Abstract
Inhaled corticosteroids (ICS) are one of the most commonly used asthma therapies and have highly variable treatment success. Polymorphisms in TBX21, a gene important for the biological action of corticosteroids, could be associated with treatment response in asthmatics. We genotyped for rs9910408 in TBX21 in 208 adult asthmatic patients, treated at least 3 years with ICS. Polymorphism rs9910408 was associated with response to ICS treatment. When treatment success was assessed by a decrease in bronchial hyperresponsiveness (BHR), the frequency of AA genotype was significantly higher in good responders (P = 0.049). This genotype related response was even more evident in the subgroups of non-smokers (P = 0.008) and in non-atopic patients (P = 0.009). AA genotype was overrepresented among good responders according to changes in FEV1 in the subgroups of non-smokers (P = 0.013) and in non-atopic patients (P = 0.048). Our results showed that treatment response to ICS, assessed as changes in BHR and FEV1, is associated with TBX21.
Asthma is a widespread disease affecting more than 300 million people worldwide. The most effective controller therapy in the treatment of asthma is considered inhaled corticosteroids (ICS). Because of large inter-individual variability and highly repeatable individual treatment response to ICS, it is reasonable to postulate a genetic basis for this heterogeneity1. It would be helpful to identify patients at risk for poor response because they and patients with possible adverse reactions to ICS would be candidates for novel, target-specific therapies. Several studies have evaluated the associations of longitudinal change in lung function or bronchial hyperresponsiveness (BHR) with single nucleotide polymorphisms (SNPs) in the candidate genes important for the biological action of corticosteroids2,3,4,5,6.
Characteristics of asthma are airway inflammation and hyperresponsiveness, reversible airway obstruction, and airway remodeling. Airways in asthma are infiltrated by Th2 lymphocytes. Conversely, in T cells from the airways of asthmatic patients, reduced expression of the Th1 transcription factor T-bet (TBX21, T-box 21) has been observed in comparison with those from airways of nonasthmatics7. TBX21 serves as a regulator of Th1 development both by inducing IFN-γ production and by inhibiting Th2 cytokines interleukin IL-4, IL-5, and IL-138. Gene knockout mice lacking TBX21 spontaneously develop histological and physiological features of asthma, including bronchial hyperresponsiveness (BHR)7. Because BHR is moderated by the use of ICS in asthma, it is conceivable that genetic variation in TBX21 may alter asthma phenotypes and treatment response to ICS2.
The question of possible involvement of genetic variants in the TBX21 gene on bronchial hyperresponsiveness and response to ICS treatment has been addressed in only a few studies. Raby et al9. reported on an association of SNP rs9910408 (c.-7947) in TBX21 with bronchial hyperresponsiveness in children. The association between rs9910408 and BHR was replicated in a cohort of adults: older men with BHR9.
Furthermore, Tantisira et al2. demonstrated significant improvement of PC20 for methacholine with ICS treatment of asthmatic children that had a functional variation in TBX21, coding for replacement of histidine 33 with glutamine. However, no association between H33Q and changes in forced expiratory volume in 1 second (FEV1) was observed after either 1 year or 4 years of therapy. A limitation of this study was that the minor allele frequency of this variant was only 4.5% and no minor homozygotes were observed.
The aim of our study was to further determine whether improvements in lung function (change in FEV1), decrease of BHR, and subjective assessment of therapy success or changes in the Asthma Control Test (ACT)10 and Asthma Quality of Life Questionnaire (AQLQ)11 scores are associated with polymorphism rs9910408 in the TBX21 gene in adult patients after at least 3 years of treatment with ICS.
Results
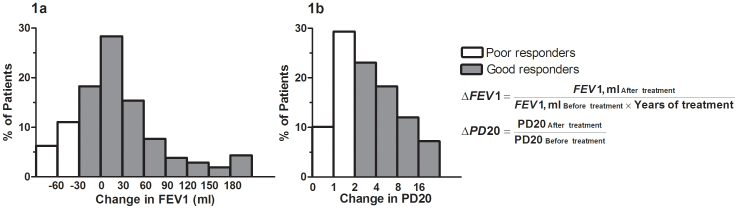
When analyzing the entire group of asthmatic patients, all parameters of asthma treatment outcome that were monitored (change in FEV1, change in PD20 for methacholine, and also in ACT and AQLQ scores after at least 3 years of treatment) showed a significant improvement (P < 0.0001, Table 1). However, large inter-individual variation in response to ICS was observed (Figure 1).
Table 1. Clinical characteristics of patients with asthma.
| CHARACTERISTICS | ALL PATIENTS | ATOPIC | NON-ATOPIC | EVER - SMOKERS | NON-SMOKERS | ICS ALONE recipients | ICS + LABA recipients | p-value4 |
|---|---|---|---|---|---|---|---|---|
| Subjects, n (%) | 208 | 90 (43) | 118 (57) | 53 (25.4) | 155 (74.6) | 121(58) | 87(42) | |
| Age, years, mean (SD) | 43 (14) | 36 (13) | 49 (12) | 44 (14) | 43 (15) | 42 (15) | 45 (13) | |
| Male sex, n (%) | 70 (43) | 41 (46) | 29 (24) | 25 (47) | 45 (29) | 31 (26) | 39 (45) | |
| %PREDICTED FEV1, median (IQR) | ||||||||
| At baseline | 84 (18) | 88 (19) | 83 (20) | 79 (17) | 87 (19) | 88 (15) | 76 (21) | |
| Change after 3 months of treatment 1 | 5 (9) | 5 (10) | 6 (9) | 5 (7) | 5 (9) | 4 (9) | 6 (9) | <0.0001 |
| Change after > 3 years of treatment 1 | 4 (9) | 4 (11) | 4 (8) | 4 (9.5) | 4 (9) | 4 (6.5) | 5 (11) | <0.0001 |
| PD20 mg at baseline, median (IQR) | 0.65 (1.09) | 0,56 (1.10) | 0.69 (1.09) | 0.48 (0.87) | 0.71 (1.20) | 1.00 (1.41) | 0.44 (0.73) | |
| PD20 mg change after > 3 years of treatment, median (IQR) 2 | 1.20 (2.35) | 1.27 (0.52) | 1.16 (2.13) | 1.07 (2.48) | 1.21 (2.36) | 1.27 (2.05) | 0.91 (2.83) | <0.0001 |
| ACT at baseline, median (IQR) | 13 (7) | 15 (8) | 12 (5) | 11.5 (6.25) | 13 (8) | 14 (8) | 12 (7) | |
| ACT, change after > 3 years of treatment, median (IQR) 3 | 7 (6) | 7 (6.5) | 8 (5) | 7 (5) | 7 (5) | 7 (6) | 7 (6) | <0.0001 |
| AQLQ at baseline, median (IQR) | 117 (61.5) | 125 (67) | 111 (54) | 110 (59.5) | 119 (61) | 118 (67) | 112 (58) | |
| AQLQ, change after > 3 years of treatment, median (IQR) 3 | 51 (46.5) | 50 (53.5) | 51 (40) | 52 (54.5) | 51 (44) | 51 (46) | 51 (52) | <0.0001 |
1The change from baseline in the % predicted FEV1 after 3 months and after > 3 years of treatment.
2The change from baseline PD20 in mg after > 3 years of treatment.
3The change in ACT questionnaire and in AQLQ questionnaire scores after > 3 years of treatment.
4Significance of changes from baseline for the entire group of 208 patients (Wilcoxon matched-pairs signed rank test).
Figure 1. Distribution of changes in FEV1 (a) and PD20 (b) in response to ICS therapy, showing large inter-individual variations.
Among the genotyped patients for rs9910408 in the TBX21 gene, 73 (35.1%) were major AA homozygotes, 111 (53.4%) were heterozygotes, and 24 (11.5%) were minor GG homozygotes, similar to the CEU population. The genotype distribution of the polymorphism analyzed was in Hardy–Weinberg equilibrium.
In the pharmacogenetic analysis, the patients were first stratified according to changes in BHR into good and poor responders after at least 3 years of ICS treatment (alone or in combination with LABA), as described in the Methods section. When the additive genetic model was assessed, genotype AA in TBX21 was associated with a greater decrease of BHR compared to the GG genotype (P = 0.049, OR = 2.74, 95% CI 1.06–7.06) in the entire group of patients (N = 208; Figure 2). The AA genotype frequency in good responders was 40% compared to 27% in patients with poor response, and there were no differences in BHR between patients with the AA and GG genotypes when therapy was first applied.
Figure 2. The distribution of genotypes in TBX21 gene in patients stratified to good and poor responders according to changes in PD20 after at least 3 years of ICS treatment in the entire group, in non-smokers and in non-atopic patients.
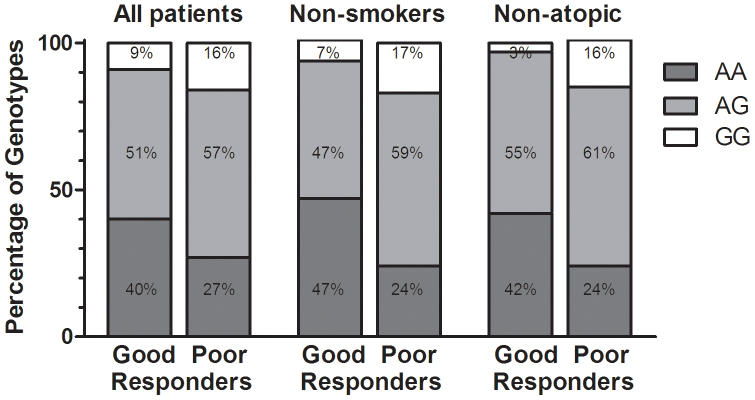
The association between the decrease in BHR and rs9910408 in TBX21 was even stronger in the subgroup of asthmatic never-smokers, using the additive genetic model (AA vs. GG; P = 0.008, OR = 5.26, 95% CI 1.65–16.69, Figure 2) as well as the recessive model (AA vs. AG + GG; P = 0.004, OR = 2.81, 95% CI 1.38–5.71) or dominant model (AG + AA vs. GG; P = 0.039, OR = 0.33, 95% CI 0.12–0.95). Furthermore, this association was also evident in the subgroup of non-atopic asthmatic patients, in which the AA genotype was overrepresented among good responders in comparison to the GG genotype (P = 0.009, OR = 9.33, 95% CI 1.72–50.63, Figure 2). The association between BHR and rs9910408 in TBX21 in non-atopic patients was also evident when using the recessive model (AA vs. AG + GG; P = 0.049, OR = 2.33, 95% CI 1.04–5.24) or dominant model (GG vs. AG + AA; P = 0.019, OR = 0.17, 95% CI 0.033–0.82).
Next we addressed the question of a possible association of TBX21 polymorphism rs9910408 and treatment outcome as monitored by changes in FEV1. When we stratified the patients into groups with good and poor response according to changes in FEV1 after at least 3 years of ICS treatment (alone or in combination with LABA), no differences in genotype distribution were evident when the entire group of patients was analyzed. However, similar to the analysis of BHR, in the subgroup of never-smokers as well as in the subgroup of non-atopic asthmatics, the AA genotype was overrepresented in the group of good responders. Patients without a smoking history with rs9910408 in the AA genotype had a greater improvement of FEV1 after at least 3 years of therapy in comparison to patients with the GG genotype (P = 0.013, OR = 5.78, 95% CI 1.49–22.37, Figure 3). The association was also statistically significant when using the recessive genetic model (AA vs. AG + GG; P = 0.019, OR = 3.30, 95% CI 1.18–9.22).
Figure 3. The distribution of genotypes in TBX21 gene in patients stratified to good and poor responders according to changes in FEV1 after at least 3 years of ICS treatment in the entire group, in non-smokers and in non-atopic patients.
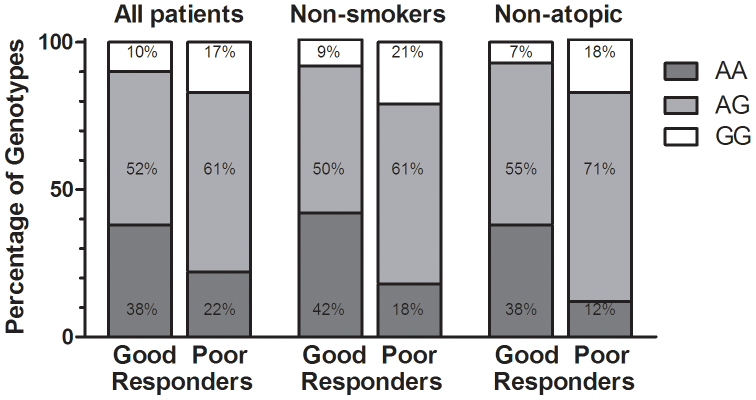
In non-atopic patients, the association between improvement in FEV1 and polymorphism in TBX21 was significant using the additive genetic model (AA vs. GG; P = 0.048, OR = 8.14, 95% CI 1.14–57.98, Figure 3) and marginally significant when using a recessive model (AA vs. AG + GG; P = 0.051, OR = 4.52, 95% CI 0.98, 20.89).
When stratifying patients into good and poor responders according to the improvement in questionnaire scores (ACT and AQLQ), no association between improvement in the ACT score and SNP rs9910408 in TBX21 was evident. However, the only association between improvement in the AQLQ score and rs9910408 in TBX21 was in the subgroup of non-atopic asthmatics, where this association was statistically significant using the additive genetic model (AA vs. GG; P = 0.037, OR = 23.82, 95% CI 1.05–542.70) as well as the recessive genetic model (AA vs. AG + GG; P = 0.050, OR = 9.77, 95% CI 0.55–173.80).
Discussion
To our knowledge, this is the first report describing the association of rs9910408 polymorphism in the TBX21 gene with improvement of bronchial hyperresponsiveness, FEV1, and quality of life in adult asthmatics treated with ICS. Altogether, our results indicate that adult asthmatic patients with the AA genotype in rs9910408 in TBX21 benefit considerably from ICS treatment. Those patients had a better therapeutic response compared to GG homozygotes as well as heterozygotes, suggesting that asthmatic airway inflammation in homozygotic AA patients is more effectively reduced by ICS treatment.
ICS are one of the most important asthma therapies, although the treatment success is highly variable. The reason for poor response in some patients is likely to be multifactorial, depending on phenotype and environmental exposure, and investigation of potential genetic determinants for ICS responsiveness is also recommended8. TBX21 is one of the potential pharmacogenetic candidate genes, encoding a transcription factor important for regulation of Th1 differentiation12, and could have important role in asthma pathogenesis because gene knockout mice develop features of asthma, including BHR7. Reduced TBX21 expression in CD4 lymphocytes in asthmatic airways leads to Th2 cytokine production (IL-4, IL-5, and IL-13)7. Our results support observations that these cytokine profiles are similar among patients with atopic and non-atopic (or “intrinsic”) asthma13,14. IL-13 seems to be the most important of them because of mediation of eosinophil recruitment, BHR, and mucus hypersecretion, even in the absence of IL-4 and IL-5. A local blockade of IL-13 by intranasal administration of blocking antibodies in T-bet–deficient mice reduces airway inflammation, BHR, and subsequent airway remodeling15. Tantisira et al. observed decreased IL-13 production in cellular models with polymorphism in TBX21 coding for H33Q, a genetic variant associated with better responses to ICS2. Different therapeutic responses to ICS regarding polymorphism rs9910408 in the TBX21 gene in our study may be mediated mostly by these effects on IL-13 cytokine production.
When analyzing the entire group of asthmatic patients, all parameters of asthma treatment outcome monitored (change in FEV1 (after 3 months and after at least 3 years of treatment), change in PD20 for methacholine, ACT and AQLQ scores after at least 3 years of treatment), showed significant improvement (p < 0.0001, Table 1). However, large inter-individual variation in response to corticosteroid treatment was observed, which could at least partly be attributed to a genetic basis. Further pharmacogenetic analysis confirmed our expectations and revealed that adult asthmatic patients with the AA genotype in rs9910408 in TBX21 benefit considerably from ICS treatment. We found a statistically significant decrease of BHR after at least 3 years of ICS treatment in adult patients with AA genotype compared to the GG genotype. The genotype related improvement in PD20 was even more evident in the subgroups of non-smokers and non-atopic patients.
Patients with the AA genotype also had higher improvement of FEV1 after at least 3 years of therapy with ICS in the subgroup of non-smokers and in non-atopic patients. Furthermore, in the subgroup of non-atopic patients, those with the AA genotype had a markedly greater improvement in AQLQ score compared to the GG genotype.
Although rs9910408 is located in the intergenic region, the studies performed showed similar results of its contribution to asthma. Our results are in line with a previous study, in which the AA genotype was found to be associated with BHR in children and adult men9.
The levels of airway hyperresponsiveness and airway obstruction are primarily related to the prognosis of asthma16. As observed by Tantisira et al2. genetic variations in TBX21 may alter treatment response to ICS. They described a 3.5-fold greater mean increase in log-transformed PC20 for methacholine in asthmatic children with glutamine variants compared to those homozygous for histidine 33 TBX21 rs2240017 after 4 years of treatment with ICS. However, the allele frequency of the minor allele H33Q was only 4.5% and no minor homozygotes were detected. Our association study analyzed the single nucleotide polymorphism rs9910408 in TBX21 with minor allele frequency of 43% in the white population9.
Previous studies have shown that smoking asthmatics had a decreased response to ICS treatment regarding changes in lung function and BHR17,18. Increased production of Th2 inflammatory cytokines in smokers, such as IL-4, observed in a study by Byron et al. could be associated with this corticosteroid resistance19. In our patients, increase of FEV1 from baseline after 3 years of treatment in the entire group was similar in non-smokers and in (ever–)smokers. However, non-smokers with the AA genotype in TBX21 had a significantly better therapeutic response in FEV1, as well as in PD20 increase (compared to those with the GG or AG genotype), suggesting that the heritability of the treatment response can be strongly modified by environmental factors such as smoking exposure, thus making it difficult to determine the genetic basis of treatment response. It also remains unclear whether smoking affects the treatment's success itself or whether it only modifies the disease progression.
Three previous studies found no evidence of association between TBX21 polymorphisms and atopy among asthmatics9,20,21. These findings were also confirmed in our study. However, in our pharmacogenetic study, non-atopic patients with the AA genotype had an even better response to ICS than atopic patients.
In conclusion, we have shown a TBX21 rs9910408 genotype-specific treatment response in adult asthmatics after inhaled corticosteroid therapy. Our results suggest that rs9910408 genotypes may allow the identification of patients that are more or less likely to respond well to ICS therapy. However, in the future functional studies and investigation of several SNPs, among which rs9910408 in TBX21 is a good candidate, will be needed to confirm the actual efficacy of such a predictive test.
Methods
This study is a prospective study involving 208 adult (>18 years) patients with atopic and non-atopic, mild to moderate persistent asthma that attended pneumological care at the outpatient pneumological practice. At their first visit, lung function and methacholine challenge tests were performed. All patients showed a positive methacholine test defined as a decrease of baseline FEV1 of 20% with a cumulative dose of methacholine (PD20) less than 4 mg, and the great majority of them had normal or near-normal spirometry testing results. All patients also underwent skin prick tests for common allergens and completed the Asthma Control Test (ACT) and Asthma Quality of Life Questionnaire (AQLQ). Detailed clinical and laboratory parameters are listed in Table 1.
After the diagnosis was established, all patients started treatment with inhaled corticosteroids (alone or in combination with long-acting beta agonists (LABA), according to achieved asthma control). Follow-up visits with spirometry testing were made every 3 or 6 months, with the last visit after at least 3 years of treatment (mean 4.6, SD 1.3 years). At this last time point, we also repeated the ACT, AQLQ, and methacholine test. All spirometry and methacholine challenge testing was performed by the same technician, with the same (dosimeter) method and equipment (Spirojet, Provojet nebulizer, Ganshorn, Germany) to avoid bias. Patients were seated and wearing nose clips. In atopic patients sensitized for pollens, final methacholine challenge tests were performed in the same season of the year as the initial tests.
Definition of response
According to the response to ICS therapy (alone or in combination with LABA), patients were divided into “poor” and “good” responders in accordance with the American Thoracic Society (ATS) and European Respiratory Society (ERS) interpretation of changes in PD20 and FEV1 and data from other studies evaluating treatment response in asthma22,23,24,25,26,27,28,29,30.
Bronchial hyperresponsiveness: When stratifying patients according to change in BHR, poor response was defined as an increase of PD20 for methacholine that was smaller than one doubling dose compared to the initial PD20.
Lung function: Poor response according to changes in FEV1 was defined as a decrease in FEV1 by more than 30 ml/year.
Asthma control: Poor response was defined as less than a three-point increase in the ACT score after at least 3 years of treatment31.
Asthma-related quality of life: Poor response was defined as less than a 16-point increase from the initial AQLQ score32.
We also analyzed the therapeutic response in the subgroups of different asthma phenotypes (atopic and non-atopic asthma) and to smoking history ((ever-) smokers and non-smokers). Therapeutic responses according to phenotype are listed in Table 2.
Table 2. TBX21 rs9910408 genotype frequencies in good and poor responders after > 3 years of treatment.
| Response | TBX21 rs9910408 genotype | ALL PATIENTS (n, %) | ATOPIC (n, %) | NON-ATOPIC(n, %) | SMOKERS (n, %) | NON-SMOKERS (n, %) | ICS ALONE recipients (n, %) | ICS + LABA recipients (n, %) | |
|---|---|---|---|---|---|---|---|---|---|
| % FEV1 change | good | AA | 65 (37.8) | 27 (38.0) | 38 (37.6) | 12 (26.7) | 53 (41.7) | 37 (39.8) | 28 (35.4) |
| AG | 89 (51.7) | 33 (46 5) | 56 (55.4) | 26 (57.8) | 63 (49.6) | 45 (48.4) | 44 (55.7) | ||
| GG | 18 (10.5) | 11 (15.5) | 7 (7.0) | 7 (15.5) | 11 (8.7) | 11 (11.8) | 7 (8.9) | ||
| poor | AA | 8 (22.2) | 6 (31.6) | 2 (11.8) | 3 (37.5) | 5 (17.9) | 7 (25.0) | 1 (12.5) | |
| AG | 22 (61.1) | 10 (52.6) | 12 (70.6) | 5 (62.5) | 17 (60.7) | 16 (57.1) | 6 (75.0) | ||
| GG | 6 (16.7) | 3 (15.8) | 3 (17.6) | 0 (0.0) | 6 (21.4) | 5 (17.9) | 1 (12.5) | ||
| PD20 change | good | AA | 51 (40.5) | 23 (39.0) | 28 (41.8) | 8 (23.5) | 43 (46.7) | 28 (41.8) | 23 (39.0) |
| AG | 64 (50.8) | 27 (45.8) | 37 (55.2) | 21 (61.8) | 43 (46.7) | 31 (46.3) | 33 (55.9) | ||
| GG | 11 (8.7) | 9 (15.2) | 2 (3.0) | 5 (14.7) | 6 (6.6) | 8 (11.9) | 3 (5.1) | ||
| poor | AA | 22 (26.8) | 10 (32.3) | 12 (23.5) | 7 (36.9) | 15 (23.8) | 16 (29.6) | 6 (21.4) | |
| AG | 47 (57.3) | 16 (51.6) | 31 (60.8) | 10 (52.6) | 37 (58.7) | 30 (55.6) | 17 (60.7) | ||
| GG | 13 (15.9) | 5 (16.1) | 8 (15.7) | 2 (10.5) | 11 (17.5) | 8 (14.8) | 5 (17.9) | ||
| ACT score change | good | AA | 61 (34.3) | 24 (32.9) | 37 (35.2) | 12 (26.1) | 50 (37.9) | 37 (34.9) | 24 (33.3) |
| AG | 98 (55.0) | 37 (50.7) | 61 (58.1) | 29 (63.0) | 68 (51.5) | 55 (51.9) | 43 (59.7) | ||
| GG | 19 (10.7) | 12(16.4) | 7 (6.7) | 5 (10.9) | 14 (10.6) | 14 (13.2) | 5 (7.0) | ||
| Poor | AA | 12 (40.0) | 9 (52.9) | 3 (23.1) | 3 (42.8) | 9 (39.1) | 7 (46.7) | 5 (33.3) | |
| AG | 13 (43.3) | 6 (35.3) | 7 (53.8) | 2 (28.6) | 11 (47.8) | 6 (40.0) | 7 (46.7) | ||
| GG | 5 (16.7) | 2 (11.8) | 3 (23.1) | 2 (28.6) | 3 (13.1) | 2 (13.3) | 3 (20.0) | ||
| AQLQ score change | good | AA | 68 (36.4) | 28 (36.4) | 40 (36.4) | 13 (27.1) | 55 (39.6) | 41 (36.9) | 27 (35.5) |
| AG | 100 (53.5) | 38 (49.3) | 62 (56.4) | 30 (62.5) | 70 (50.3) | 56 (50,5) | 44 (57.9) | ||
| GG | 19 (10.1) | 11 (14.3) | 8 (7.2) | 5 (10.4) | 14 (10.1) | 14 (12.6) | 5 (6.6) | ||
| poor | AA | 5 (23.8) | 5 (38.5) | 0 (0.0) | 2 (40.0) | 3 (18.7) | 3 (30.0) | 2 (18.2) | |
| AG | 11 (52.4) | 5 (38.5) | 6 (75.0) | 1 (20.0) | 10 (62.6) | 5 (50.0) | 6 (54.5) | ||
| GG | 5 (23.8) | 3 (23.0) | 2 (25.0) | 2 (40.0) | 3 (18.7) | 2 (20.0) | 3 (27.3) |
The study was approved by the Slovenian national medical ethics committee and all patients gave their informed written consent.
DNA isolation and single nucleotide polymorphism (SNP) genotyping
Genomic DNA was extracted from EDTA-containing whole blood samples, using a QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The genotypes of the SNP analyzed were determined using a 5′-nuclease allelic discrimination assay in a 96-well format.
Primers and probes were purchased from Applied Biosystems (Foster City, CA, USA) for SNP genotyping assay rs9910408 in TBX21. Allelic discrimination assays were performed in 5 μL reaction volumes, using approximately 5 ng of DNA as a template, 2× TaqMan Fast Advanced Master Mix, and the predesigned SNP genotyping assay provided by Applied Biosystems. The temperature conditions for the PCR were set at 50°C for 2 minutes and 95°C for 20 seconds, followed by 40 cycles at 95°C for 3 seconds and at 60°C for 30 seconds. Genotyping of the amplified PCR products was determined on the basis of the differences in VIC and FAM fluorescent levels, using the ABI Prism 7500 Fast Real-Time PCR system (system instrument equipped with SDS v2.0.5 software; Applied Biosystems).
Statistical analysis
The Hardy–Weinberg equilibrium was tested using the chi-squared test for the goodness-of-fit (one degree of freedom) model. Data distribution was evaluated by the D'Agostino–Pearson test. Parametric statistics (paired and unpaired t-tests) were used on normally distributed data, and non-parametric statistics (the Mann–Whitney, Wilcoxon, and Kruskal–Wallis tests) were used if the distribution deviated from normal.
Genotypic distribution and allelic frequencies in “poor” and “good” responders (with regard to change from baseline in FEV1, changes in PD20 for methacholine, and in ACT and AQLQ scores after at least 3 years of therapy) were compared using the chi-squared test calculated on contingency tables. Odds ratios (ORs) with 95% confidence intervals (95% CIs) were calculated using the same test. We used GraphPad Prism software (version 6.0 for Windows; GraphPad Software, San Diego, CA, USA). A p-value of less than 0.050 was accepted as statistically significant.
Author Contributions
R.M., K.P. and F.M. designed the experiments. L.A., Ž.M. and R.M. performed the experiments. L.A. and R.M. wrote the main manuscipt text. K.P. and F.M. supervised the work. All authors reviewed the manuscript.
Acknowledgments
This research was supported by grant no. P3-0360 from the Slovenian Research Agency and we are grateful to all patients that participated in this study.
References
- Weiss S. T. New approaches to personalized medicine for asthma: Where are we? J. Allergy Clin. Immunol. 129, 327–334 (2012). [DOI] [PubMed] [Google Scholar]
- Tantisira K. G. et al. TBX21: a functional variant predicts improvement in asthma with the use of inhaled corticosteroids. Proc. Natl. Acad. Sci. USA 101, 18099–18104 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantisira K. G. et al. Corticosteroid Pharmacogenetics: Association of sequence variants in CRHR1 with improved lung function in asthmatics treated with inhaled corticosteroids. Hum. Mol. Genet. 13, 1353–1359 (2004). [DOI] [PubMed] [Google Scholar]
- Tantisira K. G. et al. Genomewide association between GLCCI1 and response to glucocorticoid therapy in asthama. N. Engl. J. Med. 365, 1173–1183 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeachie M. J. et al. Predicting inhaled corticosteroid response in asthma with two associated SNPs. Pharmacogenomics J. 1–6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balantic M. et al. Asthma treatment outcome in children is associated with vascular endothelial growth factor A (VEGFA) polymorphisms. Mol. Diagn. Ther. 16, 173–180 (2012). [DOI] [PubMed] [Google Scholar]
- Finotto S. et al. Development of spontaneous airway changes consistent with human asthma in mice lacking T-bet. Science 295, 336–338 (2002). [DOI] [PubMed] [Google Scholar]
- Glimcher L. H. & Murphy K. M. Lineage commitment in the immune system: the T helper lymphocyte grows up. Genes Dev. 14, 1693–1711 (2000). [PubMed] [Google Scholar]
- Raby B. A. et al. T-bet polymorphisms are associated with asthma and bronchial hyperresponsiveness. Am. J. Respir. Crit. Care Med. 173, 64–70 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan R. A. et al. Development of the Asthma Control Test: a survey for assessing asthma control. J. Allergy Clin. Immunol. 113, 59–65 (2004). [DOI] [PubMed] [Google Scholar]
- Juniper E. F. et al. Evaluation of impairment of health related quality of life in asthma: development of a questionnaire for use in clinical trials. Thorax 47, 76–83 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaga M., Zervas E. & Chanez P. Update on severe asthma: what we know and what we need. Eur Respir. Rev. 18, 58–65 (2009). [DOI] [PubMed] [Google Scholar]
- Humbert M. et al. Elevated expression of messenger ribonucleic acid encoding IL-13 in the bronchial mucosa of atopic and nonatopic subjects with asthma. J. Allergy Clin. Immunol. 99, 657–665 (1997). [DOI] [PubMed] [Google Scholar]
- Humbert M. et al. IL-4 and IL-5 mRNA and protein in bronchial biopsies from patients with atopic anf nonatopic asthma: evidence against »intrinsic« asthma being distinct immunopathologic entity. Am. J. Respir. Crit. Care Med. 154, 1497–1504 (1996). [DOI] [PubMed] [Google Scholar]
- Finotto S. et al. Asthmatic changes in mice lacking T-bet are mediated by IL-13. Int. Immunol. 17, 993–1007 (2005). [DOI] [PubMed] [Google Scholar]
- Kerstjens H. A. M. et al. Airways hyperresponsiveness, bronchodilator response, allergy and smoking predict improvement in FEV1 during long-term inhaled corticosteroid treatment. Eur. Respir. J. 6, 868–876 (1993). [PubMed] [Google Scholar]
- Chauduri R. et al. Cigarette smoking impairs the therapeutic response to oral corticosteroids in chronic asthma. Am. J. Respir. Crit. Care Med. 168, 1308–1311 (2003). [DOI] [PubMed] [Google Scholar]
- Chalmers G. W. et al. Influence of cigarette smoking on inhaled corticosteroid treatment in mild asthma. Thorax 57, 226–230 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byron K. A., Varigos K. A. & Wootton A. M. Il-4 production is increased in cigarette smoker. Clin. Exp. Immunol. 95, 333–336 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H. T. et al. Association analysis of novel TBX21 variants with asthma phenotypes. Hum. Mutat. 22, 257 (2003). [DOI] [PubMed] [Google Scholar]
- Ylikoski E. et al. Association study of 15 novel single-nucleotide polymorphisms of the T-bet locus among Finnish asthma families. Clin. Exp. Allergy 34, 1049–1055 (2004). [DOI] [PubMed] [Google Scholar]
- Crapo R. O. et al. Guidelines for methacholine and exercise challenge testing-1999. Am. J. Respir. Crit. Care Med. 161, 309–329 (2000). [DOI] [PubMed] [Google Scholar]
- Pellegrino R. et al. Interpretative strategies for lung function tests. ATS/ERS task force: Standardisation of lung function testing. Eur. Respir. J. 26, 948–968 (2005). [DOI] [PubMed] [Google Scholar]
- Sont J. K. et al. Clinical control and histopathologic outcome of asthma when using bronchial hyperresponsiveness as an aditional guide to long-term tratment. Am. J. Respir. Crit. Care Med. 159, 1043–1051 (1999). [DOI] [PubMed] [Google Scholar]
- Masuko H. et al. Lower FEV1 in non-COPD, nonasthmatic subjects: association with smoking, annual decline in FEV1, total IgE levels, and TSLP genotypes. Int. J. Chron. Obstruct. Pulmon. Dis. 6, 181–189 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imboden M. et al. Glutathione S-transferase genotypes modify lung function decline in the general population:SAPALDIA cohort study. Respir. Res. 8, 1–17 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett W. S., Pace P. A., Sferlazza S. J., Carey V. J. & Weiss S. T. Annual variability in metacholine responsiveness in nonasthmatic working adults. Eur. Respir. J. 10, 2515–2521 (1997). [DOI] [PubMed] [Google Scholar]
- Ulrik C. S. Outcome of asthma: longitudinal changes in lung function. Eur. Respir. J. 13, 904–918 (1999). [DOI] [PubMed] [Google Scholar]
- Reddel H. K. et al. An Official American Thoracic Society/European Respiratory Society Statement: Asthma control and Exacerbations. Am. J. Respir. Crit. Care Med. 180, 59–99 (2009). [DOI] [PubMed] [Google Scholar]
- van Grunsven P. M., van Schayck C. P., Molema J., Akkermans R. P. & van Weel C. Effect of inhaled corticosteroids on bronchial responsiveness in patients with corticosteroid naive mild asthma: a meta-analysis. Thorax 54, 316–322 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz M. et al. The minimally important difference of the Asthma Control Test. J Allergy Clin. Immunol. 124, 719–723 (2009). [DOI] [PubMed] [Google Scholar]
- Riccioni G. et al. Bronchial hyperresponsiveness and quality of life in asthmatics. Respiration 70, 496–499 (2003). [DOI] [PubMed] [Google Scholar]