Abstract
Rapid development of broad regional and international DNA barcode libraries have brought new insights into the species diversity of many areas and groups. Many new species, even within well-investigated species groups, have been discovered based initially on differences in DNA barcodes. We barcoded 437 collection specimens belonging to 40 pre-identified Palearctic species of the Elachista bifasciella group of moths (Lepidoptera, Elachistidae). Although the study group has been a subject of several careful morphological taxonomic examinations, an unexpectedly high number of previously undetected putative species is revealed, resulting in a 34% rise in species number in the study area. The validity of putative new species was subsequently supported with diagnostic morphological traits. We show that DNA barcodes provide a powerful method of detecting potential new species even in taxonomic groups and geographic areas that have previously been under considerable morphological taxonomic scrutiny.
Estimates of the number of species on Earth vary from 3 to 100 million, the most recent survey concluding that there are about 8.7 million (±1.3 million SE) species based on a quantitative extrapolation of current taxonomic knowledge1. About 1.1 million of the thus far described ~1.9 million eukaryotic species are arthropods, predominantly insects2, and the vast majority of arthropod species remains to be described. The huge number of species on Earth and the shortage of taxonomic expertise attending to them have led to the predicament often called the ‘taxonomic crisis’ or ‘taxonomic impediment’3,4. It has been suggested that, by accelerating species discovery and making species delimitation more straightforward, DNA based tools might be powerful in overcoming the crisis5,6.
Animal DNA barcode is a short, standardized fragment of the mitochondrial COI gene designed to enable the rapid accurate identification of species and to accelerate species discovery5. Similar barcodes, but from different regions of the genome, have been developed both for fungi7 and plants8. COI has proven efficient in species identification in many species-rich animal groups9,10, revealing cryptic diversity11,12, while in others the results have been less promising, with even a large portion of species showing interspecific overlap with closely related species13,14. DNA barcodes have also been criticized for the shortcomings of the commonly used methodology15 and potential problems caused by anomalies of mitochondrial DNA16,17.
Several factors may have caused considerable biases in estimations of DNA barcode performance. First, in most investigations of species-rich animal groups only a fraction of the species concerned have been sampled. Second, most studies have not examined specimens exhaustively across the distribution ranges, thereby underestimating the level of geography-related intraspecific variability. Third, the sample sizes within populations have often been small, giving an unreliable picture of the distinction between species. While all these factors are likely to produce overoptimistic estimations of the power of barcodes in species identification, other factors may have biased other estimations in the reverse direction. Many papers have examined the power of the barcoding concept in cases which are known a priori to be especially difficult (e.g.18,19). In such cases, barcodes are often compared to the existing morphology-based taxonomy, either explicitly or implicitly considered to represent the “true taxonomy”. A mismatch between barcodes and existing species delineation has been suggested as providing evidence that barcodes do not perform well. Some other studies examining the utility of barcodes have been based largely on data uploaded from GenBank, which is known to include a high degree of misidentifications20,21. Some of those investigations are among the ones that have presented the lowest identification success and the highest rates of intraspecific variability of barcodes13,19,22.
Tests of the efficacy of barcodes in species identification always reflect the opinions of investigators (or the taxonomists behind the reference taxonomy) and are prone to some subjectivity, at least in the treatment of allopatric populations (cf.23), and to a variety of existing species definitions. Therefore, even identical data sets might easily lead to different conclusions. An example of this is the Astraptes fulgerator complex of skipper species, in which the same data led different investigators to deviant estimations of the number of species involved11,24. We recognize that barcodes have their limitations, as differences in the mitochondrial DNA sequences are not the cause, but rather a result, of a speciation event, and the formation of discrete haplotype clusters between the species generally requires time. Thus, the existence of even considerably different haplotypes per se is not sufficient evidence for basing taxonomic decisions25. Young species are even unlikely to be differentiated by barcoding alone, as it can be suggested that they share more or less the same haplotype pool as the ancestral taxon. Thus, in addition to a discrete barcode, a taxon can only be recognized as a distinct species with the support of other evidence, be that molecular (e.g. nuclear markers), morphological, ecological, or the proven absence of successful interbreeding as shown by the absence of panmictism. Still, there is mounting evidence that generally, at least on a local scale, different, established species usually possess their own cluster of barcode haplotypes22,23,24,25,26,27,28,29,30,31.
Wide adoption of molecular techniques in taxonomic research has significantly increased the recognition of cryptic species, i.e., species that have remained previously undetected because of close relatedness and morphological similarity32. Many other species first detected by DNA differences have retrospectively been shown to be morphologically distinguishable, representing cases of cryptic or morphologically closely similar species33,34,35,36,37,38,39,40,41,42 (whether a species is similar enough to be ‘cryptic’ or not is a fine line). A remarkable amount of DNA barcode data has been generated during recent years in association with the campaigns of the International Barcode of Life Project (iBOL, http://ibol.org/). While it is evident that massive barcoding campaigns of regional biotas will further accelerate species discoveries especially in poorly investigated areas and groups, several case studies have demonstrated unexpected discoveries among characteristic species and in well-known regions34,36,37.
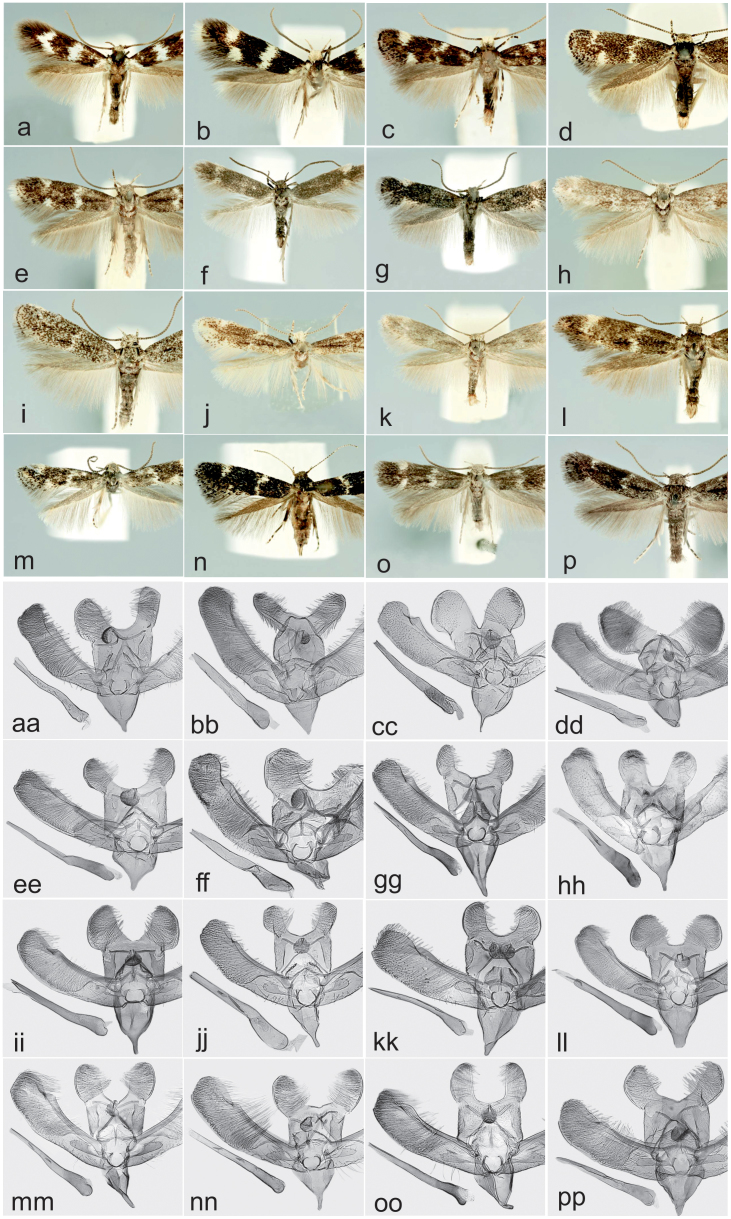
We used moths of the genus Elachista (Lepidoptera: Gelechioidea: Elachistidae) as an exemplar group. The genus worldwide comprises approximately 700 named species43. We targeted the Elachista bifasciella group of the Palearctic region. These are difficult to identify. This is because of the large number of species (130 species described worldwide), small size (wingspan typically 5–12 mm), drab coloration, generally little differentiation in external morphology, and subtle differences in genitalia (Figure 1, where a maximum of variation in appearance and morphology within the group is shown). They have nevertheless been the subject of thorough morphology-based taxonomic investigations during the past decades44,45,46,47,48,49,50,51,52,53,54,55,56 covering the fauna of the whole Palearctic region, although the European part of the region is evidently better investigated than that of other areas. As species of E. bifasciella group show close similarity in their morphology, an assumption that the species have diverged quite recently seems plausible. DNA barcodes are expected to show low resolution in such circumstances, because loci of young species may not have had sufficient time to fully diverge (often called incomplete lineage sorting) and may have been subjected to mitochondrial introgression57.
Figure 1. Habitus and male genitalia of selected N. European species of Elachista bifasciella group (habitus in comparable scale).
(a), aa: E. bifasciella; (b), bb: E. dimicatella; (c), cc: E. albifrontella; (d), dd: E. griseella; (e), ee: E. kilmunella; (f), ff: E. irenae; (g), gg: E. humilis; (h), hh: E. herrichii; (i), ii: E. orstadii; (j), jj: E. canapennella; (k), kk: E. krogeri; (l), ll: E. atricomella; (m), mm: E. deriventa; (n), nn: E. elegans; (o), oo: E. nielswolffi; (p), pp: E. poae. The photographs were taken by LK.
We examined the performance of COI barcodes in differentiating morphologically similar and allegedly young species of the Palearctic bifasciella group. We also examined whether broad sampling of collection specimens would reveal genetically distinct lineages, potentially representing new species. We went still further and examined whether these genetically distinct lineages bear unique morphological characteristics that would further support their validity as new species. While large-scale analyses of DNA barcodes have often documented intraspecific genetic splits10,30,31,58, few of them have been extended to systematically examine whether such lineages represent entities that gain integral support by independent lines of evidence, such as nuclear DNA, morphology or life-history traits, although there are notable exceptions30,59,60). We used morphology-based research of the group as a backbone hypothesis, without making presumptions as to whether they are correct in all details.
Results
The original analysis of data revealed several species having considerable (>1%) intraspecific variation and likewise clusters of unidentified specimens showing a distinct gap (>1%) from any pre-identified specimens. Altogether, twenty-five such cases were detected. All these were subjected to an in-depth morphological examination. This resulted in the detection of sixteen putative new species whose species integrity was independently supported by genetic and morphological uniqueness. Eight of these species were from Russia, three from Kyrgyzstan, two from Greece and one each from Slovenia, Turkey and Ukraine (Figure S1). Formal descriptions of these taxa have been prepared and will be published in another context (L. Kaila, in prep). Detailed morphological diagnoses for the new species will be provided in a forthcoming paper. A summary of key diagnostic features with regards of all other species is presented in Table S1. In a few cases of species showing a remarkable (>1%) intraspecific split, notably E. maculicerusella and E. canapennella, the morphological examination failed to support the presence of multiple distinct lineages.
The mean of minimum K2P distances between the genetically closest species, with both described and newly discovered putative species included, was 3.82% (range 0%–7.41%) (Figure 2, Table 1). The same measure was 3.98% (range 0–7.41%) and 3.42% (range 1.08–6.9%) in the described species and putative new species, respectively. The maximum intraspecific variation across species (singletons excluded, newly discovered putative species included) ranged from 0% to 3.76%, with an average across the species of 0.74% in described species (35 species) and 0.61% with newly discovered species included (45 species). The maximum intraspecific variation exceeded 1% in 10 species and 2% in 4 species.
Figure 2. A Neighbor-Joining tree, generated under the K2P nucleotide substitution model, of the study taxa.
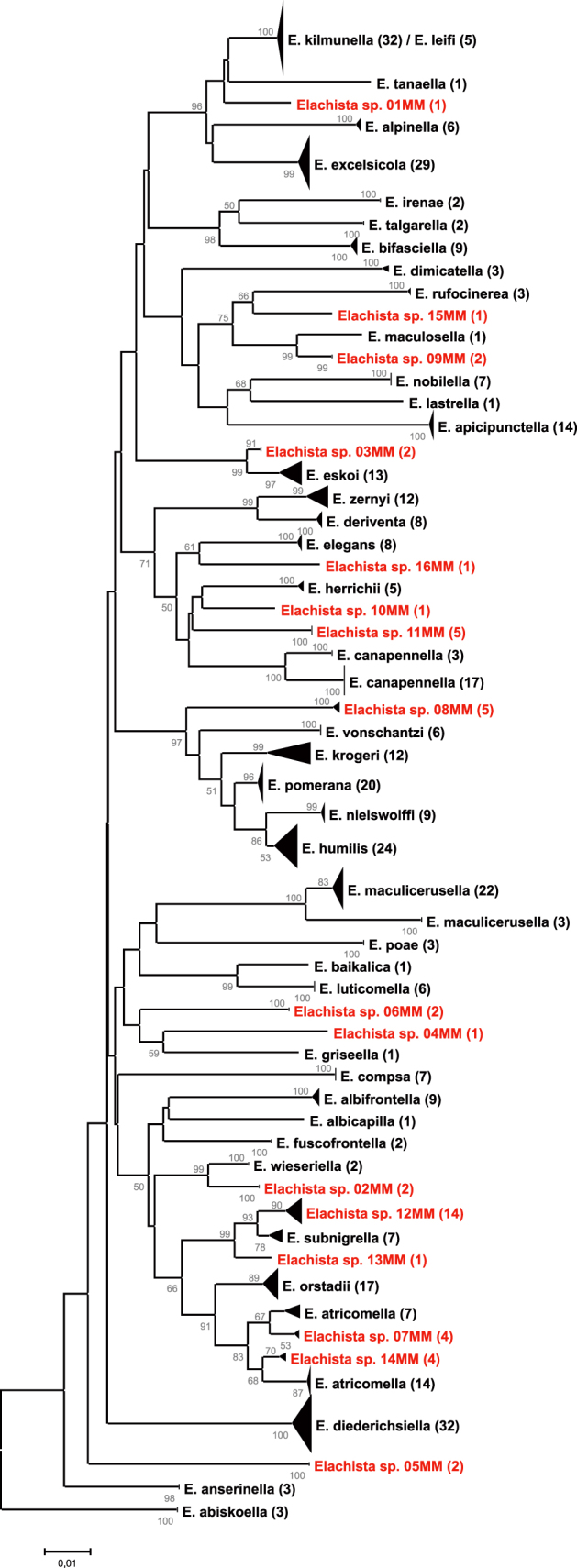
Node bootstrap support values of >50% are indicated. Sample size for each species is indicated in parentheses after species names. The height of terminal triangles is proportional to sample size, the depth to degree of intraspecific variation. Deep (>2%) intraspecific splits are shown separated. Newly discovered species are highlighted in red.
Table 1. Distance statistics (numbers in per cents, implemented under K2P nucleotide substitution model) of Elachista bifasciella group of species with observed putative species included in the comparison.
| Species | Mean intraspecific | Max intraspecific | Nearest species | Min distance to Nearest Species |
|---|---|---|---|---|
| Elachista albicapilla | N/A | N/A | Elachista orstadii | 4.53 |
| Elachista albifrontella | 0.2 | 0.31 | Elachista orstadii | 5.59 |
| Elachista alpinella | 0.1 | 0.32 | Elachista sp. 01 MM | 4.45 |
| Elachista anserinella | 0 | 0 | Elachista orstadii | 4.52 |
| Elachista apicipunctella | 0.02 | 0.15 | Elachista sp. 09 MM | 7.23 |
| Elachista atricomella | 1.2 | 2.7 | Elachista sp. 07 MM | 1.08 |
| Elachista baikalica | N/A | N/A | Elachista luticomella | 3.15 |
| Elachista bifasciella | 0.14 | 0.32 | Elachista talgarella | 5.85 |
| Elachista canapennella | 0.63 | 2.41 | Elachista elegans | 5.4 |
| Elachista compsa | 0 | 0 | Elachista pomerana | 6.94 |
| Elachista deriventa | 0.12 | 0.32 | Elachista zernyi | 2.13 |
| Elachista diederichsiella | 0.47 | 1.4 | Elachista orstadii | 5.65 |
| Elachista dimicatella | 0.21 | 0.24 | Elachista nobilella | 6.75 |
| Elachista elegans | 0.07 | 0.15 | Elachista sp. 16 MM | 4.94 |
| Elachista eskoi | 0.38 | 1.08 | Elachista sp. 03 MM | 1.39 |
| Elachista excelsicola | 0.23 | 1.24 | Elachista kilmunella | 3.62 |
| Elachista fuscofrontella | 0 | 0 | Elachista sp. 02 MM | 4.72 |
| Elachista griseella | N/A | N/A | Elachista sp. 14 MM | 6.39 |
| Elachista herrichii | 0.12 | 0.31 | Elachista sp. 10 MM | 3.78 |
| Elachista humilis | 0.52 | 1.4 | Elachista nielswolffi | 1.01 |
| Elachista irenae | 0 | 0 | Elachista talgarella | 5.88 |
| Elachista kilmunella | 0.18 | 0.92 | Elachista leifi | 0 |
| Elachista krogeri | 0.55 | 1.97 | Elachista pomerana | 2.68 |
| Elachista lastrella | N/A | N/A | Elachista nobilella | 6.57 |
| Elachista leifi | 0 | 0 | Elachista kilmunella | 0 |
| Elachista luticomella | 0 | 0 | Elachista baikalica | 3.15 |
| Elachista maculicerusella | 0.94 | 5.29 | Elachista lastrella | 7.1 |
| Elachista maculosella | N/A | N/A | Elachista sp. 09 MM | 2.18 |
| Elachista nielswolffi | 0.07 | 0.25 | Elachista humilis | 1.01 |
| Elachista nobilella | 0 | 0 | Elachista lastrella | 6.57 |
| Elachista orstadii | 0.34 | 0.77 | Elachista atricomella | 2.06 |
| Elachista poae | 0 | 0 | Elachista sp. 14 MM | 7.41 |
| Elachista pomerana | 0.08 | 0.31 | Elachista humilis | 1.56 |
| Elachista rufocinerea | 0.05 | 0.15 | Elachista sp. 15 MM | 4.75 |
| Elachista subnigrella | 0.27 | 0.61 | Elachista sp. 12 MM | 1.24 |
| Elachista talgarella | 0 | 0 | Elachista bifasciella | 5.85 |
| Elachista tanaella | N/A | N/A | Elachista kilmunella | 4.17 |
| Elachista vonschantzi | 0 | 0 | Elachista pomerana | 3.59 |
| Elachista wieseriella | 0 | 0 | Elachista sp. 02 MM | 2.02 |
| Elachista zernyi | 0.4 | 1.26 | Elachista deriventa | 2.13 |
| Elachista sp. 01 MM | N/A | N/A | Elachista kilmunella | 2.66 |
| Elachista sp. 02 MM | 0 | 0 | Elachista wieseriella | 2.02 |
| Elachista sp. 03 MM | 0 | 0 | Elachista eskoi | 1.39 |
| Elachista sp. 04 MM | N/A | N/A | Elachista griseella | 6.53 |
| Elachista sp. 05 MM | 0 | 0 | Elachista anserinella | 6.9 |
| Elachista sp. 06 MM | 0 | 0 | Elachista fuscofrontella | 6.56 |
| Elachista sp. 07 MM | 0.21 | 0.41 | Elachista atricomella | 1.08 |
| Elachista sp. 08 MM | 0.15 | 0.31 | Elachista humilis | 3.62 |
| Elachista sp. 09 MM | 0 | 0 | Elachista maculosella | 2.18 |
| Elachista sp. 10 MM | N/A | N/A | Elachista herrichii | 3.78 |
| Elachista sp. 11 MM | 0 | 0 | Elachista herrichii | 4.44 |
| Elachista sp. 12 MM | 0.33 | 1.24 | Elachista subnigrella | 1.24 |
| Elachista sp. 13 MM | N/A | N/A | Elachista subnigrella | 1.87 |
| Elachista sp. 14 MM | 0.12 | 0.41 | Elachista atricomella | 1.39 |
| Elachista sp. 15 MM | N/A | N/A | Elachista sp. 09 MM | 4.14 |
| Elachista sp. 16 MM | N/A | N/A | Elachista elegans | 4.94 |
BIN algorithms split the data into 49 distinct categories (Figure S2). In two cases, a single species contained two BINs (E. maculicerusella, E. canapennella), while five BINs contained two species, and two BINs three species. Of the 16 newly detected putative species, 11 had unique BINs, whereas 5 newly detected putative species, although having clearly distinct barcodes, were included in a BIN containing one or two additional species.
Discussion
Our study provides insights into the two fundamental features of DNA barcoding. First, the study serves as a test of the utility of DNA barcodes in recognizing species. Second, we show that broad sampling of museum specimens aids the discovery of new species in a remarkable way. Species of the test group belong to a diverse group of moths that include species that generally have overall similar morphology (Figure 1), but which have been under unusually comprehensive morphological taxonomic scrutiny. The former factor is likely to increase the proportion of previously undetected species, while the latter is expected to show the reverse effect. Provided that few groups of small-sized moths, let alone most other insect groups, have been under taxonomic scrutiny comparable to that of Elachista of the study region, we assume that surveys conducted with other insect groups would similarly lead to the discovery of many new species. Some former large-scale investigations have similarly yielded the discovery of several apparent new species61,62.
Based initially on gaps in genetic variation of DNA barcode regions, we discovered twenty-five lineages that were either not closely associated with identified specimens or showed considerable (>1%) intraspecific variation. Fourteen of these cases were subsequently shown to represent two distinct lineages and one other (E. atricomella) three distinct lineages, as demonstrated by independent sources of evidence, notably differences in external appearance and in genital structures, which generally evolve rapidly and divergently, hence providing cues for the separation of young species63. Following traditionally used criteria of species delimitation in the study group and insect taxonomy in general, these lineages can be seen to represent valid species. However, species delineation is inherently subjective as it depends, for example, on the species concept applied. Two additional species are genetically split into two widely different clusters and separate BINs. One of these splits (E. maculicerusella) concern sympatric populations and has previously been suspected of comprising two species based on possibly differing external appearance and life history traits, while in the second case (E. canapennella) the split is correlated with a wide geographic gap. Whether these species truly contain cryptic diversity should preferably be examined with other lines of evidence. As we were not able to find constant morphological differences between these clusters, the sequencing of nuclear markers might provide further insights into the question (cf.40). Mitochondrial and nuclear genomes evolve independently, and different nuclear genes may vary in their gene genealogies. Therefore, nuclear genes have a great potential for resolving whether mitochondrial haplotypes represent either panmictic populations or truly separate, reproductively isolated lineages. This is most applicable when the taxa in question occur in sympatry, because in such circumstances nuclear genes are not expected to show consistent differences as compared to mitochondrial haplogroups within a species.
The Lepidoptera in the study area have been much more intensively studied than in any other area of the world, suggesting that a similar survey conducted elsewhere with a comparable effort would reveal even more new species. Furthermore, while our survey increased the number of known species by one-third, we recognize that a still broader sampling of material from collections would almost inevitably lead to the discovery of still more undetected species. Our sample comprised only a small fraction of specimens deposited in zoological museums and private collections and does not include samples from wide areas, including sites known to host endemic taxa in other insect groups. Although the new species were retrospectively observed to be unique in their morphology, we are convinced that several of them would have remained undetected for some time without DNA barcodes. Numerous recent taxonomic studies have demonstrated the usefulness of DNA barcoding in initial detection of taxa that have been recognized as new after closer examination33,34,35,36,37,38,39,40,41,42. Our study is rare in the breadth of sampling and the independent validation of genetic results with morphological evidence, including the large-scale examination of genital structures. An example of a different pattern is reported in a study including an in-depth morphological examination of eight genetically highly variable Romanian butterfly species. This study did not yield evidence for any additional species30. European butterflies are, however, an exception among all insect groups in the extent of previous morphology-based investigation and hence generally lower levels of new species discoveries are expected in this group than in most other insect groups.
All previously named species, except E. kilmunella and E. leifi which share the same haplotypes, could unequivocally be identified based on barcodes alone, equating to 95% identification success. This provides evidence not only of the efficiency of DNA barcodes in separating species, but also of the general validity of traditionally used criteria for delimiting species in the study group. The validity of E. leifi has been under dispute, as it shows only minor morphological differences to E. kilmunella47. E. leifi males swarm only at dawn, while E. kilmunella males are day- and dusk-active. There is yet the possibility that E. leifi is a northern, yet sympatric form of E. kilmunella, perhaps adapted to a two-year life cycle, a phenomenon known from many lepidopteran species. This alternative does not, however, explain the behavioural difference. Genetic identifiers may, however, also fail to discriminate species. Especially with young species, the time elapsed since speciation may have not been sufficient for lineage sorting to be completed16,61. Young species may also show introgression16. We tested the utility of DNA barcodes in multiple cases of conditions where both of those phenomena were anticipated to occur, but found the pair E. kilmunella/E. leifi to be the only one in which these effects are possibly involved.
E. atricomella was found to constitute three genetically and morphologically distinct species, two of which were given a status of putative new species. This split rendered the remaining specimens of E. atricomella as forming a polyphyletic species. While incomplete lineage sorting or introgression may result in gene-level para- or polyphyly, we cannot rule out that this species includes a fourth, morphologically indistinguishable species, pending the examination of other genetic markers.
Two or three species were lumped together by BIN algorithms in seven cases. In each of these, except with E. kilmunella and E. leifi, the ability of DNA barcodes to discriminate species remained. BINs are deliberately designed to be relatively conservative in highlighting species having high potential of cryptic diversity and in minimizing the over-splitting of species62. We consider the conservativeness of BINs to be a meaningful property, as the unjustified splitting of species with accumulated synonyms is seen as having a more confusing impact in taxonomy than the opposite.
In summary, our results support many earlier observations that DNA barcodes effectively differentiate closely related species. We demonstrate that a comprehensive sampling of collection material is an efficient way to discover hidden portions of biodiversity and that by accelerating taxonomic workflow DNA barcoding provides an important tool that, when widely used, might substantially help to overcome the taxonomic impediment. Lepidoptera represents one of the most thoroughly investigated groups of insects, and our focal group has been under considerable previous taxonomic investigation. Our results therefore suggest that the assessment of insect species number may often be underestimated by the overlooking of morphologically similar species. Along with growing DNA barcoding activity, we assume an increasing rate of discoveries of new species across all insect groups and areas.
Methods
We aimed at sampling all Palearctic species of the E. bifasciella group, excluding those of Japan and the Russian Far East (Sakhalin, Primorsk Region), from where we could not obtain samples. A total of 49 named species are known from this region. We obtained 40 of these for study (Table S2). Besides a focused sampling of identified specimens of described species, we performed a bulk sampling of unidentified museum and other collection specimens, paying attention particularly to samples from areas whose fauna has been less exhaustively studied. Our sampling is geographically biased towards northern Europe, where 31 out of 40 sampled species occur (of these only E. herrichii was barcoded outside this region). We supplemented this sampling with available fresh specimens from Central and South Europe, from the Ural region and several areas of Central Asia and eastern Siberia. Given the vast area of the Palearctic region, our sampling cannot be considered exhaustive in geographic terms. Similarly, in most species specimens were not comprehensively sampled from different areas of their known range. Localities for the specimens included in this study are indicated in the Figure S1. The map was created using an online tool SimpleMappr64.
All specimens subjected to DNA sequencing were given a label with a unique sample ID. One or two legs of each specimen were deposited in microplate wells with 30 μL of absolute alcohol in each well. The sequencing was carried out at the Canadian Centre for DNA Barcoding following laboratory protocols used routinely in the Canadian Centre for DNA Barcoding (CCDB) as explained in detail in DeWaard et al.65. In short, this involves the Chelax-based Dry-Release DNA extraction done for an aliquot of 30–110 μL. A volume of 0.5–2 μL of aliquot is used for the PCR. CCDB uses a wide set of PCR primers for DNA amplification, depending, e.g., on the taxon group and specimen condition. The primers used, as with all other laboratory protocol details, are available at the sequence page of each record at the BOLD database. In Lepidoptera, the primer pair used is usually LepF1-LepR1. PCR is done in a volume of about 11–13 μL. PCR products are checked using Invitrogen E-gel 96 system with precast and bufferless agarose gels. Since sequencing is done bi-directionally, the PCD-products are not purified, but sequencing reactions are set up directly from PCR products. Depending on the intensity of gel bands, 0.5–2 μL of PCR product is used for sequencing reactions. The sequencing follows a routinely used recipe of ingredients with Dye terminator mix v3.1 and each reaction is done in a volume of 10 μL. Sequence reaction cleanup is done using the Sephadex column method. Sequence alignments are done using programs (Sequencer, SeqScape, Lasergene) permitting the assembling of bidirectional reads and trace file edits. Sequences were carefully checked for the detection of COI pseudogenes (NUMTS) and contaminations. Consequently, a sample of E. anserinella contaminated by another species was discarded.
A total of 437 specimens yielded at least a partial barcode sequence, with 95% of the barcodes being over 600 bp in length (Table S3). In only six cases were barcodes of less than 400 bp included, none of which were the sole representative of their species. The average sample size per species with sixteen discovered putative new species assessed as separate species was 7.8 (range 1–32). Distance statistics were calculated using BOLD (Barcode of Life Data Systems) tools, accessible at http://www.boldsystems.org66. The Barcode Index Number (BIN) grouping of haplotypes into Operational Taxonomic Units (OTUs)62, a recent option of BOLD, was used in the interpretation of results. The BIN delimitation of sequences into OTUs includes two steps. In the first stage, sequences are clustered based on the single linkage clustering method. In the second step, this preliminary clustering is refined using graph-based Markov Clustering (MCL). Based on eight independent datasets, BINs were shown to have very high correspondence with pre-existing assignments of species62. In this study, we used BIN assignments only for advisory purposes. We did not base our species delimitation on them nor on other quantitative delimitation algorithms, because in addition to molecular data, we incorporated morphological, though not quantitative morphometric data, in our analyses.
Sequences were aligned using BOLD alignment, which performs the alignment against a wide variety of animal barcodes deposited in BOLD. A few resulting gaps were subsequently removed. Sequence alignments were manually checked in Mega 567, resulting in the detection of a misalignment of a single codon in two cases. A Neighbor-Joining tree was constructed using Mega 5 under the Kimura 2 Parameter (K2P) model, with pairwise deletion of missing data and 500 bootstrap pseudoreplicates in order to test node robustness, especially for nodes leading to species. The full specimen collection details, voucher photographs, sequence data with trace files and GenBank accession numbers are available under the public dataset DS-ELABIF in BOLD. Information on specimens is also provided in the Table S3.
All species showing clear discontinuities (over 1% intraspecific gap; also including one case having a 0.99% gap) or an unusual extent of more continuous intraspecific variation (over 1% maximum intraspecific variation) were subjected to an in-depth morphological examination. Likewise, specimens not directly linked to any named taxa were examined morphologically. Morphological examination was based primarily on male and female genital characteristics, but also wing patterns were examined. This follows traditional guidelines of species delimitation in Elachista, where typically slight or moderate differences in genitalia have been used as criteria in species delimitation. This seems generally well-justified as in well-investigated species morphological delimitation criteria have been shown to correlate with different life-history properties, such as larval food plants. Species of the study group typically show narrow diets and high level of niche specialization.
For the study of the morphology of the genitalia, the abdomen was macerated in 10% aqueous solution of potassium hydroxide, rinsed thoroughly in clean water, then put to an object slide where the water was replaced by absolute EtOH. Excessive scales were removed to improve clarity. The abdominal pelt, as well as the female genitalia, were stained using chlorazol black, and the male genitalia using yellow eosin. The genitalia were severed from the abdomen by cutting the abdomen along the pleurum and along intersegment 7 and 8. The abdomen and the genitalia were transferred to another slide, to which a drop of Euparal™ was applied. In the male genitalia, the phallus was gently severed from the genital capsule. The valvae were gently opened, and the uncus, vinculum and valvae were slightly pressed using forceps to align them in the horizontal plane, and the phallus positioned in a lateral position, sometimes dorsoventrally, so as to better allow the examination of the shape of the often bifurcate apex. The genitalia were then covered by a cover glass and incubated in 40°C for two weeks. The female genitalia were positioned ventral-side upwards.
The delimitation of species in Elachistidae, as usually in animal taxonomy, assumes that the species are ‘natural’ units, the underlying hypothesis being that the species are populations or clusters of populations that may interbreed in nature, i.e., they form cohesive genealogical units. This is, of course, next to impossible to directly observe. Therefore indirect information, derived from morphology and life history traits, is routinely used for obtaining an approximation of species delimitation. Each ‘species’ recognized is therefore a hypothesis that can, and should, be subjected to further testing. The most obvious methods for testing are comparing samples collected in ‘new’ localities, acquiring further biological knowledge by rearing of larvae, and studying genomic traits (cf.43)
As criteria for the evaluation of possible morphological distinction, special attention was paid to the following traits, following the general tradition, and morphological particulars of the focal group (terminology follows Traugott-Olsen & Schmidt Nielsen 197745 and Kaila 199953): in external appearance, the wing pattern and shape, the thickness of antennae and length of the labial palpi, and the colour of the head and the neck tuft; in male genitalia, the relative size and shape of the uncus, gnathos, valva, digitate process, phallus and cornuti; in the female genitalia, the size, shape and position of the ostium bursae, the shape of the antrum, the inception position and shape of the ductus seminalis, the length and sclerotisations of the ductus bursae, the shape of the corpus bursae, and the size and shape of the signum.
Author Contributions
M.M. and L.K. designed the study. All authors took part in material acquisition and preparing samples for DNA analyses. M.M. performed analyses of molecular data and L.K. analysed morphological data. M.M. and L.K. prepared the figures and tables. M.M and L.K. wrote the manuscript and J.T. commented it.
Supplementary Material
Supplementary information
Acknowledgments
We thank Bengt Å. Bengtsson, Sami Haapala, Juhani Itämies, Jari Junnilainen, Petri Hirvonen, Jari Kaitila, Ali Karhu, Erkki and Leena Laasonen, Kari Nupponen, Esko Saarela, Leo Sippola, Ingvar Svensson (deceased), Panu Välimäki and Bo Wikström for providing specimens for sequence analysis. We cordially thank Peter Huemer for providing access to two specimens of E. wieseriella and Andreas Segerer for access to a record of E. lastrella. We are grateful for support provided by the Government of Canada through Genome Canada and the Ontario Genomics Institute to the International Barcode of Life Project. This funding enabled the Canadian Centre for DNA Barcoding (University of Guelph) to carry out the sequence analysis on our specimens. We also thank the Ontario Ministry of Economic Development and Innovation for funding the ongoing development of BOLD. Alex Borisenko provided crucial help with geographical coordinates and map construction, and James Robertson helped with making a public dataset of records. We thank Kone foundation and Finnish Cultural Foundation for their financial support to the Finnish Barcode of Life Project. We are also grateful to staff of BIO for their efforts and help. Kent Tankersley checked the English language.
References
- Mora C., Tittensor D. P., Adl S., Simpson A. G. B. & Worm B. How Many Species Are There on Earth and in the Ocean? PLoS Biol 9(8), e1001127 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman A. D. Numbers of living species in Australia and the world. Report for the Australian Biological Resources Study, Canberra (2009).[Retrieved from http://www.environment.gov.au/biodiversity/abrs/publications/other/species-numbers/2009/pubs/nlsaw-2nd-complete.pdf. Accessed August 10, 2013]. [Google Scholar]
- Godfray H. C. J. Challenges for taxonomy. Nature 417, 17–19 (2002). [DOI] [PubMed] [Google Scholar]
- Wheeler Q. D., Raven P. H. & Wilson E. O. Taxonomy: impediment or expedient? Science 303, 285 (2004). [DOI] [PubMed] [Google Scholar]
- Hebert P. D. N., Cywinska A., Ball S. L. & deWaard J. R. Biological identifications through DNA barcodes. Proc R. Soc. Lond. B. 270, 313–321 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D., Arctander P., Minelli A., Thomas R. H. & Vogler A. P. A plea for DNA taxonomy. Trends Ecol. Evol. 18, 70–74 (2003). [Google Scholar]
- Schoch C. L. et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl Acad. Sci. USA 109, 6241–6246 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- CBOL Plant Working Group. A DNA barcode for land plants. Proc. Natl Acad. Sci. USA 106, 12794–12797 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr K. et al. Filling the gap - COI barcode resolution in eastern Palearctic birds. Front. Zool. 6, 29 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P. D. N., Dewaard J. R. & Landry J. F. DNA barcodes for 1/1000 of the animal kingdom. Biol. Lett. 6, 359–362 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P. D. N., Penton E. H., Burns J. M., Janzen D. H. & Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl Acad. Sci. USA 101, 14812–14817 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. A., Woodley N. E., Janzen D. H., Hallwachs W. & Hebert P. D. N. DNA barcodes reveal cryptic host-specificity within the presumed polyphagous members of a genus of parasitoid flies (Diptera: Tachinidae). Proc. Natl Acad. Sci. USA 103, 3657–3662 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier R., Shiyang K., Vaidya G. & Ng P. K. DNA barcoding and taxonomy in Diptera: a tale of high intraspecific variability and low identification success. Syst. Biol. 55, 715–728 (2006). [DOI] [PubMed] [Google Scholar]
- Elias M. et al. Limited performance of DNA barcoding in a diverse community of tropical butterflies. Proc. R. Soc. Lond. B. 274, 2881–2889 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will K. W., Mishler B. D. & Wheeler Q. D. The perils of DNA barcoding and the need for integrative taxonomy. Syst. Biol. 54, 844–851 (2005). [DOI] [PubMed] [Google Scholar]
- Rubinoff D., Cameron S. & Will K. A genomic perspective on the shortcomings of mitochondrial DNA for "barcoding" identification. J. Hered. 97, 581–594 (2006). [DOI] [PubMed] [Google Scholar]
- Hurst G. D. D. & Jiggins F. M. Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: the effects of inherited symbionts. Proc R. Soc. Lond. B. 272, 1525–1534 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias M. et al. Limited performance of DNA barcoding in a diverse community of tropical butterflies. Proc. R. Soc. Lond. B 274, 2881–2889 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemers M. & Fiedler K. Does the DNA barcoding gap exist? - a case study in blue butterflies (Lepidoptera: Lycaenidae). Front. Zool. 4, 8 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris D. J. Can you bank on GenBank? Trends Ecol. Evol. 18, 317–319. [Google Scholar]
- Bidartondo M. I. et al. Preserving accuracy in GenBank. Science 319, 1616 (2008). [DOI] [PubMed] [Google Scholar]
- Hickerson M. J., Meyer C. P. & Moritz C. DNA barcoding will often fail to discover new animal species over broad parameter space. Syst. Biol. 55, 729–739 (2006). [DOI] [PubMed] [Google Scholar]
- Mutanen M. et al. Allopatry as a Gordian knot for taxonomists: patterns of barcode divergences in arctic-alpine Lepidoptera. PLoS ONE 7, e47214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower A. V. Z. Problems with DNA barcodes for species delimitation: ‘ten species’ of Astraptes fulgerator reassessed (Lepidoptera: Hesperiidae). Syst. Biodivers. 4, 127–132 (2006). [Google Scholar]
- Dasmahapatra K. K., Elias M., Hill R. I., Hoffman J. I. & Mallet J. Mitochondrial DNA barcoding detects some species that are real, and some that are not. Mol. Ecol. Res. 10, 264–273 (2010). [DOI] [PubMed] [Google Scholar]
- Hebert P. D. N., Ratnasingham S. & deWaard J. R. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. Lond. B 270, Suppl 1, S96–99 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P. D. N., Stoeckle M. Y., Zemlak T. S. & Francis C. M. Identification of Birds through DNA Barcodes. PLoS Biol. 2, e312 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett R. D. H. & Hebert P. D. N. Identifying spiders through DNA barcodes. Canadian J. Zool. 83, 481–491 (2005). [Google Scholar]
- Lukhtanov V. A., Sourakov A., Zakharov E. V. & Hebert P. D. N. DNA barcoding Central Asian butterflies: increasing geographical dimension does not significantly reduce the success of species identification. Mol. Ecol. Res. 9, 1302–1310 (2009). [DOI] [PubMed] [Google Scholar]
- Dinca V., Zakharov E. V., Hebert P. D. N. & Vila R. Complete DNA barcode reference library for a country's butterfly fauna reveals high performance for temperate Europe. Proc. R. Soc. Lond. B 278, 347–355 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann A., Haszprunar G. & Hebert P. D. N. DNA Barcoding the Geometrid Fauna of Bavaria (Lepidoptera): Successes, Surprises, and Questions. PLoS ONE 6, e17134 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickford D. et al. Cryptic diversity as a window on diversity and conservation. Trends Ecol. Evol. 22, 148–155 (2006). [DOI] [PubMed] [Google Scholar]
- Vaglia T., Haxaire J., Kitching I. J., Meusnier I. & Rougerie R. Morphology and DNA barcoding reveal three cryptic species within the Xylophanes neoptolemus and loelia species-groups (Lepidoptera: Sphingidae). Zootaxa 1923, 18–36 (2008). [Google Scholar]
- Hausmann A. et al. Revision of the Australian Oenochroma vinaria Guenee, 1858 species-complex (Lepidoptera: Geometridae, Oenochrominae): DNA barcoding reveals cryptic diversity and assesses status of type specimen without dissection. Zootaxa 2239, 1–21 (2009). [Google Scholar]
- Huemer P. & Hebert P. D. N. Cryptic diversity and phylogeography of high alpine Sattleria- a case study combining DNA barcodes and morphology (Lepidoptera: Gelechiidae). Zootaxa 2981, 1–22 (2011). [Google Scholar]
- Segerer A. H., Haslberger A. & Gruenewald T. Occurrence of Olethreutes subtilana (Falkovitsh, 1959) in Central Europe uncovered by DNA barcoding (Tortricidae: Olethreutinae). Nota lepid. 33, 209–218 (2011). [Google Scholar]
- Wilson J. J. et al. Identity of the ailanthus webworm moth (Lepidoptera, Yponomeutidae), a complex of two species: evidence from DNA barcoding, morphology and ecology. ZooKeys 46, 41–60 (2011). [Google Scholar]
- Mutanen M. et al. DNA barcodes reveal that the widespread European tortricid moth Phalonidia manniana (Lepidoptera: Tortricidae) is a mixture of two species. Zootaxa 3262, 1–21 (2012). [Google Scholar]
- Kaila L. & Mutanen M. DNA barcoding and morphology support the division of Elachista nuraghella sensu auct. (Lepidoptera: Elachistidae: Elachistinae) into two vicariant species. Zootaxa 3343, 57–68 (2012). [Google Scholar]
- Huemer P. & Mutanen M. Taxonomy of spatially disjunct alpine Teleiopsis albifemorella sensu auctt. revealed by molecular and morphological data - how many species are there? Zootaxa 3580, 1–23 (2012). [Google Scholar]
- Mutanen M., Aarvik L., Landry J.-F., Segerer A. & Karsholt O. Epinotia cinereana (Haworth, 1811) bona sp., a Holarctic tortricid distinct from E. nisella (Clerck, 1759) (Lepidoptera: Tortricidae: Eucosmini) as evidenced by DNA barcodes, morphology and life history. Zootaxa 3318, 1–25 (2012). [Google Scholar]
- Yang Z. et al. DNA barcoding and morphology reveal three cryptic species of Anania (Lepidoptera: Crambidae: Pyraustinae) in North America, all distinct from their European counterpart. Syst. Entomol. 37, 686–705 (2012). [Google Scholar]
- Kaila L. Elachistine Moths of Australia: (Lepidoptera: Gelechioidea: Elachistidae). (Csiro Publishing, Collingwood) (2011). [Google Scholar]
- Svensson I. Six new species of Microlepidoptera from northern Europe. Entomol. Scand. 7, 195–206 (1976). [Google Scholar]
- Traugott-Olsen E. & Schmidt Nielsen E. The Elachistidae (Lepidoptera) of Fennoscandia and Denmark. Fauna Entomol. Scand. 6, 1–299 (Klampenborg, Denmark 1977). [Google Scholar]
- Kyrki J. & Karvonen J. Elachista eskoi sp. n., a new species of Elachistidae from Finland (Lepidoptera). Entomol. Scand. 15, 521–525 (1985). [Google Scholar]
- Kaila L. & Kerppola S. Elachista leifi sp. n. from northern Finland (Lepidoptera, Elachistidae). Entomol. Fennica 3, 155–158 (1992). [Google Scholar]
- Kaila L. The Elachistidae of southern Siberia and Central Asia, with descriptions of five new species (Lepidoptera). Entomol. Fennica 3, 177–194 (1992). [Google Scholar]
- Sruoga V. A. & Puplesis R. K. New species of gramineal Elachistid moths (Lepidoptera, Elachistidae) from Middle Asia and Kazakhstan. Entomologicheskoe Obozrenie, 71, 428–441 (1992). [Google Scholar]
- Sinev S. Yu. & Sruoga V. A. New species of the mining moths (Lepidoptera, Elachistidae) from Russian Far East. Entomologicheskoe Obozrenie, 74, 120–137 (1995). [Google Scholar]
- Kaila L. Two new Elachista species (Lepidoptera, Elachistidae) from the Polar Urals region, Russia. Entomol. Fennica 9, 219–223 (1998). [Google Scholar]
- Kaila L. A revision of the Nearctic species of the genus Elachista s. l. III. The bifasciella, praelineata, saccharella and freyerella groups (Lepidoptera, Elachistidae). Acta Zool. Fennica, 211, 1–235 (1999). [Google Scholar]
- Kaila L. Phylogeny and classification of the Elachistidae s.s. (Lepidoptera: Gelechioidea). Syst. Entomol. 24, 139–169 (1999). [Google Scholar]
- Aarvik L. & Berggren K. Description of Elachista tanaella sp. n. (Elachistidae) from Arctic Norway. Nota lepid. 26, 83–87 (2004). [Google Scholar]
- Sugisima K. Japanese Elachista studied by Parenti (1983) (Lepidoptera, Elachistidae): the subgenus Aphelosetia and the gleichenella-, tetragonella-, and bifasciella-groups. Tijdschrift voor Entomologie 148, 225–246 (2005). [Google Scholar]
- Kaila L. et al. Elachista deriventa sp. n. (Lepidoptera, Elachistidae: Elachistinae), a new species from southern Finland. Entomol. Fennica 19, 184–192 (2008). [Google Scholar]
- Funk D. J. & Omland K. E. Species-level paraphyly and polyphyly: Frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annu Rev. Ecol. Evol. S. 34, 397–423 (2003). [Google Scholar]
- deWaard J. R., Hebert P. D. & Humble L. M. A comprehensive DNA barcode library for the looper moths (Lepidoptera: Geometridae) of British Columbia, Canada. PLoS ONE 6, e18290 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekrem T., Stur E. & Hebert P. Females do count: Documenting Chironomidae (Diptera) species diversity using DNA barcoding. Org. Divers. Evol. 10, 397–408 (2010). [Google Scholar]
- Strutzenberger P., Brehm G. & Fiedler K. DNA barcoding-based species delimitation increases species count of Eois (Geometridae) moths in a well-studied tropical mountain forest by up to 50%. Insect Science 18, 349–362 (2010). [Google Scholar]
- Kaila L. & Ståhls G. DNA barcodes: Evaluating the potential of COI to diffentiate closely related species of Elachista (Lepidoptera: Gelechioidea: Elachistidae) from Australia. Zootaxa 1170, 1–26 (2006). [Google Scholar]
- Ratnasingham S. & Hebert P. D. N. A DNA-based registry for all animal species: The Barcode Index Number (BIN) System. PLoS ONE 8, e66213 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnqvist G. Comparative evidence for the evolution of genitalia by sexual selection. Nature 393, 784–786 (1998). [Google Scholar]
- Shorthouse D. P. SimpleMappr, an online tool to produce publication-quality point maps (2010). [Retrieved from http://www.simplemappr.netAccessed August 22, 2013].
- deWaard J. R., Ivanova N. V., Hajibabaei M. & Hebert P. D. N. Assembling DNA Barcodes: Analytical Protocols. [Cristofre, M., (ed.). Methods in Molecular Biology: Environmental Genetics. Pp. 275–293] (Humana Press Inc., Totowa) (2008). [DOI] [PubMed] [Google Scholar]
- Ratnasingham S. & Hebert P. D. N. The Barcode of Life Data Systems (http://www.barcodinglife.org) Mol. Ecol. Notes 7, 355–364 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Method. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information