Abstract
The phylogenetic relationships among species of Triatoma matogrossensis subcomplex ( T. baratai, T. guazu, T. matogrossensis, T. sordida, T. vandae, and T. williami) was addressed by using fragments of cytochrome oxidase I (COI), 16S rDNA (16S), and cytochrome b (Cytb) through Bayesian and parsimony analyses. We did not recover a monophyletic T. matogrossensis subcomplex, and their members were found clustered in three strongly supported clades, as follows: i) T. jurbergi + T. matogrossensis + T. vandae + T. garciabesi + T. sordida; ii) with T. guasayana as the sister group of clade (i); and iii) T. williami + T. guazu, however not closely related to the clade formed by the previously mentioned species. The other two endemic species from Central-Western Brazil, T. baratai and T. costalimai, were not recovered with strong clade support as related to other members of this subcomplex. Results call for a further revision in the classification of the subcomplexes within the genus Triatoma.
Introduction
Carlos Chagas 1 described Trypanosoma cruzi, the causative agent of American trypanosomiasis, in 1909 and noted that this protozoan was transmitted by triatomines. After the first reported human cases of the disease, studies on the vector species reservoirs and description of new triatominae species started to increase. Galvão and others 2 included 19 genera and 137 species into the subfamily Triatominae, but currently it consists of 145 valid species belonging to 18 genera, of which 65 species occur in Brazil. 3– 11
Some Triatoma species are grouped into complexes, and subcomplexes based on morphological similarities, geographical distribution, epidemiological importance, phylogenetic relationships, and others. At the moment, there is no consensus about the features that define complexes. 11– 14 Thirteen Triatoma species have been found in Central-Western Brazil, but only seven are considered endemic: T. baratai, T. costalimai, T. deanorum, T. jurbergi, T. matogrossensis, T. vandae, and T. williami. 2, 13, 15, 16 Based on morphological similarities, Carcavallo and others, 17 grouped these species into the T. oliveirai complex together with T. klugi and T. oliveirai, which are found in Southern Brazil. We must stress that T. oliveirai, T. baratai, and T. deanorum are rare species, difficult to collect 18; and for this reason studies about them are scarce on all grounds. The epidemiological importance related to Chagas disease vectors is continually changing. Triatoma sherlocki, for example, was described in 2002 as a sylvatic species, 19 and was recently found invading and colonizing human domiciles in Bahia State 20, 21; it highlights the importance of understanding the phylogenetic relationships of sylvatic species. Several other species can be mentioned in this context. 14
The latest classification scheme, proposed by Schofield and Galvão, 13 summarizing results from different analyses, placed members previously considered belonging to the T. oliveirai complex into two subcomplexes (within the T. infestans complex): i) T. baratai, T. costalimai, T. deaneorum, T. guazu, T. jurbergi, T. matogrossensis, T. vandae, and T. williami, forming the T. matogrossensis subcomplex; and ii) T. oliveirai and T. klugi, included in the T. rubrovaria subcomplex, along with four other species from the Southern region of Brazil: T. carcavalloi, T. circummaculata, T. limai, and T. rubrovaria. Of additional interest to this work, Triatoma sordida was placed in the T. sordida subcomplex, which included T. garciabesi, T. guasayana, and T. patagonica.
Triatominae species that overlap in geographic distribution are sometimes subjected to the same ecological pressures, and it remains a question whether the evident morphological similarities among them are a result of ecological convergences or of their phylogenetic relationships. On the other hand, distinct ecological forces might also promote morphological differentiation among sympatric phylogenetically related species. 22 In sylvatic species, for which samples are difficult to be obtained, species complex definition has been chiefly based on morphological and ecological features; and for this reason, cytochrome b (Cytb), cytochrome oxidase I (COI), and 16S rDNA (16S) sequences of mitochondrial DNA were used to evaluate the phylogenetic relationships among seven Triatoma species occurring in Central-Western Brazil.
Material and Methods
Multiple specimens of seven sylvatic species of Triatoma were used for gene sequencing: T. baratai ( N = 4), T. costalimai ( N = 6), T. guazu ( N = 6), T. matogrossensis ( N = 5), T. vandae ( N = 4), T. williami ( N = 6), and T. sordida ( N = 5) ( Table 1, Figure 1 ). Except for T. sordida, all of the previously mentioned species belong to the T. matogrossensis subcomplex. However, T. sordida was included in the analysis because of its epidemiological importance and because some studies have shown this species as related to T. matogrossensis. 12, 23, 24 According to Forattini 25 and Galvão and others, 2 Central-Western Brazil is the center of dispersion of these seven species. The specimens studied were randomly obtained from colonies maintained at the Triatominae Insectarium of the Department of Biological Sciences, School of Pharmaceutical Sciences, São Paulo State University (Araraquara, Brazil). Because sequences of the mitochondrial genes studied herein of T. vandae and T. baratai were obtained for the first time, representative species of other groups of Latin America triatomines, such as the T. brasiliensis species complex and T. rubrovaria subcomplex, were also included for comparison.
Table 1.
Colony number (CTA - from the Triatominae Insectarium, São Paulo State University), origin and start date of colonies studied
| Species | Colony number | Initiated | Origin | Coordinates |
|---|---|---|---|---|
| Triatoma baratai | CTA 247 | 08/2007 | Nioaque, Mato Grosso do Sul (MS) | 55.83°W 21.13°S |
| Triatoma costalimai | CTA 191 | 03/1996 | Mambaí, Goiás (GO) | 46.11°W 14.48°S |
| Triatoma guazu | CTA 232 | 07/2001 | Barra do Garça, Mato Grosso (MT) | 52.25°W 15.89°S |
| Triatoma matogrossensis | CTA 248 | 05/2007 | Rio Verde de Mato Grosso, Mato Grosso do Sul (MS) | 54.84°W 18.91°S |
| Triatoma sordida | CTA 028 | 08/1982 | Brasilândia, Mato Grosso do Sul (MS) | 52.03°W 21.25°S |
| Triatoma vandae | CTA 231 | 07/2001 | Itiquira, Mato Grosso (MT) | 54.15°W 17.20°S |
| Triatoma williami | CTA 184 | 12/1995 | Barra do Garça, Mato Grosso (MT) | 52.25°W 15.89°S |
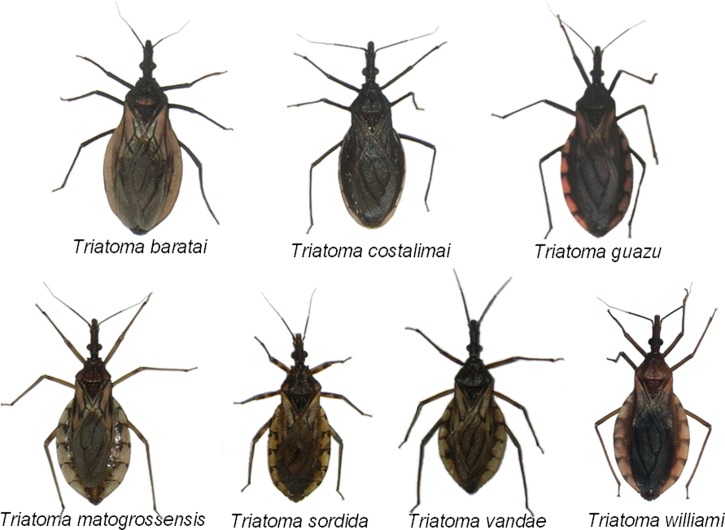
Figure 1.
Dorsal view of specimens from the colonies studied: T. baratai, T. costalimai, T. guazu, T. matogrossensis, T. sordid, T. vandae, and T. williami. Pictures of Sueli Gardim.
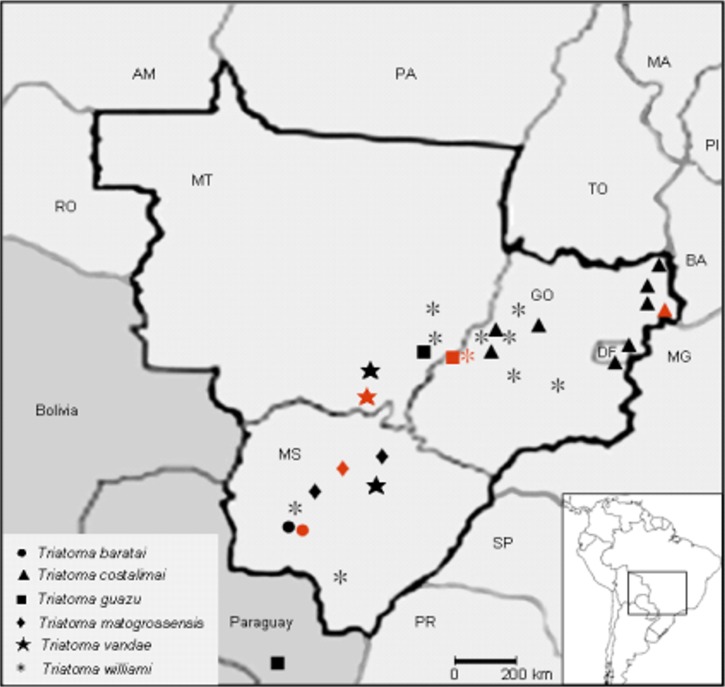
The origin of the colonies of T. matogrossensis subcomplex maintained at the insectarium and their published distributional records were used in the construction of a map of the known geographic distribution ( Figure 2). 2, 12, 13, 16– 18, 23– 32 In Brazil, T. sordida overlaps the geographic distribution of all species herein studied (also occurring in the states of Bahia, Goiás, Mato Grosso, Mato Grosso do Sul, Maranhão, Minas Gerais, Paraná, Pernambuco, Piauí, Rio Grande do Sul, Santa Catarina, and São Paulo). This species is also found in the Chaco eco-region of other countries: Argentina, Bolivia, Paraguay, and Uruguay. 2, 25, 33 Regarding the members of T. matogrossensis subcomplex, only T. deaneorum has not been included in this phylogenetic study because of its rarity, which has been also addressed by others authors. 18.
Figure 2.
The known geographic distribution of species of the T. matogrossensis subcomplex in the Central-Western Region of Brazil. 2, 13– 16, 22– 28 In red spots where the specimens herein studied originated. For details, see Table 1. The geographic distribution of T. sordida overlaps all spots for the members of T. matogrossensis subcomplex, and the Chaco eco-region of Argentina, Bolivia, Paraguay, and Uruguay. 2, 25, 33
Genomic DNA extraction was performed according to the protocol described by Sambrook and Russell. 34 From the extracted DNA, 16S and COI fragments were amplified as described by Sainz and others, 23 and for Cytb as by Lyman and others, 35 and purified using the Illustra GFX PCR DNA and Gel Band Purification Kit (GE Healthcare Life Sciences, Piscataway, NJ). Purified products were subjected to a sequencing reaction using BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) and were analyzed in the ABI PRISM 377 DNA Sequencer (Applied Biosystems). Additional sequences of Cytb and 16S and COI deposited at GenBank were added to the analysis. Rhodnius prolixus was used as an outgroup ( Table 2). 12, 20, 23, 24, 35– 43
Table 2.
Accession codes from GeneBank sequences of Triatoma and outgroup species used in this study
| Subcomplex | Species | 16S | Cytb | COI |
|---|---|---|---|---|
| T. brasiliensis | T. brasiliensis | EU827222 | AY336524 | AF021186 |
| T. juazeirensis (haplotype F) | – | AY494168 | – | |
| T. melanica (haplotype H) | – | AY336527 | – | |
| T. sherlocki | EU489057 | EU489058 | KC608987 | |
| T. infestans | T. infestans | EU143699 | AY702023 | AF021199 |
| T. maculata | T. maculata | EU827231 | KC608977 | AF449139 |
| T. pseudomaculata | EU827225 | KC608979 | KC608986 | |
| T. matogrossensis | T. baratai | KC571991 | KC608974 | – |
| T. costalimai | KC571993 | KC608975 | KC608983 | |
| T. guazu | KC571994 | KC608976 | KC608984 | |
| T. jurbergi | AY035456 | – | – | |
| T. matogrossensis | KC571995 | KC608978 | KC608985 | |
| T. vandae | KC571997 | – | KC608989 | |
| T. williami | KC571998 | KC608981 | KC608990 | |
| T. rubrovaria | T. carcavalloi | KC571992 | – | KC608982 |
| T. circummaculata | AF021188 | – | AF021191 | |
| T. klugi | AY035463 | * | – | |
| T. rubrovaria | AF021203 | GQ398003* | AF021204 | |
| T. sordida | T. garciabesi | AY185835 | – | – |
| T. guasayana | AF021194 | * | AF021193 | |
| T. patogonica | AY035464 | – | – | |
| T. sordida | KC571996 | KC608980 | KC608988 | |
| outgroup | P. megistus | AF045701 | AF045722 | AF021179 |
| R. prolixus | AF045707 | EF011726 | AF449138 |
Sequences were edited with BioEdit 7.0.5 and aligned with ClustalW. 44 Nucleotide data for Cytb and COI were transformed into amino acid sequences to check the alignment. Separate and combined phylogenetic analyses of 16S, Cytb, and COI sequences were run under a Bayesian framework in MrBayes 3.1 (2 independent runs, 4 chains, and 1 M gens) 45 and the maximum parsimony criterion in PAUP ( hsearch, 1,000 random addition replicates with TBR branch swap, gaps treated as “?”). 46 The following evolutionary models were chosen for the three partitions in the mixed-model Bayesian analysis using the Akaike information criterion in MrModeltest 47, 48: HKY+I+G for 16S rDNA; HKY+I+G for Cytb; for COI was used GTR+I+G. Clade support was estimated by Bayesian posterior probabilities (BPP) and 1,000 pseudoreplicates of non-parametric parsimony bootstrap (PB).
Results
Twenty-five new sequences were obtained (8 for 16S, 8 for Cytb, and 9 for COI; Table 2). These sequences were aligned with other sequences available at GenBank and cropped according to the shorter sequences. The alignment constructed for the phylogenetic analysis included 256 basepairs (bp) of 16S (83 variable sites, including 70 parsimony informative), 313 bp of Cytb (109 variable sites, including 84 parsimony informative), and 202 bp of COI (75 variable sites, including 58 parsimony informative), totalizing 771 bp evaluated.
For the Triatoma species a single haplotype for each gene was found, except for two Cytb haplotypes for T. matogrossensis and T. baratai, and three haplotypes of Cytb for T. guazu. In cases of two haplotypes, p-distances (all < 0.6%) were within the expected for intraspecific variation and after pilot phylogenic reconstructions, they were always clustered together. Therefore, a single haplotype was chosen as representative. Two of the sequenced Cytb haplotypes of T. guazu showed stop codons, and most probably represent pseudogenes, therefore were excluded from the analysis (see discussion below). Triatoma costalimai showed a large deletion of position 182 to 220 of Cytb sequence referring to a string of 13 amino acids. If this fragment is the homologous mitochondrial copy, apparently this deletion is not deleterious because it was confirmed in six adult specimens. However, because of the absence of stop codons, this sequence was maintained in the analysis, although there is a possibility it might be a pseudogene like as in T. guazu.
Interestingly, finding intraspecific variation in only these three species is consistent with the date of the start of the colony. These younger colonies have been maintained from 5 to 11 years, whereas most other species have been maintained from 15 to 30 years and are possibly highly inbred. Triatoma baratai and T. vandae did not have any sequences of 16S deposited in GenBank, and also T. baratai, T. costalimai, T. guazu, T. matogrossensis, T. vandae, and T. williami of Cytb.
Interspecific pairwise divergence (uncorrected p-distances) within Triatoma herein focused varied in 16S sequences from 0.4% (between T. pseudomaculata– T. maculata) to 8.7% ( T. garciabesi– T. brasiliensis and T. sordida–T. infestans), in Cytb sequences from 0.7% ( T. pseudomaculata– T. maculata) to 21.3% ( T. sordida– T. sherlocki), and in COI sequences from 1.5% ( T. williami– T. guazu) to 22.9% ( T. sordida–T. infestans) ( Supplemental files).
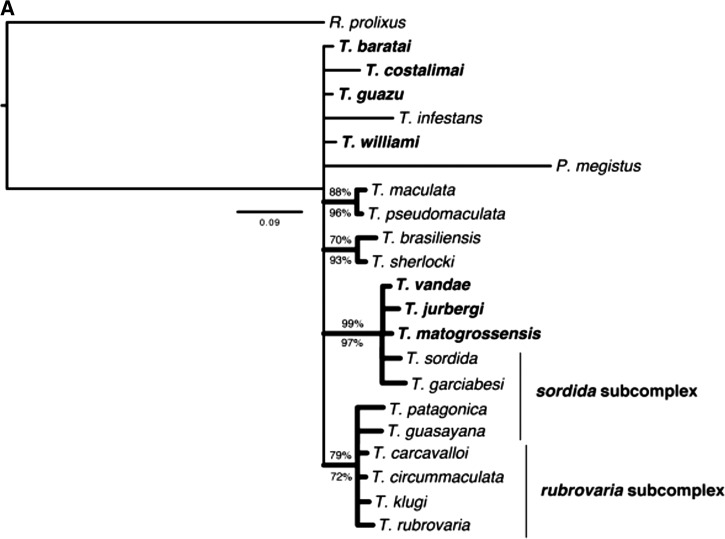
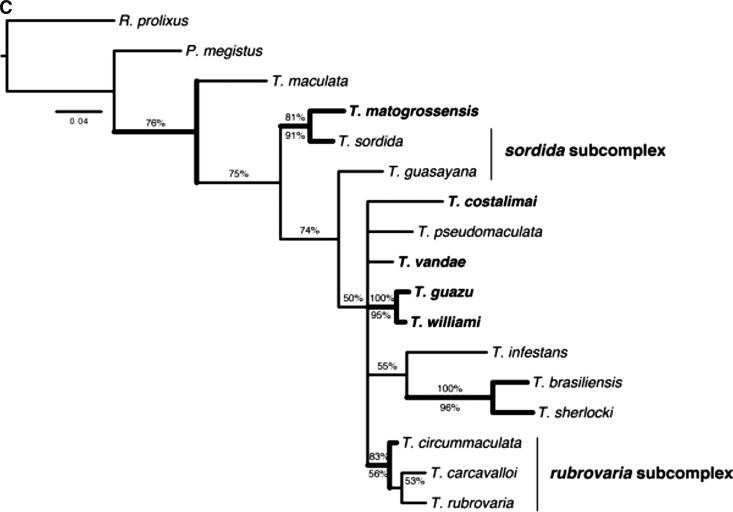
Figure 3 shows the contribution of each gene for the phylogenetic reconstruction of members of T. matogrossensis subcomplex. In the analysis of 16S ( Figure 3A), a clade including T. jurbergi, T. vandae, T. sordida, T. garciabesi, and T. matogrossensis was recovered with high support (PB = 97% and BPP = 99%). However, for this gene, T. guasayana was not included in this group, being placed in T. rubrovaria subcomplex. The analysis of COI ( Figure 3C) gene by itself recovered T. sordida and T. matogrossensis as closely related (PB = 91% and BPP = 81%), a relationship also recovered by the Cytb analysis ( Figure 3B), but in this case, T. sordida is found to be sister to T. guasayana + T. matogrossensis (PB = 98% and BPP = 100%). Except for the analyses based on the 16S gene, all others recovered T. williami and T. guazu as sister species with high clade support (All PB ≥ 95% and BPP ≥ 99%; Figures 3B and C and Figure 4 ).
Figure 3.
Bayesian inference consensus for each molecular marker of Triatoma species: ( A) 16S (HKY+I+G), ( B) Cytb (HKY+I+G), and ( C) COI (GTR+I+G). Thick clades represent those also recovered by parsimony. Percentages above the nodes indicate Bayesian posterior probabilities (BPP), whereas those below indicate parsimony bootstrap percentages (PB). Taxa in bold are current members of the T. matogrossensis subcomplex.
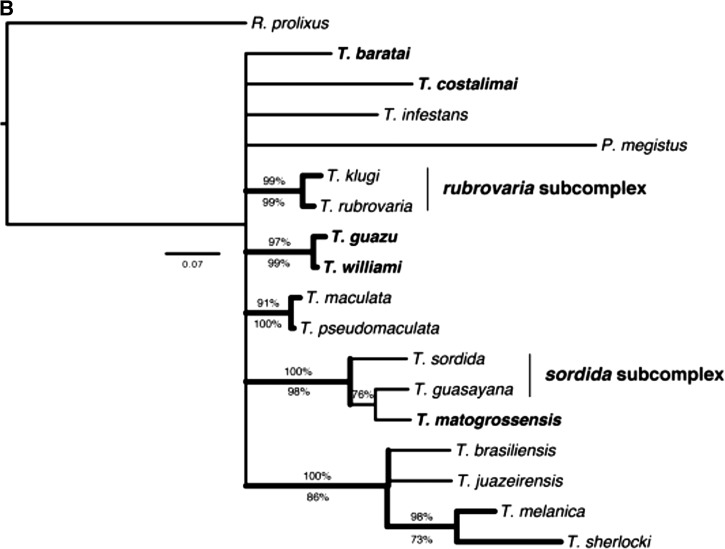
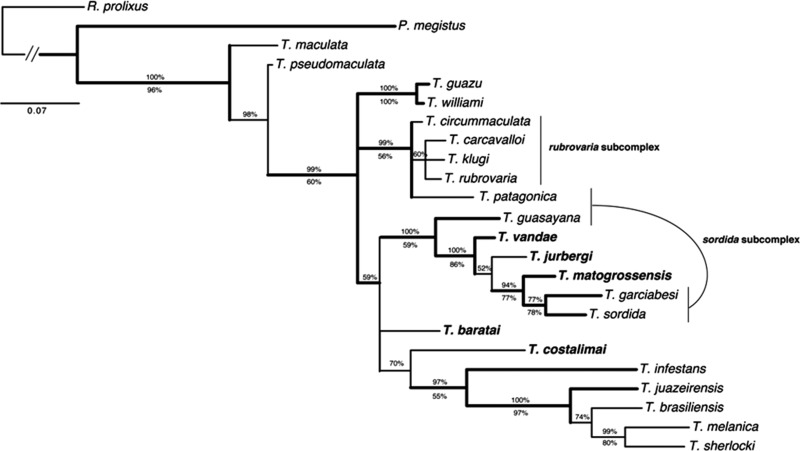
Figure 4.
Bayesian inference consensus of the combined analysis of sequences of Triatoma species occurring in Central-Western Brazil. Molecular evolution models for each partition were HKY+I+G for 16S, GTR+I+G for COI, and HKY+I+G for Cytb. Thick clades represent those also recovered by parsimony. Percentages above the nodes indicate Bayesian posterior probabilities (BPP), whereas those below indicate parsimony bootstrap percentages (PB). Taxa in bold are current members of the T. matogrossensis subcomplex.
Both Bayesian and parsimony analyses resulted in a similar topology ( Figure 4) for the analyses of the combined genes, with a markedly higher branch support for certain clades, as compared with separate analyses ( Figure 3). Focusing on the Central-Western species, three strongly supported clades were recovered as follows: i) one including the species T. jurbergi, T. matogrossensis, T. vandae, T. garciabesi, and T. sordida (PB = 83% and BPP = 100%); ii) one with T. guasayana as sister to this previously mentioned clade (PB = 59% and BPP = 100%); and iii) another with T. williami + T. guazu (PB = 99% and BPP = 100%). Based on the results of the combined analysis presented herein, it is not possible to say whether clade (iii) is related to the other previously mentioned Central-Western species. However, T. williami and T. guazu were recovered (without strong support) as related to each other and, also to the remaining Central-Western species, T. baratai and T. costalimai, in both 16S and Cytb separate analyses.
According to our results, the T. matogrossensis subcomplex as currently defined by Schofield and Galvão 13 was not found to be a monophyletic unit, because other species, currently placed in the T. sordida subcomplex, including one of the main Chagas disease vectors in Brazil, T. sordida, were recovered as strongly related to some species from this group, especially T. matogrossensis, T. jurbergi, and T. vandae.
Discussion
Traditionally, the phylogeny of newly described or strictly sylvatic triatomine species has been inferred based on morphological comparisons. 16, 49, 50 In this study, the phylogenetic relationships among Triatoma species ( T. baratai, T. costalimai, T. guazu, T. matogrossensis, T. sordida, T. vandae, and T. williami) occurring in Central-Western Brazil were analyzed using molecular information (mtDNA fragments of 16S ribosomal RNA, Cytb, and COI) through Bayesian and parsimony analysis. Both analyses provided satisfactory information for species, which had never been studied, addressing the phylogenetic reconstruction of these groups of triatomines found in Central-Western Brazil. Overall, not a single gene was particularly great in recovering these relationships, showing low clade support in most clades, however the combined analysis showed higher clade support for the clades of interest.
The use of insects from colonies for several kinds of approaches remains controversial because of genetic drift and loss of genetic variability 51; an interesting observation of this study is the apparent homogeneity of mtDNA from bugs maintained in colonies for several generations. Garcia and Powell 36 sequenced 16S (AF021212) and COI (AF021213) from T. sordida from the same colony used herein when it was 14 years old. In this study, 16 years later, the same haplotypes obtained by these authors were also found for both genes. Considering that no selective pressure appears to be affecting and driving mtDNA variation, we concluded that mtDNA is a suitable source of information for exploring phylogenetic relationships, even when obtained from individuals maintained in laboratory conditions over decades.
Sequences of 16S were overall more informative for the combined analysis between species, whereas Cytb and COI were possibly more useful for recovering relationships among more closely related species, because of its faster rate of evolution (in average 3.3 times faster), which can be observed by comparing the pairwise divergences.
Aside from the poor resolution obtained by the combined analyses, at least for separate analyses of the molecular markers, this study does not recover a strict monophyletic group for the seven aforementioned species placed in the T. matogrossensis subcomplex, mostly because members of the T. sordida subcomplex, such as T. guasayana, T. garciabesi, and T. sordida, tend to be recovered as related to them. This proximity found among members of the T. sordida subcomplex to members of the T. matogrossensis subcomplex is in agreement with previously published analyses, although this was previously recovered only with 16S and 12S sequences. 12, 13, 23, 24 With the herein newly added Cytb and COI sequences of species belonging to the T. matogrossensis subcomplex, the need for a future revision for both T. matogrossensis and T. sordida subcomplexes to better define the limits of the current classification is reinforced.
The placement of T. guasayana in our analyses disagrees in part with previous published analyses. In none of the analyses conducted herein, T. guasayana was recovered as sister to T. sordida, which was expected because of isoenzyme electrophoresis genetic distances, morphological and ecological similarities, and overlapping distribution in northern Argentina and part of the Bolivian Chaco and Paraguay. 52 Furthermore, it also disagrees with Sainz and others, 23 Garcia and others, 24 and Almeida and others, 53 which reconstructed the phylogeny of selected Triatoma species based on fragments of the mtDNA genes: 12S +16S + COI and placed this species as more related to members of the T. rubrovaria subcomplex. In fact, by using only 16S, T. guasayana was clustered with members of the T. rubrovaria subcomplex; however, by using the COI and Cytb, T. guasayana (both obtained in this study) was clustered with members of T. sordida and T. matogrossensis subcomplexes ( Figure 3).
The position of T. costalimai was also incongruent in each of the separate analyses, but usually it was recovered as related to a member of the T. matogrossensis subcomplex. This is not a surprising result considering that Hypsa and others 12 based on a parsimony analysis of 16S sequences and Sainz and others 23 based on a parsimony analysis of 16S + 12S, found this species as the sister species to all other Central-Western Brazil species. A maximum likelihood reanalysis of Hypsa and others 12 data set by Schofield and Galvão, 13 however placed this species at an even more distant position, relating it to a clade composed of members of some other subcomplexes like T. brasiliensis, T. maculata, and T. rubrovaria, in addition to T. matogrossensis and T. sordida.
The other two recovered clades of Triatoma from Central-Western Brazil actually corroborated morphometric and isoenzymatic phylogenetic analyses by Noireau and others 16 focusing on the called “ T. oliveirai complex” (actually T. matogrossensis subcomplex, according to Carcavallo and others 54 and Schofield and Galvão 13). Both distance analyses of morphological measurements and 20 isoenzyme loci support a close relationship between T. vandae and T. jurbergi as a sister clade of T. matogrossensis, whereas T. guazu and T. williami with a significant distance from T. klugi. These relationships are in complete agreement with the present analysis. In fact, T. guazu and T. williami showed only 1.2% sequence divergence of 16S, representing the lowest found herein, concerning the sylvatic species from Central-Western Brazil. The close phylogenetic relationship between these two species had already been addressed by Noireau and others 16 and De Paula and others, 55 and was confirmed herein with high node support for the Bayesian analysis (BPP = 100). Low values of p-distances associated with an overlapping geographic distribution have also been found by Almeida and others 53 for species occurring in Southern Brazil, which might be a result of recent speciation events.
Triatoma sordida is currently one of the most important Chagas disease vectors in some states of Brazil (e.g., Minas Gerais). 56, 57 According to Forattini, 25 its dispersion center was the Central-Western Region of Brazil and it stands out from the others species because it is a predominant species in Mato Grosso do Sul, Goiás and other states in Brazil and it forms large colonies in human dwellings, especially in the peridomicile. 58, 59 This distribution was confirmed by Gurgel-Gonçalves and others. 33 The epidemiologic scenario related to new sylvatic vectors that have been invading and colonizing homes are continually increasing. 14, 53, 60 Therefore, the close relationship between T. sordida, T. jurbergi, T. matogrossensis, T. vandae, and T. garciabesi calls the attention for the epidemiologic potential of these species; as such T. guasayana, has already been found invading and colonizing homes. 52
Two of the Cytb sequences for T. guazu obtained, but not used in this analysis, most likely represent pseudogenes, possibly a nuclear copy ( numts) that can be very common in insect genomes 61 and not uncommon to amplify using direct polymerase chain reaction (PCR) methods. 62 The assumption that the recovered sequences were paralogous copies, was a result of being impossible to properly align both haplotypes of Cytb to other sequences. When aligned based on nucleotide similarity, it required three indel events, two insertion events of a single nucleotide and a downstream deletion event of two nucleotides, and it shifted the open reading frame in the area between these events, resulting in stop codons being observed in all sequences. Previously, amplification of pseudogenes in Triatominae was reported for ND1, ND2, and COI 63 and is herein reported for Cytb. Considering that the density of numts in insect genomes is apparently lineage-specific and, as far as it is known, there is little information in Hemiptera, 61, 63 future sequencing by direct PCR of Cytb for phylogenetic reconstruction should be done with caution.
Conclusion
Considering species from Central-Western Brazil, which had some genes sequenced for the first time, T. baratai and T. vandae were analyzed phylogenetically based on DNA sequences. The analyses confirmed the relationship of both T. vandae and T. baratai as members of the T. matogrossensis subcomplex, however the monophyly of this subcomplex was not recovered because of the close relationship of some members to members of the T. sordida subcomplex. This added phylogenetic evidence calls for a re-evaluation of the current classification of triatomines. This re-evaluation should be based on additional multiple molecular markers, especially nuclear genes, as mitochondrial protein-coding genes used so far are apparently too variable, and possibly saturated, to recover these relationships with strong support.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dayse da Silva Rocha, José Jurberg, Ionizete Garcia da Silva, José Maria Soares Barata, Paulo Silva de Almeida, and Sebastião Aldo da Silva Valente for providing the first specimens that gave rise to the colonies of Triatoma baratai, T. costalimai, T. guazu, T. matogrossensis, and T. vandae used herein.
Footnotes
Financial support: Financial support includes grants from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brazil), Fundação de Amparo à Pesquisa do Estado de São Paulo (process numbers 2010/50.355-1, 2010/17027-0, and 2011/22378-0 - Fapesp, Brazil), and Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ, Brazil).
Authors' addresses: Sueli Gardim and Cláudia S. Rocha, UNESP, Biological Sciences, Araraquara, São Paulo, Brazil, E-mails: sugardim@gmail.com and clausolanorocha@yahoo.com.br. Carlos E. Almeida and Regina M. B. Cicarelli, Faculdade de Ciências Farmacêuticas/UNESP, Ciências Biológicas, Araraquara, São Paulo, Brazil, E-mails: almeida_ce@hotmail.com and cicarell@fcfar.unesp.br. Daniela M. Takiya, Federal University of Rio de Janeiro, Department of Zoology, Rio de Janeiro, RJ, Brazil, E-mail: takiya@gmail.com. Marco T. A. da Silva, University of São Paulo (USP), Physics Institute of São Carlos, São Carlos, São Carlos, Brazil, E-mail: tulio@ursa.ifsc.usp.br. Daniela L. Ambrósio, University of Connecticut Health Center, Department of Genetics and Developmental Biology, Farmington, CT, E-mail: dlambrosio@yahoo.com.br.
References
- 1.Chagas CR. Nova tripanozomiaze humana. Estudos sobre a morfologia e o ciclo evolutivo do Schizotrypanum cruzi, n. gen. n. esp, agente etiológico de nova entidade mórbida do homem. Mem Inst Oswaldo Cruz. 1909;1:159–218. [Google Scholar]
- 2.Galvão C, Carcavallo R, Rocha DS, Jurberg J. Checklist of the current valid species of the subfamily Triatominae Jeannet, 1919 (Hemíptera, Reduviidae) and their geographical distribution, with nomenclatural and taxonomic notes. Zootaxa. 2003;202:1–36. [Google Scholar]
- 3.Poinar JR. Triatoma dominicana sp. n. (Hemiptera: Reduviidae: Triatominae) and Trypanosoma antiquus sp. n. (Stercoraria: Trypanosomatidae), the first fossil evidence of a Triatomine-Trypanosomatid vector association. Vector Borne Zoonotic Dis. 2005;5:72–81. doi: 10.1089/vbz.2005.5.72. [DOI] [PubMed] [Google Scholar]
- 4.Costa J, Argolo AM, Felix M. Redescription of Triatoma melanica Neiva and Lent, 1941, new status (Hemiptera: Reduviidae: Triatominae) Zootaxa. 2006;1385:47–52. [Google Scholar]
- 5.Galvão C, Angulo VM. Belminus corredori, a new specie of Bolboderini (Hemiptera: Reduviidae: Triatominae) form Santander, Colômbia. Zootaxa. 2006;1241:61–68. [Google Scholar]
- 6.Bérenger JM, Blanchet D. A new species of the genus Panstrongylus from French Guiana (Heteroptera; Reduviidae; Triatominae) Mem Inst Oswaldo Cruz. 2007;102:733–736. doi: 10.1590/s0074-02762007005000088. [DOI] [PubMed] [Google Scholar]
- 7.Costa J, Felix M. Triatoma juazeirensis sp. nov. from the state of Bahia, northeastern Brazil (Hemiptera: Reduviidae: Triatominae) Mem Inst Oswaldo Cruz. 2007;102:87–90. doi: 10.1590/s0074-02762007000100015. [DOI] [PubMed] [Google Scholar]
- 8.Martínez E, Chávez T, Sossa D, Aranda R, Benigno V, Vidaurre P. Triatoma boliviana sp. n. de los valles subandinos de La Paz, Bolívia (Hemiptera: Reduviidae: Triatominae), similar a Triatoma nigromaculata Stål, 1859. Bol Inst Investig Salud y Desarrollo. 2007;3:1–10. [Google Scholar]
- 9.Sandoval CM, Pabon E, Jurberg J, Galvão C. Belminus ferroae n. sp. from the Colombian north-east, with a key to the species of the genus (Hemiptera: Reduviidae: Triatominae) Zootaxa. 2007;1443:55–64. [Google Scholar]
- 10.Jurberg J, Rocha DS, Galvão C. Rhodnius zeledoni sp. nov. afim de Rhodnius paraensis Sherlock, Guitton and Miles, 1977 (Hemiptera, Reduviidae, Triatominae) Biota Neotrop. 2009;9:123–128. [Google Scholar]
- 11.Rosa JA, Rocha CS, Gardim S, Pinto MP, Mendonça VJ, Ferreira Filho JC, Carvalho EO, Camargo LM, Oliveira J, Nascimento JD, Cilense M, Almeida CE. Description of Rhodnius montenegrensis sp.n. (Hemiptera: Reduviidae: Triatominae) from the state of Rondônia, Brazil. Zootaxa. 2012;3478:62–76. [Google Scholar]
- 12.Hypsa V, Tietz D, Zrzavy J, Rego RO, Galvão C, Jurberg J. Phylogeny and biogeography of Triatominae (Hemiptera: Reduviidae): molecular evidence of a New World origin of the Asiatic clade. Mol Phylogenet Evol. 2002;23:447–457. doi: 10.1016/s1055-7903(02)00023-4. [DOI] [PubMed] [Google Scholar]
- 13.Schofield CJ, Galvão C. Classification, evolution, and species groups within the Triatominae. Acta Trop. 2009;110:88–100. doi: 10.1016/j.actatropica.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 14.Costa J, Lorenzo M. Biology, diversity and strategies for the monitoring and control of triatomines - Chagas disease vectors. Mem Inst Oswaldo Cruz. 2009;104:46–51. doi: 10.1590/s0074-02762009000900008. [DOI] [PubMed] [Google Scholar]
- 15.Usinger RL, Wygodzinsky P, Ryckman RE. The biosystematics of Triatominae. Ann Rev Ent. 1966;11:309–330. doi: 10.1146/annurev.en.11.010166.001521. [DOI] [PubMed] [Google Scholar]
- 16.Lent H, Wygodzinsky P. Revision of the triatominae (Hemiptera, Reduviidae), and their significance as vectors of Chagas's disease. Bull Am Mus Nat Hist. 1979;163:123–520. [Google Scholar]
- 17.Carcavallo RU, Jurberg J, Rocha DS, Galvão C, Noireau F, Lent H. Triatoma vandae n. sp. do complexo oliveirai encontrada no Estado de Mato Grosso, Brasil (Hemiptera: Reduviidae: Triatominae) Mem Inst Oswaldo Cruz. 2002;97:649–654. doi: 10.1590/s0074-02762002000500011. [DOI] [PubMed] [Google Scholar]
- 18.Noireau F, Santos SM, Gumiel M, Dujardin JP, Soares MS, Carcavallo RU, Galvão C, Jurberg J. Phylogenetic relationships within the oliveirai complex (Hemiptera, Reduviidae, Triatominae) Infect Genet Evol. 2002;2:1–17. doi: 10.1016/s1567-1348(02)00059-x. [DOI] [PubMed] [Google Scholar]
- 19.Papa AR, Jurberg J, Carcavallo RU, Cerqueira RL, Barata JM. Triatoma sherlocki sp. n. coletada na Bahia, Brasil (Hemiptera, Reduviidae, Triatominae) Entomología y Vectores. 2002;9:133–146. [Google Scholar]
- 20.Almeida CE, Folly-Ramos E, Peterson AT, Lima-Neiva V, Gumiel M, Duarte R, Lima MM, Locks M, Beltrão M, Costa J. Could Triatoma sherlocki be vectoring Chagas disease in small mining communities in Bahia, Brazil? Medical and veterinary entomology. Med Vet Entomol. 2009;23:410–417. doi: 10.1111/j.1365-2915.2009.00822.x. [DOI] [PubMed] [Google Scholar]
- 21.Almeida CE, Oliveira HL, Correia N, Dornak LL, Gumiel M, Neiva VL, Harry M, Mendonça VJ, Costa J, Galvão C. Dispersion capacity of Triatoma sherlocki, Triatoma juazeirensis and laboratory-bred hybrids. Acta Trop. 2012;122:71–79. doi: 10.1016/j.actatropica.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 22.Futuyma DJ. Evolution. Second edition. Sunderland, MA: Palgrave Macmillan Sinauer Associates; 2009. [Google Scholar]
- 23.Sainz AC, Mauro LV, Moriyama EN, García BA. Phylogeny of triatomine vectors of Trypanosoma cruzi suggested by mitochondrial DNA sequences. Genetica. 2004;121:229–240. doi: 10.1023/b:gene.0000039842.82574.02. [DOI] [PubMed] [Google Scholar]
- 24.García BA, Moriyama EN, Powell JR. Mitochondrial DNA sequences of Triatomines (Hemiptera: Reduviidae): phylogenetic relationships. J Med Entomol. 2001;38:675–683. doi: 10.1603/0022-2585-38.5.675. [DOI] [PubMed] [Google Scholar]
- 25.Forattini OP. Biogeography, origin, and distribution of triatominae domiciliarity in Brazil. Rev Saude Publica. 1980;14:265–299. doi: 10.1590/s0034-89101980000300002. [DOI] [PubMed] [Google Scholar]
- 26.Mello DA, Borges MM. Primeiro encontro do Triatoma costalimai naturalmente infectado pelo Trypanosoma cruzi: estudo de aspectos biológicos da amostra isolada. Mem Inst Oswaldo Cruz. 1981;76:61–69. doi: 10.1590/s0074-02761981000100007. [DOI] [PubMed] [Google Scholar]
- 27.Mello DA. Roedores, marsupiais e triatomíneos silvestres capturados no município de Mambaí-Goiás. Infecção natural pelo Trypanosoma cruzi. Rev Saude Publica. 1982;16:282–291. doi: 10.1590/s0034-89101982000500003. [DOI] [PubMed] [Google Scholar]
- 28.Lent H, Jurberg J, Galvão C. Descrição do alótipo (macho) de Triatoma guazu Lent, Wygodzinsky, 1979 proveniente do Estado do Mato Grosso, Brasil (Hemiptera, Reduviidae) Mem Inst Oswaldo Cruz. 1996;91:313–315. [Google Scholar]
- 29.Carcavallo RU, Galvão C, Lent H. T. jurbergi sp.n. do norte do Estado do Mato Grosso, Brasil (Hemiptera, Reduviidae, Triatominae) com uma atualização das sinonímias e outros táxons. Mem Inst Oswaldo Cruz. 1998;93:459–464. doi: 10.1590/s0074-02761998000400007. [DOI] [PubMed] [Google Scholar]
- 30.Carcavallo RU, Jurberg J. T. baratai sp.n. do Estado do Mato Grosso do Sul, Brasil (Hemiptera, Reduviidae, Triatominae) Entomol. Vectores. 2000;7:373–387. [Google Scholar]
- 31.Oliveira AW, Silva IG. Distribuição geográfica e indicadores entomológicos de triatomíneos sinantrópicos capturados no Estado de Goiás. Rev Soc Bras Med Trop. 2007;40:204–208. doi: 10.1590/s0037-86822007000200011. [DOI] [PubMed] [Google Scholar]
- 32.Almeida PS, Ceretti-Júnior W, Obara MT, Santos HR, Barata JM, Faccenda O. Survey of Triatominae (Hemiptera: Reduviidae) fauna in domestic environments and natural infection by Trypanosomatidae in the State of Mato Grosso do Sul. Rev Soc Bras Med Trop. 2008;41:374–380. doi: 10.1590/s0037-86822008000400010. [DOI] [PubMed] [Google Scholar]
- 33.Gurgel-Gonçalves R, Galvão C, Costa J, Peterson AT. Geographic distribution of Chagas disease vectors in Brazil based on ecological niche modeling. J Trop Med. 2012 doi: 10.1155/2012/705326. doi:10.1155/2012/705326. Epub 2012 Feb 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Third edition. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 35.Lyman ED, Monteiro FA, Escalante AA, Cordon-Rosales C, Wesson DM, Dujardin JP, Beard CB. Mitochondrial DNA sequence variation among triatomine vectors of Chagas' disease. Am J Trop Med Hyg. 1999;60:377–386. doi: 10.4269/ajtmh.1999.60.377. [DOI] [PubMed] [Google Scholar]
- 36.Garcia BA, Powell JR. Phylogeny of species of Triatoma (Hemiptera: Reduviidae) based on mitochondrial DNA sequences. J Med Entomol. 1998;35:232–238. doi: 10.1093/jmedent/35.3.232. [DOI] [PubMed] [Google Scholar]
- 37.Gaunt MW, Miles MA. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol Biol Evol. 2002;19:748–761. doi: 10.1093/oxfordjournals.molbev.a004133. [DOI] [PubMed] [Google Scholar]
- 38.Giordano R, Cortez JCP, Paulk S, Stevens L. Genetic diversity of Triatoma infestans (Hemiptera: Reduviidae) in Chuquisaca, Bolivia based on the mitochondrial cytochrome b gene. Mem Inst Oswaldo Cruz. 2005;100:753–760. doi: 10.1590/s0074-02762005000700014. [DOI] [PubMed] [Google Scholar]
- 39.Monteiro FA, Donnelly MJ, Beard CB, Costa J. Nested clade and phylogeographic analyses of the Chagas disease vector Triatoma brasiliensis in northeast Brazil. Mol Phylogenet Evol. 2004;32:46–56. doi: 10.1016/j.ympev.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 40.Pavan MG, Monteiro FA. A multiplex PCR assay that separates Rhodnius prolixus from members of the Rhodnius robustus cryptic species complex (Hemiptera: Reduviidae) Trop Med Int Health. 2007;12:751–758. doi: 10.1111/j.1365-3156.2007.01845.x. [DOI] [PubMed] [Google Scholar]
- 41.Ceretti-Junior W, Vendrami DP, Gil JM, Barata JM, Marrelli MT. Análise das relações taxonômicas e sistemáticas entre espécies de triatomíneos (Hemiptera, Reduviidae) de colônias mantidas pelo Serviço Especial de Saúde de Araraquara, inferida de seqüências do 16S rDNA mitocondrial. Rev Bras Entomol. 2008;52:455–462. [Google Scholar]
- 42.Mendonça VJ, Silva MT, Araujo RF, Junior JM, Junior MB, Almeida CE, Costa J, Graminha MA, Cicarelli RM, Rosa JA. Phylogeny of Triatoma sherlocki (Hemiptera: Reduviidae: Triatominae) inferred from two mitochondrial genes suggests its location within the Triatoma brasiliensis complex. Am J Trop Med Hyg. 2009;81:858–864. doi: 10.4269/ajtmh.2009.08-0664. [DOI] [PubMed] [Google Scholar]
- 43.Segura EL, Torres AG, Fusco O, García BA. Mitochondrial 16S DNA variation in populations of Triatoma infestans from Argentina. Med Vet Entomol. 2009;23:34–40. doi: 10.1111/j.1365-2915.2008.00749.x. [DOI] [PubMed] [Google Scholar]
- 44.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 45.Huelsenbeck JP, Ronquist F. MrBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 46.Swofford DL. Sunderland, MA: Sinauer Associates; 2002. PAUP. Phylogenetic analysis using parsimony (*and other methods), Version 4. [Google Scholar]
- 47.Akaike H. A new look at the statistical model identification. IEEE Trans Automat Contr. 1974;19:716–723. [Google Scholar]
- 48.Nylander JA. MrModeltest v2. Evolutionary Biology Centre; Uppsala University, Sweden: 2004. Program distributed by the author. [Google Scholar]
- 49.China WE, Miller NC. Check list and keys to the families and sub-families of the Hemiptera-Heteroptera. Bull Brit Mus (Nat Hist). Ent. 1959;8:3–45. [Google Scholar]
- 50.Ryckman RE. Biosystematics and hosts of Triatoma protracta complex in North America (Hemiptera: Reduviidae) (Rodentia: Cricetidae) Univ California Publ Entomol. 1962;27:93–240. [Google Scholar]
- 51.Gómez-Sucerquia LJ, Triana-Chávez O, Jaramillo-Ocampo N. Quantification of the genetic change in the transition of Rhodnius pallescens Barber, 1932 (Hemiptera: Reduviidae) from field to laboratory. Mem Inst Oswaldo Cruz. 2009;104:871–877. doi: 10.1590/s0074-02762009000600009. [DOI] [PubMed] [Google Scholar]
- 52.Noireau F, Gutierrez T, Flores R, Brenière F, Bosseno MF, Wisnivesky-Colli C. Ecogenetics of Triatoma sordida and T. guasayana (Hemiptera: Reduviidae) in the Bolivian Chaco. Mem Inst Oswaldo Cruz. 1999;94:451–457. doi: 10.1590/S0074-02761999000400004. [DOI] [PubMed] [Google Scholar]
- 53.Almeida CE, Marcet PL, Gumiel M, Takiya DM, Cardozo-de-Almeida M, Pacheco RS, Lopes CM, Dotson EM, Costa J. Phylogenetic and phenotypic relationships among Triatoma carcavalloi (Hemiptera: Reduviidae: Triatominae) and related species collected in domiciles in Rio Grande do Sul State. Brazil. J Vector Ecol. 2009;34:164–173. doi: 10.1111/j.1948-7134.2009.00023.x. [DOI] [PubMed] [Google Scholar]
- 54.Carcavallo RU, Jurberg J, Lent H, Galvão C, Steindel M, Pinto CJ. Nova Espécie do complexo oliveirai (Nova Denominação para o Complexo matogrossensis) (Hemiptera, Reduviidae, Triatominae) do Estado do Rio Grande do Sul, Brasil. Mem Inst Oswaldo Cruz. 2001;96:71–79. [PubMed] [Google Scholar]
- 55.De Paula AS, Diotaiuti L, Schofield CJ. Testing the sister-group relationship of the Rhodniini and Triatomini (Insecta: Hemiptera: Reduviidae: Triatominae), Northeastern Brazil (Hemiptera: Reduviidae: Triatominae) Mol Phylogenet Evol. 2005;35:712–718. doi: 10.1016/j.ympev.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 56.Diotaiuti L, Loiola CF, Falcão PL, Dias JC. The ecology of Triatoma sordida in natural environments in two different regions of the State of Minas Gerais, Brazil. Rev Bras Med Trop. 1993;35:237–245. doi: 10.1590/s0036-46651993000300004. [DOI] [PubMed] [Google Scholar]
- 57.Diotaiuti L, Azeredo BV, Busek SC, Fernandes AJ. Controle do Triatoma sordida no peridomicílio rural do município de Porteirinha, Minas Gerais, Brasil. Rev Panam Salud Publica. 1998;3:21–25. doi: 10.1590/s1020-49891998000100004. [DOI] [PubMed] [Google Scholar]
- 58.Diotaiuti L, Paula OR, Falcão PL, Dias JC. Avaliação do programa de controle vetorial da doença de Chagas em Minas Gerais, Brasil, com referência especial ao Triatoma sordida. Bol Oficina Sanit Panam. 1995;118:211–219. [Google Scholar]
- 59.Argolo AM, Felix M, Pacheco R, Costa J. Doença de Chagas e seus Principais Vetores no Brasil. Rio de Janeiro, Brazil: Imperial Novo Milênio Gráfica e Editora Ltda; 2008. [Google Scholar]
- 60.Almeida CE, Vinhaes MC, Almeida JR, Silveira AC, Costa J. Monitoring the domiciliary and peridomiciliary invasion process of Triatoma rubrovaria in the State of Rio Grande do Sul, Brazil. Mem Inst Oswaldo Cruz. 2000;95:761–768. doi: 10.1590/s0074-02762000000600003. [DOI] [PubMed] [Google Scholar]
- 61.Pamilo P, Viljakainen L, Vihavainen A. Exceptionally high density of NUMTs in the honeybee genome. Mol Biol Evol. 2007;24:1340–1346. doi: 10.1093/molbev/msm055. [DOI] [PubMed] [Google Scholar]
- 62.Song H, Buhay JE, Whiting MF, Crandall KA. Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proc Natl Acad Sci USA. 2008;105:13486–13491. doi: 10.1073/pnas.0803076105. (PNAS) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mas-Coma S, Bargues MD. Populations, hybrids and the systematic concepts of species and subspecies in Chagas disease triatomine vectors inferred from nuclear ribosomal and mitochondrial DNA. Acta Trop. 2009;110:112–136. doi: 10.1016/j.actatropica.2008.10.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.