Abstract
Dermatophytes belonging to the Trichophyton and Arthroderma genera cause skin infections in humans and animals. From genome sequencing data, we mined a conserved gene cluster among dermatophytes that are homologous to one that produces an immunosuppressive polyketide in Aspergillus fumigatus. Using a recombination-based cloning strategy in yeast, we constructed fungal heterologous expression vectors that encode the cryptic clusters. When integrated into the model Aspergillus nidulans host, a structurally related compound neosartoricin B was formed, suggesting a possible role of this compound in the pathogenesis of these strains.
Keywords: natural products, polyketide, heterologous expression, prenyltransferase
Secondary metabolites (SMs) play important roles in the pathogenesis of microorganisms. In pathogenic fungi, polyketide and nonribosomal peptides have been shown to be the potential virulence factors and immunosuppressants1-5. Identification of these factors is therefore highly important towards understanding the molecular basis of host-pathogen interactions6, 7. Genome sequencing of numerous dermatophytes belonging to the Trichophyton and Arthroderma genera revealed that each genome encodes numerous SM clusters1,8. Unfortunately, under laboratory conditions many of the gene clusters in fungi are silent and hence mask the products encoded in them 9. The difficulties involved in manipulating these fungi, such as long doubling times and lack of genetic tools, further impedes the establishment of metabolite-cluster correlations.
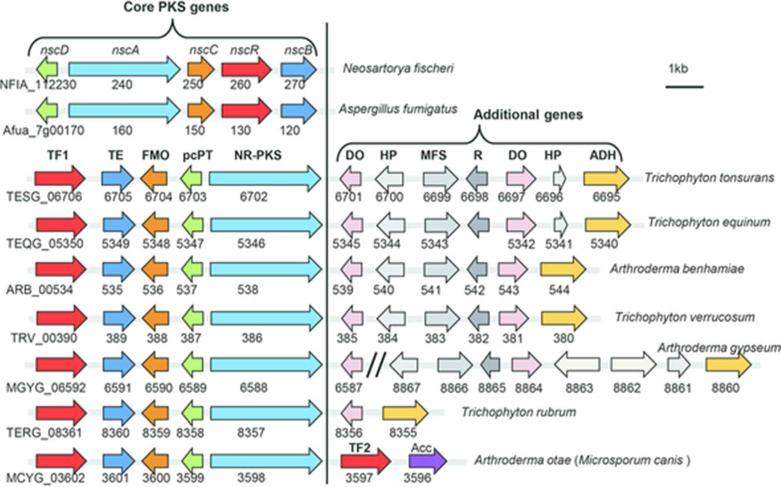
When analyzing the polyketide gene clusters from the sequenced dermatophyte genomes1, 8, we found an orthologous gene cluster, which is also conserved in pathogenic fungi Aspergillus fumigatus and Neosartorya fischeri 10, 11(Figure 1). In each genome, the cluster contains a set of four genes that includes polyketide synthase (PKS), a β-lactamase like thioesterase (TE), a flavin-dependent monooxygenase (FMO) and a polycyclic prenyltransferase (pcPT) 10, 11. In addition to these four well-conserved “core PKS genes”, the neighboring regions encode additional genes, some of which are highly syntenic and conserved between genomes (Figure 1). We recently showed that when activated, these core PKS genes from A. fumigatus and N. fischeri can synthesize a prenylated, tricyclic polyketide neosartoricin11. Neosartoricin was shown to inhibit T-cell proliferation with an IC50 of 3 μM and was proposed to possibly play a role in suppressing host adaptive immune response during infection11. The conservation of the core PKS genes in the dermatophytes (>90% identity between them) (Table S3, Supplementary Information), and the lower sequence homology to those in A. fumigatus and N. fischeri (~70% identity, Table S3), raise an interesting question as to whether these fungi produce similar compounds, and whether the presence of additional conserved genes may lead to further tailoring of the neosartoricin scaffold.
Figure 1.
Comparison of conserved gene clusters across different dermatophyte and pathogenic fungi. All of the fungi shown contain a conserved core PKS gene cassette that consists of a nonreducing polyketide synthase (NR-PKS), β-lactamase thioesterase (TE), flavin-dependent monooxygenase (FMO) and polycyclic prenyltransferase (pcPT). A fungal transcription factor (TF) is also found immediately adjacent to the core PKS genes. In dermatophytes, additional genes are also found adjacent to the core PKS gene cassette, among which many are conserved across different fungi. Some of the genes include dioxygenase (DO), hypothetical protein (HP), amino acid racemase (R), major facilitator superfamily transporters (MFS) and alcohol dehydrogenase (ADH). In A. otae, an additional C6 transcriptional factor (TF2) is adjacent to the core PKS gene cassette.
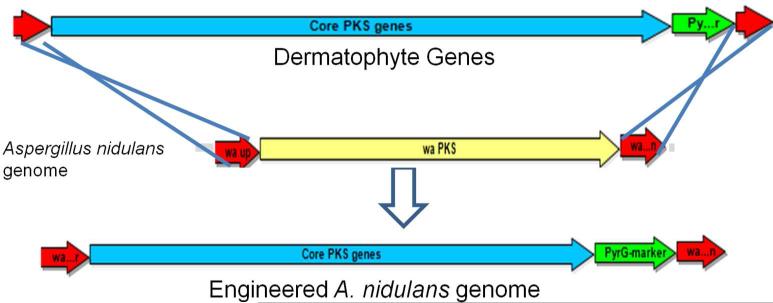
To examine the activities of the conserved core PKS genes from dermatophytes, we cloned the clusters using yeast recombination12 followed by expression in Aspergillus nidulans (Figure S1, Supplementary Information). We initiated the cloning strategy by creating an E. coli-yeast-Aspergillus shuttle vector, pYH-wA-pyrG, which consists of a ColE1 origin of replication from SuperCos1, a yeast centromere sequence (CEN) and an autonomously replicating sequence (ARS) 13, 14 (Table S1, Materials and Methods). The choice of the low copy number in Saccharomyces cerevisiae allows stable maintenance of the subsequent large (>20 kb) plasmids containing cloned gene clusters. For fungal heterologous expression, we used the well-studied A. nidulans containing the ΔnkuA deletion (Table S1), which minimizes non-homologous recombination events15, 16. The wA locus in A. nidulans, which encodes a pigment-encoding and nonessential PKS17, was chosen as the site for homologous integration of the gene clusters (Figures 2 and S1B). This allowed the facile, initial screening of correct recombination through visual evaluation of the resulting white conidia.
Figure 2.
The strategy for integrating dermatophyte gene cluster into A. nidulans. The dermatophyte genes were cloned between A. nidulans wA flanking regions and then integrated into wA locus by replacing the wAPKS gene.
Using in vivo yeast recombination cloning (Figure S1A), the core PKS genes (TESG_6702-6705), along with a putative pathway specific fungal transcription factor (TF, TESG_6706) (Figure 1) from Trichophyton tonsurans was cloned into the pYH-wA-pyrG vector adjacent to the A. fumigatus pyrG (AfpyrG) selection marker. The entire cassette was flanked by upstream and downstream sequences of the wA gene (Figure S1A). The native promoter of the TF (TESG_6706) gene was replaced with the A. nidulans gpdA promoter. The use of the constitutive promoter is to ensure expression of the foreign gene cluster in A. nidulans, since neosartoricin-like compounds are not isolated from the dermatophytes under a variety of culture conditions (data not shown). The resultant plasmid containing the four core PKS genes was then linearized and introduced into A. nidulans. A number of the nutrition-complemented transformants displayed the loss of the green spore pigments associated to the wA mutant phenotype. In addition, a yellow pigmentation was observed for these mutants, which is similar to the N. fischeri strain overexpressing neosartoricin in our previous study 11 (Figure S2).
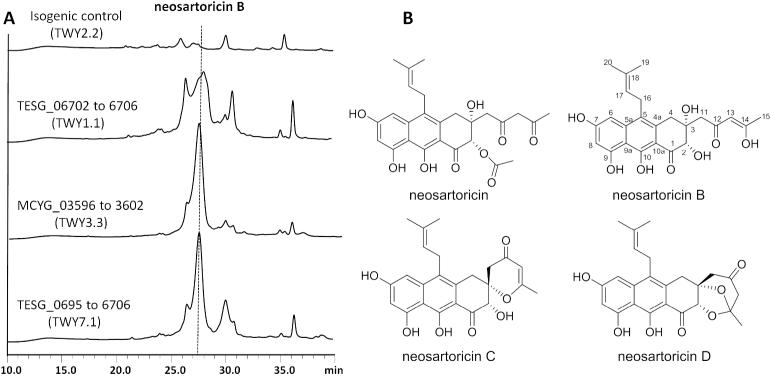
Following PCR verification of the integration of all four genes, the strain TWY1.1 (TESG_06702-6706) was cultivated in GMM media with 0.5 μM pyridoxine HCl; and the metabolites were extracted and analyzed with LC-MS (Figure 3). Compared to the isogenic control TWY2.2 (ΔwAPKS) in which the wA locus was disrupted with the pyrG cassette alone, a new metabolite (yield 10 mg/L) was observed with UV absorption identical to that of neosartoricin. Mass analysis showed that the compound has m/z = 443 [M+H]+ (Figure S3C), which is 42 mass units (mu) lower than that of neosartoricin. The new metabolite (hereafter named neosartoricin B) was purified from the crude extract and fully characterized by one and two-dimensional NMR analyses (Table S4 and Supplementary NMR spectra). Neosartoricin B is structurally similar to neosartoricin, with the loss of acetyl group at the C2 hydroxyl. All other structural features, including the polyhydroxylated aromatic scaffold, C5 prenylation and 1,3-diketo substituent at C3 were all verified to be identical. The C2 acetyl group in neosartoricin was attributed to the action of an unidentified and likely endogenous acetyltransferase in A. fumigatus. The absence of this group in neosartoricin B may be attributed to the lack of this activity in A. nidulans.
Figure 3.
The production of neosartoricins from synthetic mutants in A. nidulans. (A) HPLC traces (400 nm) of organic extracts from different strains. Trace 1: isogenic control TWY2.2; Trace 2: TWY1.1 (TESG_06702 to 6706); Trace 3: TWY7.1 (TESG_0695 to 6706); and Trace 4: TWY3.3 (MCYG_03596 to 3602). (B) The structures of neosartoricin, neosartoricins B, C and D.
During purification of neosartoricin B, we observed that under slightly acidic conditions, neosartoricin B can be converted into two additional compounds neosartoricins C and D (Figure 3B), both of which have the molecular weight of 424 (Figure S4). Full structural characterizations were performed to reveal their structures as shown in Figure 3B. Interestingly, neosartoricin C is a spirocyclic compound that is cyclized through the attack of C3 hydroxyl on C14, followed by dehydration. On the other hand, neosartoricin D is a further cyclized compound in which attack of C2 on C14 in neosartoricin C resulted in the formation of the acetal-containing dioxabicyclo-octanone ring. Both of these compounds are novel and possibly represent related metabolites of the gene cluster. Indeed, analysis of the organic extract of TWY1.1 also revealed small amounts of neosartoricins C and D, although it is not known whether these are formed during culturing or during the extraction process.
Other highly identical and syntenic genes were found immediately adjacent to most of the core PKS gene ensembles in some of the dermatophytes, including T. tonsurans, T. equinum, T. verrucosum, and A. benhamiae (Figure 1). Among them include enzymes that one may anticipate to associate with the modification of secondary metabolites, including putative dioxygenase (DO), amino acid racemase (R) and alcohol dehydrogenase (ADH). Hence it appears tantalizingly possible that among these organisms, neosartoricin B may be further morphed into a different product that may have additional biological properties. To assess the possible roles of these enzymes in tailoring of neosartoricin B, we used a similar strategy to insert different combinations of these genes from T. tonsurans into the neosartoricin B-producing host TWY1.1 (Figure S1B). Specifically, we added TESG06695-6701, TESG06695-6701 (without 6696) and TESG_06701 to yield TWY7.1, TWY8.1 and TWY9.1, respectively. However, follow-up analysis of the extracts from these strains yielded metabolic profiles identical to that of TWY1.1 (Figures 3 and S3A). This suggests that although highly conserved, the genes either may not express or do not co-transcribe with the core PKS genes. Indeed, RT-PCR analysis of the genes TESG6698 and TESG6701 in TWY7.1 showed that the genes included in this cassette are not co-transcribed with the PKS-associated genes with the constitutively expressed TF (Figure S5), hence are likely not involved in the neosartoricin B pathway. Lastly, six contiguous genes in Arthroderma otae (Microsporum canis) (MCYG_03596-3602) including the four core PKS genes, a TF gene and a putative aminocyclopropane carboxylic acid synthase (ACCS) gene were also introduced into A. nidulans to create TWY3.3, which was confirmed to produce neosartoricin B as well (Figures 3 and S2).
Our discovery of neosartoricin B demonstrates that the highly conserved core PKS genes (PKS, TE, FMO and pcPT) in dermatophytes can synthesize nearly the same prenylated aromatic polyketide as in A. fumigatus and N. fischeri, of which we showed have notable immunosuppressive activities. Neosartoricin B may therefore mediate immunomodulatory interactions with the host during infection and colonization of the pathogenic fungi. The biosynthetic mechanism of neosartoricin B is expected to follow that proposed for neosartoricin11. Hence the neosartoricin compounds may represent conserved SM shared by pathogenic fungal strains. Our approach shows the potential to use the model fungus to analyze the products of biosynthetic pathways from difficult to handle organisms.
MATERIALS AND METHODS
Strain, media and growth conditions
The fungal strains used in this study are listed in the Supplementary Information Table S1. All strains were grown at 37°C on glucose minimum medium (GMM) 18 and when appropriate were supplemented with 0.56 g uracil L−1, 1.26 g uridine L−1, 0.5 μM pyridoxine HCl, and maintained as glycerol stocks at −80°C. Escherichia coli strains XL-1 Blue (Stratagene) was used for DNA manipulation.
Gene cloning, plasmid construction and genetic manipulation
The plasmids utilized in this work are listed in Table S1. The oligonucleotide sequences for PCR primers are given in Table S2. PCR reactions were performed with Phusion high-fidelity DNA polymerase (New England Biolabs). PCR screening for transformants and mutants were carried out with Quick-Load® Taq 2X Master Mix (New England Biolabs).
The basic strategy for assembling of large PCR fragments is splicing by overlapping extension (SOE)-PCR and yeast homologous recombination as described in Figure S1. Saccharomyces cerevisiae strain BJ5464-NpgA (MATα ura3-52 his3-Δ200 leu2-Δ1 trp1 pep4::HIS3 prb1Δ1.6R can1 GAL) was used as the host12. The vector pYH-WA-pyrG is constructed by in vivo yeast recombination of three linear DNA fragments consisting of 1) the ColE1 origin of replication and ampR from SuperCos1 vector (Stratagene), 2) a yeast centromere sequence (CEN), autonomously replicating sequence (ARS) and a URA3 marker from pXP742 19, and 3) a 5′wA-AfpyrG-3′wA cassette synthesized by fusion PCR. The pyrG auxotrophy marker was amplified from A. fumigatus genomic DNA, while the 5′ and 3′ regions of the wA gene were amplified from A. nidulans genomic DNA. All three DNA fragments for in vivo yeast recombination contained a minimum of 35 bp overlapping bases with the flanking fragments. Primers used for construction of the plasmid are listed in Table S2.
Using the same strategy, all plasmids pWY13.2, 14.1, 15, 16 and 17.1 (Table S1) were constructed in this study. Overlapping regions between two flanking segments of TESG_06695-6706 and MCYG_03596-3602 ranged from 60-150 bp and overlapping regions between the segments and the vector pYH-WA-pyrG were 40 bp. Briefly, four PCR fragments (ca. 4 kb/ each fragment) of TESG_06706-6702 were amplified with T. tonsurans genomic DNA by using designated primers (Table S2) respectively. The gpdA promoter was amplified by using A. nidulans genomic DNA and fused with transcription factor (TF) TESG_06706 using designated primers (Table S2). Then, all the fragments, NheI digested vector were gel purified and transformed into S. cerevisiae BJ5464-NpgA by using S. c. EasyCompTM Transformation Kit (Invitrogen). The obtained yeast colonies were characterized by PCR. Yeast plasmids were isolated by using Zymoprep™ (D2001) Kit (Zymo Research) and transformed into E. coli XL1 Blue. The fragments for MCYG_03596-3602 from A. otae were assembled to create plasmid pWY14.2. For the construction of plasmids pWY15, 16 and 17.1, the selectable marker AfpyroA was amplified from A. fumigatus and assembled with NotI digested pYH-WA-pyrG, and amplified genomic DNA segments from the fungal hosts (Figure 1). All plasmids were confirmed by restriction enzyme digestion and sequencing.
Transformation of A. nidulans
A. nidulans strain RJMP1.49 was used as the recipient host. Fungal protoplast preparation and transformation were modified as the description from Bok and Keller20. The modifications are the following: 1) culture medium for conidia germination is reduced to 20-50 mL and germination time is shortened. Inoculate 20-50 mL of sterile liquid Minimal Medium (LMM, containing the appropriate supplements) with about 5 × 108 fresh spores (1 × 107 conidia mL−1) and shake at 37°C and 280 rpm for approximately 5-6 h for spore germination. The germination time can be shortened 1-2 h if yeast extract is added to medium. 2) Digestion temperature with lysing enzyme is adjusted to 37°C which can reduce digestion time 1-2 h. Optionally, digestion can also be performed at room temperature overnight to obtain better protoplasts. 5 to 15 micrograms of plasmids were linearized and transformed into RJMP1.49. Transformation with linearized pYH-WA-pyrG was used as control. Transformants were verified by using diagnostic PCR with appropriate primers (Table S2).
RNA extraction and reverse transcriptase PCR (RT-PCR)
106 spores from TWY2.1 (Isogenic control), TWY1.1, TWY3.3 and TWY7.1 were inoculated into 10 mL LMM with pyridoxine (0.5 μM) and cultivated at 37°C for 2 days under dark conditions with two replicates each. Then, the mycelia were harvested and total RNA was extracted using the Ambion RiboPure™-Yeast Kit according to the instructions (Invitrogen, Carlsbad, CA). For transcription assessment of dermatophyte clusters in A. nidulans, the single strand cDNAs from TWY2.1, TWY1.1 TWY3.3 and TWY7.1 were synthesized by using ImProm-II™ Reverse Transcription System (Promega, Madison, WI). Genes coding for putative TF1 (TESG_06706), NR-PKS (TESG_06702), dioxygenase (TESG_06701) and asp/glu racemase (TESG_06698) were used for the transcription assessment of TESG_06695-6706 from T. tonsurans. Genes coding for putative TF1 (MCYG_03602) and NR-PKS (MCYG_03598) were used for the assessment of MCYG_03596-3602 transcription from A. otae.
Fermentation and LC-MS analysis
A. nidulans strains were cultivated at 37°C in liquid GMM (supplemented with 0.5 μM pyridoxine HCl) at 1.0 × 105 spores per 10 cm plate in the dark. After 2 days, 700 μL cultures were taken from each strain and transferred to a 1.5 mL eppendorf tube. The cultures were extracted with 700 μL of ethyl acetate (EtOAc)/methanol (MeOH)/acetic acid (AcOH) (89:10:1). The organic phase was evaporated to dryness and re-dissolved in 100 μL MeOH. 10 μL of dissolved extract was injected for high performance liquid chromatography-photodiode array detection- mass spectrometry (HPLC-DAD-MS) analysis. LC-MS spectra were obtained on a Shimadzu 2010 EV liquid chromatography mass spectrometer using positive and negative electrospray ionization and a Phenomenex Luna 5 μm, 2.0 mm X 100 mm C18 reverse-phase column. Samples were separated on a linear gradient of 5-95% acetonitrile (CH3CN) in water (0.1% formic acid) for 30 min at a flow rate of 0.1 mL/min followed by isocratic 95% CH3CN in water (0.1% formic acid) for another 15 min.
Neosartoricin B extraction and purification
Neosartoricin B was purified from 2 day TWY1.1 culture in stationary liquid GMM culture (100 mL/per plate, 10 plates) with 0.5 μM pyridoxine HCl. The compound was extracted from the cultures using equal volume of EtOAc/MeOH/AcOH (89:10:1) twice. After evaporation of the organic phase, the crude extracts were separated by a chromatographic step on a Sephadex LH-20 column using MeOH/chloroform (CHCl3) (9:1) as the mobile-phase. Further purification was carried out by reverse phase HPLC using a Beckman Coulter System Gold LC with a Phenomenex Luna 250 × 10 mm 5 micron C18 column. The compounds were separated using a solvent gradient of 50-80% solvent B (CH3CN) at a flow rate of 2.5 mL/min over 30 min. Neosartoricins C and D were purified using the same procedures as described above except 0.1 % trifluoroacetic acid was added to the HPLC solvents. The HPLC fractions containing pure compounds were pooled and dried completely under vacuum before NMR analysis.
NMR characterization
All 1H, 13C and 2D (1H-13C HSQC and 1H-13C HMBC) NMR were performed on a Bruker DRX-500 spectrometer at the UCLA Department of Chemistry and Biochemistry NMR facility. CDCl3 was used as the solvent for neosartoricin B, C and D.
Supplementary Material
Acknowledgement
We thank Prof. Nancy Keller (University of Wisconsin Madison) for the gift of A. nidulans strain RJMP1.49. We thank Wei Xu, Hsiao-Ching Lin for the discussion of NMR spectra. This work is supported by NIH 1DP1GM106413 to YT and NIH R21AI081235 to TCW. NMR instrumentation was supported by the NSF equipment grant CHE-1048804 to UCLA.
Footnotes
Author Contributions
W.B.Y. designed and carried out the experiments; created plasmids used, analyzed the data and wrote the manuscript. R.A.C. and Y.H. analyzed NMR data. A.R.S. and T.C.W. prepared the dermatophyte cultures. Y.H.C. and Y.T. designed the experiments, analyzed the data and wrote the manuscript.
Supporting Information
Supporting figures, tables, and NMR spectra. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Note added during revision
During the review the manuscript, a study using a similar genetic approach to study Aspergillus metabolites was reported21.
REFERENCES
- 1.Burmester A, Shelest E, Glockner G, Heddergott C, Schindler S, Staib P, Heidel A, Felder M, Petzold A, Szafranski K, Feuermann M, Pedruzzi I, Priebe S, Groth M, Winkler R, Li W, Kniemeyer O, Schroeckh V, Hertweck C, Hube B, White TC, Platzer M, Guthke R, Heitman J, Wostemeyer J, Zipfel PF, Monod M, Brakhage AA. Comparative and functional genomics provide insights into the pathogenicity of dermatophytic fungi. Genome Biol. 2011;12:R7. doi: 10.1186/gb-2011-12-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perrin RM, Fedorova ND, Bok JW, Cramer RA, Wortman JR, Kim HS, Nierman WC, Keller NP. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 2007;3:508–517. doi: 10.1371/journal.ppat.0030050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rivera ZS, Losada L, Nierman WC. Back to the future for dermatophyte genomics. MBio. 2012;3 doi: 10.1128/mBio.00381-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yin WB, Baccile JA, Bok JW, Chen Y, Keller NP, Schroeder FC. A nonribosomal peptide synthetase-derived iron(III) complex from the pathogenic fungus Aspergillus fumigatus. J. Am. Chem. Soc. 2013;135:2064–2067. doi: 10.1021/ja311145n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Survase SA, Kagliwal LD, Annapure US, Singhal RS. Cyclosporin A--a review on fermentative production, downstream processing and pharmacological applications. Biotechnol. Adv. 2011;29:418–435. doi: 10.1016/j.biotechadv.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 6.Heinekamp T, Thywissen A, Macheleidt J, Keller S, Valiante V, Brakhage AA. Aspergillus fumigatus melanins: interference with the host endocytosis pathway and impact on virulence. Front. Microbiol. 2012;3:440. doi: 10.3389/fmicb.2012.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Achterman RR, White TC. Dermatophyte virulence factors: identifying and analyzing genes that may contribute to chronic or acute skin infections. Int. J. Microbiol. 2012;2012:358305. doi: 10.1155/2012/358305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez DA, Oliver BG, Graser Y, Goldberg JM, Li W, Martinez-Rossi NM, Monod M, Shelest E, Barton RC, Birch E, Brakhage AA, Chen Z, Gurr SJ, Heiman D, Heitman J, Kosti I, Rossi A, Saif S, Samalova M, Saunders CW, Shea T, Summerbell RC, Xu J, Young S, Zeng Q, Birren BW, Cuomo CA, White TC. Comparative genome analysis of Trichophyton rubrum and related dermatophytes reveals candidate genes involved in infection. MBio. 2012;3:e00259–00212. doi: 10.1128/mBio.00259-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brakhage AA. Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 2013;11:21–32. doi: 10.1038/nrmicro2916. [DOI] [PubMed] [Google Scholar]
- 10.Chooi YH, Wang P, Fang J, Li Y, Wu K, Tang Y. Discovery and characterization of a group of fungal polycyclic polyketide prenyltransferases. J. Am. Chem. Soc. 2012;134:9428–9437. doi: 10.1021/ja3028636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chooi YH, Fang J, Liu H, Filler SG, Wang P, Tang Y. Genome mining of a prenylated and immunosuppressive polyketide from pathogenic fungi. Org. Lett. 2013;15:780–783. doi: 10.1021/ol303435y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayorga ME, Timberlake WE. Isolation and molecular characterization of the Aspergillus nidulans wA gene. Genetics. 1990;126:73–79. doi: 10.1093/genetics/126.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu J, Bian X, Hu S, Wang H, Huang F, Seibert PM, Plaza A, Xia L, Muller R, Stewart AF, Zhang Y. Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat. Biotechnol. 2012;30:440–446. doi: 10.1038/nbt.2183. [DOI] [PubMed] [Google Scholar]
- 15.Maiya S, Grundmann A, Li SM, Turner G. The fumitremorgin gene cluster of Aspergillus fumigatus: identification of a gene encoding brevianamide F synthetase. ChemBioChem. 2006;7:1062–1069. doi: 10.1002/cbic.200600003. [DOI] [PubMed] [Google Scholar]
- 16.Yeh HH, Chang SL, Chiang YM, Bruno KS, Oakley BR, Wu TK, Wang CC. Engineering fungal nonreducing polyketide synthase by heterologous expression and domain swapping. Org. Lett. 2013;15:756–759. doi: 10.1021/ol303328t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujii I, Watanabe A, Sankawa U, Ebizuka Y. Identification of Claisen cyclase domain in fungal polyketide synthase WA, a naphthopyrone synthase of Aspergillus nidulans. Chem. Biol. 2001;8:189–197. doi: 10.1016/s1074-5521(00)90068-1. [DOI] [PubMed] [Google Scholar]
- 18.Shimizu K, Keller NP. Genetic involvement of a cAMP-dependent protein kinase in a g protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics. 2001;157:591–600. doi: 10.1093/genetics/157.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen MW, Fang F, Sandmeyer S, Da Silva NA. Development and characterization of a vector set with regulated promoters for systematic metabolic engineering in Saccharomyces cerevisiae. Yeast. 2012;29:495–503. doi: 10.1002/yea.2930. [DOI] [PubMed] [Google Scholar]
- 20.Bok JW, Keller NP. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell. 2004;3:527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiang YM, Oakley CE, Ahuja M, Entwistle R, Schultz A, Chang SL, Sung CT, Wang CC, Oakley BR. An efficient system for heterologous expression of secondary metabolite genes in Aspergillus nidulans. J. Am. Chem. Soc. 2013;135:7720–7731. doi: 10.1021/ja401945a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.