Abstract
The pathogenic fungus Cryptococcus neoformans synthesizes a complex family of glycosylinositolphosphoceramide (GIPC) structures. These glycosphingolipids (GSLs) consist of mannosylinositolphosphoceramide (MIPC) extended by β1-6-linked galactose, a unique structure that has to date only been identified in basidiomycetes. Further extension by up to five mannose residues and a branching xylose has been described. In this study, we identified and determined the gene structure of the enzyme Ggt1, which catalyzes the transfer of a galactose residue to MIPC. Deletion of the gene in C. neoformans resulted in complete loss of GIPCs containing galactose, a phenotype that could be restored by the episomal expression of Ggt1 in the deletion mutant. The entire annotated open reading frame, encoding a C-terminal GT31 galactosyltransferase domain and a large N-terminal domain of unknown function, was required for complementation. Notably, this gene does not encode a predicted signal sequence or transmembrane domain. The demonstration that Ggt1 is responsible for the transfer of a galactose residue to a GSL thus raises questions regarding the topology of this biosynthetic pathway and the function of the N-terminal domain. Phylogenetic analysis of the GGT1 gene shows conservation in hetero- and homobasidiomycetes but no homologs in ascomycetes or outside of the fungal kingdom.
Keywords: basidiomycete, fungal glycans, galactosyltransferase, GIPC, glycolipids
Introduction
Cryptococcus neoformans is a basidiomycete fungal pathogen that primarily causes systemic infection in immunocompromised patients but can also cause disease in immunocompetent individuals. The incidence of cryptococcosis has risen significantly with the occurrence of the HIV pandemic and increases in other immunocompromised patient populations. In these patients, the fungus can spread from the lungs to the central nervous system to cause life-threatening meningoencephalitis. A well-established virulence factor of C. neoformans is its polysaccharide capsule consisting of glucuronoxylomannan and glucoronoxylomannogalactan (GXMGal; sometimes termed galactoxylomannan; Kumar et al. 2011; Heiss et al. 2013). Besides these capsular polysaccharides, the glycome of C. neoformans includes protein-linked N- and O-glycans and glycosphingolipids (GSLs).
GSLs consist of a carbohydrate unit covalently linked to a ceramide and are found in all eukaryotic cells. The ceramide consists of the long-chain base sphingosine and a fatty acid. Sphingolipid synthesis in the endoplasmic reticulum yields chemically different types of sphingoid bases and fatty acid chains, varying in length, saturation and hydroxylation (Schnaar et al. 2009). Rather than being simply membrane structural components GSLs cluster in membrane microdomains and function in the regulation of signal transduction (van Meer et al. 2008; Wymann and Schneiter 2008). In addition, GSLs have been shown to mediate cell–cell interactions, modulate protein activities and participate in protein sorting (Schnaar et al. 2009). In multicellular organisms GSLs mediate and modulate intercellular coordination and are essential for development, although they are dispensable for viability at a single-cell level.
Two types of GSLs have been identified in fungi. Neutral ceramide monohexosides (cerebrosides) have been found in most fungi, including fungal pathogens (other than Candida glabrata) and a variety of Saccharomyces species (Takakuwa et al. 2002), although they are absent from S. cerevisiae (Nimrichter and Rodrigues 2011). Most fungal cerebrosides contain a single glucose or galactose residue linked to a ceramide composed of 9-methyl-4,8-sphingadienine and C18 or C16 α-hydroxy fatty acids. More extended neutral GSLs have been described in Neurospora crassa (tetraglycosylated neurosporaside; Dickson and Lester 1999; Costantino et al. 2011), Mucor hiemalis (Aoki et al. 2004) and Magnaporthe grisea (Maciel et al. 2002).
The second class of fungal GSLs comprises complex acidic glycosylinositolphosphoceramides (GIPCs). Inositolphosphoceramide (IPC), which does not occur in mammalian cells, consists of inositol linked via a phosphodiester bond to ceramide composed of phytosphingosine and a saturated, usually hydroxylated, long-chain fatty acid (Lester and Dickson 1993). IPC is essential for fungal growth: Saccharomyces and Aspergillus mutants that do not synthesize IPC are not viable (Nagiec et al. 1997; Cheng et al. 2001; Dickson and Lester 2002; Hu et al. 2007) and pathogenic fungi are killed by treatment with inhibitors of IPC synthase, the enzyme that catalyzes the transfer of phosphoinositol to phytoceramide (Gutierrez et al. 2007).
Addition of a carbohydrate moiety to IPC typically yields three types of core structures, modified with either α1-2- or α1-6-linked mannose or α1-2-linked glucosamine (Simenel et al. 2008); IPC bearing α1-4-linked mannose has only been reported in basidiomycetes (Jennemann et al. 2001). The core structures can be further extended in a species-specific manner (Simenel et al. 2008) with additional mannose, fucose, xylose, galactofuranose or choline-phosphate. Substitution of the first mannose with a β-galactopyranose at position 6 has been described for several fruiting body-forming homobasidiomycetes (Jennemann et al. 2001) and for the heterobasidomycete C. neoformans (Heise et al. 2002; Gutierrez et al. 2007).
Biosynthesis and function of fungal GIPCs have been extensively studied in the ascomycete S. cerevisiae. This organism does not further glycosylate mannosylinositolphosphoceramide (MIPC), although it generates M(IP)2C by addition of a second phosphoinositol group (Dickson and Lester 1999). MIPC biosynthesis in S. cerevisiae is initiated by the transfer of a mannose to IPC, mediated by the redundant enzymes Csg1 and Csh1 acting with the Ca2+-dependent regulator Csg2 (Uemura et al. 2003, 2007). The only other enzyme involved in fungal GIPC biosynthesis identified to date is the xylosyltransferase Cxt1 from C. neoformans, which was shown to add a branching xylose to GIPC. Mutants lacking Cxt1 do not elongate the glycan moiety with mannoses beyond the xylose branch (Castle et al. 2008). This is reminiscent of lipid-linked oligosaccharide biosynthesis, where high substrate specificity guarantees a strict sequence of enzyme action to ensure the generation of a single product (Burda et al. 1999). Interestingly, the absence of Cxt1 also affected capsule biosynthesis by reducing its xylose content, suggesting a dual activity of Cxt1 (Klutts and Doering 2008).
Several roles have been proposed for fungal GIPCs. Studies of S. cerevisiae mutants depleted in sphingolipids indicated that these lipids are required for microdomain formation and secretion of certain proteins to the plasma membrane (Bagnat et al. 2000). Analysis of lipid distribution within yeast cells also showed that non-glycosylated IPC occur predominantly in the Golgi membrane, whereas MIPC and M(IP)2C localize to the plasma membrane. These findings led to the suggestion that mannosylation of IPC is a potential branch point in sphingolipid trafficking that may affect protein traffic (Hechtberger et al. 1994). Identification and deletion of the relevant mannosyltransferases, however, showed that glycosylation of sphingolipids is dispensable for protein secretion in yeast (Lisman et al. 2004). The role of sphingolipid glycosylation thus remains elusive.
Significant questions remain about the enzymes involved in fungal GIPC biosynthesis and the biological role of GIPCs. As a first step in addressing this gap in knowledge, we set out to identify additional GIPC glycosyltransferases in C. neoformans. Below, we demonstrate our discovery of a galactosyltransferase, Ggt1 (GIPC-galactosyltransferase), which is responsible for the biosynthesis of the basidiomycete-specific galactosylated GIPCs.
Results
Identification and phylogenetic analysis of a candidate MIPC-galactosyltransferase in C. neoformans
The Carbohydrate-Active Enzyme database classifies glycosyltransferases into families according to sequence similarities (Cantarel et al. 2009). According to the stereochemistry of the substrates and reaction products, glycosyltransferases can be designated as retaining or inverting enzymes (Sinnott 1990). We wished to identify the enzyme that transfers galactose from its activated precursor uridine diphosphate-galactose (UDP-Gal) to MIPC in a β1-6 linkage, thus exhibiting an inverting mechanism. As the correlation between transferase family and stereochemistry of the catalyzed reaction is fairly reliable, we excluded families with a retaining mechanism from our search. From the remaining families with an inverting mechanism, we further selected those comprising β-galactoside or glycolipid forming members.
Several candidate genes in the annotated genome of C. neoformans (Jec21; serotype D) met our selection criteria: CNB01840/CNB05490, CNG04310, CNG01130, CND01800 and CND04970. These candidates included homologs of Alg13/14 (both CNB01840/CNB05490 are required for enzymatic activity), a UDP glucosesteroltransferase (CNG04310) from glycosyltransferase family 1 (GT1), a hypothetical protein of the GT47 family (CNG01130), and two members of the GT31 family: a putative galactosyltransferase (CND01800) and a hypothetical protein (CND04970). Interestingly, homologs of the last candidate could be found only in basidiomycetes (Figure 1), corresponding to the occurrence of the synthetic process of interest. For this reason, this sequence was chosen for further investigation.
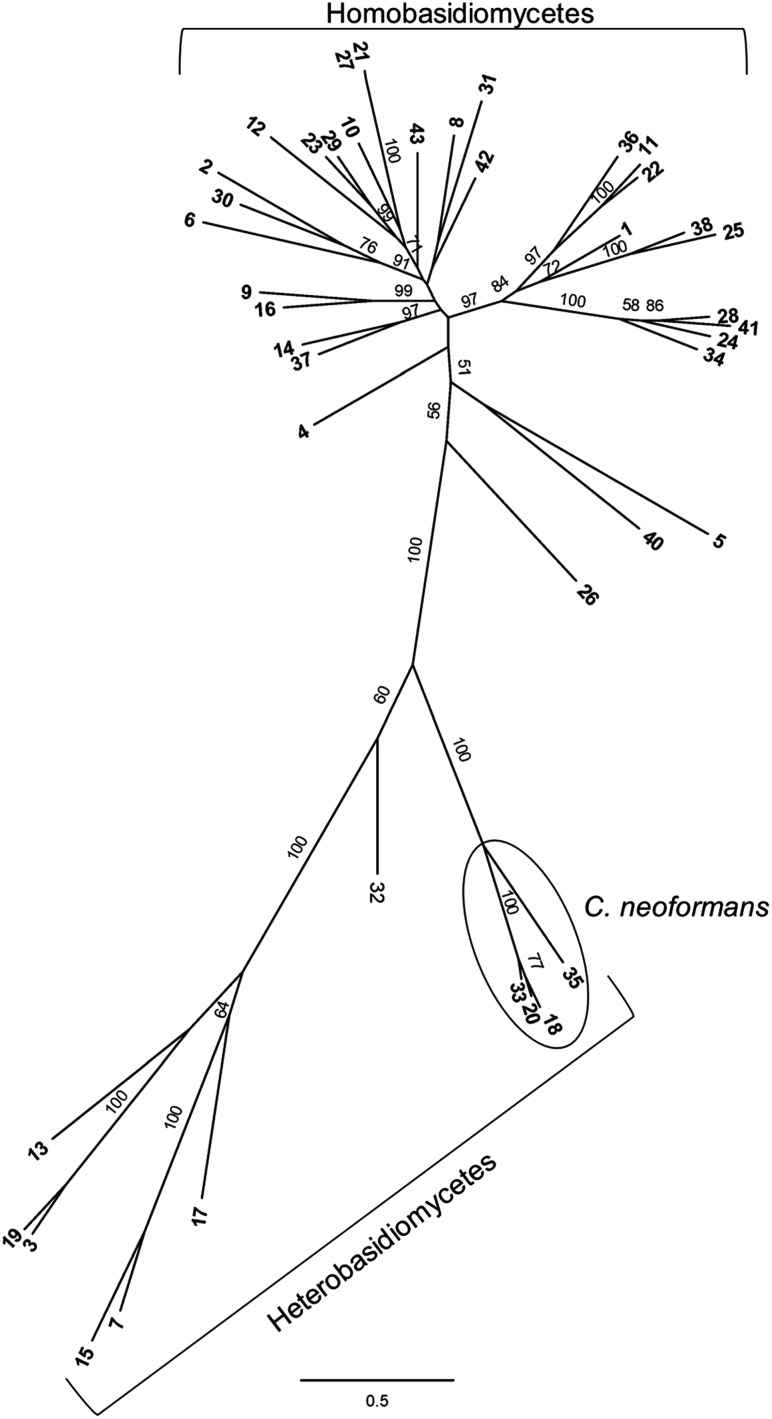
Fig. 1.
Phylogenetic analysis of the Ggt1 involved in GIPC biosynthesis in basidiomycetes. Shown is a radial tree with highest likelihood score from maximum likelihood phylogenetic analysis of 42 Ggt1 homologs from basidiomycetes. Bootstrap values of >50 are shown. Proteins used in this study are listed in Supplementary data, Table S1.
The hydrophilic N-terminal part of the protein encoded by CND04970 had minimal homology to homologs in non-Cryptococcus species and contained a high proportion of charged residues; it was therefore omitted in the phylogenetic analysis. The remainder of the open reading frame (ORF), consisting of a putative galactosyltransferase domain and a C-terminal part of unknown function, was found to be distributed across basidiomycetes, including both hetero- and homobasidiomycetes (Figure 1). In contrast, no homologs were present in ascomycetes or outside the fungal kingdom.
Determination of the gene structure
CNAG01385 of C. neoformans H99 (CND04970 of C. neoformans Jec21) is predicted to encode a protein of 867 amino acids containing a galactosyltransferase domain of the GT31 family including the characteristic DXD-motif (not shown; Wiggins and Munro 1998). To predict the length of the galactosyltransferase domain, we compared the candidate amino acid sequence to previously characterized members of the GT31 family. The only protein structure available in the GT31 family is that of the mouse O-fucosylpeptide β-1,3-GlcNAc transferase (PDB 2J0A). This protein of 321 amino acids comprises a 270-residue catalytic domain adopting a GT-B fold, an N-terminal transmembrane domain and a short stem region. These features are conserved among other GT31 family members such as the galactomannan β-1,3-galactosyltransferase (Pvg3;SPBC1921.06c; GenBank CAB58972.1) of Schizosaccharomyces pombe, where the GT-B fold catalytic domain starts at about residue 60 and is ∼310 amino acids long. Alignment of the S. pombe sequence with that encoded by C. neoformans CNAG01385 suggests that the catalytic domain of the latter protein most likely starts at position 380. The exact end of the galactosyltransferase domain is difficult to determine due to low sequence similarity, but assuming a length of 270–310 residues, it would be at positions 650–690.
In addition to the conserved galactosyltransferase domain comprising 270–310 residues, we observed two unexpected features of the candidate sequence. First, the predicted ORF contains stretches of ∼380 amino acids at the N- and 177–217 amino acids at the C terminus that lack any conserved domain. Due to these regions of low homology, we questioned the annotation of the ORF and speculated that the actual protein might be shorter. Second, we expected this protein to occur in the secretory pathway, based on the known localization of MIPC biosynthesis in S. cerevisiae, where ceramide made in the endoplasmic reticulum is converted to IPC and further modified to MIPC and M(IP)2C in the Golgi (Nakase et al. 2010). Interestingly, the candidate ORF did not encode a signal peptide or an N-terminal transmembrane domain that would be consistent with secretory pathway localization. To confirm the function of the putative MIPC-galactosyltransferase, we therefore needed to determine the structure of the gene and the actual ORF.
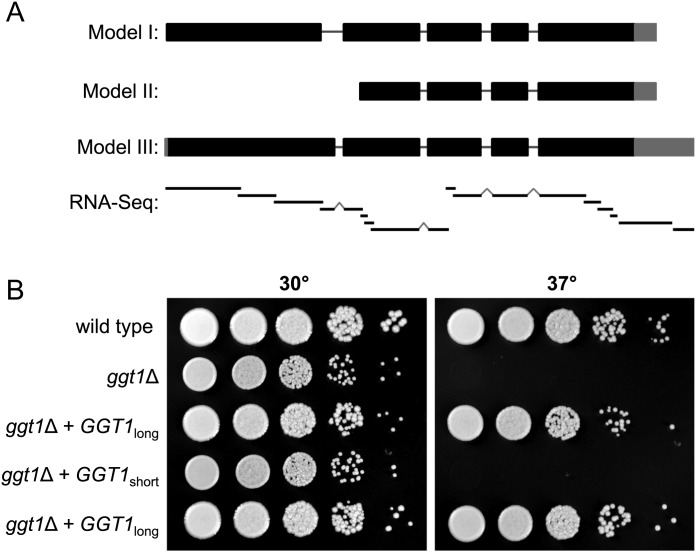
We used RNA sequencing to investigate the genomic region implicated in encoding the Ggt1 (CNAG_01385). For this purpose, RNA was isolated from C. neoformans strain KN99α, which is a serotype A strain derived from the sequenced reference strain H99. Short-read sequencing on cDNA made from these RNA preparations (RNA-Seq) was performed, and the 282 million short reads obtained were assembled into contigs. Alignment of the contigs to the H99 reference sequence supported a gene model similar to the initial annotation (Figure 2A, model I), although it differed at the first intron and extended the 5′- and 3′-untranslated regions (Figure 2A, model III). cDNA was polymerase chain reaction (PCR)-amplified using primers designed to span the first intron and the resulting amplicon was sequenced (not shown). This analysis confirmed that the first intron is 90 bp shorter than originally annotated, lengthening the coding sequence but maintaining the reading frame. Together, these studies defined the gene structure of this locus, which will be referred to as GGT1, and supported the model that an unusually large protein catalyzes the galactosyltransferase activity.
Fig. 2.
Structure determination of the predicted gene CND04970 and growth defect of a C. neoformans ggt1Δ mutant. (A) Gene models of the locus encoding Ggt1. Coding regions are depicted as thick black bars, introns by thin black lines and untranslated regions by thick gray bars. Model I shows the CNAG01385 annotation from the Broad Institute. Model II shows the annotation suggested by homology, existing EST evidence and preliminary polymerase chain reaction (PCR) studies (see text). Model III shows our revised annotation, as confirmed by PCR studies (not shown) and short read assemblies of cDNA. The contigs derived from RNA-Seq data are depicted below the gene models as thin black bars with intron-bridging indicated by grey peaks (RNA-Seq). Importantly, the predicted galactosyltransferase domain lies in the region that is common to all three models. (B) Growth defect and complementation of a C. neoformans ggt1Δ mutant. Wild-type and ggt1Δ mutant strains carrying empty vector, episomal GGT1short or GGT1long were spotted in serial 10-fold dilutions on YPD plates and incubated at the indicated temperatures for 3 days. Two independent transformants are shown for the plasmid containing the long form of the gene.
Deletion of C. neoformans CSG1 and GGT1 and effect on growth
To investigate GIPC biosynthesis in C. neoformans, genes involved in early GIPC glycosylation steps were deleted. Two redundant enzymes are responsible for the mannosylation of IPC in S. cerevisiae, one of which has a homolog in C. neoformans that was accordingly named CnCSG1. To confirm the roles of the putative C. neoformans GGT1 (CnGGT1) and CnCSG1 in the synthesis of GIPCs, deletion strains were created by replacing each locus by a cassette encoding an antibiotic resistance gene (NAT) flanked by 1 kb of homologous sequence. Both mutant strains showed normal growth on yeast extract-peptone-dextrose (YPD) plates at 30°C, but reduced growth upon temperature shift to 37°C. This temperature sensitivity could partially be restored by supplementing YPD plates with 1 M sorbitol (Figure 2B and Supplementary data, Figure S1).
Effect of CSG1 and GGT1 deletion on GIPC biosynthesis
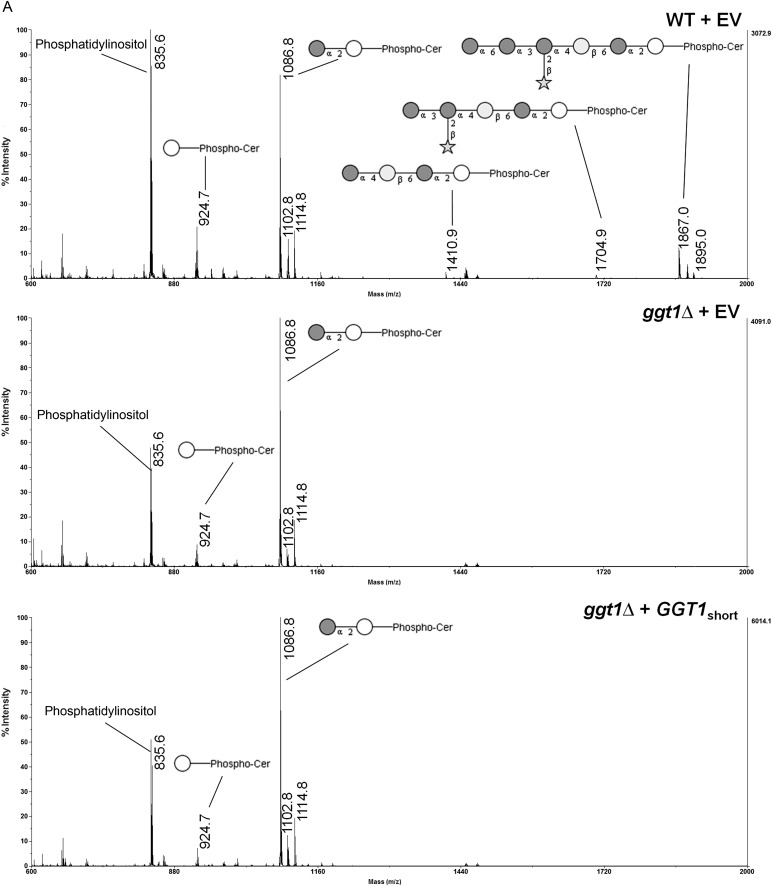
To confirm the transferase activity proposed for CnCsg1 and CnGgt1, GIPCs of wild-type and both mutant strains (csg1Δ and ggt1Δ) were analyzed. Strains were grown in liquid medium to midlog phase, cells were lysed and lipids were extracted and analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS). Wild-type extracts contained abundant phosphatidylinositol, IPC and MIPC (Figure 3A, top panel), as well as further glycosylated sphingolipids with the most prominent of these being the hexasaccharide-IPC (1867 Da) described previously (Heise et al. 2002; Gutierrez et al. 2007). As expected, no MIPC was generated in the Cncsg1Δ strain, which displayed a corresponding accumulation of its IPC precursor (Supplementary data, Figure S1). MIPC was present in the ggt1Δ strain, although further glycosylated sphingolipids were not detected (Figure 3A, second panel). This strongly supports the hypothesis that CnGgt1 mediates the addition of the galactose residue to MIPC and is required for further elaboration of this structure.
Fig. 3.
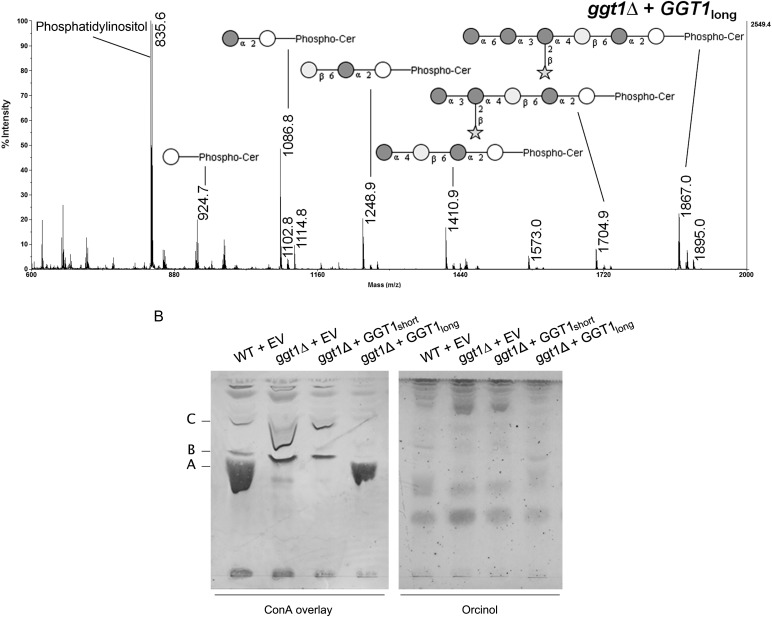
Analysis of GIPCs from different Cryptococcus transformants. (A) Lipid extracts from C. neoformans wild-type (WT)/empty vector (EV), ggt1Δ/EV, ggt1Δ/GGT1short and ggt1Δ/GGT1long were analyzed by MALDI-TOF-MS in the negative ion mode. Masses (Da) of phospholipids and GIPC structures are as follows: 835.6: phosphatidylinositol (PI); 924.7: IPC; 1086.8: mannosyl-IPC; 1248.9: IPC-disaccharide; 1410.9: IPC-trisaccharide; 1704.9: IPC-pentasaccharide; 1867.0: IPC-hexasaccharide. Accompanying peaks represent derivatives with an additional hydroxyl group (+16) or an extended fatty acid moiety (C2: +28) as previously reported (Heise et al. 2002; Guan and Wenk 2006). White circle, inositol; dark gray circle, mannose; light gray circle, galactose; star, xylose. (B) ConA overlay and orcinol stain of TLCs of GIPCs from different Cryptococcus strains. Lipid extracts from the indicated C. neoformans strains were separated by TLC and probed with biotinylated ConA or stained with orcinol. Band A corresponds to mannose-containing IPC-hexasaccharide in WT and ggt1Δ complemented with GGT1long strains. Bands B and C correspond to shorter mannose-containing GIPC structures.
Episomal expression of GGT1 in C. neoformans
Based on the domain structure discussed in Determination of the gene structure, we wondered whether the gene region containing the predicted galactosyltransferase domain alone lacking the N-terminal sequence of low homology would confer Ggt1 activity. To test this, we PCR-amplified N-terminally truncated (“short”) and complete (“long”) versions of the gene (Figure 2A, models II and III, respectively), confirmed the amplicons by sequencing and cloned them into a vector carrying a G418 resistance marker. Each of the resulting plasmids was then transformed into the ggt1Δ mutant strain and the empty vector was transformed into both the mutant strain and the wild-type parent (H99) as controls. Growth assays of the transformants revealed that the entire (“long”) GGT1 gene was required to complement the temperature sensitivity of the ggt1Δ strain (Figure 2B). Interestingly, the use of the G418 resistance marked plasmid revealed a modest growth defect in the ggt1Δ strain at 30°C in the presence of this compound (Supplementary data, Figure S2), which is more subtle on medium without G418 (Figure 2B).
Analysis of GIPCs from ggt1Δ complementation
GIPCs of the ggt1Δ strain complemented with either the short or the long version of ggt1 were analyzed by MALDI-TOF-MS (Figure 3A). Only complementation with full-length GGT1 restored galactosylated GIPCs, whereas complementation with the short version yielded results equivalent to transformation with the empty vector. This supports the conclusion that the entire GGT1 ORF is required for galactosylation of GIPCs. It appears that galactosylation is the limiting step in the biosynthesis pathway as overexpression of GGT1 (driven by the actin promoter in the plasmid) yielded more downstream products than were observed in the wild type. In addition to MS analysis, lipid extracts were separated by thin layer chromatography (TLC) and probed with the mannose-binding plant lectin ConA (Figure 3B). ConA binding confirmed that mannose-containing IPC-hexasaccharide (band A) was present in the ggt1Δ strain complemented with full-length GGT1 but absent in the ggt1Δ strain transformed with the empty vector or the short version of GGT1. Again, mannosylation appeared to be more complete in ggt1Δ cells complemented with the long form of the gene, as shown by a reduction in shorter mannose-containing GIPC structures (bands B and C) compared with the wild type.
Discussion
Cryptococcus neoformans generates GIPCs with a characteristic β1-6-linked galactose. Using bioinformatics, we identified a basidiomycete-specific putative galactosyltransferase, Ggt1. Indeed, the deletion of GGT1 in C. neoformans H99 led to the absence of galactosylated GIPC. MIPC was the most abundant glycolipid species in the ggt1Δ strain and no further glycosylated GIPCs were observed. In addition, glycolipid analysis of a C. neoformans csg1Δ mutant showed the absence of MIPC, M(IP)2C and further extended GIPC, suggesting that only one mannosyltransferase is involved in MIPC biosynthesis in C. neoformans. This finding is in agreement with the fact that, in contrast to S. cerevisiae, no homolog of the redundant mannosyltransferase Csh1 is present in C. neoformans.
Csg1 and Csh1 were found to be necessary for growth of S. cerevisiae at elevated temperature and increased calcium concentration (Cid et al. 1995). Further analysis showed that IPC accumulation rather than the absence of MIPC caused this conditional lethal phenotype in an S. cerevisiae csg1Δ mutant (Beeler et al. 1997). Similarly, C. neoformans csg1Δ showed normal growth at 30°C but did not grow at 37°C. This temperature sensitivity could partially be restored by addition of the osmotic stabilizer sorbitol, suggesting that this growth inhibition might be caused by a cell wall defect. Interestingly, C. neoformans ggt1Δ also displayed temperature sensitivity that could be restored by sorbitol. Thus, not only accumulation of IPC but also that of MIPC appears to affect cell wall integrity. Notably, the presence of the aminoglycoside G418 induced a growth defect in the ggt1Δ mutant. Aminoglycosides are known to interact with phospholipids (Mingeot-Leclercq et al. 1995), raising the intriguing possibility that G418-binding to phosphoinositides may exacerbate the glycosylated-IPC deficiency or possibly bind directly to extra/intracellular pools of IPC/GIPC.
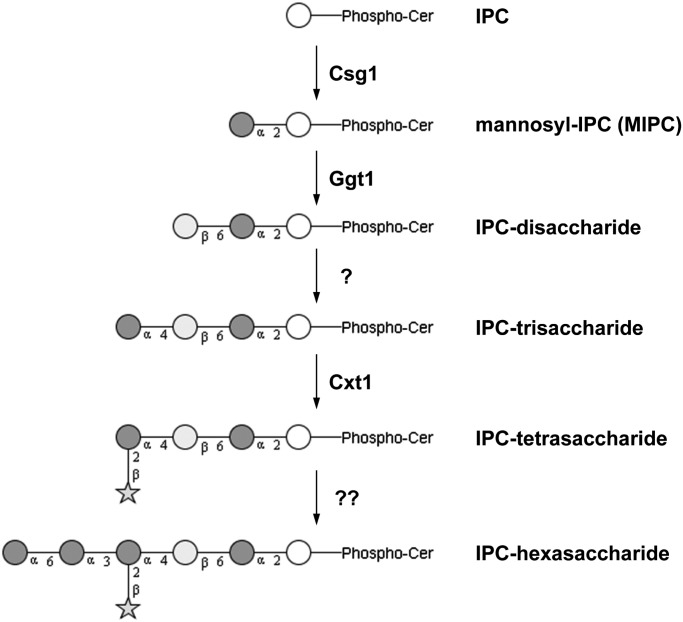
A putative schematic pathway for GIPC biosynthesis in C. neoformans is shown in Figure 4. Glycosyltransferases acting downstream of Ggt1 in the GIPC biosynthesis machinery of C. neoformans remain to be identified. Candidates for such transferases are found in the GT32 family that also comprises the redundant mannosyltransferases Csg1 and Csh1 of S. cerevisiae. Within this family, four additional homologs exist in C. neoformans that are absent in S. cerevisiae, potentially reflecting the increased complexity of GIPC in Cryptococcus. Deletion of these candidate genes and characterization of the mutants with regard to temperature sensitivity as well as GIPC structure will be required to clarify a possible role in GIPC biosynthesis.
Fig. 4.
Putative schematic pathway for GIPC biosynthesis in C. neoformans. GIPC structures are shown in black and white using the notation recommended by the Consortium for Functional Glycomics (CFG). White circle, inositol; dark gray circle, mannose; light gray circle, galactose; star, xylose. Enzyme names are indicated next to the catalyzed reaction. For references, see text.
Complementation analysis showed that the N-terminal region of the Ggt1 protein is required for Ggt1 activity, even though it lacks obvious glycosyltransferase homology. Due to the lack of any sequence or structural homology predicted by the PHYRE 2 server (Kelley and Sternberg 2009), it is currently not possible to assign a possible function to this domain. One possibility is that this domain may be involved in correct targeting of the enzyme, given the absence of other localization signals. Such a targeting mechanism may involve the interaction of this domain with other components of the basidiomycete-specific GIPC biosynthesis machinery. The lack of a signal peptide for classical secretion or a transmembrane domain in GGT1 suggest that the enzyme is either secreted to the Golgi lumen in a non-classical way or remains in the cytoplasm of the cell. The latter case would challenge the model of MIPC biosynthesis known from S. cerevisiae in terms of topology: both the IPC synthase Aur1p and the mannosyltransferases Csg1 and Csh1 reside in the Golgi lumen (Levine et al. 2000; Uemura et al. 2007). The localization of the catalytic site of the mannosyldiphosphoinositolceramide synthase Ipt1p is still under debate, but its C terminus is cytosolic (Kim et al. 2006).
To test whether GGT1 was not only necessary but also sufficient for Ggt1 activity, the complete GGT1-encoding ORF was cloned into a low copy yeast vector under the control of the glyceraldehyde-3-phosphate dehydrogenase gene promoter. Although heterologous expression of GGT1 in an S. cerevisiae ipt1Δ strain was confirmed by immunoblotting, no galactosylated MIPC was detected in MALDI-TOF analysis of lipid extracts from these cells (Supplementary data, Figure S3). It may be that the protein is not properly localized due to the lack of other components of the targeting machinery. Alternatively, a sufficient amount of UDP-Gal—if this is the reaction substrate—may not be available in this organism. In this regard, we were unable to demonstrate enzymatic activity of an N-terminally truncated version of Ggt1 comprising the galactosyltransferase domain in vitro, using recombinant protein produced in Escherichia coli or insect cells with UDP-Gal and MIPCs extracted from C. neoformans as substrates (data not shown).
Several glycoconjugates of C. neoformans share structural motifs. One intriguing example occurs between GIPCs and the capsule polysaccharide GXMGal. This polysaccharide, which has a α1-6 galactan backbone, is branched with an oligosaccharide highly similar to the one extending MIPC (βGal4-αMan [2-βXyl] 3-αMan), although this branch may be further substituted with xylose and glucuronic acid (Heiss et al. 2009, 2013). Interestingly, the same xylosyltransferase is required for synthesis of both glycans (Castle et al. 2008; Klutts and Doering 2008). Whether the shared motif is a consequence of dual activity of the enzyme or whether GIPC and capsule biosynthesis are themselves physically interconnected is an interesting question that remains to be addressed.
Sphingolipid metabolism plays an important role in virulence of C. neoformans by regulating signaling events leading to the production of virulence factors. It further impacts the pathogenesis of cryptococcal infection, by influencing internalization of C. neoformans by macrophages and fungal growth in these environments (Henry et al. 2011). Although the full role of C. neoformans GIPCs during host–pathogen interactions is still unknown, the mutants constructed in this work will move us forward in answering this question.
Materials and methods
Materials
Unless otherwise noted, all chemicals were from Sigma-Aldrich (Buchs, Switzerland), primers were from Invitrogen (Basel, Switzerland) and Microsynth (Balgach, Switzerland), and restriction enzymes were from New England Biolabs (Ipswich, MA) and Fermentas (Wohlen, Switzerland). All reagents were used according to the manufacturer recommendations.
Strains and growth conditions
Cryptococcus neoformans serotype A strains used in this study are listed in Supplementary data, Table S2. Mutants were generated by replacing the corresponding locus of H99 with a disruption cassette by means of homologous recombination as described (Davidson et al. 2002). The resistance cassette was generated by overlap PCR between the 5′- and 3′-flanking regions of the target gene and a nourseothricin-resistance gene (NAT). Primers used for the generation of csg1Δ and ggt1Δ strains are shown in Supplementary data, Table S3. Constructs were transformed by biolistics as described previously (Toffaletti et al. 1993). Yeast were grown with continuous shaking (230 rpm) in liquid culture in DMEM or YPD (1% w/v yeast extract, 2% w/v peptone, 2% w/v glucose) or on solid medium (YPD with 2% w/v agar) at the temperatures indicated in the text. Escherichia coli DH5α cells were grown at 37°C on Luria-Bertani agar plates (1% w/v tryptone, 0.5% w/v yeast extract, 1% w/v NaCl, 1.5% w/v agar). As appropriate, media were supplemented with 100 μg/mL of ampicillin, 60 µg/mL of kanamycin or 100 μg/mL of Geneticin® (G418; from Invitrogen).
RNA sequencing and assembly
Twenty-five independent cultures of KN99α cells were grown overnight in YPD at 30°C and incubated for 90 min in either capsule-inducing (DMEM, 37°C, 5% CO2) or capsule non-inducing (DMEM, 30°C, room air) conditions prior to the isolation of total RNA as in Haynes et al. (2011). PolyA+ RNA was purified from total RNA using the Invitrogen Dynabeads mRNA Purification Kit, and each sample was resuspended in 2 µL of 100 mM zinc acetate and heated at 60°C for 3 min to fragment the RNA by hydrolysis. The reaction was quenched by the addition of 2 µL volumes of 200 mM ethylenediaminetetraacetic acid and purified with an Illustra Microspin G25 column (from GE Healthcare). First-strand cDNA was made using hexameric random primers and SuperScript III Reverse Transcriptase (from Invitrogen), and the product was treated with E. coli DNA ligase, DNA polymerase I and RNase H to prepare double-stranded cDNA using the standard methods. The cDNA libraries were end-repaired with a Quick Blunting kit (from New England BioLabs) and A-tailed using Klenow exo- and dATP. Illumina adapters with four base barcodes were ligated to the cDNA and fragments ranging from 150 to 250 bp in size were selected using gel electrophoresis. The resulting libraries were enriched in a 10-cycle PCR with Phusion Hot Start II High-Fidelity DNA Polymerase (from Finnzymes Reagents) and pooled in equimolar ratios for multiplex sequencing. Single read 36-cycle and 42-cycle runs were completed on the Illumina Genome Analyzer IIx and Illumina Hi-Seq, respectively, producing a total of 282 million reads. De novo assembly of the short reads was performed with Velvet (Zerbino and Birney 2008) using a 21 k-mer hash length. Assembled contigs were aligned to the H99 reference sequence using BLAST (Zhang et al. 2000).
Cloning of CnGGT1 and mutant complementation
“Long” and “short” forms of the GGT1 sequence were expressed in the ggt1Δ mutant under control of an actin promoter. Genomic DNA was first isolated from KN99Δ cells as in Nelson et al. (2001) and used as the template for PCRs with AM87 (Supplementary data, Table S3) as the reverse primer and either AM93 or AM94 as the forward primer to form the long and short forms of the gene, respectively. The actin promoter was amplified from the vector GMC200 using primers AM84 and AM92. The three amplicons were cloned into TOPO® vectors (Invitrogen), transformed into E. coli DH5α, and kanamycin-resistant colonies were screened by colony PCR. DNA from confirmed transformants was isolated and sequenced. Each GGT1 insert was next released from the corresponding plasmid by digestion with AscI and ApaI, ligated to the similarly cut plasmid containing the actin promoter so as to be downstream of it, and the resulting plasmid used to transform E. coli. Plasmids from confirmed transformants were isolated, digested with SpeI and NcoI (for GGT1long) or Spe1 and MscI (for GGT1short), treated with Klenow fragment, digested with ApaI and the resulting segments containing the fused promoter and GGT1 sequence were ligated to the G418 marked plasmid pIBB103 (Bose and Doering 2011) that had been previously digested with PsiI and ApaI and treated with calf intestinal phosphatase. The final plasmids were transformed into E. coli and screened as above, and DNA from confirmed transformants was isolated and checked by restriction digestion. Each GGT1 plasmid and pIBB103 alone was then linearized with I-SceI and transformed into the H99α ggt1Δ strain by electroporation (500 V, 25 µF, 1000 Ω) as in reference (Skowyra and Doering 2012), with G418-resistant transformants recovered for further analysis.
Growth assay
Cells to be tested were grown overnight at 30°C in YPD medium containing 100 µg/mL G418, washed twice in phosphate buffered saline (PBS), counted, adjusted to 2 × 106 cells/mL in PBS and then 10-fold serially diluted four times in PBS. 5 µL of each dilution was spotted onto solid YPD medium or solid YPD medium supplemented with 1 M sorbitol. The plates were incubated for 3 days at the indicated temperatures.
GIPC analysis
50 mL of yeast culture was harvested at OD600 = 1 and washed with 30 mL of water. The cell suspension was adjusted with ddH2O to a volume of 210 μL and lysed with glass beads by shaking for 20 min at 4°C. 700 μL each of methanol and chloroform were added to the lysate to obtain an organic extraction solution (C:M:W) with the ratio 10:10:3 (v/v/v), and samples were vortexed for 20 min at room temperature. The supernatant was collected after centrifugation and the pellet was re-extracted twice more with C:M:W 10:10:3. The supernatants of the three extractions were pooled and dried in a SpeedVac. The dried pellet was partitioned with water saturated 1-butanol and ddH2O in a 1:1 (v/v) mixture and the butanol phases of three extractions were pooled and dried in a SpeedVac. For MS analysis, dried lipids were resuspended in 70% MeOH, spotted on a MALDI plate and covered with matrix [ATT (6-aza-2-thio-thymine) 20 mg/mL in 70% MeOH with 10 mM Ammonium citrate]. MALDI-TOF-MS was performed in the negative ion mode on an ABI 4700 MALDI TOF/TOF™ (Applied Biosystems Inc.). For TLC analysis, lipid extracts were resolved on an aluminum-backed silica-60 high-performance TLC plate using a solvent mixture of 10:10:3 (v/v/v) [chloroform/methanol/ammonium hydroxide (1 M)]. Orcinol and TLC overlay staining were performed as described (Barrows et al. 2006) using biotinylated ConA (Sigma-Aldrich) at a concentration of 650 nM in PBS, pH 7.0, containing 1 mg/mL each Ca2+ and Mn2+.
Phylogenetic analysis
Amino acid sequences of Ggt1 homologs from basidiomycetes were retrieved from NCBI, the JGI genome portal and the Broad Institute. Sequences were aligned using NCBI BLAST (Altschul and Lipman 1990) and a phylogenetic tree was constructed by RAxML (Stamatakis et al. 2008) with 1000 bootstrap replicates under the BLOSUM 62 model (Henikoff and Henikoff 1992). Bootstrap values were mapped onto the tree with highest likelihood score and the tree was formatted for presentation using FigTree 1.3.1.
Supplementary data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Funding
This work was supported by the European Commission Marie Curie Program (EuroGlycoArrays ITN) and ETH Zürich awards to MA and by National Institutes of Health (R01 GM071007) to TLD.
Abbreviations
GGT1, GIPC-galactosyltransferase; GIPC, glycosylinositolphosphoceramide; CSG1, IPC-mannosyltransferase; GSL, glycosphingolipid; GT1, glycosyltransferase family 1; GXMGal, glucoronoxylomannogalactan; IPC, inositolphosphoceramide; MIPC, mannosylinositolphosphoceramide; M(IP)2C, mannosyl-diinositolphosphoceramide; ORF, open reading frame; TLC, thin layer chromatography; UDP-Gal, uridine diphosphate-galactose.
Supplementary Material
Acknowledgements
We thank Dr. Megan McDonald (Institute of Integrative Biology, ETH Zurich) for assistance in phylogenetic analysis, Dr. Peter Gehrig (Functional Genomics Center, University and ETH Zürich) for support in MALDI-MS analysis and Christine Opelz (Institute of Microbiology, ETH Zürich) for cloning His-tagged GGT1. We also thank Alyssa L Marulli for assistance with plasmid construction and complementation of C. neoformans and Stacey Gish for plasmid construction for expression in S. cerevisiae.
References
- Altschul SF, Lipman DJ. Protein database searches for multiple alignments. Proc Natl Acad Sci USA. 1990;87:5509–5513. doi: 10.1073/pnas.87.14.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Uchiyama R, Yamauchi S, Katayama T, Itonori S, Sugita M, Hada N, Yamada-Hada J, Takeda T, Kumagai H, et al. Newly discovered neutral glycosphingolipids in aureobasidin A-resistant zygomycetes: Identification of a novel family of Gala-series glycolipids with core Gal alpha 1-6Gal beta 1-6Gal beta sequences. J Biol Chem. 2004;279:32028–32034. doi: 10.1074/jbc.M312918200. [DOI] [PubMed] [Google Scholar]
- Bagnat M, Keranen S, Shevchenko A, Simons K. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc Natl Acad Sci USA. 2000;97:3254–3259. doi: 10.1073/pnas.060034697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrows BD, Griffitts JS, Aroian RV. Caenorhabditis elegans carbohydrates in bacterial toxin resistance. Methods Enzymol. 2006;417:340–358. doi: 10.1016/S0076-6879(06)17021-4. [DOI] [PubMed] [Google Scholar]
- Beeler TJ, Fu D, Rivera J, Monaghan E, Gable K, Dunn TM. SUR1 (CSG1/BCL21), a gene necessary for growth of Saccharomyces cerevisiae in the presence of high Ca2+ concentrations at 37 degrees C, is required for mannosylation of inositolphosphorylceramide. Mol Gen Genet. 1997;255:570–579. doi: 10.1007/s004380050530. [DOI] [PubMed] [Google Scholar]
- Bose I, Doering TL. Efficient implementation of RNA interference in the pathogenic yeast Cryptococcus neoformans. J Microbiol Methods. 2011;86:156–159. doi: 10.1016/j.mimet.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burda P, Jakob CA, Beinhauer J, Hegemann JH, Aebi M. Ordered assembly of the asymmetrically branched lipid-linked oligosaccharide in the endoplasmic reticulum is ensured by the substrate specificity of the individual glycosyltransferases. Glycobiology. 1999;9:617–625. doi: 10.1093/glycob/9.6.617. [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle SA, Owuor EA, Thompson SH, Garnsey MR, Klutts JS, Doering TL, Levery SB. Beta1,2-xylosyltransferase Cxt1p is solely responsible for xylose incorporation into Cryptococcus neoformans glycosphingolipids. Eukaryot Cell. 2008;7:1611–1615. doi: 10.1128/EC.00458-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Park TS, Fischl AS, Ye XS. Cell cycle progression and cell polarity require sphingolipid biosynthesis in Aspergillus nidulans. Mol Cell Biol. 2001;21:6198–6209. doi: 10.1128/MCB.21.18.6198-6209.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cid VJ, Duran A, del Rey F, Snyder MP, Nombela C, Sanchez M. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol Rev. 1995;59:345–386. doi: 10.1128/mr.59.3.345-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino V, Mangoni A, Teta R, Kra-Oz G, Yarden O. Neurosporaside, a tetraglycosylated sphingolipid from Neurospora crassa. J Nat Prod. 2011;74:554–558. doi: 10.1021/np1009493. [DOI] [PubMed] [Google Scholar]
- Davidson RC, Blankenship JR, Kraus PR, de Jesus Berrios M, Hull CM, D'Souza C, Wang P, Heitman J. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology. 2002;148:2607–2615. doi: 10.1099/00221287-148-8-2607. [DOI] [PubMed] [Google Scholar]
- Dickson RC, Lester RL. Yeast sphingolipids. Biochim Biophys Acta. 1999;1426:347–357. doi: 10.1016/s0304-4165(98)00135-4. [DOI] [PubMed] [Google Scholar]
- Dickson RC, Lester RL. Sphingolipid functions in Saccharomyces cerevisiae. Biochim Biophys Acta. 2002;1583:13–25. doi: 10.1016/s1388-1981(02)00210-x. [DOI] [PubMed] [Google Scholar]
- Guan XL, Wenk MR. Mass spectrometry-based profiling of phospholipids and sphingolipids in extracts from Saccharomyces cerevisiae. Yeast. 2006;23:465–477. doi: 10.1002/yea.1362. [DOI] [PubMed] [Google Scholar]
- Gutierrez AL, Farage L, Melo MN, Mohana-Borges RS, Guerardel Y, Coddeville B, Wieruszeski JM, Mendonca-Previato L, Previato JO. Characterization of glycoinositolphosphoryl ceramide structure mutant strains of Cryptococcus neoformans. Glycobiology. 2007;17:1–11C. doi: 10.1093/glycob/cwm030. [DOI] [PubMed] [Google Scholar]
- Haynes BC, Skowyra ML, Spencer SJ, Gish SR, Williams M, Held EP, Brent MR, Doering TL. Toward an integrated model of capsule regulation in Cryptococcus neoformans. PLoS Pathog. 2011;7:e1002411. doi: 10.1371/journal.ppat.1002411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hechtberger P, Zinser E, Saf R, Hummel K, Paltauf F, Daum G. Characterization, quantification and subcellular localization of inositol-containing sphingolipids of the yeast, Saccharomyces cerevisiae. Eur J Biochem. 1994;225:641–649. doi: 10.1111/j.1432-1033.1994.00641.x. [DOI] [PubMed] [Google Scholar]
- Heise N, Gutierrez AL, Mattos KA, Jones C, Wait R, Previato JO, Mendonca-Previato L. Molecular analysis of a novel family of complex glycoinositolphosphoryl ceramides from Cryptococcus neoformans: Structural differences between encapsulated and acapsular yeast forms. Glycobiology. 2002;12:409–420. doi: 10.1093/glycob/cwf053. [DOI] [PubMed] [Google Scholar]
- Heiss C, Klutts JS, Wang Z, Doering TL, Azadi P. The structure of Cryptococcus neoformans galactoxylomannan contains beta-D-glucuronic acid. Carbohydr Res. 2009;344:915–920. doi: 10.1016/j.carres.2009.03.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss C, Skowyra ML, Liu H, Klutts JS, Wang Z, Williams M, Srikanta D, Beverley SM, Azadi P, Doering TL. Unusual galactofuranose modification of a capsule polysaccharide in the pathogenic yeast Cryptococcus neoformans. J Biol Chem. 2013;288:10994–11003. doi: 10.1074/jbc.M112.441998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. Amino-acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry J, Guillotte A, Luberto C, Del Poeta M. Characterization of inositol phospho-sphingolipid-phospholipase C 1 (Isc1) in Cryptococcus neoformans reveals unique biochemical features. FEBS Lett. 2011;585:635–640. doi: 10.1016/j.febslet.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Sillaots S, Lemieux S, Davison J, Kauffman S, Breton A, Linteau A, Xin C, Bowman J, Becker J, et al. Essential gene identification and drug target prioritization in Aspergillus fumigatus. PLoS Pathog. 2007;3:e24. doi: 10.1371/journal.ppat.0030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennemann R, Geyer R, Sandhoff R, Gschwind RM, Levery SB, Grone HJ, Wiegandt H. Glycoinositolphosphosphingolipids (basidiolipids) of higher mushrooms. Eur J Biochem. 2001;268:1190–1205. doi: 10.1046/j.1432-1327.2001.01963.x. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Kim H, Melen K, Osterberg M, von Heijne G. A global topology map of the Saccharomyces cerevisiae membrane proteome. Proc Natl Acad Sci USA. 2006;103:11142–11147. doi: 10.1073/pnas.0604075103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klutts JS, Doering TL. Cryptococcal xylosyltransferase 1 (Cxt1p) from Cryptococcus neoformans plays a direct role in the synthesis of capsule polysaccharides. J Biol Chem. 2008;283:14327–14334. doi: 10.1074/jbc.M708927200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Yang M, Haynes BC, Skowyra ML, Doering TL. Emerging themes in cryptococcal capsule synthesis. Curr Opin Struct Biol. 2011;21:597–602. doi: 10.1016/j.sbi.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester RL, Dickson RC. Sphingolipids with inositolphosphate-containing head groups. Adv Lipid Res. 1993;26:253–274. [PubMed] [Google Scholar]
- Levine TP, Wiggins CA, Munro S. Inositol phosphorylceramide synthase is located in the Golgi apparatus of Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:2267–2281. doi: 10.1091/mbc.11.7.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman Q, Pomorski T, Vogelzangs C, Urli-Stam D, de Cocq van Delwijnen W, Holthuis JC. Protein sorting in the late Golgi of Saccharomyces cerevisiae does not require mannosylated sphingolipids. J Biol Chem. 2004;279:1020–1029. doi: 10.1074/jbc.M306119200. [DOI] [PubMed] [Google Scholar]
- Maciel DM, Rodrigues ML, Wait R, Villas Boas MH, Tischer CA, Barreto-Bergter E. Glycosphingolipids from Magnaporthe grisea cells: Expression of a ceramide dihexoside presenting phytosphingosine as the long-chain base. Arch Biochem Biophys. 2002;405:205–213. doi: 10.1016/s0003-9861(02)00365-x. [DOI] [PubMed] [Google Scholar]
- Mingeot-Leclercq MP, Brasseur R, Schanck A. Molecular parameters involved in aminoglycoside nephrotoxicity. J Toxicol Environ Health. 1995;44:263–300. doi: 10.1080/15287399509531960. [DOI] [PubMed] [Google Scholar]
- Nagiec MM, Nagiec EE, Baltisberger JA, Wells GB, Lester RL, Dickson RC. Sphingolipid synthesis as a target for antifungal drugs. Complementation of the inositol phosphorylceramide synthase defect in a mutant strain of Saccharomyces cerevisiae by the AUR1 gene. J Biol Chem. 1997;272:9809–9817. doi: 10.1074/jbc.272.15.9809. [DOI] [PubMed] [Google Scholar]
- Nakase M, Tani M, Morita T, Kitamoto HK, Kashiwazaki J, Nakamura T, Hosomi A, Tanaka N, Takegawa K. Mannosylinositol phosphorylceramide is a major sphingolipid component and is required for proper localization of plasma-membrane proteins in Schizosaccharomyces pombe. J Cell Sci. 2010;123:1578–1587. doi: 10.1242/jcs.059139. [DOI] [PubMed] [Google Scholar]
- Nelson RT, Hua J, Pryor B, Lodge JK. Identification of virulence mutants of the fungal pathogen Cryptococcus neoformans using signature-tagged mutagenesis. Genetics. 2001;157:935–947. doi: 10.1093/genetics/157.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimrichter L, Rodrigues ML. Fungal glucosylceramides: From structural components to biologically active targets of new antimicrobials. Front Microbiol. 2011;2:212. doi: 10.3389/fmicb.2011.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaar RL, Suzuki A, Stanley P. Glycosphingolipids. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2009. [PubMed] [Google Scholar]
- Simenel C, Coddeville B, Delepierre M, Latge JP, Fontaine T. Glycosylinositolphosphoceramides in Aspergillus fumigatus. Glycobiology. 2008;18:84–96. doi: 10.1093/glycob/cwm122. [DOI] [PubMed] [Google Scholar]
- Sinnott ML. Catalytic mechanisms of enzymatic glycosyl transfer. Chem Rev. 1990;90:1171–1202. [Google Scholar]
- Skowyra ML, Doering TL. RNA interference in Cryptococcus neoformans. Methods Mol Biol. 2012;845:165–186. doi: 10.1007/978-1-61779-539-8_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Takakuwa N, Kinoshita M, Oda Y, Ohnishi M. Existence of cerebroside in Saccharomyces kluyveri and its related species. FEMS Yeast Res. 2002;2:533–538. doi: 10.1111/j.1567-1364.2002.tb00120.x. [DOI] [PubMed] [Google Scholar]
- Toffaletti DL, Rude TH, Johnston SA, Durack DT, Perfect JR. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J Bacteriol. 1993;175:1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura S, Kihara A, Inokuchi J, Igarashi Y. Csg1p and newly identified Csh1p function in mannosylinositol phosphorylceramide synthesis by interacting with Csg2p. J Biol Chem. 2003;278:45049–45055. doi: 10.1074/jbc.M305498200. [DOI] [PubMed] [Google Scholar]
- Uemura S, Kihara A, Iwaki S, Inokuchi J, Igarashi Y. Regulation of the transport and protein levels of the inositol phosphorylceramide mannosyltransferases Csg1 and Csh1 by the Ca2+-binding protein Csg2. J Biol Chem. 2007;282:8613–8621. doi: 10.1074/jbc.M606649200. [DOI] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW. Membrane lipids: Where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggins CA, Munro S. Activity of the yeast MNN1 alpha-1,3-mannosyltransferase requires a motif conserved in many other families of glycosyltransferases. Proc Natl Acad Sci USA. 1998;95:7945–7950. doi: 10.1073/pnas.95.14.7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wymann MP, Schneiter R. Lipid signalling in disease. Nat Rev Mol Cell Biol. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.