Summary
In bloodstream-form Trypanosoma brucei (the causative agent of African sleeping sickness) the glycosylphosphatidylinositol (GPI) anchor biosynthetic pathway has been validated genetically and chemically as a drug target. The conundrum that GPI anchors could not be in vivo labelled with [3H]-inositol led us to hypothesize that de novo synthesis was responsible for supplying myo-inositol for phosphatidylinositol (PI) destined for GPI synthesis. The rate-limiting step of the de novo synthesis is the isomerization of glucose 6-phosphate to 1-d-myo-inositol-3-phosphate, catalysed by a 1-d-myo-inositol-3-phosphate synthase (INO1). When grown under non-permissive conditions, a conditional double knockout demonstrated that INO1 is an essential gene in bloodstream-form T. brucei. It also showed that the de novo synthesized myo-inositol is utilized to form PI, which is preferentially used in GPI biosynthesis. We also show for the first time that extracellular myo-inositol can in fact be used in GPI formation although to a limited extent. Despite this, extracellular inositol cannot compensate for the deletion of INO1. Supporting these results, there was no change in PI levels in the conditional double knockout cells grown under non-permissive conditions, showing that perturbation of growth is due to a specific lack of de novo synthesized myo-inositol and not a general inositol-less death. These results suggest that there is a distinction between de novo synthesized myo-inositol and that from the extracellular environment.
Introduction
Trypanosoma brucei, the causative agent of African sleeping sickness, expresses a dense cell-surface coat consisting of approximately 5 × 106 dimers of variant surface glycoprotein which are attached to the plasma membrane by a covalent linkage to a glycosylphosphatidylinositol (GPI)1 membrane anchor. This coat protects the parasite from the alternative complement pathway of the host and through antigenic variation, from specific immune responses (Cross, 1996). The biosynthesis of this conserved GPI anchor has previously been genetically (Nagamune et al., 2000; Chang et al., 2002) and chemically validated (Smith et al., 2004) as a drug target in bloodstream-form T. brucei.
The structure and biosynthesis of GPI membrane anchors and related molecules have been recently reviewed (Ferguson et al., 1999; Kinoshita and Inoue, 2000; McConville and Menon, 2000; Morita et al., 2000a; de Macedo et al., 2003). The basic GPI core structure attached to protein comprises NH2CH2CH2PO4H-6Manα1-2Manα1-6Manα1-4GlcNα1-6d-myo-inositol-1-HPO4-lipid (EtN-P-Man3GlcN-PI), where the lipid can be diacylglycerol, alkylacylglycerol or ceramide (Ferguson et al., 1999).
The sequence of events underlying GPI biosynthesis has been studied in T. brucei (Masterson et al., 1989; Masterson et al., 1990; Menon et al., 1990a,b; Güther and Ferguson, 1995; Morita et al., 2000b), Trypanosoma cruzi (Heise et al., 1996), Toxoplasma gondii (Striepen et al., 1999), Plasmodium falciparum (Gerold et al., 1999), Leishmania (Smith et al., 1997; Ralton and McConville, 1998; Ralton et al., 2002), Saccharomyces cerevisiae (Sutterlin et al., 1998; Flury et al., 2000) and mammalian cells (Hirose et al., 1992; Puoti and Conzelmann, 1993; Chen et al., 1998). In all cases, GPI biosynthesis is initiated by the addition of GlcNAc from UDP-GlcNAc to phosphatidylinositol (PI) to give GlcNAc-PI, which is then de-N-acetylated to form GlcN-PI to be processed further in a species-specific manner. To date all eukaryotes synthesize PI by the exchange of the CMP moiety of CDP-diacylglycerol with inositol.
myo-Inositol is a six-carbon cyclitol, which is an essential metabolite in all eukaryotes. The metabolism of myo-inositol plays a vital role in growth regulation, signal transduction, membrane biogenesis, osmotolerance and other essential biochemical processes, as well as the formation of GPI anchors. In prokaryotes inositol is found sparingly, an example of an exception is mycobacteria, where it is essential for biogenesis of mycothiol, PI, phosphatidylinositol mannosides (PIM) and GPIs (Besra and Chatterjee, 1994; Haites et al., 2005, and references containing therein).
Cellular myo-inositol can arise from three sources: first, myo-inositol can be taken up from the extracellular environment; second, myo-inositol can be released by dephosphorylation of inositol phosphates produced from the turnover of inositol phospholipids. Finally, myo-inositol can be de novo synthesized via the concerted action of two enzymes: 1-d-myo-inositol-3-phosphate synthase (INO1) (E.C.5.5.1.4) and myo-inositol monophosphatase (IMPase) (E.C.3.1.3.25). The rate-limiting step of the de novo synthesis of myo-inositol is the NAD+-dependent conversion of d-glucose 6-phosphate to 1-d-myo-inositol-3-phosphate by INO1 via a three stage reaction involving: (i) oxidation of d-glucose 6-phosphate to 5-keto-d-glucose 6-phosphate, (ii) cyclization of myo-2-inosose1-phosphate and (iii) reduction to 1-d-myo-inositol-3-phosphate (Majumder et al., 1997).
INO1s have been characterized from few prokaryotic and various eukaryotic organisms such as Porteresia coarctata (Majee et al., 2004), Arabidopsis thaliana (Johnson and Sussex, 1995), Mycobacterium tuberculosis (Norman et al., 2002; Movahedzadeh et al., 2004), Mycobacterium smegmatis (Haites et al., 2005), Drosophila melanogaster (Park and Kim, 2004), S. cerevisiae (Donahue and Henry, 1981), Entamoeba histolytica (Lohia et al., 1999), Leishmania mexicana (Ilg, 2002) and Homo sapiens (Guan et al., 2003; Ju et al., 2004). Genetic studies in the two pathogenic organisms, L. mexicana (Ilg, 2002) and M. tuberculosis (Movahedzadeh et al., 2004), have revealed that INO1 is important in regulating their intracellular myo-inositol levels, and thus INO1 is a potential drug target in these organisms.
In this study, we begin to investigate the de novo synthesis and metabolism of myo-inositol in T. brucei. Here we show the molecular cloning and expression in Escherichia coli of the T. brucei myo-inositol-3-phosphate synthase and characterization of the recombinant enzyme. We also report the creation of a T. brucei conditional null mutant which demonstrates that the de novo synthesis of myo-inositol is essential for the bloodstream form of the parasite, even in the presence of excess myo-inositol. Our results also suggest that de novo synthesized myo-inositol is preferentially used for GPI anchor biosynthesis and that there is differentiation of the de novo synthesized myo-inositol and myo-inositol taken up from the extracellular environment.
Results
Cloning and sequencing T. brucei INO1
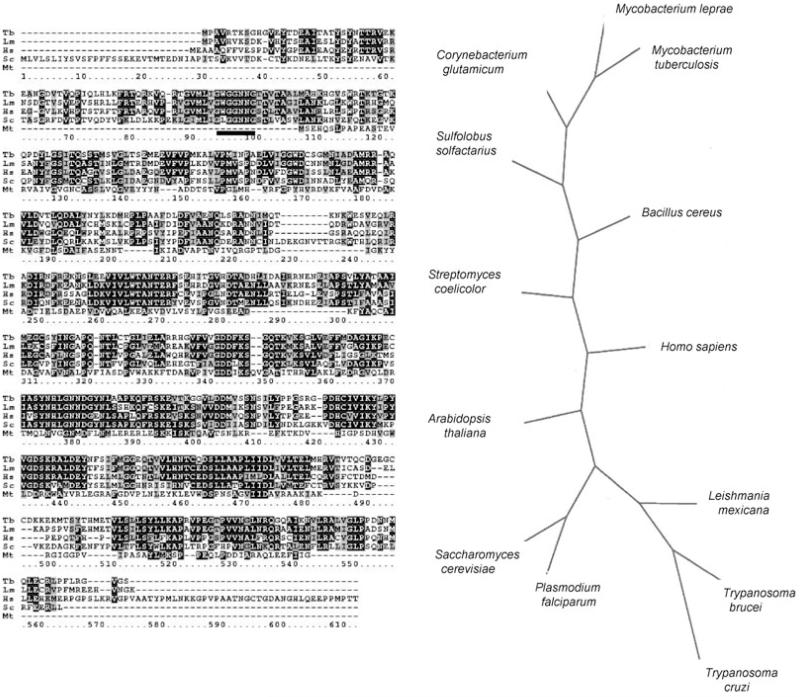
An INO1 was identified in the T. brucei genome database (Sanger Centre) using tblastn; this putative open reading frame (ORF) was PCR-amplified, cloned and sequenced. The sequence has been submitted to GenBank, Accession No. AJ86670. An alignment of the predicted translated sequence with INO1s from other organisms is shown in Fig. 1A and an unrooted phylogenetic tree in Fig. 1B. The predicted TbINO1 has a perfect copy of the motif GWGGNNG (underlined in Fig. 1A), which is involved in NAD+ binding, via a Rossmann fold typical of an oxidoreductase (Majumder et al., 2003). It also contains three other motifs which have been previously identified in other eukaryotic INO1s, LWTANTERY, NGSPQNTFVPGL and SYNHLGNNDG (Majumder et al., 2003). Although TbINO1 shows considerable similarity to INO1s from a variety of organisms, it is most closely related to those from two other kinetoplastids, T. cruzi and L. mexicana (Fig. 1B). The size of the predicted TbINO1 protein is 58 kDa, which is in good agreement with other INO1s which generally fall within the range of 58–67 kDa (Majumder et al., 1997; 2003).
Fig. 1.
A. clustalw alignment of the prediced amino acid sequence of the Trypanosoma brucei (Tb, AJ866770) INO1 with those of Leishmania mexicana (Lm, AJ344543), Saccharomyces cerevisiae (Sc, A30902), Homo sapiens (Hs, AAF26444) and Mycobacterium tuberculosis (Mt, P71703). Identical residues are shown in reverse type with black background, and conserved residues are shown in reverse type with grey background. The NAD+-binding motif GXGGXXG (residues 95–101) identified previously (Majumder et al., 2003) is shown by a solid line.
B. An unrooted phylogenetic tree of deduced amino acid sequences of INO1 proteins, showing relationship between those from protozoan parasites (T. brucei, T. cruzi, L. mexicana, P. falciparum) and others from yeast, plant, bacterial and human origins.
Overexpression of INO1 in E. coli
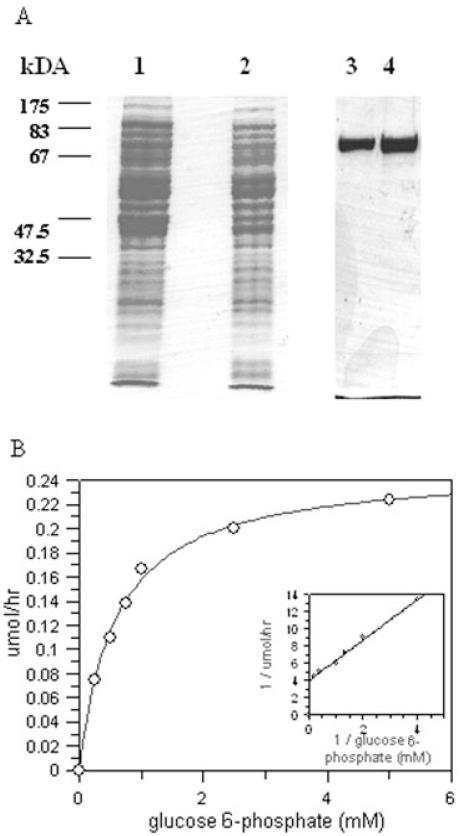
To enable biochemical characterization the T. brucei INO1 was overexpressed in E. coli using pBAD TA vector, which encodes a C-terminal hexa-His tag. This His-tagged recombinant protein was purified using Ni2+ and eluted using increasing concentration of imidazole. This purification protocol resulted in a pure protein preparation, as shown in Fig. 2A. The apparent molecular weight of this recombinant protein was approximately 75 kDa by SDS-PAGE analysis, higher than the predicted size of 64 kDa, which includes the short leader sequence and the hexa-His tag. However, the molecular weight of purified recombinant protein was shown to be approximately 64 kDa by MALDI analysis (data not shown).
Fig. 2.
Expression and purification of recombinant TbINO1 in E. coli.
A. TbINO1 was cloned into the expression vector pBAD TA (C-terminal hexa-His tag) and transformed into TOP10 E. coli competent cells, and production of recombinant TbINO1 was induced with 0.2% arabinose. After cell disruption, the soluble TbINO1 was purified by affinity chromatography using a chelating column charged with Ni2+, and proteins were separated on a SDS-PAGE gel and stained with Coomassie brilliant blue. Lane 1, total E. coli cellular protein after induction; lane 2, soluble E. coli protein after induction, which was loaded onto affinity chromatography column; lane 3, purified recombinant TbINO1 after affinity chromatography; lane 4, purified recombinant TbINO1 after dialysis.
B. Kinetics of recombinant TbINO1 for glucose 6-phosphate. Enzyme activity was measured as described in Experimental procedures, NAD+ concentration was held constant (1 mM) and glucose 6-phosphate concentration varied. Insert shows Lineweaver-Burk plot of data.
This recombinant protein was shown to be a catalytically active inositol-3-phosphate synthase through the use of a coupled assay, whereby the product of INO1 activity, inositol-3-phosphate, was used as a substrate for an IMPase, and the phosphate released by this IMPase measured colorimetrically. To study some kinetic parameters of glucose 6-phosphate, all components of the standard assay were held constant and the glucose 6-phosphate concentration varied between 0 and 5 mM. Saturation kinetics were shown by the enzyme, with an apparent Km of 0.58 mM and Vmax of 0.25 μmole h−1 (Fig. 2B) and the specific activity of the recombinant TbINO1 was found to be 756 U mg−1. Like other studied INO1 proteins, the recombinant TbINO1 activity is dependent on NAD+ no activity was observed in the absence of NAD+, or when NAD+ was replaced with either NADH or NADPH. Some activity (5% of normal) was observed when NAD+ was replaced with NADP+, suggesting that NADP+ can also act as hydrogen acceptor/donor during the reaction but only to a limited degree. Interestingly, even with NAD+ concentrations as low as 0.1 μmole, 100% of activity was observed compared with 1 mM, suggesting an extremely efficient binding of NAD+. The enzyme activity of the T. brucei INO1 was stimulated by the presence of NH+4 in the reaction mixture; there was very little activity observed in the absence of NH4+ (10 U mg−1), when compared with the activity in the presence of 2 mM NH4Ac (756 U mg−1), and no further enhancement of activity was observed when the NH4Ac concentration was increased to 10 mM.
INO1 is an essential gene in T. brucei bloodstream-form cells
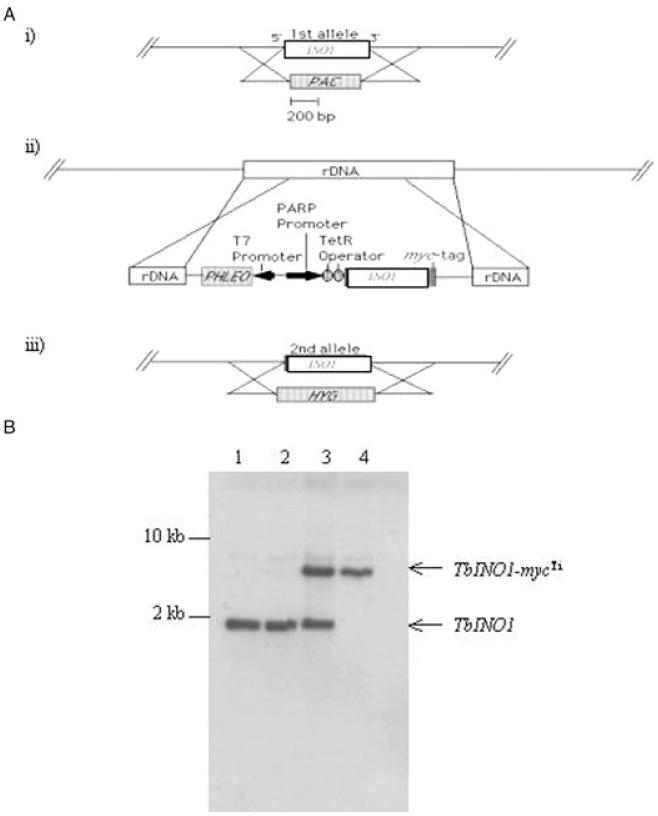
Southern blot analysis of T. brucei genomic DNA showed that INO1 is present as a single-copy gene per haploid genome (Fig. S1 in Supplementary material). A schematic of the construction of the conditional double knockout is shown in Fig. 3A. One allele of INO1 was replaced by the puromycin drug resistance gene by homologous recombination and selection with puromycin, creating the ΔINO1::PAC cell line (Fig. 3B, lane 2). Attempts to create a null mutant by homologous replacement of the second allele with the hygromycin resistance gene were unsuccessful, even when inositol level in the media was increased from 40 μM (normal HMI-9 media) to 100 mM. Therefore, as the ‘wild-type’ cell line used here constitutively expresses the T7 RNA polymerase and the tetracycline repressor protein, it was decided to introduce a tetracycline-inducible (Ti) myc-tagged ectopic copy of the INO1 into the ΔINO1::PAC cell line prior to deletion of the second allele, allowing the creation of a conditional double knockout which is under the control of tetracycline. The ectopic copy was integrated via the pLew 100 vector, in which the INO1 had been inserted downstream of the trypanosome procyclin promoter and two tetracycline operators. Integration of the pLew vector is known to occur in the rDNA locus, and it also encodes the phleomycin resistance protein (Wirtz et al., 1999). After drug selection with puromycin and phleomycin, several INO1-mycTi ΔINO1::PAC clones were obtained, and the integration of the pLew 100 ectopic copy was confirmed by PCR using primers specific to the pLew 100 vector (data not shown) and later by Southern analysis (Fig. 3B, lane 3). The second allele was replaced with the hygromycin resistance gene in the presence of tetracycline. One INO1-mycTi ΔINO1::PAC/ΔINO1::HYG clone was obtained and the genotype confirmed by Southern blotting (Fig. 3B, lane 4).
Fig. 3.
Construction of INO1 conditional double knockout cell line.
A. Schematic representation of construction strategy used. (i) One allele of INO1 was replaced by puromycin resistance gene (PAC) by homologous recombination generating ΔINO1::PAC cell line. (ii) A tetracycline-inducible ectopic copy of INO1 was introduced into the rDNA, generating INO1-mycTi ΔINO1::PAC cell line. (iii) While tetracycline induced the expression of the ectopic copy, the remaining allele was replaced by hygromycin resistance gene by homologous recombination, resulting in conditional double knockout cell line INO1-mycTi ΔINO1::PAC/ΔINO1::HYG.
B. Confirmation of genotype of T. brucei INO1 conditional double knockout cell line. Southern blot analysis of NcoI-digested genomic DNA (~2 μg) from wild-type T. brucei cells (lane 1), ΔINO1::PAC (lane 2), INO1-mycTi ΔINO1::PAC (lane 3) and INO1-mycTiINO1 ΔINO1::PAC/ΔINO1::HYG (lane 4); the INO1 ORF probe shows allelic TbINO1 at 1.9 kb and the ectopic copy TbINO1-mycTi at ~7 kb.
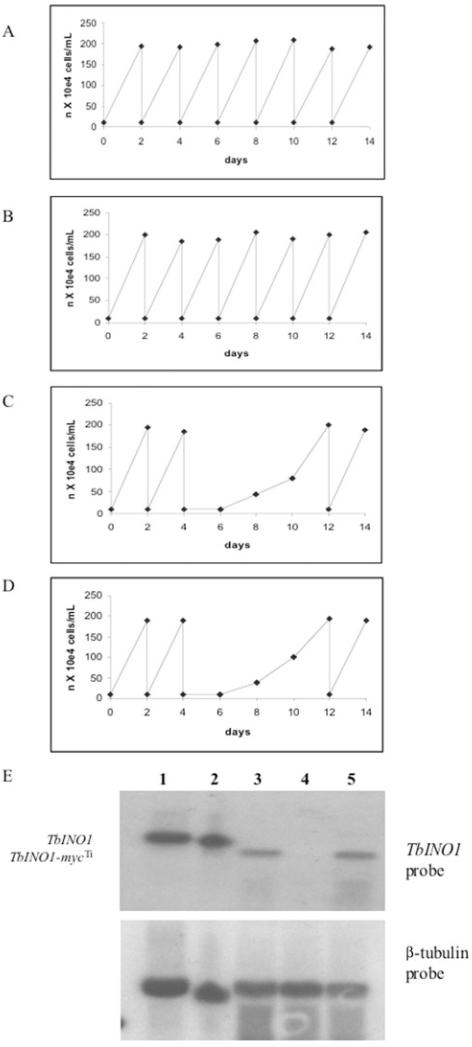
To establish whether INO1 was an essential gene, the growth of the INO1-mycTi ΔINO1::PAC/ΔINO1::HYG cell line was monitored in tetracycline containing and tetracycline-free HMI-9 which contained 40 μM myo-inositol. In the presence of tetracycline the INO1-mycTi ΔINO1::PAC/ΔINO1::HYG cell line showed normal growth rates (Fig. 4B) when compared with wild-type cells (Fig. 4A). However, in the absence of tetracycline cells grew normally for the first 2 days, cell numbers then declined to below the limits of detection by light microscopy and increasing cellular debris was observed suggesting the cell death (Fig. 4C). Even when this medium was supplemented with an additional 100 mM myo-inositol, the INO1 conditional knockouts were unable to survive in tetracycline-free media (Fig. 4D). However, in this tetracycline-free HMI-9 (with either 40 μM or 100 mM myo-inositol) from c. day 8 some live cells were visible by light microscopy, and after day 12 the cells resumed normal growth kinetics, shown in Fig. 4C. Northern blot analysis (Fig. 4E) showed that the transcript level of the ectopic INO1-mycTi was slightly lower than that of the endogenous copy seen for both bloodstream-form wild-type cells and the insect-form procyclic cells. In the absence of tetracycline for 48 h the transcript of INO1-mycTi was undetectable (Fig. 4E, lane 4), whereas in the cells which had spontaneously resumed normal growth after the absence of tetracycline for 12 days, the transcript of the INO1-mycTi is clearly visible (Fig. 4E, lane 5). This suggests that these revertant cells were able to overcome the tetracycline control, a phenomena that has been described for other essential genes in T. brucei (Krieger et al., 2000; Milne et al., 2001; Chang et al., 2002; Roper et al., 2002; 2005; Martin and Smith, 2006).
Fig. 4.
Growth curves and Northern blot of the TbINO1 conditional double knockout cell line.
A–D. Cells were washed with tetracycline-free HMI-9, and transferred to tetracycline-free media and counted daily. Growth curves are shown for wild-type cells in this medium (A), TbINO1 conditional double knockout with added tetracycline (B), TbINO1 conditional double knockout without tetracycline (C) and TbINO1 conditional double knockout without tetracycline, with added myo-inositol (100 mM) (D).
E. A Northern blot of total RNA from procyclic cells (lane 1), wild-type cells (lane 2), TbINO1 conditional double knockout without tetracycline on day 0 (lane 3), TbINO1 conditional double knockout without tetracycline on day 2 (lane 4) and TbINO1 conditional double knockout without tetracycline on day 12 (lane 5). The blot was either probed with TbINO1 ORF or β-tubulin, as detailed in Experimental procedures.
In vivo labelling of wild-type cells
Wild-type bloodstream-form T. brucei were labelled with [3H]-mannose, [3H]-glucose or [3H]-inositol in either pulse or pulse-chase experiments. After labelling for 1 h (pulse experiments) or 2 h (pulse-chase experiments) samples were taken for protein and lipid analysis.
When the cells were pulse labelled with [3H]-mannose the mature GPI glycolipids A and C were observed (Fig. 5A, lane 3), as well as low levels of galactosylated versions of glycolipid A, all of which have been described previously (Menon et al., 1988). As expected the pulse chase labelling showed decreased amounts of [3H]-labelled glycolipids A and C (Fig. 5, lane 4). This decrease is due to the addition of an equal volume of HMI-9 media to the labelling at the start of the chase and the longer labelling time of a pulse-chase experiment, allowing the labelled glycolipids A and C to be used in GPI-VSG attachment. Protein analysis confirmed that the [3H]-labelled glycolipds A and C were incorporated into mature GPI-anchored VSG and an equivalent signal was observed for the pulse and the pulse-chase experiments (Fig. 5C, lanes 5 and 6).
Fig. 5.
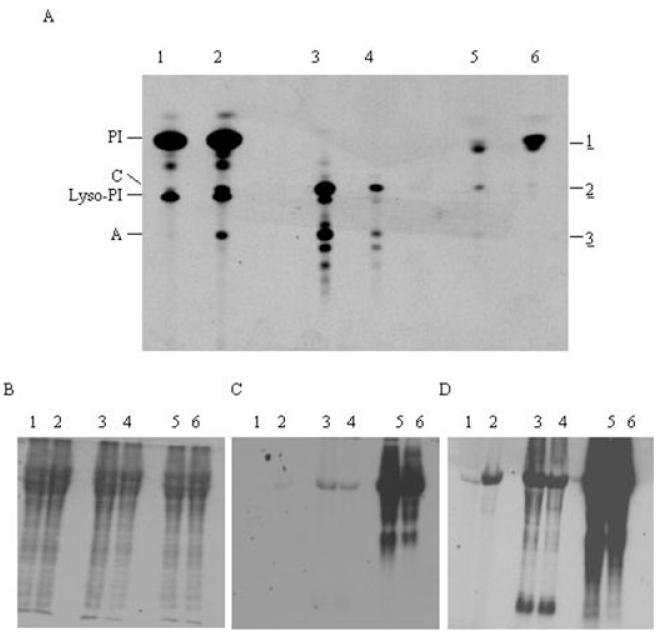
In vivo labelling of wild-type cells.
A. Wild-type cells were either pulse or pulse chase labelled and lipids were extracted, desalted, separated by HPTLC and detected by fluorography. Lane 1, [3H]-inositol pulse; lane 2, [3H]-inositol pulse chase; lane 3, [3H]-mannose pulse; lane 4, [3H]-mannose pulse chase; lane 5, [3H]-glucose pulse; lane 6, [3H]-glucose pulse chase.
B–D. From the labellings described for (A) samples were taken for protein analysis. Proteins were separated by SDS-PAGE and detected by Coomassie blue staining (B), or fluorography with either 2-day (C) or 2-week (D) exposure.
When labelled with [3H]-glucose either in the presence or in the absence of myo-inositol, wild-type cells produced three main lipid species (1, 2, 3) (Fig. 5A, lane 5). Lipid species 1 had an identical Rf to PI; digests showed it to be sensitive to PI-PLC and PLD, indicative of a PI containing phospholipid. Further results from base treatment, hydrogen fluoride (HF) treatment and deamination are all consistent with the [3H]-label being on the head group and not the diacylglycerol portion. To confirm the identity of the head group the aqueous phase from HF treatment was desalted, concentrated and analysed by high performance thin-layer chromatography (HPTLC); the Rf of the [3H]-labelled species was identical to an inositol standard, clearly showing that the head group is inositol (data not shown). These results clearly show the existence of a functional INO1 in bloodstream-form T. brucei, as the [3H]-glucose was converted into [3H]-inositol. This [3H]-inositol was subsequently used by the cell to form PI (lipid species 1).
Lipid species 2 from [3H]-glucose labelling had an identical Rf to a glycolipid C standard and to glycolipid C obtained by [3H]-mannose labellings (lanes 3 and 4). Lipid species 2’s insensitivity to PI-PLC and sensitivity to PLD are consistent with it being glycolipid C. Lipid species 3 had an identical Rf to glycolipid A standard and to glycolipid A from the [3H]-mannose labelling (lanes 3 and 4). These results and the identification lipid species 2 as glycolipid C suggest that lipid species 3 is glycolipid A. Furthermore, lipids from [3H]-glucose labelling were subjected to deamination and butanol-water partioning. HPTLC autoradiography of the butanol phase revealed that only PI was detected, and glycolipids C and A had been deaminated, thus losing their glycan head group, confirming their identities as GPIs (Fig. S2 in Supplementary material). Moreover, to confirm that the [3H]-label was on the myo-inositol head group (like PI) and not in the glycan head group as [3H]-GlcN or [3H]-Man, the relative amounts of [3H]-label in the butanol and aqueous phases after deamination were measured. As shown in Table S1 in Supplementary material, deamination of the [3H]-glucose labelling lipids resulted in almost all of the [3H]-label remaining in the butanol phase, showing that [3H]-label has remained bound to a lipid moiety. If significant [3H]-label had been detected in the aqueous phase it would be an indication that the [3H]-label had been released as part of the glycan head group upon deamination. As a control, [3H]-mannose-labelled glycolipids A and C were deaminated and as expected almost all of the [3H]-label was partitioned into the aqueous phase as the [3H]-mannose glycan head group (Table S1 in Supplementary material), indicating the deamination process was working efficiently. These results confirm that the majority of the [3H]-glucose is incorporated into GPI precursors as [3H]-myo-inositol and not as [3H]-GlcN or [3H]-mannose. Together these results show that [3H]-glucose is converted to [3H]-inositol, which is used to form [3H]-PI, and this is subsequently utilized by the GPI biosynthetic pathway to form [3H]-glycolipids A and C. Similar to the results observed for the pulse-chase [3H]-mannose labelling, the amount of [3H]-glucose-labelled glycolipids A and C decreased during the pulse chase, indicative of the [3H]-glycolipid species being utilized for GPI-VSG attachment (Fig. 5A, lane 6).
Analysis of the protein samples from [3H]-glucose labelling showed that the [3H]-labelled glycolipids A and C are incorporated into mature GPI-anchored VSG (Fig. 5C). A similar intensity of signal was observed for both pulse and pulse-chase experiments (Fig. 5C, lanes 3 and 4). Although analysis of lipid species 1 and 2 had clearly shown that the [3H]-label was on an inositol head group, we wanted to confirm that [3H]-label observed was on the GPI-anchored VSG and was not due to incorporation into N-linked oligosaccharides present on the VSG. This was confirmed by two methods. First, after labelling with [3H]-glucose, sVSG (VSG with the soluble portion of the GPI anchor attached) was purified and an aliquot was deglycosylated with PNGaseF. A clear difference in the molecular weight of the sVSG was observed by SDS-PAGE analysis, suggesting that the N-linked oligosaccharides had been successfully removed (Fig. S3A in Supplementary material, lanes 1 and 2). Autoradiography revealed that there was no significant decrease in the intensity of the [3H] signal after deglycosylation (Fig. S3A in Supplementary material, lanes 3 and 4). Second, when we conducted [3H]-glucose and [3H]-mannose labellings in the presence of tunicamycin (an inhibitor of N-glycosylation) the [3H]-label remained on the GPI-anchored VSG (Fig. S3B and C in Supplementary material). Together these results confirm that the signal observed on sVSG when labelling with [3H]-glucose is not due to the N-glycosylation. We also checked that the incorporation of the [3H]-label into mature GPI-anchored VSG was not due to an exchange mechanism as observed previously for the incorporation of myristate (Buxbaum et al., 1996). Labelling was performed in the presence or absence of cycloheximide and the incorporation of the [3H]-label into the GPI-anchored VSG determined. We could not detect any incorporation of the [3H]-glucose into the GPI-VSG in the presence of cycloheximide showing that incorporation is not due to an exchange reaction which uses the GPI-VSG as a substrate (Fig. S4 in Supplementary material). The expected incorporation of the [3H]-myristate was observed in the presence of cycloheximide and has been described previously (Buxbaum et al., 1996).
Not surprisingly when cells were pulse labelled with [3H]-inositol, PI and lyso-PI were observed (Fig. 5A, lane 1). However, when the cells were pulse chase labelled with [3H]-inositol two additional lipid species were observed with identical Rf values to glycolipids A and C respectively (Fig. 5A, lane 2). The identity of these new two lipid species as glycolipids A and C was confirmed by digests and by the incorporation of the [3H]-inositol on the GPI-anchored VSG (Fig. 5C and D), which could only be from the inositol head group. The appearance or increase of glycolipids A and C during the pulse-chase experiment with [3H]-inositol was opposite to what was observed when labelling with [3H]-glucose or [3H]-mannose, where the levels of glycolipids A and C decreased. In parallel there was an increase in the incorporation of the [3H]-labelled glycolipids A and C into the GPI-anchored VSG during pulse-chase experiments when compared with pulse alone (compare Fig. 5D lanes 1 and 2). In fact during a pulse labelling the incorporation of [3H]-inositol into mature GPI-anchored VSG was less than 5% than the incorporation of [3H]-glucose under the same conditions (Fig. 5C and D).
Biochemical phenotype of the INO1 conditional knockout
The biochemical phenotype of the INO1 conditional knockout cells was investigated by in vivo pulse-labelling experiments with [3H]-mannose and [3H]-glucose or pulse-chase experiments with [3H]-inositol after the cells had been grown in either the presence or absence of tetracycline for 2 days and compared with those undertaken with wild-type cells. After labelling, lipids were extracted, desalted, separated by HPTLC and detected by fluorography. As expected wild-type and INO1 conditional knockout cells grown in the presence of tetracycline showed identical incorporation of the [3H]-lipids (see earlier for identification of [3H]-lipids).
Labelling of the INO1 conditional knockout cells grown in the absence of tetracycline with [3H]-glucose showed a decrease in the amount of [3H]-PI formed (Fig. 6, lane 3), confirming that the deleted gene is a functional INO1. There is also a concomitant decrease in the amount of glycolipid C and glycolipid A, suggesting there is a direct knock-on effect to the GPI biosynthetic pathway due to the lack of PI in which the [3H]-inositol is derived from [3H]-glucose. This knock-on effect to the GPI biosynthetic pathway was confirmed by [3H]-mannose labelling (Fig. 6). There was a significant and reproducible decrease in the amounts of glycolipids A and C when the INO1 conditional knockout cells were grown in the absence of tetracycline (Fig. 6, lane 7). Again this suggests that the GPI biosynthetic pathway had slowed significantly to affect the dynamic pools of glycolipids A and C.
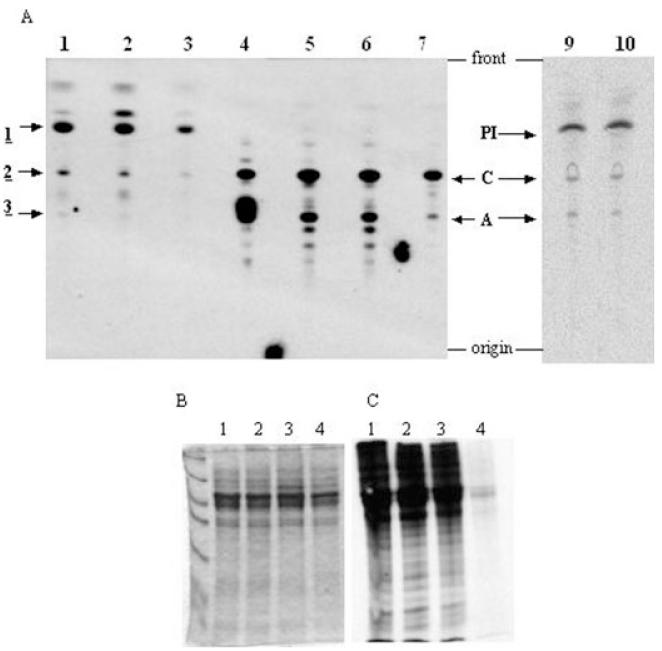
Fig. 6.
Phenotype analysis of the INO1 conditional knockout by in vivo labelling. A. Lipids from in vivo labelling with either [3H]-glucose, [3H]-mannose or [3H]-inositol were extracted, desalted and separated by HPTLC; radiolabelled lipids were detected by fluorography. Lipids from [3H]-glucose labelling are in lane 1 wild-type cells, lane 2 INO1 conditional knockout cells grown in the presence of tetracycline and lane 3 INO1 conditional knockout cells grown in the absence of tetracycline. Lipids from [3H]-mannose labelling are in lane 5 wild-type cells, lane 6 INO1 conditional knockout cells grown in the presence of tetracycline and lane 7 INO1 conditional knockout cells grown in the absence of tetracycline. Lane 4 contains glycolipids A and C standards. Lipids from [3H]-inositol labelling are in lane 9 wild-type cells and lane 10 INO1 conditional knockout cells grown in the absence of tetracycline.
B and C. Cells were labelled with 35S-methionine, and proteins were separated by SDS-PAGE and detected by either Coomassie blue staining or fluorography. Lane 1, wild-type cells; lane 2, TbINO1 conditional double knockout cells grown in the presence of tetracycline; lane 3, TbINO1 conditional double knockout cells grown in the absence of tetracycline; and lane 4 wild-type cells pre-incubated for 5 min with 60 μg ml−1 cycloheximide prior to labelling.
When the conditional double knockout cells were grown in the absence of tetracycline for 2 days and pulse chase labelled with [3H]-inositol, there was no significant change in the levels of PI, lyso-PI, glycolipids A or C observed when compared with either wild-type cells or conditional double knockout cells grown in the presence of tetracycline (Fig. 6). From these [3H]-inositol labellings it was inferred that the cells were still viable at the point of labelling as they were still able to synthesis lipids. To further confirm this we investigated the cells’ ability to synthesize new proteins. In parallel with the [3H]-glucose, [3H]-mannose and [3H]-inositol labellings described, the cells were labelled with [35S]-methionine. After the labelling was quenched, proteins were separated by SDS-PAGE and labelled proteins detected by autoradiography. All three cells lines (wild type, conditional double knockout with tetracycline and conditional double knockout without tetracycline) were able to incorporate significant and almost identical amounts of [35S]-methionine into protein (Fig. 6C, lanes 1, 2 and 3). These results clearly illustrate that at the point of labelling the INO1 conditional knockout cells grown under non-permissive conditions were still viable by their ability to synthesize protein at a level comparable to both wild-type cells or conditional double knockout cells grown in the presence of tetracycline. Therefore, at the time of labelling the cells are still viable so any biochemical phenotype observed is a direct result of the deletion of INO1.
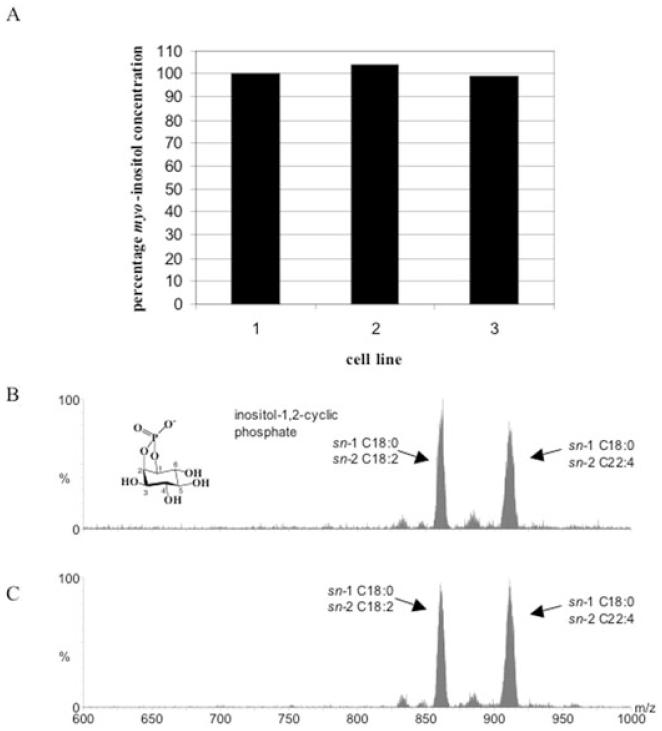
The amount of myo-inositol containing lipids in all the cell lines (wild-type cells, conditional double knockout cells grown in the presence and absence of tetracycline) was quantified by GC-MS. There was no significant difference in the amount of lipid-bound myo-inositol between the three cell lines (Fig. 7A), suggesting deletion of INO1 has not perturbed the overall cellular PI levels. We also qualitatively analysed inositol containing phospholipids by ES-MS-MS using parent-ion scanning of m/z 241 in negative ion mode (indicative collision induced fragment of all PI species) and did not detect any significant difference between the three cell lines (Fig. 7B).
Fig. 7.
Phosphatidylinositol analyses of TbINO1 conditional double knockout cells.
A. Lipid myo-inositol levels were quantified by GC-MS wild-type cells (cell line 1), TbINO1 conditional double knockout cells grown in the presence of tetracycline (cell line 2) and TbINO1 conditional double knockout cells grown in the absence of tetracycline (cell line 3). One hundred per cent represents 55 pmol myo-inositol per 5 × 104 cell equivalents.
B and C. Inositol phospholipids were qualitatively analysed by ES-MS-MS using parent-ion scanning of m/z 241 (the collision induced fragment of PI, inositol-1,2-cyclic phosphate) in negative ion mode either from (B) wild-type cells or (C) TbINO1 conditional double knockout cells grown in the absence of tetracycline.
INO1 is a cytosolic protein in bloodstream-form T. brucei
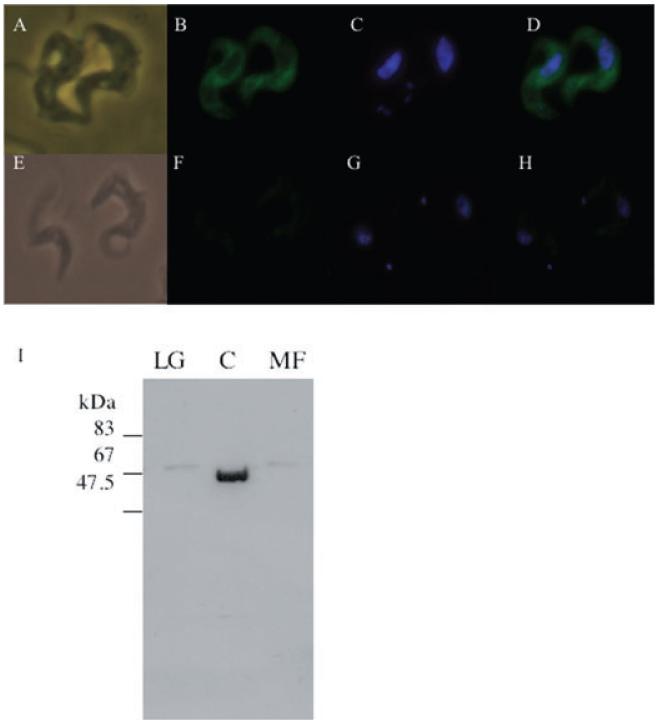
The cellular location of INO1 in T. brucei bloodstream-form cells was investigated using a tetracycline-inducible overexpression vector, pLew82 containing the TbINO1 and encoding a C-terminal haemagglutinin (HA) tag. This construct was introduced into bloodstream-form T. brucei cells, generating the cell line INO1-HATi and integration confirmed by PCR using primers specific to the vector (data not shown). Transcription of this ectopic INO1-HATi when induced by tetracycline was confirmed by Northern blotting (data not shown). Tetracycline-induced INO1-HATi cells were fixed, allowed to adhere to poly lysine slides and permeablized. The INO1-HATi protein was detected by immunofluorescence using a primary antibody against the HA tag, and the secondary (which was FITC conjugated) against the primary antibody. The FITC signal was observed throughout the cell body, and not seen in the flagella, nucleus or kinetoplast (Fig. 8A–D), suggesting that TbINO1 is cytoplasmic. This result was confirmed by subcellular fractionation studies (Fig. 8I), where the INO1-HA protein was detected in the soluble cytosolic fraction after differential centrifugation.
Fig. 8.
Subcellular localization of TbINO1-HATi in bloodstream-form T. brucei cells.
A–H. Wild-type cells (E–H) and TbINO1-HATi cells (A–D) were incubated with rat anti-HA antibodies, rabbit anti-rat FITC-conjugated antibodies (B and F), and DNA stained with 4′,6-diamidino-2-phenylindole (DAPI) (C and G). Phase images are shown in (A) and (E); FITC and DAPI merged images are shown in (D) and (H).
I. Western blot analysis of fractions from differential centrifugation, large granular (LG), cytosol (C) and microsomal fractions (MF) of TbINO1-HATi cells were prepared as described in Experimental procedures.
Discussion
Prior to the results presented in this study, it had been accepted in the field that although the GPI anchors contain inositol, GPI intermediates or mature GPIs could not be in vivo labelled with [3H]-inositol in bloodstream-form T. brucei, despite being able to efficiently label PI. This paradox could be explained if the myo-inositol used in GPIs was de novo synthesized from glucose. The results presented here support this hypothesis, not only demonstrating the existence of a functional myo-inositol de novo synthesis pathway in bloodstream-form T. brucei, but also showing that de novo synthesis is essential to the survival of the parasite and disruption of the pathway has detrimental effects on GPI anchor biosynthesis, which can not be overcome by the use of extracellular myo-inositol. These results raise several interesting questions about the distinction between de novo synthesized myo-inositol and that obtained from extracellular sources.
Most eukaryotic cells are able to de novo synthesize myo-inositol from glucose via INO1 and an IMPase. Recombinant expression of the putative TbINO1 in E. coli demonstrated that it is a catalytically active INO1 by its ability to convert glucose 6-phosphate to inositol-3-phosphate, also its dependence on NAD+ and stimulation by NH4+ are consistent with INO1s from other organisms (e.g. Donahue and Henry, 1981; Johnson and Sussex, 1995; Ju et al., 2004). Saturation kinetics were shown by the enzyme, with an apparent Km of 0.58 mM; this is very similar to the human INO1 which had a Km of 0.5 mM (Ju et al., 2004). Interestingly, the specific activity of the recombinant TbINO1 (756 U mg−1) is approximately 10-fold higher than that of the human recombinant INO1 (70 U mg−1) (Ju et al., 2004), which may be an indication of the importance of TbINO1 to T. brucei.
It has previously been shown that GPI anchor biosynthesis is essential for the survival of bloodstream-form T. brucei (Nagamune et al., 2000; Chang et al., 2002; Smith et al., 2004); we surmized that in light of this, INO1 and hence the de novo synthesis of myo-inositol would also be essential if they were responsible for providing myo-inositol for synthesis of PI for GPI anchors. We were unable to replace both allelic copies of INO1 to create a null mutant, suggesting that TbINO1 may be an essential gene and surprisingly, the addition of extra myo-inositol (from 40 μM to 100 mM) to the media did not enable the creation of a null mutant. Through the creation of a conditional double knockout and the cessation of growth under non-permissive conditions, it has been clearly demonstrated that INO1 is an essential gene in bloodstream-form T. brucei. Interestingly, even a substantial increase in the myo-inositol concentration in the growth media (40 μM to 100 mM) did not enable the conditional knockout cells to overcome the deletion of INO1 when grown in the absence of tetracycline (Fig. 4D). This inability to overcome the deletion of INO1 was surprising and unexpected as it is in contrast with all other ΔINO1 mutants to date, e.g. S. cerevisiae (Henry et al., 1977), L. mexicana (Ilg, 2002), and M. tuberculosis (Movahedzadeh et al., 2004) require 27 μM, 200 μM and 77 mM extracellular myo-inositol respectively. This clear difference between the T. brucei INO1 conditional mutant and null INO1 mutants from other organisms was intriguing and was probed further by biochemical phenotyping of INO1 conditional double knockout cells when grown under non-permissive conditions.
To our surprise in vivo labelling of wild-type cells show for the first time that is it possible to label both glycolipids A and C with [3H]-inositol. Moreover, we were able to show that this labelled myo-inositol was incorporated into GPI-anchored VSG, although at a significantly lower level than either [3H]-mannose or [3H]-glucose. However, we were only able to observe this labelling when the cells were pulse chase labelled. This increase in the labelling of GPIs and intermediates during the chase of a pulse chase labelling is quite surprising and it is opposite to what is observed when labelling with either mannose or glucose, where the labelling of glycolipids A and C actually decrease during the chase as they are used for GPI-VSG attachment. These results suggest that extracellular myo-inositol is entering the GPI biosynthetic pathway at a much slower rate than either the de novo synthesized myo-inositol or mannose. This latency may be explained by recycling, where extracellular inositol or PI synthesized from extracellular inositol is relocated within the cell and results in the mixing of the two types of myo-inositol/PI. These results also suggest that de novo synthesized myo-inositol is the primary source for GPI synthesis and extracellular inositol is a secondary source. This observation is supported twofold: first, the in vivo labelling showed that although GPI biosynthesis has slowed in the conditional double knockout cells grown under non-permissive conditions due to a lack of de novo synthesized inositol/PI, there was no concomitant increase in the levels of extacellular inositol feeding into the GPI pathway. Second, there was no significant change in the total cellular levels of lipid-based myo-inositol between wild-type cells or conditional double knockout cells grown under non-permissive conditions. Together, the results presented clearly demonstrate that deletion of INO1 is not causing an ‘inositol-less death’, but more intriguingly the cells are dying due to the decrease in the de novo synthesized myo-inositol, whose particular function appears to be supplying the GPI pathway with myo-inositol/PI. Moreover, these results explain why the additional myo-inositol in the media did not allow the creation of a null mutant and did not enable the conditional double knockout cells to overcome the deletion of INO1 when grown in the absence of tetracycline.
The clear preference for use of de novo synthesized for GPI anchors biosynthesis suggests a distinction or compartmentalization of the two types of myo-inositol (extracellular and de novo synthesized). To probe this idea further we have started investigating the location/s of enzymes of the de novo synthesis pathway in bloodstream-form T. brucei. Previously we have shown that a sole PI synthase (PIS) is responsible for synthesis of bulk cellular PI as well as PI destined for GPI anchor synthesis and is located in both the Golgi and endoplasmic reticulum (ER) (Martin and Smith, 2006) and we have shown here that TbINO1 is localized to in the cytosol, which is consistent with INO1s from other organisims. Based on these observations we propose the following: first, extracellular myo-inositol is utilized by the Golgi PIS to make bulk cellular PI. Second, TbINO1 catalyses the conversion of glucose 6-phosphate to inositol-3-phosphate in the cytoplasm, the presence of this phosphate group distinguishes de novo synthesized myo-inositol from that acquired from extracellular sources. This inositol-3-phosphate is transported to the ER, dephosphorylated by an IMPase and used by the ER PIS to form PI, which is subsequently used by the GPI biosynthetic machinery also located within the ER (see Ferguson, 1999, and references contained therein). Further evidence supporting this hypothesis is the identification of a putative IMPase (GeneDB annotation) which appears to be located in the ER (K.L. Martin and T.K. Smith, unpublished). To our knowledge, this is the first report from any organism of differentiation between de novo synthesized and extracellular myo-inositol and our current efforts are focused on understanding the complex mechanisms of this phenomena further. The physiological reason behind this preference for de novo synthesized myo-inositol remains unclear; it may be an evolutionary adaptation to guarantee that the parasites can sustain the high level of GPI anchor production, ensuring the integrity of its cell surface protective coat of VSG, enabling evasion of the hosts’ immune system.
In summary, the results presented here clearly demonstrate that bloodstream-form T. brucei is dependent on the de novo synthesis of myo-inositol. This dependence has been confirmed genetically by the perturbation of growth of INO1 conditional double mutants when grown in tetracycline-free media. Furthermore, our results suggest compartmentalization of PI synthesis, which has not been reported in any other organism to date. Surprisingly, there appears that there are no compensatory mechanisms whereby the extracellular myo-inositol can replace de novo synthesized myo-inositol when de novo synthesis is compromised, strongly validating both INO1 and the de novo synthesis of myo-inositol in bloodstream-form T. brucei as potential therapeutic drug targets. Currently, we are investigating compounds that interfere with the de novo synthesis of inositol as drug leads against African sleeping sickness and other related neglected diseases.
Experimental procedures
Cloning of the T. brucei INO1 gene and ligation into pLew vectors
A putative INO1 gene was identified in the T. brucei genome database (Sanger Centre) using tblastn search with the S. cerevisiae INO1 genomic sequence as the query. The ORF was amplified from T. brucei genomic DNA using the forward and reverse primers 5′-GAGGAGAAGCTTATGCCAGCCGTCCGTACG-3′ and 5′-TCGTTAATTAAACTTCCCACGCCGCGAAG-3′ containing HindIII and PacI restriction sites respectively (underlined). A band of the expected size of ~1.5 kb was amplified using Pfu polymerase, purified using a QIAquick PCR purification kit (Qiagen) and cloned into pCR-Blunt II TOPO (Invitrogen). The TbINO1 ORF was excised using HindIII and PacI and subsequently ligated into the tetracycline-inducible expression vectors pLew82 and pLew100 (Wirtz et al., 1999) via the HindIII and PacI restriction sites.
Construction of T. brucei gene replacement cassettes
The 5′ and 3′ untranslated regions (UTRs) immediately adjacent to the INO1 ORF were amplified from T. brucei genomic DNA using Pfu polymerase. The primers 5′-ATAAGAATGCGGCCGCAATGGCTGTTGTGGCACAAGG-3′ and 5′-GGATCCGTTTAAACTTACGGACCGTCAAGCTTGTACCACAACACGAATCTTCAGG-3′ were used for the 5′ UTR, amplifying the expected 398 bp product and primers 5′-AAGCTTTAAACGGATCCGTAAGTTTAAACGGATCCTGAATGTAACCGTCTACTTCGTTCCGC-3′ and 5′-ATAAGAATGCGGCCGCATGAAAAGAAAACATCGGGGAAT-3′ resulted in the correct 356 bp product of the 3′ UTR. These amplified products were used in a knitting PCR, resulting in a 744 bp product in which the 5′ UTR was joined to the 3′ UTR via a short BamHI–HindIII linker region contained within the described primers (italics) and a NotI site (underlined) at each end. This PCR product was ligated into pGEM-5Zf(+) (Promega) via the NotI sites and the hygromycin phosphotransferase (HYG) or puromycin acetyltransferase (PAC) resistance genes were ligated between the BamHI and HindIII restriction sites. Plasmid DNA was prepared using a QIAprep Miniprep Plasmid Kit (Qiagen); after digestion with NotI, it was precipitated with sodium acetate/ethanol and redissolved in sterile water to a final concentration of 1 μg μl−1.
Expression of T. brucei INO1 in E. coli
The INO1 ORF was amplified from the TbINO1-TOPO plasmid with Taq polymerase using the primers 5′-CGG GATCCCGGGATGCCAGCCGTCCGTACGAAAAGT-3′ and 5′-CCTTTCCTTCGCGGCGTGGGAAGTAGGCCGCTCGA GCGGC-3′ resulting in a single band of ~1.5 kb. The amplified product was purified using a QIAquick PCR purification kit (Qiagen) and ligated into pBAD TOPO TA (Invitrogen) vector and sequence confirmed. This construct was transformed into TOP10 competent cells (Invitrogen) for expression. A single colony was used to inoculate 200 ml of Luria–Bertani media containing 100 μg ml−1 ampicillin and grown at 37°C until OD (600 nm) was 0.5. The cells were induced with 0.2% arabinose and grown at 37°C for a further 3 h, after which the cells were harvested by centrifugation and stored at −20°C.
Purification of recombinant INO1
Frozen pellets were resuspended in lysis buffer (20 mM Tris pH 8.0, 300 mM NaCl) and incubated on ice with 0.2 mg ml−1 lysozyme (Sigma) for 30 min. Cells were disrupted by sonication (3 × 30 s) on ice and the supernatant was cleared by centrifugation (50 000 g, 30 min, 4°C). This supernatant was filtered (0.45 μm) prior to loading onto a Hi-Trap Chelating Sepharose HP column (Amersham) which had been charged with Ni2+ and equilibrated with lysis buffer. The column was connected to an AKTA FPLC (Amersham) and bound protein eluted in the same buffer containing 0.5 M imidazole, using a linear gradient of 0–0.25 M, a flow rate of 1 ml min−1 and collecting 1 ml fractions. Fractions containing the TbINO1 protein were pooled and dialysed against 20 mM Tris pH 8.0, 0.05 mM DTT for 24 h with regular buffer changes. The purity of the recombinant INO1 was checked by SDS-PAGE (10% gel). The dialysed sample was then adjusted to 15% glycerol and stored at −80°C.
Enzyme assays
The assay used for INO1 activity was based on that described by Eisenberg and Parthasarathy, (1987). Briefly, the standard assay reaction consisted of 50 mM Tris-Ac pH 8.0, 5 mM glucose 6-phosphate, 1 mM DTT, 1 mM NAD+, 2 mM NH4Ac and 5 μg of purified protein in a total volume of 150 μl. The assay was incubated at 37°C for 1 h, and the assay stopped by heating at 100°C for 5 min. To determine the amount of glucose 6-phosphate which had been converted to inositol-1-phosphate, 1.25 mU of IMPase from bovine brain (Sigma) and 4 mM MgCl2 were added in a total of 50 μl and incubated at 37°C for 1 h; the assay was stopped by heating at 100°C for 5 min. As this IMPase has negligible activity against glucose 6-phosphate, this resulted in the quantitative release of phosphate from inositol-1-phosphate which had been formed solely due to the action of TbINO1. For each assay a substrate blank and enzyme blank were also incubated with IMPase to account for any free phosphates already present in the sample and any non-specific activity of IMPase. The free phosphate was determined by the malachite green method of Itaya and Ui (1966) and compared with a standard curve of KH2PO4. Specific activity is defined as units (U) per milligram of protein, where 1 U is the amount of enzyme catalysing the formation of 1 nmole of inositol-1-phosphate per min at 37°C.
Southern and Northern blots
The INO1 ORF was PCR-amplified using the same primers described previously for ligation into pLew vectors and gel-purified with a QIAquick gel extraction kit (Qiagen). This fragment was then labelled with either fluorescein (Gene ImagesRandom prime module, Amersham) for Southern blotting or [α-32P]-dCTP (RediprimeII random prime labelling system, Amersham) for Northern blotting.
For Southern blots genomic T. brucei DNA (2 μg) was digested with various restriction enzymes, and the digestion products were separated on a 0.8% agarose gel and transferred to Hybon-N membrane (Amersham). The membrane was hybridized overnight in ULTRA-HYB (Ambion) at 42°C with the fluorescein labelled INO1 ORF probe. Stringency washes were carried out at 42°C, and consisted of two washes at low stringency for 15 min each (2× SSC, 0.1% SDS) and two washes at high stringency again for 15 min each (0.2× SSC, 0.1% SDS). Bound probe was detected using a CDP-STAR detection module (Amersham) and autoradiography.
For Northern blots total RNA was purified using RNeasy Mini Kit (Qiagen) and separated on a formaldehyde gel and transferred to Hybon N+ (Amersham). Hybridization conditions and stringency washes were as described for Southern blotting, using the 32P-labelled INO1 ORF as the probe. Bound probe was detected by autoradiography.
Cultivation and genetic modification of T. brucei
Bloodstream-form T. brucei strain 427, which had been previously modified to express both T7 polymerase and the tetracycline repressor protein (Wirtz et al., 1999), are referred to here as wild-type cells for convenience. Cells were grown in HMI-9 media supplemented with G418 (2.5 μg ml−1), at 37°C with 5% CO2 as described elsewhere (Wirtz et al., 1999; Milne et al., 2001; Chang et al., 2002; Roper et al., 2002). Transformation conditions and subsequent drug selection were also described elsewhere (Wirtz et al., 1999; Milne et al., 2001; Chang et al., 2002; Roper et al., 2002). For experiments requiring tetracycline-free conditions, Tet system approved fetal calf serum (Clonetech) was used. When tetracycline was added to the media, a final concentration of 1 μg ml−1 was used. For tetracycline-free experiments, cells were washed three times in tetracycline-free HMI-9 and resuspended in the same tetracycline-free media at 5 × 104 cells ml−1. Cells were counted each day and were passaged only when the density was between 2 × 106 and 3 × 106 (normally every second day).
Subcellular localization studies
Mid-log T. brucei INO1-HATi bloodstream-form cells which had been grown in the presence of tetracycline for 2 days were harvested by centrifugation (800 g, 10 min) and used for either immunofluorescence or differential centrifugation. For immunofluorescence, the cells were fixed with 4% paraformaldehyde (4°C, overnight). An aliquot of the fixed cells was placed on a poly lysine slide and allowed to air dry. Cells were rehydrated with PBS and washed with PBS-glycine (0.1 M) before permeablization with Triton X-100 (0.1%). The cells were blocked with 1% BSA in PBS before incubation with the primary monoclonal antibody; rat anti-HA (Roche) and the secondary antibody; FITC-conjugated rabbit anti-rat immunoglobulins (DakoCytomation).
For differential centrifugation, cells (1 × 108) were lysed in ice-cold water (100 μl) containing leupeptin (2 μg ml−1) and TLCK (0.1 μM) for 15 min on ice, before mechanical disruption of the cells by the addition of glass beads (425–600 μm, Sigma), and vortexing vigorously for 3 × 30 s, with 2 min intervals on ice. Glass beads were removed by centrifugation (500 g, 5 min). Differential centrifugation was a described previously (Sarkar et al., 2003). Briefly, unbroken cells and nuclei were removed by centrifugation at 1000 g for 10 min. The supernatant was removed and centrifuged at 14 500 g for 10 min to remove the large granular fraction. The resulting supernatant was removed and centrifuged at 140 000 g for 1 h, the supernatant contained the cytosolic fraction and pellet contained the small granular fraction. All pellets were resuspended in trypanosome dilution buffer (TDB) (25 mM KCl, 400 mM NaCl, 5 mM MgSO4, 100 mM Na2HPO4, NaH2PO4, 100 mM glucose) to an equal volume to that of the cytosolic fraction. Proteins were separated by SDS-PAGE and transferred to ECL-Nylon membrane by Western blotting. After blocking overnight in PBS-5% skim milk powder, protein was detected using the primary monoclonal antibody; rat anti-HA (Roche) and secondary antibody; horseradish peroxidase-conjugated rabbit anti-rat immunoglobulins (Jackson) and ECL western detection reagents.
In vivo T. brucei metabolic labelling
For each metabolic labelling, 2 × 107 mid-log cells were centrifuged (800 g, 10 min), washed in glucose-free minimal essential media, before being resuspended in the same media at a final concentration of 1 × 107 cells ml−1. Cells were labelled for 1 h (pulse) or 2 h (pulse chase) at 37°C with 50 μCi ml−1 of either d-[2-3H]-glucose (30 Ci mmol−1, ARC), d-[2-3H]-mannose (14 Ci mmol−1, Amersham) or d-[2-3H]-inositol (30 Ci mmol−1, Amersham) in a shaking water bath. For pulse-chase experiments, after 1 h of labelling an equal volume of normal media (HMI-9) was added and the labelling allowed to continue for another hour. The cells were collected by centrifugation (800 g, 10 min) and samples taken for either lipid or protein analysis. Lipids were extracted using chloroform:methanol:water (10:10:3 v/v) for 1 h, the supernatant was removed and the pellet was re-extracted with chloroform:methanol (2:1 v/v) for 1 h. The supernantants were pooled and dried before samples were desalted using butanol/water partioning. Lipids were separated by HPTLC using silica 60 HPTLC plates and chloroform:methanol:water (10:10:3 v/v) as the solvent. Radiolabelled lipids were either detected by fluorography at −70°C, after spraying with En3hance™ and using Kodak XAR-5 film with an intensifying screen, or when quantification was required the HPTLC plates were imaged using a Fuji FLA-2000 phosphorimager. To analyse proteins SDS-PAGE sample buffer was added to the cell pellet and heated at 95°C for 5 min. Proteins were separated on a 10% SDS-PAGE gel and visualized by Coomassie blue staining. To detect [3H]-labelled proteins the destained gel was soaked in En3hance™ (NEN) for 30 min, washed with water twice, soaked in 10% glycerol and dried and exposed to XAR-5 film at −70°C. Small-scale sVSG purification and PNGase F (Calbiochem) was as performed as described previously (Jones et al., 2005).
When labelling with [35S]-methionine, 1 × 107 mid-log cells were collected by centrifugation, washed in methionine-free minimal essential media and resuspended in the same media at a final concentration of 1 × 107 cells ml−1. The cells were labelled for 30 min with 20 μCi [35S]-methionine (MP Biomedicals, 1175 Ci mmol−1) at 37°C. To quench the labelling, the cells were diluted in 20 ml of cold TDB containing 1 mM l-methionine and centrifuged (800 g, 10 min, 4°C). After the supernatant was removed, the cells were resuspended in TDB and an equal volume of 2× SDS-PAGE sample buffer was added prior to heating at 100°C for 5 min. Proteins were separated on a 10% SDS-PAGE gel and visualized by Coomassie blue staining. To detect [35S]-labelled proteins the destained gel was soaked in En3hance™ (NEN) for 30 min, washed with water twice, soaked in 10% glycerol and dried. The dried gel was then exposed to XAR-5 film overnight at −70°C.
Enzymatic digests and chemical characterization of radiolabelled lipid/glycolipid species
Digestion with GPI-specific phospholipase D (GPI-PLD, Glyco) and PI-specific phospholipase C (PI-PLC, Glyco), deamination, base hydrolysis and HF treatment were as described previously (Güther et al., 1994; Güther and Ferguson, 1995; Smith et al., 1996). After deamination, reactions were subjected to butanol/water partitioning and total [3H] cpm in these fractions was determined by scintillation spectrometry using a Beckman LS6000E with formula 989 scintillation fluid (Packard Biosciences). HPTLC and detection were the same as described for in vivo T. brucei metabolic labelling.
Inositol analysis
Mid-log cells were collected by centrifugation (800 g, 10 min) washed with TDB and stored at −20°C. Lipids were extracted from these samples by the addition of 500 μl of chloroform methanol mixture (2:1 v/v) and incubated at room temperature for 1 h. The supernatant was removed and the pellet re-extracted with chloroform:methanol:water mixture (1:2:0.8 v/v). Pooled supernatants were dried under nitrogen prior to desalting by biphasic partitioning using 2:1 butanol:water (v/v). An internal standard of d6-myo-inositol was added to samples prior to hydrolysis by strong acid (6 M HCl, 110°C), derivitization with TMS and analysis by GC-MS, according to the method of Ferguson (1993). myo-Inositol was quantified and the means of three separate analyses were determined.
Supplementary Material
Acknowledgements
This work was supported by a Wellcome Trust Senior Research Fellowship 067441.
Footnotes
Supplementary material
The following supplementary material is available for this article online:
Fig. S1. Southern blot analysis of T. brucei genomic DNA, using TbINO1 ORF as a probe.
Fig. S2. Deamination of lipid samples from in vivo labelling.
Fig. S3. sVSG and deglycosylation analysis of samples from in vivo labelling.
Fig. S4. Analysis of exchange reaction with GPI-VSG as substrate.
Table S1. Deamination of lipid samples from in vivo labelling. Cells were labelled with [3H]-glucose or [3H]-mannose, lipids extracted, deaminated and subjected to butanol water partitioning. [3H]-Levels in the butanol and water phases were determined by scintillation spectrometry and are expressed as a percentage of the total cpm.
This material is available as part of the online article from http://www.blackwell-synergy.com
References
- Besra GS, Chatterjee D. Lipids and carbohydrates of Mycobacterium tubercolosis. In: Broom BR, editor. Tuberculosis: Pathogenesis, Protection and Control. American Society for Microbiology; Washington, DC: 1994. pp. 285–306. [Google Scholar]
- Buxbaum LU, Milne KG, Werbovetz KA, Englund PT. Myristate exchange on the Trypanosoma brucei variant surface glycoprotein. Proc Natl Acad Sci USA. 1996;93:1178–1183. doi: 10.1073/pnas.93.3.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T, Milne KG, Güther MLS, Smith TK, Ferguson MAJ. Cloning of the Trypansoma brucei and Leishmania major genes encoding the GlcNAc-phoshatidylinositol de-N-acetylase of glycosylphosphatidylinositol biosynthesis that is essential to the African sleeping sickness parasite. J Biol Chem. 2002;277:50176–50182. doi: 10.1074/jbc.M208374200. [DOI] [PubMed] [Google Scholar]
- Chen R, Walter EI, Parker G, Lapurga JP, Millan JL, Ikehara Y, et al. Mammalian glycophosphatidylinositol anchor transfer to proteins and posttransfer deacylation. Proc Natl Acad Sci USA. 1998;95:9512–9517. doi: 10.1073/pnas.95.16.9512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross GAM. Antigenic variation in trypanosomes: secrets surface slowly. Bioessays. 1996;18:283–287. doi: 10.1002/bies.950180406. [DOI] [PubMed] [Google Scholar]
- Donahue TF, Henry SA. Myo-inositol-1-phosphate synthase. J Biol Chem. 1981;256:7077–7085. [PubMed] [Google Scholar]
- Eisenberg FJ, Parthasarathy R. Measurement of biosynthesis of myo-inositol from glucose-6-phosphate. Methods Enzymol. 1987;141:127–143. doi: 10.1016/0076-6879(87)41061-6. [DOI] [PubMed] [Google Scholar]
- Ferguson MAJ. GPI membrane anchors: isolation and analysis. In: Fukuda M, Kobata A, editors. Glycobiology: A Practical Approach. IRL Press; Oxford: 1993. pp. 349–383. [Google Scholar]
- Ferguson MAJ. The structure, biosynthesis and functions of glycosylphosphatidylinositol anchors, and the contributions of trypanosome research. J Cell Sci. 1999;112:2799–2809. doi: 10.1242/jcs.112.17.2799. [DOI] [PubMed] [Google Scholar]
- Ferguson MAJ, Brimacombe JS, Brown JR, Crossman A, Dix A, Field RA, et al. The GPI biosynthetic pathway as a therapeutic target for African sleeping sickness. Biochim Biophys Acta. 1999;1455:327–340. doi: 10.1016/s0925-4439(99)00058-7. [DOI] [PubMed] [Google Scholar]
- Flury I, Benachour A, Conzelmann A. L031c belongs to a novel family of membrane proteins involved in the transfer of ethanolaminephosphate onto the core structure of glycosylphosphatidylinositol anchors in yeast. J Biol Chem. 2000;275:24458–24465. doi: 10.1074/jbc.M003844200. [DOI] [PubMed] [Google Scholar]
- Gerold P, Jung N, Azzouz N, Freiberg N, Kobe S, Schwarz RT. Biosynthesis of glycosylphosphatidylinositols of Plasmodium falciparum in a cell-free incubation system: inositol acylation is needed for mannosylation of glycosylphosphatidylinositols. Biochem J. 1999;344:731–738. [PMC free article] [PubMed] [Google Scholar]
- Guan G, Dai P, Shechter I. cDNA cloning and gene expression analysis of human myo-inositol 1-phosphate synthase. Arch Biochem Biophys. 2003;417:251–259. doi: 10.1016/s0003-9861(03)00388-6. [DOI] [PubMed] [Google Scholar]
- Güther MLS, Ferguson MAJ. The role of inositol acylation and inositol deacylation in GPI biosynthesis in Trypanosoma brucei. EMBO J. 1995;14:3080–3093. doi: 10.1002/j.1460-2075.1995.tb07311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güther MLS, Masterson WJ, Ferguson MAJ. The effects of phenylmethylsulfonyl fluoride on inositol-acylation and fatty acid remodelling in African trypanosomes. J Biol Chem. 1994;269:18694–18701. [PubMed] [Google Scholar]
- Haites RE, Morita YS, McConville MJ, Billman-Jacobe H. Function of phosphatidylinositol in mycobacteria. J Biol Chem. 2005;280:10981–10987. doi: 10.1074/jbc.M413443200. [DOI] [PubMed] [Google Scholar]
- Heise N, Raper J, Buxbaum LU, Peranovich TM, de Almeida ML. Identification of complete precursors for the glycosylphosphatidylinositol protein anchors of Trypanosoma cruzi. J Biol Chem. 1996;271:16877–16887. doi: 10.1074/jbc.271.28.16877. [DOI] [PubMed] [Google Scholar]
- Henry SA, Atkinson KD, Kolat AL, Culbertson MR. Growth and metabolism of inositol-starved Saccharomyces cerevisiae. J Bacteriol. 1977;68:2888–2898. doi: 10.1128/jb.130.1.472-484.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose S, Prince GM, Sevlever D, Ravi L, Rosenberry TL, Ueda E, Medof ME. Characterization of putative glycoinositol phospholipid anchor precursors in mammalian cells. Localization of phosphoethanolamine. J Biol Chem. 1992;267:16968–16974. [PubMed] [Google Scholar]
- Ilg T. Generation of myo-inositol-auxotrophic Leishmania mexicana mutants by targeted replacement of the myo-inositol-1-phosphate synthase gene. Mol Biochem Parasitol. 2002;120:151–156. doi: 10.1016/s0166-6851(01)00435-2. [DOI] [PubMed] [Google Scholar]
- Itaya K, Ui M. A new micromethod for the colorimetric determination of inorganic phosphate. Clinica Chimica Acta. 1966;14:361–366. doi: 10.1016/0009-8981(66)90114-8. [DOI] [PubMed] [Google Scholar]
- Johnson MD, Sussex IM. 1l-myo-inositol 1-phosphate synthase from Arabidopsis thaliana. Plant Physiol. 1995;107:613–619. doi: 10.1104/pp.107.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DC, Mehlert AM, Güther ML, Ferguson MAJ. Deletion of the glucosidase II gene in Trypanosoma brucei reveals novel N-glycosylation mechanisms in the biosynthesis of variant surface glycoprotein. J Biol Chem. 2005;280:35929–35942. doi: 10.1074/jbc.M509130200. [DOI] [PubMed] [Google Scholar]
- Ju S, Shaltiel G, Shamir A, Agam G, Greenberg ML. Human 1d-myo-inositol 3-phosphate synthase is functional in yeast. J Biol Chem. 2004;279:21759–21765. doi: 10.1074/jbc.M312078200. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Inoue N. Dissecting and manipulating the pathway for glycosylphosphatidylinositol-anchor biosynthesis. Curr Opin Chem Biol. 2000;4:632–638. doi: 10.1016/s1367-5931(00)00151-4. [DOI] [PubMed] [Google Scholar]
- Krieger S, Schwarz W, Ariyanayagam MR, Fairlamb AH, Krauth-Siegel RL, Clayton C. Trypanosomes lacking trypanothione reductase are avirulent and show increased sensitivity to oxidative stress. Mol Microbiol. 2000;35:542–552. doi: 10.1046/j.1365-2958.2000.01721.x. [DOI] [PubMed] [Google Scholar]
- Lohia A, Hait NC, Majumder AL. 1-myo-Inositol 1-phosphate synthase from Entamoeba histolytica. Mol Biochem Parasitol. 1999;98:67–79. doi: 10.1016/s0166-6851(98)00147-9. [DOI] [PubMed] [Google Scholar]
- de Macedo CS, Shams-Eldin H, Smith TK, Schwarz RT, Azzouz N. Inhibitors of glycosyl-phosphatidylinositol anchor biosynthesis. Biochimie. 2003;85:465–472. doi: 10.1016/s0300-9084(03)00065-8. [DOI] [PubMed] [Google Scholar]
- McConville MJ, Menon AK. Recent developments in the cell biology and biochemistry of glycosylphosphatidylinositol anchors. Mol Membr Biol. 2000;17:1–16. doi: 10.1080/096876800294443. [DOI] [PubMed] [Google Scholar]
- Majee M, Maitra S, Dastidar KD, Pattnaik S, Chatterjee A, Hait NC, et al. A novel salt-tolerant 1-myo-inositol-1-phosphate synthase form Porteresia coarctata (Roxb.) Tateoka, a halophytic wild rice. J Biol Chem. 2004;279:28539–28552. doi: 10.1074/jbc.M310138200. [DOI] [PubMed] [Google Scholar]
- Majumder AL, Johnson MD, Henry SA. Chapter XIX 1l-myo-Inositol-1-phosphate synthase. Biochim Biophys Acta. 1997;1348:245–256. doi: 10.1016/s0005-2760(97)00122-7. [DOI] [PubMed] [Google Scholar]
- Majumder AL, Chatterjee A, Dastidar KD, Majee M. Diversification and evolution of 1-myo-inositol 1-phosphate synthase. FEBS Lett. 2003;553:3–10. doi: 10.1016/s0014-5793(03)00974-8. [DOI] [PubMed] [Google Scholar]
- Martin KL, Smith TK. Phosphatidylinositol synthesis is essential in bloodstream form T. brucei. Biochem J. 2006;396:287–295. doi: 10.1042/BJ20051825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masterson WJ, Doering TL, Hart GW, Englund PT. A novel pathway for glycan assembly: biosynthesis of the glyosyl-phosphatidylinositol anchor of the trypanosome variant surface glycoprotein. Cell. 1989;56:793–800. doi: 10.1016/0092-8674(89)90684-3. [DOI] [PubMed] [Google Scholar]
- Masterson WJ, Raper J, Doering TL, Hart GW, Englund PT. Fatty acid remodeling: a novel reaction sequence in the biosynthesis of trypanosome glycosyl-phosphatidylinositol membrane anchors. Cell. 1990;62:73–80. doi: 10.1016/0092-8674(90)90241-6. [DOI] [PubMed] [Google Scholar]
- Menon AK, Major S, Ferguson MAJ, Duszenko M, Cross GAM. Candidate glycophospholipid precursor for the glycosylphosphatidylinositol membrane anchor of Trypanosoma brucei variant surface glycoproteins. J Biol Chem. 1988;263:1970–1977. [PubMed] [Google Scholar]
- Menon AK, Mayor S, Schwarz RT. Biosynthesis of glycosyl-phosphatidylinositol lipids in Trypanosoma brucei: involvement of mannosyl-phosphoryldolichol as the mannose donor. EMBO J. 1990a;9:4249–4258. doi: 10.1002/j.1460-2075.1990.tb07873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon AK, Schwarz RT, Mayor S, Cross GAM. Cell-free synthesis of glycosyl-phosphatidylinositol precursors for the glycolipid membrane anchor of variant surface glycoproteins. Structural characterization of putative biosynthetic intermediates. J Biol Chem. 1990b;265:9033–9042. [PubMed] [Google Scholar]
- Milne KG, Güther MLS, Ferguson MAJ. Acyl-CoA binding protein is essential in bloodstream form T. brucei. Mol Biochem Parasitol. 2001;112:301–304. doi: 10.1016/s0166-6851(00)00369-8. [DOI] [PubMed] [Google Scholar]
- Morita YS, Acosta-Serrano A, Buxbaum LU, Englund PT. Glycosyl phosphatidylinositol myristoylation in African trypanosomes. New intermediates in the pathway for fatty acid remodeling. J Biol Chem. 2000a;275:14147–14154. doi: 10.1074/jbc.275.19.14147. [DOI] [PubMed] [Google Scholar]
- Morita YS, Acosta-Serrano A, Englund PT. The biosynthesis of GPI enchors. In: Ernst P, Sinay P, Hart G, editors. Oligosaccharides in Chemistry and Biology – A Comprehensive Handbook. Wiley-VCH; Weinheim, Germany: 2000b. pp. 417–433. [Google Scholar]
- Movahedzadeh F, Smith DA, Norman RA, Dinadayala P, Murray-Rust J, Russell DG, et al. The Mycobacterium tuberculosis ino1 gene is essential for growth and virulence. Mol Microbiol. 2004;51:1003–1014. doi: 10.1046/j.1365-2958.2003.03900.x. [DOI] [PubMed] [Google Scholar]
- Nagamune K, Nozaki T, Maeda Y, Ohishi K, Fukuma T, Hara T, et al. Critical roles of glycosylphosphatidylinositol for Trypanosoma brucei. Proc Natl Acad Sci USA. 2000;97:10336–10341. doi: 10.1073/pnas.180230697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman RA, McAlister MSB, Murray-Rust J, Movahedzadeh F, Stoker NG, McDonald NQ. Crystal structure of inositol 1-phosphate synthase from Mycobacterium tuberculosis, a key enzyme in phosphatidylinositol synthesis. Structure. 2002;10:393–402. doi: 10.1016/s0969-2126(02)00718-9. [DOI] [PubMed] [Google Scholar]
- Park S, Kim J. Characterization of recombinant Drosophila melanogaster myo-inositol-1-phosphate synthase expressed in Escherichia coli. J Microbiol. 2004;42:20–24. [PubMed] [Google Scholar]
- Puoti A, Conzelmann A. Characterization of abnormal free glycophosphatidylinositols accumulating in mutant lymphoma cells of classes B, E, F, and H. J Biol Chem. 1993;268:7215–7224. [PubMed] [Google Scholar]
- Ralton JE, McConville MJ. Delineation of three pathways of glycosylphosphatidylinositol biosynthesis in Leishmania mexicana. Precursors from different pathways are assembled on distinct pools of phosphatidylinositol and undergo fatty acid remodeling. J Biol Chem. 1998;273:4245–4257. doi: 10.1074/jbc.273.7.4245. [DOI] [PubMed] [Google Scholar]
- Ralton JE, Mullin KA, McConville MJ. Intracellular trafficking of glycosylphosphatidylinositol (GPI)-anchored proteins and free GPIs in Leishmania mexicana. Biochem J. 2002;363:365–375. doi: 10.1042/0264-6021:3630365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper JR, Güther MLS, Milne KG, Ferguson MAJ. Galatose metabolism is essential for the African sleeping sickness parasite Trypanosoma brucei. Proc Natl Acad Sci USA. 2002;99:5884–5889. doi: 10.1073/pnas.092669999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper JR, Güther MLS, MacRae JI, Prescott AR, Hallyburton I, Acosta-Serrano A, Ferguson MAJ. The suppression of galactose metabolism in procyclic form Trypanosoma brucei causes cessation of cell growth and alters procyclin glycoprotein structure and copy number. J Biol Chem. 2005;280:19728–19736. doi: 10.1074/jbc.M502370200. [DOI] [PubMed] [Google Scholar]
- Sarkar M, Hamilton CJ, Fairlamb AH. Properties of phosphoenolpyruvate mutase, the first enzyme in the aminoethylphophonate biosynthetic pathway in Trypanosoma cruzi. J Biol Chem. 2003;278:22703–22708. doi: 10.1074/jbc.M302419200. [DOI] [PubMed] [Google Scholar]
- Smith TK, Cottaz S, Brimacombe JS, Ferguson MAJ. Substrate specifities of the dolicol phosphate mannose: glucosaminyl phospatidylinositol a1-4-mannosyltransferase of the glycosylphosphatidylinositol biosynthetic pathway of African trypanosomes. J Biol Chem. 1996;271:6476–6482. doi: 10.1074/jbc.271.11.6476. [DOI] [PubMed] [Google Scholar]
- Smith TK, Milne FC, Sharma DK, Crossman A, Brimacombe JS, Ferguson MAJ. Early steps in glycosylphosphatidylinositol biosynthesis in Leishmania major. Biochem J. 1997;326:393–400. doi: 10.1042/bj3260393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TK, Crossman A, Brimacombe JS, Ferguson MAJ. Chemical validation of GPI biosynthesis as a drug target against African sleeping sickness. EMBO J. 2004;23:4701–4708. doi: 10.1038/sj.emboj.7600456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striepen B, Dubremetz J-F, Schwarz RT. Glucosylation of glycosylphosphatidylinositol membrane anchors: identification of uridine diphosphate-glucose as the direct donor for side chain modification in Toxoplasma gondii using carbohydrate analogues. Biochemistry. 1999;38:1478–1487. doi: 10.1021/bi981884q. [DOI] [PubMed] [Google Scholar]
- Sutterlin C, Escribano MV, Gerold P, Maeda Y, Mazon MJ, Kinoshita T, et al. Saccharomyces cerevisiae GPI10, the functional homologue of human PIG-B, is required for glycosylphosphatidylinositol-anchor synthesis. Biochem J. 1998;332:153–159. doi: 10.1042/bj3320153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GAM. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.