Abstract
In this study, we used microarray analysis to investigate the biogenesis and progression of intervertebral disc degeneration. The gene expression profiles of 37 disc tissue samples obtained from patients with herniated discs and degenerative disc disease collected by the National Cancer Institute Cooperative Tissue Network were analyzed. Differentially expressed genes between more and less degenerated discs were identified by significant analysis of microarray. A total of 555 genes were significantly overexpressed in more degenerated discs with a false discovery rate of < 3%. Functional annotation showed that these genes were significantly associated with membrane-bound vesicles, calcium ion binding and extracellular matrix. Protein-protein interaction analysis showed that these genes, including previously reported genes such as fibronectin, COL2A1 and β-catenin, may play key roles in disc degeneration. Unsupervised clustering indicated that the widely used morphology-based Thompson grading system was only marginally associated with the molecular classification of intervertebral disc degeneration. These findings indicate that detailed, systematic gene analysis may be a useful way of studying the biology of intervertebral disc degeneration.
Keywords: genes, intervertebral disc degeneration, molecular classification, protein-protein interaction
Introduction
The intervertebral disc (IVD) is a cartilaginous structure that shows degenerative and age-related changes earlier than other connective tissues in the body (Urban and Roberts, 2003). Disc degeneration occurs when its cells die or become dysfunctional, especially in an acidic environment. During degeneration, the IVD becomes dehydrated and vascularized, and there is an ingrowth of nerves (Hughes et al., 2012). As a multi-factorial process, disc degeneration is influenced by factors such as genetic predisposition, lifestyle conditions, including obesity, occupation, smoking and alcohol consumption (Samartzis et al., 2012), aging phenomena (Gruber and Hanley, 2007) and other health factors, such as diabetes. Disc degeneration is important clinically because of a strong association between disc degeneration and back pain (Luoma et al., 2000), which is a major public health problem in modern industrialized societies. In addition, the altered physiology of the IVD during degeneration is associated with many other clinical symptoms or conditions, such as lower limb pain, paraesthesia, spinal stenosis and disc herniation (Hughes et al., 2012).
Previous studies have identified some molecular markers associated with disc degeneration. For example, asporin was found to be expressed at higher levels in degenerated human IVD (Gruber et al., 2009). The mRNA expression levels of ADAMTS-7 and ADAMTS-12 were significantly higher in endplate cells of degenerative discs compared with those in non-degenerative discs (Zhang et al., 2012). In addition, genetic polymorphisms in collagen I, collagen IX, collagen XI, aggrecan, extracellular matrix-degrading enzymes, inflammatory cytokines, such as IL (interleukin)-1, IL-6 and TNFα (tumor necrosis factors α), Fas/FasL and vitamin D receptors have been associated with the development of intervertebral disc degeneration (IVDD) (Kalb et al., 2012).
In this study, we compared the gene expression profile data of IVDD patients with that of healthy controls. The gene profiles were downloaded from the gene expression omnibus (GEO) database and the intervertebral disc data were classified into two groups according to the Thompson grading system (Thompson et al., 1990).
Materials and Methods
Microarray data analysis
Microarray datasets were downloaded from the GEO database (accession numbers GSE15227 (Gruber et al., 2009) and GSE23130 (Gruber et al., 2012)). The signal intensities were re-calculated with the robust multi-array average (RMA) (Irizarry et al., 2003) using the Entrez Gene-based center custom Chip Description File (CDF) developed by University of Michigan (Dai et al., 2005). The single sample classified as Thompson grade I was considered as healthy (Gruber et al., 2009) and ignored before merging the GSE15227 and GSE23130 data. ComBat (Johnson et al., 2007) was applied to the merged dataset to remove potential unwanted batch effects. The samples were divided into two groups based on the Thompson grading system. Grade IV and V samples were considered as more degenerated (Gruber et al., 2009). Significant analysis of microarray (SAM) (Tusher et al., 2001) was applied to identify differentially expressed genes using a delta of 1. Functional annotation was done on DAVID (Huang da et al., 2009) using significantly differentially expressed genes.
Protein-protein interaction analysis
Differentially expressed genes were submitted to STRING 9.0 (Szklarczyk et al., 2011) to detect possible protein-protein interactions. EntrezGene IDs were translated into official gene symbols by DAVID before submitting. A threshold score of 0.4 was applied as default. Only those interactions that were proven experimentally were considered. A simple Perl script was used to exclude genes/proteins not in the query gene list.
Cluster analysis
The data for each gene was median-centered before clustering. Centered correlation was applied as a similarity metric and average linkage as a clustering method. The analysis was done in Cluster 3.0 and a heatmap was generated by TreeView. A one-sided Fisher’s exact test was used to assess the consistence of the molecular and Thompson grade classifications.
Results
Differentially expressed genes in IVD degeneration
Eleven disc samples (eight Grade IV and three Grade V) were classified as the more degenerated group while the remaining 26 samples (10 Grade II and 16 Grade III) were classified as the less degenerated group. One thousand sample permutations were used to estimate the false discovery rate (FDR). When delta was set to 1,555 genes were over-expressed in the more degenerated disc samples (Table 1) and 98 genes were underexpressed in the more degenerated disc samples (Table 2). Both groups had an FDR < 3%. The overexpressed gene list was referred to as IVDD_UP and the underexpressed gene list as IVDD_DN. Several of the IVDD_UP genes have been reported before. For example, asporin, which is present in the cartilage extracellular matrix (ECM) (Henry et al., 2001) and may be genetically associated with osteoarthritis (Loughlin, 2005), showed higher expression levels in Thompson Grade IV human discs compared to lower grade discs (Gruber et al., 2009). A highly conserved secreted serine protease, HTRA1, which degrades numerous extracellular matrix proteins, was also found in IVDD_UP. HTRA1 mRNA and protein levels are significantly elevated in degenerated disc tissue (Tiaden et al., 2012). The gene expression of cathepsin K, a cysteine protease that cleaves the triple helical domains of collagen types I and II, was significantly greater in more degenerated discs (grades III and IV) compared to healthier discs (grades I and II) (Gruber et al., 2011).
Table 1.
Top 30 IVDD_UP genes with p < 0.05 and FDR < 3%.
| Gene name | Score | Fold change | q-value |
|---|---|---|---|
| BCAS3 | 5.525645110 | 2.015911968 | 0 |
| AP2S1 | 4.993647193 | 2.695484514 | 0 |
| RTN3 | 4.831739884 | 1.906392737 | 0 |
| MAT2B | 4.795801549 | 2.661280400 | 0 |
| SMOC2 | 4.722070979 | 3.529131297 | 0 |
| TMEM214 | 4.609155146 | 2.397158886 | 0 |
| SETD3 | 4.553095283 | 1.580288009 | 0 |
| RCAN1 | 4.515015896 | 2.901977015 | 0 |
| NUDT15 | 4.387565829 | 1.611085005 | 0 |
| PDGFRA | 4.352020487 | 2.013095239 | 0 |
| HOXC6 | 4.312467370 | 4.046628242 | 0 |
| DAXX | 4.270628387 | 1.822846390 | 0 |
| SLC40A1 | 4.255751181 | 5.142171070 | 0 |
| RYFY1 | 4.250428720 | 2.629236672 | 0 |
| IFITM1 | 4.237500892 | 2.139999154 | 0 |
| CHMP1A | 4.220758109 | 2.250578926 | 0 |
| SF4 | 4.213669108 | 1.548679838 | 0 |
| GJA1 | 4.184343713 | 5.752785110 | 0 |
| ZFYVE26 | 4.095399931 | 1.536943270 | 0 |
| CHT3L1 | 4.080014011 | 5.398762712 | 0 |
| C3ORF1 | 4.069293869 | 2.752723264 | 0 |
| CPN2 | 4.067581072 | 4.689686574 | 0 |
| POLR2G | 4.050051549 | 2.546570992 | 0 |
| LGALS1 | 4.027157407 | 4.896502415 | 0 |
| ITM2B | 4.026549335 | 3.916969611 | 0 |
| PTTG1 | 4.006722096 | 4.387834233 | 0 |
| CDK2AP1 | 3.968504177 | 6.601587442 | 0 |
| TMEM9 | 3.967576485 | 1.540136100 | 0 |
| CLU | 3.947513637 | 7.942443254 | 0 |
| RHOC | 3.942703220 | 3.185787316 | 0 |
Table 2.
Top 30 IVDD_DN genes with p < 0.05 and FDR < 3%.
| Gene name | Score | Fold change | q-value |
|---|---|---|---|
| KRTAP4-11 | −4.465760325 | 0.478119384 | 0 |
| OR5E1P | −4.306673904 | 0.598139097 | 0 |
| TTTY14 | −4.266771085 | 0.697225016 | 0 |
| PPY | −4.230640542 | 0.327213756 | 0 |
| C100RF25 | −4.209818145 | 0.58437279 | 0 |
| DPCR1 | −4.101123182 | 0.700400802 | 0 |
| LOC1002893 | −4.052210058 | 0.560921078 | 0 |
| DNAH8 | −3.978399529 | 0.708059442 | 0 |
| CSMD2 | −3.948145754 | 0.685029482 | 0 |
| KRTAP4-6 | −3.870547452 | 0.669765942 | 0 |
| HOXD11 | −3.810598074 | 0.706993483 | 0.417452925 |
| TMEM102 | −3.798006024 | 0.71228122 | 0.417452925 |
| KRTAP4-8 | −3.76590512 | 0.625274173 | 0.417452925 |
| KRTAP4-4 | −3.745342914 | 0.733361857 | 0.417452925 |
| ZNF440 | −3.7332524 | 0.644960742 | 0.417452925 |
| OR2M4 | −3.721057881 | 0.656210145 | 0.417452925 |
| TKTL1 | −3.659388232 | 0.704826703 | 0.538764031 |
| KRTAP-7 | −3.657243192 | 0.692592119 | 0.538764031 |
| KRT76 | −3.633206767 | 0.533949119 | 0.538764031 |
| C80RF12 | −3.629644489 | 0.677570179 | 0.538764031 |
| LOC1001312 | −3.562150167 | 0.721969038 | 0.538764031 |
| TFE3 | −3.55931541 | 0.789925298 | 0.538764031 |
| FRS3 | −3.540271969 | 0.622015836 | 0.538764031 |
| HHIP | −3.525281501 | 0.717589029 | 0.538764031 |
| HTR3C | −3.522633996 | 0.603942904 | 0.538764031 |
| ELAC1 | −3.519001164 | 0.785612099 | 0.538764031 |
| LOC401097 | −3.492245688 | 0.766487773 | 1.000561772 |
| TRAV24 | −3.487013115 | 0.656667894 | 1.000561772 |
| FMO1 | −3.457758868 | 0.658106046 | 1.000561772 |
| WDR53 | −3.456389239 | 0.626137741 | 1.000561772 |
Functional annotation of differentially expressed genes
Differentially expressed genes were submitted to DAVID (Huang da et al., 2009) for functional annotation. Databases for disease information, functional categories, gene ontology, curated pathways and protein domains were included to provide a comprehensive knowledge source. For the IVDD_UP genes, 195 functional annotation clusters were identified. The top three clusters ranked by the enrichment score were mainly associated with membrane-bounded vesicles, calcium ion binding and extracellular matrix, with enrichment scores of 4.7, 3.61 and 3.25, respectively. Noticeably, these enriched genes were also related to the biological process of skeletal system development, including cartilage development. Among the IVDD_DN genes, there were only 19 clusters with the highest enrichment score being only 0.87; this score was for a cluster involving mainly zinc fingers and the Krueppel-associated box.
Protein-protein interactions among differentially expressed genes
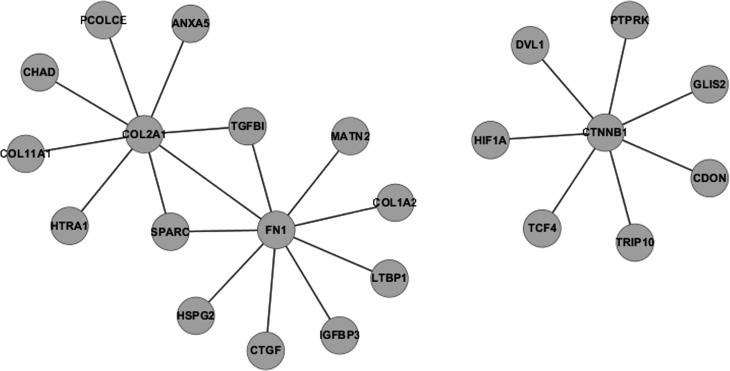
STRING showed that 159 experimentally proven protein-protein interactions were formed by 175 genes in IVDD_UP (Figure 1). Fibronectin 1, COL2A1 (collagen, type II, α1) and β-catenin were the three genes/proteins that had the most interacting partners (Figure 2). Of these 175 genes, 107 had only one interacting partner in IVDD_UP and thus resembled the phenomenon of scale-free biological networks (Barabasi and Albert, 1999). Surprisingly, no interactions were observed amongst IVDD_DN genes.
Figure 1.
Protein-protein interaction network for IVDD_UP genes. The larger the font size, the greater the number of interacting partners per node. Fibronectin 1 (FN1), COL2A1 and CTNNB1 had the most interacting partners in this network.
Figure 2.
Protein-protein interactions involving the three most connected proteins. Fibronectin 1 (FN1), COL2A1 and CTNNB1 were the first, second and third most connected proteins, with nine, eight and seven edges, respectively.
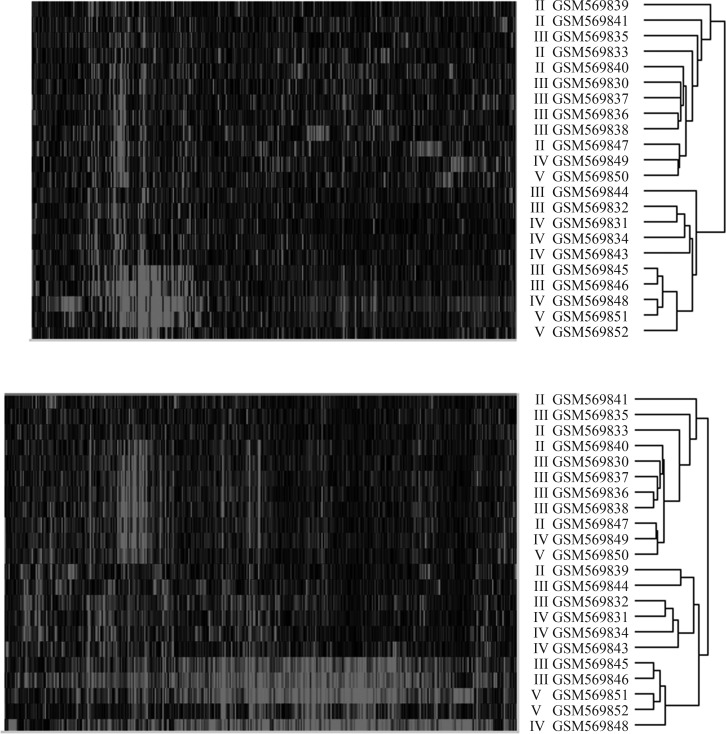
Cluster analysis of IVD tissues
Unsupervised hierarchical clustering was done on 22 disc tissues of GSE23130. The experimental results obtained using all 18,818 genes or only 1,984 genes with a minimum sample standard deviation of 1 were highly similar (Figure 3). A one-tailed Fisher’s exact test showed a marginally significant association between the Thompson-based classification and the unsupervised molecular classification (p = 0.04799 for the 18,818-gene case and 0.09133 for the 1,984-gene case).
Figure 3.
Unsupervised hierarchical clustering of 22 disc tissues of GSE23130. The pattern shown on the left was obtained by using all 18,818 genes while the result obtained by using only 1,984 genes with a minimum sample standard deviation of 1 is shown on the right.
Discussion
Our microarray analysis of the gene expression profile of degenerated IVD detected hundreds of differentially expressed genes that may be associated with IVD degeneration. Functional annotation supported the biological relevance of our findings. Protein-protein interaction analysis of IVDD_UP revealed genes that may have key roles in IVD degeneration. Fibronectin 1 (FN1), a molecular marker for fibrosis (Leask and Abraham, 2004), was up-regulated in punctured mouse tail discs that showed progressive degenerative changes and were fibrocartilaginous (Yang et al., 2009). Earlier studies reported that the degradation fragments of fibronectin (Fn-f) accumulate in the disc during degeneration (Oegema et al., 2000) and induce IVD degeneration in rabbits (Greg Anderson et al., 2003). Given that HTRA1 can induce fibronectin proteolysis and one of the resultant fragments was found to be increased in HTRA1-treated IVD cell cultures as well as in disc tissue from patients with IVD degeneration, it was hypothesized that HTRA1 promoted IVD degeneration through the proteolytic cleavage of fibronectin and subsequent activation of resident disc cells (Tiaden et al., 2012). The over-expression of these two genes was consistent with this hypothesis. Interestingly, fibronectin mRNA and protein levels were significantly down-regulated following the up-regulation of HTRA1 in rhesus monkey choroid-retina endothelial cells (RF/6A) and human umbilical vein endothelial cells (HUVECs) (Jiang et al., 2012), suggesting that some other unknown factors may be involved in the interaction between HTRA1 and fibronectin in IVD tissues.
The IVDD_UP group of genes contained four collagen genes: COL11A1, COL1A2, COL3A1 and COL2A1. COL2A1 interacted with eight other proteins encoded by genes in IVDD_UP. Mutations in COL2A1 give rise to a spectrum of phenotypes that predominantly affecting cartilage and bone. These chondrodysplasias are typically characterized by a disproportionately short stature, eye abnormalities, cleft palate and hearing loss. Increasing evidence has also implicated COL2A1 mutations in individuals with isolated degenerative joint disease (Kannu et al., 2010). The relationship between COL2A1 gene polymorphisms and knee osteoarthritis were also investigated in Han Chinese women (Xu et al., 2011).
Mutations in the COL11A1 gene (also known as STL2) have been identified in some people with Stickler syndrome (Martin et al., 1999). Other mutations in this gene cause segments of DNA to be skipped when the protein is being made, resulting in an abnormally short pro-alpha chain. Alterations in COL11A1 impair collagen function and can lead to hearing loss, tearing of the lining of the eye, and bone and joint abnormalities (Rodriguez et al., 2004). COL1A2 is a protein found in most connective tissues. Mutations in this gene are associated with asteo-genesis imperfecta, Ehlers-Danlos syndrome, idiopathic osteoporosis and atypical Marfan syndrome (Vasan et al., 1991; Ward et al., 2001; Hartikka et al., 2004). However, the symptoms associated with mutations in this gene tend to be less severe than with mutations in the gene for α1 type I collagen since the α2 form is less abundant (Bou-Gharios et al., 2004). Multiple messages for this gene result from multiple polyadenylation signals, a feature shared with most of the other collagen genes (Zuo et al., 2012).
COL3A1 is a protein that, in humans, is encoded by the COL3A1 gene located on chromosome 2 (Cutting et al., 1990). COL3A1 is a fibrillar collagen found in extensible connective tissues such as skin, lung and the vascular system, frequently in association with type 1 collagen. Mutations in this gene often lead to the exclusion of multi-exons (Superti-Furga et al., 1988). Other studies have shown that (1) IVD cells respond strongly to changes in the osmotic environment by altering their mRNA expression, e.g., human cells cultured over five days showed increased expression of aggrecan and collagen II in the nucleus and annulus cells in the presence of elevated osmolarity (Wuertz et al., 2007), and (2) there is an increase in collagen-II, aggrecan and Sox-9 protein expression in the nucleus pulposus (NP) and anulus fibrosus (AF) regions of discs from running exercised rats compared with non-exercised controls (Brisby et al., 2010). Our result may indicate an as yet unknown role for COL2A1 in degenerated IVD.
Proper regulation of Wnt/β-catenin signaling is required for development and organization of the IVD (Kondo et al., 2011). Furthermore, β-catenin was overexpressed in IVD extracted from patients with IVD degeneration and X-ray and micro-CT analyses revealed osteophyte formation and narrowing of the disc space in three-month-old β-catenin conditional activation (cAct) mice (Tang et al., 2012). Our results may provide clues for studying the biological mechanism of β-catenin in human disc tissue.
Unsupervised clustering has provided insights into the molecular heterogeneity of complex diseases such as cancer and is useful in cancer diagnosis and therapy. The application of this analysis in our study showed that the widely used morphology-based Thompson grading system was only marginally associated with the molecular classification of IVDD. This interesting finding indicates that there is scope for improving the clinical assessment of the progress of IVDD.
Footnotes
Associate Editor: Houtan Noushmer
References
- Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- Bou-Gharios G, Ponticos M, Rajkumar V, Abraham D. Extra-cellular matrix in vascular networks. Cell Prolif. 2004;37:207–220. doi: 10.1111/j.1365-2184.2004.00306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisby H, Wei AQ, Molloy T, Chung SA, Murrell GA, Diwan AD. The effect of running exercise on intervertebral disc extracellular matrix production in a rat model. Spine (Phila Pa 1976) 2010;35:1429–1436. doi: 10.1097/BRS.0b013e3181e0f5bc. [DOI] [PubMed] [Google Scholar]
- Cutting GR, McGinniss MJ, Kasch LM, Tsipouras P, Antonarakis SE. Physical mapping by PFGE localizes the COL3A1 and COL5A2 genes to a 35-kb region on human chromosome 2. Genomics. 1990;8:407–410. doi: 10.1016/0888-7543(90)90302-b. [DOI] [PubMed] [Google Scholar]
- Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greg Anderson D, Li X, Tannoury T, Beck G, Balian G. A fibronectin fragment stimulates intervertebral disc degeneration in vivo. Spine (Phila Pa 1976) 2003;28:2338–2345. doi: 10.1097/01.BRS.0000096943.27853.BC. [DOI] [PubMed] [Google Scholar]
- Gruber HE, Hanley EN. Do we need biomarkers for disc degeneration? Biomark Insights. 2007;1:131–133. [PMC free article] [PubMed] [Google Scholar]
- Gruber HE, Hoelscher G, Loeffler B, Chow Y, Ingram JA, Halligan W, Hanley EN., Jr Prostaglandin E1 and misoprostol increase epidermal growth factor production in 3D-cultured human annulus cells. Spine J. 2009a;9:760–766. doi: 10.1016/j.spinee.2009.04.024. [DOI] [PubMed] [Google Scholar]
- Gruber HE, Ingram JA, Hoelscher GL, Zinchenko N, Hanley EN, Jr, Sun Y. Asporin, a susceptibility gene in osteoarthritis, is expressed at higher levels in the more degenerate human intervertebral disc. Arthritis Res Ther. 2009b;11:R47. doi: 10.1186/ar2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber HE, Ingram JA, Hoelscher GL, Zinchenko N, Norton HJ, Hanley EN., Jr Constitutive expression of cathepsin K in the human intervertebral disc: New insight into disc extracellular matrix remodeling via cathepsin K and receptor activator of nuclear factor-kappaB ligand. Arthritis Res Ther. 2011;13:R140. doi: 10.1186/ar3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber HE, Hoelscher GL, Ingram JA, Hanley EN., Jr Genome-wide analysis of pain-, nerve- and neurotrophin-related gene expression in the degenerating human annulus. Mol Pain. 2012;8:63. doi: 10.1186/1744-8069-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartikka H, Kuurila K, Körkkö J, Kaitila I, Grénman R, Pynnönen S, Hyland JC, Ala-Kokko L. Lack of correlation between the type of COL1A1 or COL1A2 mutation and hearing loss in osteogenesis imperfecta patients. Hum Mutat. 2004;24:147–154. doi: 10.1002/humu.20071. [DOI] [PubMed] [Google Scholar]
- Henry SP, Takanosu M, Boyd TC, Mayne PM, Eberspaecher H, Zhou W, de Crombrugghe B, Hook M, Mayne R. Expression pattern and gene characterization of asporin, a newly discovered member of the leucine-rich repeat protein family. J Biol Chem. 2001;276:12212–12221. doi: 10.1074/jbc.M011290200. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hughes SP, Freemont AJ, Hukins DW, McGregor AH, Roberts S. The pathogenesis of degeneration of the intervertebral disc and emerging therapies in the management of back pain. J Bone Joint Surg Br. 2012;94:1298–1304. doi: 10.1302/0301-620X.94B10.28986. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jiang J, Huang L, Yu W, Wu X, Zhou P, Li X. Overexpression of HTRA1 leads to down-regulation of fibronectin and functional changes in RF/6A cells and HUVECs. PLoS One. 2012;7:e46115. doi: 10.1371/journal.pone.0046115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Kalb S, Martirosyan NL, Kalani MY, Broc GG, Theodore N. Genetics of the degenerated intervertebral disc. World Neurosurg. 2012;77:491–501. doi: 10.1016/j.wneu.2011.07.014. [DOI] [PubMed] [Google Scholar]
- Kannu P, Bateman JF, Randle S, Cowie S, du Sart D, McGrath S, Edwards M, Savarirayan R. Premature arthritis is a distinct type II collagen phenotype. Arthritis Rheum. 2010;62:1421–1430. doi: 10.1002/art.27354. [DOI] [PubMed] [Google Scholar]
- Kondo N, Yuasa T, Shimono K, Tung W, Okabe T, Yasuhara R, Pacifici M, Zhang Y, Iwamoto M, Enomoto-Iwamoto M. Intervertebral disc development is regulated by Wnt/beta-catenin signaling. Spine (Phila Pa 1976) 2011;36:E513–E518. doi: 10.1097/BRS.0b013e3181f52cb5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leask A, Abraham DJ. TGF-beta signaling and the fibrotic response. FASEB J. 2004;18:816–827. doi: 10.1096/fj.03-1273rev. [DOI] [PubMed] [Google Scholar]
- Loughlin J. Polymorphism in signal transduction is a major route through which osteoarthritis susceptibility is acting. Curr Opin Rheumatol. 2005;17:629–633. doi: 10.1097/01.bor.0000176687.85198.49. [DOI] [PubMed] [Google Scholar]
- Luoma K, Riihimaki H, Luukkonen R, Raininko R, Viikari-Juntura E, Lamminen A. Low back pain in relation to lumbar disc degeneration. Spine (Phila Pa 1976) 2000;25:487–492. doi: 10.1097/00007632-200002150-00016. [DOI] [PubMed] [Google Scholar]
- Martin S, Richards AJ, Yates J, Scott JD, Pope M, Snead MP. Stickler syndrome: further mutations in COL11A1 and evidence for additional locus heterogeneity. Eur J Hum Genet. 1999;7:807–814. doi: 10.1038/sj.ejhg.5200377. [DOI] [PubMed] [Google Scholar]
- Oegema TR, Jr, Johnson SL, Aguiar DJ, Ogilvie JW. Fibronectin and its fragments increase with degeneration in the human intervertebral disc. Spine (Phila Pa 1976) 2000;25:2742–2747. doi: 10.1097/00007632-200011010-00005. [DOI] [PubMed] [Google Scholar]
- Rodriguez R, Seegmiller R, Stark M, Bridgewater L. A type XI collagen mutation leads to increased degradation of type II collagen in articular cartilage. Osteoarthritis Cartilage. 2004;12:314–320. doi: 10.1016/j.joca.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Samartzis D, Karppinen J, Chan D, Luk KD, Cheung KM. The association of lumbar intervertebral disc degeneration on magnetic resonance imaging with body mass index in overweight and obese adults: a population-based study. Arthritis Rheum. 2012;64:1488–1496. doi: 10.1002/art.33462. [DOI] [PubMed] [Google Scholar]
- Superti-Furga A, Gugler E, Gitzelmann R, Steinmann B. Ehlers-Danlos syndrome type IV: a multi-exon deletion in one of the two COL3A1 alleles affecting structure, stability, and processing of type III procollagen. J Biol Chem. 1988;263:6226–6232. [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Xu G, Hou W, Ye X, Shi Q, Chen D, Wang Y. β-Catenin plays a key role in lumbar intervertebral disc degeneration. Spine (Phila Pa 1976) 2012. pp. 18–37. [DOI] [PubMed]
- Thompson JP, Pearce RH, Schechter MT, Adams ME, Tsang IK, Bishop PB. Preliminary evaluation of a scheme for grading the gross morphology of the human intervertebral disc. Spine (Phila Pa 1976) 1990;15:411–415. doi: 10.1097/00007632-199005000-00012. [DOI] [PubMed] [Google Scholar]
- Tiaden AN, Klawitter M, Lux V, Mirsaidi A, Bahrenberg G, Glanz S, Quero L, Liebscher T, Wuertz K, Ehrmann M, et al. Detrimental role for human high temperature requirement serine protease A1 (HTRA1) in the pathogenesis of intervertebral disc (IVD) degeneration. J Biol Chem. 2012;287:21335–21345. doi: 10.1074/jbc.M112.341032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JP, Roberts S. Degeneration of the intervertebral disc. Arthritis Res Ther. 2003;5:120–130. doi: 10.1186/ar629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasan NS, Kuivaniemi H, Vogel BE, Minor R, Wootton J, Tromp G, Weksberg R, Prockop D. A mutation in the pro alpha 2 (I) gene (COL1A2) for type I procollagen in Ehlers-Danlos syndrome type VII: Evidence suggesting that skipping of exon 6 in RNA splicing may be a common cause of the phenotype. Am J Hum Genet. 1991;48:305–317. [PMC free article] [PubMed] [Google Scholar]
- Ward L, Lalic L, Roughley P, Glorieux F. Thirty-three novel COL1A1 and COL1A2 mutations in patients with osteogenesis imperfecta types I–IV. Hum Mutat. 2001;17:434–434. doi: 10.1002/humu.1124. [DOI] [PubMed] [Google Scholar]
- Wuertz K, Urban JP, Klasen J, Ignatius A, Wilke HJ, Claes L, Neidlinger-Wilke C. Influence of extracellular osmolarity and mechanical stimulation on gene expression of intervertebral disc cells. J Orthop Res. 2007;25:1513–1522. doi: 10.1002/jor.20436. [DOI] [PubMed] [Google Scholar]
- Xu P, Yao J, Hou W. Relationships between COL2A1 gene polymorphisms and knee osteoarthritis in Han Chinese women. Mol Biol Rep. 2011;38:2377–2381. doi: 10.1007/s11033-010-0371-0. [DOI] [PubMed] [Google Scholar]
- Yang F, Leung VY, Luk KD, Chan D, Cheung KM. Injury-induced sequential transformation of notochordal nucleus pulposus to chondrogenic and fibrocartilaginous phenotype in the mouse. J Pathol. 2009;218:113–121. doi: 10.1002/path.2519. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Huang M, Wang X, Xu X, Ni M, Wang Y. Negative effects of ADAMTS-7 and ADAMTS-12 on endplate cartilage differentiation. J Orthop Res. 2012;30:1238–1243. doi: 10.1002/jor.22069. [DOI] [PubMed] [Google Scholar]
- Zuo C, Wen F, Li M, Zhang X, Chen H, Wu K, Zeng R. COL1A2 polymorphic markers confer an increased risk of neovascular age-related macular degeneration in a Han Chinese population. Mol Vision. 2012;18:1787–1793. [PMC free article] [PubMed] [Google Scholar]