Abstract
Lynch syndrome is an autosomal dominant cancer predisposition syndrome classically caused by germline mutations of the mismatch repair genes, MLH1, MSH2, MSH6 and PMS2. Constitutional epimutations of the MLH1 gene, characterized by soma-wide methylation of a single allele of the promoter and allelic transcriptional silencing, have been identified in a subset of Lynch syndrome cases lacking a sequence mutation in MLH1. We report two individuals with no family history of colorectal cancer who developed that disease at age 18 and 20 years. In both cases, cancer had arisen because of the de novo occurrence of a constitutional MLH1 epimutation and somatic loss-of-heterozygosity of the functional allele in the tumors. We show for the first time that the epimutation in one case arose on the paternally inherited allele. Analysis of 13 tumors from seven individuals with constitutional MLH1 epimutations showed eight tumors had lost the second MLH1 allele, two tumors had a novel pathogenic missense mutation and three had retained heterozygosity. Only 1 of 12 tumors demonstrated the BRAF V600E mutation and 3 of 11 tumors harbored a mutation in KRAS. The finding that epimutations can originate on the paternal allele provides important new insights into the mechanism of origin of epimutations. It is clear that the second hit in MLH1 epimutation-associated tumors typically has a genetic not epigenetic basis. Individuals with mismatch repair–deficient cancers without the BRAF V600E mutation are candidates for germline screening for sequence or methylation changes in MLH1.
Keywords: colorectal cancer, Lynch syndrome, MLH1 epimutation, microsatellite instability, BRAF
Lynch syndrome (OMIM 120435) is an autosomal dominant cancer susceptibility syndrome characterized by the early onset of colorectal and endometrial cancers as well as other extra-colonic cancers.1 Lynch syndrome is caused by heterozygous germline sequence mutations of the mismatch repair genes MLH1, MSH2, MSH6 or PMS2, most frequently affecting MLH1 and MSH2.2 These are typically loss-of-function point mutations, identified by direct sequencing of the exons,3 as well as deletions and rearrangements of MLH1 and MSH2, identified by multiplex ligation-dependent probe amplification (MLPA).4,5
Although the genetic predisposition to cancer development is transmitted in an autosomal dominant fashion, somatic loss of the remaining functional allele is required for tumor development, consistent with Knudson’s “two-hit” model.6 Lynch syndrome–associated tumors demonstrate microsatellite instability (MSI) as a direct consequence of defective mismatch repair activity.7,8 Immunohistochemical analysis of the mismatch repair proteins in the tumor can also be instrumental in determining which of the mismatch repair genes should be the focus of germline screening.9–11 However, sporadic MSI colorectal cancers that are associated with somatic biallelic methylation of MLH1 can be a confounding factor in identifying tumors that have arisen in the context of Lynch syndrome.12,13 Although sporadic colorectal cancers demonstrating MSI and MLH1 methylation tend to occur in females over the age of 65 years, these are also closely associated with presence of the BRAF V600E mutation and the CpG island methylator phenotype (CIMP+), whereas Lynch syndrome–associated tumors are usually BRAF wild type.14–16 Thus, determination of the mutation status of BRAF has been proposed as a means of identifying those individuals with mismatch repair–deficient colorectal cancer who should be offered germline testing.17–19
Pathogenic sequence mutations of the mismatch repair genes remain unidentified in a number of cases meeting the clinical criteria for Lynch syndrome. Recently, deletions of the 3′ terminus of the EPCAM gene have been identified in a small number of families with Lynch syndrome whose tumors demonstrate loss of MSH2.20 In these cases, a failure in transcriptional termination of EPCAM results in the generation of fusion transcripts with the adjacent MSH2 gene downstream, giving rise to methylation of the MSH2 promoter, particularly in epithelial tissues where EPCAM is expressed at high levels.20 Constitutional epimutations of the MLH1 gene have also been identified in mutation-negative individuals with a clinical diagnosis of Lynch syndrome.21–31 This defect is characterized by soma-wide promoter methylation and transcriptional silencing of a single allele of the MLH1 gene.22–25 The coregulated EMP2AIP1 gene, which lies head-to-head with MLH1 and is transcribed in the opposite direction,32 is also subject to allelic inactivation in epimutation cases.25 The clinical phenotype associated with this defect appears to be consistent with Lynch syndrome,33 although the molecular and histopathological characteristics of epimutation-associated tumors have not been comprehensively investigated. The salient difference in cases with a constitutional MLH1 epimutation from their counterparts with sequence mutations is the lack of a remarkable family history, as MLH1 epimutations tend to arise spontaneously and are reversible between generations. Nevertheless, maternal transmission to the successive generation has been demonstrated in two families, with non-Mendelian inheritance clearly demonstrated in one of these.25,27 Here, the epimutation was transmitted from the affected mother, Patient A, to just one of her three sons who had each inherited the same maternal allele, with reversal of the epimutation in the other two.25 The mechanism by which MLH1 epimutations may be reversed in some cases but not in others remains to be defined, although the involvement of a fully penetrant genetically linked defect is unlikely.25 To date, MLH1 epimutations have been shown to either arise spontaneously on the maternally inherited allele or have been inherited through the maternal germline, thus demonstrating a maternal bias in origin.24,25,27 The absence of MLH1 methylation in the spermatozoa of two male carriers of a soma-wide constitutional MLH1 epimutation suggests that this epigenetic defect is not transmitted through the male germline with its epigenetic modifications intact.25,34 However, the potential for paternal transmission remains if epimutations are genetically predetermined, as the epimutation could be reinstated in the somatic cells of offspring postfertilization.33
In this study, we describe two new individuals with constitutional MLH1 epimutations, both of whom developed colorectal cancer at a very young age. We demonstrate for the first time that the epimutation arose spontaneously on the paternally inherited allele in one case. In addition, we have determined the molecular features of 15 tumors arising in seven individuals with constitutional epimutations. These investigations further elucidate the phenotype associated with constitutional MLH1 epimutations and their potential for inheritance.
Material and Methods
Patients and specimens
Individuals referred to family cancer clinics in Australia and the USA who (i) met the revised Bethesda criteria for hereditary nonpolyposis colorectal cancer, (ii) had no identifiable pathogenic sequence mutation of the key mismatch repair genes and (iii) whose tumors demonstrated MSI and immunohistochemical loss of MLH1 and PMS2 were included in this study. This study was approved by the St Vincent’s Human Research Ethics Committee (Approval number H04/024) and by the Baylor Research Institute Review Board (Approval number 003-180). Each of the probands and family members provided their informed consent to join the study.
Extraction of DNA and RNA
Genomic DNA was isolated from EBV-transformed lymphoblastoid cell lines (LCLs) and peripheral blood lymphocytes (PBL) by standard phenol-chloroform extraction and QIAamp DNA Mini Kit (Qiagen). DNA was extracted from buccal mucosa and hair follicles using the BuccalAmp™ DNA extraction kit (Epicentre). DNA was extracted from multiple 10-µm sections of formalin-fixed paraffin-embedded (FFPE) blocks of tumor and matched normal mucosa using the QIAamp DNA Mini Kit (Qiagen). DNA and total RNA from saliva samples were collected and extracted using the Oragene-DNA and Oragene-RNA kits (DNA Genotek). Total RNA was extracted from PBLs or LCLs using the Purelink Micro-Midi total RNA isolation kit (Invitrogen).
Methylation analyses
Genomic DNA (1 µg) was converted with sodium bisulfite using the EZ DNA Methylation-Gold kit (Zymo Research, CA). The converted DNA (50–100 ng) was tested for the presence of methylation in the “Deng-C” region35 of the MLH1 promoter by quantitative methylation-specific PCR followed by temperature denaturation analysis (qMSP), as previously described,25 or by bisulfite pyrosequencing. The pyrosequencing assay quantified the levels of methylation at five individual CpG sites within the amplicon. Amplification was conducted using forward primer 5′-GGTATTTTTGTTTTTATTGGTT-3′ and biotinylated reverse primer 5′-biotin-ACTCTATAAATTACTAAATCTCTT-3′ with annealing at 47°C. Amplicons were rendered single stranded, and the biotinylated strand captured on streptavidin-coated sepharose beads (Biotage, Uppsala, Sweden), on a Pyrosequencing workstation according to the manufacturer’s instructions (Pyrosequencing AB, Uppsala, Sweden). The pyrosequencing reaction was performed using sequencing primer 5′-AAAAAGAATTAATAGGAA-3′ on a PSQ 96MA system with the PyroGold SQA reagent kit (Pyrosequencing), with nucleotide dispensation order AGATGTCGATATGTCGACTTGATCAGTCGTATGTCGTA. The sequence interrogated was GAGYGGATAGYGATTTTTAAYGYGTAAGYGTATATTTTTTTAGGTAG. The relative levels of the C (methylated) and T (representing unmethylated) nucleotides at the Y position of the target CpG sites were determined using the Q-CpG software V1.0.9 (Biotage).
Clonal bisulfite sequencing of the MLH1 C-region was performed to study the allelic patterns of methylation as previously described.23–25
CIMP analysis was performed on tumor DNA following sodium bisulfite conversion, as previously described.16
Allelic expression analyses
RNA samples were treated with DNase I, and cDNAs were prepared from 2-µg total RNA by reverse transcription using the First Strand Superscript III cDNA synthesis kit with oligo-dT20 primers (Invitrogen). A control with reverse transcriptase omitted was performed for each sample. For allelic expression analyses at the c.655A>G SNP (rs1799977) within MLH1 exon 8, the relative levels of the “A” and “G” alleles were determined in genomic DNA and cDNAs using quantitative pyrosequencing assays designed for genomic DNA and cDNA templates, respectively, as previously described.36 For analysis of the c.474C>T (rs4647256) SNP within MLH1 exon 6, PCR and sequencing of genomic DNA was conducted using previously designed primers.37 Amplification of cDNA templates was performed between exons 5 and 8 using primers 5′-GCAAGTTACTCAGATGGAAAAC-3′ and 5′-CGAC TAACAGCATTTCCAAAGA-3′ for 35 cycles with annealing at 56°C. The purified PCR products were sequenced across the SNP site using internal primer 5′-CTCAGATGGAAA ACTGAAAGC-3′. For allelic expression analyses of the C>A SNP rs9311149 within the intron-less EPM2AIP1 gene, the same PCR amplification primers 5′-GTCCTGTTGTAGC AGTGAATAT-3′ and 5′-GCAGCATTGGAGAATTGGTA AA-3′ flanking the SNP were used for genomic DNA and cDNA templates, with amplification for 30 cycles at an annealing temperature of 58°C. Dideoxy sequencing of the purified PCR products was performed using internal primers 5′-TAGGTCCTTACCAGTTACTG-3′ and 5′-CATCAATTA GGGAAGATCTAG-3′. All PCR products were purified using the QIAquick PCR purification kit (Qiagen) before dideoxy sequencing. Sequences were resolved on an ABI Prism 3730 Capillary DNA Analyzer.
Haplotyping and screen of MLH1 promoter for sequence changes
Haplotyping was conducted at various SNP sites within MLH1 as previously described.24,25 Screening for mutations within the critical region of the MLH1 promoter and 5′UTR was performed by PCR amplification and sequencing from nucleotide position −295 with respect to the translation start site to the boundary of intron 1, using primers 5′-AAACGAACCAATAGGAAGAGC-3′ and 5′-ACTCCCTCCGTACCAGTTCT-3′.
Loss-of-heterozygosity analysis
Loss-of-heterozygosity (LOH) analysis was performed on DNA extracted from FFPE tumor tissue and compared to DNA extracted from normal tissue at informative SNP sites within MLH1, either by direct sequence analysis or by pyrosequencing (at the rs1799977 SNP), as previously described.36
BRAF and KRAS mutation screening
DNA extracted from tumor specimens was screened for the BRAF V600E mutation and mutations within codons 12–13 of KRAS by pyrosequencing, as previously described.38
Results
Identification of two new Lynch syndrome cases with colorectal cancer at a very young age due to a constitutional MLH1 epimutation
Constitutional MLH1 epimutations were identified in two unrelated individuals, YT and BF. Each had previously undergone a negative germline screen for mutations in the mismatch repair genes through family cancer clinics in Australia and the USA.
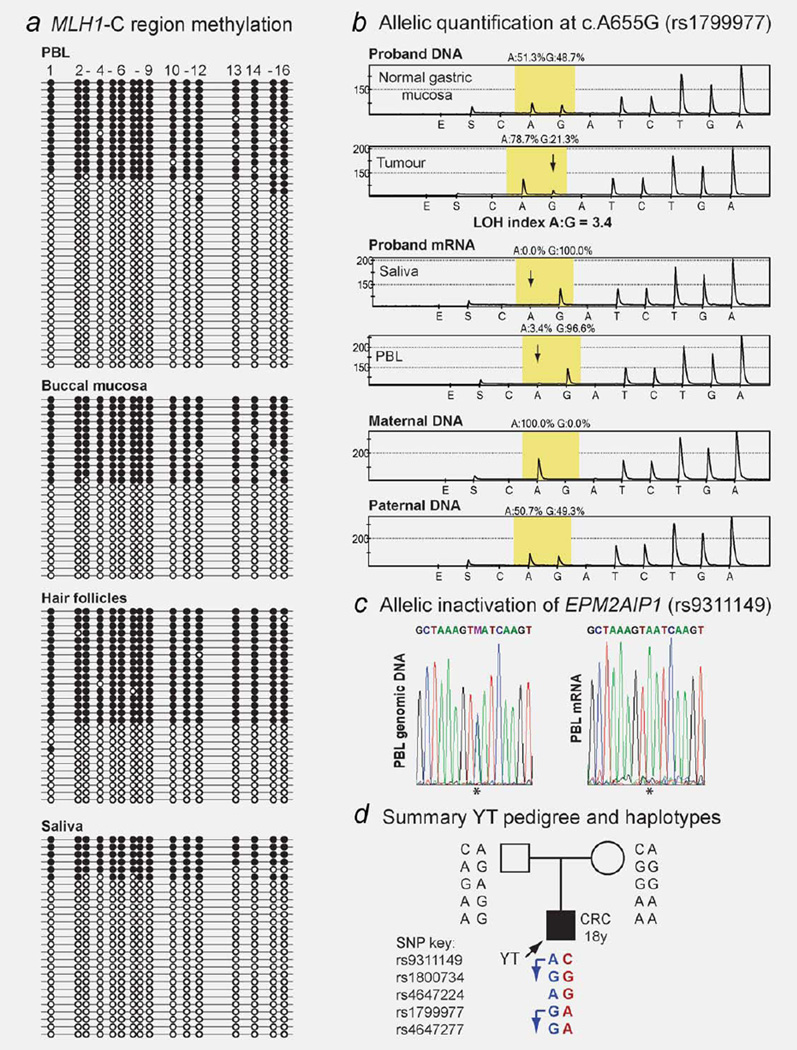
Male proband YT presented with a T3N1M0, mucinous, mismatch repair–deficient (microsatellite unstable at 6/6 markers, loss of expression of MLH1 and PMS2 protein complex but retention of MSH2 and MSH6) adenocarcinoma of the ascending colon at the age of 18 years (Table 1). The proband was of Sri Lankan heritage and had no family history of cancer. Germline screening of the MLH1 gene by direct sequencing did not detect any pathogenic sequence mutation, but revealed the proband to be heterozygous for two benign polymorphisms, c.655A>G (p.I219V) within exon 8 and c.1668A>G SNP within exon 19. No structural alterations were identified in MLH1 or MSH2 by MLPA. Testing of the “Deng-C” region of the MLH1 promoter35 in the proband’s DNA by quantitative real-time methylationspecific PCR (qMSP) was positive for methylation, with levels of 36–50% methylation detected in his PBLs, buccal mucosa, hair follicles and saliva (Supporting Information Fig. 1). Clonal bisulfite sequence analysis confirmed dense hemiallelic methylation in each tissue (Fig. 1a), consistent with a somawide MLH1 epimutation. Methylation analysis in DNA derived from the same four somatic tissue types in the proband’s parents showed no trace of methylation (Supporting Information Fig. 1), indicating the epimutation arose de novo in proband YT. Sequence analysis of the C-region of the MLH1 promoter and 5′UTR did not reveal any sequence change in the proband or his parents. Allelic expression and LOH analyses were conducted at informative exonic SNPs within MLH1 and EPM2AIP1 to determine the effect of the epimutation on transcriptional activity and tumor development. The epimutation was associated with transcriptional inactivation of a single allele of both MLH1 and EPM2AIP1 (Figs. 1b and 1c) and the opposite, active allele was lost in YT’s colorectal tumor (Fig. 1b). Genotyping of the parents revealed that the epimutation arose on the maternally inherited allele (Figs. 1b–1d).
Table 1.
Summary of the molecular features of the tumors in carriers of constitutional MLH1 epimutations
| ID | Age at diagnosis (years) |
Diagnosis | BRAF (V600 E) | KRAS mutation | Second hit |
|---|---|---|---|---|---|
| TT23 | 43 | Colon, cecum | ND | ND | LOH |
| TT | 44 | Colon, descending | No | No | ROH |
| TT | 51 | Three synchronous, small bowel | No | No | LOH |
| TT | 59 | Ampulla of Vater | No | No | LOH |
| VT | 46 | Colon, cecum | Yes | No | LOH |
| VT | 53 | Endometrium | ND | ND | ROH |
| VT | 63 | Breast | ND | ND | LOH |
| PtA25 | 63 | Colon, ascending | No | Yes, codon 13, c.38G>A | Missense |
| PtA | 67 | Rectum | No | No | Missense |
| PtA | 68 | Sarcomatoid lesion | No | No | ROH |
| PtB25 | 41 | Recto/sigmoid | No | No | ND |
| PtB | 48 | Rectum | No | No | ND |
| ST24 | 39 | Colon, transverse | No | Yes, codon 12, c.35G>A | LOH |
| YT | 18 | Colon, ascending | No | ND | LOH |
| BF | 20 | Colon, sigmoid | No | Yes, codon 12, c.35G>A | LOH |
The mechanisms resulting in the somatic loss of the normal allele in the tumors and the mutation status at the BRAF V600 site and KRAS codons 12 and 13 are listed.
Pt: Patient; LOH: loss-of-heterozygosity; ROH: retention of heterozygosity; No: not mutation identified; ND: not done.
Figure 1.
Constitutional MLH1 epimutation in “Proband YT.” (a) Allelic bisulfite sequencing demonstrated that hemiallelic methylation at the “Deng-C” region of the MLH1 promoter was widespread throughout his somatic tissues. Horizontal lines represent individual clones and dots indicate CpG dinucleotides within the sequenced fragment; black is methylated, white is unmethylated. (b) Allelic quantification of genomic DNA and mRNA samples in the proband and parents by pyrosequencing at the benign c.655A>G SNP (rs1799977) within MLH1 exon 8. The yellow-shaded region shows the peaks at the SNP site. Two peaks showing equal levels of the “A” and “G” alleles in the proband’s normal gastric mucosa DNA indicate heterozygosity. In the tumor DNA there is significant reduction of the transcriptionally active “G” allele (indicated by downward arrow), consistent with loss-of-heterozygosity (LOH) of the unmethylated allele. In the mRNA from the proband’s saliva and PBL there was loss of expression of the “A” allele (indicated by downward arrows). Genotyping of this SNP in the parents showed that the silenced “A” allele was maternally inherited. (c) Allelic expression analysis at the C/A SNP (rs9311149) within the 3′UTR of the EPM2AIP1 gene shows allelic transcriptional loss of EPM2AIP1 in proband YT as well. (d) Proband YT pedigree and summary of haplotypes from informative SNPs according to the key provided. Allele colors relate to parental origin of inheritance, with red indicating maternally derived alleles, blue showing paternally derived alleles and black indicating unknown parentage. Transcription of the paternally inherited allele was demonstrated, as indicated by arrows. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Female proband BF presented with a T3N0MO mucinous, mismatch repair–deficient (microsatellite unstable at 5/5 markers, loss of expression of MLH1, PMS2 and MSH6 but retention of MSH2) cancer of the sigmoid colon at age 20 years (Supporting Information Fig. 2). The proband was of African American, Caucasian and Eastern Indian descent and had no family history of cancer. Germline mutation analysis revealed the proband to be heterozygous for the c.474C>T SNP (rs4647256) within MLH1 exon 6, which is predominantly identified in individuals of African descent, but did not identify any pathogenic mutation of either of the three mismatch repair genes MLH1, MSH2 or MSH6.
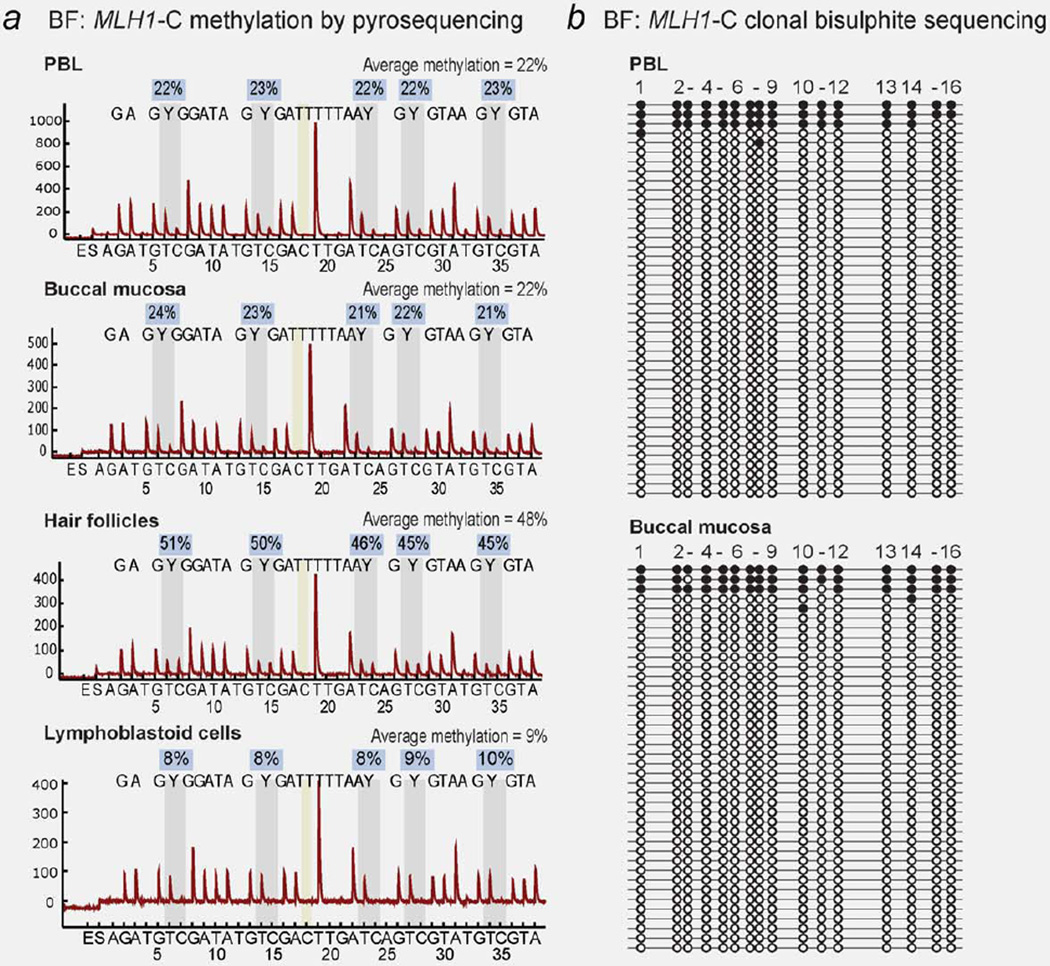
Methylation analysis of the “Deng-C” region of the MLH1 promoter by bisulfite pyrosequencing and qMSP of the proband’s PBL and LCL DNA was positive, with average methylation levels of 22% and 9% detected, respectively (Fig. 2a). Pyrosequencing in additional tissues from the proband revealed a mosaic pattern of MLH1 methylation, with buccal mucosa giving an average methylation score of 22% and hair follicles showing 48% methylation (Fig. 2a). Clonal bisulfite sequencing of PBL and buccal mucosa DNA showed that a small proportion of alleles were affected by dense methylation (Fig. 2b). Bisulfite pyrosequencing and qMSP analyses of the PBL and LCL DNA from BF’s parents and her unaffected brother showed no evidence for MLH1 methylation (Supporting Information Fig. 2), indicating the epimutation arose de novo in the proband. Screening of the C-region of the MLH1 promoter and 5′UTR did not reveal any sequence changes in the proband or any family member.
Figure 2.
Constitutional MLH1 methylation in somatic tissues of Proband BF. (a) Bisulfite pyrosequencing in peripheral blood lymphocytes (PBL) and other somatic tissues in BF show soma-wide mosaic methylation of the MLH1 “Deng-C” region. The sequence analyzed is written above the peaks within each pyrogram. Vertical gray bars indicate the position of the C/T (Y) nucleotides within the five CpG sites interrogated for methylation status. The level of methylation C:T at each site is given in the boxes above, with the average methylation score of the five CpG sites provided at the top of each pyrogram. The vertical yellow bar indicates a control for complete bisulphite conversion. (b) Clonal bisulfite sequencing within the MLH1-C region shows the allelic pattern of methylation. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
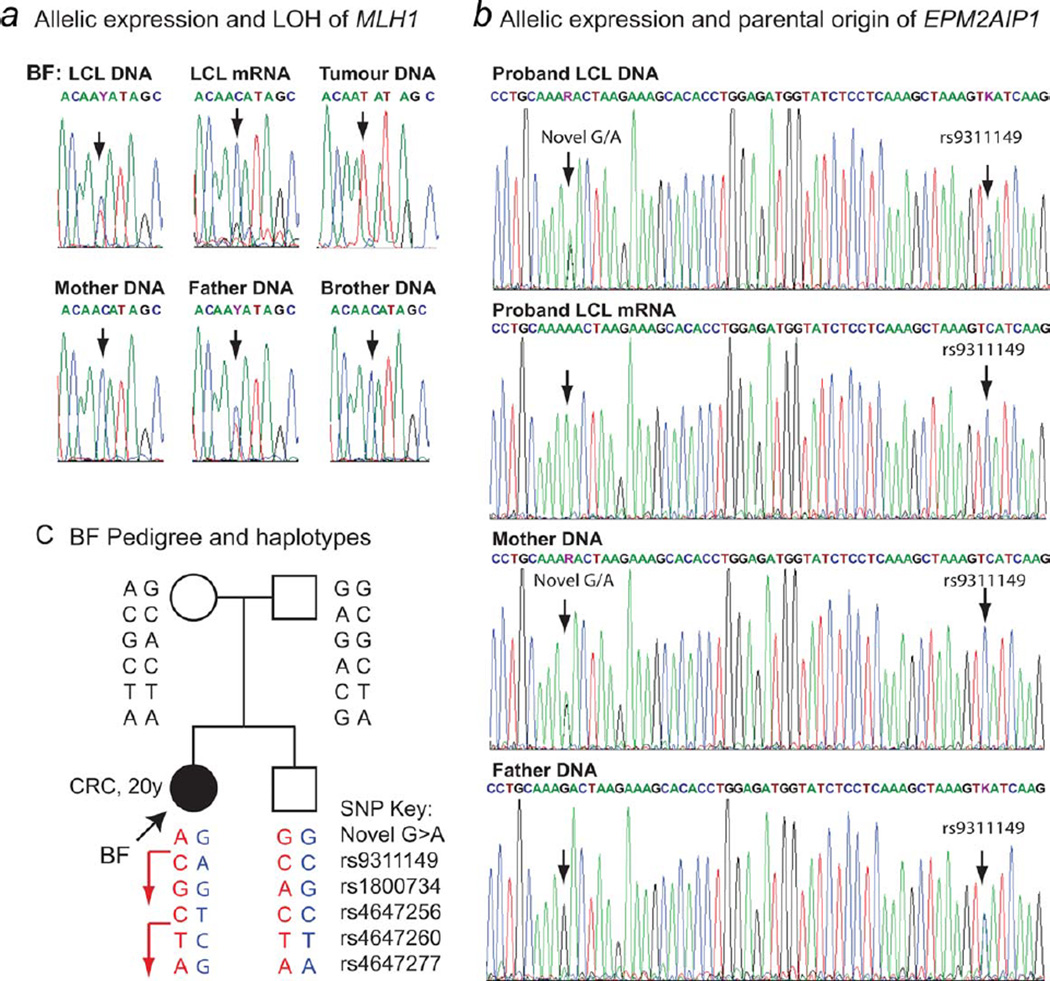
Allelic expression analyses of MLH1 and EPM2AIP1 at informative exonic SNP sites demonstrated complete loss of expression of a single allele in the proband’s LCL mRNA (Figs. 3a and 3b). LOH analysis of her tumor DNA showed a significant reduction of the opposite, functionally active allele, consistent with somatic loss of the unmethylated allele in her tumor (Fig. 3a). In contrast to YT, however, the allelic expression analyses in BF and genotyping in the pedigree indicated that the epimutation in proband BF arose on the paternally inherited allele (Figs. 3b–3d). The haplotypes across MLH1 and EPM2AIP1 (Fig. 3c), and sex determination by fragment length analysis of the X-Y Amelogenin homolog (data not shown),39 were consistent for parentage and gender of the samples obtained for each family member, confirming the origin of the epimutation on the paternal allele. As the father was devoid of methylation in his PBLs, the epimutation is likely to have arisen de novo.
Figure 3.
Transcriptional inactivation of expression of the paternal MLH1 and EPM2AIP1 alleles in Proband BF and LOH of the maternal allele in the tumor. (a) Sequence electropherograms across the MLH1 exon 6 c.474C>T (rs4647256) SNP (indicated by arrows) in Proband BF and family members. The genomic DNA and RNA extracted from the proband’s lymphoblastoid cells (LCL) show that she is heterozygous for the SNP, but only the “C” allele is expressed, indicating loss of expression of the T allele. Genotyping in the parents showed obligate inheritance of the active “C” allele from the mother, who is homozygous “C.” Therefore, the inactivated “T” allele was paternally inherited. The tumor shows considerable reduction of the functional “C” allele. (b) Electropherograms spanning a novel G/A SNP site and the C/A rs9311149 SNP in the 3′UTR of EPM2AIP1 in the proband’s genomic DNA (gDNA), mRNA and in the genomic DNA of family members. The proband was found to be heterozygous for both SNPs. The LCL mRNA shows monoallelic expression of the “A” allele of the novel G/A SNP and the “C” allele of SNP rs9311149. Both SNP sites were informative as to the parental origin of the alleles, with the mother heterozygous for the novel SNP and homozygous “C” for the rs9311149 SNP and the father homozygous “G” for the novel SNP and heterozygous for rs9311149. The expressed allele was thus maternally derived, confirming the epimutation arose on the paternal allele. (c) Proband BF pedigree and summary of haplotypes from informative SNPs according to the key provided, with red showing maternal and blue showing paternal inheritance. The expressed maternal allele is indicated by an arrow. The proband’s sibling inherited the opposite parental alleles. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Mechanism of inactivation of the second allele in epimutation carriers
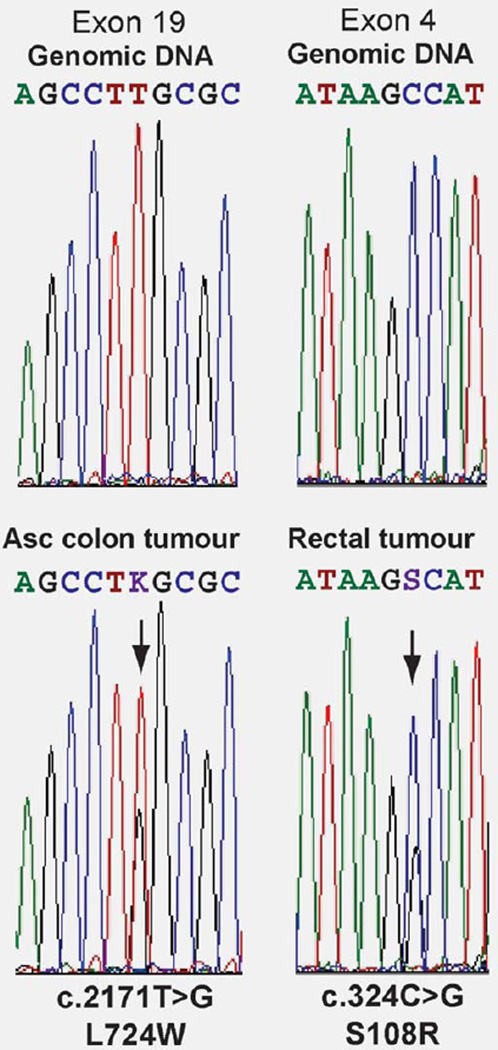
To date we have demonstrated that six tumors from five epimutation carriers have somatic loss of the nonmethylated allele as the cause of their mismatch repair deficiency (Table 1).23–25 Newly described patients YT and BF also displayed somatic loss of the nonmethylated allele in their tumors. For previously described Patient A (Table 1),25 the somatic event affecting the functional allele in her colorectal cancers was unknown. Subsequent to the initial report of Patient A, she developed an undifferentiated partially sarcomatoid skin tumor with widespread metastasis. This tumor was microsatellite unstable and also showed loss of expression of MLH1 and PMS2. We sought to determine the nature of the second hit in this tumor as well as her colorectal cancers. LOH analysis across the exon 8 c.655A>G SNP for which Patient A was informative showed retention of heterozygosity of MLH1 in Patient A’s ascending colon, rectal and skin tumor (Supporting Information Fig. 3). Clonal bisulfite sequence analysis of the MLH1 promoter, encompassing the −93G>A SNP (rs1800734) which was heterozygous in Patient A, showed methylation was confined to the epimutant “A” allele, as detected in the matched normal colonic mucosa, whereas the ‘G’ allele remained unmethylated (Supporting Information Fig. 3). Finally, exonic sequencing of the entire coding region of MLH1 in all three tumors led to the identification of novel point mutations in the ascending colon and rectal tumors, which were not present in the germline. A c.2171T>G missense mutation leading to a L724W amino acid change was identified in exon 19 in the tumor of the ascending colon, and a c.324C>G missense mutation leading to a S108R change was identified in exon 4 of the rectal tumor (Fig. 4). To assess the potential pathogenicity of these acquired mutations, in silico analysis was performed using the SIFT program (http://sift.jcvi.org), in which a SIFT score of <0.05 denotes a deleterious change. The L724W and S108R mutations gave SIFT scores of 0.01 and 0.00, respectively, predicting they are highly likely to affect protein function. Assuming the mutations arose on the wild-type allele, these are likely to account for the loss of MLH1 expression in the respective tumors. No mutation was identified in the skin sarcoma, and no further specimen was available for additional study. The nature of the second hit in some tumors remained unaccounted for because of lack of availability (data not shown) or inadequate sample for comprehensive testing.
Figure 4.
Point mutations serving as the “second hit” in the colorectal tumors of Patient A. Electropherograms showing the acquired somatic missense mutations identified in the tumors (bottom as labeled), which were not present in the germline DNA (top). The positions of each mutation are indicated by arrows. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Low rate of common BRAF V600E and KRAS mutations in epimutation-associated tumors
Of the 15 tumors from epimutation carriers, which were available for analysis, 12 were tested for the BRAF V600E mutation and only one, a cecal cancer from patient VT was found to harbor this mutation (Table 1). Of the 11 tumors tested for mutations in codons 12 and 13 of KRAS, only three carried a mutation (Table 1).
Discussion
Herein, we report the identification of two new Lynch syndrome cases whose cancer predisposition was caused by a de novo constitutional epimutation of MLH1. Both cases developed colorectal cancer at the very young ages of 18 and 20 years, representing the youngest cases amongst epimutation carriers reported to date. Consistent with the lack of a family history of Lynch syndrome–associated cancers in both cases, the epimutations in YT and BF arose spontaneously. In the case of YT, the epimutation arose on the maternally inherited allele, which has been a consistent finding amongst the previously reported cases for whom the parental origin of the epimutation was determinable. The discovery of maternal transmission of the MLH1 epimutations in two families, together with the bias in the maternal origin of epimutations in sporadic cases, led to a suggestion that MLH1 epimutations might be borne epigenetically intact specifically within the oocyte.33 Interestingly, however, we demonstrate for the first time in proband BF that the MLH1 epimutation originated on the paternally inherited allele. This has significant implications regarding the mechanistic origins of MLH1 epimutations, as previous studies demonstrated that epimutations were fully reversed in the spermatozoa of two male probands who harbored a constitutional MLH1 epimutation throughout their somatic tissues.25,34 Whilst proband BF’s father did not carry the epimutation in his blood, we were unable to obtain a spermatozoa sample to seek evidence that the epimutation may have arisen de novo in his germline. Nevertheless, we consider it more plausible that the epimutation arose postfertilization in this case. The possibility that epimutations are triggered postfertilization by a genetic cue, such as the coinheritance of a mutant trans-acting factor, is now more plausible than gametic epigenetic inheritance, though this remains to be substantiated. Lack of knowledge regarding the underlying mechanistic basis for MLH1 epimutations is a confounding factor in genetic diagnosis and counseling. In the case of BF’s brother, he inherited the opposite parental allele of MLH1 compared to his sister and had no trace of MLH1 methylation in his blood, so it appears unlikely that he has a significant risk of developing cancer as a consequence of this particular defect.
Another notable feature of the epimutation in BF was the degree of methylation mosaicism between her various somatic tissues. This varied from 22% in blood to 48% in hair follicles. Even though methylation was detected at levels of just 7–9% in her LCL using two independent methods, there was nevertheless complete transcriptional inactivation of her epimutant allele in the same cells, as opposed to a partial repression, as witnessed in a previous case displaying significant methylation mosaicism.27 It is possible that the accrual of methylation to the allele may represent a secondary consequence of inherent allelic inactivation, although no sequence changes that might underlie this were found in the critical region of the MLH1 promoter or 5′UTR.
In demonstrating the very early age of onset in probands YT and BF as well as the extensive spectrum of tumor types presented by Patient A (Table 1), our findings extend the current knowledge regarding the phenotype associated with constitutional MLH1 epimutations. Furthermore, the differing ethnic origins of these two cases provide further evidence that MLH1 epimutations are not confined to any particular ethnic group. Previously reported cases have been of Northern European, Mediterranean, Japanese and Chinese heritage.22–27,31 Taken together, these findings illustrate the importance of screening for epimutations in suspected cases of Lynch syndrome irrespective of the age of presentation with disease or ethnic origin.
We additionally aimed to better characterize the molecular pathways that lead to the development of MSI tumors lacking expression of MLH1 in individuals with constitutional MLH1 epimutations. First, we established the mode of somatic inactivation of the unmethylated allele in the tumors of the current cases and previously reported case, Patient A. In both YT and BF, the “second hit” was attributable to somatic LOH of the normally regulated MLH1 allele. However, the additional loss of expression of the MSH6 protein in BF’s tumor remains unaccounted for, although this could be a secondary consequence of MSI. No germline mutation was identified in MSH6 in this patient. In Patient A, two missense mutations, predicted to be deleterious, were identified in her colorectal tumors. Thus, it appears that the second hit leading to MSI tumors with MLH1 loss in epimutation carriers tends to be genetically mediated, with LOH the most frequent mechanism observed. It remains possible that epigenetic silencing of the second allele occurred in a proportion of epimutation-associated tumors in which the second hit remains unaccounted for, but on the basis of these cases, the presence of a constitutional epigenetic error does not appear to incur an increased susceptibility to subsequent somatic methylation of the second MLH1 allele. Rather the loss of MLH1 expression in epimutation-associated tumors appears to be consistent with those of classic Lynch syndrome carriers of germline sequence mutations. Furthermore, screening for somatic mutation of BRAF and KRAS in the tumors of the current cases and our previously reported epimutation carriers revealed a low frequency of mutations at common mutation hotspots in both genes. The BRAF V600E mutation was detected in a single early-onset colorectal tumor from one epimutation carrier, but insufficient tumor was available for further analysis to establish if this was concomitant with biallelic MLH1 methylation. Nevertheless, it appears that the molecular pathogenesis in the greater proportion of tumors in epimutation carriers parallels that of Lynch syndrome tumors associated with germline sequence mutations, as opposed to sporadic MSI colorectal cancers demonstrating MLH1 loss due to biallelic methylation.
Supplementary Material
Acknowledgements
This work was funded by NIH Grant R01 CA 72851, the NH&MRC, Australia and Cancer Council NSW. MPH is the recipient of a Career Development and Support Fellowship from the Cancer Institute NSW.
Footnotes
Additional Supporting Information may be found in the online version of this article
References
- 1.Lynch HT, Lynch JF. Lynch syndrome: history and current status. Dis Markers. 2004;20:181–198. doi: 10.1155/2004/460240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peltomaki P. Lynch syndrome genes. Fam Cancer. 2005;4:227–232. doi: 10.1007/s10689-004-7993-0. [DOI] [PubMed] [Google Scholar]
- 3.Peltomaki P, Vasen H. Mutations associated with HNPCC predisposition—update of ICG-HNPCC/INSiGHT mutation database. Dis Markers. 2004;20:269–276. doi: 10.1155/2004/305058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor CF, Charlton RS, Burn J, Sheridan E, Taylor GR. Genomic deletions in MSH2 or MLH1 are a frequent cause of hereditary non-polyposis colorectal cancer: identification of novel and recurrent deletions by MLPA. Hum Mutat. 2003;22:428–433. doi: 10.1002/humu.10291. [DOI] [PubMed] [Google Scholar]
- 5.Grabowski M, Mueller-Koch Y, Grasbon-Frodl E, Koehler U, Keller G, Vogelsang H, Dietmaier W, Kopp R, Siebers U, Schmitt W, Neitzel B, Gruber M, et al. Deletions account for 17% of pathogenic germline alterations in MLH1 and MSH2 in hereditary nonpolyposis colorectal cancer (HNPCC) families. Genet Test. 2005;9:138–146. doi: 10.1089/gte.2005.9.138. [DOI] [PubMed] [Google Scholar]
- 6.Knudson AG. Hereditary cancer: two hits revisited. J Cancer Res Clin Oncol. 1996;122:135–140. doi: 10.1007/BF01366952. [DOI] [PubMed] [Google Scholar]
- 7.Tannergard P, Liu T, Weger A, Nordenskjold M, Lindblom A. Tumorigenesis in colorectal tumors from patients with hereditary non-polyposis colorectal cancer. Hum Genet. 1997;101:51–55. doi: 10.1007/s004390050585. [DOI] [PubMed] [Google Scholar]
- 8.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 9.de Leeuw WJ, Dierssen J, Vasen HF, Wijnen JT, Kenter GG, Meijers-Heijboer H, Brocker-Vriends A, Stormorken A, Moller P, Menko F, Cornelisse CJ, Morreau H. Prediction of a mismatch repair gene defect by microsatellite instability and immunohistochemical analysis in endometrial tumours from HNPCC patients. J Pathol. 2000;192:328–335. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH701>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 10.Lindor NM, Burgart LJ, Leontovich O, Goldberg RM, Cunningham JM, Sargent DJ, Walsh-Vockley C, Petersen GM, Walsh MD, Leggett BA, Young JP, Barker MA, et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol. 2002;20:1043–1048. doi: 10.1200/JCO.2002.20.4.1043. [DOI] [PubMed] [Google Scholar]
- 11.Hendriks Y, Franken P, Dierssen JW, De Leeuw W, Wijnen J, Dreef E, Tops C, Breuning M, Brocker-Vriends A, Vasen H, Fodde R, Morreau H. Conventional and tissue microarray immunohistochemical expression analysis of mismatch repair in hereditary colorectal tumors. Am J Pathol. 2003;162:469–477. doi: 10.1016/S0002-9440(10)63841-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, Kane MF, Kolodner RD, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cunningham JM, Christensen ER, Tester DJ, Kim CY, Roche PC, Burgart LJ, Thibodeau SN. Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res. 1998;58:3455–3460. [PubMed] [Google Scholar]
- 14.Deng G, Bell I, Crawley S, Gum J, Terdiman JP, Allen BA, Truta B, Sleisenger MH, Kim YS. BRAF mutation is frequently present in sporadic colorectal cancer with methylated hMLH1, but not in hereditary nonpolyposis colorectal cancer. Clin Cancer Res. 2004;10(1 Part 1):191–195. doi: 10.1158/1078-0432.ccr-1118-3. [DOI] [PubMed] [Google Scholar]
- 15.McGivern A, Wynter CV, Whitehall VL, Kambara T, Spring KJ, Walsh MD, Barker MA, Arnold S, Simms LA, Leggett BA, Young J, Jass JR. Promoter hypermethylation frequency and BRAF mutations distinguish hereditary non-polyposis colon cancer from sporadic MSIH colon cancer. Fam Cancer. 2004;3:101–107. doi: 10.1023/B:FAME.0000039861.30651.c8. [DOI] [PubMed] [Google Scholar]
- 16.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, Koh H, Simms L, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 17.Domingo E, Laiho P, Ollikainen M, Pinto M, Wang L, French AJ, Westra J, Frebourg T, Espin E, Armengol M, Hamelin R, Yamamoto H, et al. BRAF screening as a low-cost effective strategy for simplifying HNPCC genetic testing. J Med Genet. 2004;41:664–668. doi: 10.1136/jmg.2004.020651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loughrey MB, Waring PM, Tan A, Trivett M, Kovalenko S, Beshay V, Young MA, McArthur G, Boussioutas A, Dobrovic A. Incorporation of somatic BRAF mutation testing into an algorithm for the investigation of hereditary non-polyposis colorectal cancer. Fam Cancer. 2007;6:301–310. doi: 10.1007/s10689-007-9124-1. [DOI] [PubMed] [Google Scholar]
- 19.Schofield L, Watson N, Grieu F, Li WQ, Zeps N, Harvey J, Stewart C, Abdo M, Goldblatt J, Iacopetta B. Population-based detection of Lynch syndrome in young colorectal cancer patients using microsatellite instability as the initial test. Int J Cancer. 2009;124:1097–1102. doi: 10.1002/ijc.23863. [DOI] [PubMed] [Google Scholar]
- 20.Ligtenberg MJ, Kuiper RP, Chan TL, Goossens M, Hebeda KM, Voorendt M, Lee TY, Bodmer D, Hoenselaar E, Hendriks-Cornelissen SJ, Tsui WY, Kong CK, et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nat Genet. 2009;41:112–117. doi: 10.1038/ng.283. [DOI] [PubMed] [Google Scholar]
- 21.Gazzoli I, Loda M, Garber J, Syngal S, Kolodner RD. A hereditary nonpolyposis colorectal carcinoma case associated with hypermethylation of the MLH1 gene in normal tissue and loss of heterozygosity of the unmethylated allele in the resulting microsatellite instability-high tumor. Cancer Res. 2002;62:3925–3928. [PubMed] [Google Scholar]
- 22.Miyakura Y, Sugano K, Akasu T, Yoshida T, Maekawa M, Saitoh S, Sasaki H, Nomizu T, Konishi F, Fujita S, Moriya Y, Nagai H. Extensive but hemiallelic methylation of the hMLH1 promoter region in early-onset sporadic colon cancers with microsatellite instability. Clin Gastroenterol Hepatol. 2004;2:147–156. doi: 10.1016/s1542-3565(03)00314-8. [DOI] [PubMed] [Google Scholar]
- 23.Suter CM, Martin DI, Ward RL. Germline epimutation of MLH1 in individuals with multiple cancers. Nat Genet. 2004;36:497–501. doi: 10.1038/ng1342. [DOI] [PubMed] [Google Scholar]
- 24.Hitchins M, Williams R, Cheong K, Halani N, Lin VA, Packham D, Ku S, Buckle A, Hawkins N, Burn J, Gallinger S, Goldblatt J, et al. MLH1 germline epimutations as a factor in hereditary nonpolyposis colorectal cancer. Gastroenterology. 2005;129:1392–1399. doi: 10.1053/j.gastro.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 25.Hitchins MP, Wong JJ, Suthers G, Suter CM, Martin DI, Hawkins NJ, Ward RL. Inheritance of a cancer-associated MLH1 germ-line epimutation. N Engl J Med. 2007;356:697–705. doi: 10.1056/NEJMoa064522. [DOI] [PubMed] [Google Scholar]
- 26.Valle L, Carbonell P, Fernandez V, Dotor AM, Sanz M, Benitez J, Urioste M. MLH1 germline epimutations in selected patients with early-onset non-polyposis colorectal cancer. Clin Genet. 2007;71:232–237. doi: 10.1111/j.1399-0004.2007.00751.x. [DOI] [PubMed] [Google Scholar]
- 27.Morak M, Schackert HK, Rahner N, Betz B, Ebert M, Walldorf C, Royer-Pokora B, Schulmann K, von Knebel-Doeberitz M, Dietmaier W, Keller G, Kerker B, et al. Further evidence for heritability of an epimutation in one of 12 cases with MLH1 promoter methylation in blood cells clinically displaying HNPCC. Eur J Hum Genet. 2008;16:804–811. doi: 10.1038/ejhg.2008.25. [DOI] [PubMed] [Google Scholar]
- 28.Gylling A, Ridanpaa M, Vierimaa O, Aittomaki K, Avela K, Kaariainen H, Laivuori H, Poyhonen M, Sallinen SL, Wallgren-Pettersson C, Jarvinen HJ, Mecklin JP, et al. Large genomic rearrangements and germline epimutations in Lynch syndrome. Int J Cancer. 2009;124:2333–2340. doi: 10.1002/ijc.24230. [DOI] [PubMed] [Google Scholar]
- 29.Mueller J, Gazzoli I, Bandipalliam P, Garber JE, Syngal S, Kolodner RD. Comprehensive molecular analysis of mismatch repair gene defects in suspected Lynch syndrome (hereditary nonpolyposis colorectal cancer) cases. Cancer Res. 2009;69:7053–7061. doi: 10.1158/0008-5472.CAN-09-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niessen RC, Hofstra RM, Westers H, Ligtenberg MJ, Kooi K, Jager PO, de Groote ML, Dijkhuizen T, Olderode-Berends MJ, Hollema H, Kleibeuker JH, Sijmons RH. Germline hypermethylation of MLH1 and EPCAM deletions are a frequent cause of Lynch syndrome. Genes Chromosomes Cancer. 2009;48:737–744. doi: 10.1002/gcc.20678. [DOI] [PubMed] [Google Scholar]
- 31.Zhou HH, Yan SY, Zhou XY, Du X, Zhang TM, Cai X, Lu YM, Cai SJ, Shi DR. MLH1 promoter germline-methylation in selected probands of Chinese hereditary non-polyposis colorectal cancer families. World J Gastroenterol. 2008;14:7329–7334. doi: 10.3748/wjg.14.7329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin JC, Jeong S, Liang G, Takai D, Fatemi M, Tsai YC, Egger G, Gal-Yam EN, Jones PA. Role of nucleosomal occupancy in the epigenetic silencing of the MLH1 CpG island. Cancer Cell. 2007;12:432–444. doi: 10.1016/j.ccr.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hitchins MP, Ward RL. Constitutional (germline) MLH1 epimutation as an aetiological mechanism for hereditary nonpolyposis colorectal cancer. J Med Genet. 2009;46:793–802. doi: 10.1136/jmg.2009.068122. [DOI] [PubMed] [Google Scholar]
- 34.Hitchins MP, Ward RL. Erasure of MLH1 methylation in spermatozoa-implications for epigenetic inheritance. Nat Genet. 2007;39:1289. doi: 10.1038/ng1107-1289. [DOI] [PubMed] [Google Scholar]
- 35.Deng G, Peng E, Gum J, Terdiman J, Sleisenger M, Kim YS. Methylation of hMLH1 promoter correlates with the gene silencing with a region-specific manner in colorectal cancer. Br J Cancer. 2002;86:574–579. doi: 10.1038/sj.bjc.6600148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwok C-T, Ward RL, Hawkins NJ, Hitchins MP. Detection of allelic imbalance in MLH1 expression by pyrosequencing serves as a tool for the identification of germline defects in Lynch syndrome. Fam Cancer. doi: 10.1007/s10689-009-9314-0. in press. [DOI] [PubMed] [Google Scholar]
- 37.Hegde M, Blazo M, Chong B, Prior T, Richards C. Assay validation for identification of hereditary nonpolyposis colon cancer-causing mutations in mismatch repair genes MLH1, MSH2, and MSH6. J Mol Diagn. 2005;7:525–534. doi: 10.1016/S1525-1578(10)60584-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Packham D, Ward RL, Ap Lin V, Hawkins NJ, Hitchins MP. Implementation of novel pyrosequencing assays to screen for common mutations of BRAF and KRAS in a cohort of sporadic colorectal cancers. Diagn Mol Pathol. 2009;18:62–71. doi: 10.1097/PDM.0b013e318182af52. [DOI] [PubMed] [Google Scholar]
- 39.Akane A. Sex determination by PCR analysis of the X-Y amelogenin gene. Methods Mol Biol. 1998;98:245–249. doi: 10.1385/0-89603-443-7:245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.