Abstract
Poly(ADP-ribose)polymerase-1 (PARP1) is a nuclear protein implicated in DNA repair, recombination, replication, and chromatin remodeling. The aim of this study was to evaluate possible differences between PARP1−/− and wild-type mice regarding induction and repair of DNA lesions in irradiated male germ cells. Comet assay was applied to detect DNA damage in testicular cells immediately, and two hours after 4 Gy X-ray irradiation. A similar level of spontaneous and radiation-induced DNA damage was observed in PARP1−/− and wild-type mice. Conversely, two hours after irradiation, a significant level of residual damage was observed in PARP1−/− cells only. This finding was particularly evident in round spermatids. To evaluate if PARP1 had also a role in the dynamics of H2AX phosphorylation in round spermatids, in which γ-H2AX foci had been shown to persist after completion of DNA repair, we carried out a parallel analysis of γ-H2AX foci at 0.5, 2, and 48 h after irradiation in wild-type and PARP1−/− mice. No evidence was obtained of an effect of PARP1 depletion on H2AX phosphorylation induction and removal. Our results suggest that, in round spermatids, under the tested experimental conditions, PARP1 has a role in radiation-induced DNA damage repair rather than in long-term chromatin modifications signaled by phosphorylated H2AX.
Keywords: poly(ADP-ribose)polymerase-1, DNA repair, male mouse germ cells, comet assay, H2AX phosphorylation, ionizing radiation
1. Introduction
The maintenance of DNA integrity in the paternal genome is of utmost importance for reproduction and it is well known that DNA lesions in germ cells can be transmitted to the next generation [1]. Germ cells need efficient systems to repair DNA damage to prevent inheritable mutations. Differences have been observed among DNA repair pathways in somatic and male germ cells, in which the expression or the presence of some DNA repair proteins depends on the phase of germ cell development [2–9].
Poly(ADP-ribose)polymerase-1 (PARP1) is one of the members of a family of nuclear proteins involved in several cellular processes like DNA repair, replication and transcription, chromatin structure, and intracellular calcium signaling, and is critical for the long-term maintenance of genomic stability [10]. Activation of PARP1 is one of the immediate responses of eukaryotic cells to DNA damage. It recognizes DNA strand breaks and, at the site of breakage, catalyzes the transfer of the ADP-ribose moiety from the respiratory co-enzyme NAD+ to nuclear protein acceptors [11,12]. PARP1 signals the presence of DNA lesions to downstream effectors involved in coordinating the cellular response to DNA damage, recruits repair enzymes to the damaged sites, and affects chromatin structure to allow repair factors to access DNA lesions [11,13–15]. PARP1 has been involved in the repair of various types of DNA lesions by multiple pathways [15]. The role of this enzyme and other PARP family members in DNA repair processes has been extensively studied in somatic cells, both using chemical inhibitors and mouse and cellular models genetically defective for the enzymes [13,16–24]. These studies show that inhibition or lack of PARP slows down DNA repair and increase the cytotoxicity of ionizing radiation and alkylating agents. The contribution of PARP1 to DNA repair in male germ cells has been considerably less investigated. It is known that germinal cells are characterized by a high expression level of PARP [25] and difference among testis cell subpopulations in PARP activity and production of ADP-ribose polymers was also observed [26–30]. In male germ cells poly(ADP ribose) metabolism is important for the regulation of meiotic process and seems to have a role in DNA repair and chromatin remodeling in post-meiotic cells [27,28,30–32]. A delay of radiation-induced DNA damage repair was shown in cultured rat spermatocytes and spermatids [25], and mouse spermatids in vivo [3] when poly(ADP ribose) metabolism was inhibited by chemical treatment. These studies left unanswered the question of the specific role of PARP1 in the germ cell DNA damage response because of the poor specificity of chemical inhibitors towards different PARP family members [25].
PARP1 knockout mice have been produced to investigate the specific role of PARP1 in cellular processes [24]. PARP1−/− mice are more sensitive to the lethal effects of alkylating agents and ionizing radiation, show an increased frequency of spontaneous sister chromatid exchanges in bone marrow cells and increased levels of chromatid and chromosome aberrations after exposure to genotoxic agents [24,33–35]. Although their fertility is not compromised, more subtle effects on the germ cell DNA damage response in these mice cannot be excluded. To our knowledge, no studies have been published so far to evaluate male germ cell capability of PARP1−/− mice to repair induced DNA damage.
In this study, alkaline comet assay has been applied to evaluate the level of basal and X-ray induced DNA lesions in testis cells from wild-type (WT) and PARP1−/− mice. In addition, to investigate the role of PARP1 in DNA repair in male germ cells, DNA damage was assessed at different times after irradiation. Exploiting the capacity of comet assay to identify DNA lesions in individual cells versus their ploidy [9,36], we evaluated the response to irradiation of different testis cell subpopulations. Post-meiotic early spermatids were the most affected by lack of PARP1. So, in these cells the induction of double strand breaks (DSB) was also specifically investigated by γ-H2AX immunolabeling. Finally, the persistence of γ-H2AX foci after DNA repair was evaluated to assess the role of PARP1 in long-lasting chromatin remodeling [37].
2. Results and Discussion
Cytotoxic effects induced by 4 Gy X-rays on WT and PARP1−/− testis cells were assessed by flow cytometric DNA content analysis 48 h after irradiation. The most radiosensitive testicular cell population is that of differentiating spermatogonia. Thus, shortly after irradiation, cytotoxic effects are reflected by a decrease of the S-phase flow cytometric compartment, which includes mainly proliferating spermatogonia. X-rays induced a comparable cytotoxic effect in WT and PARP1−/− mice (Table 1), the magnitude of which was in agreement with a previously published dose-effect relationship [38].
Table 1.
Percentage of testis cells in each population (standard error) as evaluated by flow cytometric DNA content analysis.
| Elongated spermatids | Round spermatids | Diploid cells | S-phase cells | Tetraploid cells | |
|---|---|---|---|---|---|
| WT control | 23.11 (1.06) | 49.85 (1.13) | 11.32 (0.52) | 2.69 (0.13) | 10.90 (0.20) |
| PARP1−/− control | 23.71 (0.31) | 48.42 (0.66) | 11.36 (0.31) | 2.46 (0.16) | 11.07 (0.27) |
| WT 4 Gy 48 h | 23.21 (0.61) | 49.32 (0.93) | 12.13 (0.91) | 1.04 (0.22) b | 10.94 (0.33) |
| PARP1−/− 4 Gy 48 h | 26.28 (0.79) a | 46.83 (0.93) | 11.57 (0.54) | 0.70 (0.11) b | 11.29 (0.27) |
p < 0.05;
p < 0.005.
Short term genotoxic damage induced by 4 Gy X-rays on WT and PARP1−/− testis cells was assessed by alkaline comet assay immediately after irradiation; removal of DNA lesions was assessed by comet analyses at 2 and 48 h after treatment. Overall data are summarized in Table 2.
Table 2.
Mean fraction tail DNA (± standard error) of unirradiated testis cells and of 4 Gy-irradiated cells sampled immediately, 2 or 48 h after exposure.
| WT Fraction tail DNA (± s.e.) | PARP1−/− Fraction tail DNA (± s.e.) | |
|---|---|---|
| Control | 0.09 (0.01) | 0.08 (0.01) |
| 4 Gy T0 | 0.14 (0.02) a | 0.13 (0.01) a |
| 4 Gy T2 | 0.08 (0.01) | 0.11 (0.01) a |
| 4 Gy T48 | 0.09 (0.01) | 0.08 (0.01) |
p < 0.05 with respect to matched controls.
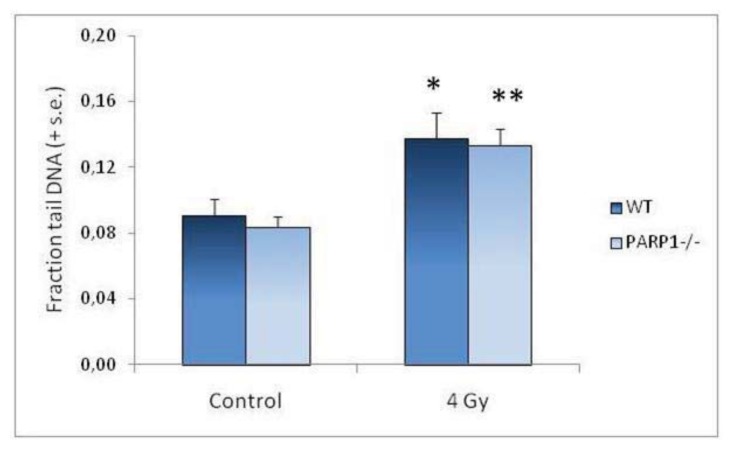
The mean fraction tail DNA values in control and irradiated testis cells evaluated in WT and PARP1−/− mice immediately after irradiation are also reported in Figure 1. No differences in the basal level of DNA strand breaks were observed; 4 Gy X-rays induced a similar significant increase of mean fraction tail DNA values in WT and PARP1−/− mice. These results suggest that, in male germ cells, lack of PARP1 does not affect the level of endogenous damage or modify chromatin structure in a way that makes it more susceptible to radiation-induced lesions.
Figure 1.
Fraction tail DNA of unirradiated testis cells and of 4 Gy-irradiated cells sampled immediately after exposure. Columns represent the mean of fraction tail DNA values (+ standard error) for each experimental group. Asterisks evidence results significantly different from matched controls (* p < 0.05; ** p < 0.01).
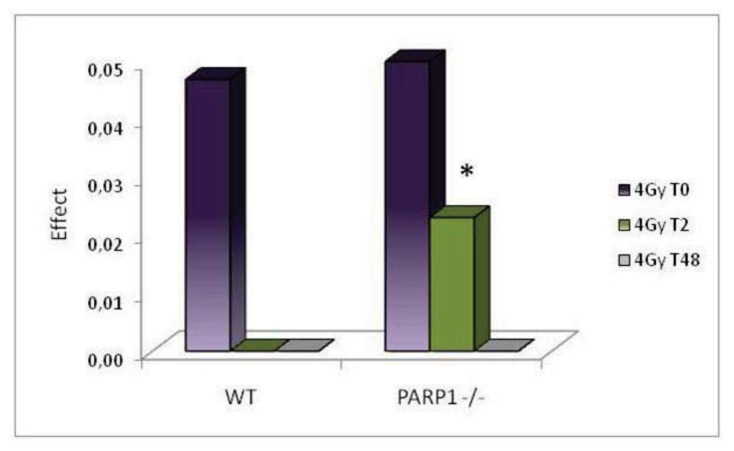
Histograms in Figure 2 report data on the level of residual damage observed at different times after irradiation expressed as differences between the mean fraction tail DNA of irradiated and unirradiated groups. WT mice showed a complete repair of DNA lesions within two hours after treatment; on the contrary, in PARP1−/− mice, although the level of damage decreased with time after irradiation, a statistically significant (p < 0.05) level of residual damage was still detected two hours after treatment. Similarly to WT animals, 48 h after irradiation no residual damage was detectable in PARP1−/− mice. These results are in agreement with previous studies in rodent testis cells reporting a significant delay of DNA repair following oxidative stress induced by chemical and physical agents when PARP activity was inhibited by chemical inhibitors [3,25,27], and suggest that PARP1 is specifically involved in DNA strand break repair after X-ray irradiation in one or more spermatogenic cell stages. The complete recovery observed 48 h after treatment indicates that, eventually, the absence of PARP1 is compensated by other proteins with a similar function and/or alternative repair pathways. Previous in vitro studies on PARP1 null cells had already suggested that lack of PARP1 decreases the efficiency of and delays DNA repair, although such effects are ultimately overcome by backup mechanisms [19,24,39].
Figure 2.
Differences between the mean fraction tail DNA of irradiated and unirradiated groups (Effect) immediately, 2 and 48 h after irradiation in testis cells of wild-type and PARP1−/− mice. Columns represent the mean values for each experimental group. The asterisk evidences results significantly different from matched controls (* p < 0.05).
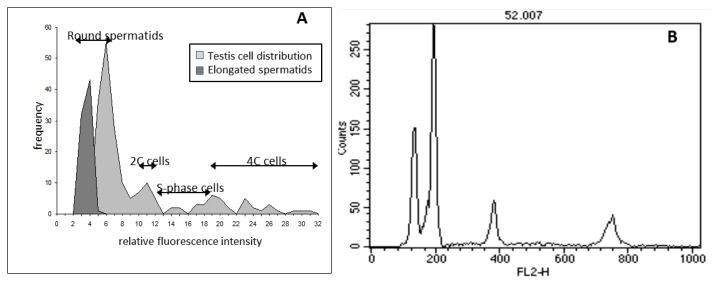
In order to investigate the response to the insult of different testis cell subpopulations we exploited the capacity of comet assay to identify DNA lesions in individual cells versus their ploidy. The cell distribution obtained by this approach (Figure 3A), that had been previously applied to evaluate DNA damage in different cell cycle phases [40] and different testicular cell types [36], was validated by comparing it with the DNA content distribution histogram obtained by flow cytometry (Figure 3B). The two distributions resulted very similar also considering that flow cytometric histogram was obtained measuring 10,000 cells, while the comet assay histogram is based on 200 cells.
Figure 3.
Comparison between DNA content histograms obtained by comet assay (A) and flow cytometry (B). The histograms were obtained by analyzing the samples of untreated testis cells. Two hundreds and 10,000 cells were analyzed by comet assay and flow cytometry respectively.
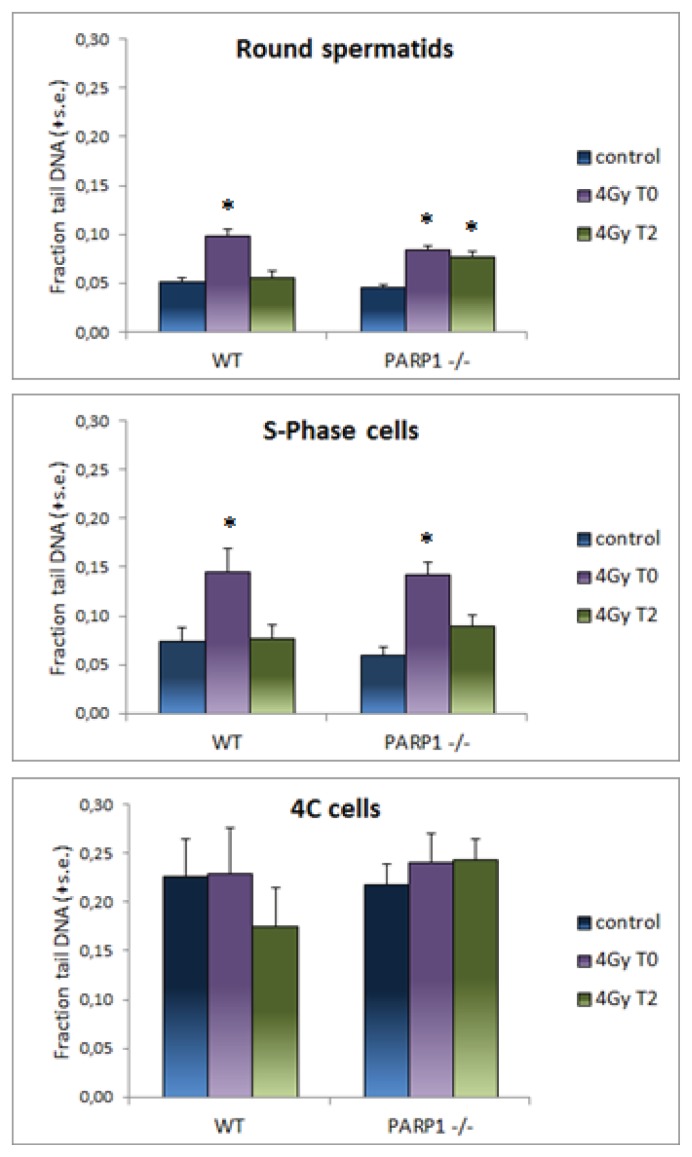
The mean fraction tail DNA values were then evaluated on cells attributed to different cell subpopulations, as described in the Experimental Section and are reported in Figure 4. The distribution of strand breaks in different cell types of unirradiated testes was similar in WT and PARP1−/− cells, with four-times higher mean fraction tail DNA values in untreated 4C cells than in the other cell types. This finding could be ascribed to DNA strand breaks generated during meiotic synapsis and recombination occurring in pachytene cells [41,42], which represent a relevant fraction of this subpopulation. Immediately after irradiation an increase of fraction tail DNA values was induced in round spermatids, 2C and S-phase cells while no effect was observed in 4C cells, indicating different sensitivity of various cell subpopulations. This result, in agreement with Zheng and Olive [9], could reflect differences in chromatin condensation and interaction of target DNA with histone and non-histone proteins during the differentiation process that make it differentially susceptible to radiation-induced lesions. It is known that radiosensitivity of testicular subpopulations varies, the mitotically active spermatogonia being the most sensitive to ionizing radiation, and spermatocytes and spermatids the most resistant cells [43]. Two hours after treatment, a significant reduction of radiation damage was observed in all cell subpopulations with the exception of PARP1−/− round spermatids, indicating that the delay in strand break rejoining observed in testis cells was mainly imputable to this cell subpopulation. This finding is consistent with the observation that round spermatids respond to genotoxic stress with a more elevated production of poly(ADPribose) than other testis cells and provides evidence that lack of PARP1, among all family members, is specifically implicated in the delay of DNA repair shown in rodent post-meiotic cells [3,25] treated with chemical inhibitors of poly(ADP) ribosylation. Considering that ADP-ribosylation of nuclear proteins releases chromatin structure and, in this way, facilitates repair of damaged regions [11,17], it is conceivable that, due to the progressive chromatin condensation occurring during post-meiotic maturation, spermatids increasingly rely upon PARP1 activity to give access to repair enzymes. It was not possible to specifically investigate the requirement for PARP1 in elongated spermatids because our experimental conditions failed to decondense their compact chromatin. However, since all DNA repair processes decrease with maturation, the role of PARP1 in mature spermatids is probably less relevant.
Figure 4.
Fraction tail DNA in different testis subpopulations, discriminated on the basis of their ploidy, after irradiation of WT and PARP1−/− mice. Mean of fraction tail DNA values (+ standard error) are shown for each experimental group. Control, untreated sample; 4 Gy T0, immediately after irradiation; 4 Gy T2, 2 h after irradiation. Asterisks evidence results significantly different from matched controls (* p < 0.05).
We had previously shown that round spermatids are peculiar also regarding the dynamics of H2AX phosphorylation, which persists long time after DNA damage induced by irradiation has been repaired [37]. To evaluate if PARP1 had also a role in the dynamics of H2AX phosphorylation in these cells, the induction and removal of γ-H2AX foci were compared in round spermatids from PARP1−/− and WT mice. H2AX phosphorylation has become a popular marker of radiation-induced DSB [44] and more recently, it has been proposed to also mark chromatin modifications independently from the presence of DNA breaks [45–47]. In unirradiated testes, H2AX is highly phosphorylated in the sex body of pachytene cells, which corresponds to the heterochromatic domain of sex chromosomes [41,48] and γ-H2AX marks sites of recombinational DSB preceding chromosome synapsis [41]. After irradiation, increased phosphorylation was found in the whole testis [49] and γ-H2AX foci were detected in A spermatogonia, pachytene spermatocytes, and round spermatids [3,37,42], as well as in neonatal male germ cells [50]. Due to the interplay between Ataxia Telangiectasia Mutated (ATM) kinase protein and poly(ADP-ribosyl)ation that is important for the phosphorylation of H2AX [51] a difference in the basal and radio-induced level of γ-H2AX foci could be expected between PARP1−/− and WT mice.
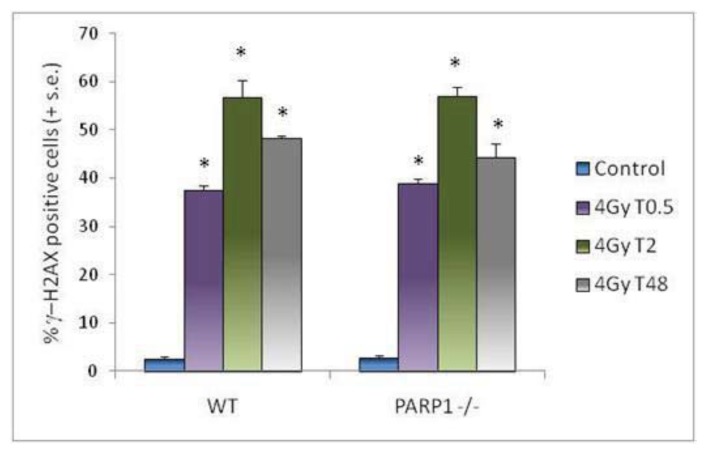
Under our experimental conditions, no difference was observed between PARP1−/− and WT mice in the background frequency of γ-H2AX-positive spermatids (Figure 5). Thirty minutes after irradiation a comparable highly significant increase (p < 0.001) of γ-H2AX-positive cells was found in both lines. The subsequent temporal evolution of γ-H2AX foci was also quantitatively similar in WT and PARP1−/− mice: 2 h after irradiation, a further increase of positive cells was observed, and 48 h after irradiation, although the number of foci was reduced, a high percentage of positive cells was still present. At the same time, the size of foci increased and they became discrete and countable. The average number was 5.7 ± 0.28 in WT and 5.0 ± 0.13 in PARP1−/− γ-H2AX positive cells, not showing significant differences between the two mouse lines. Such large foci were never detected in the few γ-H2AX positive unirradiated cells. Examples of γ-H2AX labeled testicular cells are shown in Figure 6. A number reduction and a size increase of foci had been observed in somatic and germ cells with time after irradiation and related to clustering of small foci and relocalization of repair enzymes to sites of complex unrepaired DNA lesions or to remaining scaffold structures used for DSB repair [37,52–54]. Our results suggest that PARP1 does not strongly influence the dynamics of H2AH phosphorylation in round spermatids. This finding is not completely consistent with data reported by Ahmed and co-workers [3] who showed that, after a comparable induction of foci by 1 Gy gamma-rays in mice treated or not with PARP chemical inhibitors, significantly more foci remained in PARP inhibited mice at eight hours after irradiation. Such different observations might be explained considering differences in tested doses (1 vs. 4 Gy), sampling times (8 h vs. 48 h), and analyzed endpoints (number of foci vs. percentage of positive cells), or might be actually due to a secondary role of PARP1 among PARP family members in radiation-induced γ-H2AX foci removal, or to a combination of all these factors. Additionally, it should be noted that inhibition of PARP1 is not equivalent to genetic deletion of PARP1, as also suggested by a comparison between RNAi depleted HeLa cells and the same cells treated with a PARP1 inhibitor [55]. In this study, the effects of inhibition were more serious as the PARP1 protein was still able to engage in the formation of a DNA damage chromatin complex making the shift to an alternative repair mode more difficult. Further experiments in PARP1−/− mice with lower X-ray doses might contribute to solve these issues.
Figure 5.
Percentages of γ-H2AX-positive round spermatids. Control, untreated sample; 4 Gy T0.5, 30 min after irradiation; 4 Gy T2, 2 h after irradiation; 4 Gy T48, 48 h after irradiation. Columns represent the mean values (+ standard error) for each experimental group. Asterisks evidence results significantly different from matched controls (* p < 0.001).
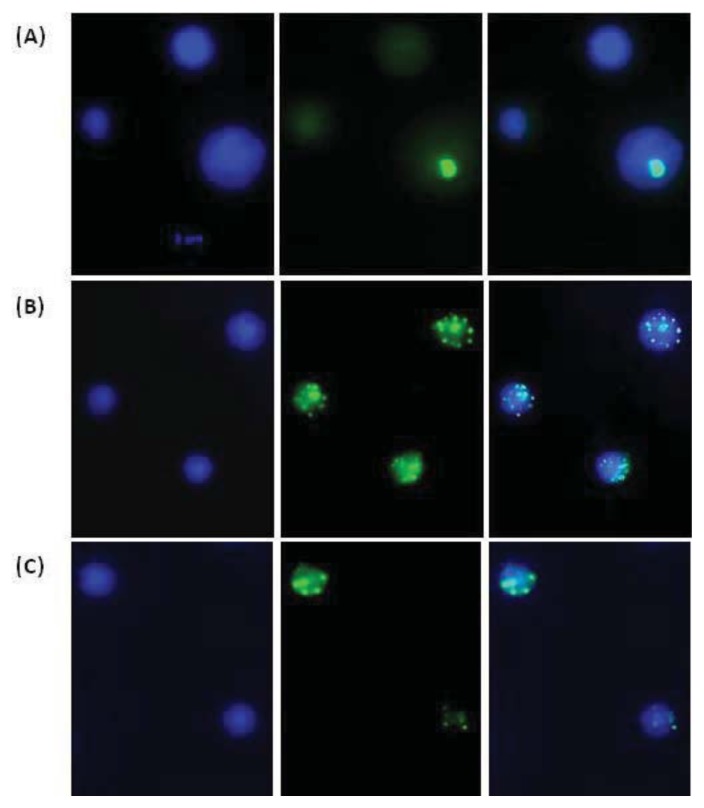
Figure 6.
Representative images of γ-H2AX immunostained testicular cells. Left: DNA fluorescence; middle: γ-H2AX fluorescence; right: merge. (A) Unirradiated cells: one pachytene spermatocyte showing a bright stained XY body and two round spermatids; (B) Three round spermatids 30 min after 4 Gy X-ray; (C) Two round spermatids 48 h after 4 Gy X-ray.
3. Experimental Section
3.1. Mice
Male C57Bl mice aged 12–14 weeks were obtained from Harlan (Udine, Italy); PARP1−/− mice were bred at ENEA from founders received from P. de Boer (Nijmegen, The Netherlands) upon licence of G. de Murcia (Strasbourg, France). Animals were maintained under standard conditions (20–22 °C, 60% relative humidity, on a 12 h light/dark cycle, with chlorinated water and feed ad libitum). All experimental protocols were reviewed and approved by the Institutional Animal Care and Use Committee.
3.2. X-Ray Irradiation
Mice were anaesthetized with Avertin (Sigma Aldrich St. Louis, MO, USA) diluted at 2.5% in sterile saline solution given intraperitoneally at a dose of 10 μL/g b.w. and only the testes region was irradiated with 4 Gy X-rays, the remainder of the body was shielded by 1-mm thick lead shield. Irradiation was carried out with a Gilardoni X-ray machine (15 mA, 250 kV; dose-rate 0.96 Gy/min). At different times after irradiation, mice were sacrificed and testes were removed and minced to obtain cellular suspensions to be analyzed.
3.3. Comet Assay of Testis Cells
Alkaline comet assay (pH > 13) was performed on testis cells from control mice and mice irradiated with 4 Gy and sacrificed immediately, 2 or 48 h after irradiation. At least four wild type (WT) and four PARP1−/− mice were used per experimental point. One testis from each mouse was minced and the cell suspensions were filtered through a 70 μm nylon mesh, centrifuged, resuspended in PBS (approximately 107 cells/mL), and mixed with low-melting point agarose (Bio-Rad, Hercules, CA, USA) to prepare slides for comet assay.
Alkaline comet assay was performed as described by Singh et al. [56] with minor modification [36]. Immediately before scoring, slides were stained with 12 μg/mL ethidium bromide (Sigma-Aldrich, St. Louis, MO, USA) and examined, at 200× magnification, with an Olympus fluorescence microscope. Slides were analyzed by a computerized image analysis system (Delta Sistemi, Rome, Italy). To evaluate the amount of DNA damage, computer generated fraction tail DNA values were used. Two-hundred cells were scored for each mouse from two different slides. Elongated spermatids, morphologically recognized, were not included in DNA damage analysis because our standard conditions failed to decondense their compact chromatin. Furthermore, the integral fluorescence intensity of each comet was taken as a measure of DNA content and used to discriminate cells with different DNA content [40]. The procedure to assign cells to the various subpopulations was adapted from the method previously described by Zheng and Olive [9]. In particular, cells were classified as haploid spermatids on the basis of DNA content distribution histograms and morphology; this subpopulation is defined “round spermatids” in the manuscript, although it is rather broad and includes different maturation stages from early round spermatids up to the elongating ones. Somatic and germinal cells, were considered 2C if their total fluorescence was comprised between one standard deviation minus and one standard deviation plus twice the mean haploid cell value. Cells with total fluorescence higher than double fluorescence of 2C cells were considered 4C. Cells included between 2C and 4C populations were considered S-phase cells. A representative histogram of relative fluorescence intensity obtained from a control sample is reported in Figure 3A and compared with the DNA distribution histogram obtained by flow cytometry on the same sample (Figure 3B).
3.4. Immunofluorescent Analysis of H2AX Histone Phosphorylation
Immunofluorescent analysis of H2AX histone phosphorylation was performed on round spermatids in groups of 4 PARP1−/− and 4 WT mice irradiated with 4 Gy X-rays and sacrificed 30 min, 2 h or 48 h later. Four unirradiated mice of each line were used as matched controls. Testis cell suspensions in PBS were mechanically prepared and filtered through 70 μm nylon mesh. The suspensions were fixed in 70% ethanol and diluted to a concentration of approximately 4 × 105 total cells/mL. Cells were cytospun onto slides (10 min, 600 rpm), air-dried, and re-fixed for 30 min in 2% paraformaldehyde in PBS. After 3 rinses in 0.05% Triton-X100 in PBS (TBS), slides were blocked in 5% nonfat dry milk in TBS for 1 h at 37 °C and incubated overnight at 4 °C with mouse monoclonal anti-γ-H2AX antibody (Upstate Biotechnology, Lake Placid, NY, USA) diluted 1:1000 in 10% goat serum, 5% nonfat dry milk in TBS. Slides were then washed 3 times in PBS, blocked in 5% nonfat dry milk in PBS for 30 min at 37 °C and finally incubated for 2 h at 37 °C with Alexa 488-conjugated goat antimouse IgG (Molecular Probes, Eugene OR, USA) diluted 1:2000 in 10% goat serum, 5% nonfat dry milk in PBS. Slides were rinsed in PBS and mounted in Vectashield mounting medium with DAPI (Vector Laboratories Inc., Burlingame, CA, USA).
Slides were viewed under 1000× magnification using an Olympus fluorescence microscope (Olymphus Optical co. Tokyo, Japan) equipped with CCD camera. Round spermatids were identified by their morphology according to Mahadevaiah et al. [41]. At 0.5 and 2 h post-irradiation time points there were tens of small foci on multiple focal planes and it was not possible to reliably count their number. For this reason, spermatids were simply classified in γ-H2AX positive and γ-H2AX negative (Figure 6). At 48 h after irradiation, few large countable foci were observed in positive cells, and their average number was also determined. At least 100 spermatids were analyzed per mouse.
3.5. Flow Cytometric DNA Content Analysis of Testis Cells
Flow cytometric (FCM) DNA content analysis was performed 48 h after irradiation on one testis of control and irradiated mice to evaluate radiation-induced cytotoxicity. The procedure used is described in detail elsewhere [57]. The DNA content of the testis cells was measured using a FACScan flow cytometer (Becton Dickinson, San Jose, CA, USA). A total of 1 × 104 events were accumulated for each measurement. Typical DNA content fluorescence intensity distribution histograms from adult mouse testicular cells (Figure 3B) are characterized by four main peaks representing elongated haploid spermatids, round haploid spermatids, cells with a 2C DNA content including G1 somatic and germ cells plus secondary spermatocytes, and cells with a 4C DNA content including G2 somatic and germ cells and primary spermatocytes; S-phase cells are included between 2C and 4C cell compartments, as described in Zante et al. [58]. The calculation of the relative frequencies of the various testicular cell types was performed automatically according to the model described in Lampariello and coworkers [59].
3.6. Statistical Analysis
Individual mouse data were considered the experimental unit. Mean values and standard errors relative to each homogeneous group were calculated. Statistical analysis was performed using STATISTICA software (StatSoft, Inc., Tulsa, OK, USA). Comparison between group means was performed by one-way ANOVA, and Duncan’s test was used for post hoc comparisons. Differences were considered due to the treatment when their probability level was lower than 5%.
4. Conclusions
In summary, our results suggest that in round spermatids lack of PARP1 delays the repair of radiation-induced DNA damage, whereas it does not seem to affect H2AX phosphorylation dynamics signaling double strand breaks or post-repair chromatin modifications. Additional experiments could clarify if these conclusions could be extended also to the radiation response of round spermatids in the low-dose range.
Acknowledgments
Partially supported by FILAS—Regione Lazio, in agreement with ENEA, Program TOP-IMPLART, 6 October 2010.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Brinkworth M.H. Paternal trasmission of genetic damage: Findings in animals and humans. Int. J. Androl. 2000;23:123–135. doi: 10.1046/j.1365-2605.2000.00222.x. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed E.A., van der Vaart A., Barten A., Kal H.B., Chen J., Lou Z., Minter-Dykhouse K., Bartkova J., Bartek J., de Boer P., et al. Differences in DNA double strand breaks repair in male germ cell types: Lessons learned from a differential expression of Mdc1 and 53BP1. DNA Repair (Amst. ) 2007;6:1243–1254. doi: 10.1016/j.dnarep.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed E.A., de Boer P., Philippensc E.P., Kal H.B., de Rooij D.G. Parp1–XRCC1 and the repair of DNA double strand breaks in mouse round spermatids. Mutat. Res. 2010;683:84–90. doi: 10.1016/j.mrfmmm.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 4.Olsen A.K., Bjortuft H., Wiger R., Holme J., Seeberg E., Bjoras M., Brunborg G. Highly efficient base excision repair (BER) in human and rat male germ cells. Nucleic Acid Res. 2001;29:1781–1790. doi: 10.1093/nar/29.8.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olsen A.K., Lindeman B., Wiger R., Duale N., Brunborg G. How do male germ cells handle DNA damage? Toxicol. Appl. Pharmacol. 2005;207:521–531. doi: 10.1016/j.taap.2005.01.060. [DOI] [PubMed] [Google Scholar]
- 6.Sathees C.R., Raman M.J. Mouse testicular extracts process DNA double-strand breaks efficiently by DNA end-to-end joining. Mutat. Res. 1999;433:1–13. doi: 10.1016/s0921-8777(98)00055-x. [DOI] [PubMed] [Google Scholar]
- 7.Srivastava N., Raman M.J. Homologous recombination-mediated double-strand break repair in mouse testicular extracts and comparison with different germ cell stages. Cell Biochem. Funct. 2007;25:75–86. doi: 10.1002/cbf.1375. [DOI] [PubMed] [Google Scholar]
- 8.Xu G., Spivak G., Mitchell D.L., Mori T., McCarrey J.R., McMahan C.A., Walter R.B., Hanawalt P.C., Walter C.A. Nucleotide excision repair activity varies among murine spermatogenic cell types. Biol. Reprod. 2005;73:123–130. doi: 10.1095/biolreprod.104.039123. [DOI] [PubMed] [Google Scholar]
- 9.Zheng H., Olive P.L. Influence of oxigen on radiation induced DNA damage in testicular cells of C3H mice. Int. J. Radiat. Biol. 1997;71:275–282. doi: 10.1080/095530097144157. [DOI] [PubMed] [Google Scholar]
- 10.Kikland J.B. Poly ADP-ribose polymerase-1 and health. Exp. Biol. Med. 2010;568:235–561. doi: 10.1258/ebm.2010.009280. [DOI] [PubMed] [Google Scholar]
- 11.Bouchard V.J., Rouleau M., Poirier G.G. PARP-1, a determinant of cell survival in response to DNA damage. Exp. Hematol. 2003;31:446–454. doi: 10.1016/s0301-472x(03)00083-3. [DOI] [PubMed] [Google Scholar]
- 12.Kim M.Y., Zhang T.W., Kim L.K. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- 13.Dantzer F., Schreiber V., Niedergang C., Trucco C., Flatter E., de La Rubia G., Oliver J., Rolli V., Menisier-de Murcia J., de Murcia G. Involvement of poly(ADP-ribose)polymerase in base excision repair. Biochimie. 1999;81:69–75. doi: 10.1016/s0300-9084(99)80040-6. [DOI] [PubMed] [Google Scholar]
- 14.Schreiber V., Ame J., Dollè P., Schultz I., Rinaldi B., Fraulob V., deMurcia J.M., deMurcia G. Poly(ADP-ribose) polimerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem. 2002;277:23028–23036. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- 15.Krishnakumar R., Kraus W.L. The PARP side of the nucleus: Molecular actions, physiological outcomes, and clinical targets. Mol. Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cardoso R.S., Espanhol A.R., Passos G.A.S., Sakamoto-Hojo E.T. Differential gene expression in γ-irradiated BALB/3T3 fibroblast under the influence of 3-aminobenzamide, an inhibitor of PARP enzyme. Mutat. Res. 2002;508:33–40. doi: 10.1016/s0027-5107(02)00141-0. [DOI] [PubMed] [Google Scholar]
- 17.D’Amours D., Desnoyers S., D’Silva I., Poirier G.G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 18.Masutani M., Nozaki T., Nakamoto K., Nakagama H., Suzuki H., Kusuoka O., Tsutsumi M., Sugimura T. The response of PARP knockout mice against DNA damaging agents. Mutat. Res. 2000;462:159–166. doi: 10.1016/s1383-5742(00)00033-8. [DOI] [PubMed] [Google Scholar]
- 19.Masutani M., Nakagama H., Sugimura T. Poly(ADP-ribose) and carcinogenesis. Genes Chromosom. Cancer. 2003;38:339–348. doi: 10.1002/gcc.10250. [DOI] [PubMed] [Google Scholar]
- 20.Meyer-Ficca M.L., Meyer R.G., Jacobson E.L., Jacobson M.K. Poly(ADP-ribose)polymerases: Managing genome stability. Int. J. Biochem. Cell Biol. 2005;37:920–926. doi: 10.1016/j.biocel.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 21.Muiras M.L. Mammalian longevity under the protection of PARP-1’s multi-facets. Ageing Res. Rev. 2003;2:129–148. doi: 10.1016/s1568-1637(02)00062-4. [DOI] [PubMed] [Google Scholar]
- 22.Petermann E., Keil C., Oei S.L. Importance of poly(ADP-ribose)polymerases in the regulation of DNA-dependent processes. Cell. Mol. Life Sci. 2005;62:731–738. doi: 10.1007/s00018-004-4504-2. [DOI] [PubMed] [Google Scholar]
- 23.Satoh M.S., Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356:356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 24.Shall S., de Murcia G. Poly(ADP-ribose)polymerase-1: What have we learned from the deficient mouse model? Mutat. Res. 2000;460:1–15. doi: 10.1016/s0921-8777(00)00016-1. [DOI] [PubMed] [Google Scholar]
- 25.Atorino L., di Meglio S., Farina B., Jones R., Quesada P. Rat germinal cells require PARP for repair of DNA damage induced by γ irradiation and H2O2 treatment. Eur. J. Cell Biol. 2001;80:222–229. doi: 10.1078/0171-9335-00153. [DOI] [PubMed] [Google Scholar]
- 26.Di Meglio S., Denegri M., Vallefuoco S., Tramontano F., Scovassi A.I., Quesada P. Poly(ADPR)polymerase-1 and poly(ADPR) glycohydrolase level and distribution in differentiating rat germinal cells. Mol. Cell. Biochem. 2003;248:85–91. doi: 10.1023/a:1024136927637. [DOI] [PubMed] [Google Scholar]
- 27.Di Meglio S., Tramontano F., Cimmino G., Jones R., Quesada P. Dual role of poly(ADP-ribose) polymerase-1 and −2 and poly(ADP-ribose) glycohydrolase as DNA-repair and pro-apoptotic factors in rat germinal cells exposed to nitric oxide donors. Biochim. Biophys. Acta. 2004;1692:35–44. doi: 10.1016/j.bbamcr.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 28.Meyer-Ficca M.L., Scherthan H., Burkle A., Meyer R.G. Poly(ADP-ribosyl)ation during chromatin remodeling steps in rat spermiogenesis. Chromosoma. 2005;114:67–74. doi: 10.1007/s00412-005-0344-6. [DOI] [PubMed] [Google Scholar]
- 29.Quesada P., Atorino L., Cardone A., Ciarcia G., Farina B. Poly(ADPribosyl)ation system in rat germinal cells at different stages of differentiation. Exp. Cell Res. 1996;226:183–190. doi: 10.1006/excr.1996.0217. [DOI] [PubMed] [Google Scholar]
- 30.Tramontano F., Malanga M., Quesada P. Differential contribution of poly(ADP-ribose)polymerase-1 and −2 (PARP-1 and −2) to the poly(ADP-ribosyl)ation reaction in rat primary spermatocytes. Mol. Hum. Reprod. 2007;13:821–828. doi: 10.1093/molehr/gam062. [DOI] [PubMed] [Google Scholar]
- 31.Meyer-Ficca M.L., Lonchar J., Credidio C., Ihara M., Li Y., Wang Z.Q., Meyer R.G. Disruption of poly(ADP-ribose) homeostasis affects spermiogenesis and sperm chromatin integrity in mice. Biol. Reprod. 2009;81:46–55. doi: 10.1095/biolreprod.108.075390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer-Ficca M.L., Ihara M., Lonchar J.D., Meistrich M.L., Austin C.A., Min W., Wang Z.Q., Meyer R.G. Poly(ADP-ribose) Metabolism is essential for proper nucleoprotein exchange during mouse spermiogenesis. Biol. Reprod. 2011;84:218–228. doi: 10.1095/biolreprod.110.087361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masutani M., Nozaki T., Nishiyama E., Shimokawa T., Tachi Y., Suzuki H., Nakagama H., Wakabayashi K., Sugimura T. Function of poly(ADP-ribose)polymerase in response to DNA damage: Gene-distruption study in mice. Mol. Cell. Biochem. 1999;193:149–152. [PubMed] [Google Scholar]
- 34.Menisier-deMurcia J., Niedergang C., Trucco C., Recoul M., Dutrillaux B., Mark M., Olivier F.J., Massan M., Dierich A., LeMeur M., et al. Requirement of poly(ADP-ribose)polymerase in recovery from DNA damage in mice and in cells. Proc. Natl. Acad. Sci. USA. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z.Q., Auer B., Stingl L., Berghammer H., Haidacher D., Schweiger M., Wagner E.F. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 1995;9:509–520. doi: 10.1101/gad.9.5.509. [DOI] [PubMed] [Google Scholar]
- 36.Cordelli E., Fresegna A.M., Leter G., Eleuteri P., Spanò M., Villani P. Evaluation of DNA damage in different stages of mouse spermatogenesis after testicular X-irradiation. Radiat. Res. 2003;160:443–451. doi: 10.1667/rr3053. [DOI] [PubMed] [Google Scholar]
- 37.Paris L., Cordelli E., Eleuteri P., Grollino M.G., Pasquali E., Ranaldi R., Meschini R., Pacchierotti F. Kinetics of γ-H2AX induction and removal in bone marrow and testicular cells of mice after X-ray irradiation. Mutagenesis. 2011;26:563–572. doi: 10.1093/mutage/ger017. [DOI] [PubMed] [Google Scholar]
- 38.Hacker-Klom U., Goehde W., Schumann J. Mammalian Spermatogenesis as a Biological Dosimeter for Ionizing Radiation. In: Eisert W.G., Mendelsohn M.L., editors. Biological Dosimetry. Springer-Verlag; Berlin/Heidelberg, Germany: 1984. pp. 127–137. [Google Scholar]
- 39.Cieslar-Pobudaa A., Saenkoa Y., Rzeszowska-Wolnya J. PARP-1 inhibition induces a late increase in the level of reactive oxygen species in cells after ionizing radiation. Mutat. Res. 2012;732:9–15. doi: 10.1016/j.mrfmmm.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 40.Villani P., Altavista P.L., Castaldi L., Leter G., Cordelli E. Analysis of DNA oxidative damage related to cell proliferation. Mutat. Res. 2000;464:229–237. doi: 10.1016/s1383-5718(99)00192-8. [DOI] [PubMed] [Google Scholar]
- 41.Mahadevaiah S.K., Turner J.M., Baudat F., Rogakou E.P., de Boer P., Blanco-Rodriguez V., Jasin M., Keeney S., Bonner W.M., Burgoyne P.S. Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 2001;27:271–276. doi: 10.1038/85830. [DOI] [PubMed] [Google Scholar]
- 42.Hamer G., Roepers-Gajadien H.L., van Duyn-Goedhart A., Gademan I.S., Kal H.B., van Buul P.P.W., de Rooij D.G. DNA double-strand breaks and γ-H2AX signaling in the testis. Biol. Reprod. 2003;68:628–634. doi: 10.1095/biolreprod.102.008672. [DOI] [PubMed] [Google Scholar]
- 43.Hamer G., Roepers-Gajadien H.L., Gademan I.S., Kal H.B., de Rooij D.G. Intracellular bridges and apoptosis in clones of male germ cells. Int. J. Androl. 2003;26:348–353. doi: 10.1111/j.1365-2605.2003.00436.x. [DOI] [PubMed] [Google Scholar]
- 44.Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 45.Baure J., Izadi A., Suarez V., Giedzinski E., Cleaver J.E., Fike J.R., Limoli C.L. Histone H2AX phosphorylation in response to changes in chromatin structure induced by altered osmolarity. Mutagenesis. 2009;24:161–167. doi: 10.1093/mutage/gen064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soutoglou E., Misteli T. Activation of the cellular DNA damage response in the absence of DNA lesions. Science. 2008;320:1507–1510. doi: 10.1126/science.1159051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ichijima Y., Sakasai R., Okita N., Asahina K., Mizutani S., Teraoka H. Phosphorylation of histone H2AX at M phase in human cells without DNA damage response. Biochem. Biophys. Res. Commun. 2005;336:807–812. doi: 10.1016/j.bbrc.2005.08.164. [DOI] [PubMed] [Google Scholar]
- 48.Fernandez-Capetillo O., Mahadevaiah S.K., Celeste A., Romanienko P.J., Camerini-Otero R.D., Bonner W.M., Manova K., Burgoyne P., Nussenzweig A. H2AX is required for chromatin remodelling and inactivation of sex chromosomes in male mouse meiosis. Dev. Cell. 2003;4:497–508. doi: 10.1016/s1534-5807(03)00093-5. [DOI] [PubMed] [Google Scholar]
- 49.Olive P.L., Banath J.P. Phosphorylation of histone H2AX as a measure of radiosensitivity. Int. J. Radiat. Oncol. Biol. Phys. 2004;58:331–335. doi: 10.1016/j.ijrobp.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 50.Forand A., Dutrillaux B., Bernardino-Sgherri J. Gamma-H2AX expression pattern in non-irradiated neonatal mouse germ cells and after low-dose gamma-radiation: Relationships between chromatid breaks and DNA double-strand breaks. Biol. Reprod. 2004;71:643–649. doi: 10.1095/biolreprod.104.027466. [DOI] [PubMed] [Google Scholar]
- 51.Haince J.F., Kozlov S., Dawson V.L., Dawson T.M., Hendzel M.J., Lavin M.F., Poirier G.G. Ataxia Telangiectasia Mutated (ATM) signaling network is modulated by a novel poly(ADP-ribose)-dependent pathway in the early response to DNA-damaging agents. J. Biol. Chem. 2007;282:16441–16453. doi: 10.1074/jbc.M608406200. [DOI] [PubMed] [Google Scholar]
- 52.Bouquet F., Muller C., Salles B. The loss of gammaH2AX signal is a marker of DNA double strand breaks repair only at low levels of DNA damage. Cell Cycle. 2006;5:1116–1122. doi: 10.4161/cc.5.10.2799. [DOI] [PubMed] [Google Scholar]
- 53.Costes S.V., Chiolo I., Pluth J.M., Barcellos-Hoff M.H., Jakob B. Spatiotemporal characterization of ionizing radiation induced DNA damage foci and their relation to chromatin organization. Mutat. Res. 2010;704:78–87. doi: 10.1016/j.mrrev.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Markova E., Schultz N., Belyaev I.Y. Kinetics and dose response of residual 53BP1/gamma-H2AX foci: Co-localization, relationship with DSB repair and clonogenic survival. Int. J. Radiat. Biol. 2007;83:319–329. doi: 10.1080/09553000601170469. [DOI] [PubMed] [Google Scholar]
- 55.Godon C., Cordelieres F.P., Biard D., Giocanti N., Megnin-Chanet F., Hall J., Favaudon V. PARP inhibition versus PARP-1 silencing: Different outcomes in terms of single-strand break repair and radiation susceptibility. Nucleic Acids Res. 2008;36:4454–4464. doi: 10.1093/nar/gkn403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh N.P., McCoy M.T., Tice R.R., Schneider E.L. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 57.Spanò M., Bartoleschi C., Cordelli E., Leter G., Segre L., Mantovani A., Fazzi P., Pacchierotti F. Flow cytometric and histological assessment of 1,2,3,4-diepoxybutane toxicity on mouse spermatogenesis. J. Toxicol. Environ. Health. 1996;47:101–119. doi: 10.1080/009841096161582. [DOI] [PubMed] [Google Scholar]
- 58.Zante J., Schumann J., Gohde W., Hacker U. DNA fluorocytometry of mammalian sperm. Histochemistry. 1977;54:1–7. doi: 10.1007/BF00493324. [DOI] [PubMed] [Google Scholar]
- 59.Lampariello F., Mauro F., Uccelli R., Spanò M. Automatic analysis of flow cytometric DNA histograms from irradiated mouse male germ cells. Cytometry. 1989;10:62–69. doi: 10.1002/cyto.990100111. [DOI] [PubMed] [Google Scholar]