Abstract
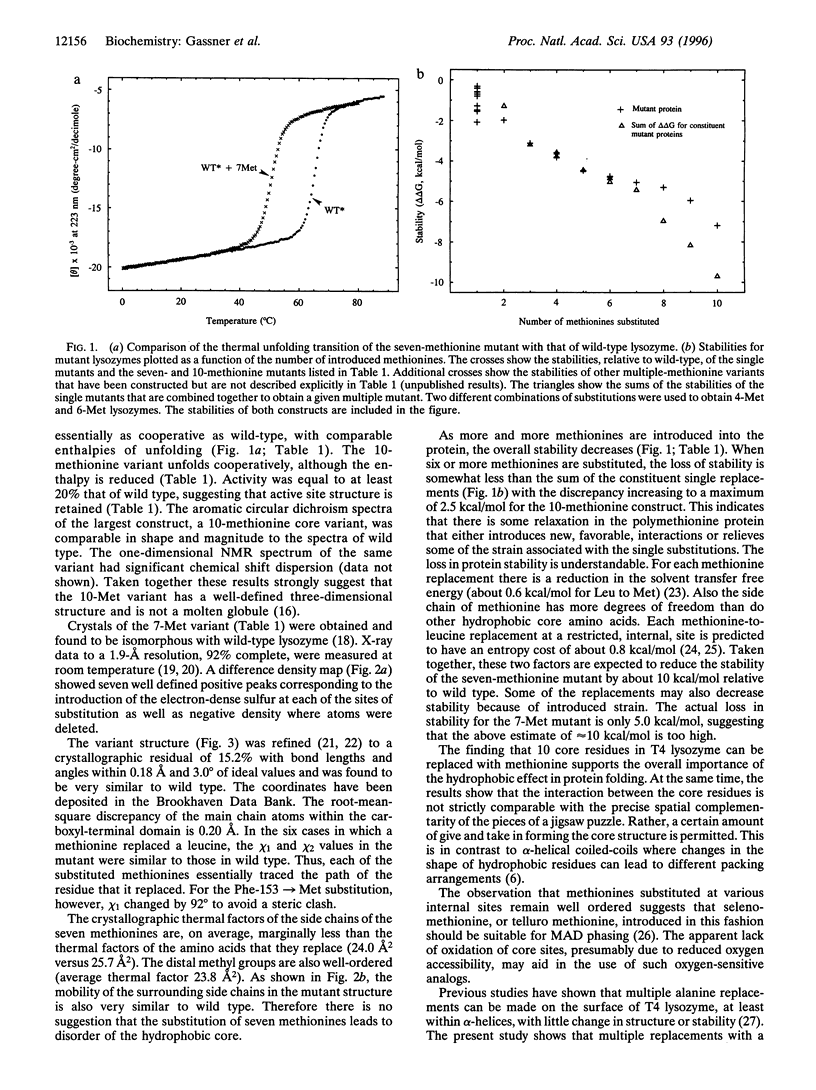
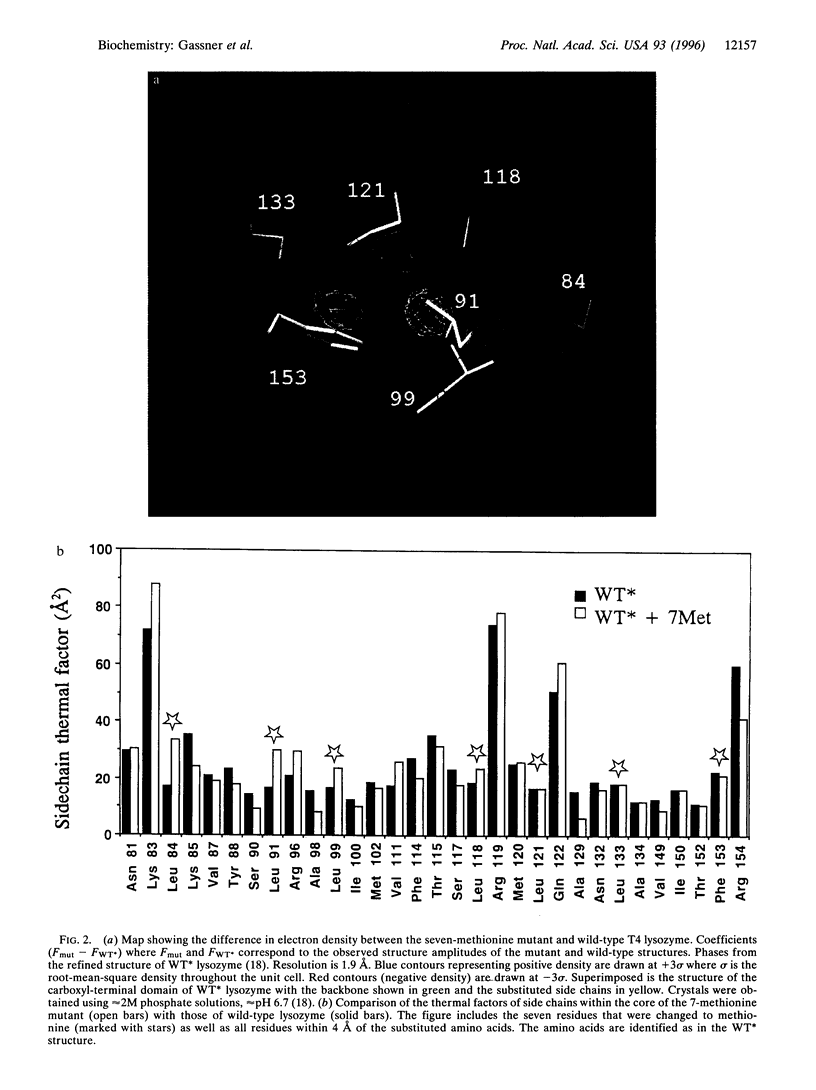
To test whether the structure of a protein is determined in a manner akin to the assembly of a jigsaw puzzle, up to 10 adjacent residues within the core of T4 lysozyme were replaced by methionine. Such variants are active and fold cooperatively with progressively reduced stability. The structure of a seven-methionine variant has been shown, crystallographically, to be similar to wild type and to maintain a well ordered core. The interaction between the core residues is, therefore, not strictly comparable with the precise spatial complementarity of the pieces of a jigsaw puzzle. Rather, a certain amount of give and take in forming the core structure is permitted. A simplified hydrophobic core sequence, imposed without genetic selection or computer-based design, is sufficient to retain native properties in a globular protein.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Axe D. D., Foster N. W., Fersht A. R. Active barnase variants with completely random hydrophobic cores. Proc Natl Acad Sci U S A. 1996 May 28;93(11):5590–5594. doi: 10.1073/pnas.93.11.5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin E. P., Hajiseyedjavadi O., Baase W. A., Matthews B. W. The role of backbone flexibility in the accommodation of variants that repack the core of T4 lysozyme. Science. 1993 Dec 10;262(5140):1715–1718. doi: 10.1126/science.8259514. [DOI] [PubMed] [Google Scholar]
- Behe M. J., Lattman E. E., Rose G. D. The protein-folding problem: the native fold determines packing, but does packing determine the native fold? Proc Natl Acad Sci U S A. 1991 May 15;88(10):4195–4199. doi: 10.1073/pnas.88.10.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaber M., Baase W. A., Gassner N., Matthews B. W. Alanine scanning mutagenesis of the alpha-helix 115-123 of phage T4 lysozyme: effects on structure, stability and the binding of solvent. J Mol Biol. 1995 Feb 17;246(2):317–330. doi: 10.1006/jmbi.1994.0087. [DOI] [PubMed] [Google Scholar]
- Chothia C., Levitt M., Richardson D. Helix to helix packing in proteins. J Mol Biol. 1981 Jan 5;145(1):215–250. doi: 10.1016/0022-2836(81)90341-7. [DOI] [PubMed] [Google Scholar]
- Creamer T. P., Rose G. D. Side-chain entropy opposes alpha-helix formation but rationalizes experimentally determined helix-forming propensities. Proc Natl Acad Sci U S A. 1992 Jul 1;89(13):5937–5941. doi: 10.1073/pnas.89.13.5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson A. E., Baase W. A., Matthews B. W. Similar hydrophobic replacements of Leu99 and Phe153 within the core of T4 lysozyme have different structural and thermodynamic consequences. J Mol Biol. 1993 Feb 5;229(3):747–769. doi: 10.1006/jmbi.1993.1077. [DOI] [PubMed] [Google Scholar]
- Eriksson A. E., Baase W. A., Zhang X. J., Heinz D. W., Blaber M., Baldwin E. P., Matthews B. W. Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science. 1992 Jan 10;255(5041):178–183. doi: 10.1126/science.1553543. [DOI] [PubMed] [Google Scholar]
- Hamlin R. Multiwire area X-ray diffractometers. Methods Enzymol. 1985;114:416–452. doi: 10.1016/0076-6879(85)14029-2. [DOI] [PubMed] [Google Scholar]
- Harbury P. B., Zhang T., Kim P. S., Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993 Nov 26;262(5138):1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- Harrison S. C., Durbin R. Is there a single pathway for the folding of a polypeptide chain? Proc Natl Acad Sci U S A. 1985 Jun;82(12):4028–4030. doi: 10.1073/pnas.82.12.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson W. A. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science. 1991 Oct 4;254(5028):51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- Kellis J. T., Jr, Nyberg K., Sali D., Fersht A. R. Contribution of hydrophobic interactions to protein stability. Nature. 1988 Jun 23;333(6175):784–786. doi: 10.1038/333784a0. [DOI] [PubMed] [Google Scholar]
- Klapper M. H. The independent distribution of amino acid near neighbor pairs into polypeptides. Biochem Biophys Res Commun. 1977 Oct 10;78(3):1018–1024. doi: 10.1016/0006-291x(77)90523-x. [DOI] [PubMed] [Google Scholar]
- Kuwajima K. The molten globule state as a clue for understanding the folding and cooperativity of globular-protein structure. Proteins. 1989;6(2):87–103. doi: 10.1002/prot.340060202. [DOI] [PubMed] [Google Scholar]
- Lim W. A., Sauer R. T. The role of internal packing interactions in determining the structure and stability of a protein. J Mol Biol. 1991 May 20;219(2):359–376. doi: 10.1016/0022-2836(91)90570-v. [DOI] [PubMed] [Google Scholar]
- Pielak G. J., Auld D. S., Beasley J. R., Betz S. F., Cohen D. S., Doyle D. F., Finger S. A., Fredericks Z. L., Hilgen-Willis S., Saunders A. J. Protein thermal denaturation, side-chain models, and evolution: amino acid substitutions at a conserved helix-helix interface. Biochemistry. 1995 Mar 14;34(10):3268–3276. doi: 10.1021/bi00010a017. [DOI] [PubMed] [Google Scholar]
- Ponder J. W., Richards F. M. Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes. J Mol Biol. 1987 Feb 20;193(4):775–791. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- Poteete A. R., Sun D. P., Nicholson H., Matthews B. W. Second-site revertants of an inactive T4 lysozyme mutant restore activity by restructuring the active site cleft. Biochemistry. 1991 Feb 5;30(5):1425–1432. doi: 10.1021/bi00219a037. [DOI] [PubMed] [Google Scholar]
- Richards F. M. The interpretation of protein structures: total volume, group volume distributions and packing density. J Mol Biol. 1974 Jan 5;82(1):1–14. doi: 10.1016/0022-2836(74)90570-1. [DOI] [PubMed] [Google Scholar]
- STREISINGER G., MUKAI F., DREYER W. J., MILLER B., HORIUCHI S. Mutations affecting the lysozyme of phage T4. Cold Spring Harb Symp Quant Biol. 1961;26:25–30. doi: 10.1101/sqb.1961.026.01.007. [DOI] [PubMed] [Google Scholar]
- Shortle D., Stites W. E., Meeker A. K. Contributions of the large hydrophobic amino acids to the stability of staphylococcal nuclease. Biochemistry. 1990 Sep 4;29(35):8033–8041. doi: 10.1021/bi00487a007. [DOI] [PubMed] [Google Scholar]
- Sternberg M. J., Chickos J. S. Protein side-chain conformational entropy derived from fusion data--comparison with other empirical scales. Protein Eng. 1994 Feb;7(2):149–155. doi: 10.1093/protein/7.2.149. [DOI] [PubMed] [Google Scholar]
- Zhang X. J., Baase W. A., Shoichet B. K., Wilson K. P., Matthews B. W. Enhancement of protein stability by the combination of point mutations in T4 lysozyme is additive. Protein Eng. 1995 Oct;8(10):1017–1022. doi: 10.1093/protein/8.10.1017. [DOI] [PubMed] [Google Scholar]