A network analysis approach to gene regulation during seed maturation of Medicago truncatula uncovers distinct temporal regulatory programs related to desiccation tolerance, longevity, and pod abscission and the key regulators governing these programs.
Abstract
In seeds, desiccation tolerance (DT) and the ability to survive the dry state for prolonged periods of time (longevity) are two essential traits for seed quality that are consecutively acquired during maturation. Using transcriptomic and metabolomic profiling together with a conditional-dependent network of global transcription interactions, we dissected the maturation events from the end of seed filling to final maturation drying during the last 3 weeks of seed development in Medicago truncatula. The network revealed distinct coexpression modules related to the acquisition of DT, longevity, and pod abscission. The acquisition of DT and dormancy module was associated with abiotic stress response genes, including late embryogenesis abundant (LEA) genes. The longevity module was enriched in genes involved in RNA processing and translation. Concomitantly, LEA polypeptides accumulated, displaying an 18-d delayed accumulation compared with transcripts. During maturation, gulose and stachyose levels increased and correlated with longevity. A seed-specific network identified known and putative transcriptional regulators of DT, including ABSCISIC ACID-INSENSITIVE3 (MtABI3), MtABI4, MtABI5, and APETALA2/ ETHYLENE RESPONSE ELEMENT BINDING PROTEIN (AtAP2/EREBP) transcription factor as major hubs. These transcriptional activators were highly connected to LEA genes. Longevity genes were highly connected to two MtAP2/EREBP and two basic leucine zipper transcription factors. A heat shock factor was found at the transition of DT and longevity modules, connecting to both gene sets. Gain- and loss-of-function approaches of MtABI3 confirmed 80% of its predicted targets, thereby experimentally validating the network. This study captures the coordinated regulation of seed maturation and identifies distinct regulatory networks underlying the preparation for the dry and quiescent states.
Seed development in higher plants is divided into embryogenesis and maturation, which includes organ expansion together with storage reserve accumulation. Besides the regulatory pathways involved in these processes, additional pathways are activated during and after seed filling that confer to the seeds the remarkable capacity to survive almost complete desiccation. In nature, anhydrobiosis (life without water) bridges periods of adverse conditions. This trait is an important factor in the preservation of seed viability and quality during dry storage and an essential parameter to ensure fast and homogenous seedling establishment to ensure high yield. Whereas the regulatory networks underlying seed development have been studied (Santos-Mendoza et al., 2008; Le et al., 2010; Peng and Weselake, 2011), little is known about the regulatory networks underlying seed survival in the dry state.
To survive in the dry state, seeds first acquire desiccation tolerance (DT; i.e. the ability to survive complete drying and rehydration), which occurs for most species during seed filling. Subsequently, during late maturation, seeds progressively acquire longevity (also known as storability or life span), which is the ability to remain alive in the dry state for certain periods of time (Hay and Probert, 1995; Sanhewe and Ellis, 1996; Sinniah et al., 1998). The longevity of seeds can be remarkably long, ranging from decades to centuries (Walters et al., 2005) and even millennia (Shen-Miller et al., 1995). The relative timing between the progressive increase in seed longevity and the end of seed reserve accumulation, seed desiccation, and fruit abscission varies between species (for review, see Probert et al., 2007). During seed maturation of Medicago truncatula, a model plant closely related to economically relevant legumes such as alfalfa (Medicago sativa), soybean (Glycine max), and pea (Pisum sativum), longevity starts once seed reserve accumulation is terminated and increases progressively over 30-fold during a 2-week maturation phase (Gallardo et al., 2007; Chatelain et al., 2012). Subsequently, a final desiccation step occurs concomitantly with pod abscission. In Arabidopsis (Arabidopsis thaliana), late maturation is associated with transcriptional and metabolic switches (Angelovici et al., 2010). More than 6,900 genes (i.e. approximately one-third of the Arabidopsis genome) were identified as regulated at this specific stage. Fait et al. (2006) showed that in Arabidopsis, seed maturation was marked by a significant decrease of sugars, organic acids, and amino acids due to their incorporation into storage molecules. Inversely, during desiccation, they observed an accumulation of these compounds to prepare for germination. However, the short maturation phase of Arabidopsis (i.e. about 3 d) makes it difficult to separate seed filling, the acquisition of DT, longevity, and abscission-related processes.
On the molecular level, a number of mechanisms have been discovered that influence survival in the dry state. They include the synthesis of protective molecules, such as nonreducing sugars (Hoekstra et al., 2001), late embryogenesis abundant (LEA) proteins (Hundertmark et al., 2011; Chatelain et al., 2012), heat shock proteins (HSPs; Prieto-Dapena et al., 2006), various other stress proteins (Sugliani et al., 2009), and a set of antioxidant defenses against oxidative stress such as glutathione (GSH; Kranner et al., 2006), tocopherols (Sattler et al., 2004), and flavonoids present in the testa (Debeaujon et al., 2000). In addition, seed longevity has been shown to be associated with DNA and protein repair systems such as DNA ligase (Waterworth et al., 2010) and protein l-isoaspartyl methyltransferase (Ogé et al., 2008). Progress on the elucidation of signaling pathways and developmental programs controlling DT has come from the isolation and analysis of mutants impaired in embryogenesis and maturation, such as the abscisic acid-insensitive3 (abi3) and leafy cotyledon (lec1) mutants of Arabidopsis. Loss-of-function mutants of ABI3 and LEC1 produce seeds that lose their viability during desiccation or during the first few weeks after harvest (Ooms et al., 1993; Sugliani et al., 2009). However, abi3 and lec1 mutants are affected in multiple processes of maturation, including storage accumulation and dormancy (Santos-Mendoza et al., 2008). In addition, together with LEC2 and FUS3, these master regulator genes have partially redundant functions.
Besides the characterization of these few transcription factors (TFs), genetic evidence suggests that other regulatory factors are likely to play a role in the complex network that regulates DT and longevity, either interacting or downstream of the four master regulators (Sugliani et al., 2009). For example, in tobacco (Nicotiana tabacum), overexpression of the sunflower (Helianthus annuus) seed-specific HEAT SHOCK TRANSCRIPTION FACTOR A9 (HSFA9) that is regulated by ABI3 leads to an increase of seed longevity and thermal tolerance of seedlings (Prieto-Dapena et al., 2006). When the sunflower HSFA9 was overexpressed conjointly with the drought-responsive TF HaDREB2, positive effects on seed longevity were observed beyond those observed with the overexpression of HaHSFA9 alone (Almoguera et al., 2009).
This study aimed at identifying the complex set of regulatory factors as well as to understand the downstream mechanisms leading to DT and longevity. Correlation of gene expression (coexpression analysis) is a powerful approach to analyze large data sets, as coexpressed genes have an increased likelihood of being involved in the same biological conditions or developmental pathways (Usadel et al., 2009; Bassel et al., 2012). In addition, conditional-dependent data sets, like those restricted to seed development, are more likely to identify gene interactions relevant to a specific biological question (Usadel et al., 2009). However, the temporal modulation of gene networks during development is limited by the availability of a sufficient number of data sets that capture specific processes over time. The large time span of the late maturation phase in M. truncatula provides an elegant model to dissect these regulatory mechanisms (Chatelain et al., 2012). Using a combination of physiology, metabolomic, and transcriptomic approaches, we built and partially validated a genetic regulatory network, which revealed several developmental modules related to the acquisition of DT, longevity, and final drying/abscission with distinct specific biological functions. A survey of seed-specific TFsidentified a set of potential transcriptional regulators involved in the acquisition of DT and longevity that will facilitate future investigations in the understanding of seed quality acquisition.
RESULTS
Physiological Characterization of M. truncatula Seed Development
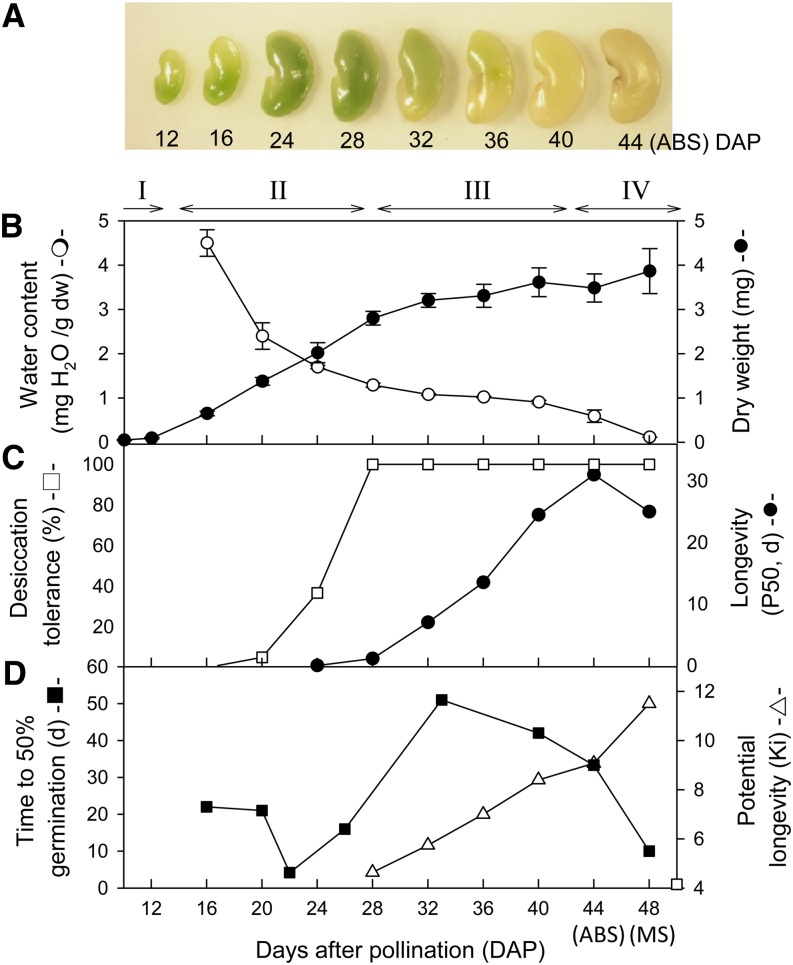
The different seed traits that were acquired during M. truncatula seed development were characterized by extending the study from Chatelain et al. (2012), which was performed on the same seed lots. Until 10 d after pollination (DAP), seeds undergo the embryogenesis program, after which seed filling (or early maturation) is initiated and lasts until about 32 DAP (Fig. 1, A and B). During seed filling, from 24 DAP onward, seeds acquire DT (i.e. the ability to survive in the dry state; Fig. 1C). From 28 DAP onward (phase III), seeds gradually acquire their longevity (i.e. the ability to remain alive in the dry state for extended periods of time). This longevity can be quantified by the time until 50% of the seed batch loses its viability during controlled aging (expressed as P50). In M. truncatula, the P50 increased over 30-fold between 28 and 44 DAP (point of abscission; Fig. 1D). The seed water content during this period remained constant and underwent a final decrease to 0.1 g water g−1 dry weight between 40 and 44 DAP, which is defined by pod abscission (phase IV; Fig. 1B). As found previously for a number of species (Probert et al., 2007), we observed the highest P50 at pod abscission (i.e. 44 DAP). Modeling longevity by the viability equation of Ellis and Roberts (1980) showed that during late maturation, the initial seed quality increased steadily (Fig. 1D).
Figure 1.
Physiological characterization of seed maturation of M. truncatula. Seed development is divided into four major phases: embryogenesis (I), seed filling (II), late maturation (III), and pod abscission (IV). A, Time course of seed development. B, Water content and dry weight (dw) changes. C, Acquisition of DT, measured as the percentage germination after rapid drying to 43% relative humidity, and longevity (P50), determined as the time to reduce viability to 50% under storage at 75% relative humidity and 35°C. D, Changes in initial viability (or potential longevity [KI]), determined from the viability equation, and germination speed or dormancy, determined as the time required to germinate 50% of seeds at 20°C. Part of the data were derived from Chatelain et al. (2012). [See online article for color version of this figure.]
Apart from the acquisition of longevity, maturation also affects germination speed (Bolingue et al., 2010). During early development, the germination capacity of freshly harvested seeds was acquired from 16 DAP onward, and the rate of germination increased from 16 to 22 DAP (Fig. 1D). From 22 DAP onward, the germination speed decreased gradually, indicating the installation of dormancy. At 32 DAP, it took 51 d for 50% of the seed population to germinate. After 6 months of storage, fully matured seeds germinated within 24 h (Fig. 1D, white square). Therefore, we interpret this strong reduction in germination speed as the acquisition of a shallow physiological dormancy (Bolingue et al., 2010). Further maturation leads to an increase in germination speed, indicating a partial release of dormancy, although the speed of seed germination is still much slower than in fully after-ripened seeds (16 versus 1 d).
Dynamic Changes in the Transcriptome of Late Seed Maturation
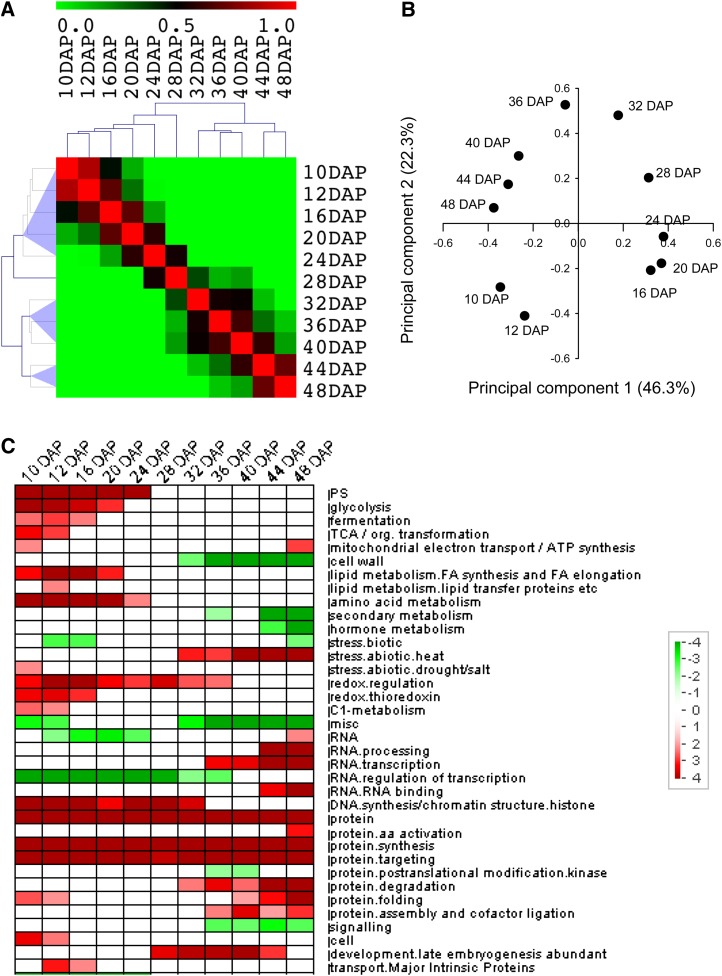
Changes in transcriptome were followed at seven stages throughout late seed maturation at 24, 28, 32, 36, and 40 DAP, at pod abscission (i.e. approximately 44 DAP), and at the final mature stage (i.e. dry seeds around 48 DAP). Total RNA was isolated in three biological replicates and used to hybridize the Affymetrix M. truncatula GeneChip containing 52,796 probe sets (Benedito et al., 2008). During the Robust Multiarray Average normalization process, we combined our data set of 21 Affymetrix GeneChips generated in this study with the data set that we previously generated for the M. truncatula gene expression atlas, covering six earlier seed stages to encompass the whole seed development (Benedito et al., 2008). From this extended data set, 19,012 probe sets were selected as differentially expressed throughout the complete seed development (i.e. with a coefficient of variation above 30% and a maximum of relative expression above 50; Supplemental Table S1). Pearson correlation coefficient (PCC) calculations and principal component analysis were performed to compare fine changes between the transcriptomes of the different developmental stages (Fig. 2, A and B). Three major transcriptional switches occurred during seed development: after 24, 32, and 40 DAP. An overrepresentation analysis of functional classes (Usadel et al., 2009) using the 11 developmental stages showed that the beginning of seed development, corresponding to embryogenesis and early maturation (i.e. 10–24 DAP), was marked by the overrepresentation of functional classes related to growth and metabolic activities such as photosynthesis, cell wall, lipid metabolism, hormone metabolism, secondary metabolism, protein synthesis, and cell cycle (Fig. 2C). Around 28 to 32 DAP, overrepresented functional classes changed to abiotic stress, RNA transcription, protein folding and degradation, as well as development (mainly LEA proteins). Underrepresented classes were related to cell wall, peroxidases, and signaling. After 40 DAP, overrepresented classes were related to mitochondrial electron transport, RNA (i.e. processing, binding, and transcription) and proteins (i.e. amino acid activation, degradation, and folding), whereas classes related to secondary and hormone metabolism and biotic stress were significantly underrepresented (Fig. 2C).
Figure 2.
Comparison of transcriptomes during seed development. A, Pairwise PCCs were used to generate the heat map. The color scale indicates the degree of correlation. B, Principal component analysis performed using median centering of the transcriptomes of seed developmental stages. C, Overrepresentation analysis of functional classes during seed development using PageMan software. Data were subjected to a bin-wise Wilcoxon test, and resulting P values were adjusted according to Bonferroni. Overrepresented and underrepresented classes are indicated in red and green, respectively. Functional classes and subclasses statistically affected are indicated according to MapMan ontology. The scale bar indicates the z-score calculated from P values (i.e. P of 0.05 represents after adjustment a z-score of 1.96).
Generation and Analysis of the Coexpression Gene Regulatory Network of Seed Development
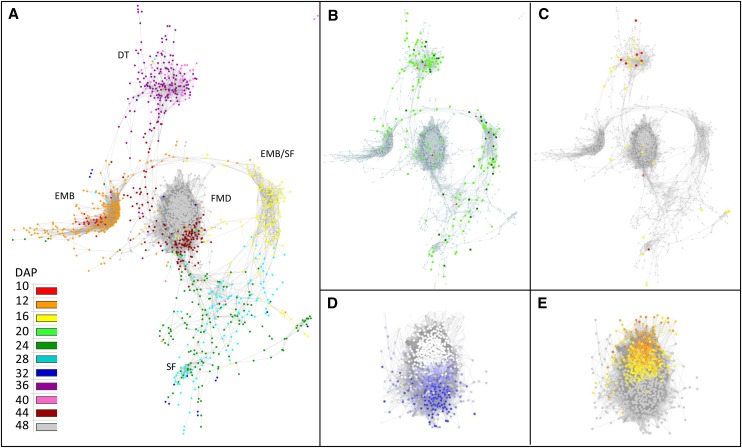
To identify the main regulatory processes that are initiated during late seed maturation together with their main regulatory factors, a weighted gene coexpression network analysis was carried out (Zhang and Horvath, 2005; Glaab et al., 2009). Using the gene coexpression network analysis module of ArrayMining (Glaab et al., 2009), a simple topological network analysis was applied to construct a weighted gene coexpression network using the graph-layout algorithm by Fruchterman and Reingold (1991). The edge adjacency threshold was set at 0.95 and visualized using an organic layout in Cytoscape. This gene regulatory network (GRN) analysis revealed several major subnetworks of gene interactions that we referred to as modules (Fig. 3). To identify the temporal expression profiles that correspond to the different coexpression modules, each node of the GRN was colored according to its maximum expression, indicated in DAP (Fig. 3A; Supplemental Table S2). By crossing this information with the developmental stages and associated physiological processes, distinct modules were found to correspond to different developmental processes: embryogenesis (red/orange), switch between embryogenesis and early seed filling (yellow), late seed filling (blue/green), acquisition of DT and dormancy (pink/purple), and final maturation drying (FMD) module encompassing genes with maximum expression at 44 DAP (brown), and genes with increased expression at the final two stages, related to pod abscission (gray; Fig. 3A).
Figure 3.
Weighted gene coexpression gene network of seed development of M. truncatula. The edge adjacency threshold was set at 0.95 and visualized using an organic layout in Cytoscape. A, Temporal analysis of network modules was obtained by coloring each gene by its maximum expression along seed development. B, Projection of seed-specific probe sets (light green), seed-specific and legume-specific probe sets (dark green), and seed specific TFs (red). C, Distribution of probe sets linked to DT and LEA genes. Genes previously identified in relation to DT were derived from Buitink et al. (2006) and are projected in yellow on the transcriptional network. Genes encoding LEA proteins are indicated in red. D, Analysis of correlation between expression profiles and P50. The color intensity of the scale ranges from 0.99 (dark blue) to 0.85 (light blue). E, Analysis of expression profiles that correlate with abscission. The scale indicates the fold increase in gene expression between 40 and 48 DAP (i.e. during abscission) and ranges from 50-fold (orange) to 2-fold (yellow).
Considering that the ability to survive the dry state in higher plants is restricted to pollen and seeds (except for resurrection plants [Craterostigma platagineum]), “seed-related” or “seed-specific” genes were projected on the regulatory network (Fig. 3B). Using data from the M. truncatula Gene Expression Atlas (MtGEA; Benedito et al., 2008), z-score values of the 19,012 probe sets were calculated and 794 probe sets were identified with a seed-specific expression or preferentially expressed in seeds in comparison with other tissues (i.e. leaf, root, petiole, stem, flower, nodule, and vegetative bud; Supplemental Table S3). Out of the 794 probe sets, 198 were present in the GRN (Fig. 3B, green and red symbols). Seed-specific genes were mainly found in the switch between embryogenesis and early seed filling module (41 of 143), in the late seed filling module (38 of 287), and in the acquisition of DT and dormancy module (72 of 218). For the acquisition of DT and dormancy module, 33% of the genes were seed specific, which is in strong contrast to the FMD module, in which the seed-specific genes represent only 1.8% (14 of 771).
Identification and Functional Analysis of Coexpression Modules in Relation to DT, Longevity, and Abscission
Since we are interested in the mechanisms that govern the capacity to survive the dry state, we investigated which module contained genes related to DT or longevity. For this purpose, a group of 191 genes, which were previously identified as being up-regulated in relation to DT (Buitink et al., 2006), were mapped to the Affymetrix probe sets, and 101 probe sets could be retrieved (Supplemental Table S2). These genes were selected based on increased transcript levels upon the acquisition of DT during maturation as well as during the reinduction of DT in germinated radicles of M. truncatula (Buitink et al., 2006). A projection of these genes on the GRN indicated that 26 probe sets were present, out of which 54% were found in the acquisition of DT and dormancy module (14 of 26; Fig. 3C; Supplemental Table S2). Six probe sets were found in the module with probe sets displaying a maximum of expression at 44 DAP (FMD module), and another six genes were found in the module with their maximum expression around 28 DAP (late seed filling module). In addition, probe sets of LEA genes were projected in red on the network and were found mainly in the acquisition of DT and dormancy module (Fig. 3C).
Overrepresented biological processes that are related to the acquisition of DT and dormancy module were investigated by analysis of the probe sets for enriched Gene Ontology (GO) terms using a χ2 statistical test method with the Yekutieli multitest adjustment method in AgriGO (Du et al., 2010; Table I). The acquisition of DT and dormancy module was enriched in genes with GO terms related to response to stresses (e.g. response to desiccation, heat, temperature, abiotic stimulus) and in GSH metabolic process (Table I; Supplemental Fig. S1). They included many LEA genes, HSPs or small heat shock proteins (sHSPs; DNAJ), universal stress proteins, aldose reductases, and abscisic acid (ABA)-induced genes (Supplemental Table S2).
Table I. GO enrichment analysis of maturation modules obtained from gene coexpression analysis.
Analysis was performed using AgriGO with the M. truncatula Affymetrix background applying a χ2 statistical test method with the Yekutieli multitest adjustment method. The graphical view of the GO enrichment analysis for the FMD module is provided in Supplemental Figures S1 and S2. BG, Number of genes annotated in the GO term in background; FDR, false discovery rate; P, biological process; F, molecular function.
| GO Term | Ontology Source | Description | No. Input List | BG | P | FDR |
|---|---|---|---|---|---|---|
| Acquisition of DT and dormancy module | ||||||
| GO:0009611 | P | Response to wounding | 6 | 124 | 4.30E-09 | 2.10E-06 |
| GO:0000272 | P | Polysaccharide catabolic process | 6 | 139 | 4.90E-08 | 1.20E-05 |
| GO:0006022 | P | Aminoglycan metabolic process | 5 | 112 | 5.80E-07 | 5.60E-05 |
| GO:0006749 | P | GSH metabolic process | 5 | 112 | 5.80E-07 | 5.60E-05 |
| GO:0009620 | P | Response to fungus | 6 | 155 | 3.90E-07 | 5.60E-05 |
| GO:0050832 | P | Defense response to fungus | 5 | 118 | 1.40E-06 | 0.00011 |
| GO:0009414 | P | Response to water deprivation | 6 | 176 | 3.40E-06 | 0.00023 |
| GO:0009408 | P | Response to heat | 5 | 126 | 3.80E-06 | 0.00023 |
| GO:0009415 | P | Response to water | 6 | 189 | 1,00E-05 | 0.00044 |
| GO:0006950 | P | Response to stress | 29 | 2,261 | 1.10E-05 | 0.00046 |
| GO:0009266 | P | Response to temperature stimulus | 7 | 319 | 0.00029 | 0.01 |
| GO:0009628 | P | Response to abiotic stimulus | 15 | 1,051 | 0.0003 | 0.01 |
| Longevity submodule | ||||||
| GO:0006364 | P | rRNA processing | 5 | 35 | 1.80E-16 | 5.00E-14 |
| GO:0016072 | P | rRNA metabolic process | 5 | 35 | 1.80E-16 | 5.00E-14 |
| GO:0006457 | P | Protein folding | 12 | 233 | 4.30E-13 | 8.20E-11 |
| GO:0034470 | P | ncRNA processing | 5 | 82 | 8.00E-07 | 0.00011 |
| GO:0034660 | P | ncRNA metabolic process | 6 | 164 | 0.00013 | 0.013 |
| GO:0006396 | P | RNA processing | 9 | 327 | 0.00013 | 0.013 |
| GO:0051082 | F | Unfolded protein binding | 12 | 125 | 9.10E-27 | 1.60E-24 |
| Abscission submodule | ||||||
| GO:0006396 | P | RNA processing | 14 | 327 | 3.70E-10 | 2.60E-07 |
| GO:0008380 | P | RNA splicing | 7 | 101 | 1.80E-09 | 6.30E-07 |
| GO:0034470 | P | ncRNA processing | 5 | 82 | 6.40E-06 | 0.0011 |
| GO:0006511 | P | Ubiquitin-dependent protein catabolic process | 9 | 235 | 6.20E-06 | 0.0011 |
| GO:0016070 | P | RNA metabolic process | 27 | 1,352 | 2.70E-05 | 0.0023 |
| GO:0044257 | P | Cellular protein catabolic process | 9 | 259 | 3.10E-05 | 0.0023 |
| GO:0006605 | P | Protein targeting | 6 | 132 | 4.40E-05 | 0.003 |
| GO:0016071 | P | mRNA metabolic process | 7 | 175 | 5.10E-05 | 0.0032 |
| GO:0006397 | P | mRNA processing | 6 | 143 | 0.00012 | 0.0071 |
| GO:0034613 | P | Cellular protein localization | 9 | 327 | 0.00076 | 0.037 |
| GO:0070727 | P | Cellular macromolecule localization | 9 | 331 | 0.00088 | 0.04 |
| GO:0008565 | F | Protein transporter activity | 5 | 104 | 0.00014 | 0.034 |
To discover modules related to longevity, 315 probe sets were identified in the network with expression profiles that correlated with P50 values above a PCC of 0.85 (Supplemental Table S2). A projection of these probe sets on the network showed that they were all located in the FMD module (Fig. 3D). In addition, 338 probe sets increasing around the point of abscission were identified by determining the difference in transcript levels between 40 and 48 DAP (more than 2-fold increase; Fig. 3E). All probe sets of this abscission module were also detected in the FMD module, but with a distinctly different topology from those related to longevity (P50). The differential topology of these two groups of probe sets suggested that they might govern different functions. For this reason, both subclusters were analyzed separately for GO enrichment. The longevity submodule (315 probe sets; PCC > 0.85) was enriched in genes with GO terms related to ribosomal RNA processing, RNA and noncoding RNA processing, and unfolded protein binding (Table I; Supplemental Fig. S2). The abscission submodule was enriched in genes related RNA processing and RNA splicing as well as in ubiquitin-dependent protein catabolic processes and protein targeting and localization (Table I; Supplemental Fig. S2).
Posttranscriptional Regulation during Late Maturation
To further characterize the processes occurring during the acquisition of longevity, GO enrichment analysis was performed on all genes with transcripts that correlated with the P50 values (i.e. corresponding to the acquisition of longevity, irrespective of whether they were part of the coexpression GRN or not [PCC > 0.90]; Supplemental Table S4). Besides the 118 genes that were present in the network, the total list of up-regulated genes comprised 1,332 probe sets. As for the longevity module in the GRN, these probe sets also displayed a significant enrichment in genes related to gene expression (194 probe sets) and translation (62 probe sets) as well as ribonucleoprotein complex biogenesis and ribosome biogenesis (51 probe sets; Supplemental Fig. S3; Supplemental Table S5). The expression profiles of these genes were already elevated during early seed development but increased gradually 2- to 5-fold from 28 to 48 DAP (Supplemental Fig. S4). This suggests that transcription and translation regulation processes might be activated in a coordinated way after the seed filling and during the acquisition of longevity.
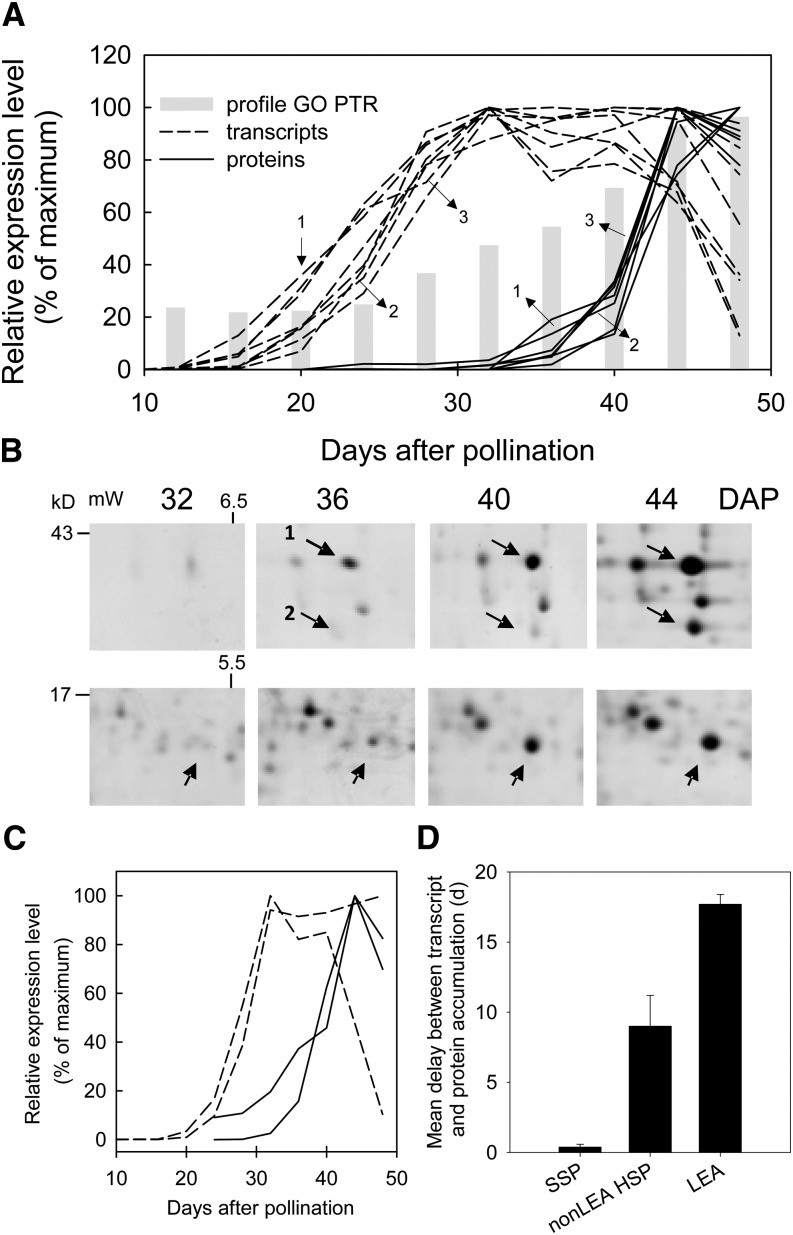
Further evidence for the regulation of translational processes during late seed maturation comes from the comparison of transcript levels and corresponding polypeptide contents of LEA proteins that appear late during seed maturation, using our proteome analysis of Chatelain et al. (2012). These data were obtained from the same seed lots as used for the transcriptome analysis. Out of the 14 LEA genes identified by these authors, eight genes showed transcript profiles that were very tightly coregulated (Fig. 4A, dashed lines). All of them were part of the acquisition of DT and dormancy module. For the eight genes, the corresponding proteins accumulated 18 to 20 d later than transcript levels (Fig. 4A, solid lines). This is visualized in Figure 4B, showing that polypeptides encoded by three of these LEA genes (DHN3, MP2, and EM) accumulated from 36 to 40 d onward. When the accumulation of transcript and protein levels of the LEA proteins is compared with the average of the expression profiles of the genes related to posttranscriptional regulation from the longevity module (Table II; Fig. 4A, gray bars; Supplemental Fig. S2), it can be seen that transcripts of genes involved in translational regulation increase 10 d later than LEA transcripts and advance the accumulation of the protein levels: transcripts have a high but steady level between 10 and 24 DAP, after which transcript levels increase by 5-fold until 44 DAP.
Figure 4.
Posttranscriptional regulation during late seed maturation. A, Transcript (dashed lines) and polypeptide (solid lines) abundance of eight LEA genes. Polypeptide abundance was derived from Chatelain et al. (2012). LEA proteins are as follows (IMGAG version 3.0): DHN3 (spot 1); TC144427, maturation polypeptide2, Medtr1g073780 (spot 2); Em protein, AJ498523 (spot 3); EM6, Medtr3g166750.1; SBP65, Medtr4g109500; PM10, Medtr8g134020; LEAM, Medtr2g017540. Gray bars indicate the average expression profiles of the genes of the longevity submodule related to GO terms of posttranscriptional regulation (PTR). B, Two-dimensional protein electrophoresis of the heat-stable protein fraction at different stages during M. truncatula seed maturation. Arrows indicate polypeptides corresponding to three genes indicated in A. C, Transcript and polypeptide abundance of sHSP20 (Medtr5g088740) and a phosphatidylethanolamine-binding protein (TC14831). D, Average delay of transcript versus protein accumulation of vicilin and legumin (data from Gallardo et al., 2007) and of seven genes from Chatelain et al. (2012): TC163177, Gly-rich protein, Mtr.44922.1.S1; Medtr5g088740, HSP20, Mtr.50164.1.S1_at; TC146236, 1-Cys-peroxiredoxin, Mtr.11099.1.S1_at; Medtr2g005480, chaperone DnaK, Mtr.50134.1.S1_s_at; TC148321, phosphatidylethanolamine-binding protein, Mtr.11203.1.S1_at; Medtr8g044470, cystatin, Mtr.49348.1.S1_s_at; Medtr4g095700.1, RNA-binding protein RPN-1, Mtr.52040.1.S1_at. Gene annotations are from IMGAG version 3.0.
Table II. Metabolite changes during maturation of M. truncatula seeds.
Metabolite contents were normalized with respect to ribitol content used as an internal standard. PCCs with P50 are indicated, as are ratios between metabolite content between 24 and 44 DAP and between 48 and 44 DAP. ABS, Abscission; DS, dry mature seeds.
|
DAP |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Compound | 24 | 28 | 32 | 36 | 40 | 44 (ABS) | 48 (DS) | PCC P50 | Ratio 24:44 | Ratio 48:44 |
| Amino acid | ||||||||||
| Allo-Thr | 0.113 | 0.069 | 0.040 | 0.023 | 0.018 | 0.010 | 0.029 | −0.88 | 10.73 | 2.74 |
| Gly | 0.470 | 0.244 | 0.153 | 0.104 | 0.069 | 0.035 | 0.202 | −0.68 | 13.27 | 5.70 |
| Homo-Ser | 5.626 | 7.720 | 7.649 | 6.482 | 5.639 | 3.892 | 4.907 | −0.97 | 1.45 | 1.26 |
| l-Ala | 2.854 | 1.554 | 1.086 | 0.759 | 0.533 | 0.078 | 0.182 | −0.97 | 36.71 | 2.34 |
| l-Arg | 3.181 | 1.482 | 0.702 | 0.419 | 0.394 | 0.426 | 0.370 | −0.80 | 7.47 | 0.87 |
| l-Asn | 10.136 | 2.477 | 1.369 | 1.307 | 0.877 | 0.764 | 0.961 | −0.90 | 13.27 | 1.26 |
| l-Asp | 4.109 | 3.814 | 2.937 | 1.692 | 0.589 | 0.195 | 0.386 | −0.98 | 21.09 | 1.98 |
| l-Glu | 2.290 | 1.466 | 1.201 | 0.601 | 0.284 | 0.109 | 0.190 | −0.98 | 20.96 | 1.74 |
| l-Gln | 2.336 | 1.202 | 0.684 | 0.259 | 0.043 | 0.003 | 0.004 | −0.94 | 902.83 | 1.45 |
| l-Homo-Ser | 0.057 | 0.068 | 0.057 | 0.054 | 0.053 | 0.039 | 0.046 | −0.93 | 1.48 | 1.19 |
| l-Ile | 0.291 | 0.146 | 0.089 | 0.061 | 0.057 | 0.025 | 0.041 | −0.93 | 11.47 | 1.61 |
| l-Leu | 0.926 | 0.730 | 0.399 | 0.242 | 0.156 | 0.087 | 0.118 | −0.92 | 10.65 | 1.36 |
| l-Lys | 1.097 | 0.616 | 0.339 | 0.195 | 0.082 | 0.033 | 0.057 | −0.94 | 33.65 | 1.74 |
| l-Met | 0.131 | 0.079 | 0.054 | 0.048 | 0.051 | 0.035 | 0.034 | −0.86 | 3.75 | 0.96 |
| l-Phe | 0.204 | 0.104 | 0.056 | 0.037 | 0.027 | 0.022 | 0.045 | −0.85 | 9.33 | 2.04 |
| l-Pro | 0.475 | 0.306 | 0.208 | 0.192 | 0.208 | 0.108 | 0.275 | −0.59 | 4.42 | 2.56 |
| l-Ser | 0.793 | 0.506 | 0.321 | 0.214 | 0.150 | 0.035 | 0.018 | −0.95 | 22.43 | 0.50 |
| l-Tyr | 0.433 | 0.272 | 0.219 | 0.735 | 1.709 | 1.362 | 1.093 | 0.90 | 0.32 | 0.80 |
| l-Val | 0.540 | 0.303 | 0.186 | 0.136 | 0.114 | 0.067 | 0.105 | −0.93 | 8.09 | 1.57 |
| Orn | 2.485 | 1.331 | 0.761 | 0.535 | 0.540 | 0.538 | 0.454 | −0.81 | 4.62 | 0.85 |
| Carboxylate | ||||||||||
| 4-O-Methyl-myoinositol | 0.460 | 0.173 | 0.136 | 0.132 | 0.092 | 0.055 | 0.092 | −0.98 | 8.41 | 1.68 |
| d-Pinitol | 0.768 | 0.417 | 0.341 | 0.255 | 0.130 | 0.057 | 0.050 | −0.98 | 13.54 | 0.89 |
| Digalactosylglycerol | 0.349 | 0.401 | 0.619 | 0.405 | 0.682 | 0.408 | 0.504 | 0.11 | 0.86 | 1.24 |
| Galactinol | 8.818 | 8.574 | 9.349 | 3.828 | 5.075 | 3.097 | 4.681 | −0.83 | 2.85 | 1.51 |
| Gal methoxyamine | 0.179 | 0.147 | 0.208 | 0.081 | 0.144 | 0.063 | 0.052 | −0.66 | 2.83 | 0.82 |
| Gentiobiose | 0.317 | 1.268 | 0.834 | 0.658 | 0.262 | 0.078 | 0.037 | −0.97 | 4.05 | 0.47 |
| Glucopyranose | 0.182 | 0.119 | 0.106 | 0.062 | 0.051 | 0.030 | 0.022 | −0.95 | 6.03 | 0.74 |
| Gulose | 0.118 | 0.077 | 0.127 | 0.180 | 0.626 | 0.621 | 1.098 | 0.82 | 0.19 | 1.77 |
| Maltose methoxyamine | 0.537 | 1.038 | 1.115 | 0.289 | 0.191 | 0.027 | 0.014 | −0.92 | 19.55 | 0.51 |
| Myoinositol | 11.632 | 6.201 | 3.936 | 2.402 | 1.852 | 1.122 | 0.894 | −0.93 | 10.36 | 0.80 |
| Raffinose | 1.342 | 1.207 | 1.805 | 1.319 | 1.803 | 0.522 | 0.401 | −0.51 | 2.57 | 0.77 |
| Ribitol | 1.643 | 1.061 | 1.434 | 0.620 | 0.946 | 0.591 | 0.705 | −0.66 | 2.78 | 1.19 |
| Rib methoxyamine | 0.427 | 0.219 | 0.186 | 0.062 | 0.065 | 0.044 | 0.048 | −0.90 | 9.63 | 1.08 |
| Suc | 67.758 | 60.156 | 55.634 | 44.710 | 29.479 | 5.706 | 3.551 | −0.94 | 11.88 | 0.62 |
In order to evaluate the importance of the delay of LEA translation compared with other seed development-regulated genes, the standard rate of delay was calculated for four seed storage protein genes expressed during seed filling (between 12 and 24 DAP) using the data set from Gallardo et al. (2007) on M. truncatula seed development. In addition, we determined the rate of delay for the other seven proteins appearing during late maturation, at the same time as LEA proteins appear (between 24 and 44 DAP), using our transcriptome data and proteome data (Chatelain et al., 2012). The transcript levels and protein accumulation are shown for two of these late maturation proteins, a sHSP and 1-Cys-peroxiredoxin (Fig. 4C). The average rate of delay for seed storage proteins and non-LEA proteins was approximately 0.4 and 9 d, respectively, compared with 18 d for LEA proteins.
Metabolome Changes during Seed Maturation
To complete the study of maturation, metabolite dynamics were analyzed at the same stages chosen for the gene expression analysis (i.e. from 24 to 48 DAP) by gas chromatography coupled to mass spectrometry (GC-MS). A total of 63 polar and 33 nonpolar metabolites were identified and characterized using different chemical libraries (Supplemental Table S6). Most of the metabolites decreased during maturation (Table II). This concurred with the GO enrichment analysis of 2,409 genes displaying a negative correlation with P50, suggesting that down-regulated biological processes were all related to metabolism (Supplemental Table S5). The first group of down-regulated genes was related to cellular amino acid and derivative metabolic processes, with many enzymes involved in Glu metabolism (e.g. Gln synthetase, Glu dehydrogenase, Pro dehydrogenase, Ala aminotransferase, dehydroascorbate reductase). The decrease in amino acid biosynthetic genes was confirmed at the metabolite level by a negative correlation between P50 and most of the amino acid content profiles (Table II), which displayed a global decrease along the seed desiccation process. The exception was Tyr, which is the only amino acid that accumulates during late seed maturation. The second and third down-regulated processes were lipid metabolic process and secondary metabolic processes, including phospholipid metabolism, isoprenoid/phenylprenoid, and anthocyanin biosynthesis. Finally, the fourth down-regulated process was related to carbohydrate metabolic processes, including hexose biosynthetic process and Glc catabolic process. Monosaccharides (Glc and Fru) and Suc correlated negatively with longevity (Table II).
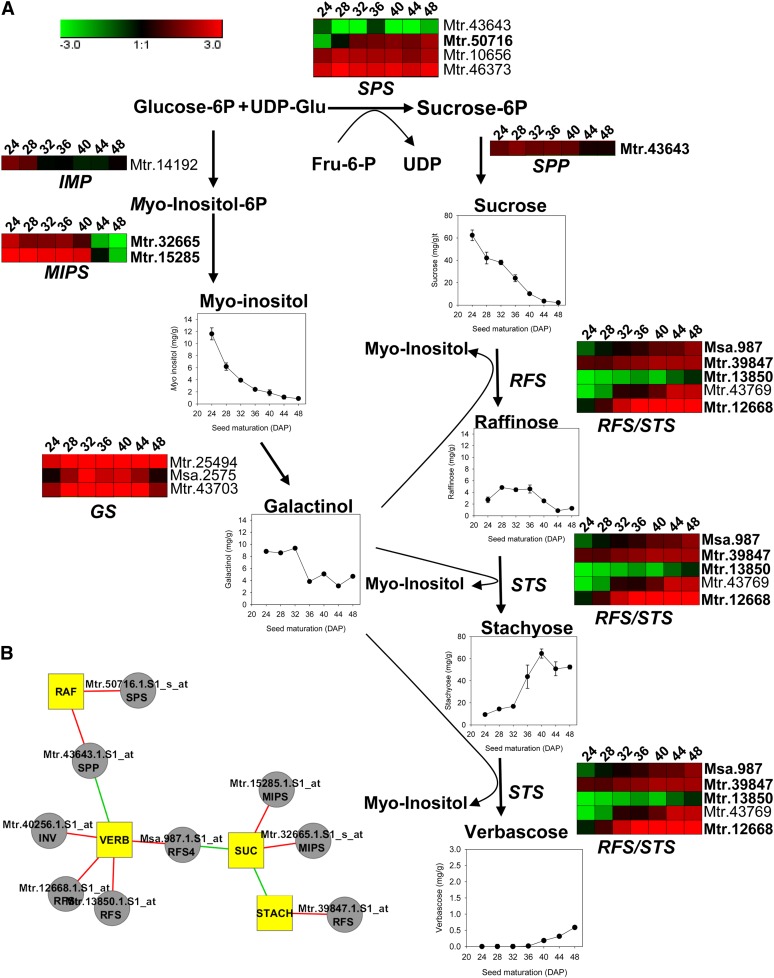
Since seeds of M. truncatula accumulate raffinose family oligosaccharides (RFO) at the expense of Suc (Rosnoblet et al., 2007), we analyzed the soluble carbohydrate composition related to RFO metabolism in more detail using Dionex-HPLC (Fig. 5A). According to these data, we proposed a model for the metabolic pathway of RFO biosynthesis (Fig. 5A) describing metabolite contents as well as transcript levels of the enzymes. Sugar analysis confirmed the decrease in monosaccharides and the strong negative correlation between Suc and longevity found by the GC-MS analysis (Table II). In addition, raffinose increased transiently from 24 to 36 DAP, whereas stachyose and verbascose increased sharply from 32 and 36 DAP onward, respectively (Fig. 5). Stachyose was the major soluble sugar in M. truncatula seed (more than 90% of the total sugars). This tetrasaccharide correlated positively with longevity (PCC = 0.92; Fig. 5A). To identify those enzymes of the RFO pathway with transcript levels that correlated with the increase in stachyose, a correlation analysis was performed (Fig. 5B). The transcript level of a stachyose synthase (Mtr.39847.1.S1_at) correlated strongly with the increase in stachyose, whereas the Suc amount correlated with the two myoinositol-3-phosphate synthase genes (Mtr.15285.1.S1_at and Mtr.32665.1.S1_s_at) and RFS4 (Msa.987.1.S1_at), another raffinose synthase.
Figure 5.
Predictive metabolic pathway of RFO biosynthesis in M. truncatula seeds. A, Sugar contents were analyzed during late seed maturation. Sugar data are means ± se of five replications using five seeds. According to their annotations, expression profiles of putative enzymes involved in the RFO pathway are represented. B, Network based on correlation between major RFO sugars and the expression of enzymes involved in the RFO pathway. Red and green lines represent positive and negative correlations, respectively. GS, Galactinol synthase; IMP, myoinositol monophosphatase; INV, invertase; MIPS, myoinositol phosphate synthase; RAF, raffinose; RFS, raffinose synthase; SPP, Suc phosphate phosphatase; SPS, Suc phosphate synthase; STACH, stachyose; STS, stachyose synthase; SUC, Suc; VERB, verbascose. Probe sets indicated in boldface are represented in the network in B.
While most metabolites decreased gradually from 24 DAP onward, a change in metabolite profile occurred around abscission between 44 and 48 DAP: many free amino acids increased around 2-fold (Table II). Likewise, l-gulose, a C-3 epimer of Gal involved in one of the three pathways leading to ascorbic acid (Dowdle et al., 2007; Wolucka and Van Montagu, 2007), increased 3.6-fold when the seeds dried from 1 to 0.1 g g−1. l-Gulose and l-Gal are intermediates of two possible biosynthetic pathways for ascorbate, but it is not yet clear what the relative contribution of these pathways in vivo is. l-Gulose is a rare sugar found in plants (Wolucka and Van Montagu, 2007) and has not yet been detected in seeds. Ascorbate was not detected in this study, but in other species its amount decreases during maturation to undetectable levels in dry seeds (Arrigoni et al., 1992).
Transcriptional Regulation of LEA, P50, and DT Genes
To elucidate transcriptional regulation of the acquisition of DT and longevity, a coexpression gene regulatory network analysis was performed using the PCC between the log2 expression of seed-specific TFsand seed-specific probe sets (Supplemental Table S7). A total of 34 seed-specific TFs were detected with transient expression profiles at different times during seed developmental (Supplemental Fig. S5). We retained 22 TFs with a maximum of expression during maturation (i.e. six with a maximum of expression between 20 and 28 DAP, 13 between 32 and 40 DAP, and three between 44 and 48 DAP; Supplemental Fig. S5).
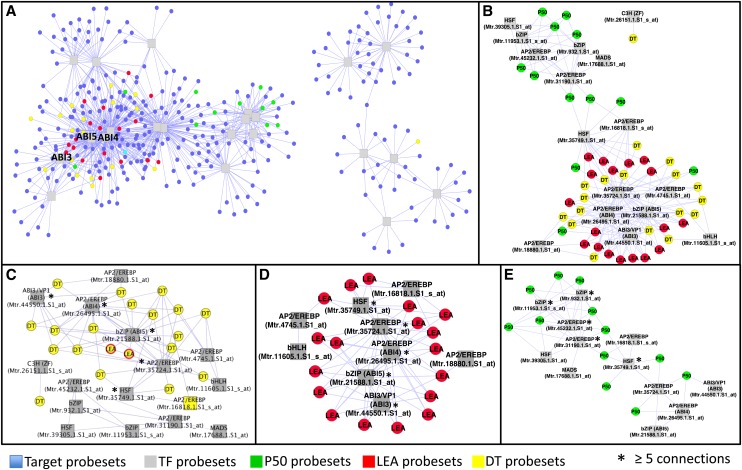
To visualize and identify key regulators of longevity, LEA and DT, we highlighted probe sets of the network that correlated strongly with P50 (Supplemental Table S2), probe sets shown to be involved in DT (Buitink et al., 2006; Supplemental Table S2), and probe sets encoding LEA proteins (Chatelain et al., 2012; Supplemental Table S2). Using a PCC threshold of 0.9, the global regulatory network was composed of two core modules: a large one containing 16 TFs and 292 putative target genes including most of the LEA, DT, and P50 probe sets and a smaller one with six TFs and 76 putative targets (Fig. 6A). Figure 6, B to E, visualizes the TFs that are connected to the LEA, DT, and P50 probe sets. Two clusters are apparent (Fig. 6B): one containing the LEA and DT probe sets and the other containing most of the P50 probe sets. Both clusters are connected by two TFs, an MtAP2/EREBP and an MtHSF TF that is a homolog of the Arabidopsis HSFA9, known to be regulated by AtABI3 (Kotak et al., 2007). An analysis of the number of connections revealed the TFs that are highly connected (i.e. at least five connections), representing interesting candidate genes controlling the different gene probe sets (Supplemental Table S7). The four highly connected TFs with DT probe sets were MtABI3, MtABI4, MtABI5, and an MtAP2/EREBP gene (Fig. 6C). These TFs also connected highly with LEA probe sets, together with the HSFA9 homolog (Fig. 6D). In the cluster of P50 probe sets (Fig. 6E), highly connected TFs were different from those for LEA and DT probe sets and included two M. truncatula basic leucine zipper TFs and two MtAP2/EREBP TFs (Fig. 6E).
Figure 6.
Coexpression gene regulatory network between the 22 seed-specific TFs and seed-specific genes of M. truncatula. A, View of the global gene regulatory network containing all probe sets showing a PCC above 0.9 between their expression profiles. B, Simplified projection of the GRN with only P50, LEA, and DT probe sets. C, Focus on the regulation of DT-related genes. D, Focus on the regulation of LEA protein genes. E, Focus on the regulation of P50-related genes. All networks were displayed using the Cytoscape organic layout.
Validation of the Gene Regulatory Network
To experimentally validate the network shown in Figure 6, we focused on MtABI3 because it is connected with 40 putative target genes (Table III), making it one of the most connected TFs in our network (Supplemental Table S7). In addition, experimental evidence on the direct targets of Arabidopsis has been recently published (Mönke et al., 2012). We first compared our M. truncatula predictive targets against this list of 98 ABI3 regulons from Arabidopsis that were validated using chromatin immunoprecipitation analysis, mutant transcriptome analysis, and transient promoter activation assay (Mönke et al., 2012). Out of the 40 M. truncatula putative target genes present in the network, 37 had putative orthologs in Arabidopsis (Supplemental Table S8). A total of 70% (26 of 37) of these M. truncatula MtABI3 putative targets were identified as ABI3 regulons in Arabidopsis and include seed storage proteins, oleosins, and LEA proteins (Table III).
Table III. List of putative MtABI3 regulons according to our GRN analysis.
The 2-kb promoter sequence analysis was performed to reveal the presence and number of RYL and GBL cis-regulatory motifs known to bind ABI3 for each predicted M. truncatula regulon. Promoter sequences used for the analysis are provided in Supplemental Table S9. Loss-of-function data from expression in Mtabi3 mutant seeds compared with wild-type seeds at 32 DAP and gain-of-function data from hairy roots transformed with MtABI3 compared with control lines are expressed as log2 ratios and are from probe sets present on the Nimblegen gene chip with their corresponding P values. Arabidopsis orthologs of the putative MtABI3 regulons identified in our network are indicated, and the methods used by Mönke et al. (2012) to validate them as targets of ABI3 in Arabidopsis are specified as follows: A, transient promoter activation assay; C, chromatin immunoprecipitation chip analysis; P, quantitative reverse transcription-PCR; T, array-based transcriptome analysis. Affymetrix identifiers in boldface represent genes displaying significant changes in expression in both gain- and loss-of function experiments.
| Probe Set Identifier of Putative ABI3 Regulons According to the GRN |
Promoter Analysis (2-kb Sequence) |
Ectopic Expression (Hairy Roots) |
abi3 Mutants 32 DAP | Targets of ABI3 in Arabidopsis (Mönke et al., 2012) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 32 DAP |
|||||||||||
| Affymetrix Identifier | Tentative Annotation | IMGAG Mt3.5.1 | No. of RYL Motifs (CATGCA) | No. of GBL Motifs (ACGTGTC) | Nimblegen Identifier | Log2 ABI3/Control | P | Log2 abi3/Wild Type | P | Arabidopsis Orthologs | Methods Used to Identify Regulons |
| Mtr.37233.1.S1_at | Oleosin | – | – | – | – | – | – | – | – | AT5G40420 | CTPA |
| Mtr.24397.1.S1_at | Cupin | Medtr1g071710 | 1 | 1 | Medtr_v1_006027 | 3.05 | 2.29E-03 | −2.86 | 6.92E-08 | AT3G22640 | CT |
| Mtr.43017.1.S1_at | Cupin | Medtr1g071710 | 1 | 1 | Medtr_v1_006027 | 3.05 | 2.29E-03 | −2.86 | 6.92E-08 | AT3G22640 | CT |
| Mtr.31203.1.S1_s_at | Legumin A | – | – | – | Medtr_v1_006092 | 2.81 | 2.61E-03 | −1.43 | 7.09E-06 | AT5G44120 | CTP |
| Mtr.21257.1.S1_at | LEA | Medtr2g014040 | 2 | 0 | Medtr_v1_009629 | 2.98 | 0.002341 | −0.93 | 0.000896 | AT4G21020 | CT |
| Mtr.21376.1.S1_at | Mitochondrial translocase | Medtr2g034550 | 1 | 0 | Medtr_v1_010795 | 3.85 | 0.000438 | −1.16 | 4.95E-05 | AT4G16160 | CTPA |
| Mtr.17433.1.S1_at | Unknown | Medtr3g096890 | 0 | 0 | Medtr_v1_019345 | 1.23 | 0.103578 | −1.94 | 1.07E-05 | AT1G27990 | TP |
| Mtr.17102.1.S1_at | Dehydrin | Medtr3g117190 | 5 | 0 | Medtr_v1_020579 | 3.67 | 0.001524 | −0.98 | 0.000286 | AT2G21490 | CTP |
| Mtr.8682.1.S1_s_at | Cupin | Medtr4g060780 | 3 | 1 | Medtr_v1_021810 | 3.08 | 9.30E-03 | −0.24 | 0.094514 | AT2G28490 | CTP |
| Mtr.37595.1.S1_s_at | Cupin | Medtr4g060780 | 3 | 1 | Medtr_v1_024309 | 3.08 | 0.009303 | −1.01 | 9.35E-05 | AT2G28490 | CTP |
| Mtr.37595.1.S1_at | Allergen Gly | Medtr4g060780 | 3 | 1 | Medtr_v1_024309 | 3.08 | 0.009303 | −1.01 | 9.35E-05 | AT2G28490 | CTP |
| Mtr.41721.1.S1_at | Oleosin | Medtr6g005040 | 1 | 0 | Medtr_v1_038147 | 3.3 | 0.030741 | −2.89 | 1.12E-07 | AT3G27660 | CP |
| Mtr.18309.1.S1_at | Tonoplast intrinsic protein | Medtr6g069040 | 2 | 1 | Medtr_v1_039779 | 2.51 | 0.009378 | −0.63 | 0.001548 | AT1G73190 | CTPA |
| Mtr.198.1.S1_s_at | Tonoplast intrinsic protein | Medtr6g069040 | 2 | 1 | Medtr_v1_039779 | 2.51 | 9.38E-03 | −0.63 | 0.001548 | AT1G17810 | CTA |
| Mtr.23887.1.S1_at | Unknown | Medtr7g080030 | 6 | 1 | Medtr_v1_044786 | 3.96 | 2.92E-05 | −2.13 | 4.87E-07 | AT5G16460 | TP |
| Mtr.12358.1.S1_at | LEA | Medtr7g093160 | 6 | 0 | Medtr_v1_045825 | 4.35 | 8.92E-05 | −1.89 | 1.82E-06 | AT5G06760 | CPA |
| Mtr.12942.1.S1_at | LEA4, LEA5 | Medtr7g093170 | 0 | 2 | Medtr_v1_045825 | 4.35 | 8.92E-05 | −1.89 | 1.82E-06 | AT5G06760 | CPA |
| Mtr.5956.1.S1_s_at | LEA | Medtr7g093170 | 0 | 2 | Medtr_v1_045826 | 4.59 | 7.89E-07 | −0.98 | 0.000197 | AT5G06760 | CPA |
| Mtr.9420.1.S1_s_at | Calcium-binding protein | – | – | – | Medtr_v1_068117 | 4.03 | 1.35E-05 | −2.51 | 2.44E-07 | AT4G26740 | CTPA |
| Mtr.43091.1.S1_s_at | LEA | Medtr7g093160 | 6 | 0 | Medtr_v1_074975 | 2.95 | 4.31E-03 | −0.97 | 0.000130 | AT5G06760 | CPA |
| Mtr.9049.1.S1_at | SBP65 | – | – | – | Medtr_v1_083614 | 4.18 | 0.003071 | −2.26 | 4.25E-06 | AT2G42560 | TP |
| Mtr.37679.1.S1_at | Tonoplast intrinsic protein | – | – | – | Medtr_v1_086805 | 3.51 | 0.007926 | −1.44 | 7.73E-06 | AT1G73190 | CTPA |
| Mtr.35625.1.S1_s_at | Unknown | – | – | – | Medtr_v1_088080 | 3.66 | 5.45E-05 | −0.78 | 0.002302 | AT5G07330 | CT |
| Mtr.37272.1.S1_at | Legumin A | – | – | – | Medtr_v1_088220 | 0.02 | 0.833278 | −1.03 | 6.69E-05 | AT5G44120 | CTP |
| Mtr.37270.1.S1_at | Legumin | – | – | – | Medtr_v1_091453 | 2.13 | 0.094826 | −1.33 | 0.000107 | AT5G44120 | CTP |
| Mtr.31204.1.S1_s_at | Oleosin | – | – | – | Medtr_v1_093522 | 3.61 | 0.000696 | −1.45 | 6.43E-06 | AT3G01570 | CTA |
| Mtr.12327.1.S1_s_at | LEA | – | – | – | Medtr_v1_085905 | 1.06 | 0.069602 | −0.87 | 0.000219 | AT1G52690 | – |
| Mtr.7496.1.S1_at | Unknown | – | – | – | Medtr_v1_083970 | 4.79 | 0.000158 | −0.87 | 0.001075 | AT5G36100 | – |
| Mtr.10877.1.S1_at | Aldehyde dehydogenase | – | – | – | Medtr_v1_072305 | 2.86 | 0.034275 | −0.61 | 0.002170 | AT1G23800 | – |
| Mtr.8651.1.S1_a_at | Dehydrin | – | – | – | Medtr_v1_066754 | 3.32 | 1.10E-05 | −0.79 | 0.000374 | AT3G50970 | – |
| Mtr.7231.1.S1_at | Defensin | Medtr8g095370 | 7 | 0 | Medtr_v1_051884 | 0.46 | 0.200574 | −0.55 | 0.004721 | AT2G02130 | – |
| Mtr.50543.1.S1_at | Unknown | Medtr8g020560 | 3 | 1 | Medtr_v1_048842 | 0.61 | 0.352299 | −0.07 | 0.720525 | AT3G13960 | – |
| Mtr.50542.1.S1_at | Unknown | Medtr8g020550 | 0 | 0 | Medtr_v1_048841 | 0.45 | 0.399151 | 0.16 | 0.247374 | AT3G13960 | – |
| Mtr.11561.1.S1_at | Unknown | Medtr5g025800 | 3 | 0 | Medtr_v1_031191 | 2.56 | 0.007446 | −0.93 | 0.000140 | AT5G45310 | – |
| Msa.3093.1.S1_at | Desiccation-induced protein | Medtr4g063500 | 2 | 0 | Medtr_v1_024482 | 2.69 | 0.000739 | −2.28 | 3.44E-07 | AT1G07645 | – |
| Mtr.17894.1.S1_at | Unknown | Medtr1g014110 | 0 | 1 | Medtr_v1_004332 | 3.53 | 0.000135 | −1.09 | 0.001158 | AT4G31830 | – |
| Mtr.38008.1.S1_at | Phosphofructokinase | AC233669_8 | 2 | 0 | Medtr_v1_002457 | 1.35 | 0.048458 | −1.09 | 5.32E-05 | AT5G47810 | – |
| Msa.1254.1.S1_at | Unknown | – | – | – | – | – | – | – | – | – | – |
| Msa.900.1.S1_s_at | Unknown | – | – | – | – | – | – | – | – | – | – |
| Mtr.8651.1.S1_at | Dehydrin | – | – | – | – | – | – | – | – | – | – |
A previous study demonstrated that Arabidopsis ABI3 regulon promoters were enriched for RY element-like (RYL) and G-box-derived ABA-responsive element (GBL) motifs (Mönke et al., 2012). To investigate this, promoter sequences were isolated from the putative target genes of M. truncatula (Supplemental Table S9). Cis-regulatory analysis of the M. truncatula promoter sequences of the 29 predicted target genes revealed the presence of at least one RYL (i.e. CATGCA) or one GBL (i.e. ACGTGTC) motif (Table III).
To obtain further experimental evidence supporting our network, we performed gain-of-function and loss-of-function experiments of MtABI3. A Mtabi3 mutant was retrieved from the Tnt1 insertion mutant population, and differential gene expression between the mutant lines and wild-type seeds was determined using the Nimblegen slides at 32 DAP. This time point was chosen because gene expression of the target genes reached their highest expression level then (Fig. 3; Supplemental Tables S1 and S7). A total of 1,955 genes were significantly down-regulated in the Mtabi3 mutant (log2 < −1, P < 0.001; Supplemental Table S8). Out of the 40 M. truncatula putative targets identified via the GRN, 36 were present on the M. truncatula Nimblegen slides used to analyze the transcriptome (Table III). Four Affymetrix probe sets turned out to be duplicates and BLASTed to the same Nimblegen probe set, resulting in 32 Nimblegen probe sets that represented putative ABI3 targets. A total of 29 out of 32 probe sets (88%) were significantly down-regulated in the Mtabi3 mutant and represent direct or indirect target genes of MtABI3 (Table III). To distinguish direct gene induction from indirect, downstream responses in the mutant seeds, composite M. truncatula transgenic hairy roots were generated, transformed with the genomic sequence of the MtABI3 gene, and subsequently used for transcriptome analysis using the M. truncatula Nimblegen slides. This ectopic expression analysis of MtABI3 identified 368 up-regulated genes displaying statistically significant changes in expression (log2 > 1, P < 0.01) in comparison with control transformant lines containing only the GUS gene (Supplemental Table S8). Eighty-four probe sets were in common between mutant (down-regulated) and MtABI3 overexpression in the hairy roots (up-regulated). Out of the 32 predictive target genes present on the Nimblegen chip used for hybridization in this experiment, 25 genes (80%) displayed both a significant up-regulation of gene expression between transformed and control lines and a significant down-regulation between mutant and wild-type lines (Table III).
DISCUSSION
To prepare for the quiescent and dry state, developing seeds need to activate regulatory pathways to install protective mechanisms to confer DT and life span. With a detailed transcriptome analysis and gene regulatory network construction of developing M. truncatula seeds, we visualized the presence of several tightly coregulated modules of gene expression that coincided with specific developmental processes, such as embryogenesis, seed filling, acquisition of DT, longevity, and final maturation drying. The coexpression analysis of this network is based on the guilt-by-association paradigm, where genes under the control of a common transcriptional regulatory mechanism have a greater probability of being involved in the same biochemical or developmental pathways (Usadel et al., 2009; Lee et al., 2010; Bassel et al., 2011). Indeed, these distinct modules were enriched with different molecular functions. The acquisition of DT and dormancy module was enriched in stress response genes, such as LEA, HSP, and oxidative stress-related genes. This observation supports the hypothesis that during the course of evolution, seed maturation resulted from the acquisition of pathways associated with ABA-mediated vegetative responses to abiotic stress (Santos-Mendoza et al., 2008; Oliver et al., 2011).
We discovered that late maturation genes display a higher rate of delay of translation compared with genes coding for seed storage proteins, which appear earlier during maturation (Table I; Supplemental Fig. S2; Supplemental Table S5). This observation confirms and extends those of Gallardo et al. (2007), who suggested posttranscriptional regulation during maturation of M. truncatula seeds, evident from an increase in transcripts between 24 and 36 DAP without any increase in corresponding protein levels at 36 DAP. These authors suggested that these transcripts presumably contribute to the stored mRNA pool used for protein synthesis during germination. Here, we show that at least for a certain number of proteins, this translation still occurs after 36 DAP. Probe sets with GO terms related to RNA processing and translation were significantly more abundant during late seed maturation and associated with the longevity module. A group of genes that show such a long delay in translation are LEA genes, for which transcripts accumulate between 16 and 24 DAP, and corresponding proteins accumulate approximately 18 d later (Fig. 4; Supplemental Table S5). Indeed, posttranscriptional regulation has been suggested before for Em proteins in Arabidopsis seeds (Bies et al., 1998) and was experimentally demonstrated for a dehydrin via interaction with an RNA-binding protein (Li et al., 2002). Possibly, the enrichment of the posttranscriptional regulation during late seed maturation that is highlighted here might be linked to the anticipation by the seed tissues of fluctuating environmental conditions that accelerate or delay the time until abscission and final maturation drying, thus ensuring the timely production of LEA proteins to improve seed storability. Several lines of evidence suggest that LEA proteins are implicated in seed longevity. In Arabidopsis, the strong down-regulation of three seed-specific dehydrins resulted in a decrease in survival during storage (Hundertmark et al., 2011), whereas in M. truncatula, the four most abundant seed LEA proteins correlated with the increase in longevity during maturation (Chatelain et al., 2012).
The seed-specific coexpression network demonstrated that the transcriptional regulation of LEA genes was highly connected to MtABI3, MtABI4, and MtABI5 as well as an uncharacterized MtAP2/EREPB TF (Fig. 6D). ABI3 is a known regulator of the response to osmotic stress in vegetative tissues as well as DT in seeds (Ooms et al., 1993; Cutler et al., 2010; Hauser et al., 2011). The regulation of LEA and DT genes by MtABI3 was experimentally verified and revealed that 80% of the predicted MtABI3 regulons according to our GRN showed more than 2-fold change of expression in both hairy roots overexpressing MtABI3 in comparison with control roots and in Mtabi3 mutant seeds compared with wild-type seeds (Table III). These data highlight the robustness of our network as an exploratory tool to decipher pathways regulating the acquisition of DT and longevity. The topology of this maturation network shows that MtABI4 and MtABI5 genes also occupy a central position and are highly connected to LEA and DT genes. Thus, these genes together with MtABI3 and an uncharacterized MtAP2/EREPB TF belonging to the DREB-A2 family are predicted to be important core components of the seed-specific ABA signaling pathway that regulates survival in the dry state. Arabidopsis abi5 and abi4 monogenic mutants display extremely weak phenotypes during development and produce desiccation-tolerant seeds (Finkelstein, 1994). According to our network topology and their high connectivity, MtABI3, MtABI4, and MtABI5 together with the MtDREBA2 TFs probably function in an intricate combination of transcriptional interactions. ABI3 interacts with ABI5 to regulate the expression of downstream genes, whereas ABI4 controls the induction of ABI5 (Bossi et al., 2009; Cutler et al., 2010). Nearly half of the ABI4-specific targets were also underexpressed in abi5 mutant seeds and vice versa (Reeves et al., 2011). Also, among the 16 common targets of ABI4 and ABI5 resulting from their overexpression in seedlings, 10 were identified as LEA genes.
The putative ortholog of the Arabidopsis HSFA9 was found at the transition of the DT and longevity modules, connecting to both gene sets (Fig. 6B). This central position suggests that this MtHSF is an integrator at the interface between DT and longevity programs, reinforcing the unique and intriguing role of this seed-specific HSF in regulating seed maturation (Prieto-Dapena et al., 2006; Kotak et al., 2007; Almoguera et al., 2009). In Arabidopsis seeds, the expression of HSFA9 is controlled by ABI3 (Kotak et al., 2007). Consistent with these data, the M. truncatula HSFA9 ortholog is embedded into the LEA gene regulatory cluster that includes ABI3 (Fig. 6D). Whereas it is not known whether MtHSFA9 regulates LEA genes, overexpression of HSFA9 leads to an increase in sHSP (Prieto-Dapena et al., 2006). Many genes encoding MtHSPs are present in the acquisition of DT and dormancy module (i.e. MtHSP70, MtHSP101, MtHSP90, the small MtHSP18.2, MtHSP17.4, MtHSP22, MtHSP17.6I, and MtHSP17.6II; Fig. 3; Supplemental Table S2). In addition, transcript levels of the major HSP players, MtHSP70 and MtHSP90, also increase during seed maturation (Supplemental Table S2), suggesting that a tight regulation is needed to ensure protein homeostasis. The central position of the MtHSFA9 ortholog in the M. truncatula network concurs with data showing that ectopic expression of a sunflower HaHSFA9 in Arabidopsis leads both to increased seed longevity as determined by controlled deterioration and to tolerance of seedlings to severe dehydration (Prieto-Dapena et al., 2006). When HaHSFA9 was conjointly overexpressed with a HaDREB2 TF in tobacco, seed longevity was increased compared with HaHSFA9 alone (Almoguera et al., 2009). Interestingly, a putative ortholog of this DREB2 TF (MtAP2/EREBP Mtr.35724.1.S1_at) is part of the DT/LEA cluster (Fig. 6, B and C). It is interesting that weak abi3 alleles are desiccation tolerant but display reduced longevity, suggesting that some of the pathways of DT and longevity are overlapping (Ooms et al., 1993). Tejedor-Cano et al. (2010) observed that the loss-of-function mutant of HSFA9 did not lead to a decreased DT in tobacco seeds. This may not be surprising considering both the M. truncatula network topology and the multiple connections of the MtHSFA9 with several MtAP2/EREBPs.
A group of genes that were overrepresented in the acquisition of DT and dormancy module are related to the biosynthesis of GSH, such as a γ-glutamyl-Cys ligase that catalyzes the first and rate-limiting step and a γ-glutamyl-Cys synthetase (Noctor et al., 2012). In Arabidopsis, gsh1 mutants confer a recessive embryo-lethal phenotype (Cairns et al., 2006). These authors concluded that the autonomous synthesis of GSH in the embryo was necessary for proper seed maturation. Our network analysis extends this conclusion by suggesting that such lethality might be conferred by the absence of the acquisition of DT in these gsh1 embryos. This hypothesis is further supported by the recent metabolomic comparison of the desiccation-tolerant plant Sporobolus stapfianus with the desiccation-sensitive Sporobolus pyramidalis (Oliver et al., 2011). During drying, the desiccation-tolerant species exhibited an important increase in GSH and γ-glutamyl amino acids. Also in our M. truncatula network, the GSH metabolic process cluster contains a GSH peroxidase and a glutathione S-transferase, two enzymes involved in separate pathways implicated in the scavenging of hydrogen peroxide and peroxide via thioredoxins and GSH, respectively (Noctor et al., 2012). Thus, GSH might represent a key antioxidant involved in survival in the dry state. GSH half-cell reduction has been shown to be a marker for seed longevity (Kranner et al., 2006). The developmentally regulated increase in GSH content during the acquisition of DT might also serve later during seed development to enhance longevity.
Among the main metabolic events concomitant with the increase in longevity are the increase in stachyose (the most abundant RFO in M. truncatula) and the decrease in the precursors Suc and myoinositol (Fig. 5). A strong correlation (r2 = 0.91) was found between transcripts of a stachyose synthase gene (RFS/STS, Mtr.39847.1.S1_at) and the accumulation of stachyose (Fig. 5B). In pea seeds, levels of galactinol synthase and stachyose synthase activity are reflected by steady-state levels of corresponding mRNAs, and an increased stachyose synthase activity correlates with an increase in stachyose (Peterbauer et al., 2001). Further research will be needed to determine whether the stachyose synthase gene that correlates with stachyose is indeed regulating stachyose accumulation in M. truncatula. There is no evidence for a causal relationship between the protective properties of RFO and the acquisition of DT or longevity (Bentsink et al., 2000; Buitink et al., 2000; Gurusinghe and Bradford, 2001). Nonetheless, a correlation has been reported between the seed longevity and the ratio of Suc to RFO (Horbowicz and Obendorf, 1994; Sinniah et al., 1998), suggesting that RFO synthesis at the expense of Suc might be part of the same regulatory pathways leading to longevity. This is confirmed by the presence of the stachyose synthase gene (Mtr.39847.1.S1) in the longevity module as a first neighbor both of MtABI5 (Mtr.21588.1.S1_at) and MtAP2/EREBP (Mtr.35724.1.S1_at; Supplemental Table S7). However, to our knowledge, the role of MtABI5 or MtAP2/EREBP TFs in the regulation of RFO metabolism is unknown. Another gene that is present in the longevity module as a first neighbor of MtABI4, MtABI5, and MtAP2/EREPB (Mtr.35724.1.S1_at) is MtSNF4b (Mtr.42144.1.S1_at; Supplemental Table S7). Seeds of M. truncatula RNA interference lines deficient in MtSNF4b displayed a decrease of stachyose accumulation associated with a loss in longevity (Rosnoblet et al., 2007).
A significant switch in the transcriptome and metabolome occurs from 40 DAP onward, when seeds start their final maturation drying related to pod abscission (Fig. 2; Table II). A total of 4,602 and 4,485 probe sets, respectively, increase or decrease 2-fold in abundance between 40 DAP and mature dry seeds (48 DAP; Supplemental Table S1). Even between the point of abscission at 44 DAP, when the water content is already relatively low (less than 0.7 g g−1), and final maturation drying (at 48 DAP), 1,687 probe sets showed an increase in transcript levels. Many of these probe sets are represented in the central, tightly coregulated expression module of the GRN corresponding to the abscission submodule (Fig. 3). The precise role of these transcripts is unknown, but the GO enrichment study revealed an increased number of probe sets related to proteolysis via the ubiquitin pathway. The activation of proteolysis processes might indicate that proteins are degraded in a coordinated way during this period. According to our metabolomics data, amino acid contents increase sharply between 44 and 48 DAP (Table II). This increase has already been observed in Arabidopsis seeds (Fait et al., 2006), and we propose that protein degradation may serve to produce a pool of free amino acids that may be stored as part of the process to prepare germination.
MATERIALS AND METHODS
Plant Material and Physiological Analyses
Plants of Medicago truncatula (A17) were grown in a sterile mix of vermiculite and soil in a growth chamber at 24°C/21°C with a 16-h photoperiod at 200 µmol m−2 s−2. Flowers were tagged, and developing seeds were removed from the pods at different time intervals. DT, fresh and dry weights, as well as water content were determined according to Rosnoblet et al. (2007), and seed longevity data were derived from Chatelain et al. (2012). Data of the survival curves were probit transformed, and initial seed quality was calculated as the intercept according to Ellis and Roberts (1980). Speed of germination was determined by incubation of freshly harvested seeds in water at different time points and was calculated as the time to obtain 50% germination. For transcriptomic analysis using Affymetrix slides, three replicates of 50 seeds harvested from different plants were rapidly frozen in liquid nitrogen and stored at −80°C prior to RNA isolation.
A M. truncatula abi3 mutant (NF6003) was isolated from a Tnt1 insertional mutant population using a nested PCR approach (Cheng et al., 2011). Tnt1 insertion in the MtABI3 gene was confirmed by purification and sequencing of PCR products using ABI3 (i.e. ABI3-F, 5′-ATGGAGTGTGGTTTAGATCTGC-3′, and ABI3-R, 5′-TCATTTTTTTCTCTTTGGTGAAG-3′) and Tnt1 primer (5′-GCATTCAAACTAGAAGACAGTGCTACC-3′). Mutant plants were grown under controlled conditions as described above and backcrossed twice with wild-type plants to remove the undesirable mutant background.
Affymetrix Microarray, Data Extraction, and Normalization
Total RNA was extracted using a modified cetyltrimethylammonium bromide method (Verdier et al., 2008), and 10 μg of total RNA from each sample was DNase treated (Turbo DNase; Ambion) and purified (RNeasy MinElute Cleanup kit; Qiagen) according to the manufacturer’s instructions. RNA was quantified and evaluated for purity using an ND-1000 Nanodrop Spectrophotometer (NanoDrop Technologies) and a Bioanalyzer 2100 (Agilent). The Affymetrix M. truncatula GeneChip Array (Affymetrix) was used for expression analysis during seed development. RNA from three independent biological replicates was analyzed for each time point. Probe synthesis/labeling was carried out from 500 ng of RNA using the GeneChip 3′ IVT express kit, according to the manufacturer’s instructions (Affymetrix). Array hybridization, scanning, and data normalization were performed as described by Benedito et al. (2008). Each Cel file from the hybridized Affymetrix array was exported from GeneChip Operating Software version 1.4 (Affymetrix) and imported into Robust Multiarray Average Express (Irizarry et al., 2003) for global normalization. Presence/absence call for each probe set to remove background noise was obtained using dCHIP (Li and Wong, 2001). Raw microarray data were deposited at Array-Express (http://www.ebi.ac.uk/arrayexpress) as E-MEXP-3719.
Transcriptomic Data Analysis of Seed Development
To identify probe sets differentially regulated during seed development, coefficient of variation (CV) was calculated as follows: CV = (SD/mean), where SD is the sd of relative expression and mean is the average of the relative expression value for each probe set across all the studied developmental stages. Distance analysis between different developmental stages was calculated using pairwise Pearson correlation using R (version 2.15) and visualized using MultiExperiment Viewer (MeV version 4, part of the microarray software suite; Saeed et al., 2003). Principal component analysis was performed using median centering mode in MeV software, and axes 1 and 2 were selected to visualize results. PageMan software was used to perform overrepresentation analysis of functional classes using a bin-wise Wilcoxon test, and resulting P values were adjusted according to Bonferroni. A color scale was used to show overrepresented and underrepresented functional classes in red and blue, respectively (Usadel et al., 2009). To identify seed-related or seed-specific probe sets, a Z score was calculated using the following formula: Z = (X − mean)/SD, where X is the relative expression value, mean is the average of relative expression of tissues available in the MtGEA (i.e. leaf, root, petiole, stem, flower, nodule, and vegetative bud [http://mtgea.noble.org/]), and SD is the sd of relative expression in all tissues available in the MtGEA. Probe sets with a Z score above 2.66 in seeds were considered as seed specific or preferentially expressed in seeds.
Network Generation and Visualization
A gene regulatory network of seed development was constructed from the data set presented in Supplemental Table S1, but keeping only those probes with a minimum of relative expression of 100 and increasing the coefficient of variation to 40% (Supplemental Table S2). Weighted gene coexpression network analysis (Zhang and Horvath, 2005) was performed using the ArrayMining Web resource (http://www.arraymining.net) with edge adjacency threshold set to 0.5 using the one-step automatic network construction approach (Glaab et al., 2009). Text tables were imported into the open-source software Cytoscape (version 2.8.1) to generate CYS files (Smoot et al., 2011). Graphs were laid out using yFiles Organic layout after applying a cutoff to the PCC between genes of greater than 0.95. Probe sets of each module were analyzed for enrichment in GO terms using the Singular Enrichment Analysis tool of AgriGO with a χ2 statistical test method and the Yekutieli multitest adjustment method (Du et al., 2010). Data lists (Supplemental Table S2) were analyzed using the M. truncatula Affymetrix background.
Ectopic Expression of MtABI3 using Agrobacterium rhizogenes Transformation
Genomic DNA was isolated from 3-week-old leaves (R108 genotype) using the NucleoSpin Plant II kit (Macherey-Nagel). MtABI3 genomic DNA was amplified using the primers ABI3-TOPO-Fw (5′-CACCATGGAGTGTGGTTTA-3′) and ABI3-TOPO-Rv (5′-TCATTTTTTTCTCTTTGGTGA-3′) with Phusion polymerase (Thermo Scientific). The full-length fragment was cloned into the binary vector pK7GW2D (Karimi et al., 2002) containing a 35S cauliflower mosaic virus promoter and GFP reporter that was used as transformation marker. Control (plasmid containing the GUS gene) and construct containing the MtABI3 gene were introduced in the ARqua1 strain of A. rhizogenes (Quandt et al., 1993). M. truncatula R108 seeds were sterilized, grown, and transformed according to Boisson-Dernier et al. (2001). Cotransformed roots were selected based on the presence of GFP marker using a fluorescence stereomicroscope after 4 weeks of culture. Roots were immediately frozen in liquid nitrogen for conservation. For each construct, three independent biological replicates were collected. Verification of MtABI3 overexpression was performed by reverse-transcription PCR according to Bolingue et al. (2010) using internal primers 5′-TCAAAATGAAGATGAAAACCCTT-3′ and 5′-TCTTTGTTCGATTTAAGCCACTC-3′.
Nimblegen Microarray, Data Extraction, and Normalization
Custom-designed M. truncatula Nimblegen arrays were designed based on the M. truncatula genome (Young et al., 2011) and containing sequences of the IMGAG Mt3.5.1 version as well as additional genomic sequences (F. Debellé, P. Gamas, and J. Gouzy, personal communication). High-density microarray slides containing 119,780 quality-filtered unigenes were designed and produced by Nimblegen. Total RNA of transgenic roots and wild-type and Mtabi3 seed tissues was extracted using the nucleospin RNAplant kit (Macherey-Nagel), and 400 ng was amplified using the Ambion messageAmp II following the manufacturer’s instructions. Five micrograms of amplified RNA was retrotranscribed with 400 units of SuperScript II reverse transcriptase (Invitrogen), labeled with 1.5 mmol of Cyanine-3 or Cyanine-5 (Interchim), and then purified using NucleoSpin Gel and PCR Clean-up column kits (Macherey-Nagel). Purified and labeled complementary DNA was quantified using NanoDrop ND-1000. Labeled samples (30 pmol) of 35S::GUS (control) roots were mixed with 30 pmol of labeled samples of 35S::ABI3 roots and cohybridized to the Medtr_v1.0 12x135K arrays. Likewise, labeled samples of wild-type (R108) seeds at 32 DAP were mixed with labeled samples of the Mtabi3 mutant at 32 DAP and cohybridized. Three biological replicates were analyzed per comparison using dye switch as described by Depuydt et al. (2009). Hybridizations were performed on a NimbleGen Hybridization System 4 (mix mode B) at 42°C overnight. Slides were washed, dried, and scanned at 2-μm resolution and high sensitivity with a Roche-NimbleGen MS200. NimbleScan Software version 2.4 was used to extract pair data files from the scanned images. Statistical analyses of the gene expression data were performed using the R language, version 2.5.1, and the Linear Models for Microarray Analysis package (Smyth et al., 2005) from the Bioconductor project. For the preprocessing step, data were normalized by the lowess method. Log ratio and log intensity were calculated before differential expression analyses were performed using the lmFit function and the Bayes-moderated Student’s t test in Linear Models for Microarray Analysis. Probes with P < 0.001 were considered as differentially expressed. Nimblegen microarray data were deposited at Gene Expression Omnibus (GSE44291).
Metabolite Analysis
Polar and nonpolar metabolite extractions were performed according to Broeckling et al. (2005). Three replicates of developing seeds (approximately 150–200 seeds) were ground in liquid nitrogen and lyophilized overnight using a freeze dryer. Six milligrams of dry tissues was ground with 1.5 mL of chloroform containing docosanol (as a nonpolar standard) and 1.5 mL of HPLC-grade water containing ribitol (as a polar internal standard). The mixture was centrifuged at 2,900g for 30 min at 4°C, and 1 mL of the separated polar and nonpolar layers were collected and dried under vacuum and nitrogen, respectively. Nonpolar samples were hydrolyzed using 1.25 m HCl and dried. Then, both polar and nonpolar samples were treated with methoxyamine-HCl in pyridine and derivatized with N-methyl-N-(trimethylsilyl)trifluoroacetamide + 1% trimethyl chlorosilane (Thermo Scientific). Samples were analyzed using an Agilent 6890 gas chromatograph coupled to a 5973 mass spectrophotometer detector (http://www.chem.agilent.com/). Different compounds were identified by Automated Mass Spectral Deconvolution and Identification System software using commercial and in-house libraries and quantified using MET-IDEA software (Broeckling et al., 2006).
Sugar Determination
Seeds at different developmental stages were harvested and directly frozen into liquid nitrogen. Sugar were extracted and analyzed by HPLC on a Carbopac PA-1 column (Dionex) as described by Rosnoblet et al. (2007). Three independent extractions and assays were performed from samples of 25 seeds.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. GO enrichment analysis of probe sets corresponding to the acquisition of DT and dormancy module using AgriGO.
Supplemental Figure S2. GO enrichment analysis of probe sets corresponding to the FMD module using AgriGO.
Supplemental Figure S3. GO enrichment analysis of probe sets correlating with longevity.
Supplemental Figure S4. Transcript profiles of 47 genes correlating with longevity.
Supplemental Figure S5. Hierarchical clustering of seed-specific TFs throughout the different stages of seed development.
Supplemental Table S1. List of 19,012 probe sets differentially expressed during seed development.
Supplemental Table S2. Classification of probe sets in the different modules of the weighted coexpression gene network.
Supplemental Table S3. List of 794 probe sets showing seed-specific expression or preferentially expressed in seeds.
Supplemental Table S4. Genes showing an expression profile positively or negatively correlated (PCC > 0.9 or PCC < −0.9) with the acquisition of longevity (P50) during seed maturation of M. truncatula.
Supplemental Table S5. GO enrichment analysis of probe sets positively or negatively correlated with longevity (P50).
Supplemental Table S6. List of metabolites identified from polar and nonpolar seed fractions using GC-MS.
Supplemental Table S7. PCC values between the 22 seed-specific TFs expressed during seed late maturation and 794 seed-specific probe sets.
Supplemental Table S8. List of probe sets showing a statistically significant increase in transcript level of at least 2-fold between hairy roots transformed with the MtABI3 genomic DNA in comparison with control lines containing only the GUS gene and/or significantly decreased transcript levels in the Mtabi3 mutant at 32 DAP compared with wild-type seeds.
Supplemental Table S9. Promoter sequences (up to 2 kb) of the predicted MtABI3 regulons presented in Table III.
Acknowledgments
We thank Yuhong Tang and Stacy Allen for assistance with Affymetrix data analysis and Mohamed Bedair and Lloyd Sumner for GC-MS analysis.
Glossary
- DT
desiccation tolerance
- LEA
late embryogenesis abundant
- HSP
heat shock protein
- DAP
days after pollination
- P50
time until 50% of the seed batch loses its viability during controlled aging
- PCC
Pearson correlation coefficient
- GRN
gene regulatory network
- FMD
final maturation drying
- MtGEA
M. truncatula Gene Expression Atlas
- GO
Gene Ontology
- sHSP
small heat shock protein
- GC-MS
gas chromatography coupled to mass spectrometry
- RFO
raffinose family oligosaccharides
- TF
transcription factor
- RYL
RY element-like
- GBL
G-box-derived ABA-responsive element
- ABA
abscisic acid
- GSH
glutathione
References
- Almoguera C, Prieto-Dapena P, Díaz-Martín J, Espinosa JM, Carranco R, Jordano J. (2009) The HaDREB2 transcription factor enhances basal thermotolerance and longevity of seeds through functional interaction with HaHSFA9. BMC Plant Biol 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelovici R, Galili G, Fernie AR, Fait A. (2010) Seed desiccation: a bridge between maturation and germination. Trends Plant Sci 15: 211–218 [DOI] [PubMed] [Google Scholar]
- Arrigoni O, De Gara L, Tommasi F, Liso R. (1992) Changes in the ascorbate system during seed development of Vicia faba L. Plant Physiol 99: 235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel GW, Glaab E, Marquez J, Holdsworth MJ, Bacardit J. (2011) Functional network construction in Arabidopsis using rule-based machine learning on large-scale data sets. Plant Cell 23: 3101–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel GW, Lan H, Glaab E, Gibbs DJ, Gerjets T, Krasnogor N, Bonner AJ, Holdsworth MJ, Provart NJ. (2011) Genome-wide network model capturing seed germination reveals coordinated regulation of plant cellular phase transitions. Proc Natl Acad Sci USA 108: 9709–9714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, et al. (2008) A gene expression atlas of the model legume Medicago truncatula. Plant J 55: 504–513 [DOI] [PubMed] [Google Scholar]
- Bentsink L, Alonso-Blanco C, Vreugdenhil D, Tesnier K, Groot SPC, Koornneef M. (2000) Genetic analysis of seed-soluble oligosaccharides in relation to seed storability of Arabidopsis. Plant Physiol 124: 1595–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bies N, Aspart L, Carles C, Gallois P, Delseny M. (1998) Accumulation and degradation of Em proteins in Arabidopsis thaliana: evidence for post-transcriptional controls. J Exp Bot 49: 1925–1933 [Google Scholar]
- Boisson-Dernier A, Chabaud M, Garcia F, Bécard G, Rosenberg C, Barker DG. (2001) Hairy roots of Medicago truncatula as tools for studying nitrogen-fixing and endomycorrhizal symbioses. Mol Plant Microbe Interact 14: 693–700 [DOI] [PubMed] [Google Scholar]
- Bolingue W, Rosnoblet C, Ly Vu B, Leprince O, Aubry C, Buitink J. (2010) The MtSNF4b subunit of the sucrose non-fermenting-related kinase complex connects after-ripening and constitutive defense responses in seeds of Medicago truncatula. Plant J 61: 792–803 [DOI] [PubMed] [Google Scholar]
- Bossi F, Cordoba E, Dupré P, Mendoza MS, Román CS, León P. (2009) The Arabidopsis ABA-INSENSITIVE (ABI) 4 factor acts as a central transcription activator of the expression of its own gene, and for the induction of ABI5 and SBE2.2 genes during sugar signaling. Plant J 59: 359–374 [DOI] [PubMed] [Google Scholar]
- Broeckling CD, Huhman DV, Farag MA, Smith JT, May GD, Mendes P, Dixon RA, Sumner LW. (2005) Metabolic profiling of Medicago truncatula cell cultures reveals the effects of biotic and abiotic elicitors on metabolism. J Exp Bot 56: 323–336 [DOI] [PubMed] [Google Scholar]
- Broeckling CD, Reddy IR, Duran AL, Zhao XJ, Sumner LW. (2006) MET-IDEA: data extraction tool for mass spectrometry-based metabolomics. Anal Chem 78: 4334–4341 [DOI] [PubMed] [Google Scholar]
- Buitink J, Hemminga MA, Hoekstra FA. (2000) Is there a role for oligosaccharides in seed longevity? An assessment of intracellular glass stability. Plant Physiol 122: 1217–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buitink J, Leger JJ, Guisle I, Vu BL, Wuillème S, Lamirault G, Le Bars A, Le Meur N, Becker A, Küster H, et al. (2006) Transcriptome profiling uncovers metabolic and regulatory processes occurring during the transition from desiccation-sensitive to desiccation-tolerant stages in Medicago truncatula seeds. Plant J 47: 735–750 [DOI] [PubMed] [Google Scholar]
- Cairns NG, Pasternak M, Wachter A, Cobbett CS, Meyer AJ. (2006) Maturation of Arabidopsis seeds is dependent on glutathione biosynthesis within the embryo. Plant Physiol 141: 446–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatelain E, Hundertmark M, Leprince O, Le Gall S, Satour P, Deligny-Penninck S, Rogniaux H, Buitink J. (2012) Temporal profiling of the heat-stable proteome during late maturation of Medicago truncatula seeds identifies a restricted subset of late embryogenesis abundant proteins associated with longevity. Plant Cell Environ 35: 1440–1455 [DOI] [PubMed] [Google Scholar]
- Cheng X, Wen J, Tadege M, Ratet P, Mysore KS. (2011) Reverse genetics in Medicago truncatula using Tnt1 insertion mutants. Methods Mol Biol 678: 179–190 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Léon-Kloosterziel KM, Koornneef M. (2000) Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol 122: 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depuydt S, Trenkamp S, Fernie AR, Elftieh S, Renou JP, Vuylsteke M, Holsters M, Vereecke D. (2009) An integrated genomics approach to define niche establishment by Rhodococcus fascians. Plant Physiol 149: 1366–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdle J, Ishikawa T, Gatzek S, Rolinski S, Smirnoff N. (2007) Two genes in Arabidopsis thaliana encoding GDP-L-galactose phosphorylase are required for ascorbate biosynthesis and seedling viability. Plant J 52: 673–689 [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang ZH, Su Z. (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RH, Roberts EH. (1980) Improved equations for the prediction of seed longevity. Ann Bot (Lond) 45: 13–30 [Google Scholar]
- Fait A, Angelovici R, Less H, Ohad I, Urbanczyk-Wochniak E, Fernie AR, Galili G. (2006) Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol 142: 839–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR. (1994) Mutations at 2 new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J 5: 765–771 [Google Scholar]
- Fruchterman T, Reingold E. (1991) Graph drawing by force directed placement. Software Practice and Experience 21: 1129–1164 [Google Scholar]
- Gallardo K, Firnhaber C, Zuber H, Héricher D, Belghazi M, Henry C, Küster H, Thompson R. (2007) A combined proteome and transcriptome analysis of developing Medicago truncatula seeds: evidence for metabolic specialization of maternal and filial tissues. Mol Cell Proteomics 6: 2165–2179 [DOI] [PubMed] [Google Scholar]
- Glaab E, Garibaldi JM, Krasnogor N. (2009) ArrayMining: a modular Web-application for microarray analysis combining ensemble and consensus methods with cross-study normalization. BMC Bioinformatics 10: 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurusinghe S, Bradford KJ. (2001) Galactosyl-sucrose oligosaccharides and potential longevity of primed seeds. Seed Sci Res 11: 121–133 [Google Scholar]
- Hauser F, Waadt R, Schroeder JI. (2011) Evolution of abscisic acid synthesis and signaling mechanisms. Curr Biol 21: R346–R355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay FR, Probert RJ. (1995) Seed maturity and the effects of different drying conditions on desiccation tolerance and seed longevity in foxglove (Digitalis purpurea L.). Ann Bot (Lond) 76: 639–647 [Google Scholar]
- Hoekstra FA, Golovina EA, Buitink J. (2001) Mechanisms of plant desiccation tolerance. Trends Plant Sci 6: 431–438 [DOI] [PubMed] [Google Scholar]
- Horbowicz M, Obendorf RL. (1994) Seed desiccation tolerance and storability: dependence on flatulence-producing oligosaccharides and cyclitols. Review and survey. Seed Sci Res 4: 385–405 [Google Scholar]
- Hundertmark M, Buitink J, Leprince O, Hincha DK. (2011) The reduction of seed-specific dehydrins reduces seed longevity in Arabidopsis thaliana. Seed Sci Res 21: 165–173 [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kotak S, Vierling E, Bäumlein H, von Koskull-Döring P. (2007) A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. Plant Cell 19: 182–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranner I, Birtić S, Anderson KM, Pritchard HW. (2006) Glutathione half-cell reduction potential: a universal stress marker and modulator of programmed cell death? Free Radic Biol Med 40: 2155–2165 [DOI] [PubMed] [Google Scholar]
- Le BH, Cheng C, Bui AQ, Wagmaister JA, Henry KF, Pelletier J, Kwong L, Belmonte M, Kirkbride R, Horvath S, et al. (2010) Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA 107: 8063–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Ambaru B, Thakkar P, Marcotte EM, Rhee SY. (2010) Rational association of genes with traits using a genome-scale gene network for Arabidopsis thaliana. Nat Biotechnol 28: 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH. (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 98: 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Kinoshita T, Pandey S, Ng CK-Y, Gygi SP, Shimazaki K-I, Assmann SM. (2002) Modulation of an RNA-binding protein by abscisic-acid-activated protein kinase. Nature 418: 793–797 [DOI] [PubMed] [Google Scholar]
- Mönke G, Seifert M, Keilwagen J, Mohr M, Grosse I, Hähnel U, Junker A, Weisshaar B, Conrad U, Bäumlein H, et al. (2012) Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon. Nucleic Acids Res 40: 8240–8254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Mhamdi A, Chaouch S, Han Y, Neukermans J, Marquez-Garcia B, Queval G, Foyer CH. (2012) Glutathione in plants: an integrated overview. Plant Cell Environ 35: 454–484 [DOI] [PubMed] [Google Scholar]
- Ogé L, Bourdais G, Bove J, Collet B, Godin B, Granier F, Boutin JP, Job D, Jullien M, Grappin P. (2008) Protein repair L-isoaspartyl methyltransferase 1 is involved in both seed longevity and germination vigor in Arabidopsis. Plant Cell 20: 3022–3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver MJ, Guo L, Alexander DC, Ryals JA, Wone BWM, Cushman JC. (2011) A sister group contrast using untargeted global metabolomic analysis delineates the biochemical regulation underlying desiccation tolerance in Sporobolus stapfianus. Plant Cell 23: 1231–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms JJJ, Leon-Kloosterziel KM, Bartels D, Koornneef M, Karssen CM. (1993) Acquisition of desiccation tolerance and longevity in seeds of Arabidopsis thaliana: a comparative study using abscisic acid-insensitive abi3 mutants. Plant Physiol 102: 1185–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng FY, Weselake RJ. (2011) Gene coexpression clusters and putative regulatory elements underlying seed storage reserve accumulation in Arabidopsis. BMC Genomics 12: 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterbauer T, Lahuta LB, Blöchl A, Mucha J, Jones DA, Hedley CL, Gòrecki RJ, Richter A. (2001) Analysis of the raffinose family oligosaccharide pathway in pea seeds with contrasting carbohydrate composition. Plant Physiol 127: 1764–1772 [PMC free article] [PubMed] [Google Scholar]
- Prieto-Dapena P, Castaño R, Almoguera C, Jordano J. (2006) Improved resistance to controlled deterioration in transgenic seeds. Plant Physiol 142: 1102–111216998084 [Google Scholar]
- Probert R, Adams J, Coneybeer J, Crawford A, Hay F. (2007) Seed quality for conservation is critically affected by pre-storage factors. Aust J Bot 55: 326–335 [Google Scholar]
- Quandt HJ, Pühler A, Broer I. (1993) Transgenic root nodules of Vicia hirsutea: a fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol Plant Microbe Interact 6: 699–706 [Google Scholar]
- Reeves WM, Lynch TJ, Mobin R, Finkelstein RR. (2011) Direct targets of the transcription factors ABA-Insensitive(ABI)4 and ABI5 reveal synergistic action by ABI4 and several bZIP ABA response factors. Plant Mol Biol 75: 347–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosnoblet C, Aubry C, Leprince O, Vu BL, Rogniaux H, Buitink J. (2007) The regulatory gamma subunit SNF4b of the sucrose non-fermenting-related kinase complex is involved in longevity and stachyose accumulation during maturation of Medicago truncatula seeds. Plant J 51: 47–59 [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Sanhewe AJ, Ellis RH. (1996) Seed development and maturation in Phaseolus vulgaris. 2. Post-harvest longevity in air-dry storage. J Exp Bot 47: 959–965 [Google Scholar]
- Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L. (2008) Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J 54: 608–620 [DOI] [PubMed] [Google Scholar]
- Sattler SE, Gilliland LU, Magallanes-Lundback M, Pollard M, DellaPenna D. (2004) Vitamin E is essential for seed longevity and for preventing lipid peroxidation during germination. Plant Cell 16: 1419–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Miller J, Mudgett MB, Schopf JW, Clarke S, Berger R. (1995) Exceptional seed longevity and robust growth: ancient sacred lotus from China. Am J Bot 82: 1367–1380 [Google Scholar]
- Sinniah UR, Ellis RH, John P. (1998) Irrigation and seed quality development in rapid-cycling Brassica: soluble carbohydrates and heat-stable proteins. Ann Bot (Lond) 82: 647–655 [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27: 431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK, Michaud J, Scott HS. (2005) Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21: 2067–2075 [DOI] [PubMed] [Google Scholar]
- Sugliani M, Rajjou L, Clerkx EJM, Koornneef M, Soppe WJJ. (2009) Natural modifiers of seed longevity in the Arabidopsis mutants abscisic acid insensitive3-5 (abi3-5) and leafy cotyledon1-3 (lec1-3). New Phytol 184: 898–908 [DOI] [PubMed] [Google Scholar]
- Tejedor-Cano J, Prieto-Dapena P, Almoguera C, Carranco R, Hiratsu K, Ohme-Takagi M, Jordano J. (2010) Loss of function of the HSFA9 seed longevity program. Plant Cell Environ 33: 1408–1417 [DOI] [PubMed] [Google Scholar]
- Usadel B, Obayashi T, Mutwil M, Giorgi FM, Bassel GW, Tanimoto M, Chow A, Steinhauser D, Persson S, Provart NJ. (2009) Co-expression tools for plant biology: opportunities for hypothesis generation and caveats. Plant Cell Environ 32: 1633–1651 [DOI] [PubMed] [Google Scholar]
- Verdier J, Kakar K, Gallardo K, Le Signor C, Aubert G, Schlereth A, Town CD, Udvardi MK, Thompson RD. (2008) Gene expression profiling of M. truncatula transcription factors identifies putative regulators of grain legume seed filling. Plant Mol Biol 67: 567–580 [DOI] [PubMed] [Google Scholar]
- Walters C, Wheeler LM, Grotenhuis JM. (2005) Longevity of seeds stored in a genebank: species characteristics. Seed Sci Res 15: 1–20 [Google Scholar]
- Waterworth WM, Masnavi G, Bhardwaj RM, Jiang Q, Bray CM, West CE. (2010) A plant DNA ligase is an important determinant of seed longevity. Plant J 63: 848–860 [DOI] [PubMed] [Google Scholar]
- Wolucka BA, Van Montagu M. (2007) The VTC2 cycle and the de novo biosynthesis pathways for vitamin C in plants: an opinion. Phytochemistry 68: 2602–2613 [DOI] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GE, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KFX, Gouzy J, Schoof H, et al. (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480: 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. (2005) A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 4: Article 17 [DOI] [PubMed] [Google Scholar]