Abstract
Isolated human immunoglobulin G (IgG) CH2 domains are promising scaffolds for novel candidate therapeutics. Unlike other human IgG domains, CH2 is not involved in strong interchain interactions and isolated CH2 is relatively stable. However, isolated single CH2 is prone to aggregation. In native IgG and Fc molecules, the N-terminal residues of CH2 from the two heavy chains interact with each other and form hinge regions. By contrast, the N-terminal residues are highly disordered in isolated CH2. We have hypothesized that removal of the CH2 N-terminal residues may not only increase its stability but also its aggregation resistance. To test this hypothesis we constructed a shortened variant of IgG1 CH2 (CH2s) where the first seven residues of the N-terminus were deleted. We found that the thermal stability of CH2s was increased by 5°C compared to CH2. Importantly, we demonstrated that CH2s is significantly less prone to aggregation than CH2 as measured by Thioflavin T (ThT) fluorescence, turbidity and light scattering. We also found that the CH2s exhibited pH-dependent binding to a soluble single-chain human neonatal Fc receptor (shFcRn) which was significantly stronger than the very weak shFcRn binding to CH2 as measured by flow cytometry. Computer modeling suggested a possible mode of CH2 aggregation involving its N-terminal residues. Therefore, deletion of the N-terminal residues could increase drugability of CH2-based therapeutic candidates. This strategy to increase stability and aggregation resistance could also be applicable to other Ig-related proteins.
Keywords: IgG, Fc, CH2 domain, stability, aggregation resistance
INTRODUCTION
Full-size engineered monoclonal antibodies (mAbs) which typically are composed of an antigen-binding fragment (Fab) and a fragment (Fc) which mediates effector functions have been highly successful biological therapeutics.[1,2,3,4,5,6,7] However, their large size (M.W.: ~150kD) may not allow efficient penetration in solid normal and diseased tissues (e.g. solid tumors) as well as unable to bind to regions on the surface of some molecules (e.g., the HIV envelope glycoprotein (Env)) that are accessible by molecules of smaller size.[8] A number of protein scaffolds based on immunoglobulin (Ig) domains (e.g.: antibody variable domains (Vs)) and non-Ig domains (e.g. the 10th type III domain of human fibronectin) have been developed to overcome these limitations[9] . A major drawback of such scaffolds and corresponding binders is that they lack full-size mAb functions conferred by the Ig Fc which can bind to Fc receptors including the neonatal Fc receptor (FcRn), and is important for extension of half-life and stability in vivo. [10]
CH2 (M.W.: ~12kD) is the penultimate constant domain of immunoglobulins (Igs) (CH2 of IgG, IgA and IgD, and CH3 of IgE and IgM). The isolated, aglycosylated CH2 is monomeric and independently folded domain and its crystal structure was already determined.[11] CH2 domain contains seven β-strands, designated as A through G, and flexible loop regions in between the strands that are similar to the complementarity-determining regions (CDRs) in the antibody Vs. However in contrast to V, CH2 also contains binding sites or portions of binding sites of Fc receptors. Therefore, it has been proposed that CH2 could be a promising scaffold for development of novel candidate therapeutics because it could be engineered to bind to specific antigens and retain its Fc binding related functions occurring, which is one of the important advantages of CH2-based scaffolds compared to other scaffolds with similar size. In a previous study, we selected a binder against the HIV-1 Env gp120 from a library based on the CH2 scaffold.[13] However, we found that most clones aggregated, which constrained the selection of binders with high affinity.[13] We have hypothesized that the N-terminal residues that are not part of the Ig fold and do not form any secondary structure, as can be seen in the crystal structures of an intact IgG1[14] , an Fc/Fc gamma receptor III complex[15] and an isolated CH2[11] , could contribute significantly to the aggregation of CH2. Therefore, we constructed a shortened CH2 variant (denoted as CH2s) by deletion of the first seven N-terminal residues of human IgG1 CH2 preceding the first β-strand as annotated in the IMGT data base.[16] Here, we report the biophysical and biochemical characterization of the CH2s in comparison to the CH2, and investigate the role of the N-terminal residues in the conformation and function of CH2. Our results indicate an important role of these residues in the stability and aggregation resistance, which could also be applicable to other candidate protein therapeutics.
MATERIALS AND METHODS
Construction and expression of CH2 and CH2s
The plasmid for expression of CH2 with C-terminal His-tag (for purification and detection) and FLAG tag (for detection) in Escherichia coli (E. coli) was described previously.[17,18] The same vector was used for construction of CH2s expression plasmid. For small scale expression, E. coli strain HB2151 cells containing the expression plasmids were grown at 37°C in 1 mL SB medium in 14-mL round-bottom tubes (BD Biosciences, CA) to an optical density of OD600 = 0.7 – 1.0. Expression was induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 30°C or 37°C for 12 – 16 hrs. Bacterial cells were harvested and re-suspened in 100 µl PBS (9.0 g/L NaCl, 144 mg/L KH2PO4, 795 mg/L Na2HPO4, pH 7.4). Then, 1 µL Polymyxin B sulfate (Sigma-Aldrich, MO) (0.5 mu/ml) was added to the suspension. After 1 h rotation at room temperature, the cell lysate was clarified by centrifugation at 18,000 × g for 30 min at 4°C. The supernatant was checked by SDS-PAGE to compare the soluble expression levels.
Purification of CH2 and CH2s
CH2 and CH2s were expressed in 2 L SB medium as described above. The bacterial cells were harvested and re-suspended in Buffer A (PBS + 0.5 M NaCl, pH 7.4) at 1:10 (volume of Buffer A: culture volume). Polymyxin B sulfate was added to the suspension (1:1000 volume of polymyxin B sulfate: culture volume). The cell lysate was subsequently clarified by centrifugation at 18,000 × g for 45 min at 4°C. The clarified supernatant was purified by using Ni-NTA agarose (QIAGEN, CA). After elution with Buffer B (Buffer A + 200 mM Imidazole), the imidazole was removed by Amicon Ultra – 15 Centrifugal Filter Devices (MILLIPORE, MA) and the purified proteins were stored in PBS. The proteins were checked for purity by SDS-PAGE and their concentrations were determined by measuring the UV absorbance at 280 nm (GE NanoVue Spectrophotometer, GE Healthcare, USA).
Secondary structures and thermo-stabilities of CH2 and CH2s
The secondary structures of CH2 and CH2s were determined by circular dichroism (CD) spectroscopy. The purified proteins were diluted in PBS (pH7.4) at the final concentration of 0.4 mg/mL, and the CD spectra were recorded on AVIV Model 202 CD Spectrometer (Aviv Biomedical, NJ). Thermo-induced unfolding was monitored at 216 nm by recording the CD signal in the temperature range of 25 – 90°C with heating rate 1°C/min to determine the transition thermal midpoint (Tm). The temperature was recorded with an external probe sensor and the temperature inside the microcuvette was calculated by calibration – it was about 2 – 3oC (range from 1.9°C to 3.8°C for temperatures from 20°C to 80°C) lower that the one measured by the external sensor. After heating, wavelength spectra were recorded at 90°C. For evaluation of the refolding, all the samples were kept at 4°C overnight and measured at 25°C again.
The Tm of each protein was also measured by using the instrument Optim 1000 (Avacta Analytical Ltd, UK). Briefly, proteins were prepared in PBS, pH 7.4 at 0.20 mg/mL. 9 µL aliquots of each sample were loaded into an Optim 1000 compatible micro-cuvette. The instrument was programmed to read fluorescence emission from the sample, excited at 266 nm. The sample temperature was increased from 30 – 85°C at 1°C/min steps and the intensity ratios between the fluorescence at 350 nm and 330 nm were recorded and analyzed to yield Tm.
Thioflavin T (ThT) binding experiment
40 µL 15 µM ThT was added into 40 µL 1 mg/ml protein prepared by filtering through 0.22 µm filter and 10-min centrifuging at 18,000 × g to make final 80 µL working solution. Assay measured the change in fluorescence intensity from 0 to 250 min by excitation at 440 nm and emission at 482 nm at room temperature.
Turbidity assay
Turbidity assay was carried out on CH2 and CH2s similarly prepared in ThT experiment after incubation at 50°C for 0, 5, 10, 20, 30, 60 and 120 min. Turbidity was quantified by recording the absorbance values at 320 nm (Pharmacia Biotech Ultrospec 2000 UV/visible Spectrophotometer, GE Healthcare, USA).
Dynamic light scattering (DLS)
For comparison of aggregation tendency, both CH2 and CH2s were concentrated and filtered through 0.22 µm filter. The concentration was adjusted to 10 mg/mL then 220 µL samples were incubated at 37°C and 4°C respectively. On day 0, day 1, day 3 and day 7, 50 µL samples were taken out and centrifuged at 18,000 × g for 10 min. The supernatants were diluted to 1 mg/mL and 400 µL samples were used for DLS measurement to determine the size of protein particles. The same samples were also used to run size exclusion chromatography (SEC) (Superdex 200 10/300GL, GE healthcare, USA).
Static light scattering
Molecular weight measurements were made using a Zetasizer Nano ZS 3600 (Malvern Instruments Limited, MA) equipped with a 633 nm laser. All manipulations of protein solutions prior to static intensity measurements were performed in a laminar flow hood to minimize the introduction of dust. Dilutions of CH2 and CH2s were made in 0.22 µm filtered PBS from protein stocks filtered with a 0.22 µm syringe filter immediately prior to use and all concentrations centrifuged at 20,800 × g at 25°C for 15 min. Supernatants were transferred to new tubes and measurements made in a low volume quartz cuvette (Hellma Analytics, Germany). Time-averaged intensities of light scatter at a fixed 173° angle were collected for each series of dilutions at 25°C in triplicate using toluene (Sigma-Aldrich) as a scattering standard. The Zetasizer software was used to solve the Rayleigh equation, KC/ΔRθ = 2A2C + 1/MW at each concentration where K is the Debye optical constant calculated using a refractive index increment dn/dC of 0.185 mL/g for proteins, C is the protein concentration in g/mL, and ΔRθ is the difference in the Rayleigh ratio between the protein solution and pure solvent. The averaged values of KC/ΔRθ were fit versus concentration as a Debye plot for each protein and molecular weights calculated from the inverse of the y-intercept.
Construction of recombinant plasmids for yeast surface expression of CH2 and CH2s
CH2 and CH2s were cloned into pYD7 vector which was a modified version from pCTCON2[19] by moving the agglutinin protein aga2p to the C-terminal of interest proteins. The clones were verified by direct sequencing. The constructs were transformed into EBY100 cells for surface expression according to the protocol described previously.[19]
Flow cytometry assay
Yeast cells containing pYD7-CH2 and pYD7-CH2s were grown in SDCAA medium, and the expression was induced in SGCAA medium according to published protocols.[19,20] For FcRn binding measurement, 5×105 yeast cells were harvested, washed by PBSA (PBS + 0.1% bovine serum albumin) (pH 6.0) and re-suspended in 50 µL PBSA (pH 6.0) containing 100 nM biotin-conjugated soluble human FcRn (shFcRn).[21] The samples was kept on ice for 2 hrs, then the cells were washed by PBSA (pH 6.0) again and still re-suspended in 50 µL PBSA (pH 6.0). 1 µL PE-streptavidin (Invitrogen) was added into the re-suspended cells. After incubation on ice for 30 min, the cells were washed by PBSA (pH 6.0) and re-suspend in 0.5 ml PBSA (pH 6.0) for flow cytometry measurement. Same samples prepared in PBSA (pH 7.4) were also measured for comparison of the fluorescence intensity shift with that in pH 6.0. PE-streptavidin was used as negative controls. To determine the expression of CH2 and CH2s on yeast cell surface, the mouse anti-human CH2 monoclonal antibody (Abd sterotec, NC) (primary antibody) and Alexa Fluor 488-conjuated goat anti-mouse IgG (Invitrogen, NY) (secondary antibody) were used for testing at pH 6.0 and pH 7.4‥
Conformational changes of CH2 and CH2s detected by an anti-CH2 antibody
Enzyme-Linked Immunosorbent Assay (ELISA) was used for measurement of the binding of CH2 and CH2s to an anit-human CH2 Fab (m01m1) targeting the conformational epitope in CH2 selected from a human naive phage display library.[20] Briefly, CH2 and CH2s were coated on 96-well plate at the concentration of 140 nM (about 2 µg/mL). After blocking by PBS + 3% Milk, the 1:5 serially diluted m01m1 was added with concentrations from 0 to 3.3 µM. Horseradish peroxidase (HRP) conjugated goat anti-human Fab polyclonal antibody (Sigma-Aldrich, MO) was used as secondary antibody.
Binding to human serum albumin (HSA)
ELISA was used to test the binding of CH2 and CH2s to HSA. HSA was coated on 96-well plate respectively with concentration of 30 nM (2 µg/mL) overnight. The plate was blocked by Pierce® Protein-Free (PBS) Blocking Buffer (Thermo SCIENTIFIC, IL) at 37°C for 1 h. The 1:3 serially diluted CH2 and CH2s in PBS from 0 to 10 µM were added and incubated at 37°C for 2 h. 1:1000 diluted HRP-anti-FLAG (Sigma-Aldrich, MO) in PBS was used as secondary antibody.
Computational sequence and structure-based analyses of CH2
We homology modeled the N-terminal region containing seven residues (APELLGG), which were not seen and disordered as evident from the lack of clear electron density in the previously determined crystal structure of an isolated unglycosylated human antibody CH2 domain at 1.7 Å resolution[11] (PDB 3D9J), using SWISS-MODEL.[22] The N-terminal region from the human IgG Fc structure[23] (PDB 3RY6) was used as a template to build the missing N-terminal residues of the CH2 structure. We used TANGO method[24] for the predication of the aggregation prone regions (APRs) in CH2. Distribution of surface charged, hydrophobic residues and crystal packing at the lattice were analyzed in detail. We computed the solvent accessible surface areas to analyze the surface-exposure of individual residues in CH2 using GETAREA.[25] Structural modeling, analysis and molecular rendering were made with PyMOL.[26]
RESULTS
Expression levels of CH2 and CH2s
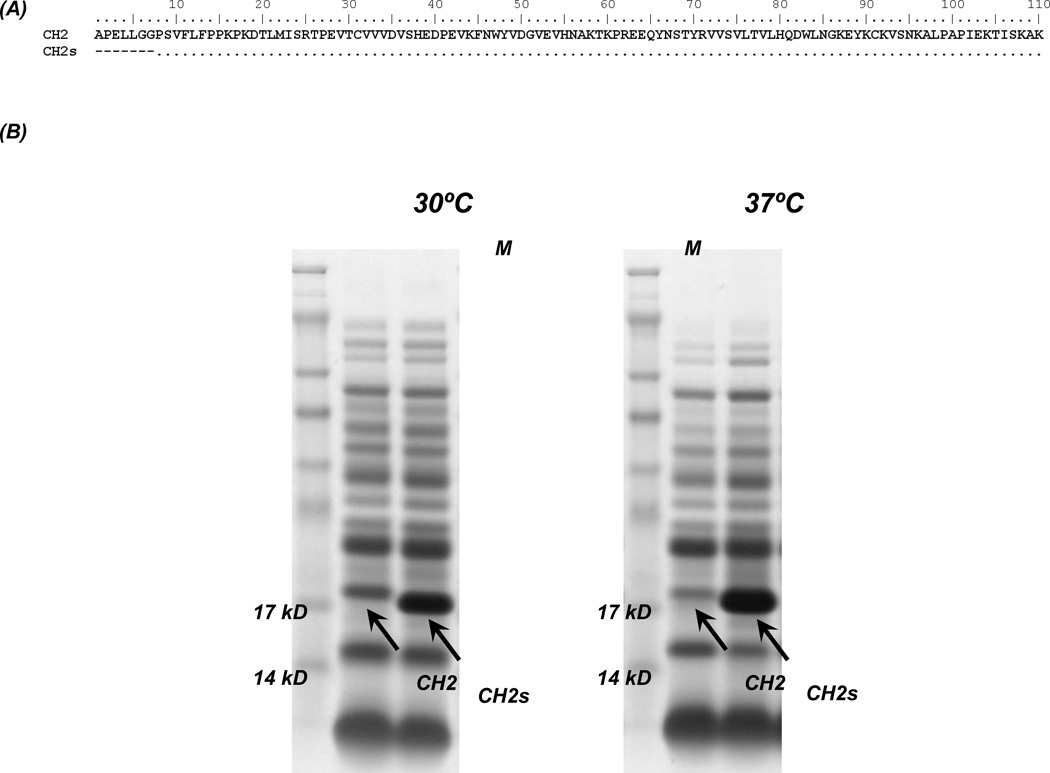
We constructed a shortened version of CH2 (CH2s) by deletion of the first seven N-terminal residues (Figure 1A). The two proteins were expressed in E. coli. We found that soluble expression of CH2s at 30°C and 37°C was much higher than that of CH2 (Figure 1B).
Figure 1.
Design and expression of CH2 and CH2s. (A) Amino acid sequence alignment of wide-type CH2 (NCB Accession No. J00228) with CH2s. (B) Comparison of the soluble expression of CH2 and CH2s at 30°C and 37°C.
Secondary structures and thermo-stabilities of CH2 and CH2s
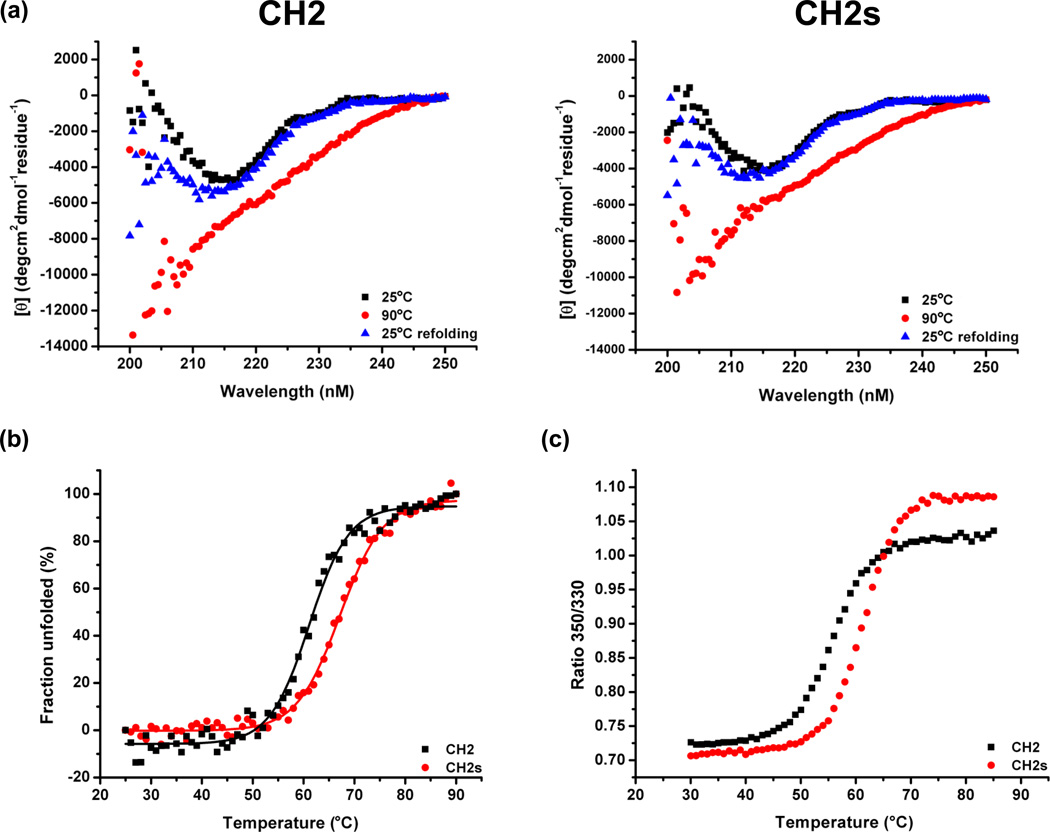
The secondary structure and thermal stability were measured by using CD. Both CH2 and CH2s showed high β-sheet contents at 25°C (Figure 2A). The β-sheet structure was gradually disrupted as the temperature increased (Figure 2B). At 90°C, both proteins were completely unfolded (Figure 2A). The sigmoid curve was fitted by a two-state model which was also previously used[17]. Notably, 50% unfolding of CH2s occurred at 62.5±1.1°C (Tm) which was significantly higher than that of CH2 (57.7±0.5 °C) (Tm values were calibrated after calculation of the raw Tm values from CD data shown in Figure 2B as described above) . The unfolding of both CH2 and CH2s was reversible (Figure 2A).
Figure 2.
Secondary structure and stability of CH2 and CH2s measured by circular dichroism (CD). (A) Folding curve at 25°C (black), unfolding at 90°C (red) and refolding (blue) at 25°C. (B) The fraction folded of the protein (ff) was calculated as ff = ([θ] -[θM])/([θT] -[θM]). [θT] and [θM] were the mean residue ellipticities at 216 nm of folded state at 25°C and unfolded state of 90°C. The Tm values (57.7°C and 62.5°C for CH2 and CH2s respectively) from were determined by the first derivative [d(Fraction folded)/dT] with respect to temperature (T). (C) Temperature induced unfolding of CH2 and CH2s. The fluorescence intensity ratio of 350/330 indicated that tryptophan residues were exposed as the CH2 and CH2s unfolded. The calculated Tm values were 56.2°C and 61.3°C for CH2 and CH2s respectively.
The temperature induced unfolding of CH2 and CH2s was also monitored in Avacta Optim1000 instrument (Figure 2C). The measurement is based on the fact that proteins that contain aromatic amino acids fluoresce when excited by ultra-violet light, and tryptophan residues that are exposed to water (on unfolded proteins) have maximal fluorescence at a wavelength of about 340–350 nm, whereas totally buried residues (inside native proteins) fluoresce at about 330 nm. Therefore, fluorescence intensity ratio of 350:330 nm reflects the change of high order protein structure during temperature ramps. We found that both proteins displayed gradual increases in the intensity ratio of 350:330 nm but with different Tms. The calculated Tm for CH2 was 56.2°C, whereas for CH2s was 61.3°C, which is consistent with that measure by CD.
Aggregation propensity
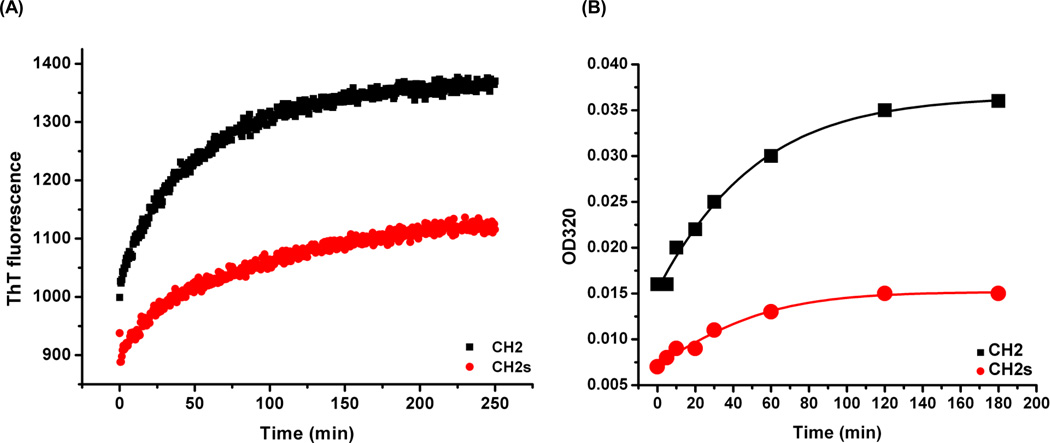
Three different methods were used to measure the aggregation propensity of CH2 and CH2s: ThT fluorescence, turbidity and DLS. It has been reported that ThT binding could be used to evaluate antibody aggregation.[27] We found that the fluorescence intensity and the rate of increase of fluorescence intensity in CH2 after binding to ThT were higher and faster than that in CH2s (Figure 3A), indicating that there are more CH2 aggregates and they form faster than that in the case of CH2s.
Figure 3.
Comparison of aggregation propensity between CH2 and CH2s. (A) ThT binding experiment at room temperature. The rate of increase of fluorescence intensity in CH2 (black) was faster than that in CH2s (red). (B) Turbidity assay of CH2 (black) and CH2s (red) after 50°C incubation at different time points. OD320 of CH2 increased faster than that for CH2s.
We also used a relatively simple turbidity assay for evaluation of aggregation propensity. OD320 was measured after different times of incubation at 50 °C (Figure 3B). The initial absorbance at OD320 in CH2 was higher than that in CH2s, which indicated larger particles have already formed after purification of CH2. The rate of increase of the absorbance at 320-nm for CH2 was also higher than that of CH2, which was consistent with the data obtained by measuring the ThT fluorescence.
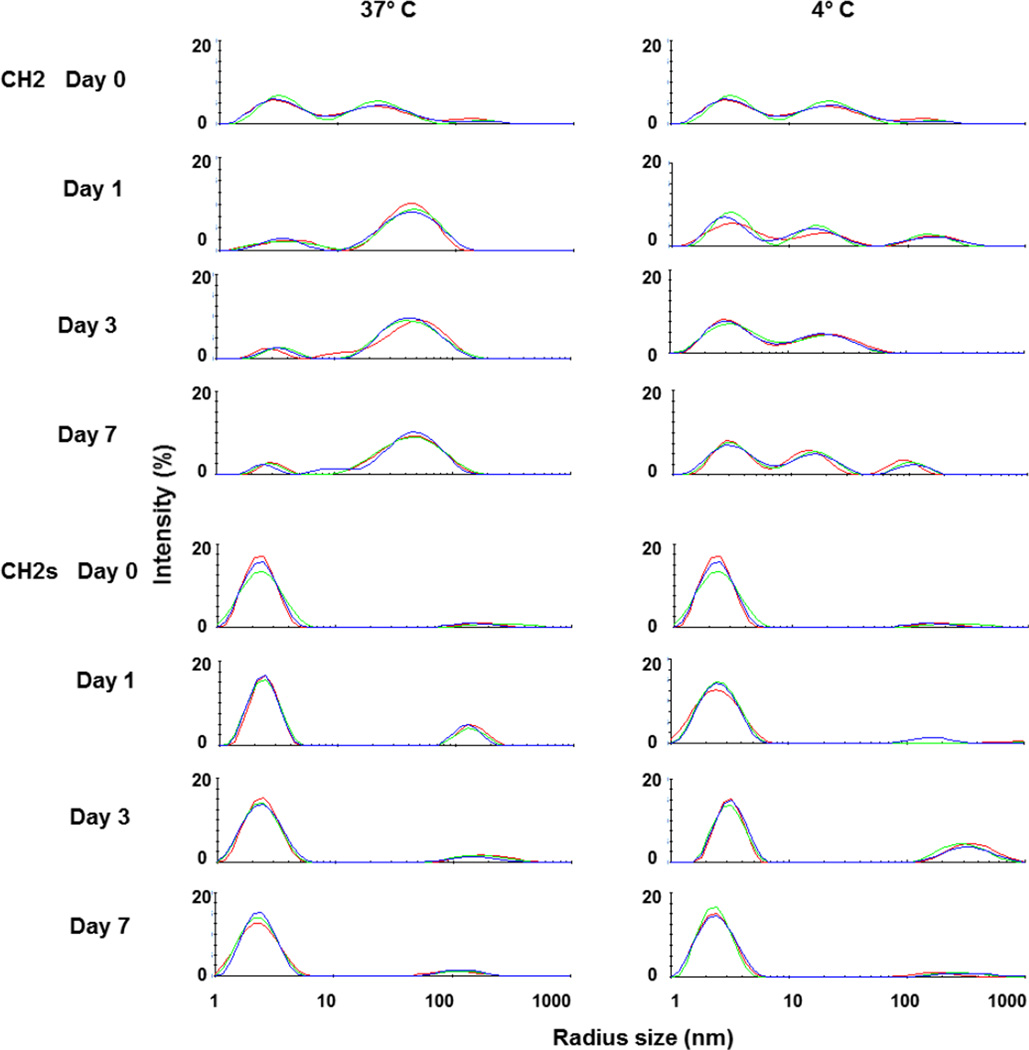
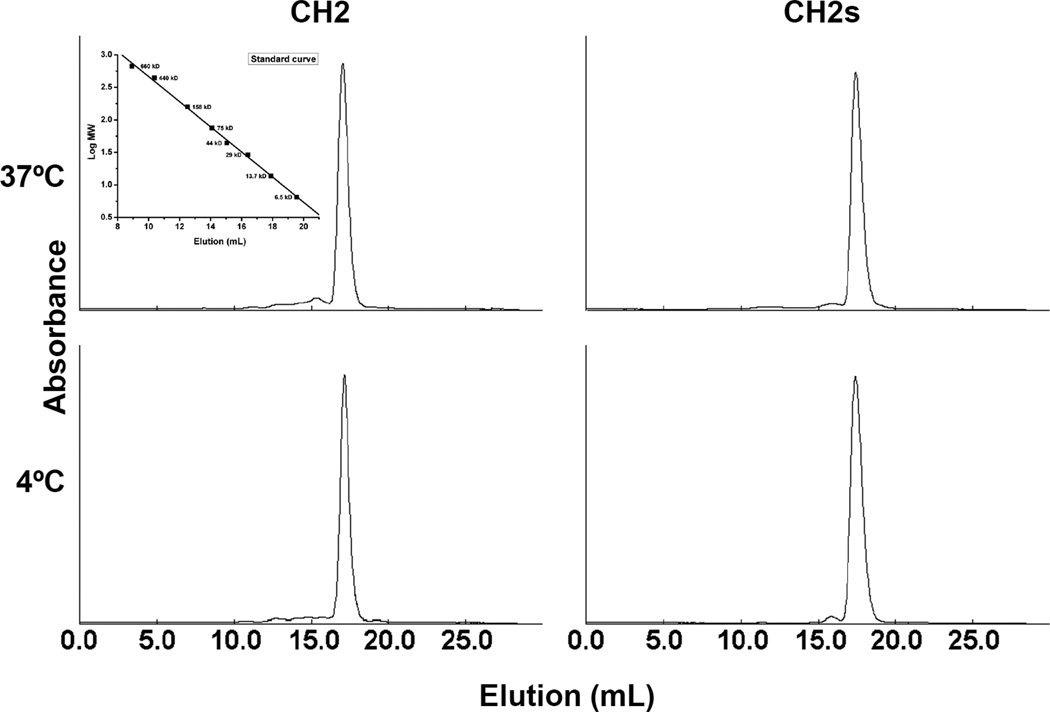
Finally, we used DLS to monitor the formation of soluble oligomers (Figure 4). We observed two major peaks at day 0 (before incubation at 37°C) for CH2 but only one for CH2s. After incubation at 37°C, the peaks from the CH2 solution shifted to larger sizes indicating formation of large soluble oligomers. In contrast to CH2, there was only slight increase in the peaks reflecting large CH2s which were very small. At day 7, there was large amount of precipitates formed in CH2 and the sample became cloudy while CH2s was still clear with no visible precipitates formed. The large particles formed more slowly at 4°C compared to 37°C. Interestingly, after 7-day incubation at 37°C and 4°C, the supernatants from CH2 and CH2s were still monomeric according to the results obtained by using SEC (Figure 5) which indicates that the formation of the soluble oligomers might be reversible and the oligomers disassociate to monomers in diluted condition when running SEC. The predicted monomer contributions based on DLS intensity measurements for CH2 and CH2s are given in Table 1.
Figure 4.
Measurement of CH2 and CH2s aggregation after incubation at 37°C and 4°C by dynamic light scattering (DLS). CH2 had two major peaks at day 0 while CH2s had only one peak indicating that the populations of CH2 molecules were not uniform and reflected possible oligomer formation after purification.
Figure 5.
Estimation of oligomer formation of CH2 and CH2s after 7-day incubation at 37°C and 4°C by SEC. Only monomeric peak was observed in both CH2 and CH2s indicating that the formation of oligomer might be reversible. The insertion is standard curve.
Table 1.
The contribution of monomer fraction predicted based on dynamic light scattering (DLS) intensity measurements for CH2 and CH2s. Numbers in parentheses denote the average size.
| Temp. | Days | Size (nm) CH2 |
Concentration (mg/mL) CH2 |
Size (nm) CH2s |
Concentration (mg/mL) CH2s |
% of Monomer | |
|---|---|---|---|---|---|---|---|
| CH2 | CH2s | ||||||
| 37° C | 0 | 7.1(17.62) | 1.0 | 4.8(9.052) | 1.0 | 45.5 | 89 |
| 1 | 6.9(54.90) | 1.5 | 4.8(42.24) | 1.1 | 17.5 | 79.8 | |
| 3 | 6.7(49.48) | 1.2 | 5.0(5.514) | 1.1 | 11.9 | 86.3 | |
| 7 | 5.1(51.20) | 1.0 | 4.9(5.726) | 1.0 | 8.1 | 84.9 | |
| 4° C | 0 | 7.1(17.62) | 1.0 | 4.8(9.052) | 1.0 | 45.5 | 89 |
| 1 | 6.5(32.96) | 1.2 | 5.3(5.264) | 1.0 | 42.9 | 89.3 | |
| 3 | 7.1(10.99) | 1.2 | 8.5(9.255) | 0.8 | 55.6 | 76.5 | |
| 7 | 7.1(75.33) | 1.3 | 5.1(5.187) | 1.0 | 51.6 | 90.4 | |
The average molecular weight (M.W.) of CH2 and CHs were measured with static light scattering method. CH2 was estimated to be 24.9 kD, which is much larger than the calculated M.W. (14.7 kD) based on its amino acid sequence. In contrast, the average M.W. of CH2s was 13.7 kD which is consistent with the calculated M.W. (14.0 kD).
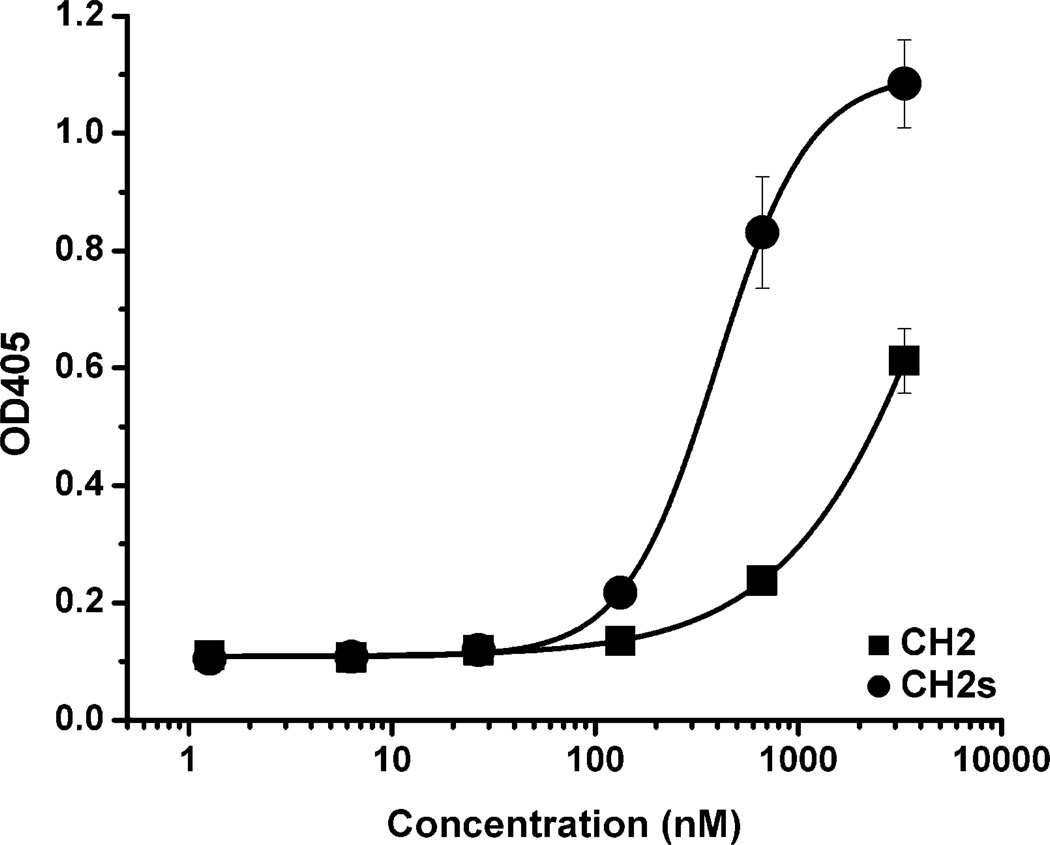
Conformational changes
To determine whether shortening of CH2 resulted in conformational changes, we used an anti-human CH2 Fab, m01m1, specific for a conformational epitope in CH2.[20] Fab m01m1 bound to CH2 with relatively low affinity (EC50 > 1400 nM) (Figure 6) as reported previously.[20] However, it bound significantly better to CH2s with an EC50 of 392 nM (Figure 6). These results indicate that there are certain differences between the conformations of CH2 and CH2s.
Figure 6.
Binding of CH2 (■) and CH2s (●) to an anti-human CH2 Fab m01m1. The EC50s of m01m1 to CH2 and CH2s were >1400 nM and 392 nM, respectively, indicating conformational differences.
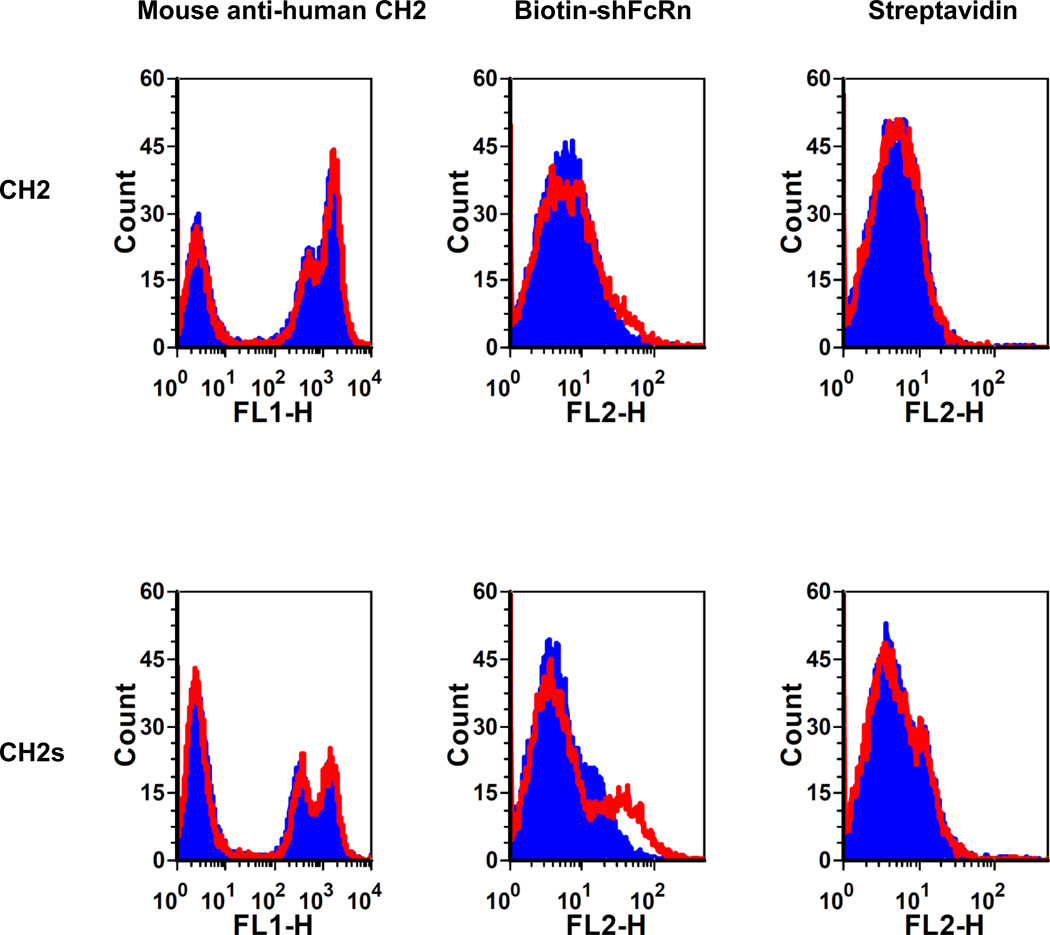
CH2s binds to shFcRn at pH 6.0 better than CH2
Recently, we developed methods to produce shFcRn in mammalian cells with high yield [28] and measure the shFcRn binding to proteins expressed on yeast cell surface[20]. We used the same methods to test CH2 and CH2s binding to shFcRn at pH 6.0 and pH7.4 (Figure 7). The fluorescence intensity shift for CH2s was larger than that for CH2, indicating stronger binding of CH2s to shFcRn than that of CH2 although both were relatively weak.
Figure 7.
Binding of yeast-expressed CH2 and CH2s to shFcRn at pH6 (red) and pH7.4 (blue). Very slight fluorescence intensity shift occurred in the case of CH2 indicating very weak binding to shFcRn. Stronger fluorescence intensity shift was observed in the case of CH2s indicating enhanced binding to shFcRn. The expression of CH2 and CH2s was tested by the corresponding antibodies. PE-streptavidin was used as negative control.
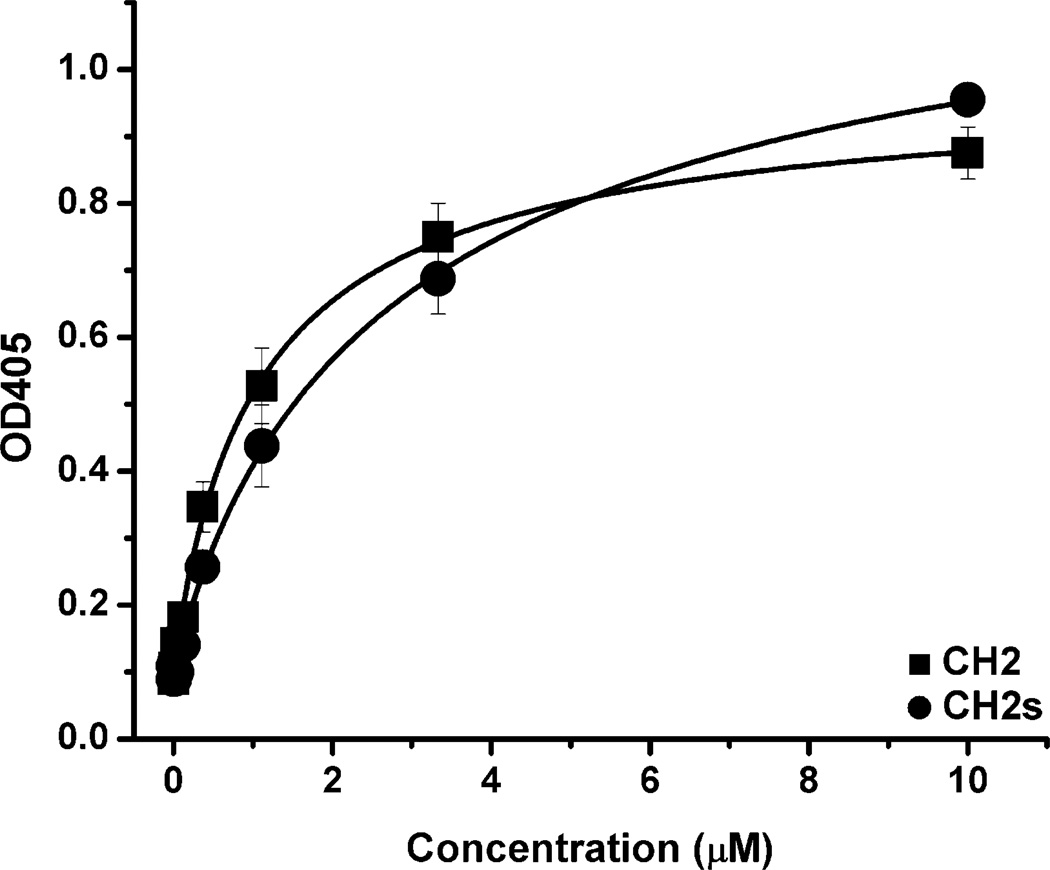
Non-specific binding of CH2 and CH2s to human serum albumin (HSA)
In order to explore the relationship between aggregation and non-specific binding, we also compared the binding of CH2 and CH2s to HSA (Figure 8). Both CH2 and CH2s could bind to HSA with low EC50. The EC50 of CH2 to HSA (0.9 µM) was slightly lower than that of CH2s (1.7 µM) indicating that decrease of aggregation could also lead to decrease of non-specific binding to HSA.
Figure 8.
Binding of CH2 and CH2s to human serum albumin (HSA) measured by ELISA. (■) binding of CH2 to HSA (EC50 ≈ 0.9 µM); (●) binding of CH2s to HSA (EC50 ≈ 1.7 µM).
Prediction of aggregation prone regions (APRs) in CH2
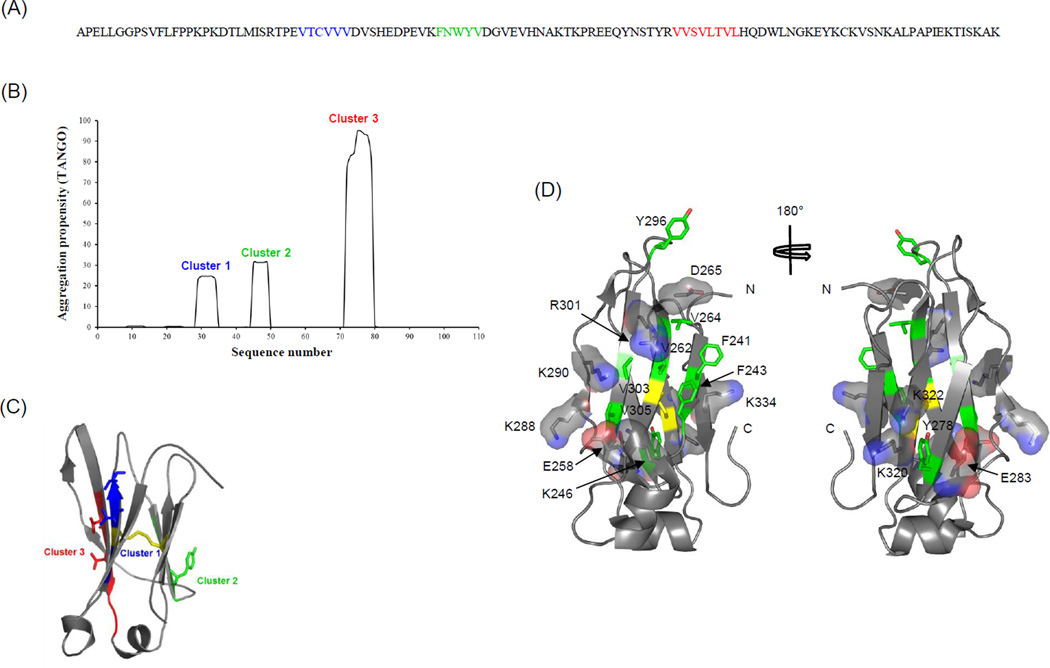
Three major APRs (cluster1–3) in CH2 predicted by TANGO method are shown in blue, green and red in Figure 9A. All the three clusters have relatively high percent of hydrophobic amino acids and the third cluster has the highest aggregation propensity of more than 90% (Figure 9B). In the three APRs, certain hydrophobic residues point out at the surface of strands B, C and E as shown in blue, green and red respectively (Figure 9C), indicating that these residues could pose potential risk of misfolding and aggregation. Cluster 3 is the biggest cluster and locates at the same side as cluster 1 while cluster 2 is at the other side (Figure 9C).
Figure 9.
Prediction of aggregation prone regions (APRs) in CH2. (A) Three major APRs, cluster 1–3, are mapped on the CH2 amino acid sequence and highlighted by blue, green and red, respectively, as predicted by TANGO method. (B) The TANGO profile for CH2 showing the sequence number on X-axis and aggregation propensity (%) on Y-axis. (C) Mapping of APRs, clusters 1–3, on the three-dimensional structure of CH2 domain. (D) Ribbon diagrams of CH2 structure illustrating the distribution of charged residues as sticks embedded in translucent surfaces, and hydrophobic residues in green sticks with atom colors are shown. The location of disulfide bond is colored in yellow.
Roles of surface charged and hydrophobic residues involving APRs in CH2
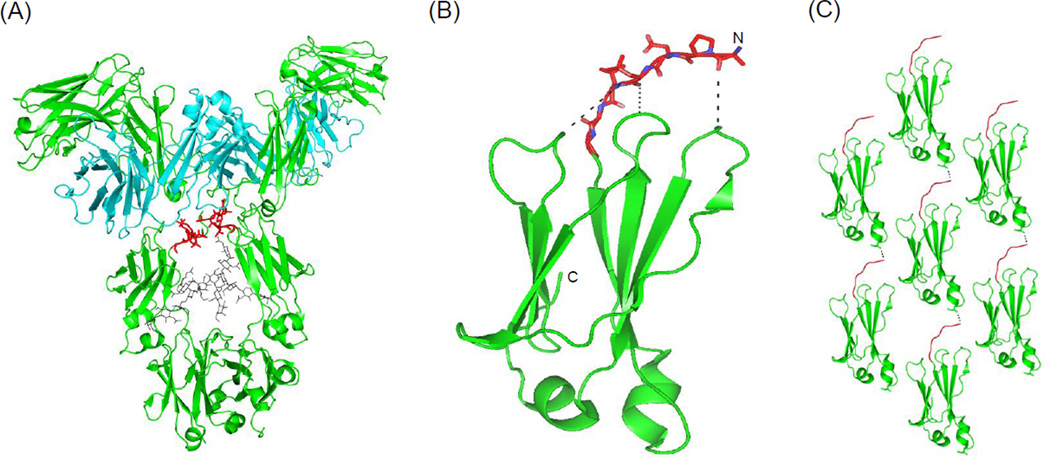
The predicted APRs were further analyzed in structural context by mapping the APRs containing residues on the crystal structure of CH2 domain. The APR clusters 1 and 3 have hydrophobic residues located at one side of the β-sheet -V262 and V264 in strand B, and V303 and V305 in strand E as shown in Figure 9D. The APR cluster 2 has a residue Y278 in strand C at the other side of the β-sheet. The analysis of CH2 structure revealed several surface-exposed and charged residues covering those hydrophobic residues in the three APRs (Figure 9D). The ARP clusters 1 and 3 were protected by positively charged residues K246, K288, K290 and R301, and a negatively charged residue E258. On the outer surface of other β-sheet, the charged residues K320, K322 and E283 surrounded the Y278. Further, we found a few hydrophobic residues, including Y296 in the CD loop, F241 and F243 in the strand A, and I253 and Leu309 at the helical regions, which are partially exposed on the surface. To understand the molecular mechanisms of the role of the N-terminal residues of CH2 in aggregation, we modeled the seven N-terminal residues (APELLGG) that are undefined in the CH2 X-ray crystal structure. We found that the modeled N-terminal residues were compatible in the crystal packing as they did not create any clashes with symmetry related molecules. Therefore, these residues could be the probable N-terminus location of the CH2 domain. We compared the conformation of the N-terminal residues of CH2 domain from an IgG structure[14] along with the modeled N-terminal residues. In the full-length IgG, the N-terminal residues of CH2 domains from the two heavy chains interact together which prevents the solvent exposure of these residues (Figure 10A). By contrast, the N-terminal residues of isolated CH2 domains were likely to be involved in intra-and inter-molecular contacts contributing to the CH2 aggregation (Figure 10B and 10C).
Figure 10.
(A) An intact IgG crystal structure is shown for highlighting the N-terminal residues of CH2 domains, colored in red sticks (PDB code 1HZH). The carbohydrates moieties are shown in gray sticks. (B) A ribbon cartoon diagram of the CH2 domain with modeled N-terminal residues (in orange sticks and colored by atoms) showing the proximity of the N-terminal residues to the loop regions of CH2. (C) A crystal packing view of CH2 monomers in the lattice indicates intermolecular contacts between the N-terminal residues and helical region of different CH2 monomers as shown by dots. Surface-exposed hydrophobic residue Leu253 at a helical region involved in the intermolecular interaction is shown as sticks.
DISCUSSION
Currently, there are a number of protein scaffolds of relatively small size (typically from 10 kD to 20 kD) under development because of their distinct advantages over conventional monoclonal antibodies against certain targets. Heavy chain variable domains (VHs) and CH2s have been extensively characterized.[8,12,29] It was found that VHs tend to aggregate[30,31] and significant amount of work has been done to reduce the VH aggregation.[32,33,34,35] CH2 is involved in Fc unfolding and aggregation.[36] Mutation of several CH2 residues could reduce aggregation of full length IgG.[37] However, aggregation of isolated CH2 domains has not been studied. Furthermore, it has been reported that removal of the N-terminal hexapeptide from human β2-microglobulin could increase protein aggregation,[38] which indicates that the N-terminal residues of isolated domain may also play a role in the aggregation. Altogether, it is important to understand the aggregation of CH2 and its prevention for developing of CH2-based therapeutics.
The isolated CH2 domains have been proposed as novel scaffolds for library construction and selection of binders against specific antigens.[8] The thermo-stability of CH2 is relatively lower than other scaffolds with similar size such as the tenth type III domain of human fibronectin,[17,39,40] and the pH-dependent binding of CH2 to shFcRn is very weak.[20] In this work, we used CH2 and a short variant (CH2s) lacking the seven residues (APELLGG) at the N-terminus to study their aggregation, stability and possible correlations to their sequence, structure and functional properties. Despite the exposure of hydrophobic residues due to lack of glycosylation in E. coli CH2, we had successfully crystallized monomeric CH2 along with 30% PEG 1500 with equal volume of protein.[11] It is known that PEG can be used as an effective additive for preventing protein aggregation. However, subsequent efforts for selection of efficient CH2-based binders with desired physicochemical properties were not as effective as selection of VH-based binders mostly due to the intrinsic aggregation propensity of the CH2 domain. We used computational prediction models of APRs including the TANGO method, which suggested several facial hydrophobic residues including mainly valine residues on the outer surfaces of the β-sheets (Figure 9C). However, close examination of the crystal structure of CH2 revealed properly placed charged residues that largely protect those hydrophobic residues predicted as APRs by covering over them (Figure 9D). The edges of the β-sheets in monomeric CH2 were also well-protected by lysine residues, K288 and K334 as nature’ design for avoiding edge-to-edge aggregation.[41] The calculated solvent accessible areas indicated contributions by some key hydrophobic residues, F241, F243 and Y296, which are partly exposed to the surface (Figure 9D). In addition, one of the residues from the cluster 3, L309, was found with multiple side-chain conformations rendering potentially a hydrophobic patch. Previously, it was clearly shown that a point mutation L309K in the CH2 domain changed the APR which led to a reduced aggregation propensity and improved thermal stability.[42] The specific L309K point mutation was consistently predicted to reduce the aggregation propensity in different methods.[43] Although most of the hydrophobic residues are covered by charged residues and not fully exposed, some of those residues might need to be engineered since any disruption of charged residues during random mutation and selection processes involved in the design of CH2-based binders could lead to aggregating themselves and/or non-specific binding to targets. In addition to the TANGO predicted residues, we noted two leucine residues in the N-terminal region which were not resolved in the crystal structure but most likely are solvent exposed in the monomer CH2. Generally, although the N-terminal region of isolated CH2 does not possess any regular secondary structure, the N-terminal regions of CH2 domains in the intact IgG or Fc molecules are found interacting with each other and form the binding site for Fcγ receptors.[14,15] We speculated that the N-terminal region of monomeric CH2 in the absence of such conserved structural and functional interactions would lead to aggregation. Therefore, to analyze the role of N-terminal residues, we modeled the missed N-terminal residues of the isolated CH2 domain that could be accommodated into the crystal lattice. We visualized the proximity of the N-terminal residues to other residues, intramolecular contacts, in the CH2 as well as intermolecular interactions observed in crystal packing (Figure 10B and 10C respectively). The assigned probable location of the N-terminal residues was found at a close proximity (within 8–12 Å) to several residues of CH2 loop regions at the top (Figure 10B). Due to the flexible nature of a couple of glycine residues in the N-terminal region, it is quite reasonable to expect that the N-terminal region could interfere with the proper folding of the CH2 loops resulting into aggregation-prone intermediates as recently observed in the folding pathways of the α-spectrin SH3 domain.[44] On the other hand, we noticed that the N-terminal residues of the CH2 monomer could make intermolecular contacts with other monomers in the crystal lattice. For instance, the N-terminal region of one CH2 monomer was found to be in close proximity with the helical region of neighboring CH2 where a solvent-exposed hydrophobic residue I253 was present as shown in sticks (Figure 10C). This could explain the possible role of the N-terminal residues in creating the hydrophobic surface interactions that would lead to aggregation. A similar observation was previously made as suggestive to amyloidogenic properties as the N-terminal region of the β-2-microglobulin monomer was found to interact with the D-strand of another monomer that lost protection in the isolated form.[45] Thus, the N-terminal residues of CH2 were most likely to play potential roles in the CH2 intra-and or inter-molecular contacts contributing to its aggregation propensity. Accordingly, this could explain the fact that after removal of the N-terminal seven residues, CH2s showed significantly increased stability and aggregation resistance compared to CH2. Furthermore, the binding of CH2s to FcRn was also enhanced.
Our results showed that the removal of the N-terminal residues of the IgG1 CH2 domains increased stability and aggregation resistance. This could not only be used for development of CH2-based therapeutics, but could be also useful for exploration of IgGs and Fc-related fusion proteins in treatment of human diseases. For example, IgG molecules with specific mutations that prevent Fc gamma receptor (FcγR) binding have proved particularly valuable in the treatment of autoimmune diseases and transplant rejection, as they retain the antigen specificity of the antibody but are unable to trigger unwanted immune-cell reactions, such as neutrophil degranulation.[46,47] Because of the absence of the N-terminal residues in CH2 that are important for FcγR[47], the CH2s and shortened Fcs could possibly lack binding to FcγR and still retain FcRn binding activity.
ACKNOWLEDGEMENTS
We thank Dr. Sergey G. Tarasov and Ms. Marzena A. Dyba of the Biophysics Resource in the Structural Biophysics Laboratory, NCI-Frederick for technical help and the Wuhan Key Laboratory on Emerging Infectious Diseases and Biosafety for support. This work was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
REFERENCES
- 1.Waldmann TA. Immunotherapy: past, present and future. Nat Med. 2003;9(3):269–277. doi: 10.1038/nm0303-269. [DOI] [PubMed] [Google Scholar]
- 2.Casadevall A, Dadachova E, Pirofski LA. Passive antibody therapy for infectious diseases. Nature reviews Microbiology. 2004;2(9):695–703. doi: 10.1038/nrmicro974. [DOI] [PubMed] [Google Scholar]
- 3.Schrama D, Reisfeld RA, Becker JC. Antibody targeted drugs as cancer therapeutics. Nature reviews Drug discovery. 2006;5(2):147–159. doi: 10.1038/nrd1957. [DOI] [PubMed] [Google Scholar]
- 4.Carter PJ. Potent antibody therapeutics by design. Nat Rev Immunol. 2006;6(5):343–357. doi: 10.1038/nri1837. [DOI] [PubMed] [Google Scholar]
- 5.Dimitrov DS, Marks JD. Therapeutic antibodies: current state and future trends--is a paradigm change coming soon? Methods Mol Biol. 2009;525(1–27):xiii. doi: 10.1007/978-1-59745-554-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dimitrov DS. Therapeutic antibodies, vaccines and antibodyomes. MAbs. 2010;2(3):347–356. doi: 10.4161/mabs.2.3.11779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu Z, Prabakaran P, Chen W, Broder CC, Gong R, Dimitrov DS. Human monoclonal antibodies as candidate therapeutics against emerging viruses and HIV-1. Virologica Sinica. 2013;28(2):71–80. doi: 10.1007/s12250-013-3313-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dimitrov DS. Engineered CH2 domains (nanoantibodies) mAbs. 2009;1(1):26–28. doi: 10.4161/mabs.1.1.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nuttall SD, Walsh RB. Display scaffolds: protein engineering for novel therapeutics. Curr Opin Pharmacol. 2008;8(5):609–615. doi: 10.1016/j.coph.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Roopenian DC, Akilesh S. FcRn: the neonatal Fc receptor comes of age. Nat Rev Immunol. 2007;7(9):715–725. doi: 10.1038/nri2155. [DOI] [PubMed] [Google Scholar]
- 11.Prabakaran P, Vu BK, Gan J, Feng Y, Dimitrov DS, Ji X. Structure of an isolated unglycosylated antibody C(H)2 domain. Acta Crystallogr D Biol Crystallogr. 2008;64(Pt 10):1062–1067. doi: 10.1107/S0907444908025274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gong R, Chen W, Dimitrov DS. Candidate antibody-based therapeutics against HIV-1. BioDrugs. 2012;26(3):143–162. doi: 10.2165/11631400-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao X, Feng Y, Vu BK, Ishima R, Dimitrov DS. A large library based on a novel (CH2) scaffold: identification of HIV-1 inhibitors. Biochem Biophys Res Commun. 2009;387(2):387–392. doi: 10.1016/j.bbrc.2009.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saphire EO, Parren PW, Pantophlet R, Zwick MB, Morris GM, Rudd PM, et al. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science. 2001;293(5532):1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- 15.Sondermann P, Huber R, Oosthuizen V, Jacob U. The 3.2-A crystal structure of the human IgG1 Fc fragment-Fc gammaRIII complex. Nature. 2000;406(6793):267–273. doi: 10.1038/35018508. [DOI] [PubMed] [Google Scholar]
- 16.Lefranc MP, Pommie C, Kaas Q, Duprat E, Bosc N, Guiraudou D, et al. IMGT unique numbering for immunoglobulin and T cell receptor constant domains and Ig superfamily C-like domains. Developmental and comparative immunology. 2005;29(3):185–203. doi: 10.1016/j.dci.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Gong R, Vu BK, Feng Y, Prieto DA, Dyba MA, Walsh JD, et al. Engineered human antibody constant domains with increased stability. The Journal of biological chemistry. 2009;284(21):14203–14210. doi: 10.1074/jbc.M900769200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong R, Chen W, Dimitrov DS. Expression, purification, and characterization of engineered antibody CH2 and VH domains. Methods Mol Biol. 2012;899:85–102. doi: 10.1007/978-1-61779-921-1_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baker R, Holloway J, Holtkamp CC, Larsson A, Hartman LC, Pearce R, et al. Effects of multi-sensory stimulation for people with dementia. J Adv Nurs. 2003;43(5):465–477. doi: 10.1046/j.1365-2648.2003.02744.x. [DOI] [PubMed] [Google Scholar]
- 20.Gong R, Wang Y, Feng Y, Zhao Q, Dimitrov DS. Shortened engineered human antibody CH2 domains: increased stability and binding to the human neonatal Fc receptor. The Journal of biological chemistry. 2011;286(31):27288–27293. doi: 10.1074/jbc.M111.254219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holtkamp M, Matzen J, Buchheim K, Walker MC, Meierkord H. Furosemide terminates limbic status epilepticus in freely moving rats. Epilepsia. 2003;44(9):1141–1144. doi: 10.1046/j.1528-1157.2003.14003.x. [DOI] [PubMed] [Google Scholar]
- 22.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 23.Ramsland PA, Farrugia W, Bradford TM, Sardjono CT, Esparon S, Trist HM, et al. Structural basis for Fc gammaRIIa recognition of human IgG and formation of inflammatory signaling complexes. J Immunol. 2011;187(6):3208–3217. doi: 10.4049/jimmunol.1101467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol. 2004;22(10):1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 25.Fraczkiewicz R, Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J Comput Chem. 1998;19(3):319–333. [Google Scholar]
- 26.Alam SM, Travers PJ, Wung JL, Nasholds W, Redpath S, Jameson SC, et al. T-cell-receptor affinity and thymocyte positive selection. Nature. 1996;381(6583):616–620. doi: 10.1038/381616a0. [DOI] [PubMed] [Google Scholar]
- 27.Kayser V, Chennamsetty N, Voynov V, Helk B, Trout BL. Conformational stability and aggregation of therapeutic monoclonal antibodies studied with ANS and Thioflavin T binding. mAbs. 2011;3(4):408–411. doi: 10.4161/mabs.3.4.15677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holthuis JC, van Meer G, Huitema K. Lipid microdomains, lipid translocation and the organization of intracellular membrane transport (Review) Mol Membr Biol. 2003;20(3):231–241. [PubMed] [Google Scholar]
- 29.Holliger P, Hudson PJ. Engineered antibody fragments and the rise of single domains. Nature biotechnology. 2005;23(9):1126–1136. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- 30.Ewert S, Huber T, Honegger A, Pluckthun A. Biophysical properties of human antibody variable domains. J Mol Biol. 2003;325(3):531–553. doi: 10.1016/s0022-2836(02)01237-8. [DOI] [PubMed] [Google Scholar]
- 31.Holt R. The new subspecialty of metabolic medicine. Diabetes, obesity & metabolism. 2003;5(5):356. [PubMed] [Google Scholar]
- 32.Holt R. Orlistat reduces features of the metabolic syndrome: the XENDOS study. Diabetes, obesity & metabolism. 2003;5(5):356. [PubMed] [Google Scholar]
- 33.Holt R. The Food and Agriculture Organization/World Health Organization expert report on diet, nutrition and prevention of chronic diseases. Diabetes, obesity & metabolism. 2003;5(5):354. [PubMed] [Google Scholar]
- 34.Holtzapffel RC, Castelli J, Finnegan PM, Millar AH, Whelan J, Day DA. A tomato alternative oxidase protein with altered regulatory properties. Biochim Biophys Acta. 2003;1606(1–3):153–162. doi: 10.1016/s0005-2728(03)00112-9. [DOI] [PubMed] [Google Scholar]
- 35.Bowmaker M, Yang MY, Yasukawa T, Reyes A, Jacobs HT, Huberman JA, et al. Mammalian mitochondrial DNA replicates bidirectionally from an initiation zone. J Biol Chem. 2003;278(51):50961–50969. doi: 10.1074/jbc.M308028200. [DOI] [PubMed] [Google Scholar]
- 36.Latypov RF, Hogan S, Lau H, Gadgil H, Liu D. Elucidation of acid-induced unfolding and aggregation of human immunoglobulin IgG1 and IgG2 Fc. J Biol Chem. 2012;287(2):1381–1396. doi: 10.1074/jbc.M111.297697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Revesz T, Holton JL. Anatamopathological spectrum of tauopathies. Movement disorders : official journal of the Movement Disorder Society. 2003;18(Suppl 6):S13–S20. doi: 10.1002/mds.10558. [DOI] [PubMed] [Google Scholar]
- 38.Wongworawat MD, Holtom P, Learch TJ, Fedenko A, Stevanovic MV. A prolonged case of Mycobacterium marinum flexor tenosynovitis: radiographic and histological correlation, and review of the literature. Skeletal Radiol. 2003;32(9):542–545. doi: 10.1007/s00256-003-0636-y. [DOI] [PubMed] [Google Scholar]
- 39.Hackel BJ, Kapila A, Wittrup KD. Picomolar affinity fibronectin domains engineered utilizing loop length diversity, recursive mutagenesis, and loop shuffling. J Mol Biol. 2008;381(5):1238–1252. doi: 10.1016/j.jmb.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feige MJ, Walter S, Buchner J. Folding mechanism of the CH2 antibody domain. J Mol Biol. 2004;344(1):107–118. doi: 10.1016/j.jmb.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 41.Richardson JS, Richardson DC. Natural beta-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc Natl Acad Sci U S A. 2002;99(5):2754–2759. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chennamsetty N, Voynov V, Kayser V, Helk B, Trout BL. Design of therapeutic proteins with enhanced stability. Proc Natl Acad Sci U S A. 2009;106(29):11937–11942. doi: 10.1073/pnas.0904191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buck PM, Kumar S, Wang X, Agrawal NJ, Trout BL, Singh SK. Computational methods to predict therapeutic protein aggregation. Methods Mol Biol. 2012;899:425–451. doi: 10.1007/978-1-61779-921-1_26. [DOI] [PubMed] [Google Scholar]
- 44.Krobath H, Estacio SG, Faisca PF, Shakhnovich EI. Identification of a Conserved Aggregation-Prone Intermediate State in the Folding Pathways of Spc-SH3 Amyloidogenic Variants. J Mol Biol. 2012;422(5):705–722. doi: 10.1016/j.jmb.2012.06.020. [DOI] [PubMed] [Google Scholar]
- 45.Trinh CH, Smith DP, Kalverda AP, Phillips SE, Radford SE. Crystal structure of monomeric human beta-2-microglobulin reveals clues to its amyloidogenic properties. Proc Natl Acad Sci U S A. 2002;99(15):9771–9776. doi: 10.1073/pnas.152337399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Proctor R, Holt JT, Allen JI, Blackford J. Nutrient fluxes and budgets for the North West European Shelf from a three-dimensional model. The Science of the total environment. 2003:314–316. 769–785. doi: 10.1016/s0048-9697(03)00083-4. [DOI] [PubMed] [Google Scholar]
- 47.Rosenwald A, Wright G, Leroy K, Yu X, Gaulard P, Gascoyne RD, et al. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J Exp Med. 2003;198(6):851–862. doi: 10.1084/jem.20031074. [DOI] [PMC free article] [PubMed] [Google Scholar]