Abstract
Upon blue light irradiation, photoactive yellow protein (PYP) undergoes a conformational change that involves large movements at the N-terminus of the protein. We reasoned that this conformational change might be used to control other protein or peptide sequences if these were introduced as linkers connecting the N and C-termini of PYP in a circular permutant. For such a design strategy to succeed, the circularly permuted PYP (cPYP) would have to fold normally and undergo a photocycle similar to that of the wild type protein. We created a test cPYP by connecting the N- and Ctermini of wild type PYP (wtPYP) with a GGSGGSGG linker polypeptide and introducing new N- and C- termini at G115 and S114 respectively. Biophysical analysis indicated that this cPYP adopts a dark state conformation much like wtPYP and undergoes wtPYP-like photoisomerization driven by blue light. However, thermal recovery of dark state cPYP is ~10-fold faster than wtPYP, so that very bright light is required to significantly populate the light state. Targeted mutations at M121E (M100 in wtPYP numbering) were found to enhance the light sensitivity substantially by lengthening the lifetime of the light state to ~10 min. NMR, circular dichroism, and UV-Vis analysis indicated that the M121E-cPYP mutant also adopts a dark state structure like that of wtPYP although protonated and deprotonated forms of the chromophore coexist giving rise to a shoulder near 380 nm in the UV-Vis absorption spectrum. Fluorine NMR studies with fluoro-tryptophan labeled M121E-cPYP show that blue-light drives large changes in conformational dynamics and leads to solvent exposure of Trp7 (Trp119 in wtPYP numbering) consistent with substantial rearrangement of the N-terminal cap structure. M121E-cPYP thus provides a scaffold that may allow a wider range of photoswitchable protein designs by replacing the linker polypeptide with a target protein or peptide sequence.
Keywords: photo-control, photoactive yellow protein, PYP, optogenetics, photoisomerization, genetically encoded, circular permutation
Introduction
Photoswitchable proteins offer the possibility for remote control of a variety of biochemical processes (1). If one wishes to photocontrol the activity of a given target protein a number of strategies may be envisaged. One design involves co-opting naturally occurring domains that undergo light driven dimerization. Fusion of one domain to the target protein of interest and of its dimerization partner to a membrane localization sequence for instance, can be used to effectively control the subcellular localization of the target (2, 3). A second strategy involves making hybrid proteins in which a photoswitchable domain is linked to a target protein in manner that results in conformational changes in the photoreceptor being transferred to functional changes in the target (4–8). As Möglich and Moffat have pointed out in a review of engineered photoreceptors, this strategy is critically dependent on the nature on the linkage between the two proteins (9).
The LOV domain has proven to be a workhorse for such engineering efforts; it has been thoroughly characterized by a range of structural methods and has the distinct advantage of having a naturally occurring co-factor (10–14). Photoactive yellow protein (PYP) is another small water soluble protein that undergoes blue-light driven photochemistry (15–18). Although the PYP cofactor p-coumaric acid is not usually present except in specialized cell types, it can be added exogenously, or the enzymes for its biosynthesis can be co-expressed with PYP (19). From the point of view of designing novel photoswitchable bioactivity, PYP has the potential advantage that it undergoes extensive conformational changes upon photoisomerization. These changes involve a large increase in protein internal dynamics and large scale rearrangements and/or detachment of the N-terminal cap domain from the rest of the protein (17, 20). The conformational change of PYP has been engineered to photocontrol other proteins, in particular the GCN4 DNA binding protein (21–23) and alpha hemolysin (24) by making targetprotein/PYP hybrids. As just described, this strategy places rather strict constraints on the target proteins that can be controlled because linkage sequences must be chosen that satisfy functional constraints of both PYP and the target protein (1, 21–23).
We reasoned that construction of a circularly permuted PYP variant might lead to a relaxation of these sequence constraints. Ramachandran et al have reported that distances between residues near the N-terminus of PYP (Ala5, Glu9) and a residue on a loop adjacent to the C-terminus (Gln99) change by up to 19 Å upon PYP isomerization to the light state* (20). We hypothesized that if the N and C-termini of PYP were linked in a circular permutant, the conformation, dynamics, and end-to-end distance of the linker would be changed significantly if the main aspects of the light triggered protein structural change were preserved. A variety of target sequences with bioactivity, but few restraints on sequence, could then be introduced as linkers and may be subject to photo-control.
Our aim in the present work was to test if circular permutation can be accomplished using an unstructured flexible linker while preserving the light driven conformational changes of PYP.
Materials and Methods
Gene synthesis and site directed mutagenesis
DNA coding for cPYP (codon optimized for E. coli) was synthesized by BioBasic Inc. (Toronto) and inserted into the pET24b(+) vector using Ndel and HindIII restriction sites. The gene for wildtype PYP was cloned into the same vector (21) . Mutations to create M100E-PYP, M121E-cPYP and M121A-cPYP were carried out using standard molecular biology protocols. Primers for the mutations were designed with PrimerX (http://www.bioinformatics.org/primerx/) following the Stratagene QuikChange protocol (Agilent, Inc.) and purchased from ACGT Corp.
Primers for M100E mutation:
-
Forward: 5' CACCTTCGATTACCAAGAGACGCCCACGAAGGTG 3'
Reverse: 5' CACCTTCGTGGGCGTCTCTTGGTAATCGAAGGTG 3'
Primers for M121E mutation:
-
Forward: 5' GAATACACCTTCGACTACCAGGAGACGCCAACTAAAGTAAAAG 3'
Reverse: 5' CTTTTACTTTAGTTGGCGTCTCCTGGTAGTCGAAGGTGTATTC 3'
Primers for M121A mutation:
-
Forward: 5' GAATACACCTTCGACTACCAGGCGACGCCAACTAAAGTAAAAG 3'
Reverse: 5' CTTTTACTTTAGTTGGCGTCGCCTGGTAGTCGAAGGTGTATTC 3'
For each mutation, 100 ng each of the forward and reverse primers were added to 50 ng of template DNA, 25 µL Pfu Turbo hotstart PCR master mix (a 2x formulation of Pfu Turbo hotstart DNA polymerase, an optimized PCR reaction buffer, magnesium, and dNTPs; Agilent Technologies), and water to a total volume of 50 µL. The reaction mixture was held at (i) 95°C for 2 minutes followed by (ii) 30 seconds at 95°C, (iii) 30 seconds at 5°C below primer Tm and (iv) 6 minutes at 72°C. Steps (ii)-(iv) were repeated 30 times, then the mixture was held at 72°C for an additional 10 minutes before being allowed to cool to room temperature. A 1 µL aliquot of DpnI was added to the reaction mixture which was then incubated at 37°C for one hour. A 0.8% agarose gel in 0.5x TBE was run to check for PCR products. A PCR purification kit (Invitrogen) was subsequently used and the product was transformed into MAX Efficiency® DH5α™ competent cells (Invitrogen™). A midiprep (Invitrogen) was carried out and the plasmids were sequenced in both directions (ACGT Corp.) to confirm the mutations.
Protein expression and purification
Expression and reconstitution of the PYP constructs was adapted from the work of Devanathan et al. (25) The protocol was the same for all constructs. DNA (0.2 ng) was transformed into BL21*(DE3) competent cells and plated onto agar plates containing 30 µg/mL kanamycin. The following day, a single colony was used to inoculate 25 mL of Luria-Bertani (LB) broth that had been supplemented with kanamycin (30 µg/mL). The 25 mL overnight culture was used to inoculate 1 L of LB supplemented with 30 µg/mL kanamycin. Cells were grown at 37°C until an OD600 of 0.6 was reached and then induced with 1 mM IPTG. The temperature was adjusted to 25°C and the cells grown for a further 1.5 hrs before 25 mg of activated chromophore dissolved in 1 mL of ethanol was added to the media. The synthesis of the activated chromophore, 4-hydroxycinnamic acid S-thiophenyl ester, was carried out as detailed by Changenet-Barret et al. (26) except that the product was not recrystallized. The cells were grown for a further 6 hours before centrifugation to separate the media from the protein-containing cell pellet.
The pellet was resuspended in lysis buffer containing 50 mM sodium phosphate, pH 8.0, 300 mM sodium chloride and 5 mM magnesium chloride and frozen at −20°C until purification. The resuspended cell pellet was sonicated in pulses on ice for 5 min and then centrifuged at 12K rpm for 1 hr to separate the supernatant from the pellet. The protein was purified on a Ni-NTA column that was equilibrated with the lysis buffer. The protein-containing supernatant was loaded onto the column, and the yellow color became associated with the resin. The resin was washed with 10 column volumes (CV) of the lysis buffer. The resin was subsequently washed with 5 CVs of high salt buffer (i.e. lysis buffer supplemented with 2 M NaCl) followed by a further 5 CVs of lysis buffer. To elute non-specifically bound proteins, the resin was subjected to 5 CVs of lysis buffer supplemented with 5 mM imidazole. The protein was eluted by increasing the concentration of imidazole to 200 mM.
The eluted protein was dialyzed extensively against 40 mM Tris-OAc, 1 mM EDTA, and 100 mM NaCl (pH 7.5) (1x TAE, 100 mM NaCl, pH 7.5) at 4°C. The dialyzed protein was concentrated to ~1.5 mL using an Amicon ultracentrifugal device (10,000 Da NMWL (nominal molecular weight limit) (Millipore). The protein was then applied to a Superdex 75 10/300 GL column (GE Healthcare) running in 1x TAE, 100 mM NaCl, pH 7.5 buffer. UV-Vis absorbance spectroscopy was used to determine which eluted fractions had the highest ratios of absorbance at 446 nm to that at 278 nm. A value of ~2 was typical for pure wtPYP or cPYP holoprotein and 1.4 for M100E-PYP and M121E-CPYP. The purity and identity of samples was confirmed using 12.5% SDS-PAGE and electrospray ionization mass spectrometry (ESI-MS). The expression levels of all three constructs was similar; a 1 L culture routinely gave >25 mg of purified protein.
UV/Vis spectra and photoisomerization
UV-Vis spectra and kinetic measurements were performed using a PE Lambda 35 or 25 spectrophotometer or using a diode array UV–Vis spectrophotometer (Ocean Optics Inc., USB4000) in each case, coupled to a temperature controlled cuvette holder (Quantum Northwest, Inc.). Protein concentrations were determined using an extinction coefficient at λmax (~446 nm) of 45 × 103 M−1cm−1 for wtPYP and cPYP and 29 × 103 for M100E-PYP and M121E-cPYP. Irradiation of the protein sample was carried out by using a Luxeon III Star LED Royal Blue (455 nm) Lambertian operating at approximately 700 mA (~50 mW/cm2). To determine the rate constants for thermal relaxation, changes in the absorbance spectrum at 350 nm were monitored. Data were fitted to single exponential functions to extract rate constants.
pH effects
Studies of pH effects were carried out in a universal buffer (25 mM sodium acetate, MES, Tris, and CAPSO, 100 mM NaCl) in a 1.0 cm path length quartz cuvette using a PE Lambda 25 spectrophotometer. To obtain different pHs, the sample was titrated with 1M HCl or 1M NaOH followed by 1 minute stirring. The rate constants for thermal relaxation were obtained by irradiating the sample for 1 minute using a Luxeon III Star LED Royal Blue (455 nm) Lambertian operating at approximately 700 mA (~50 mW/cm2) and changes in the absorbance at 350 nm were monitored at 20°C. Data was analyzed as described above.
Thermal stability
Samples were prepared in 1x TAE, 100 mM NaCl, pH 7.5. Samples were fully dark adapted before they were scanned in the UV-Vis region from 250–550 nm. At each temperature, the samples were allowed to equilibrate for 5 minutes before spectra were collected. The contribution from the buffer was subtracted from the protein spectra.
Salt effects
All studies were performed in 1x TAE buffer, 100 mM NaCl, pH 7.5 having 0.5/1/2 M (NH4)2SO4 or NH4Cl at 20°C. Samples were fully dark adapted before they were scanned in the UV-Vis region from 250–550 nm.
Guanidinium denaturation studies
Guanidinium induced denaturation of wtPYP, cPYP and M121E-cPYP was monitored by tryptophan (Trp) fluorescence using an Aviv Associates (Lakewood, NJ) model ATF 105 automated titrating spectrofluorometer. Protein samples (~5 µM) were prepared in 1x TAE, 100 mM NaCl, pH 7.5 with or without 5 M GdnHCl (ThermoFisher) and allowed to incubate overnight to fully dark-adapt. Both folding and unfolding titrations were performed. Fluorescence was acquired of protein samples using excitation at 280 nm and emission at 360 nm with a bandwidth of 2 nm. During the fluorescence experiment, the sample volume was kept constant at 2 mL, with a 1.0 cm path length quartz cuvette and the temperature was maintained at 20°C. The sample was stirred after each addition for 1 minute and then stirring was halted before the fluorescence measurement. Fluorescence data were fitted using the equation:
where αN is F360nm in the absence of guanidinium, βN is the slope (F360nm vs. [Gdn]) at the beginning of the curve, αD is the value of F360nm for the fully denatured state and βD the slope at the end of the transition. The quantity [D]50% is the Gdn concentration at the point where the protein is 50% unfolded and term mD–N is a constant that reflects the degree of unfolding of the protein in guanidinium (27).
CD measurements
CD experiments were carried out on an Olis RSM100 CD instrument. Samples were prepared at a final concentration of 15 µM in 1 mM sodium phosphate buffer pH 7.5. A cylindrical quartz cell of 0.1 cm path length was used for all measurements. Samples were fully dark adapted before they were scanned in the far-UV region from 260–190 nm at 20°C. The integration time was 2 s and three scans were averaged to give a final spectrum. For the irradiated state, samples were irradiated using a Luxeon III Star LED Royal Blue (455 nm) Lambertian operating at approximately 700 mA (50 mW/cm ) for 1 minute and five individual sets were averaged to get the final spectrum. Buffers were also scanned under the same conditions and subtracted from the protein spectra. The spectra were converted to mean residue ellipticities and smoothed using a binomial algorithm in IgorPro software (Wavemetrics).
Sedimentation equilibrium
Samples of M121E-cPYP were prepared at 5.5, 11.1, 22.2 µM concentrations in 1x TAE, 100 mM NaCl, pH 7.5 buffer and analyzed by analytical ultracentrifugation (AUC facility, Dept. Biochemistry, University of Toronto). Absorbance at 446 nm was monitored at 4°C and speeds of 20000 and 24000 rpm.
NMR
Labeled protein samples (15N, 15N and 13C, and 19F labeled cPYP and M121E-cPYP) were prepared as follows: An expression plasmid was transformed into BL21*(DE3) chemically competent cells, and these were plated on LB agar plates supplemented with 30 µg/ml kanamycin. On the following day, a single colony from the agar plate was used to inoculate 25 mL of LB broth/kanamycin media, and the culture was incubated overnight at 37°C and 250 rpm. On the following day, 6 mL of the overnight culture was centrifuged for 15 min at 4000 rpm, and the pellet was resuspended in 50 mL of M9 media containing a natural abundance of 15N. The 50-mL culture was grown at 37°C and 250 rpm until it had reached mid-log phase (OD600 ~ 0.5–0.7), then it was centrifuged as described above. The pellet was used to inoculate 1 L of 99% 15N-enriched M9 minimal media, supplemented with 0.3% Dglucose, 0.1% 15NH4Cl (Cambridge Isotope Laboratories, Inc.), 30 mg/L kanamycin, 10 mg/L thiamine, 10 mg/L biotin, 1 mM MgSO4, of 0.6, protein expression was induced by the addition of 1mM IPTG. The temperature was adjusted to 25°C, and the cells were grown for a further 1.5 hr, after which 25 mg of activated chromophore (26) dissolved in 1 mL of ethanol was added. The temperature was then adjusted to 18°C, and the culture was grown overnight. On the following day, the cells were harvested by centrifugation at 4000 rpm and 4°C for 1 hr. For doubly (15N and 13C) labeled protein samples, 13C-enriched D-glucose ( growth media enriched in both 15N and 13C was added in place of only 15N enriched. To introduce fluoro-tryptophan into M100E-PYP and M121E-cPYP, 19F-labeled 5-fluoro-L,D-Trp (Aldrich) was added to the bacterial culture at a concentration of 60 mg/L, 1 h prior to induction (28). Here, activated chromophore addition was done together with induction of protein expression by addition of 1mM IPTG and growth was halted 2 hr after induction. The protein was purified in the same manner as unlabeled samples (as described above). The mass of the proteins was confirmed by electrospray ionization mass spectrometry.
Samples were placed in a Shigemi tube (BMS-005TV, Shigemi, Inc.) with a multimode 1-mm fiber optic (Thorlabs) inserted through the top. The other end of the fiber was coupled to a LED Royal Blue LED (LXH-LR5C (700 mW); LED Supply) through a focusing lens (WPI, Inc.). Tests outside the NMR magnet and M121EcPYP into their colorless light states.
NMR experiments were recorded at the Québec/Eastern Canada High Field NMR Facility or at CSICOMP (Dept. of Chemistry, University of Toronto). Experiments for backbone assignments were performed using a 500 MHz Varian Inova spectrometer equipped with a Z-gradient HCN 5 mm cold probe. All pulse sequences were used as obtained from the Varian ‘Biopack’ sequence library. HSQC spectra were acquired with 40 increments spanning 1520 Hz in the 15N dimension. Backbone assignments were determined from HNCO (29) (32 13C increments spanning 1510 Hz), HNCACB (30) (64 13C increments spanning 8300 Hz), CBCA(CO)NH (31) (48 13C increments spanning 8300 Hz), and C(CO)NH (32) (80 13C increments spanning 9040 Hz) spectra, all acquired with 32 increments spanning 1520 Hz in the 15N dimension. For the C(CO)NH, mixing of 13C magnetization was achieved using a FLOPSY8 (33) mixing scheme applied for 12.7 ms.
Spectra were processed using the NMRPipe (34) processing suite. Typically, FID signals were zero filled to double the original data size and apodized using a squared-cosine window function prior to Fourier transformation. In the indirect dimension(s), linear prediction was used to double the original data size prior to zero-filling and apodization as above. Assignment of backbone resonances was performed using standard methods (35), aided by the NMRViewJ software program (One Moon Scientific).
19F one-dimensional NMR experiments were performed at different temperatures on a 700 MHz Agilent DD2 spectrometer, using a H-F{13C,15N} triple-resonance cold probe, in which the highfrequency channel could be tuned to 19F. At least 1024 to 16384 scans were used to obtain the 19F NMR spectrum in dark and irradiated states (typically, spectra acquired at lower temperatures required more scans). A 350 µL sample volume was used in 3 mm sample tube, with a fiber optic inserted, inserted into a 5 mm sample tube containing a reference solution of trifluoroacetic acid TFA. Spectra were referenced to the chemical shift of TFA (−78.5 ppm) and processed using 50 Hz line broadening. Lorentzian fitting was used to obtain chemical shifts. Photoswitching was accomplished in the same manner as described above.
Results & Discussion
Protein design
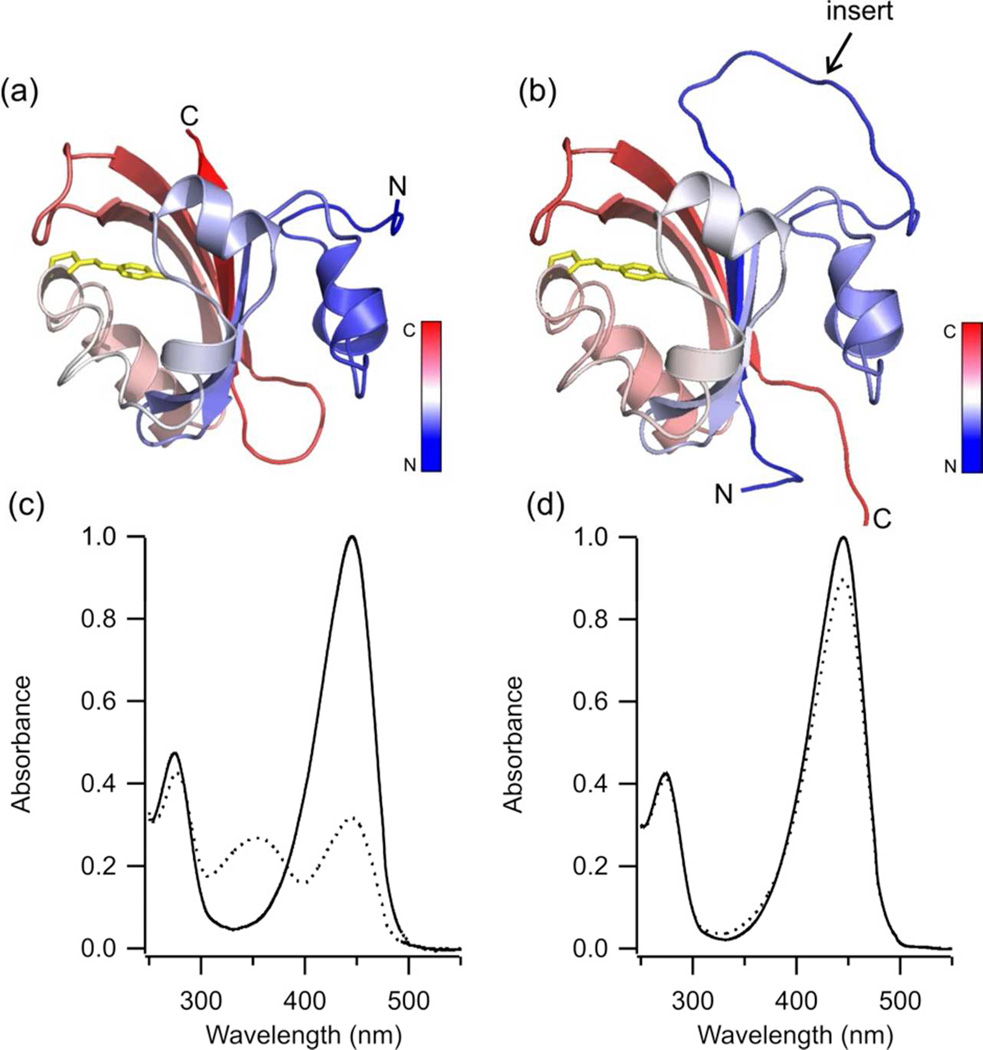
To test if a circularly permuted PYP would fold normally and undergo a native photocycle, we decided to introduce a simple flexible linker (GGSGGSGG) long enough to link the N and C termini but chosen to be minimally perturbing to the PYP scaffold. We opted to create new N and C termini at G115 and S114, since this is the loop most distant from the chromophore and mutations here, unlike other loops, have not been found to cause significant changes in the properties of PYP (15). A model of this circularly permuted PYP (cPYP) protein is shown in Fig 1.
Figure 1.
Models of (a) wtPYP and (b) cPYP in the dark-adapted state colored in a gradient from N-terminal to C-terminal (N-blue, C-red) based on the high resolution X-ray structure of dark-adapted PYP (1NWZ) (36). The chromophore is shown in yellow sticks. The GGSGGSGG insert is modeled as a flexible loop. (c) UV-Vis spectrum of wtPYP (dark-adapted (—), irradiated with 450 nm light (⋯)(pH 7.5 Tris-acetate EDTA buffer, 100 mM NaCl, 20°C) (d) UV-Vis spectrum of cPYP (dark-adapted (—), irradiated with 450 nm light (⋯)(pH 7.5 Tris-acetate EDTA buffer, 100 mM NaCl, 20°C).
Structural characterization of dark-adapted cPYP
The circularly permuted PYP expressed well and was highly soluble. UV-Vis spectra of the dark adapted states of cPYP and wtPYP at pH 7.5 are shown in Fig. 1c,d. The spectra are virtually superimposable and this was found to be the case at pH 5.0 and pH 9.0 as well (not shown). Constant intensity (~50 mW/cm2) 450 nm the steady-state (irradiated) spectra shown as dotted lines in Fig. 1c,d. Whereas the peak at 446 nm is greatly decreased and a peak appears at 350 nm for wtPYP, as expected for significant conversion to the cis, protonated form of the protein, the changes are much less pronounced for the cPYP species. As detailed below, this observation is a consequence of significantly faster thermal relaxation rate of the cPYP cis species to the dark-adapted, trans isomer. We focus first on the structure of the dark-state cPYP protein and on the extent to which this resembles wtPYP.
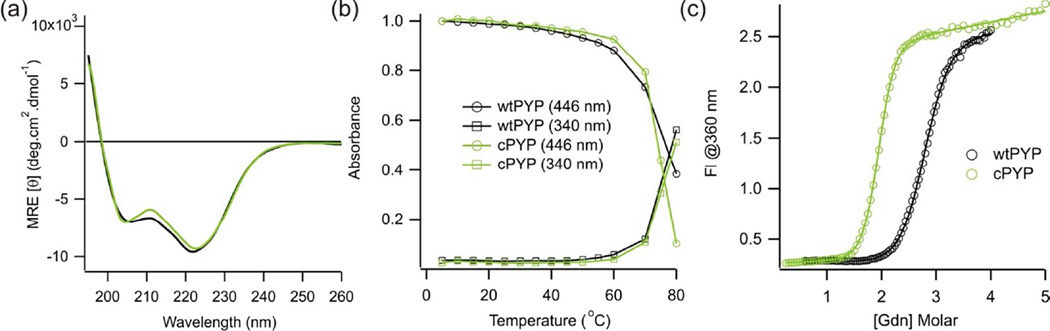
CD spectra of wtPYP and cPYP at pH 7.5 are shown in Fig. 2a. These are very similar indicating the overall secondary structures of the two proteins are very similar. A temperature denaturation experiment indicated cPYP was nearly as stable as wild type (Fig. 2b). Reversible denaturation by guanidinium was also measured (Fig. 2c) and cPYP was found to undergo a cooperative folding/unfolding transition although it was distinctly less stable than wtPYP as judged by the concentration of guanidinium at the midpoint of the denaturation curve. This result is typical for circularly permuted proteins (37).
Figure 2.
(a) Circular dichroism spectra of dark-adapted wtPYP (−) and cPYP ( ) (pH 7.5 5 mM sodium phosphate buffer, 20°C) (b) Temperature denaturation titrations of dark-adapted wtPYP (−) and cPYP (
) (pH 7.5 5 mM sodium phosphate buffer, 20°C) (b) Temperature denaturation titrations of dark-adapted wtPYP (−) and cPYP ( ) (pH 7.5 Tris-acetate EDTA buffer, 100 mM NaCl) (c) Guanidinium refolding curve for dark-adapted wtPYP (−) and cPYP (
) (pH 7.5 Tris-acetate EDTA buffer, 100 mM NaCl) (c) Guanidinium refolding curve for dark-adapted wtPYP (−) and cPYP ( ). (pH 7.5 Tris-acetate EDTA buffer, 100 mM NaCl, 20°C).
). (pH 7.5 Tris-acetate EDTA buffer, 100 mM NaCl, 20°C).
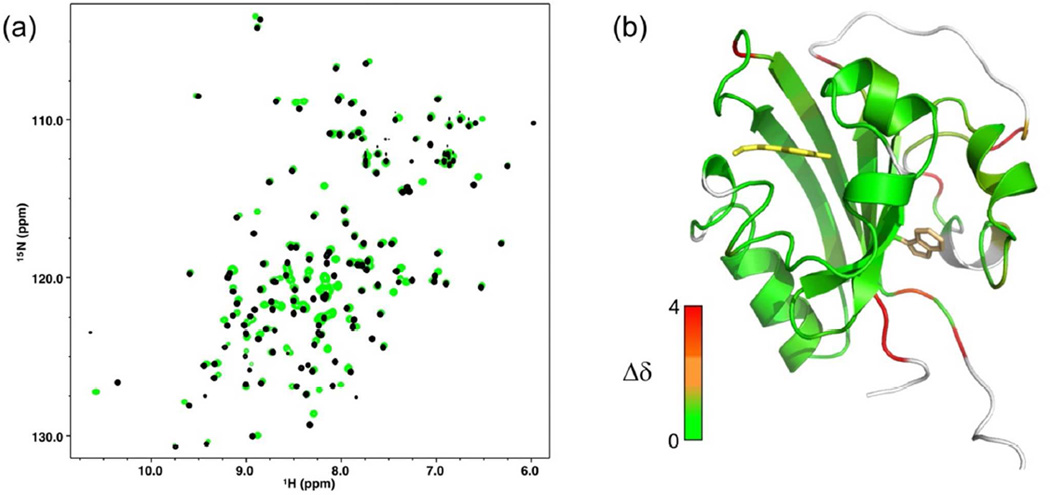
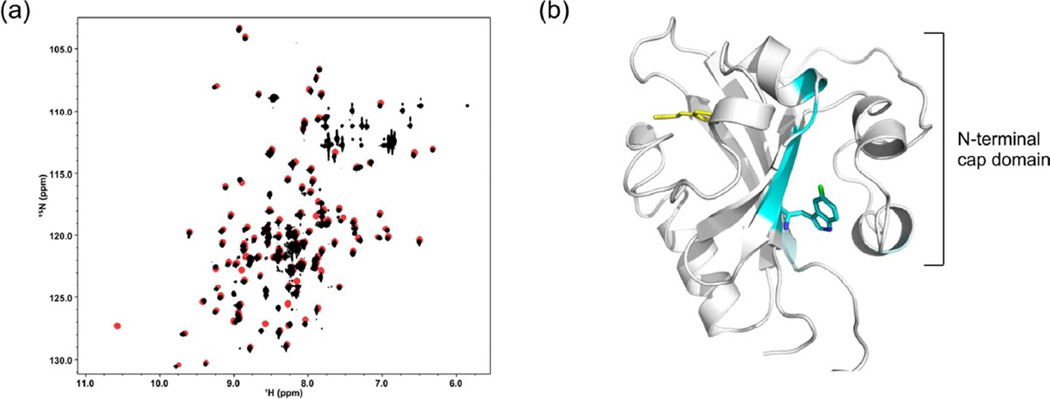
To obtain a more detailed assessment of the similarity of the dark state structure of cPYP to that of wtPYP, we expressed 15N,13C labeled protein and assigned the backbone chemical shifts using standard protein NMR methods (see supporting information). The 15N,1H-HSQC spectra of wtPYP and cPYP are shown overlaid in Figure 3 together with a model of the cPYP structure based on the X-ray structure of wtPYP (as in Fig. 1b) with residues colored according to chemical shift similarity to wtPYP. There is very close correspondence of chemical shifts across the chromophore binding domain. There are large changes in chemical shifts at the loop where the new N- and C-termini were created as expected. There are smaller changes in parts of the N-terminal cap domain including the Trp indole NH signal. The glycine rich linker connecting the wild type N and C-termini could not be assigned presumably due to overlap of resonances and/or mobility of the segment. Overall these data indicate that the dark-adapted structure of cPYP and wtPYP are very similar although local changes to structure and/or dynamics in the N-terminal cap are likely.
Figure 3.
(a)15N,1H-HSQC spectrum of wtPYP (black) and cPYP (green). For assignments see supporting information (b). Modeled structure of cPYP (based on wt PYP 1NWZ) (36) with residues colored according to chemical shift difference from wtPYP (BMRB code: 18122) (38). The chemical shift difference was calculated as The Trp side chain is shown in orange sticks. Note that the indole NH peak moves relative to wtPYP. (pH 7.5 Tris-acetate EDTA buffer, 100 mM NaCl, 20°C). Model produced using PyMol.
Rapid photoswitching of cPYP
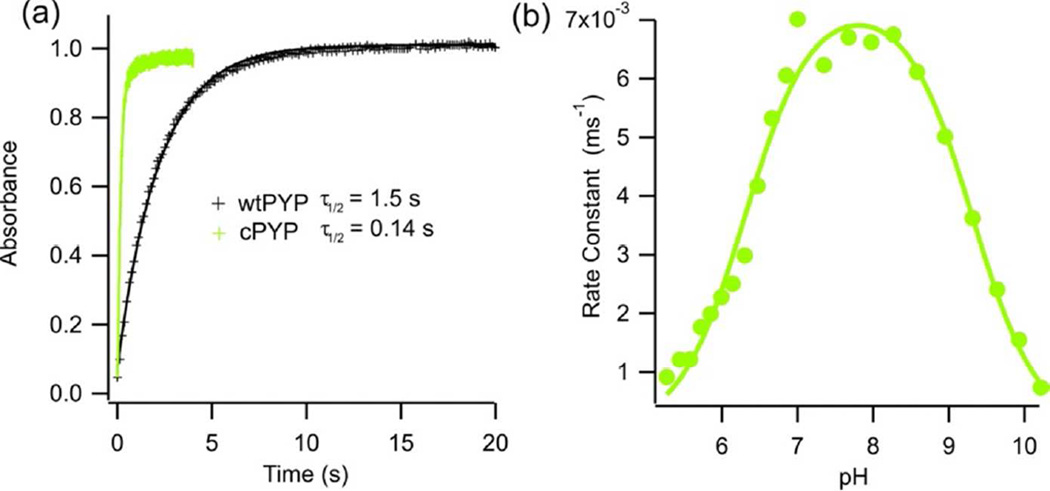
We next investigated the photoswitching characteristics of cPYP. Figure 4 shows the recovery of the dark state structure as monitored by UV-Vis spectroscopy after removal of blue light irradiation. The rate constant for cis-to-trans recovery of cPYP is seen to be ~ 10x faster than that of wtPYP at pH 7.5, 20°C. This enhanced recovery rate is also seen at lower and higher temperatures (not shown). The enhanced recovery rate explains the smaller change in steady state spectra observed under constant low intensity blue light irradiation (Fig. 1c,d). We measured the relaxation rate of cPYP as a function of pH (Fig. 4b) and found a bell shaped dependence of the relaxation rate with a maximum near pH 7.8 and apparent pKas of 6.3 and 9.3. This pH dependence is very similar to that of wild type PYP for which apparent pKas of 6.4 and 9.4 were reported (39).
Figure 4.
(a) Recovery of dark-adapted wtPYP (−) and cPYP ( ) after irradiation with 450 nm light (pH 7.5 Tris-acetate EDTA buffer, 100 mM NaCl, 20°C) (b) pH dependence of dark-state recovery of cPYP as a function of pH (100 mM NaCl, 20°C). The fitted curve is given by: kobs = kmax /(1 + 10(pK1 − pH) + 10(pH − pK2)) with pK1 = 6.3 ± 0.1 and pK2 = 9.3 ± 0.1
) after irradiation with 450 nm light (pH 7.5 Tris-acetate EDTA buffer, 100 mM NaCl, 20°C) (b) pH dependence of dark-state recovery of cPYP as a function of pH (100 mM NaCl, 20°C). The fitted curve is given by: kobs = kmax /(1 + 10(pK1 − pH) + 10(pH − pK2)) with pK1 = 6.3 ± 0.1 and pK2 = 9.3 ± 0.1
The rapid thermal recovery of the dark state structure of cPYP is rather unusual; to our knowledge, although PYP homologues from other species are known that undergo rapid thermal recovery, all mutations or modifications of wtPYP form H. halophila reported to date have resulted in a slowing of the recovery rate to varying degrees (15, 18). From the point of view of using cPYP as a photoswitch to control biomolecular function, the very rapid thermal recovery is problematic since it means that only a small fraction of the cis-state can be populated under steady state conditions with common light sources (Fig. 1d). We therefore wished to modify cPYP to slow down its thermal recovery rate.
A number of modifications to wtPYP result in a dramatic slowing of the thermal recovery rate (15). The best characterized of these are deletions to the N-terminal domain (1–25) and point mutations at M100. Since we wished to maintain the conformational changes at the N-terminus we chose to mutate M100 (M121 in cPYP sequence numbering).
M121E-cPYP shows a slower photocycle
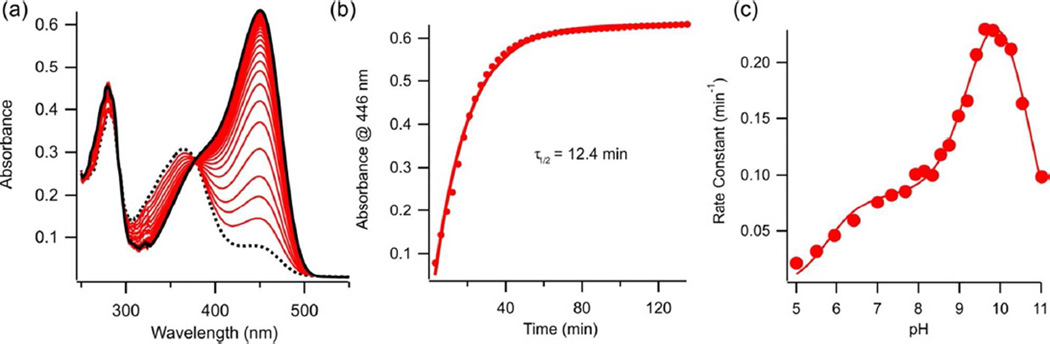
The point mutations M121A-cPYP and M121E-cPYP were created, expressed and purified. The two proteins behaved very similarly so that only one (M121E-cPYP) was studied in detail. Figure 5 shows UV-Vis spectra of this species in dark-adapted conditions together with steady state spectra observed under 450 nm irradiation. Thermal recovery of the dark state was found to occur with a half life of 12.4 min at pH 7.5, ~500 times longer than with cPYP (Fig. 5b). The pH dependence of the reisomerization rate was found to be more complex than for cPYP with a maximum near pH 10 and three apparent pKas. Throughout the entire pH range however, the recovery rate was slow enough that complete isomerization to the cis form could be easily achieved under steady state blue light illumination.
Figure 5.
(a) UV-Vis spectrum of M121E-cPYP (dark-adapted (−), irradiated with 450 nm light (⋯)(pH 7.5 Tris-acetate EDTA buffer, 100 mM NaCl, 20°C) (b) Recovery of dark-adapted M121E-cPYP after irradiation with 450 nm light (pH 7.5 Tris-acetate EDTA buffer, 100 mM NaCl, 20°C (c) pH dependence of dark-state recovery rate of M121E-cPYP as a function of pH (100 mM NaCl, 20°C). The fitted curve is given by: kobs = kmax1 /(1 + 10(pK1 − pH) + 10(pH − pK2)) + kmax 2 /(1 + 10(pK2 − pH) + 10(pH − pK3)) with pK1 = 5.8 ± 0.1, pK2 = 9.3 ± 0.1 and pK3 = 10.6 ± 0.1
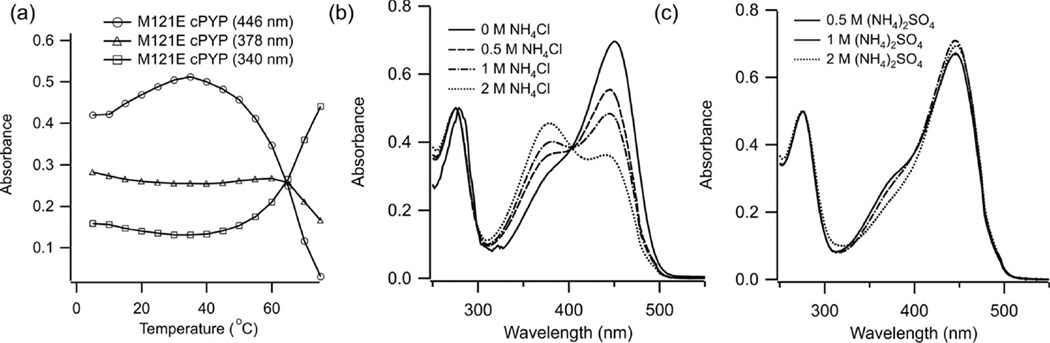
An examination of the dark state UV-Vis spectrum of M121E-cPYP (Fig. 5a) compared to cPYP (Fig. 1d) reveals the presence of a shoulder at ~380 nm in addition to the main absorbance band at 446 nm expected for the deprotonated trans form of the chromophore. The 380 band, called an “intermediate spectral from” (40) has been observed previously in M100E, M100A, M100L mutants of wtPYP as well as in the Y42F wtPYP mutant. The band has been ascribed to the presence of a fraction of protonated trans-chromophore in the dark-adapted protein (40–42) In wtPYP (and by extension cPYP) the pCA phenolic proton is completely transferred to Glu46 so that only the anionic form of the chromophore is observed. Mutations such as M100E (M121E in cPYP) or Y42F appear to alter the balance of pKas so that two distinct H-bonding arrangements co-exist, one of which involves protonated chromophore. Since the H-bonding equilibrium involves two sites on the protein, not an equilibrium between solvent and protein, changing the bulk pH does not substantially affect the UV-Vis spectrum (see supporting information). The ratio of protonated to anionic chromophore is affected by temperature however. Figure 6 shows that the fraction of anionic chromophore is maximal at 35°C, and decreases below this temperature. Above 35°C, the fraction of anionic chromophore also decreases and ultimately the protein unfolds near 60°C. Chaotropic salts such as ammonium chloride stabilize the protonated form of the chromophore whereas kosmotropic salts stabilize the anionic form (Fig. 6b,c).
Figure 6.
(a) Measured absorbance of dark-adapted M121E-cPYP (dark-adapted in pH 7.5 Tris-acetate EDTA buffer, 100 mM NaCl) at a series of temperatures (b) UV-Vis spectrum of dark-adapted M121E-cPYP (pH 7.5 Tris-acetate EDTA buffer, 100 mM NaCl) with the addition of the indicated concentration of ammonium chloride. (c) UV-Vis spectrum of dark-adapted M121E-cPYP (pH 7.5 Tris-acetate EDTA buffer, 100 mM NaCl) with the addition of the indicated concentration of ammonium sulfate.
Structural characterization of dark-adapted M121E-cPYP
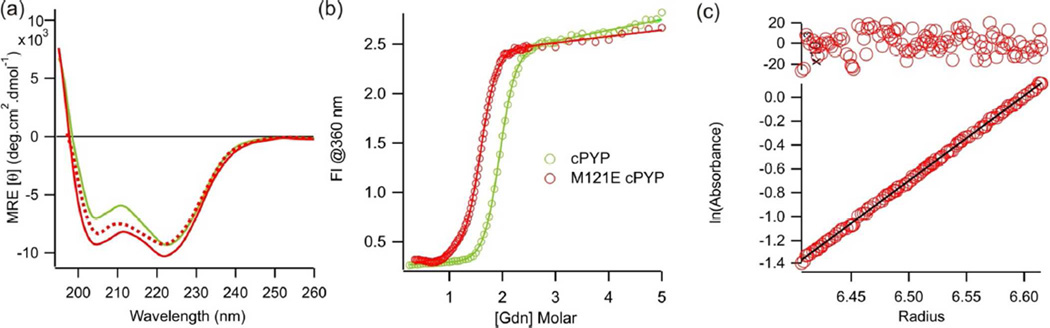
We wished to test whether the dark state structure and the light driven conformational changes were preserved in M121E-cPYP compared to cPYP and wtPYP. We first compared the CD spectrum of M121E-cPYP to cPYP and wtPYP (Fig. 7a). These spectra were normalized using the absorbance of the protonated pCA chromophore at 350 nm after denaturation of the proteins in 2% SDS. The CD spectrum of M121E-cPYP exhibits slightly more negative ellipticity near 208 nm relative to cPYP and wtPYP. It is unclear if this represents a slightly larger fraction of helical secondary structure than cPYP or whether the optical changes associated with the presence of a fraction of protonated chromophore produce alterations in this region of the CD spectrum.
Figure 7.
(a) Circular dichroism spectra of dark-adapted M121E-cPYP ( ), irradiated (450 nm) M121E-cPYP (
), irradiated (450 nm) M121E-cPYP ( ), and cPYP (
), and cPYP ( ) (pH 7.5, 5 mM sodium phosphate buffer, 20°C) (b) Guanidinium refolding curve spectra of dark-adapted M121E-cPYP (
) (pH 7.5, 5 mM sodium phosphate buffer, 20°C) (b) Guanidinium refolding curve spectra of dark-adapted M121E-cPYP ( ) and cPYP (
) and cPYP ( ). (pH 7.5 Tris-acetate EDTA buffer, 100 mM NaCl, 20°C). (c) Sedimentation equilibrium data (linear fit and residuals) indicating that M121E-cPYP (pH 7.5 Tris-acetate EDTA buffer, 100 mM NaCl) behaves as a monomer.
). (pH 7.5 Tris-acetate EDTA buffer, 100 mM NaCl, 20°C). (c) Sedimentation equilibrium data (linear fit and residuals) indicating that M121E-cPYP (pH 7.5 Tris-acetate EDTA buffer, 100 mM NaCl) behaves as a monomer.
Reversible denaturation by guanidinium was also measured (Fig. 7b) and M121E-cPYP was found to undergo a cooperative folding/unfolding transition. The M121E-cPYP mutant was somewhat less stable than cPYP as judged by the concentration of guanidinium at the midpoint of the denaturation curve.
Since circularly permuted proteins can sometimes form domain-swapped dimers (43, 44), we checked for self association of M121E-cPYP by analytical ultracentrifugation (Fig. 7c). These data indicated that the protein behaves as a monomer under the solution conditions tested.
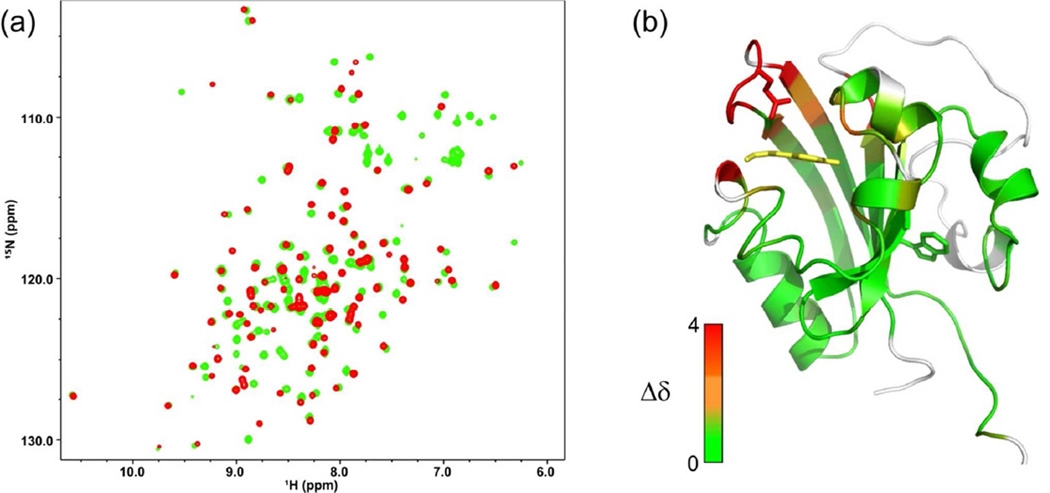
To obtain a more detailed assessment of the similarity of the dark state structure of M121E-cPYP to cPYP and wtPYP, we again compared backbone chemical shifts obtained using standard protein NMR methods (see supporting information). The 15N,1H-HSQC spectra of M121E-cPYP and cPYP are shown overlaid in Figure 8 together with a model colored according to chemical shift similarity. There is very close correspondence of chemical shifts throughput the central beta sheet as well as in those parts of the N-terminal domain that can be assigned (again the flexible linker residues could not be assigned). This overlap in chemical shifts includes the Trp indole side chain NH resonance. There are large changes in chemical shifts at the loop where the M121E mutation occurs. There are smaller changes in chemical shifts of residues near this mutation site but also near the phenolic group of the chromophore consistent with co-existence of distinct patterns of H-bonding around the chromophore and Glu67 (Glu46 in wild type numbering). Taken together, these data indicate that the M121E-cPYP mutant adopts much the same overall dark state structure as cPYP except that the mutation alters the details of the H-bonding patterns near the chromophore. The same conclusion was reached by Sasaki et al regarding the dark-sate structure of the M100L mutant of wtPYP (41) and by Devanathan et al for the M100A mutant (45).
Figure 8.
(a)15N,1H-HSQC spectrum of cPYP (green) and M121E-cPYP (red). For assignments see supporting information (b). Modeled structure of M121E-cPYP (based on wt PYP 1NWZ) with residues colored according to chemical shift difference from cPYP. The chemical shift difference was calculated as The Trp side chain is shown in green sticks; the indole NH has identical chemical shifts in cPYP and M121E-cPYP. Model produced using PyMol.
Light driven conformational change of M121E-cPYP
Joshi et al (40) have shown that the photocycle undergone by both the protonated and deprotonated Y42F-PYP mutant are essentially the same. We therefore expected that, although the switching efficiency may be lower, the conformational changes that ensue should be unaffected by the presence of the intermediate spectral form in the M121E-cPYP construct.
To characterize the nature of the conformational change, we first measured the CD spectrum of M121E-cPYP after exposure to blue light (Fig. 7a). Only a small change in the CD spectrum was observed. A similarly small CD change was seen by Lee et al (46) for wtPYP who interpreted this as indicating a molten globule like state was formed upon irradiation in which secondary structural elements were preserved but tertiary contacts were lost. Interestingly, a larger CD change was seen by Sasaki et al (41) in the M100L PYP mutant. These authors suggested that the molten globule state was in equilibrium with a structure in which helical secondary structure had been lost. Since the N-terminal cap only contains short helical sections its rearrangement may not lead to large changes in overall secondary structure.
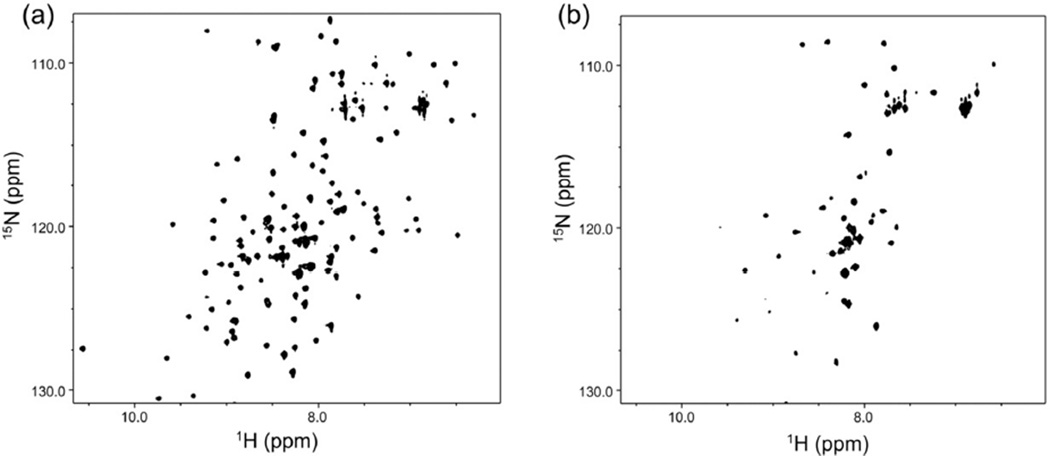
The 15N,1H-HSQC spectrum of M121E-cPYP in dark-adapted and irradiated states (Fig. 9) shows the changes expected based on the wild type protein with large changes in chemical shifts, an increase in signal intensity in the region of the spectrum associated with disordered structure, and a large decrease in the total number of peaks indicating extensive conformational exchange on a millisecond timescale (47–52).
Figure 9.
15N,1H-HSQC spectrum M121E-cPYP in dark-adapted (a) and irradiated (b) states.
Numerous pieces of evidence have led to the conclusion that blue light irradiation of wtPYP leads to large scale rearrangements and/or detachment of the N-terminal cap domain from the rest of the protein. These data include NMR studies on wtPYP and mutants, H/D exchange measurements, DEER measurements and, most recently, pump–probe X-ray solution scattering data (17, 20, 49, 50, 53–55). A central question in the present study is whether the light driven rearrangement of the N-terminal domain seen in the wild type also occurs in the M121E-cPYP construct.
To address this question directly, we opted to use the single Trp residue as a probe. As shown in Figure 10, Trp7 (119 in wtPYP numbering) is buried in the dark state structure (as calculated by the program Surface Racer (56)) as a consequence of packing of the N-terminal cap against the rest of the protein. If the N-terminal cap becomes detached or substantially reorganized, the Trp residue is expected to become solvent exposed. Indeed fluorescence measurements indicate this Trp residue becomes exposed in the light state of Δ25 PYP mutant lacking the N-terminal cap. (21, 57). Rather than using Trp fluorescence methods, which can be complicated by energy transfer to the chromophore in PYP (58), we opted to use 19F NMR measurements of a 19F-Trp substituted analogue of M121E-cPYP. 19F NMR spectra of fluorinated proteins often reveal dramatic changes in chemical shifts and line shapes between different folded states (59). Moreover, changing the solvent from D2O for H2O is known to yield an easily measured chemical shift perturbation of 19F nuclei that are solvent-exposed thus providing a quantitative measure of the degree of solvent exposure of the 19F-Trp site (28).
Figure 10.
15N,1H-HSQC spectra M121E-cPYP with (black) and without (red) 19F-Trp incorporation. (b) Chemical shifts showing perturbation are clustered in space around the fluorine atom (shown in green) on the Trp indole side chain. Model produced using PyMol.
We first measured the 15N,1H-HSQC spectrum of 19F-Trp labeled M121E-cPYP and M100EPYP to assess the degree of structural perturbation introduced by the fluorine label. Figure 10a shows an overlay of this spectrum with that of the unlabeled protein for cPYP. Spectra for M100E-PYP are included in the supporting information. Chemical shift perturbations are mapped onto the modeled M121E-cPYP structure in Fig. 10b. Only small changes are seen at sites close in space to the 19F atom confirming the non-perturbing nature of this single atom substitution. The HSQC data, together with UV-Vis spectra data and measurements of thermal relaxation rates (supporting information), indicate the fluorine substitution introduces minor local perturbations in the M100E-PYP protein also.
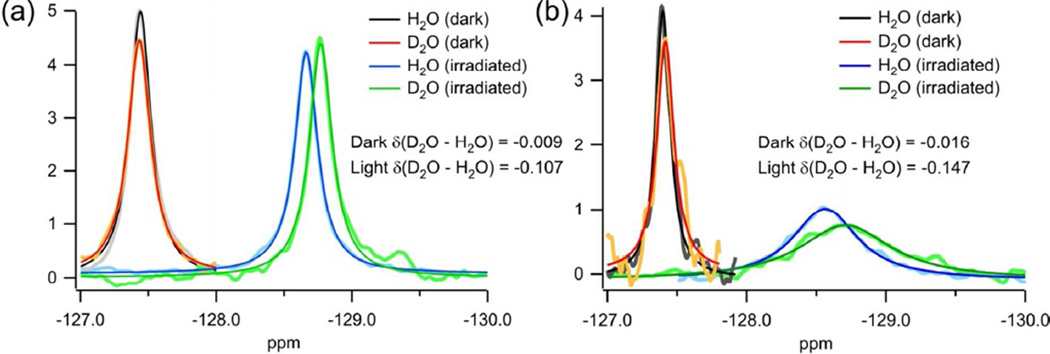
We then measured 19F NMR signals from the M121E-cPYP and M100E-PYP labeled proteins in both dark and irradiated conditions in both H2O and D2O at a series of temperatures. Figure 11 shows these data for 30°C; lower and higher temperatures are included in the supporting information. The dark adapted state of both M100E-PYP and M121E-cPYP gave sharp signals for the fluorine label near −127.4 ppm and these showed very small solvent isotope shifts (<0.016 ppm) consistent with a buried location for the Trp residue. Under similar conditions, the solvent isotope shift Δδ(D2O-H2O) for 5F-Trp free in solution is −0.109 ppm (28). At higher temperatures the solvent isotope shifts became larger (see supporting information) consistent with some temperature induced loosening of the structure. Irradiation caused the 19F signals to move upfield for both M100E-PYP and M121E-cPYP (Fig. 11, supporting information), consistent with enhanced solvent exposure (28). Irradiation also produced broadening of the 19F signal particularly for the circular permutant (Fig. 11). The degree of broadening was highly temperature sensitive with light state signals showing linewidths (FWHM values) of 400 Hz at 20°C and 130 Hz at 40°C. This behavior is consistent with a large degree of conformational exchange of the Trp residue in the light state of M121E-cPYP (60). The observation of large solvent isotope shifts Δδ(D2O-H2O) (Fig. 11, supporting information) for the 19F signal upon irradiation provides direct evidence that the Trp indole side chain is highly exposed in the light state of both M100E-PYP and M121E-cPYP.
Figure 11.
19F spectra of 5F-Trp labeled M100E-PYP (a) and M121E-cPYP (b) at 30°C under the conditions indicated in the legend.
Prospects for photo-control of target sequences
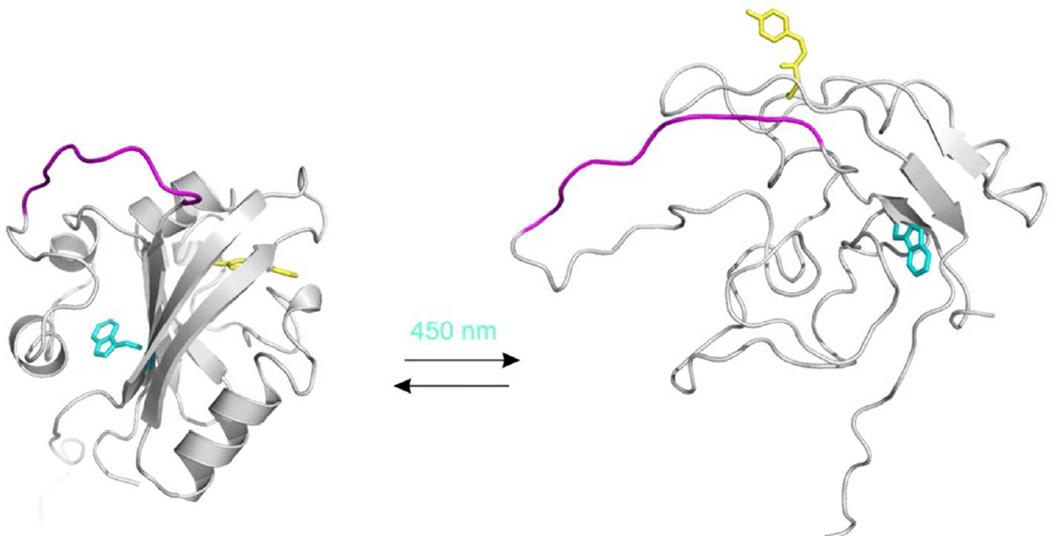
Overall the data presented here are consistent with a picture in which M121E-cPYP adopts a well folded dark state that is structurally very similar to that of wtPYP except for small perturbations in the N-terminal cap domain. Irradiation then causes a transition to a globally more flexible structure undergoing extensive conformational exchange in which the N-terminal cap that normally prevents solvent exposure of the Trp residue has moved to permit essentially complete solvent exposure. A picture of this light driven conformational change is shown in Figure 12. The dark state structure is modeled on that determined by X-ray diffraction of wtPYP (1NWZ) by Genick et al (36). The light state structure is based on a single structure taken from the ensemble of structures (2KX6) reported by Ramachandran et al for the light state of wtPYP (20). The GGSGGSGG insert sequence is highlighted in each model. Based on the data described here, it is likely that the conformational dynamics of this insert and particularly its accessibility will be affected by the large changes occurring in PYP. Based on previous experience with loop insertion, a wide range of peptide sequences and lengths are likely to be tolerated at this site (37, 44). Of course, inserts with defined conformational preferences may couple to PYP in a manner quite distinct from the short flexible linker studied here. It is unclear at present if the N-to-C distance change seen by Ramachandran et al (20) would have an associated free energy change large enough to drive folding/unfolding of a given insert sequence. Nevertheless, the light triggered conversion of the PYP domain to a globally flexible structure may permit uncovering of a binding surface or active site on an insert sequence even without the insert undergoing a conformational change. We are currently exploring the effectiveness of circularly permuted PYP to serve as a scaffold for photoswitching a range of insert sequences.
Figure 12.
Models of dark-adapted (left) and blue-light irradiated (right) M121E-cPYP based on 1NWZ (36) and 2KX6 (20) respectively. The insert sequence is colored magenta, the chromophore is shown as yellow sticks and the Trp residue is shown as cyan sticks. Models produced using PyMol.
Supplementary Material
Acknowledgements
We would like to thank Prof. Scott Prosser for advice on fluorine NMR. We would like to thank Prof. Alan Davidson for use of his titrating fluorometer.
This work has been supported by NSERC, CIHR and NIH R01 MH086379. NMR instrumentation at the Centre for Spectroscopic Investigation of Complex Organic Molecules and Polymers was supported by the CFI (Project number: 19119) and the Ontario Research Fund.
Footnotes
We use the term “light state” here to refer to the relatively long-lived putative signaling state of PYP. This state has also been called the I(2)' state and the pB state (20. Ramachandran, P. L., Lovett, J. E., Carl, P. J., Cammarata, M., Lee, J. H., Jung, Y. O., Ihee, H., Timmel, C. R., and van Thor, J. J. (2011) The short-lived signaling state of the photoactive yellow protein photoreceptor revealed by combined structural probes, J. Am. Chem. Soc. 133, 9395–9404.).
Supporting Information.
Further spectroscopic details are provided including pH dependence of UV Vis spectra for M121EcPYP and characterization of M100E-PYP. 19F spectra of 5F-Trp labeled M121E-cPYP at a series of temperatures. Backbone NMR assignments of cPYP and M121E-cPYP. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ha JH, Loh SN. Protein conformational switches: From Nature to design. Chem. Eur. J. 2012;18:7984–7999. doi: 10.1002/chem.201200348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strickland D, Lin Y, Wagner E, Hope CM, Zayner J, Antoniou C, Sosnick TR, Weiss EL, Glotzer M. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat. Methods. 2012;9:379–U392. doi: 10.1038/nmeth.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou XX, Chung HK, Lam AJ, Lin MZ. Optical control of protein activity by fluorescent protein domains. Science. 2012;338:810–814. doi: 10.1126/science.1226854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strickland D, Yao X, Gawlak G, Rosen MK, Gardner KH, Sosnick TR. Rationally improving LOV domain-based photoswitches. Nat. Methods. 2010;7:623–626. doi: 10.1038/nmeth.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Airan RD, Thompson KR, Fenno LE, Bernstein H, Deisseroth K. Temporally precise in vivo control of intracellular signalling. Nature. 2009;458:1025–1029. doi: 10.1038/nature07926. [DOI] [PubMed] [Google Scholar]
- 6.Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lungu OI, Hallett RA, Choi EJ, Aiken MJ, Hahn KM, Kuhlman B. Designing photoswitchable peptides using the AsLOV2 domain. Chem. Biol. 2012;19:507–517. doi: 10.1016/j.chembiol.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moglich A, Ayers RA, Moffat K. Addition at the Molecular Level: Signal Integration in Designed Per-ARNT-Sim Receptor Proteins. J. Mol. Biol. 2010;400:477–486. doi: 10.1016/j.jmb.2010.05.019. [DOI] [PubMed] [Google Scholar]
- 9.Moglich A, Moffat K. Engineered photoreceptors as novel optogenetic tools. Photochem. Photobiol. Sci. 2010;9:1286–1300. doi: 10.1039/c0pp00167h. [DOI] [PubMed] [Google Scholar]
- 10.Christie JM, Gawthorne J, Young G, Fraser NJ, Roe AJ. LOV to BLUF: Flavoprotein contributions to the optogenetic toolkit. Mol. Plant. 2012;5:533–544. doi: 10.1093/mp/sss020. [DOI] [PubMed] [Google Scholar]
- 11.Losi A, Gartner W. Old chromophores, new photoactivation paradigms, trendy applications: Flavins in blue light-sensing photoreceptors. Photochem. Photobiol. 2011;87:491–510. doi: 10.1111/j.1751-1097.2011.00913.x. [DOI] [PubMed] [Google Scholar]
- 12.Woolley GA. Designing chimeric LOV photoswitches. Chem. Biol. 2012;19:441–442. doi: 10.1016/j.chembiol.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 13.Nash AI, Ko WH, Harper SM, Gardner KH. A conserved glutamine plays a central role in LOV domain signal transmission and its duration. Biochemistry. 2008;47:13842–13849. doi: 10.1021/bi801430e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 15.Kumauchi M, Hara MT, Stalcup P, Xie A, Hoff WD. Identification of six new photoactive yellow proteins--diversity and structure-function relationships in a bacterial blue light photoreceptor. Photochem. Photobiol. 2008;84:956–969. doi: 10.1111/j.1751-1097.2008.00335.x. [DOI] [PubMed] [Google Scholar]
- 16.Lee BC, Pandit A, Croonquist PA, Hoff WD. Folding and signaling share the same pathway in a photoreceptor. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9062–9067. doi: 10.1073/pnas.111153598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hellingwerf KJ, Hendriks J, Gensch T. Photoactive Yellow Protein, a new type of photoreceptor protein: Will this "yellow lab" bring us where we want to go? J. Phys. Chem. A. 2003;107:1082–1094. [Google Scholar]
- 18.Meyer TE, Kyndt JA, Memmi S, Moser T, Colon-Acevedo B, Devreese B, Van Beeumen JJ. The growing family of photoactive yellow proteins and their presumed functional roles. Photochem. Photobiol. Sci. 2012;11:1495–1514. doi: 10.1039/c2pp25090j. [DOI] [PubMed] [Google Scholar]
- 19.Kyndt JA, Vanrobaeys F, Fitch JC, Devreese BV, Meyer TE, Cusanovich MA, Van Beeumen JJ. Heterologous production of Halorhodospira halophila holophotoactive yellow protein through tandem expression of the postulated biosynthetic genes. Biochemistry. 2003;42:965–970. doi: 10.1021/bi027037b. [DOI] [PubMed] [Google Scholar]
- 20.Ramachandran PL, Lovett JE, Carl PJ, Cammarata M, Lee JH, Jung YO, Ihee H, Timmel CR, van Thor JJ. The short-lived signaling state of the photoactive yellow protein photoreceptor revealed by combined structural probes. J. Am. Chem. Soc. 2011;133:9395–9404. doi: 10.1021/ja200617t. [DOI] [PubMed] [Google Scholar]
- 21.Morgan SA, Al-Abdul-Wahid MS, Woolley GA. Structure-based design of a photocontrolled DNA binding protein. J. Mol. Biol. 2010;399:94–112. doi: 10.1016/j.jmb.2010.03.053. [DOI] [PubMed] [Google Scholar]
- 22.Fan HY, Morgan SA, Brechun KE, Chen YY, Jaikaran AS, Woolley GA. Improving a designed photocontrolled DNA-binding protein. Biochemistry. 2011;50:1226–1237. doi: 10.1021/bi101432p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan SA, Woolley GA. A photoswitchable DNA-binding protein based on a truncated GCN4-photoactive yellow protein chimera. Photochem. Photobiol. Sci. 2010;9:1320–1326. doi: 10.1039/c0pp00214c. [DOI] [PubMed] [Google Scholar]
- 24.Ui M, Tanaka Y, Araki Y, Wada T, Takei T, Tsumoto K, Endo S, Kinbara K. Application of photoactive yellow protein as a photoresponsive module for controlling hemolytic activity of staphylococcal alpha-hemolysin. Chem. Comm. 2012;48:4737–4739. doi: 10.1039/c2cc18118e. [DOI] [PubMed] [Google Scholar]
- 25.Devanathan S, Genick UK, Getzoff ED, Meyer TE, Cusanovich MA, Tollin G. Preparation and properties of a 3,4-dihydroxycinnamic acid chromophore variant of the photoactive yellow protein. Arch. Biochem. Biophys. 1997;340:83–89. doi: 10.1006/abbi.1997.9902. [DOI] [PubMed] [Google Scholar]
- 26.Changenet-Barret P, Espagne A, Katsonis N, Charier S, Baudin JB, Jullien L, Plaza P, Martin MM. Excited-state relaxation dynamics of a PYP chromophore model in solution: influence of the thioester group. Chem. Phys. Lett. 2002;365:285–291. [Google Scholar]
- 27.Clarke J, Fersht AR. Engineered disulfide bonds as probes of the folding pathway of barnase: increasing the stability of proteins against the rate of denaturation. Biochemistry. 1993;32:4322–4329. doi: 10.1021/bi00067a022. [DOI] [PubMed] [Google Scholar]
- 28.Evanics F, Bezsonova I, Marsh J, Kitevski JL, Forman-Kay JD, Prosser RS. Tryptophan solvent exposure in folded and unfolded states of an SH3 domain by 19F and 1H NMR. Biochemistry. 2006;45:14120–14128. doi: 10.1021/bi061389r. [DOI] [PubMed] [Google Scholar]
- 29.Ikura M, Kay LE, Bax A. A novel approach for sequential assignment of 1H, 13C, and 15N spectra of proteins: heteronuclear triple-resonance three-dimensional NMR spectroscopy. Application to calmodulin. Biochemistry. 1990;29:4659–4667. doi: 10.1021/bi00471a022. [DOI] [PubMed] [Google Scholar]
- 30.Wittekind M, Mueller L. HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the alpha-carbon and beta-carbon resonances in proteins. J. Mag. Res. Ser. B. 1993;101:201–205. [Google Scholar]
- 31.Grzesiek S, Bax A. Correlating backbone amide and side-chain resonances in larger proteins by multiple relayed triple resonance NMR. J. Am. Chem. Soc. 1992;114:6291–6293. [Google Scholar]
- 32.Grzesiek S, Anglister J, Bax A. Correlation of backbone amide and aliphatic sidechain resonances in C-13/N-15-enriched proteins by isotropic mixing of C-13 magnetization. J. Mag. Res. Ser. B. 1993;101:114–119. [Google Scholar]
- 33.Mohebbi A, Shaka AJ. Improvements in C-13 broad-band homonuclear crosspolarization for 2D and 3D NMR. Chem. Phys. Lett. 1991;178:374–378. [Google Scholar]
- 34.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 35.Kanelis V, Forman-Kay JD, Kay LE. Multidimensional NMR methods for protein structure determination. IUBMB Life. 2001;52:291–302. doi: 10.1080/152165401317291147. [DOI] [PubMed] [Google Scholar]
- 36.Getzoff ED, Gutwin KN, Genick UK. Anticipatory active-site motions and chromophore distortion prime photoreceptor PYP for light activation. Nat. Struct. Biol. 2003;10:663–668. doi: 10.1038/nsb958. [DOI] [PubMed] [Google Scholar]
- 37.Butler JS, Mitrea DM, Mitrousis G, Cingolani G, Loh SN. Structural and thermodynamic analysis of a conformationally strained circular permutant of barnase. Biochemistry. 2009;48:3497–3507. doi: 10.1021/bi900039e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pool TJ, Oktaviani NA, Kamikubo H, Kataoka M, Mulder FAA. H-1, C-13, and N-15 resonance assignment of photoactive yellow protein. Biomol. NMR Assign. 2013;7:97–100. doi: 10.1007/s12104-012-9387-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Genick UK, Devanathan S, Meyer TE, Canestrelli IL, Williams E, Cusanovich MA, Tollin G, Getzoff ED. Active site mutants implicate key residues for control of color and light cycle kinetics of photoactive yellow protein. Biochemistry. 1997;36:8–14. doi: 10.1021/bi9622884. [DOI] [PubMed] [Google Scholar]
- 40.Joshi CP, Otto H, Hoersch D, Meyer TE, Cusanovich MA, Heyn MP. Strong hydrogen bond between glutamic acid 46 and chromophore leads to the intermediate spectral form and excited state proton transfer in the Y42F mutant of the photoreceptor photoactive yellow protein. Biochemistry. 2009;48:9980–9993. doi: 10.1021/bi9012897. [DOI] [PubMed] [Google Scholar]
- 41.Sasaki J, Kumauchi M, Hamada N, Oka T, Tokunaga F. Light-induced unfolding of photoactive yellow protein mutant M100L. Biochemistry. 2002;41:1915–1922. doi: 10.1021/bi011721t. [DOI] [PubMed] [Google Scholar]
- 42.Kumauchi M, Hamada N, Sasaki J, Tokunaga F. A role of methionine 100 in facilitating PYP(M)-decay process in the photocycle of photoactive yellow protein. J. Biochem. 2002;132:205–210. doi: 10.1093/oxfordjournals.jbchem.a003211. [DOI] [PubMed] [Google Scholar]
- 43.Szilagyi A, Zhang Y, Zavodszky P. Intra-chain 3D segment swapping spawns the evolution of new multidomain protein architectures. J. Mol. Biol. 2012;415:221–235. doi: 10.1016/j.jmb.2011.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cutler TA, Mills BM, Lubin DJ, Chong LT, Loh SN. Effect of interdomain linker length on an antagonistic folding-unfolding equilibrium between two protein domains. J. Mol. Biol. 2009;386:854–868. doi: 10.1016/j.jmb.2008.10.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Devanathan S, Genick UK, Canestrelli IL, Meyer TE, Cusanovich MA, Getzoff ED, Tollin G. New insights into the photocycle of Ectothiorhodospira halophila photoactive yellow protein: photorecovery of the long-lived photobleached intermediate in the Met100Ala mutant. Biochemistry. 1998;37:11563–11568. doi: 10.1021/bi9803776. [DOI] [PubMed] [Google Scholar]
- 46.Lee BC, Croonquist PA, Sosnick TR, Hoff WD. PAS domain receptor photoactive yellow protein is converted to a molten globule state upon activation. J. Biol. Chem. 2001;276:20821–20823. doi: 10.1074/jbc.C100106200. [DOI] [PubMed] [Google Scholar]
- 47.Fuentes G, Nederveen AJ, Kaptein R, Boelens R, Bonvin AM. Describing partially unfolded states of proteins from sparse NMR data. J. Biomol. NMR. 2005;33:175–186. doi: 10.1007/s10858-005-3207-9. [DOI] [PubMed] [Google Scholar]
- 48.Bernard C, Houben K, Derix NM, Marks D, van der Horst MA, Hellingwerf KJ, Boelens R, Kaptein R, van Nuland NA. The solution structure of a transient photoreceptor intermediate: Delta25 photoactive yellow protein. Structure. 2005;13:953–962. doi: 10.1016/j.str.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 49.Derix NM, Wechselberger RW, van der Horst MA, Hellingwerf KJ, Boelens R, Kaptein R, van Nuland NA. Lack of negative charge in the E46Q mutant of photoactive yellow protein prevents partial unfolding of the blue-shifted intermediate. Biochemistry. 2003;42:14501–14506. doi: 10.1021/bi034877x. [DOI] [PubMed] [Google Scholar]
- 50.Craven CJ, Derix NM, Hendriks J, Boelens R, Hellingwerf KJ, Kaptein R. Probing the nature of the blue-shifted intermediate of photoactive yellow protein in solution by NMR: hydrogen-deuterium exchange data and pH studies. Biochemistry. 2000;39:14392–14399. doi: 10.1021/bi001628p. [DOI] [PubMed] [Google Scholar]
- 51.Dux P, Rubinstenn G, Vuister GW, Boelens R, Mulder FA, Hard K, Hoff WD, Kroon AR, Crielaard W, Hellingwerf KJ, Kaptein R. Solution structure and backbone dynamics of the photoactive yellow protein. Biochemistry. 1998;37:12689–12699. doi: 10.1021/bi9806652. [DOI] [PubMed] [Google Scholar]
- 52.Rubinstenn G, Vuister GW, Mulder FA, Dux PE, Boelens R, Hellingwerf KJ, Kaptein R. Structural and dynamic changes of photoactive yellow protein during its photocycle in solution. Nat. Struct. Biol. 1998;5:568–570. doi: 10.1038/823. [DOI] [PubMed] [Google Scholar]
- 53.Khan JS, Imamoto Y, Harigai M, Kataoka M, Terazima M. Conformational changes of PYP monitored by diffusion coefficient: effect of N-terminal alpha-helices. Biophys. J. 2006;90:3686–3693. doi: 10.1529/biophysj.105.078196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng G, Cusanovich MA, Wysocki VH. Properties of the dark and signaling states of photoactive yellow protein probed by solution phase hydrogen/deuterium exchange and mass spectrometry. Biochemistry. 2006;45:11744–11751. doi: 10.1021/bi0608663. [DOI] [PubMed] [Google Scholar]
- 55.Kim TW, Lee JH, Choi J, Kim KH, van Wilderen LJ, Guerin L, Kim Y, Jung YO, Yang C, Kim J, Wulff M, van Thor JJ, Ihee H. Protein structural dynamics of photoactive yellow protein in solution revealed by pump-probe X-ray solution scattering. J. Am. Chem. Soc. 2012;134:3145–3153. doi: 10.1021/ja210435n. [DOI] [PubMed] [Google Scholar]
- 56.Tsodikov OV, Record MT, Jr., Sergeev YV. Novel computer program for fast exact calculation of accessible and molecular surface areas and average surface curvature. J. Comput. Chem. 2002;23:600–609. doi: 10.1002/jcc.10061. [DOI] [PubMed] [Google Scholar]
- 57.Gensch T, Hendriks J, Hellingwerf KJ. Tryptophan fluorescence monitors structural changes accompanying signalling state formation in the photocycle of photoactive yellow protein. Photochem. Photobiol. Sci. 2004;3:531–536. doi: 10.1039/b401600a. [DOI] [PubMed] [Google Scholar]
- 58.Hoersch D, Otto H, Cusanovich MA, Heyn MP. Distinguishing chromophore structures of photocycle intermediates of the photoreceptor PYP by transient fluorescence and energy transfer. J. Phys. Chem. B. 2008;112:9118–9125. doi: 10.1021/jp801174z. [DOI] [PubMed] [Google Scholar]
- 59.Danielson MA, Falke JJ. Use of 19F NMR to probe protein structure and conformational changes. Annu. Rev. Biophys. Biomol. Struct. 1996;25:163–195. doi: 10.1146/annurev.bb.25.060196.001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoeltzli SD, Frieden C. 19F NMR spectroscopy of [6-19F]tryptophan-labeled Escherichia coli dihydrofolate reductase: equilibrium folding and ligand binding studies. Biochemistry. 1994;33:5502–5509. doi: 10.1021/bi00184a019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.