Abstract
MicroRNAs (miRNAs) are endogenous ~23-nt RNAs that can play important gene-regulatory roles in animals and plants by pairing to the mRNAs of protein-coding genes to direct their posttranscriptional repression. This review outlines the current understanding of miRNA target recognition in animals and discusses the widespread impact of miRNAs on both the expression and evolution of protein-coding genes.
The miRNAs
Characterization of C. elegans genes that control the timing of larval development revealed two small regulatory RNAs, known as the lin-4 and let-7 RNAs (Lee et al., 1993; Reinhart et al., 2000). let-7 homologs, soon recognized in other bilateral animals, including mammals, exhibited temporal expression resembling that observed in C. elegans, suggesting that let-7 and perhaps other small temporal RNAs might be playing orthologous roles in diverse metazoan lineages (Pasquinelli et al., 2000). Soon thereafter, lin-4 and let-7 RNAs were reported to represent a very populous class of small endogenous RNAs found in worms, flies and mammals — a few expressed temporally, but most not — which were named microRNAs (miRNAs) (Lagos-Quintana et al., 2001; Lau et al., 2001; Lee and Ambros, 2001). miRNAs have since been found in plants, green algae, viruses, and more deeply branching animals (Griffiths-Jones et al., 2008).
Meanwhile other types of small RNAs have been found in animals, plants, and fungi. These include endogenous small interfering RNAs (siRNAs)(Reinhart and Bartel, 2002; Ambros et al., 2003) and Piwi-interacting RNAs (piRNAs)(Aravin et al., 2007). Like miRNAs, many of these other RNAs function as guide RNAs within the broad phenomenon known as RNA silencing, but miRNAs differ from these other classes of small RNAs in their biogenesis: miRNAs derive from transcripts that fold back on themselves to form distinctive hairpin structures (Bartel, 2004), whereas the other types of endogenous small RNAs derive either from much longer hairpins that give rise to a greater diversity of small RNAs (siRNAs), or from bimolecular RNA duplexes (siRNAs), or from precursors without any suspected double-stranded character (piRNAs).
Once processed from the hairpin (Grishok et al., 2001; Lee et al., 2003) and loaded into the Argonaute protein of the silencing complex (Hutvagner and Zamore, 2002; Mourelatos et al., 2002), the miRNAs pair with mRNAs to direct posttranscriptional repression. At sites with extensive pairing complementarity, metazoan miRNA can direct Argonaute-catalyzed mRNA cleavage (Hutvagner and Zamore, 2002; Song et al., 2004; Yekta et al., 2004). More commonly, though, the metazoan miRNAs direct translational repression, mRNA destabilization, or a combination of the two (Lee et al., 1993; Wightman et al., 1993; Lim et al., 2005). The various molecular processes at the heart of miRNA-directed translational repression and mRNA destabilization, which include inhibition of translation initiation and poly(A) shortening, are reviewed elsewhere (Filipowicz et al., 2008).
The number of confidently identified miRNA genes has surpassed 110 in C. elegans, 140 in D. melanogaster, and 400 in humans—numbers that approach about 1–2% the number of protein-coding genes in these respective species (Ruby et al., 2006; Landgraf et al., 2007; Ruby et al., 2007). These numbers will undoubtedly increase as high-throughput sequencing continues to be applied both to miRNA discovery and to the validation of some of the many additional candidates proposed.
Seeds of miRNA target prediction
The discovery of the abundance of miRNAs in diverse multicellular species raised many questions, including, perhaps most intriguingly, what are all these tiny noncoding RNAs doing? Key to answering this question has been to learn how to find their regulatory targets. Initial clues to miRNA target recognition came from the observation that the lin-4 RNA had some sequence complementarity to multiple conserved sites within the lin-14 mRNA (Lee et al., 1993; Wightman et al., 1993), within a region of the 3' untranslated region (UTR) that earlier molecular genetic analyses had shown was required for the repression of lin-14 by lin-4 (Wightman et al., 1991). Similarly, lin-4 and let-7 RNAs were found to have complementarity to UTR sites of lin-28 and lin-41, respectively, which are targets that were also found with help of genetic analyses (Moss et al., 1997; Reinhart et al., 2000).
What about the hundreds of miRNAs identified by cloning and computation, most of which correspond to loci without previously identified functions? In plants, many targets can be predicted with confidence simply by searching for messages with extensive complementarity to the miRNAs (Rhoades et al., 2002). In animals extensive complementarity, with consequent cleavage of the targeted message, occasionally occurs, but is much more unusual (Yekta et al., 2004; Davis et al., 2005). Thus, for metazoan miRNAs the challenge has been to devise a genomewide computational search that captures most of the regulatory targets without also bringing in too many false predictions.
Initial attempts generated algorithms and sets of predictions that were difficult for experimentalists to evaluate, which was exacerbated by the poor overlap between sets of predictions from the same organism (Enright et al., 2003; Lewis et al., 2003; Stark et al., 2003; John et al., 2004; Kiriakidou et al., 2004). Nonetheless, some of these efforts have provided methods and insights that helped set the stage for our current understanding of metazoan miRNA recognition. A key methodological advance was the use of preferential evolutionary conservation to evaluate the ability of an algorithm to distinguish miRNA target sites from the multitude of 3'-UTR segments that otherwise would score equally well with regard to the quality of miRNA pairing (Lewis et al., 2003). To the extent that sites are conserved more than would be expected by chance, they are judged to be under selective pressure and therefore biologically functional. Thus, by summing the net yield of conserved sites after correcting for the number expected by chance, features and refinements of the algorithm can be evaluated computationally. For example, short subsegments of the miRNA can be individually screened to learn which ones are subject to preferentially conserved pairing. In this way, common features of target recognition can be distinguished from those that seem equally plausible but are rarely if ever used, thereby enabling the principles of target recognition to be elucidated and algorithms to be developed without resorting to training on a known set of targets (Lewis et al., 2003; Lewis et al., 2005). Developing the algorithm without consideration of known targets avoids biases from sites that are more easily found experimentally and was particularly useful for mammalian miRNAs, for which no targets were known.
Current prediction methods are diverse, both in approach and performance (Table 1, Baek et al., 2008; Selbach et al., 2008), and all have room for improvement. Nonetheless, agreement is emerging on three conclusions, which are each reassuringly consistent with a growing body of experimental data. And, as further relief for the non-computational biologist, the most critical concepts for computational target prediction can be distilled down to a few simple guidelines that can be implemented by anyone with access to the UC Santa Cruz Genome Browser and the Find function on their word processor.
Table 1.
Tools for Predicting Metazoan miRNA Targets
| Tool (Refs)* | Clades** | Criteria for Prediction and Ranking | Website URL |
|---|---|---|---|
| Site conservation considered | |||
| TargetScan (1) | m | Stringent seed pairing, site number, site type, site context (which includes factors that influence site accessibility); option of ranking by likelihood of preferential conservation rather than site context | targetscan.org |
| f,w | Stringent seed pairing, site number, site type | targetscan.org | |
| EMBL (2) | f | Stringent seed pairing, site number, overall predicted pairing stability | russell.embl-heidelberg.de/miRNAs/ |
| PicTar (3) | m,f,w | Stringent seed pairing for at least one of the sites for the miRNA, site number, overall predicted pairing stability | pictar.mdc-berlin.de |
| EIMMo (4) | m,f,w | Stringent seed pairing, site number, likelihood of preferential conservation | mirz.unibas.ch/ElMMo2 |
| Miranda (5) | m,f,w,+ | Moderately stringent seed pairing, site number, pairing to most of the miRNA | microRNA.org |
| miRBase Targets (6) | m,f,w,+ | Moderately stringent seed pairing, site number, overall pairing | microrna.sanger.ac.uk |
| PITA Top (7) | m,f,w | Moderately stringent seed pairing, site number, overall predicted pairing stability, predicted site accessibility | genie.weizmann.ac.il/pubs/mir07/mir07_data.html |
| mirWIP (8) | w | Moderately stringent seed pairing, site number, overall predicted pairing stability, predicted site accessibility | 146.189.76.171/query |
| Site conservation not considered | |||
| TargetScan All (9) | m | Stringent seed pairing, site number, site type, site context (which includes factors that influence site accessibility) | targetscan.org |
| PITA All (7) | m,f,w | Moderately stringent seed pairing, site number, overall predicted pairing stability, predicted site accessibility | genie.weizmann.ac.il/pubs/mir07/mir07_data.html |
| RNA22 (10) | m,f,w | Moderately stringent seed pairing, matches to sequence patterns generated from miRNA set, overall predicted pairing and predicted pairing stability | cbcsrv.watson.ibm.com/rna22 |
References (refs) for current versions/releases: 1 (mammals, Friedman et al., 2008; flies, Ruby et al., 2007; worms, Ruby et al., 2006); 2 (Stark et al., 2005); 3 (Lall et al., 2006); 4 (Giadatzis et al., 2007); 5 (Betel et al., 2008); 6 (Griffiths-Jones et al., 2008); 7 (Kertesz et al., 2007); 8 (Hammell et al., 2008); 9 (Grimson et al., 2007); 10 (Miranda et al., 2006). Tools are listed according to criteria for prediction and ranking, which for those tools assessed using recent proteomics results corresponds to their overall performance (Baek et al., 2008).
Letters indicate predictions provided for the mammalian/vertebrate (m), fly (f), worm (w), or additional (+) clades.
The first major conclusion is that requiring conserved Watson–Crick pairing to the 5' region of the miRNA centered on nucleotides 2–7, which is called the miRNA ‘seed’ (Figure 1) markedly reduces the occurrence of false-positive predictions (Lewis et al., 2003; Brennecke et al., 2005; Krek et al., 2005; Lewis et al., 2005). The discovery that perfect seed pairing substantially improves prediction reliability implied that it was also important for miRNA target recognition (Lewis et al., 2003). This assertion dovetailed nicely with previous reports that the 5' region is the most conserved portion of the metazoan miRNAs (Lim et al., 2003) and the 5' region of certain Drosophila miRNAs perfectly matches 3'-UTR elements that mediate mRNA decay and translational repression (Lai, 2002), as well as subsequent experiments showing that miRNA-like regulation was most sensitive to nucleotide substitutions that disrupt seed pairing (Doench and Sharp, 2004; Kloosterman et al., 2004; Brennecke et al., 2005; Lai et al., 2005).
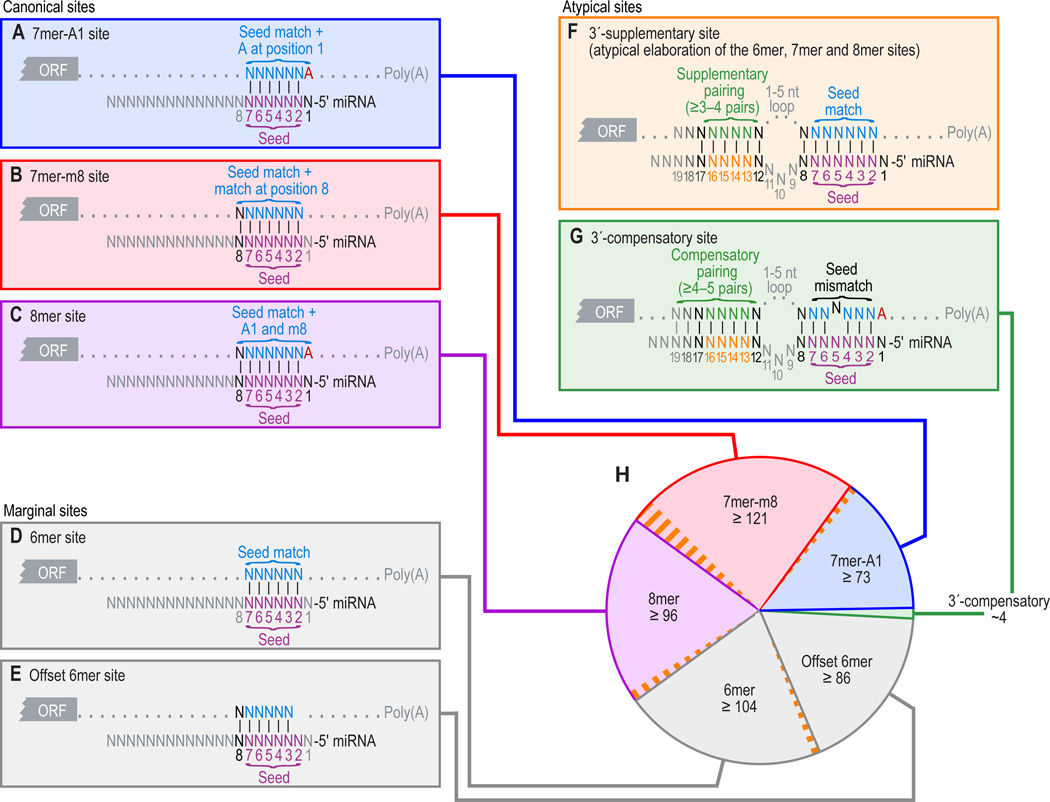
Figure 1. Types of miRNA target sites.
(A–C) Canonical, 7–8-nt seed-matched sites. Vertical dashes indicate contiguous Watson–Crick pairing.
(D–E) Marginal, 6-nt sites matching the seed region. These 6mer sites typically have reduced efficacy (Figure 4A) and are conserved by chance more frequently than the larger sites. Therefore, when prioritizing site efficacy and prediction specificity, prediction algorithms with stringent seed-pairing criteria disregard 6mer sites.
(F–G) Sites with productive 3´ pairing. For 3´-supplementary sites (F), Watson–Crick pairing usually centering miRNA nucleotides 13–16 (orange) supplements a 6–8-nt site (A–E). At least 3–4 well-positioned contiguous pairs are typically required for increased efficacy, which explains why 3´-supplementary sites are atypical. For 3´-compensatory sites (G), Watson–Crick pairing usually centering on miRNA nucleotides 13–16 (orange) can compensate for a seed mismatch and thereby create a functional site.
(H) Number of preferentially conserved mammalian sites matching a typical highly conserved miRNA (Friedman et al., 2008). For each site matching the seed region, orange-hatched subsectors indicate the fraction of conserved sites with preferentially conserved 3´-supplementary pairing. Analysis was performed using the 87 miRNA families highly conserved in vertebrates. A 7mer site is counted only if it is not part of an 8mer site, and a 6mer site is counted only if it is not part of a larger site. Values plotted are the number of preferentially conserved sites confidently detected above background, calculated as the average number of conserved sites minus the upper 95% confidence limit on the sites estimated to be conserved by chance. Thus for each site type of panels A–E, there is 95% confidence that the actual average of preferentially conserved sites is higher than that plotted.
The second conclusion is that conserved pairing to the seed region can also be sufficient on its own for predicting conserved targets above the noise of false-positive predictions (Brennecke et al., 2005; Krek et al., 2005; Lewis et al., 2005). For example, mammalian targets can be predicted by simply searching for conserved 7-nt matches in aligned regions of vertebrate 3' UTRs (Lewis et al., 2005). Prediction specificity increases when requiring an 8-nt match or multiple matches to the same miRNA, but systematic analysis of preferentially conserved features indicates that most targets of a given miRNA have only a single 7-nt match to that miRNA seed region. Fortunately, enough genomes have been sequenced and aligned such that these targets with single sites can now be predicted with confidence that most are authentic; when assessing the evolutionary conservation of 7-nt motifs that match miRNAs compared to those that do not match miRNAs but are of equal abundance in the UTRs, the ratio of predicted targets to estimated false positives is 3.5:1 in a five-genome analysis that extends to chicken (Lewis et al., 2005).
Hence, a simple three-step protocol can predict evolutionarily conserved targets for a metazoan miRNA: 1) Identify the two 7-nt matches to the seed region (Figure 1A and B). For example, miR-1, with sequence 5'-UGGAAUGUAAAGAAGUAUGUA, would recognize the CAUUCCA match and the ACAUUCC match. 2) Use available whole-genome alignments (Karolchik et al., 2008) to compile orthologous 3' UTRs. 3) Search within the orthologous UTRs for conserved occurrence of either 7-nt match. These are predicted regulatory sites. Note that members of the same miRNA family (i.e., miRNAs with the same sequence at nucleotides 2–8) all share the same predicted targets. A search for conserved 8-nt sites comprised of both 7-nt motifs (e.g., ACAUUCCA, in the case of miR-1, Figure 1C) yields greater prediction specificity, whereas a search for conserved 6-nt seed matches (Figure 1D) yields greater sensitivity. When only a few genomes are available, those sites present at orthologous positions in all genomes examined are the ones considered conserved. When more genomes are available, more sophisticated measures of conservation increase the information gleaned from the alignments (Gaidatzis et al., 2007; Kheradpour et al., 2007; Friedman et al., 2008).
As might be expected, searching for conserved instances of either of two 7-mers yields many predicted targets—hundreds of messages for each miRNA family. The surprise is that after subtracting the number of sites expected to be conserved by chance, the number of predicted targets remains very high. This leads to the third major conclusion: highly conserved miRNAs have very many conserved targets (Brennecke et al., 2005; Krek et al., 2005; Lewis et al., 2005; Xie et al., 2005). An updated analysis of preferential conservation of 7–8-nt sites reveals that the mammalian miRNAs conserved through vertebrates have an average of 300 conserved targets per miRNA family, a number that exceeds 400 if 6-mer sites are also included (Figure 1H, Friedman et al., 2008). In sum, more than half of the human protein-coding genes appear to have been under selective pressure to maintain 3´-UTR pairing to miRNAs (Friedman et al., 2008). As a result, miRNA targeting can explain a sizable fraction of the conserved motifs in mammalian 3´ UTRs (Xie et al., 2005).
As computational studies were uncovering evidence of widespread targeting, an experimental approach reached the same conclusion. After introducing miRNAs into HeLa cells, microarray analyses revealed modest effects on the levels of hundreds of mRNAs (Lim et al., 2005). Messages that decrease in response to each miRNA tend to have corresponding seed matches at a propensity indicating that most of the down-regulated messages are directly targeted by the miRNA. Because the miRNAs are introduced into cells that normally do not express them, some of the interactions observed in HeLa cells probably do not occur in the animal. Nonetheless, introduction of the miRNAs causes the expression profile of HeLa to shift towards that of the organ that normally expresses the introduced miRNA, thereby indicating that much of the observed targeting reflects, either directly or indirectly, that within the animal (Lim et al., 2005). In addition to providing experimental support for both the importance of seed pairing and the conclusion that miRNAs repress many messages, these results overturned the prevailing notion that miRNAs down-regulate protein output without influencing message levels. Indeed, re-evaluation of the effects of the lin-4 miRNA on lin-14 mRNA levels indicated substantial mRNA destabilization of this classical miRNA target (Bagga et al., 2005). Subsequent studies inhibiting or ablating endogenous miRNAs conclusively demonstrate the widespread transcriptomic effects on seed-matched messages (Krutzfeldt et al., 2005; Giraldez et al., 2006; Rodriguez et al., 2007). Identities of the mRNAs that co-immunoprecipitate with silencing complexes and the proteins that change following miRNA introduction or disruption further confirm the widespread targeting of seed-matched mRNAs (Beitzinger et al., 2007; Easow et al., 2007; Karginov et al., 2007; Baek et al., 2008; Selbach et al., 2008).
Comparison of target-prediction tools
Although initial target-prediction efforts yielded largely non-overlapping predictions, this changed with the developing consensus on the importance of seed pairing for miRNA:target recognition (Table 1). When evaluated based on proteomic changes following miRNA addition or deletion, tools that stringently require Watson–Crick seed pairing perform better than those that do not (Baek et al., 2008; Selbach et al., 2008). Tools with more moderately stringent cutoffs that allow a mismatch or wobble to the miRNA seed generally do perform better than expected by chance, but this observed efficacy can be explained by the strong response of a subset of predictions with perfect seed matches. The inability of these tools to find many functional sites with seed mismatches does not necessarily mean that mammalian messages lack such sites — it only means that, of the tools developed thus far, none are able to find many functional seed-mismatched sites.
The current predictions by TargetScan, PicTar, EMBL, and ElMMo have a high degree of overlap because they now all require stringent seed pairing. However, they are not 100% identical. Some reasons for imperfect overlap can be traced to alignment artifacts, the use of slightly different UTR databases, or the use of different miRNA sequences. Other reasons are intrinsic to the prediction algorithms themselves, such as the treatment of the target nucleotide opposite the first miRNA nucleotide. TargetScan rewards an A across from position 1 (Figure 1A, C), whereas the other algorithms with stringent seed pairing reward a Watson–Crick match across from this position (Krek et al., 2005; Lewis et al., 2005; Stark et al., 2005; Gaidatzis et al., 2007). This is not a factor for the many miRNAs that begin with a U; for such miRNAs the 7-nt matches used by each of the four tools are identical. However, for a miRNA beginning with A, C, or G, about half of the predicted targets differ because for such a miRNA one of the two 7-nt matches (the 7mer-m8, Figure 1B) is the same for all four tools, whereas the other one differs (the 7mer-A1, Figure 1A).
Several lines of evidence support the non-Watson–Crick recognition of target position 1. The first is the site-conservation analyses that originally detected this preference in vertebrates (Lewis et al., 2005). The second is array and proteomics data in mammalian cells showing that for miRNAs (and siRNAs) that do not begin with U, the 7mer-A1 sites out-perform sites with a Watson–Crick match to position 1 (Nielsen et al., 2007; Baek et al., 2008). Whether or not this preference extends beyond mammals has not yet been reported, but a biochemical preference for a mismatch at position 1 and crystallographic studies indicating that the 5´-most nucleotide of an Argonaute-bound guide RNA is not paired to the target strand both suggest that it could extend beyond mammals (Haley and Zamore, 2004; Ma et al., 2005; Parker et al., 2005).
Another difference between the prediction sets is their rank ordering of the targets, designed to help biologists focus on predictions more likely to be authentic or responsive. When assessed using available proteomics results, the higher-ranked predictions of several tools trend toward better performance, with the most robust discrimination observed for Targetscan rankings (Baek et al., 2008). When evaluated independently, each of the parameters used to rank TargetScan predictions — site conservation, site number, site type (with 8mer > 7mer-m8 > 7mer-A1) and site context (described below) — correlate with targeting efficacy (Grimson et al., 2007; Nielsen et al., 2007; Baek et al., 2008; Friedman et al., 2008; Selbach et al., 2008). Examples of top-ranked targets for well-studied miRNA families in nematodes, flies, and mammals are shown (Figure 2A).
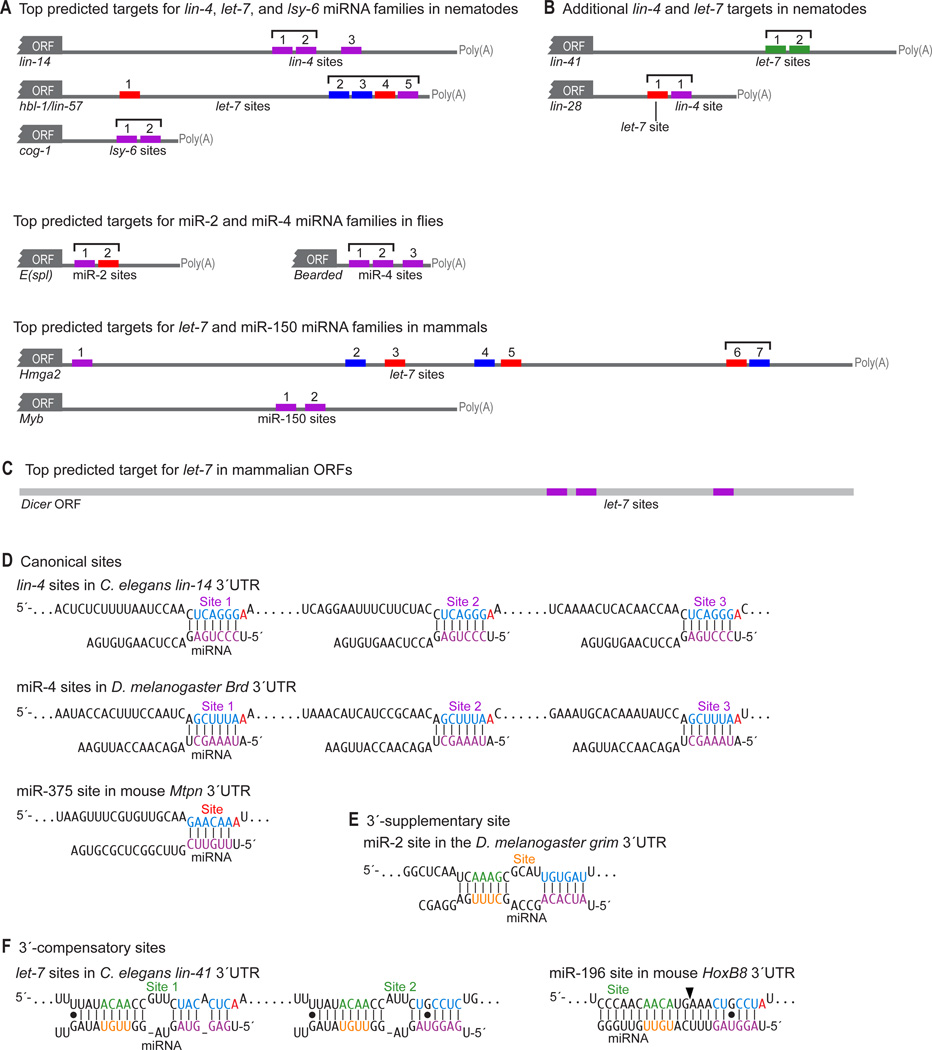
Figure 2. Examples of conserved miRNA targets and sites.
Coloring of site types and residues within the miRNAs and targets are as in Figure 1.
(A) Correspondence between the top predicted targets and genetically implicated interactions (Wightman et al., 1991; Abrahante et al., 2003; Johnston and Hobert, 2003; Lai et al., 2005; Mayr et al., 2007; Xiao et al., 2007). The nematode and mammalian predictions are the top Targetscan (release 4.2) predictions for the respective miRNAs, whereas the fly predictions are among several top but essentially equivalent predictions. Many are also among the top three predictions for the respective miRNAs at the PicTar (lin-14, hbl-1, Brd, Hmga2), EMBL (E(Spl), Brd), and EIMMo (hbl-1, Brd, Hmga2) web sites. Sites within cooperative distance of each other are indicated (square brackets).
(B) Additional examples of nematode targets originally identified with assistance from genetics (Moss et al., 1997; Reinhart et al., 2000).
(C) Top ORF target candidate of mammalian let-7 (Lewis et al., 2005).
(D) Examples of canonical miRNA target sites (Lee et al., 1993; Wightman et al., 1993; Poy et al., 2004; Lai et al., 2005), with sites and pairing reflecting current understanding of target recognition.
(E) A 3´-supplementary site with experimental support (Brennecke et al., 2005).
(F) 3´-compensatory sites with experimental support (Vella et al., 2004; Yekta et al., 2004). The position of miRNA-directed cleavage within the HoxB8 3´ UTR is indicated (arrowhead).
Supplementary and compensatory pairing
Even after recognition of the importance of seed pairing, a reasonable assumption has been that pairing to the remainder of the miRNA usually supplements seed pairing to enhance binding specificity and affinity. However, experimental evidence for frequent function of such 3´-supplementary pairing has not yet materialized (Doench and Sharp, 2004; Brennecke et al., 2005; Lim et al., 2005). Similarly, comparative analysis in flies indicates that the majority of sites under selection have no more 3´-supplementary pairing than expected by chance (Brennecke et al., 2005), and parallel analyses in mammals motivated a disregard of 3´-supplementary pairing altogether (Lewis et al., 2005). Indeed, the early pairing and energy-based rubrics initially designed to identify and rank 3´-supplementary pairing (Lewis et al., 2003; John et al., 2004; Krek et al., 2005) lack predictive value (Grimson et al., 2007; Baek et al., 2008).
Complicating genomewide attempts to evaluate the potential contribution of 3´-supplementary pairing are the numerous potential pairing possibilities involving the 3´ portion of the miRNA and the UTR, some of which might be more favored than others, perhaps because they are more compatible with the location and configuration of the miRNA in the silencing complex. Systematic analyses of pairing possibilities, searching for those associated with preferential conservation and increased efficacy on the arrays, has revealed a type of 3´-supplementary pairing that is both productive and associated with a sufficient number of sites to be supported by microarray datasets (Grimson et al., 2007). This 3´ pairing optimally centers on miRNA nucleotides 13–16 and the UTR region directly opposite this miRNA segment (Figure 1F). Such sites in which 3´ pairing productively augments seed pairing are called ‘3´-supplementary sites’. Like seed pairing, 3´ pairing appears relatively insensitive to predicted thermostability and instead is sensitive to pairing geometry, preferring at least 3–4 contiguous Watson–Crick pairs uninterrupted by bulges, mismatches or wobbles. Sites with conserved supplementary pairing of this type are predicted with significantly greater specificity, but such sites are atypical (Figure 1H) and tend to be only slightly more effective than those without the supplemental pairing (Grimson et al., 2007), suggesting that supplemental 3´ pairing plays a modest role in target recognition.
Pairing to the 3´ portion of the miRNA can not only supplement a 7–8mer match, it can also compensate for a single-nucleotide bulge or mismatch in the seed region, as illustrated by the let-7 sites in lin-41 and the miR-196 site in Hoxb8 (Figure 2F). These are called ‘3´-compensatory sites’ (Figure 1G). In all experimentally validated examples of 3´-compensatory sites, the pairing centered on miRNA nucleotides 13–17 extends to at least 9 contiguous Watson–Crick pairs — substantially more than the number needed to observe effective supplementary pairing. Indeed, for the miR-196 site in Hoxb8, the pairing is so extensive that the miRNA directs the Argonaute-mediated cleavage of the message (Yekta et al., 2004).
The let-7 sites in C. elegans lin-41 happened to be some of the first proposed and definitively validated sites in animals (Reinhart et al., 2000; Vella et al., 2004), and therefore had a particularly strong influence on early concepts of miRNA targeting. Indeed, the let-7 sites in lin-41, which had some resemblance to a few of the proposed (but not individually validated) lin-4 sites in lin-14, were originally thought to exemplify typical miRNA target sites. However, systematic examination of site conservation indicates that mismatched seed sites with 3´-compensatory pairing are only rarely under selective pressure to be conserved (Brennecke et al., 2005; Lewis et al., 2005; Friedman et al., 2008). Perhaps because such sites with extensive pairing to the 3´ portion of the miRNA possess much more informational complexity than do the 7–8mer perfect matches and therefore emerge much less frequently and are harder to maintain in evolution, these 3´-compensatory sites appear to be used only rarely for biological targeting, comprising ~1% of the conserved sites in mammals (Figure 1H). Thus to achieve prediction specificity, algorithms can either omit the prediction of 3´-compensatory sites (Lewis et al., 2005; Gaidatzis et al., 2007) or predict them at high stringency, requiring such extensive pairing to the miRNA that the 3´-compensatory sites do not substantially increase the total number of predictions (Krek et al., 2005; Stark et al., 2005; Friedman et al., 2008).
If 3´-compensatory sites are indeed rare, then the question arises as to why lin-41 would have two highly conserved 3´-compensatory sites for let-7. An answer to this question becomes apparent when considering the other let-7 family members in worms, which include let-7, miR-48, miR-84, and miR-241, all of which have the same seed region but differ in their remaining sequence (Lau et al., 2001; Lim et al., 2003). Because these three paralogs are expressed earlier than is let-7, their repression of lin-41 would presumably cause cells to precociously assume adult cell fates (Reinhart et al., 2000; Abbott et al., 2005). To prevent this undesired outcome, both lin-41 sites have two important features: 1) imperfect seed pairing to the let-7 family, which prevents regulation by most family members, and 2) extensive compensatory pairing involving the unique 3´ portion of let-7 RNA, which enables later repression by let-7 (Brennecke et al., 2005; Lewis et al., 2005). Presumably other situations requiring regulation by a specific member of a miRNA family would favor the emergence and retention of 3´-compensatory pairing, and the consequent lack of redundant function among miRNA family members might make such targets particularly sensitive to the loss of a single miRNA (and thereby more readily identified through forward genetics). Moreover, a few key 3'-compensatory sites per miRNA could explain the preferential conservation of miRNA nucleotides sometimes observed outside the seed region.
For the many sites that lack evidence for consequential 3´ pairing, the 3´ region of the miRNA might still interact with the message, but in a way that does not favor matches over mismatches and therefore does not add detectably to targeting specificity. A common practice is to show the 3´ region of the miRNA paired to the message, depicting short regions of potential pairing of the type that generally can be found between any two arbitrarily chosen RNA fragments. A reasonable alternative is to avoid proposing such pairing for a majority of miRNA sites (e.g., Figure 2D) and to depict pairing to the miRNA 3´ region only in cases for which there is reason to believe that it adds to targeting specificity (Figure 2E and F).
Suggested basis for the specialized roles of each region
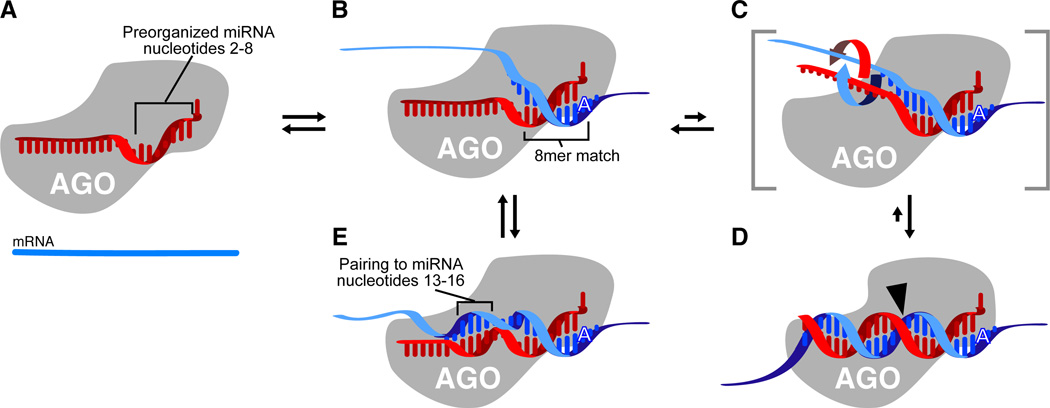
One mechanistic model for explaining the primacy of seed pairing proposes that the protein of the silencing complex presents the 5´ region of the miRNA (or siRNA) preorganized to favor Watson–Crick pairing to the mRNA (Bartel, 2004; Mallory et al., 2004) (Figure 3A). Presentation of nucleotides 2–8 prearranged in a geometry resembling an A-form helix would enhance both the affinity and specificity for matched mRNA segments, enabling 7–8-nt sites to suffice for most targeting functions (Figure 3B). Presentation of a preformed helical segment longer than ~7 nt would impose topological challenges and would not increase the effective nucleation surface because too many nucleotides would face opposing directions, whereas a shorter preformed segment would have both lower affinity and lower specificity. Consistent with this model, a co-structure with a target mimic reveals protein contacts to the guide-strand backbone that might also preorganize the seed region prior to target binding (Ma et al., 2005; Parker et al., 2005), and biochemical studies indicate that affinity to the seed is stronger than that to other regions of the RNA (Haley and Zamore, 2004; Ameres et al., 2007).
Figure 3. A speculative model explaining the roles of each miRNA region.
(A) MicroRNA (red) bound by Argonaute (AGO) such that nucleotides 2–8 are preorganized to favor efficient pairing. Nucleotide 1 is twisted away from the helix and permanently unavailable for pairing; nucleotides 9–11 are facing away from an incoming mRNA and unavailable for nucleation; the remainder of the miRNA is bound in a configuration that has not been preorganized for efficient pairing.
(B) Recognition of an 8mer site by the preformed binding pocket. An A at position 1 of the site presumably is recognized directly by the protein of the silencing complex.
(C) Massive conformational accommodation of extensively paired sites. At very rare sites of endogenous miRNAs (and the intended sites of exogenous siRNAs) pairing is extensive. In this model, pairing anchored and nucleated at the seed extends to the central region of the miRNA causing the protein to loosen its grip on the 3´ region of the miRNA and thereby allowing the miRNA and mRNA to wrap around each other.
(D) Accommodated pairing suitable for mRNA cleavage. The Argonaute protein locks down on the extensively paired duplex, which places the active site (black arrowhead) in position to cleave the mRNA.
(E) 3´-supplementary pairing. The message can pair to nucleotides 13–16, incorporating them into a short helical segment without major perturbation of the Argonaute protein or the remainder of the miRNA. Importantly, in this mode of target recognition, the miRNA and mRNA are not wrapped around each other.
Although pairing to the seed region is often sufficient for functional binding specificity, some sites involve additional pairing outside the seed region (Figure 1F–H). A disproportional importance of seed pairing nonetheless is observed at cleavage sites, suggesting that pairing might still nucleate at the seed match of these cleavage sites and then spread to the central and 3´ regions of the miRNA (or siRNA) (Bartel, 2004; Haley and Zamore, 2004; Mallory et al., 2004; Ameres et al., 2007). In this seed-nucleation model, a substantial conformational accommodation must occur to achieve extensive Watson–Crick pairing: Prior to target recognition, the miRNA is likely bound along its entire length to protein, presumably to the Argonaute protein (Figure 3A); otherwise the miRNA sugar-phosphate backbone would be accessible to cellular RNases. After nucleating at the seed (Figure 3B), pairing cannot spread much further without the protein releasing its grip on the remainder of the miRNA. Perhaps successive pairs to the more central miRNA residues helps trigger a transient release of the central and 3´ miRNA regions, allowing these regions to wrap around the message to complete the two helical turns (Figure 3C). Following this large conformational change, the protein can complete the accommodation by re-binding the central and 3´ regions of the miRNA in a mode that cleaves the message (Figure 3D). In this model, some binding energy gained in forming the central pairs would be offset by the disruption of miRNA:Argonaute interactions, causing contiguous pairing outside the seed to contribute less affinity than might have otherwise been expected. This lower contribution to affinity could explain why the sites that do not require central pairing for function (3´-supplementary and 3´-compensatory sites) tend to skip contiguous pairing to the central residues (Figure 1F–G). Pairing to these residues might be neutral or even disfavored in terms of binding energy and would begin to induce the accommodation, whereas starting a fresh pairing region at residues 13–16 (without the miRNA actually wrapping around the mRNA) would enable additional favorable interactions without incurring the cost of the large conformational change (Figure 3E).
Beyond conserved targeting
Target recognition that relies heavily on 7-nt matches to the seed region creates the possibility for a lot of nonconserved targeting, given that poorly conserved 7-nt sites outnumber preferentially conserved ones by about ten-to-one. Are a substantial fraction of these sites functional? After all, the cell cannot evaluate evolutionary conservation when choosing which of the sites should mediate repression.
Heterologous reporter assays show that when present in the same cell as the miRNA, a large fraction of nonconserved sites can indeed function (Farh et al., 2005), raising the question of how often messages with nonconserved sites are present in the same tissues as their cognate miRNAs. Analyses of mRNA and miRNA expression profiles indicate that 3´ UTRs with nonconserved sites are most often found in genes primarily expressed in tissues where the cognate miRNA is absent (Farh et al., 2005). This observation has a simple evolutionary explanation, known as selective avoidance. Over the course of evolutionary drift, sites for miRNAs that are absent in the cells where the mRNA is expressed can accumulate without consequence, whereas sites that emerge for a miRNA that is highly expressed in the same cell where the message functions will often impart a selective disadvantage and thus fail to be fixed in the population (Farh et al., 2005; Stark et al., 2005). Despite this trend, because so many messages have nonconserved 7-nt sites for each miRNA, the minority of messages that are co-expressed with the miRNA still constitutes a large number, creating the possibility for much nonconserved targeting (Farh et al., 2005). Identities of messages and proteins that increase upon inhibition or removal of an endogenous miRNA demonstrate that nonconserved targeting is even more widespread than conserved targeting (Krutzfeldt et al., 2005; Giraldez et al., 2006; Rodriguez et al., 2007; Baek et al., 2008; Selbach et al., 2008). Although much of this nonconserved targeting could represent inconsequential, evolutionarily neutral dampening of gene expression, some presumably represents important species-specific repression. With this in mind, targets have been predicted without considering site conservation (Miranda et al., 2006; Grimson et al., 2007; Kertesz et al., 2007). PITA and TargetScan predictions have been reported both with and without conservation cutoffs. Although disregarding conservation compromises overall performance, the highest ranked predictions of both tools perform at least as well as do their respective highest ranked conserved predictions (Baek et al., 2008).
The depletion of sites in messages co-expressed with the miRNA is particularly striking in messages most highly and preferentially expressed in the same tissue as the miRNA. 3´-UTRs of these mRNAs have about half as many nonconserved 7-nt sites for that miRNA as expected by chance, a depletion attributed in large part to selective avoidance (Farh et al., 2005). The evolutionary pressure to avoid emergence of fortuitous miRNA sites is also detected in ‘housekeeping genes’ and might explain the shorter 3´ UTRs of these genes in animals when compared to 3´ UTRs of orthologs in plants and fungi, which lack abundant 3´ UTR targeting (Stark et al., 2005). Messages selectively avoiding targeting to a miRNA are called the ‘antitargets’ of that miRNA (Bartel and Chen, 2004). When considering the thousands of messages avoiding targeting to particular miRNAs together with those avoiding targeting to all miRNAs, the phenomenon of selective avoidance clearly has had a widespread impact on UTR evolution, with the estimated number of antitargets comparable to the number of conserved targets (Farh et al., 2005).
When considering conserved targeting, nonconserved targeting, and antitargeting, miRNAs likely influence the expression or evolution of nearly all mammalian mRNAs. Indeed, these effects are so widespread that the spatial and temporal specificities of highly expressed miRNAs can be revealed by finding those 7-nt motifs that are underrepresented in messages preferentially expressed in particular tissues or developmental stages (Farh et al., 2005). The reason that this unusual approach of searching for the absence of nonconserved sites (rather than the presence of conserved sites) is productive lies in the fact that 7-nt sites—conserved ones as well as nonconserved ones—frequently have what it takes to mediate biological repression. Indeed, the most important implication of the selective avoidance phenomenon concerns its ramifications for endogenous target recognition. Selective avoidance shows that when 7mer sites emerge in 3´ UTRs, they often appear in contexts suitable for biological repression—otherwise, depletion of sites could not be observed.
The influence of UTR context
Although 7–8-nt matches to the seed region, coupled with whether or not the mRNA is coexpressed with the miRNA, can explain much of targeting specificity, they cannot explain all of it. Selective avoidance leads to ~50% site depletion, not the near-100% depletion that might have been expected if 7-nt sites mediated repression regardless of their sequence context. Moreover, reporter assays show that the identical site can mediate repression in some UTRs but not in others (e.g., Brennecke et al., 2005; Farh et al., 2005; Giraldez et al., 2006). Because pairing to the 3´ portion of the miRNA is rarely consequential, additional features of UTR context must influence site efficacy. Additional features recently found to boost site efficacy include 1) positioning within the 3´ UTR at least 15 nt from the stop codon, 2) positioning away from the center of long UTRs, 3) AU-rich nucleotide composition near the site or other measures of site accessibility, and 4) proximity to sites for coexpressed miRNAs. These features are discussed below.
Although most investigation into metazoan miRNA function has been for sites in 3´ UTRs, experiments using artificial sites show that targeting can occur in 5´ UTRs and open reading frames (ORFs) (Kloosterman et al., 2004; Lytle et al., 2007), and computational and experimental genome-wide analyses indicate that a significant amount of targeting, involving thousands of mRNAs, occurs in ORFs (Farh et al., 2005; Lewis et al., 2005; Lim et al., 2005; Easow et al., 2007; Grimson et al., 2007; Stark et al., 2007; Baek et al., 2008) (Figure 2C). Overall, endogenous ORF targeting appears less frequent and less effective than 3´-UTR targeting but still much more frequent than 5´-UTR targeting.
One reason that 5´ UTRs and ORFs may be less hospitable for targeting is that silencing complexes bound to these regions would be displaced by the translation machinery as it translocates from the cap-binding complex through the ORF (Bartel, 2004). Support for this notion comes with the observation that the transition to more effective and more selectively conserved sites is not at the stop codon but instead occurs ~15 nt into the 3´ UTR, precisely as expected if the first 15 nt of the 3´ UTR were cleared of silencing complexes when they enter the ribosome as it approaches the stop codon (Grimson et al., 2007). Targeting of sites perfectly complementary to artificial siRNAs is not hampered by ribosome interference, presumably because the ribosome has more difficulty disrupting extensive pairing or because sites that are cleaved need not remain associated as long to the mRNA.
The apparent interference by the ribosome along its entire path of translation implies that most messages under detectable miRNA control experience at least one round of translation prior to or concurrent with their repression; if an appreciable number of molecules were repressed prior to translation of the full-length protein, then the strong effects of ribosome interference could not be observed (Grimson et al., 2007). By similar reasoning, ORF targeting is expected to be much more effective in messages that are already inefficiently translated, because fewer ribosomes passing through the site would allow for greater residency time of the silencing complex. In this way, ORF targeting could provide an important mechanism for amplifying the effects of both miRNA 3´-UTR targeting and other types of translational repression.
Genome-wide analyses of site conservation, site efficacy, and site depletion all indicate that 7–8-nt sites within the 3´ UTR and out of the path of the ribosome tend to be most effective if they do not fall in the middle of long UTRs (Grimson et al., 2007). One explanation for these results is that sites in the middle of long UTRs might be less accessible to the silencing complex because they would have opportunities to form occlusive interactions with segments from either side, whereas sites near the UTR ends would not. These same types of analyses show that even more important than site position is the nucleotide composition in the immediate vicinity of the site, with those sites within high local AU content performing best (Grimson et al., 2007). When the site has a match to position 8, an A or U across from position 9 is particularly favorable, suggesting non-Watson–Crick recognition of this nucleotide resembling that opposite position 1 (Lewis et al., 2005; Nielsen et al., 2007). The remaining benefit of local AU composition might be, as suggested for site position, to place the site within a more accessible UTR context. Indeed, several methods have been proposed for predicting accessible UTR secondary structure favorable for miRNA targeting (Robins et al., 2005; Zhao et al., 2005; Kertesz et al., 2007; Long et al., 2007; Hammell et al., 2008). Although some of these methods have predictive value, when evaluated by monitoring the impact of the miRNA on both mRNA destabilization and protein output, they are less successful in predicting responsive targets than is scoring local AU content (Grimson et al., 2007; Baek et al., 2008). Perhaps because of RNA-binding proteins, RNA tertiary structure, and multiple competing RNA pairing conformations, the details of intracellular UTR structures might differ substantially from predicted structures, such that scoring local AU content is more reliable for predicting site accessibility.
When analyzing orthologous 3´ UTRs, conserved 7-mers in general, not just those matching miRNAs, are preferentially found in local AU-rich contexts, in predicted accessible secondary structure, away from the first 15 nt of the 3´ UTR, and away from the centers of long UTRs (Gaidatzis et al., 2007; Grimson et al., 2007; Majoros and Ohler, 2007). This result would be expected if these UTR context features found to boost miRNA effectiveness do so by enhancing site accessibility and therefore generalize to protein-binding elements. Systematic analyses of the number of sites conserved above the number expected by chance, i.e., analyses of the signal above background, rather than the signal divided by background, prevents this general effect from confounding interpretation of miRNA site conservation (Grimson et al., 2007).
The proposal of multiple lin-4 sites in the lin-14 3´ UTR (Lee et al., 1993; Wightman et al., 1993) and of sites for both lin-4 and let-7 RNAs in both the lin-14 and lin-28 UTRs (Reinhart et al., 2000), led to the notion that multiple sites for the same or different miRNAs might function cooperatively. Quantitative analyses of array data have shown that with most site configurations the increased response observed for messages with multiple sites is nearly the same as that expected if each site contributes independently to repression; that is, the response for a gene with two sites matches that anticipated by multiplying the responses from each site working on its own (Grimson et al., 2007; Nielsen et al., 2007). This multiplicative effect, a hallmark of independent and noncooperative action, was observed previously using reporter assays (Doench et al., 2003). Although not cooperative in a biochemical sense, such independent action can add up to substantial repression. For instance, a message with eight sites to co-expressed miRNAs would be repressed by ~25 fold if each site independently decreased protein output by a third [(0.67)8 = 0.04].
A notable exception to the overall tendency of independent action has been found: two sites that are close together (within 40 nt, but no closer than 8 nt) tend to act cooperatively, leading to marked enhancement in repression over that expected from the independent contributions of the two sites (Grimson et al., 2007; Saetrom et al., 2007). By analogy to transcription factors, cooperative miRNA function provides a mechanism by which repression can become more sensitive to small changes in miRNA expression levels, and it greatly enhances the regulatory effect and utility of combinatorial miRNA expression. Although not the norm, hundreds if not thousands of sites fall within a cooperative configuration with a site for the same or a co-expressed miRNA. Such cases would likely be more responsive to the miRNA, which may make them easier to identify genetically. Indeed, the miR-2:E(Spl) and miR-4:Brd interactions both involve conserved sites with intersite spacing suitable for cooperative action (Figure 2A). The same holds for the lin-4:lin-14, let-7:lin-41, let-7:hbl-1 and lsy-6:cog-1 interactions, and the lin-4 site within lin-28, which is the another classical interaction identified genetically in worms, falls within cooperative distance of a site for the co-expressed let-7 family members (Figure 2A–B).
Relative contributions of site type and context
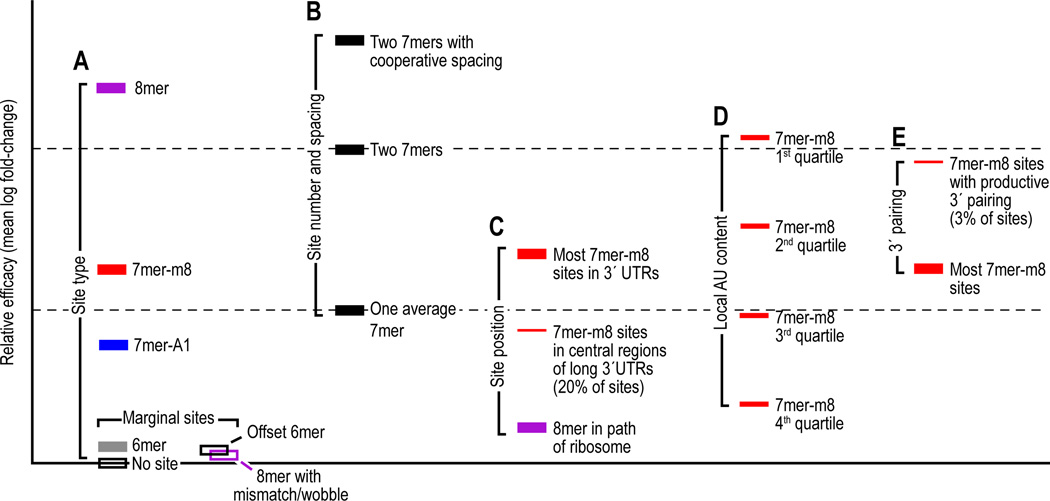
Quantitative analyses of array data provides the opportunity to compare the relative contributions of different types of 6–8-nt sites, 3´ pairing, and each of the four features of UTR context that boost site efficacy (Figure 4). The hierarchy of site efficacy is as follows: 8mer >> 7mer-m8 > 7mer-A1 >> 6mer > no site, with the 6mer differing only slightly from no site at all (Grimson et al., 2007; Nielsen et al., 2007) (Figure 4A). This hierarchy reflects average efficacies—within each site type efficacy varies widely, depending on the context of each individual site. Regarding the known context features, dual 7mers with cooperative intersite spacing tend to out-perform a single 8mer, whereas those at non-cooperative spacing do not (Figure 4B); an 8mer in the path of the ribosome tends to be less effective than a 7mer in the UTR (Figure 4A and C), and local AU content, site position, and supplemental 3´ pairing are all influential, but local AU content more commonly distinguishes sites because relatively few sites are positioned in the middle of long UTRs or proximal to residues that can provide the extra ~4 bp needed for consequential 3´ pairing (Figure 4C–E; Grimson et al., 2007).
Figure 4. Relative mean efficacy of miRNA sites.
Efficacy is plotted on a log scale as mean destabilization of messages possessing the indicated sites corresponding to an introduced miRNA, as monitored on mRNA expression arrays (Grimson et al., 2007). Site-conservation analyses, site-depletion analyses, reporter-assay results, and proteomic results all indicate that these differences measured at the mRNA level correspond to similar relative differences in protein output (Grimson et al., 2007; Baek et al., 2008). The four seed-matched sites are colored as in Figure 1. Nonconserved sites are considered together with conserved sites, and unless noted otherwise, only messages with single sites within the 3´ UTR are considered. The Y-axis is marked off in units corresponding to the mean efficacy of a single 7mer, considering both types of 7mers in aggregate (dashed lines).
(A) Mean efficacy of sites differentiated by their type. Efficacies of the four seed-matched sites (Figure 1A–D) are plotted. To illustrate the contribution of seed pairing, also plotted are efficacies of 8mer sites with a single mismatch or wobble at nucleotides 3, 4, 5, 6 or 7 (open purple bar) and of the offset 6mer match (Figure 1E, open black bar).
(B) Mean efficacies of a single 7mer and dual 7mers. Both types of 7mer sites are considered in aggregate (black filled bars).
(C) Mean efficacy of sites differentiated based on their position in the message.
(D) Mean efficacy of sites differentiated based on their local AU content.
(E) Mean efficacy of sites differentiated based on potential for 3´-supplementary pairing.
The quantitative estimates of Figure 4 were calculated from the mRNA destabilization effects of introducing a miRNA into cultured cells, leading to the possibility that these estimates might miss or underestimate the relative importance of those features that exert more of an influence on translational repression or have a greater influence in the animal than in cell culture. Helping allay these concerns is the performance of a model that quantitatively integrates these parameters into “context scores” for predicting site efficacy (Grimson et al., 2007). Proteomics data and reporter assays support the predictive value of the model for protein down-regulation, and proteomics and array data monitoring endogenous responses following miRNA knockout show that the model applies to endogenous miRNA:target interactions in fish and mice (Grimson et al., 2007; Baek et al., 2008). The model, with its context scores, is used to rank both conserved and nonconserved Targetscan predictions (Table 1). Because the model predicts site efficacy without considering site conservation, it also predicts siRNA off-targets, which appear to be repressed through the same mechanisms as endogenous miRNA targets.
Remaining recognition challenges
Although much is known about the principles of miRNA target recognition and some targets can be predicted with high confidence, much remains to be learned. A limitation of comparative sequence analysis is that some preferentially conserved sites are difficult to distinguish from those that are conserved by chance, and thus prediction sensitivity requires permissive conservation cutoffs. For example, the cutoffs used currently by TargetScan are chosen such that only about half of the conserved sites listed are thought to be preferentially conserved, some of which are indistinguishable from the others that are included as a consequence of their fortuitous conservation (Friedman et al., 2008). Probabilities of preferentially conserved targeting and context scores are both provided to guide biologists wanting to focus on the more confident predictions, but because these values imperfectly reflect the preferential conservation and efficacy of each site, using them to increase prediction specificity eliminates some biological targets without eliminating all false positives. Some high-scoring sites do not respond at all to the miRNA in reporter and proteomic experiments (Grimson et al., 2007; Beak et al., 2008), and overall, context scores explain ~60% of the variability observed in reporter assays for the same 7-nt site in different UTR contexts (A. Grimson, D.B., unpublished data). Further illustrating the room for improvement, conserved sites tend to perform better than do nonconserved sites with identical context scores, presumably because a higher fraction of the conserved sites have been under selection to fall in more favorable contexts, and current methods only partially capture features of these more favorable contexts. Thus, some determinants of targeting specificity remain to be discovered or more accurately quantified, a conclusion congruent with a recent reporter-mutagenesis experiments in worms (Didiano and Hobert, 2008).
Another challenge concerns sites that are functional despite lacking both perfect seed pairing and 3´-compensatory pairing. Many such sites have been proposed, and in a few cases the messages or their UTRs are reported to respond to changes in the levels of the miRNA. However, only very rarely has function been tested by observing the effect of mutating the site; in the other cases the responsive UTRs could be indirect targets of the miRNA, regulated downstream of the direct targets. One experimentally confirmed site, found in the human LIN28 3´ UTR, involves a mismatch at the first seed nucleotide and then perfect pairing to miRNA nucleotides 3–8 (Wu and Belasco 2005). Genomewide, such “offset 6mer” sites (Figure 1E) do have a detectable tendency to be conserved (Figure 1H), but they typically mediate very limited repression (Figure 4A, Friedman et al., 2008). Understanding what makes the LIN28 offset 6-mer site so effective could yield important new insights into miRNA targeting.
Adding to the challenge of fully understanding targeting specificity are cases in which other regulatory processes counteract miRNA-directed regulation of particular targets. For example, alternative cleavage and polyadenylation can eliminate regulatory sites from the message—a phenomenon that appears widespread in proliferating cells, in which the shorter UTRs have only half the number of conserved miRNA sites as observed in the longer isoforms that dominate in non-proliferating cells (Sandberg et al., 2008). In another example, miR-430-directed repression of nanos1 is observed in the zebrafish soma but is blocked by the binding of Dead-end RNA-binding protein in the germline, thereby allowing germline-specific expression (Mishima et al., 2006; Kedde et al., 2007). Moreover, within the same cell the miRNA-mediated repression can be modulated in response to different conditions. For example, under stress conditions miR-122-directed repression of human CAT-1 mRNA is relieved through the binding of HuR to AU-rich elements in the CAT-1 3´ UTR (Bhattacharyya et al., 2006). These examples illustrate that in addition to the general features that influence site efficacy, other mechanisms, some involving cell-type-specific UTR-binding cofactors, can influence site accessibility and subcellular message localization. Indeed, under some circumstances, in particular those that induce cells to become quiescent, miRNA targeting of UTRs is reported to enhance rather than repress translation (Vasudevan et al., 2007, 2008).
Perhaps the most important caveat of the current understanding of miRNA targeting is that virtually everything known about metazoan targeting has been learned from the minority of miRNAs that are the most highly expressed. Expression-array, protoemic, and reporter assays are all conducted with miRNAs introduced or expressed at high levels, and site-depletion analysis is informative only for the miRNAs with highly differential expression. A similar caveat applies to analyses of site conservation. The 87 mammalian miRNA families conserved throughout most vertebrates have an average of >400 preferentially conserved sites per family (Figure 1H). By contrast, the 53 families that are conserved throughout mammals but appear to have emerged well after the divergence of chicken have far fewer preferentially conserved sites; after subtracting the estimated number of sites conserved by chance, an average of only 11 sites remain per miRNA family—a number high enough to explain the conservation of these mammalian-only miRNAs but too low to contribute to our understanding of target recognition (Friedman et al., 2008). A lower number of conserved sites is expected for mammalian-only miRNAs because messages have had a relatively short time between the emergence of these miRNAs and the divergence of mammals to acquire beneficial sites, whereas for older miRNAs, they had much more time to acquire beneficial sites. Another reason for this dramatic difference might be that mammalian-only miRNAs are often expressed at lower levels and in more narrow domains than are the more broadly conserved miRNAs, which would provide fewer opportunities for evolutionary acquisition of targets. Regardless of the reasons for this difference, one ramification is that the predicted targets listed for these 53 mammalian-only families must be viewed with caution; because site conservation is barely above that expected by chance, the observation that a site is conserved provides little evidence of biological relevance.
Although found and studied largely in the context of highly expressed miRNAs, seed pairing and favorable UTR context presumably are relevant also for more modestly expressed miRNAs. However, features normally sufficient for targeting by highly expressed miRNAs might not be sufficient for targeting by more modestly expressed miRNAs. Perhaps additional determinants, such as binding of a protein cofactor or extensive 3´-supplementary pairing, would be required to concentrate a limiting amount of miRNA to a small subset of the sites that would otherwise function with higher miRNA concentration. Because many mammalian miRNAs are not highly expressed, this more restrictive (and still uncharacterized) recognition mode presumably applies to a majority of mammalian miRNAs. In sum, the state of knowledge does not sound good when expressed in terms of miRNAs: we do not understand targeting for most miRNAs. Nonetheless, it sounds better when expressed in terms of functional miRNA:target interactions: we likely understand most of miRNA targeting. This is because each of the 87 highly conserved miRNA families has so many conserved and nonconserved targets that are repressed through 7–8-nt sites, and thus the canonical, less restrictive recognition mode presumably applies to the vast majority of metazoan miRNA targeting interactions.
The biological functions of miRNAs
Since the discovery that lin-4 and let-7 play roles in the timing of C. elegans larval development, specific miRNAs have been implicated in many other biological processes. With more than a half of mammalian messages under selective pressure to maintain pairing to miRNAs (Friedman et al., 2008), it may prove difficult to find a biological function or process that is not influenced at least to some degree, in some cell type, by miRNAs.
Some miRNAs have predicted propensity to target genes with related functions, which can provide insight into biological roles of these miRNAs (Stark et al., 2003; Grun et al., 2005; Lall et al., 2006; Gaidatzis et al., 2007). For example, the vertebrate miRNAs of the miR-17~92 cluster tend to target genes involved in growth control (Lewis et al., 2005), consistent with their oncogenic properties (He et al., 2005). However, the conserved targets of particular miRNAs, even those miRNAs with very striking expression specificities, are not always statistically enriched for specific functions or processes, and even in those cases for which statistical enrichment is found, the enrichment involves only a minority of the conserved targets. Although noise in the predictions and flaws in the gene-ontology databases contribute to this low signal, it is difficult to escape the conclusion that most of the broadly conserved miRNAs each represses genes with a wide variety of biological and molecular functions.
Binary switches and much more
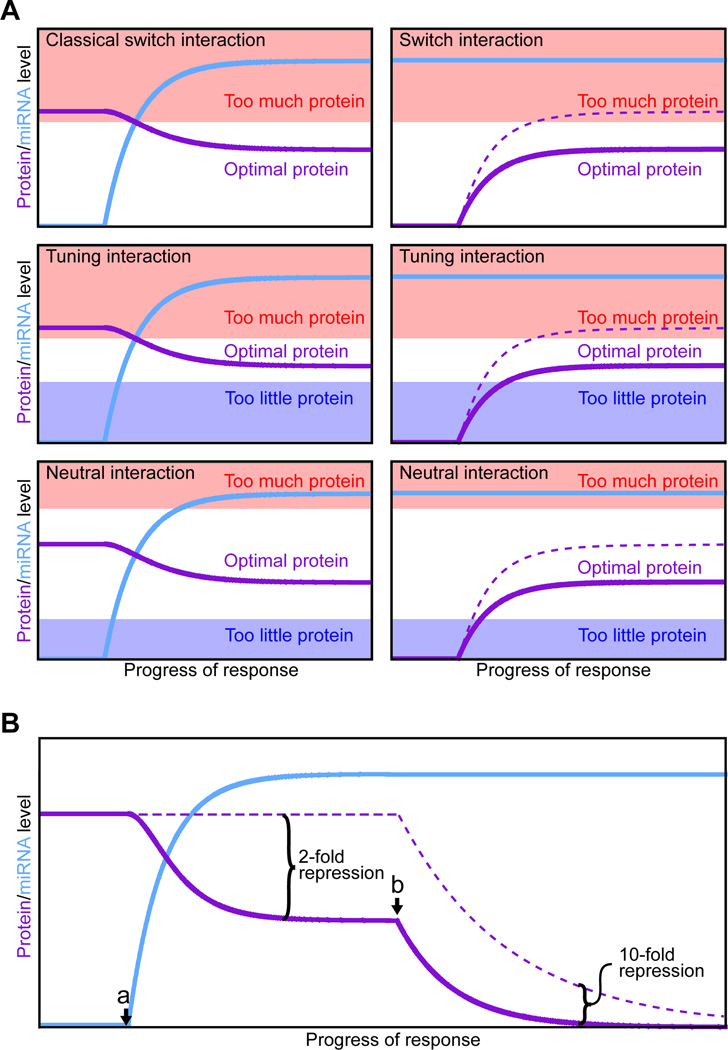
The diversity of conserved miRNA targets is rationalized in a model of miRNA function proposed as the abundance, differential expression, and targeting promiscuity of metazoan miRNAs were coming into focus (Bartel and Chen, 2004). In this model, the different expression profiles of miRNAs in different cell types constitutes a miRNA milieu, unique to each cell type, that dampens the expression of thousands of mRNAs and provides important context for the evolution of all metazoan mRNA sequences. As the UTR sequences drift over the course of evolution, they are continuously sampling matches to co-expressed miRNAs. Depending on whether the dampening of protein output is beneficial, inconsequential, or harmful, the sites are either selectively conserved, neutral, or selectively avoided during evolution (with the messages classified as conserved targets, neutral targets, and antitargets, respectively, of the miRNA). In muscle, for example, genes involved in many functions and processes might be detrimental if expressed at too high a level, and thus a wide variety of messages would be expected to accrue conserved complementary sites to muscle-specific miRNAs. For genes that should not be expressed in a particular cell type, the cell can come to depend on its miRNAs to act as binary off-switches to help to repress target protein output to inconsequential levels. Examples include the lin-4 targeting of lin-14 and lin-28, and let-7 targeting of lin-41. These classical switch interactions embodied the initial paradigm of miRNA targeting (summarized in Reinhart et al., 2000), whereby miRNA induction turns off expression of a pre-existing target. The current model expands this paradigm to include other types of switch interactions, tuning and neutral interactions, as well as instances in which messages selectively avoid miRNA targeting (described earlier in “Beyond conserved targeting”).
In contrast to the originally identified lin-4 and let-7 interactions, switch interactions can also include those in which a miRNA is already present when the target is first expressed (Figure 5A). In this scenario, the miRNA sets a more stringent threshold for consequential transcriptional activity, which can help quiet stochastic cell-to-cell noise during developmental fate decisions (Cohen et al., 2006). For example, targeting of senseless by miR-9a in fly epithelial cells prevents sporadic production of extra neuronal precursor cells (Li et al., 2006). At their extreme, such switch interactions can be regarded as failsafe interactions. For both classical switch interactions and failsafe interactions the miRNA represses protein output to inconsequential levels, but failsafe interactions differ because protein output falls below functional levels even in the absence of the miRNA. For failsafe interactions, miRNA repression adds an additional, functionally redundant layer of repression, helping ensure that aberrant transcripts do not give rise to a consequential amount of protein. Proposed examples of failsafe targeting include the miR-1 repression of non-muscle Tropomyosin isoforms and non-muscle V-ATPase subunits in the developing muscle (Stark et al., 2005).
Figure 5. MicroRNA-mediated regulatory effects.
(A) Classes of miRNA regulatory interactions (Bartel and Chen, 2004). Left panels: in response to a developmental or environmental cue, cells induce miRNA expression (blue), which in turn dampens protein production (purple) from the targeted message. Right panels: protein production from analogous targets also can be dampened by pre-existing miRNAs; accumulation in the absence of the miRNA is indicated (dashed line). All panels depict 33% repression, which illustrates that classification does not rely on the magnitude of repression but instead depends on the properties of each target, namely its threshold(s) for optimal protein output. These thresholds are depicted as the boundaries between the clear (optimal) and colored (suboptimal) regions of the graphs.
(B) Modest destabilization transitions to much greater repression after the mRNA is no longer transcribed. A miRNA is induced (point a) and mediates modest (2-fold) mRNA destabilization. After transcription of the mRNA halts (point b), the same modest destabilization quickly yields substantial (≥10-fold) repression of protein output. If the miRNA also mediates translational repression (not considered in this example), the transition to the off state is accelerated further.
Tuning interactions are those for which the miRNA acts as a rheostat rather than a binary off-switch to dampen protein output to a more optimal level but one that is still functional in the cell (Figure 5A), thereby enabling more customized expression in different cell types as well as more uniform expression within each cell type. A recent example is the Drosophila miR-8 regulation of atrophin, which reduces protein output to a level that prevents neurodegeneration but not so low as to compromise viability (Karres et al., 2007). Another likely example is the miR-375 targeting of Myotrophin (Mtpn), which dampens Mtpn output in pancreatic islets to a more optimal level but one that remains functional for insulin secretion (Poy et al., 2004). In the case of Mtnp targeting, the miRNA level remains constant in the adult animal, illustrating that the regulation need not be dynamic to be classified as tuning.
Neutral interactions dampen protein output but this repression is tolerated or offset by feedback mechanisms such that the regulatory sites are under no selective pressure to be retained or lost during the course of evolution (Figure 5A). Neutral interactions comprise cases in which biological targeting (i.e., targeting occurring in the animal) has no biological function. Because 7–8-nt sites so frequently fall in contexts suitable for repression in the animal, many ‘bystander’ messages that fortuitously pair to co-expressed miRNAs are likely subject to neutral repression. Indeed, when endogenous miRNAs are inhibited or removed, most derepressed messages have nonconserved sites (Krutzfeldt et al., 2005; Giraldez et al., 2006; Rodriguez et al., 2007; Baek et al., 2008), raising the possibility that neutral repression might be the most frequent type of biological repression. However, this possibility is difficult to confirm because tuning or switch interactions can have useful lineage-specific functions, and antitargets can have detrimental sites that have yet to be lost.
Comparing the expression of the miRNAs with that of their predicted targets can provide important clues to the more prevalent regulatory effects of metazoan miRNAs. In Drosophila, expression of miRNAs and their conserved targets usually appears ‘mutually exclusive,’ as judged by in situ hybridization (Stark et al., 2005). Microarray data from mammals paints a similar picture of a mutually exclusive tendency when considering messages with nonconserved sites (discussed above). However, the array data, which have greater dynamic range than do in situ hybridization data, suggest a different picture for messages with conserved sites, indicating that although the conserved mRNA targets tend to be expressed higher in tissues that lack the miRNA, they are still usually detected, albeit at lower levels, in tissues that express the miRNA (Farh et al., 2005; Sood et al., 2006). When miRNA-expressing cells are purified to cellular resolution, this tendency for some overlap between miRNA and target expression domains is retained, which indicates that the observed overlap is not an artifact of mixing cell types (Farh et. al., 2005; A. Shkumatava, A. Stark, H. Sive, D.B., unpublished data). The tendency of conserved targets to be present at low levels in the same tissues as the miRNA suggests that, rather than performing failsafe functions, miRNAs more frequently function to actively sculpt expression domains through a combination of tuning and classical switch targeting (Farh et al., 2005; Sood et al., 2006; A. Shkumatava, A. Stark, H. Sive, D.B., unpublished data). Still, with so many conserved targets, each highly conserved miRNA likely performs each type of regulatory function, and the proportions of classical switch, tuning, and failsafe interactions could vary widely from one miRNA to the next. Moreover, a single miRNA:target relationship could vary in different tissues or over the course of development, with, for example, active repression transitioning to failsafe repression as transcriptional output of the message declines.
The degree of target repression could also provide clues to the more prevalent functions of metazoan miRNAs. However, very little is known about the influence of miRNAs in their endogenous context on the protein output of their many of targets. Large-scale proteomic analysis has been performed for only one miRNA, miR-223, in only one biologically relevant cell type, murine neutrophils (Baek et al., 2008). This analysis revealed that although some detected proteins are repressed by 50–80%, miR-223 typically has more modest effects on its endogenous targets (even those targets that are conserved), with individual sites usually reducing protein output by less than a half and often by less than a third. Perhaps other miRNAs in their endogenous contexts have many more targets for which protein output is dramatically repressed. Even allowing for this possibility, it seems reasonable to presume that for each highly conserved miRNA, a minority of the preferentially conserved targets (much less than 150 for most miRNAs) are repressed >50% by that miRNA, whereas the hundreds of remaining preferentially conserved targets (particularly those with only 6mer sites) are repressed more modestly.
Those interactions conferring the greatest repression presumably would be enriched in switch interactions (classical or failsafe), whereas those with more modest repression would tend to be tuning interactions. Nonetheless, some targets that respond more modestly to the miRNA are likely to run counter to this tendency. For example, when target expression falls at the razor edge of efficacy, a 30% knockdown could provide switch function (Figure 5A). Alternatively, the miRNA and target can fall within a mutually repressive regulatory loop that amplifies small changes in target output to achieve switch function (Johnston et al., 2005; Li and Carthew, 2005; Yoo and Greenwald, 2005; Li et al., 2006). Another scenario in which modest miRNA-directed regulation cooperates with transcriptional regulation to achieve classical switch function occurs when a miRNA targets an mRNA that lingers after the gene has been shut off transcriptionally. In this case, depending on the threshold level for protein function, the mRNA decay rate, and the protein decay rate, modest miRNA-mediated repression can lead to substantially reduced protein at later timepoints, with a much more rapid transition to the off state (Figure 5B). Indeed this type of switch targeting applies to hundreds of maternal messages whose expression is damped by miR-430 in zebrafish [and perhaps analogous miRNAs in mammals (Farh et al., 2005)] to facilitate the transition to the zygotic gene-expression program (Giraldez et al., 2006). Nonetheless, many conserved mammalian interactions involve targets that are not strongly repressed by the miRNA, are not expressed at the razor edge of function, are not gene-regulatory molecules and thus cannot participate in amplifying regulatory loops, and are not transcriptionally shut off concurrently with miRNA repression. Most of these are unlikely to be classical switch interactions, and because modest repression would seem to impart less selective advantage for failsafe interactions than for tuning interactions, most are presumed tuning interactions.
Roles of specific miRNA:target interactions
The vast number of predicted targets, often with quite disparate functions, presents biologists with the challenge of choosing which is worthy of experimental follow-up. In some cases, known properties of a predicted target will suggest that the biological process of interest might be particularly sensitive to changes in its expression, making it especially promising for follow-up. Another way to choose targets to investigate is to assume that those messages with multiple conserved sites and particularly favorable sites might be among the most responsive to the miRNA. Examination of the predictions illustrates the utility of this approach. Among the predicted targets of C. elegans lin-4 miRNA, the one with the highest number of conserved sites is lin-14 (Figure 2A). Thus, for those interested in investigating the molecular etiology of the lin-4 phenotype, the target-prediction results suggest that lin-14 would be the top candidate for experimental follow-up, which turns out to be right on the mark (Lee et al., 1993; Wightman et al., 1993). For the other two genetically identified worm miRNAs, let-7 and lsy-6, similar correspondence is observed between targets revealed using genetic information and the top genome-wide target predictions (Figure 2A), presumably because such targets are more responsive to the miRNA than the multitude of other conserved targets and thus most easily implicated genetically. With this correspondence in mind, Hmga2, the top predicted target of the let-7 family in mammals (Figure 2A), has been investigated. As anticipated, its expression is highly responsive to changes in let-7 levels, and disrupting this regulation has phenotypic consequences in cell culture (Mayr et al., 2007). Likewise, Myb, the top predicted target of miR-150 (Figure 2A), has been investigated, and the effects of too little or too much miR-150 are largely explained by Myb repression (Xiao et al., 2007).
Once a miRNA:target interaction is chosen for study, how can the function of that specific interaction be assigned with confidence? Monitoring protein changes following miRNA knockout or knockdown is a useful starting point, but with so many targets for each miRNA, the possibility of indirect effects can be difficult to rule out. Thus, an attractive approach is to disrupt only that interaction and observe the phenotypic consequences. An expedient method to disrupt a single miRNA targeting interaction is to use antisense reagents that hybridize to the target site thereby preventing miRNA pairing. This approach reveals the importance of miR-430 regulation in balancing the expression of Nodal agonist (Squint) and antagonist (Lefty) during zebrafish mesoderm development (Choi et al., 2007). Another approach is to mutate the miRNA sites within a transgene expressing the target mRNA, ideally under the same transcriptional control as the endogenous gene. Transgene experiments performed flies even before miRNAs were known to regulate the sites can now be interpreted as evidence that miRNA regulation of Enhancer of split (E(Spl)) and Bearded (Brd) is needed for proper development of the peripheral nervous system (Lai and Posakony, 1997; Lai et al., 1998; Lai et al., 2005). In plants this approach reveals the biological importance of specific miRNA:target interactions throughout every stage of development, and because conserved miRNAs of plants fall within gene families whose members have largely redundant functions, this approach provides more information on plant miRNA functions than does disrupting miRNA loci (Jones-Rhoades et al., 2006). The transgene approach has also been fruitful in mammalian systems (Mayr et al., 2007; Teng et al., 2008).
The cleanest way to specifically disrupt a miRNA targeting interaction is to perturb pairing at an endogenous site through homologous recombination. Such an experiment shows that miR-155 repression of activation-induced cytidine deaminase (AID) production in mice helps prevent a potentially oncogenic translocation (Dorsett et al., 2008). Mapping naturally occurring variation can also uncover consequential targeting interactions. A point substitution creating a single miR-1 regulatory site in the sheep Myostatin 3' UTR dramatically impacts musculature (Clop et al., 2006), and a polymorphism that optimizes the context of a pre-existing miR-189 site in the human SLITRK1 3' UTR is associated with Tourette’s syndrome (Abelson et al., 2005).
Micromanagers of protein output
Although the biological characterization of miRNA-mediated regulatory interactions is in its infancy, the emerging picture is that the phenotypic consequences of the vast majority of conserved interactions would be very challenging to detect in the lab. Indeed, simultaneously disrupting all the interactions of a miRNA by knocking out the miRNA locus often does not have dramatic phenotypic consequences. Of the 95 C. elegans miRNA genes tested, only a few have grossly abnormal phenotypes when individually knocked out (Miska et al., 2007).
Knockouts in flies and vertebrates are likely to have more easily discernable phenotypic consequences than those in worms. Compared to worm miRNAs, far fewer conserved fly miRNAs fall into multigene families, thereby lessening opportunities for redundant functions. Moreover, tissue-specific expression patterns, which can inform more focused phenotypic analyses, are more easily determined in flies and vertebrates because these species are more amenable to dissection and in situ hybridization (Kosman et al., 2004; Wienholds et al., 2005). The lsy-6 miRNA mutant, which scores as wild-type in the assays of Miska et al. (2007) but lacks the ability to discriminate between certain chemosensory inputs, illustrates the utility of a more focused assay (Johnston and Hobert, 2003). Nonetheless, gene-knockout phenotypes in flies and vertebrates still can be subtle, or largely attributable to derepression of only a few conserved targets (Li and Carthew, 2005; Li et al., 2006; Xiao et al., 2007), thereby reinforcing the idea that individually disrupting most conserved targeting interactions would have phenotypic consequences that are difficult to detect in the lab.
At least three factors explain why the phenotypic consequences of disrupting single miRNA:target interactions are expected to be subtle. One is that >90% of the conserved miRNA:target interactions involve only a single site to the miRNA, and therefore most of these targets would be expected to be down-regulated by less than 50%. Because most messages with a conserved site to one miRNA have at least one other conserved site to an unrelated miRNA (Brennecke et al., 2005; Krek et al., 2005; Lewis et al., 2005), interactions with multiple miRNAs might need to be disrupted before the de-repression of that message had perceptible consequences.
A second factor is that miRNAs have conserved interactions with targets possessing many types of functions, and for the vast majority of these targeted genes protein output can vary by twofold without detectable consequences, as evidenced by the rarity of haplo-insufficient phenotypes. One of the more interesting reasons that such perturbations are so frequently tolerated, even for miRNA targets that are themselves gene-regulatory proteins, is the phenomenon of regulatory network buffering. Many regulatory interactions, including many miRNA:target interactions, presumably fall within complex regulatory networks with bifurcating pathways and feedback control that enable accurate response despite a defective node in the network. With this ability to buffer the effects of losing a node, such networks must be perturbed elsewhere before the lost miRNA interaction has discernable phenotypic consequences. Reciprocally, perturbing the miRNA node would be expected to sensitize the network to reveal the importance of other regulatory nodes. Such experiments are beginning to reveal important miRNA functions that otherwise would be missed (Li and Carthew, 2005).
A third reason for the subtle phenotypes is that lab conditions have been optimized to preserve and propagate mutant lines, whereas conditions that better simulate the stresses and competition that have shaped the evolution of each species would uncover many more instances in which disrupting miRNA regulation of a target has discernable phenotypic consequences. Indeed, to the extent that a miRNA-target interaction has been under selective pressure to be preserved, it must be a biologically meaningful regulatory relationship with phenotypic consequences, albeit potentially mild ones. By this criterion, comparative sequence analyses, which measures the results not of a laboratory experiment but of The Big Experiment (otherwise known as evolution), indicates a vast scope of consequential regulatory targeting. However, because a very subtle fitness disadvantage can prevent alleles with mutant sites from being the ones that become fixed in a population, purifying selection is exquisitely sensitive in retaining consequential interactions. A challenge of the next decade will be to design the laboratory experiments with the sensitivity needed to uncover the functional roles of most of these biological interactions.
When combining the observation that most mammalian messages are under selective pressure to maintain sites to miRNAs together with the observation that each conserved targeting interaction typically imparts only a modest reduction in protein output, it is difficult to escape the conclusion that the precise levels of most individual proteins impact animal fitness. That these levels should be so precise for so many proteins, with the tight tolerances so often retained through evolution, is one of the more fascinating biological conclusions arising from miRNA research of the past few years.
The emerging picture of miRNA regulation in animals is far richer and more complex than the crisp linear pathways of the previous decade, with miRNAs participating in executive decisions but also performing much of the grunt work to micromanage protein output. However, the situation is not so complicated so as to make the reductionistic approach futile. Initial experiments to characterize individual regulatory interactions in the lab will still reveal much by focusing on those with the more easily scored consequences. With help of target-predictions, expression data, and biological knowledge, candidates for such interactions can now be found by focusing on those messages that are predicted to be most responsive to the miRNA, coexpressed with the miRNA in relevant cells, and at interesting and vulnerable nodes in regulatory networks. With so many biologists now cognizant of miRNAs and how they recognize their targets, the stage is set for rapid progress in learning the functions of these more accessible miRNA:target interactions.
Acknowledgements
I thank D. Baek, C. Burge, K. Farh, R. Friedman, A Grimson, B. Lewis, L. Lim and other colleagues for their input and stimulating discussions, and members of the lab for helpful comments on this manuscript. Work on miRNAs in my lab is supported by grants from the NIH.
References
- Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V. The let-7 microRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans . Dev Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abelson JF, Kwan KY, O'Roak BJ, Baek DY, Stillman AA, Morgan TM, Mathews CA, Pauls DL, Rasin MR, Gunel M, et al. Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4:625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans . Curr Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 2007;4:76–84. doi: 10.4161/rna.4.2.4640. [DOI] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibe B, Bouix J, Caiment F, Elsen JM, Eychenne F, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38:813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- Cohen SM, Brennecke J, Stark A. Denoising feedback loops by thresholding--a new role for microRNAs. Genes Dev. 2006;20:2769–2772. doi: 10.1101/gad.1484606. [DOI] [PubMed] [Google Scholar]
- Davis E, Caiment F, Tordoir X, Cavaille J, Ferguson-Smith A, Cockett N, Georges M, Charlier C. RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr Biol. 2005;15:743–749. doi: 10.1016/j.cub.2005.02.060. [DOI] [PubMed] [Google Scholar]
- Didiano D, Hobert O. Molecular architecture of a miRNA-regulated 3' UTR. RNA. 2008;14:1297–1317. doi: 10.1261/rna.1082708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett Y, McBride KM, Jankovic M, Gazumyan A, Thai TH, Robbiani DF, Di Virgilio M, San-Martin BR, Heidkamp G, Schwickert TA, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easow G, Teleman AA, Cohen SM. Isolation of microRNA targets by miRNP immunopurification. RNA. 2007;13:1198–1204. doi: 10.1261/rna.563707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila . Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2008;27:2008. doi: 10.1101/gr.082701.108. published online October. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidatzis D, van Nimwegen E, Hausser J, Zavolan M. Inference of miRNA targets using evolutionary conservation and pathway analysis. BMC Bioinformatics. 2007;8:69. doi: 10.1186/1471-2105-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, Dongen SV, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Grun D, Wang YL, Langenberger D, Gunsalus KC, Rajewsky N. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput Biol. 2005;1:e13. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nat Struct Mol Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- Hammell M, Long D, Zhang L, Lee A, Carmack CS, Han M, Ding Y, Ambros V. mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nat Methods. 2008;9:813–819. doi: 10.1038/nmeth.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans . Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Jr, Chang S, Etchberger JF, Ortiz CO, Hobert O. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc Natl Acad Sci USA. 2005;102:12449–12454. doi: 10.1073/pnas.0505530102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, Hannon GJ. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci USA. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karres JS, Hilgers V, Carrera I, Treisman J, Cohen SM. The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila . Cell. 2007;131:136–145. doi: 10.1016/j.cell.2007.09.020. [DOI] [PubMed] [Google Scholar]
- Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Orom UA, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Kheradpour P, Stark A, Roy S, Kellis M. Reliable prediction of regulator targets using 12 Drosophila genomes. Genome Res. 2007;17:1919–1931. doi: 10.1101/gr.7090407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, Hatzigeorgiou A. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Wienholds E, Ketting RF, Plasterk RH. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res. 2004;32:6284–6291. doi: 10.1093/nar/gkh968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman D, Mizutani CM, Lemons D, Cox WG, McGinnis W, Bier E. Multiplex detection of RNA expression in Drosophila embryos. Science. 2004;305:846. doi: 10.1126/science.1099247. [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lai EC. Micro RNAs are complementary to 3' UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- Lai EC, Burks C, Posakony JW. The K box, a conserved 3' UTR sequence motif, negatively regulates accumulation of Enhancer of split Complex transcripts. Development. 1998;125:4077–4088. doi: 10.1242/dev.125.20.4077. [DOI] [PubMed] [Google Scholar]
- Lai EC, Posakony JW. The Bearded box, a novel 3' UTR sequence motif, mediates negative post-transcriptional regulation of Bearded and Enhancer of split Complex gene expression. Development. 1997;124:4847–4856. doi: 10.1242/dev.124.23.4847. [DOI] [PubMed] [Google Scholar]
- Lai EC, Tam B, Rubin GM. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 2005;19:1067–1080. doi: 10.1101/gad.1291905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lall S, Grun D, Krek A, Chen K, Wang YL, Dewey CN, Sood P, Colombo T, Bray N, Macmenamin P, et al. A genome-wide map of conserved microRNA targets in C. elegans . Curr Biol. 2006;16:460–471. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans . Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans . Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14 . Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Li X, Carthew RW. A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell. 2005;123:1267–1277. doi: 10.1016/j.cell.2005.10.040. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila . Genes Dev. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The microRNAs of Caenorhabditis elegans . Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long D, Lee R, Williams P, Chan CY, Ambros V, Ding Y. Potent effect of target structure on microRNA function. Nat Struct Mol Biol. 2007;14:287–294. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5' UTR as in the 3' UTR. Proc Natl Acad Sci USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5'-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majoros WH, Ohler U. Spatial preferences of microRNA targets in 3' untranslated regions. BMC Genomics. 2007;8:152. doi: 10.1186/1471-2164-8-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5' region. EMBO J. 2004;23:3356–3364. doi: 10.1038/sj.emboj.7600340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Mishima Y, Giraldez AJ, Takeda Y, Fujiwara T, Sakamoto H, Schier AF, Inoue K. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr Biol. 2006;16:2135–2142. doi: 10.1016/j.cub.2006.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM, Bartel DP, Ambros VR, Horvitz HR. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen CB, Shomron N, Sandberg R, Hornstein E, Kitzman J, Burge CB. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA. 2007;13:1894–1910. doi: 10.1261/rna.768207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Bartel DP. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297:1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans . Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Robins H, Li Y, Padgett RW. Incorporating structure to predict microRNA targets. Proc Natl Acad Sci USA. 2005;102:4006–4009. doi: 10.1073/pnas.0500775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U–RNAs and additional microRNAs and endogenous siRNAs in C. elegans . Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saetrom P, Heale BS, Snove O, Jr, Aagaard L, Alluin J, Rossi JJ. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 2007;35:2333–2342. doi: 10.1093/nar/gkm133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci USA. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal microRNAs confer robustness to gene expression and have a significant impact on 3'UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila microRNA targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Lin MF, Kheradpour P, Pedersen JS, Parts L, Carlson JW, Crosby MA, Rasmussen MD, Roy S, Deoras AN, et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng G, Hakimpour P, Landgraf P, Rice A, Tuschl T, Casellas R, Papavasiliou FN. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Cell-cycle control of microRNA-mediated translation regulation. Cell Cycle. 2008;7:1545–1549. doi: 10.4161/cc.7.11.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3'UTR. Genes Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wightman B, Burglin TR, Gatto J, Arasu P, Ruvkun G. Negative regulatory sequences in the lin-14 3'-untranslated region are necessary to generate a temporal switch during Caenorhabditis elegans development. Genes Dev. 1991;5:1813–1824. doi: 10.1101/gad.5.10.1813. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans . Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- Yoo AS, Greenwald I. LIN-12/Notch activation leads to microRNA-mediated down-regulation of Vav in C. elegans . Science. 2005;310:1330–1333. doi: 10.1126/science.1119481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]