Abstract
Sirtuins are a highly conserved family of nicotinamide adenine dinucleotide (NAD+)-dependent protein lysine modifying enzymes with deacetylase, adenosine diphosphateribosyltransferase and other deacylase activities. Mammals have seven sirtuins, namely SIRT1-7. They are key regulators for a wide variety of cellular and physiological processes such as cell proliferation, differentiation, DNA damage and stress response, genome stability, cell survival, metabolism, energy homeostasis, organ development, aging, and cancer. Here we present an extensive literature review of the roles of mammalian sirtuins, particularly SIRT1 as that is the most studied sirtuin, in human epithelial, neuronal, hematopoietic, and mesenchymal malignancies, covering breast, prostate, lung, thyroid, liver, colon, gastric, pancreatic, ovarian, and cervical cancers, tumors of the central nervous system, leukemia and lymphoma, and soft tissue sarcomas. Collective evidence suggests sirtuins are involved in both promoting and suppressing tumorigenesis depending on cellular and molecular contexts. We discuss the potential use of sirtuin modulators, especially sirtuin inhibitors, in cancer treatment.
Keywords: sirtuin, cancer, sirtuin modulator, deacetylation, acetylation
Introduction
The sirtuins are a family of proteins homologous to yeast silent information regulator 2 (Sir2) that was cloned and characterized in 1984 as a gene required for maintaining silent chromatin in yeast.1 The discovery of the longevity-promoting effect of Sir2 in yeast in 19992 and subsequently in higher eukaryotes nematode worm in 20013 and fruit fly in 20044 has stimulated extensive research interest in the biology of sirtuins. Although the effect of Sir2 and the most-studied mammalian Sir2 homolog, SIRT1, on longevity has recently been questioned,5–9 sirtuin family proteins appear to play important roles in many physiological and pathological processes.
There are seven sirtuin genes, SIRT1-7, in mammals.10,11 Biochemically, they are a class of proteins that possess nicotinamide adenine dinucleotide (NAD+)-dependent lysine deacetylase (SIRT1, SIRT2, SIRT3, SIRT5, SIRT6, and SIRT7) and monoribosyltransferase (SIRT4 and SIRT6) activities.12–19 Recently, SIRT5 was shown to be a NAD+-dependent protein lysine demalonylase and desuccinylase.20 Sirtuin family members share a conserved NAD+-binding and catalytic core domain. Sirtuins are also known as class III histone deacetylases (HDACs), and their unique NAD+-dependency distinguishes sirtuins from other (classes I, II, and IV) HDACs.
Brief overview of physiological functions of sirtuins
SIRT1 is primarily a nuclear deacetylase.21 It contains at least two nuclear localization signals and two nuclear export signals, and can shuttle between the nucleus and cytoplasm under certain conditions.22 SIRT1 removes the acetyl group from the ɛ-amino group of lysine residues in histones and non-histone proteins, and regulates target gene expression and protein activities that control various cellular processes such as cell proliferation, differentiation, apoptosis, metabolism, DNA damage and stress response, genome stability, and cell survival in complex matters (Table 1). SIRT1−/− mice in C57/B6 background typically die within 1 month after birth, but in some other background they may survive through adulthood with smaller body size, closed eyelids, infertility, and autoimmune-like conditions.23–26
Table 1.
Examples of SIRT1 substrates and functions
| SIRT1 substrates | SIRT1 functions |
|---|---|
| AceCS1 | Promotes AceCS1 activity and metabolism183 |
| Akt, PDK1 | Enhances their PIP3 binding and membrane localization during tumorigenesis and cardiac hypertrophy184 |
| Androgen receptor | Represses dihydrotestosterone-induced androgen receptor signaling185 |
| APE1 | Promotes base excision repair activity186 |
| ATG (autophagy genes Atg5, Atg7, and Atg8) | Promotes autophagy187 |
| β-catenin | Suppresses its ability to activate transcription and to drive cell proliferation125 |
| BMAL1 | Modulates CLOCK-mediated chromatin remodeling and circadian control188 |
| CIITA (class II transactivator) | Augments MHC II transcription by shielding CIITA from proteasomal degradation and promoting nuclear accumulation and target binding189 |
| c-MYC | Stabilizes89,190 or destabilizes c-MYC oncoprotein132 |
| CRTC1 (CREB-regulated transcription coactivator 1, or TORC1) | Activates TORC1 by promoting its dephosphorylation and its interaction with CREB for neuroprotection191 |
| CRTC2 (TORC2) | Attenuates CRTC2 activity and glucose output during fasting192 |
| DNMT1 | Deacetylation of different lysines on DNMT1 has different effects on the activities for DNA methylation193 |
| EVI1 | Triggers EVI1 degradation194 |
| eNOS | Stimulates eNOS activity, increases endothelial nitric oxide, promotes endothelium-dependent vascular relaxation195 |
| ERα | Represses its DNA binding and transcriptional activity196 |
| FOXO1 | Potentiates FOXO1-mediated transcription through its deacetylase activity197 |
| FOXO3 | Increases FOXO3’s ability to induce cell cycle arrest and resistance to oxidative stress but inhibits FOXO3’s ability to induce cell death198,199 |
| FOXp3 | Promotes its degradation, inhibits Treg functionality200 |
| FXR (nuclear bile acid receptor) | Decreases its stability but enhances transactivation activity in lipid and glucose metabolism regulation201 |
| HIF1α | Inactivates HIF-1 alpha under hypoxia130 |
| HIF2α | Promotes HIF-2 signaling during hypoxia202 |
| Histone H1(K26), H3(K9, K56), H4(K16) | Transcription regulation and chromatin function12 |
| HSF1 | Prolongs HSF1 binding to the heat shock promoter Hsp70203 |
| Ku70 | Promotes DNA repair activity204 |
| LXR (Liver X receptor) | Positively regulates its function for cholesterol and lipid homeostasis205 |
| MeCP2 (Methyl-CpG binding protein 2) | Promotes MeCP2-mediated BDNF expression206 |
| MMP2 | Enhances MMP2 protein stability81 |
| MyoD | inhibits myogenesis207 |
| NBS1 | Maintains NBS1 in a hypoacetylated state, which is required for ionizing radiation-induced NBS1 Ser343 phosphorylation208 |
| NF-κB p65 | Reduces NF-κB transcriptional activity, augments apoptosis in response to TNFα209 |
| NHLH2 | Activates MAO-A to mediate anxiety and exploratory drive210 |
| N-MYC | Promotes protein stability111 |
| NoRC | Leads to enhanced promoter-associated RNA binding and an increase in heterochromatic histone marks211 |
| NiCD (Notch1 intracellular domain) | Acts as a negative modulator of Notch signaling in endothelial cells212 |
| p300 | Represses its transactivation activity213 |
| p53 | Promotes cell survival under stress214,215 |
| PARP1 | Promotes cell survival under stress216 |
| PER2 | Promotes PER2 degradation to regulate circadian clock gene expression217 |
| PGC1α | Positively and negatively controls gene expression for glucose homeostasis218 |
| PIP5 Kγ | Regulates thyroid-stimulating hormone release by enhancing PIP5 Kgamma activity219 |
| PTEN | Modulates PTEN interaction with PDZ domain-containing proteins220 |
| RARβ | Activates alpha-secretase gene ADAM10, suppresses beta-amyloid production221 |
| Rb (Retinoblastoma tumor suppressor protein) | Inactivates retinoblastoma tumor suppressor protein222 |
| Smad7 | Inhibits transforming growth factor beta-induced apoptosis in glomerular mesangial cells223 |
| SREBP-1C | Inhibits SREBP-1C activity in regulation of hepatic lipid metabolism224 |
| STAT3 | Suppresses the inhibitory effect of STAT3 on gluconeogenesis225 |
| Survivin | Suppresses survivin thus inhibits cell survival226 |
| SUV39H1 | increases SUV39H1 activity during heterochromatin formation227 |
| Tat | Facilitates the recycling of Tat228 |
| Tip60, hMOF | Inhibits their acetyltransferase activity and promotes their degradation in DNA damage response229 |
| WRN | Increases its helicase and exonuclease activities, promotes its translocation from nucleoplasm to nucleoli after DNA damage230 |
| XBP1s | Inhibits its transcriptional activity231 |
| Xpa, Xpc | Promotes nucleotide excision repair activity232,233 |
Abbreviations: AceCS1, acetyl-coenzyme A synthetase 1; APE1, apurinic/apyrimidinic endonuclease-1; ATG, autophagy genes; BDNF, brain-derived neurotrophic factor; CREB, cyclic adenosine monophosphate response element-binding protein; DNMT1, DNA methyltransferase 1; eNOS, endothelial nitric oxide synthase; ERα, estrogen receptor alpha; EVI1, ecotropic viral integration site I; FOXO1, forkhead box protein O1; FOXO3, forkhead box protein O3; FOXp3, forkhead box P3; HIF1α, hypoxia-inducible factor 1-alpha; HIF2α, hypoxia-inducible factor 2-alpha; HSF1, heat shock factor protein 1; LXR, liver X receptor; MAO-A, monoamine oxidase A; MeCP2, methyl-CpG binding protein 2; MHC II, major histocompatibility complex class II; MMP2, matrix metalloproteinase-2; NBS1, nijmegen breakage syndrome protein 1; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; NICD, Notch1 intracellular domain; NoRC, nitric oxide reductase; PARP1, poly(adenosine diphosphate ribose) polymerase 1; PDK1, phosphoinositide-dependent kinase-1; PGC1α, peroxisome proliferator-activated receptor gamma coactivator 1-alpha; PIP3, phosphatidylinositol (3,4,5)-triphosphate; PTEN, phosphatase and tensin homolog; RARβ, retinoic acid receptor beta; SREBP-1C, sterol regulatory element-binding protein; STAT3, signal transducer and activator of transcription 3; TNF, tumor necrosis factor; WRN, Werner syndrome; XBP1, X-box binding protein 1; Xpa, xeroderma pigmentosum group A; Xpc, xeroderma pigmentosum group C.
SIRT2 is mainly localized to the cytoplasm, but can shuttle to the nucleus during mitosis.21,27,28 It deacetylates many substrates such as histone H4K16, H3K56, α-tubulin, PR-Set7, phosphoenolpyruvate carboxykinase 1, NF-κB subunit p65, FOXO, and RIP1 (receptor-interacting protein 1) (Table 2). SIRT2 regulates several cell functions including cell cycle progression, cell death, and stress response. SIRT2 knockout female mice develop mammary tumors, whereas males develop hepatic and intestinal tumors.29
Table 2.
Examples of SIRT2 substrates and functions
| SIRT2 substrates | SIRT2 functions |
|---|---|
| Alpha-tubulin | Abrogates resistance to axonal degeneration14,234 |
| CDH1, CDC20 | Promotes their degradation and cell cycle exit29 |
| FOXO1 | Inhibits adipocyte differentiation235 |
| FOXO3a | Leads to Skp2-mediated FOXO3 ubiquitination and degradation76 |
| Histone H3K56 | Inhibits its assembly into chromatin in response to DNA damage157 |
| Histone H4K16 | Promotes cell cycle G2/M transition28 |
| Keratin 8 (K207) | Increases its solubility236 |
| NF-κB p65 (K310) | Suppresses NF-κB dependent gene expression237 |
| p300 | Restores its binding to the pre-initiation complex, thereby promoting transcription238 |
| PAR-3 | Decreases the activity of the polarity complex signaling component atypical protein kinase C, thereby regulating myelin formation239 |
| PEPCK1 (Phosphoenolpyruvate carboxykinase 1) | Stabilizes PEPCK1 for gluconeogenesis240 |
| PR-Set7 | Increases its chromatin localization241 |
| RIP1 | Is required for programmed necrosis242 |
Abbreviations: FOXO1, forkhead box protein O1; FOXO3a, forkhead box protein O3a; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; PEPCK1, phosphoenolpyruvate carboxykinase 1; RIP1, receptor-interacting protein 1; PAR-3, protease-activated receptor-3.
SIRT3 is present in mitochondria,21,30,31 but is also detected in the nucleus.32,33 It is a major protein deacetylase within the mitochondrial matrix,34 and plays a crucial role in cellular energy metabolism and redox regulation by deacetylating key mitochondrial proteins, including acetyl-coenzyme A synthetase 2, isocitrate dehydrogenase 2 (IDH2), glutamate dehydrogenase (GDH), manganese superoxide dismutase (MnSOD) (Table 3). SIRT3-null mice exhibit reduction of respiration and adenosine triphosphate levels, defect of fatty acid oxidation, metabolic syndrome, and development of mammary tumors.35–37
Table 3.
Examples of mitochondrial sirtuin substrates and functions
| Sirtuins | Sirtuin substrates | Sirtuin functions |
|---|---|---|
| SIRT3 | AceCS2 | Promotes AceCS2 activity183,243 |
| Cyclophilin D | Induces dissociation of hexokinase II from the mitochondria,244 and suppresses age-related cardiac hypertrophy245 | |
| FOXO3a | Promotes its nuclear localization and mitochondrial biogenesis246 | |
| GDH | Increases its activity34,247 | |
| Histone H4K16 | Gene transcription33 | |
| IDH2 | Activates its activity, protects cells from oxidative stress, prevents age-related hearing loss under caloric restriction146,147,247 | |
| Ku70 | Protects cells from stress-mediated cell death248 | |
| LCAD | Increases its enzymatic activity249 | |
| LKB1 | Activates LKB1, thus augmenting the activity of the LKB1-AMPK pathway250 | |
| HMGCS2 | Enhances its enzymatic activity251 | |
| MnSOD | Enhances its enzymatic activity252 | |
| MRPL10 | Suppresses translational activity of mitochondrial ribosomes253 | |
| NDUFA9 | Augments complex I activity of the electron transport chain254 | |
| OTC | Stimulates its activity255 | |
| SDH (Succinate dehydrogenase) | Promotes its enzyme activity256,257 | |
| SIRT4 | GDH | Mono-ADP-ribosylates it, reduces GDH activity16 |
| MCD | Deacetylates and inhibits the activity of MCD258 | |
| SIRT5 | CPS1 | Deacetylates and upregulates its activity to regulate the urea cycle42 |
| Pyruvate dehydrogenase and succinate dehydrogenase | Desuccinylates and represses their biochemical activity and thus cellular respiration44 |
Notes: There are a large and growing number of mitochondrial sirtuin substrates. The list only shows some examples that have been studied in more detail.
Abbreviations: AceCS2, acetyl-coenzyme A synthetase 2; ADP, adenosine diphosphate; AMPK, adenosine monophosphate-activated protein kinase; CPS1, carbamoyl phosphate synthetase I; FOXO3a, forkhead box protein O3a; FOXp3, forkhead box P3; GDH, glutamate dehydrogenase; HMGCS2, mitochondrial 3-hydroxy-3-methylglutaryl coenzyme A synthase 2; IDH2, isocitrate dehydrogenase 2; LCAD, long-chain acyl coenzyme A dehydrogenase; LKB1, liver kinase B1; MCD, malonyl coenzyme A decarboxylase; MnSOD, manganese superoxide dismutase; MRPL10, mitochondrial ribosomal protein L10; NDUFA9, nicotinamide adenine dinucleotide dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial; OTC, ornithine transcarbamoylase; SDH, succinate dehydrogenase.
SIRT4 is localized to mitochondria,21 and is a NAD+-dependent protein adenosine diphosphate (ADP)-ribosyl transferase, which catalyzes the transfer of ADP-ribosyl groups onto target proteins, such as GDH.16 SIRT4 regulates cellular metabolic functions like insulin secretion and fatty acid oxidation.16,38–40 Following genotoxic stress, SIRT4 has also exhibited an anti-apoptotic function by maintaining mitochondrial NAD+ levels together with SIRT3.41 SIRT4-depleted mice develop hyperinsulinemia and lung tumors.16,40
SIRT5 is also localized to mitochondria.21 It can deacetylate carbamoyl phosphate synthetase 1 and activate its catalytic activity in the initial step of the urea cycle for ammonia detoxification and disposal.42 It also possesses NAD+-dependent lysine demalonylase and desuccinylase activities that remove malonyl and succinyl groups on target proteins including GDH, carbamoyl phosphate synthetase 1, pyruvate dehydrogenase, succinate dehydrogenase, and many other substrates impacting diverse metabolic pathways.20,43,44 Interestingly, a proteomics study by Park et al44 showed significant cytoplasmic activity of SIRT5, in line with a previous study by Matsushita et al45 showing that there are two isoforms of human SIRT5 differing in the C-terminal sequence, with the shorter isoform (SIRT5iso2) mainly localized in mitochondria and the longer form (SIRT5iso1) localized in both cytoplasm and mitochondria. SIRT5-null mice exhibit urea cycle defect and hyperammonemia after fasting.42
SIRT6 is a nuclear protein having both deacetylase and ADP-ribosyltransferase activity.17,46 Recently SIRT6 was shown to be able to remove long-chain fatty acyl group from lysine to regulate tumor necrosis factors (TNF)-α secretion.47 SIRT6 has been implicated in the regulation of transcription, genome stability, metabolism, and lifespan. Its substrates include histone H3K9, H3K56, C-terminal binding protein interacting protein, poly(ADP-ribose) polymerase 1, DNA-dependent protein kinase, and GCN5 (Table 4). SIRT6 deficient mice die around 4 weeks after birth, showing premature aging phenotypes, hypoglycemia, increased glucose uptake, cardiac hypertrophy and heart failure, hypersensitivity to DNA damage, and genomic instability. The observed lethal hypoglycemia directly results from its histone H3K9 deacetylase function that controls the expression of glycolytic genes.48–62
Table 4.
Examples of nuclear SIRT6 and SIRT7 substrates and functions
| Sirtuins | Sirtuin substrates | Sirtuin functions |
|---|---|---|
| SIRT6 | CtIP | Deacetylates it to promote DNA end resection53 |
| DNA-PK | Deacetylates and stabilizes it at chromatin for DNA double-strand break repair52 | |
| GCN5 | Interacts with and modifies GCN5, enhancing GCN5’s activity to acetylate PGC1α and suppresses hepatic glucose production59 | |
| Histone H3K56 | Deacetylates H3K56 to promote genomic stability19,156 | |
| Histone H3K9 | Deacetylates H3K9: maintains telomeric chromatin;46,57 represses NF-κB50 and HIF1α51,60 dependent gene expression; attenuates IGF-Akt signaling58 | |
| PARP1 | Mono-ADP-ribosylates it, thereby stimulating its poly-ADP-ribosylase activity and enhancing DSB repair under oxidative stress56 | |
| TNF-α | Removes the fatty acyl modification on K19 and K20 of TNF-α to promote its secretion47 | |
| SIRT7 | Histone H3K18 | Maintains the transformation phenotype of cancer cells18 |
Abbreviations: ADP, adenosine diphosphate; CtIP, C-terminal binding protein interacting protein; DNA-PK, DNA-dependent protein kinase; HIF1α, hypoxia-inducible factor 1-alpha; IGF, insulin-like growth; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; PARP1, poly(adenosine diphosphate ribose) polymerase 1; PGC1α, peroxisome proliferator-activated receptor gamma coactivator 1-alpha; TNF, tumor necrosis factor.
SIRT7 is localized to the nucleolus.21 It exhibits high selectivity for histone H3K18, and functions to maintain the transformed phenotypes of cancer cells.18 SIRT7 is a positive regulator of RNA polymerase I transcription and therefore ribosome biogenesis, and its knockdown induces apoptosis in human cells, indicating that SIRT7 is required for cell survival.63,64 SIRT7-deficient mice die around 1 year, showing premature aging phenotypes (kyphosis and loss of subcutaneous fat), and enhanced inflammatory cardiomyopathy as well as enhanced cardiomyocyte apoptosis.65 Some available mouse models for sirtuin research are summarized in Table 5.
Table 5.
Available mouse models for sirtuins research
| Sirtuin | Mouse models | Phenotypes |
|---|---|---|
| SIRT1 | KO (whole body) | In C57/B6 background, mice die within 1 month after birth. In BALB/c background or mixed background, mice can survive through adulthood with smaller body size, closed eyelids, infertility, and autoimmune-like conditions |
| KO (brain) | Memory defect, no adaptive feeding response to calorie restriction, less serum insulin-like growth factor 1 | |
| KO (liver) | Defect in circadian gene oscillation, develop hepatic steatosis and inflammation | |
| KO (macrophage) | Increased inflammation, glucose intolerance and insulin resistance induced by high fat diet | |
| Tg (whole body) | Protected against various metabolic disorders (fatty liver and type 2 diabetes) induced by high fat diet, protected against age-induced cancer, osteoporosis and glucose intolerance | |
| Tg (brain) | enhanced memory formation and feeding behavior, protected against Alzheimer’s disease | |
| Tg (heart) | Cardioprotection (mild expression), cardiac hypertrophy (high expression) | |
| Tg (gut) | Protected against colon cancer | |
| Tg (kidney) | Protected against acute renal failure | |
| SIRT2 | KO (whole body) | SiRT2 knockout female mice develop mammary tumors, whereas males develop liver and intestinal tumors |
| SIRT3 | KO (whole body) | Defect in fatty acid oxidation, cancer prone, their oocytes exhibit developmental arrest after in vitro fertilization, accumulation of hyperacetylated mitochondrial proteins, reduced respiration and adenosine triphosphate levels |
| Tg (heart) | Protected against cardiac hypertrophy | |
| SIRT4 | KO (whole body) | Developed hyperinsulinemia and lung tumors |
| SIRT5 | KO (whole body) | Defect in urea cycle, hyperammonemia after fasting |
| Tg (liver) | Increased urea cycle activity, increased urea production | |
| SIRT6 | KO (whole body) | Died around 4 weeks showing premature aging phenotype (lymphopenia, loss of subcutaneous fat), hypoglycemia, increased glucose uptake, genomic instability |
| KO (liver) | Increased glycolysis, triglyceride synthesis, reduced β oxidation and fatty liver formation | |
| Tg (whole body) | Protected against metabolic disorder induced by high fat diet | |
| SiRT7 | KO (whole body) | Died around 1 year showing premature aging phenotypes (kyphosis, loss of subcutaneous fat, degenerative cardiac hypertrophy), increased apoptosis |
Note: Adapted with permission from J Cell Sci. 2011;124(Pt 6):833–838. Nakagawa T, Guarente L. Sirtuins at a glance.259
Abbreviations: KO, knockout; Tg, transgenic.
Roles of sirtuins in cancer
All mammalian sirtuins except SIRT5 have been reported to be involved in tumorigenesis. But the roles of sirtuins in cancer are complex and may contribute to either tumor promotion or suppression depending on cellular and molecular contexts as reviewed recently.66
SiRT1 in cancer
In the past decade, numerous substrates of SIRT1 have been identified, including many important regulators for cancer cell proliferation, DNA damage repair, and survival under various stress conditions (Table 1). SIRT1 plays a dual role in cancer promotion and suppression, depending on tissue contexts and the temporal and spatial distribution of SIRT1 upstream and downstream factors (Figure 1). This section will review SIRT1 functions in several types of cancer.
Figure 1.
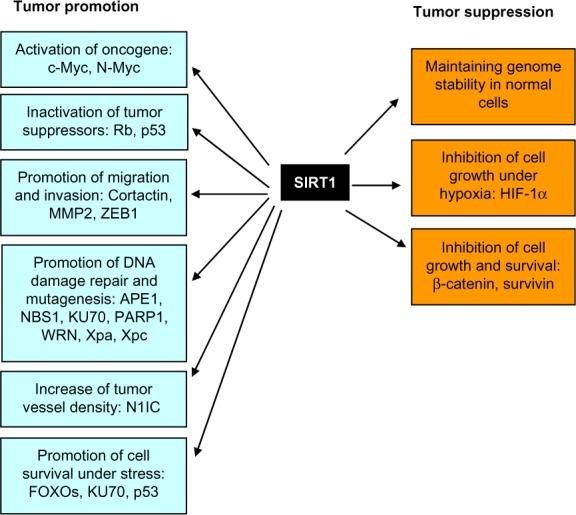
Bifurcated roles of SIRT1 in tumor promotion and suppression.
Abbreviations: APE1, apurinic/apyrimidinic endonuclease-1; FOXO, forkhead box protein O; HIF-1 α, hypoxia-inducible factor 1-al pha; MMP2, matrix metalloproteinase-2; NBS1, nijmegen breakage syndrome protein 1; NIC, Notch 1 intracellular domain; PARP 1, poly(adenosine diphosphate ribose) polymerase 1; WRN, Werner syndrome; Xpa, xeroderma pigmentosum group A; Xpc, xeroderma pigmentosum group C; ZEB1, zinc finger E-box-binding homeobox 1.
Breast cancer
The expression of SIRT1 protein was seen in most human breast cancer specimens, and its expression was significantly associated with distant metastasis and poor prognosis.67–69 SIRT1 upregulation in breast cancer cells is associated with inactivation of tumor suppressor hypermethylated in cancer 1 (HIC1) by DNA hypermethylation.70 SIRT1 promotes cell survival after DNA damage through inactivation of the p53 pathway. SIRT1 upregulation is also associated with decreased miR-200a in breast cancer samples, which targets the three prime untranslated region of SIRT1 messenger RNA (mRNA) and promotes epithelial–mesenchymal transition (EMT)-like transformation in mammary epithelial cells.71 SIRT1 is essential for oncogenic signaling of estrogen/estrogen receptor α (ERα) in breast cancer. SIRT1 inactivation suppresses estrogen/ERα-induced cell growth and tumor development, and induces apoptosis. Compared to adjacent normal tissue, SIRT1 is found to be significantly upregulated in the invasive ductal carcinoma, and positively regulates the expression of aromatase, an enzyme responsible for a key step in the biosynthesis of estrogen in breast cancer.72 In addition, SIRT1 can promote cell migration by directly interacting and deacetylating cortactin,73 and promote the expression of multidrug resistance-associated protein 2 in tamoxifen-resistant breast cancer cells for chemoresistance by deacetylating FOXO1.74 SIRT1 activator SRT1720 promotes the migration and pulmonary metastasis of subcutaneously-implanted breast cancer cells in mice, further supporting the cancer promoting effect of SIRT1 in breast cancer.75
Prostate cancer
SIRT1 is significantly overexpressed in human prostate cancer cell lines and tissues, compared with normal prostate epithelial cells and adjacent normal prostate tissues. SIRT1 inhibition via nicotinamide, sirtinol, short hairpin RNAs, or mutation of the 25 amino acid C-terminal SIRT1 activator sequence, results in a significant inhibition in the cell growth, viability, and chemoresistance.76–80 SIRT1 is highly expressed in advanced prostate cancer tissues and could promote prostate cancer cell invasion, migration, and metastasis through matrix metalloproteinase-2,81 EMT inducing transcription factor ZEB1,82 and cortactin.73,83 In the transgenic mouse model, SIRT1 expression promotes murine prostate carcinogenesis initiated by phosphatase and tensin homolog deficiency.84
Lung cancer
Positive SIRT1 and cortactin expression was observed in 67% (96 of 144) and 58% (84 of 144) of patients with invasive non-small-cell lung cancer, respectively.85 SIRT1 and cortactin expression are significantly associated with unfavorable clinical factors, including high pathological T stage, lymph node metastasis, and advanced tumor invasion.85 Deregulation of the HIC1-SIRT1-p53 regulation loop was confirmed in 118 non-small-cell lung cancer patients. The patients with low p53 acetylation and high SIRT1 expression mostly showed low HIC1 expression and worse prognosis compared to other patients.86 SIRT1 could facilitate endothelial cell branching and proliferation to increase vessel density and promote lung tumor growth through downregulation of DLL4/Notch signaling and deacetylation of Notch1 intracellular domain.87 Conversely, SIRT1/2 inhibition by short interfering RNA (siRNA) or a small molecule inhibitor Salermide, causes apoptosis in human non-small-cell lung cancer cells by upregulating death receptor 5 expression.88
Colon cancer
Highly-expressed c-MYC correlates with increased SIRT1 protein level in colorectal cancer.89 c-MYC, nicotinamide phosphoribosyltransferase, deleted in breast cancer protein 1, and SIRT1 form a positive feedback regulatory loop.89 In 121 colorectal serrated lesions, the higher expression of c-MYC and SIRT1 proteins is strongly associated with higher grades of malignancy.90 In another study with a total of 485 colorectal cancer patients, SIRT1 overexpression was detected in 180 (37%) tumors.91 SIRT1 expression is associated with microsatellite instability and CpG island methylator phenotype, although not patient prognosis.91 Reduced expression of miR-34a, a negative regulator of SIRT1 mRNA, is observed in drug-resistant DLD-1 colon cancer cells, and introduction of miR-34a induces apoptosis by downregulating SIRT1.92
Thyroid cancer
SIRT1 is overexpressed in human thyroid cancers and it is positively correlated with c-MYC protein levels. Transgenic SIRT1 expression promotes murine thyroid carcinogenesis initiated by phosphatase and tensin homolog deficiency. SIRT1 increases c-MYC transcription and stabilizes c-MYC protein in thyroid cancers from SIRT1 transgenic mice or cultured thyroid cancer cells.84
Gastric cancer
SIRT1 protein expression in gastric cardiac carcinoma is significantly higher than that in normal gastric cardiac tissues and is associated with lymphatic metastasis, TNM (the extent of tumor [T], the extent of spread to lymph nodes [N], and the presence of distant metastasis [M]) stage, survival rate, and mean survival time.93 In another study, positive expression of SIRT1 was seen in 73% (130 of 177) of gastric cancer patients.94 SIRT1 expression is also significantly associated with shorter overall survival and relapse-free survival.94 SIRT1 is required for activating-transcription-factor-4-induced multidrug resistance in gastric cancer cells. Activating transcription factor 4 facilitates multidrug resistance in gastric cancer cells through direct binding to SIRT1 promoter and activating SIRT1 expression. Significantly, inhibition of SIRT1 by RNA interference or a specific inhibitor (EX-527) sensitizes gastric cancer cells to therapeutic treatment.95
Liver cancer
SIRT1 expression is significantly elevated in hepatocellular carcinoma (HCC) compared to non-tumor tissues, and the expression levels correlate with tumor grades and predict poor prognosis. SIRT1 promotes tumorigenesis and chemoresistance in HCC, and inhibition of SIRT1 consistently suppresses the proliferation of HCC cells in vitro or in vivo via the induction of cellular senescence or apoptosis.96–100 SIRT1 expression also positively correlates with c-MYC levels in HCC. SIRT1 and c-MYC regulate each other via a positive feedback loop and act synergistically to promote cell proliferation of both mouse and human liver tumor cells.101 Accordingly, expression of microRNA (miRNA)-34a is reduced in HCC, and the reduced expression of miRNA-34a is associated with worse outcome of HCC patients. Treatment of established HCC xenograft with miR-34a-expressing adenovirus in a mouse model results in complete tumor regression without recurrence.102 In addition, miRNA-29c also functions as a tumor suppressor by directly targeting oncogenic SIRT1 in HCC.103
Pancreatic cancer
SIRT1 overexpression was observed in pancreatic cancer tissues at both mRNA and protein levels.104 Increased SIRT1 positivity is associated with patients’ age (over 60 years old), larger tumor size (larger than 4 cm), and higher TNM stage. SIRT1 knockdown induces apoptosis and senescence, inhibits invasion and enhances chemosensitivity in pancreatic cancer cells.104,105 In pancreatic cancer, SIRT1 regulates acinar-to-ductal metaplasia and supports cancer cell viability through deacetylating pancreatic transcription factor-1a and β-catenin. Inhibition of SIRT1 is effective in suppression of acinar-to-ductal metaplasia and in reducing cell viability in established pancreatic ductal adenocarcinoma.106 In addition, SIRT1 promotes EMT ability as well as invasion of pancreatic cancer cells by forming a complex with Twist and MBD1, thus suppressing E-cadherin transcription activity.107
Ovarian and cervical cancers
Expression of SIRT1 protein was significantly increased in 90 cases of malignant ovarian epithelial tumors compared to 40 cases of benign and 36 cases of borderline epithelial tumors.108 In granulosa cells, SIRT1 suppresses the activity of transcriptional factor FOXL2 on targets involved in cell cycle and DNA repair. Conversely, inhibition of SIRT1 by nicotinamide limits granulosa cell proliferation by increasing FOXL2 expression.109 In human SiHa cervical cancer cells, SIRT1 is upregulated by oncogenic viral protein human papillomavirus E7, and may mediate the pro-survival function of human papillomavirus E7 through attenuating p53 activity.110
Tumors of the central nervous system
SIRT1 and N-MYC form a positive feedback regulation loop during the tumorigenesis of neuroblastoma, and preventive treatment with the SIRT1 inhibitor Cambinol significantly reduces tumorigenesis in N-MYC transgenic mice.111 SIRT1 regulates tyrosine hydroxylase expression and differentiation of neuroblastoma cells via FOXO3a. SIRT1 inhibition by siRNA or nicotinamide inhibits all trans-retinoic acid induced upregulation of tyrosine hydroxylase and differentiation.112 In glioblastoma, SIRT1 is highly expressed in tumor-derived CD133+ progenitor cells compared to CD133− cells and knockdown of SIRT1 expression enhances the radio-sensitivity and radiation-induced apoptosis in the CD133+ cells in vitro and in vivo.113 Also, casein kinase-2 inhibitors could sensitize glioblastoma cells to TNF-α-induced apoptosis through a mechanism involving SIRT1 inhibition.114 SIRT1 is also frequently expressed (64.2%, 77/120 patients) in human medulloblastomas relative to surrounding noncancerous cerebellar tissues and its expression is correlated with the formation and prognosis of medulloblastomas. Inhibition of SIRT1 by siRNA or nicotinamide arrests medulloblastoma cell UW228-3 in the G1 phase and induces apoptosis, suggesting SIRT1 as a potential therapeutic target in this type of tumor.115
Lymphoma and leukemia
In adult T-cell leukemia cells, overexpression of SIRT1 was observed and its inhibition by sirtinol induced apoptosis.116 In 104 diffuse large B-cell lymphoma patients, positive expression of SIRT1 protein was seen in 74% (77/104) of patients, and was significantly associated with shorter overall survival.117 SIRT1 is also overexpressed (greater than two-fold) in acute myelogenous leukemia (AML) samples.118 Increased SIRT1 expression appears critical for cell survival. Inhibition of SIRT1/2 by Cambinol induces apoptosis in Burkitt lymphoma cells.119 In a large cohort of primary AML (n=12) and B-cell chronic lymphocytic leukemia (n=36) samples and leukemia cell lines, a combination of sirtuin inhibitors such as sirtinol, cambinol, or EX-527 with HDAC inhibitors led to a synergistic anti-leukemic effect.120
In chronic myelogenous leukemia (CML), a crucial role of SIRT1 in CML development and chemoresistance has recently been demonstrated.121–123 SIRT1 is activated by oncogenic breakpoint-cluster region-Abelson-murine-leukemia (BCR-ABL) in part via STAT5 signaling in hematopoietic progenitor cells. SIRT1 inhibition efficiently impairs the growth of human CML cells and sensitizes leukemia stem cells to the BCR-ABL inhibitor imatinib both in vitro and in vivo. SIRT1 knockout robustly inhibits BCR-ABL-mediated transformation of mouse bone marrow cells and development of CML-like myeloproliferative disease. Moreover, in a CML chemoresistance model that faithfully recapitulates many features of human CML response to imatinib treatment, SIRT1 inhibition prevents BCR-ABL mutagenesis through inhibiting Ku70-mediated DNA repair pathway and blocks CML cell relapse upon imatinib treatment.121–123
Soft tissue sarcomas
SIRT1 is frequently expressed in soft tissue neoplasms with myoid differentiation including angiomyolipoma (four out of five patients), glomus tumor (five out of five patients), leiomyoma (nine out of ten patients), leiomyosarcoma (76.5% of 51 patients), and rhabdomyosarcoma (87% of 24 patients), and thus could be a potential immunohistochemical marker and therapeutic target in these tumors.124
SIRT1 in tumor suppression
The above studies support roles of SIRT1 in cancer promotion; however, there is also a body of evidence, particularly from mouse model studies, pointing to a tumor suppressor role of SIRT1. SIRT1 transgenic mice exhibit a reduced incidence of spontaneous carcinomas and sarcomas, and a reduced susceptibility to carcinogen-induced liver cancer.6 Ectopic induction of SIRT1 in an APCmin/+ (adenomatous polyposis coli) mouse colon cancer model reduces tumor formation and proliferation.125 SIRT1+/−p53+/− mice have a higher incidence of tumors than wild-type, SIRT1+/− and p53+/− mice.126
SIRT1 expression is significantly downregulated in human head and neck squamous cell carcinoma (HNSCC). High SIRT1 expression is associated with good prognosis for HNSCC patients.127 In colorectal adenocarcinoma, SIRT1 overexpression was observed in approximately 25% of stage I/II/III tumors but rarely in advanced stage IV tumors and approximately 30% of carcinomas showed lower SIRT1 expression than normal tissues.128 In another clinical observation, SIRT1 protein expression gradually decreased during the normal-adenoma-adenocarcinoma-metastasis sequence in colorectal cancers, with positivity of 100%, 80.8%, 41.9%, and 35.7%, respectively.129
SIRT1 may suppress tumor growth through distinct mechanisms. SIRT1 deacetylates and inactivates hypoxia-inducible factor 1α, thus inhibits the expression of genes targeted by hypoxia-inducible factor 1α in certain tumors.130 In HMLER breast cancer cells, SIRT1 was found to suppress EMT, and reduced SIRT1 expression increases metastasis of these cells in nude mice.131 In one study, it was shown that although c-MYC induces SIRT1 expression, SIRT1 deacetylates c-MYC to reduce c-MYC protein stability and thus cellular transformation.132 SIRT1 inhibits proliferation of pancreatic cancer cells expressing oncogenic pancreatic adenocarcinoma upregulated factor, by suppression of β-catenin and cyclin-D1.133 In hepatitis-B-virus-X-protein-overexpressed Hep3B hepatocellular carcinoma cells, SIRT1 inhibits proliferation and enhances the sensitivity of the cells to doxorubicin or oxidative stress through destabilization of β-catenin or inhibition of c-Jun N-terminal kinase, respectively.134,135 These studies further underline the distinct roles of SIRT1 in cancer cells under different conditions.
SIRT2 in cancer
Similar to SIRT1, SIRT2 may have both tumor suppression and promotion function. SIRT2 expression is reduced in gliomas, and SIRT2 inhibits colony formation of glioma cell lines.136 SIRT2 expression is also reduced in esophageal adenocarcinomas, gastric adenocarcinomas, and HNSCC.137 The direct evidence that SIRT2 may act as a tumor suppressor came from a SIRT2 knockout mouse study.29 SIRT2 deficient male mice develop HCC whereas females develop mammary tumors.29 Mechanistically, SIRT2 regulates the anaphase-promoting complex/cyclosome activity through deacetylation of its coactivators, anaphase-promoting complex (CDH1) and CDC20. SIRT2 deficiency causes increased levels of mitotic regulators, including Aurora-A and Aurora-B that direct centrosome amplification, aneuploidy, and mitotic cell death. Moreover, SIRT2 level is reduced in human breast cancer and HCC.29
On the other hand, SIRT2 knockdown leads to both necrotic and apoptotic cell death in C6 glioma cells.138 Similarly, cervical carcinoma HeLa cells undergo apoptosis in response to SIRT2 downregulation.139 SIRT2 promotes bladder cancer cell migration and invasion by targeting cortactin together with HDAC6.140 SIRT2 is upregulated in neuroblastoma cells by N-MYC and in pancreatic cancer cells by c-MYC; and in turn, SIRT2 stabilizes N-MYC and c-MYC protein by downregulating ubiquitin-protein ligase NEDD4 expression.141 In AML cells, SIRT2 and NAD+ salvage enzyme nicotinamide phosphoribosyltransferase are upregulated and involved in the aberrant proliferation and survival of leukemic cells.142 The results from these studies indicate a tumor promotion role of SIRT2.
SIRT3 in cancer
The mitochondrial sirtuin SIRT3 plays crucial roles in metabolism and oxidative stress response, and is considered as a mitochondrial tumor suppressor. SIRT3 levels are reduced in human breast and colon carcinoma,37 HNSCC,143 HCC, and osteosarcoma.144,145 About 20% of all human cancer samples and 40% of breast and ovarian cancer samples contain deletions of SIRT3.37 Mechanistically, SIRT3 may inhibit tumor growth by reducing production of reactive oxygen species (ROS) through regulating electron transport, superoxide dismutase, mitochondrial IDH2, and FOXO3a.66 Notably, SIRT3 deacetylates IDH2 at lysine 413 and activates its activity, leading to increased nicotinamide adenine dinucleotide phosphate levels and an increased ratio of reduced-to-oxidized glutathione in mitochondria, and thus reducing ROS.146,147 SIRT3 promotes antioxidant activity of superoxide dismutase MnSOD via direct deacetylation, and loss of SIRT3 increases acetylation of MnSOD and thereby increases cellular ROS. Increased ROS stabilizes hypoxia-inducible factor (HIF) 1-alpha, resulting in metabolic reprogramming toward glycolysis and thus facilitating tumor development.35,37,148 Recently, it has been shown that SIRT3 deacetylates and destabilizes the proto-oncogene product S-phase kinase-associated protein 2 (Skp2), and inactivation of SIRT3 leads to Skp2 acetylation and thereby increased Skp2 stability and cytoplasmic retention, resulting in enhanced cellular proliferation, migration, and tumorigenesis in vivo.149
However, potential roles of SIRT3 in tumor promotion have also been reported. SIRT3 expression is higher in human lymph-node-positive breast cancer150 and oral squamous cell carcinoma (OSCC).151 Inhibition of SIRT3 in OSCC cells inhibits cell growth and anoikis (a form of programmed cell death) resistance, lowers tumor burden and incidence, and sensitizes OSCC cells to radiation and cisplatin treatments in vitro.151,152 The tumor suppressor p53 is deacetylated by SIRT3, and SIRT3 rescues p53-induced growth arrest in human bladder-tumor-derived EJ-p53 cells.153 SIRT3 deacetylates mitochondrial matrix protein IDH2 to protect cells from oxidative stress; but in cancer, IDH2 activation by SIRT3 may have a pro-survival effect on cancer cells. IDH2 activity has been demonstrated to be a major factor in cancer, and as such, SIRT3 is a potential regulator of IDH2-dependent functions in cancer cell metabolism.147
SIRT4 in cancer
The roles of SIRT4 in cancer have been unclear until two recent studies revealing that it is a potential tumor suppressor.40,154 SIRT4 expression is found to be significantly lower in human bladder, breast, colon, gastric, ovarian, and thyroid carcinomas, relative to normal tissues. In cancer cells, the mammalian target of rapamycin complex 1 pathway promotes glutamine anaplerosis by repressing SIRT4, thus activating GDH. SIRT4 overexpression reduces cell proliferation and transformation, and delays tumor development in a Tsc2−/− (tuberous sclerosis complex 2) mouse embryonic fibroblast xenograft model.154 Consistently, in another study, the loss of SIRT4 led to increased glutamine-dependent cell proliferation and stress-induced genomic instability, resulting in tumorigenic phenotypes.40 SIRT4 knockout mice spontaneously develop lung tumors.40 These studies indicate a crucial role of SIRT4 in linking glutamine metabolism with tumorigenesis.
SIRT6 in cancer
There is a growing body of evidence showing SIRT6 as a tumor suppressor. SIRT6 is downregulated in several human cancers such as pancreatic cancer, colorectal cancer, and HCC, and its expression is associated with clinical outcomes in cancer patients.51,143,155 SIRT6 deacetylates histones H3K9 and H3K56.19,46,156 H3K56 has been shown to be hyperacetylated in breast, liver, skin, thyroid, and colon cancers.157 Loss of SIRT6 leads to transformation of immortalized mouse embryonic fibroblasts, and the transformed SIRT6-deficient cells display increased glycolysis. In a conditional SIRT6 knockout mouse model, SIRT6 deletion increased the number, size, and aggressiveness of tumors.60 In a genetic mouse model specific for liver cancer initiation, SIRT6 represses Survivin expression by reducing histone H3K9 acetylation and NF-κB activation, and the increased SIRT6 expression at the liver cancer initiation stage markedly impairs liver cancer development.158 Overexpression of SIRT6 leads to massive apoptosis in a variety of cancer cell lines but not in non-transformed cells.159
However, there is also some evidence inconsistent with its tumor suppression function. Compared to 17 normal volunteer controls, SIRT6 mRNA levels were significantly increased in 32 chronic lymphocytic leukemia patients, although its relationship with clinical prognosis was not clear.160 SIRT6 protein levels are elevated in paclitaxel- and epirubicin-resistant MCF-7 cells compared to the parental cells. SIRT6 depletion sensitizes cells to both paclitaxel and epirubicin treatment, whereas SIRT6 overexpression leads to increased resistance. Consistently, the stronger immunostaining of SIRT6 in 118 breast cancer patient samples was significantly associated with poorer overall survival.161 In pancreatic cancer cells, SIRT6 enhances Ca(2+) responses by activating Ca(2+) channel transient receptor potential cation channel, subfamily M, member 2 via modulating levels of ADP-ribose, which increases the expression of proinflammatory cytokines/chemokines, such as interleukin 8 and TNF, and promotes cell migration.162
SIRT7 in cancer
SIRT7 mRNA expression is increased in breast and thyroid cancer, compared to their normal counterparts.163 SIRT7 knockdown inhibits proliferation and induces apoptosis in U2OS cells.64 SIRT7 specifically deacetylates histone H3K18, which is necessary for maintaining tumor phenotypes of human cancer cells, including anchorage-independent growth and the escape from contact inhibition.18 Moreover, SIRT7 depletion markedly reduces the growth of human U251 cancer cell xenografts in mice.18 Very recently, both mRNA and protein levels of SIRT7 were shown to be increased in HCC, and knockdown of its expression efficiently suppressed tumor growth in vitro and in vivo.164
However, in HNSCC, SIRT7 mRNA expression level is lower.143 An antiproliferative role of SIRT7 has been demonstrated by using SIRT7 knockout or overexpressing cells, and an inverse correlation with tumorigenic potential has been shown in several murine cell lines.165 In HeLa, Hep3B, MDA-MB-231, and HEK293T cells, a negative transcriptional regulation of HIF1 and HIF2 by SIRT7 was established, suggesting that SIRT7 may function as a tumor suppressor through HIF signaling.166
Potential clinical implications of sirtuins in human malignancies
Apparently, sirtuins have complex roles in human malignancies. Several factors should be taken into consideration regarding some contradictory laboratory observations. 1) Species difference. Mouse studies provide crucial in vivo evidence for tumor suppressor functions of several sirtuins. But the tumorigenesis process in mice is not identical to that in humans, in spite of similarity between the two processes. 2) Genes crucial for inhibiting tumor initiation may not necessarily play the same role in the later stages of cancer development. In fact, opposing roles of tumor promotion and suppression have been observed for many genes including telomerase reverse transcriptase, transforming growth factor beta, and DNA methyltransferases.66 3) Tissue difference. Genes may play different roles in different tissues, which in turn affects their functions in cancers of different tissue origins. In-depth understanding of the roles of individual sirtuins in a particular type of cancer would thus be necessary to better guide a therapeutic strategy with sirtuin modulation. Sirtuin modulators could possibly be used as a single agent for cancer treatment if a specific sirtuin is found to be crucial for a specific type of cancer.
Although much research is stilled needed to understand SIRT2-7 in human malignancies, a consensus theme has now emerged from SIRT1 studies that it is crucial for cancer drug resistance. SIRT1 mediates multiple aspects of cancer drug resistance, by decreasing drug penetration, conferring proliferation and anti-apoptotic survival advantages to cancer cells, facilitating acquired resistance through genetic mutations, promoting survival of cancer stem cells, and changing the tumor microenvironment for resistance, as described above and in a recent review.167 Inhibition of SIRT1 in combination with a cancer-cell-specific agent would greatly benefit cancer treatment. For example, SIRT1 inhibition plus a BCR-ABL kinase inhibitor would likely eradicate CML stem cells and prevent them from acquisition of resistant mutations, which may ultimately lead to a cure of the disease. However, there may be potential side effects of SIRT1 inhibition given that SIRT1 has complex roles in regulating a wide variety of cellular and physiological functions. Intriguingly, developmental defects observed in SIRT1 homozygous knockout mice are typically far more severe in inbred mouse strains than in outbred mouse strains.23 Besides, heterozygous SIRT1 knockout has little impact on mouse development and physiology. Given that human populations are genetically heterogeneous and SIRT1 inhibition by small molecules is unlikely 100% as complete as by homozygous gene knockout, we speculate that SIRT1 inhibition by small molecules might have only mild side effects on humans, in particular, if such drugs would not be intended for life-long use. The side effects could be further reduced in adult patients without risk of developmental impact. Therefore, SIRT1 is a promising target for cancer treatment.
Numerous SIRT1/sirtuin activators and inhibitors have been developed in the past decade.168–171 Sirtuin inhibitors have been explored for cancer treatment. These inhibitors have diverse chemical scaffolds (Figure 2) and can be broadly classified into several categories: 1) Naphthol based inhibitors: for example, Sirtinol, Cambinol, Splitomicin, Salermide, AGK2. 2) Indol/indolinone based inhibitors: for example, EX-527, bisindolylmaleimide, tryptamide. 3) Nicotinamide based inhibitors: for example, carbanicotinamide adenine dinucleotide. 4) Urea and thiourea based inhibitors: for example, Tenovin and Suramin. 5) Polyphenol based inhibitors: for example, biphenylpolyphenol, benzoic acid derivative rottlerin, erbstatin. 6) Peptide based inhibitors: for example, thioacetyl-lysine peptides and H3K9TSu peptide 5. Among these inhibitors, tenovin-6111,121,122,172 and cambinol108,119 have been shown to have encouraging in vivo effect against cancers in animal studies. In a recent clinical trial, the pan-sirtuin inhibitor niacinamide was reported to improve therapeutic outcome when in combination with HDAC inhibition for treatment of human aggressive B-cell lymphomas.173 However, these inhibitors are neither potent enough nor specific enough, and their in vivo effect is also limited.122 In addition, a specific SIRT1 inhibitor EX-527 is in a Phase II clinical trial for treating Huntington’s disease,174 although its effect on cancer remains unclear. Future efforts will be needed towards developing more selective and potent SIRT1 or other individual sirtuin inhibitors.
Figure 2.
Various scaffolds of known sirtuin inhibitors.
Crystal structures of SIRT2,175 SIRT3,176 SIRT5,177 SIRT6,178 and most recently, the SIRT1 catalytic core179 have been determined. These crystal structures are expected to facilitate development of a new generation of selective sirtuin inhibitors. The conserved catalytic core of sirtuins consists of three structural parts: an NAD+ binding domain based on a large Rossmann fold, a small zinc-binding domain, and an extended cleft between the two domains where the substrate binds. Most of current sirtuin inhibitors target either the substrate binding cleft or the NAD+ binding domain according to docking studies.169,170 The crystal structure of SIRT5 bound to suramin shows that suramin interacts with both the nicotinamide binding pocket (C-pocket) within the NAD+ binding domain and the substrate cleft.177 The C-pocket is also targeted by several other inhibitors such as an analog of EX-527 on the SIRT1 catalytic core.179 Thieno[3,2-d] pyrimidine-6-carboxamides, a new class of sirtuin inhibitors that have been most recently developed using encoded library technology, also bind to the C-pocket of SIRT3 but extend through the substrate cleft on the co-crystal structure.180 Despite these progresses, certain difficulties remain, particularly for SIRT1, in that long and unstructured N-terminal and C-terminal sequences may influence catalytic core functions.79,181 A small rigid N-terminal region (amino acids 190–244) appears to mediate the interaction of sirtuin activating compounds with SIRT1.182 However, the large portions of unstructured sequences may play regulatory roles under cellular settings when SIRT1 is in complex with other proteins, and they may influence the functions of small molecule modulators and add some uncertainty to how the drugs act. A combination of biochemical, structural, and cell-based assays is thus necessary for drug development and will help improve the selectivity and specificity of candidate sirtuin inhibitors.
Conclusion
Sirtuins have diverse functions in mammalian physiology and research of these genes is continuing to grow rapidly. More research findings are expected in cancer and other age-related diseases, particularly for those less-understood sirtuin members. Future results will not only shed new insight on their biological functions, but also help devise more rational application of sirtuin inhibitors or activators for treatment of cancer and other diseases. Generation of more potent and individual sirtuin-selective inhibitors will further accelerate the endeavor to improve the management of human malignancies.
Acknowledgments
The authors would like to acknowledge the research support from the National Cancer Institute of the National Institutes of Health under award number R01 CA143421, and the State of California Tobacco Related Disease Research Program (TRDRP) award 20XT-0121 to WenYong Chen. The contents are solely the responsibility of the authors and do not represent the official views of the National Institutes of Health.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Shore D, Squire M, Nasmyth KA. Characterization of two genes required for the position-effect control of yeast mating-type genes. EMBO J. 1984;3(12):2817–2823. doi: 10.1002/j.1460-2075.1984.tb02214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13(19):2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410(6825):227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 4.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101(45):15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herranz D, Serrano M. SIRT1: recent lessons from mouse models. Nat Rev Cancer. 2010;10(12):819–823. doi: 10.1038/nrc2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herranz D, Munoz-Martin M, Canamero M, et al. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat Commun. 2010;1:3. doi: 10.1038/ncomms1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Viswanathan M, Guarente L. Regulation of Caenorhabditis elegans lifespan by sir-2.1 transgenes. Nature. 2011;477(7365):E1–E2. doi: 10.1038/nature10440. [DOI] [PubMed] [Google Scholar]
- 8.Rizki G, Iwata TN, Li J, et al. The evolutionarily conserved longevity determinants HCF-1 and SIR-2.1/SIRT1 collaborate to regulate DAF-16/FOXO. PLoS Genet. 2011;7(9):e1002235. doi: 10.1371/journal.pgen.1002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burnett C, Valentini S, Cabreiro F, et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477(7365):482–485. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273(2):793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 11.Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene:Sir2-like proteins(sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun. 1999;260(1):273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 12.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403(6771):795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 13.Landry J, Sutton A, Tafrov ST, et al. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci U S A. 2000;97(11):5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11(2):437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 15.Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci U S A. 2000;97(26):14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haigis MC, Mostoslavsky R, Haigis KM, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126(5):941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 17.Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280(22):21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- 18.Barber MF, Michishita-Kioi E, Xi Y, et al. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature. 2012;487(7405):114–118. doi: 10.1038/nature11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michishita E, McCord RA, Boxer LD, et al. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle. 2009;8(16):2664–2666. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du J, Zhou Y, Su X, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334(6057):806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16(10):4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem. 2007;282(9):6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 23.McBurney MW, Yang X, Jardine K, et al. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol. 2003;23(1):38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng HL, Mostoslavsky R, Saito S, et al. Developmental defects and p53 hyperacetylation in Sir2 homolog(SIRT1)-deficient mice. Proc Natl Acad Sci U S A. 2003;100(19):10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sequeira J, Boily G, Bazinet S, et al. sirt1-null mice develop an autoimmune-like condition. Exp Cell Res. 2008;314(16):3069–3074. doi: 10.1016/j.yexcr.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Lee SM, Shannon S, et al. The type III histone deacetylase Sirt1 is essential for maintenance of T cell tolerance in mice. J Clin Invest. 2009;119(10):3048–3058. doi: 10.1172/JCI38902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.North BJ, Verdin E. Interphase nucleo-cytoplasmic shuttling and localization of SIRT2 during mitosis. PLoS One. 2007;2(8):e784. doi: 10.1371/journal.pone.0000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaquero A, Scher MB, Lee DH, et al. SirT2 is a histone deacetylase with preference for histone H4Lys 16 during mitosis. Genes Dev. 2006;20(10):1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim HS, Vassilopoulos A, Wang RH, et al. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell. 2011;20(4):487–499. doi: 10.1016/j.ccr.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Onyango P, Celic I, McCaffery JM, Boeke JD, Feinberg AP. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci U S A. 2002;99(21):13653–13658. doi: 10.1073/pnas.222538099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol. 2002;158(4):647–657. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura Y, Ogura M, Tanaka D, Inagaki N. Localization of mouse mitochondrial SIRT proteins:shift of SIRT3 to nucleus by co-expression with SIRT5. Biochem Biophys Res Commun. 2008;366(1):174–179. doi: 10.1016/j.bbrc.2007.11.122. [DOI] [PubMed] [Google Scholar]
- 33.Scher MB, Vaquero A, Reinberg D. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev. 2007;21(8):920–928. doi: 10.1101/gad.1527307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lombard DB, Alt FW, Cheng HL, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27(24):8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim HS, Patel K, Muldoon-Jacobs K, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17(1):41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirschey MD, Shimazu T, Jing E, et al. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol Cell. 2011;44(2):177–190. doi: 10.1016/j.molcel.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finley LW, Carracedo A, Lee J, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell. 2011;19(3):416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nasrin N, Wu X, Fortier E, et al. SIRT4 regulates fatty acid oxidation and mitochondrial gene expression in liver and muscle cells. J Biol Chem. 2010;285(42):31995–32002. doi: 10.1074/jbc.M110.124164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahuja N, Schwer B, Carobbio S, et al. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J Biol Chem. 2007;282(46):33583–33592. doi: 10.1074/jbc.M705488200. [DOI] [PubMed] [Google Scholar]
- 40.Jeong SM, Xiao C, Finley LW, et al. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell. 2013;23(4):450–463. doi: 10.1016/j.ccr.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang H, Yang T, Baur JA, et al. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130(6):1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137(3):560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peng C, Lu Z, Xie Z, et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics. 2011;10(12):M111.012658. doi: 10.1074/mcp.M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park J, Chen Y, Tishkoff DX, et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell. 2013;50(6):919–930. doi: 10.1016/j.molcel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsushita N, Yonashiro R, Ogata Y, et al. Distinct regulation of mitochondrial localization and stability of two human Sirt5 isoforms. Genes Cells. 2011;16(2):190–202. doi: 10.1111/j.1365-2443.2010.01475.x. [DOI] [PubMed] [Google Scholar]
- 46.Michishita E, McCord RA, Berber E, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452(7186):492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang H, Khan S, Wang Y, et al. SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature. 2013;496(7443):110–113. doi: 10.1038/nature12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mao Z, Tian X, Van Meter M, et al. Sirtuin 6 (SIRT6) rescues the decline of homologous recombination repair during replicative senescence. Proc Natl Acad Sci U S A. 2012;109(29):11800–11805. doi: 10.1073/pnas.1200583109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim HS, Xiao C, Wang RH, et al. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. 2010;12(3):224–236. doi: 10.1016/j.cmet.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawahara TL, Michishita E, Adler AS, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136(1):62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhong L, D’Urso A, Toiber D, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140(2):280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCord RA, Michishita E, Hong T, et al. SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging (Albany NY) 2009;1(1):109–121. doi: 10.18632/aging.100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaidi A, Weinert BT, Choudhary C, Jackson SP. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329(5997):1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Xiao C, Kim HS, Lahusen T, et al. SIRT6 deficiency results in severe hypoglycemia by enhancing both basal and insulin-stimulated glucose uptake in mice. J Biol Chem. 2010;285(47):36776–36784. doi: 10.1074/jbc.M110.168039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwer B, Schumacher B, Lombard DB, et al. Neural sirtuin 6 (Sirt6) ablation attenuates somatic growth and causes obesity. Proc Natl Acad Sci U S A. 2010;107(50):21790–21794. doi: 10.1073/pnas.1016306107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mao Z, Hine C, Tian X, et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332(6036):1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tennen RI, Bua DJ, Wright WE, Chua KF. SIRT6 is required for maintenance of telomere position effect in human cells. Nat Commun. 2011;2:433. doi: 10.1038/ncomms1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sundaresan NR, Vasudevan P, Zhong L, et al. The sirtuin SIRT6 blocks IGF-Akt signaling and development of cardiac hypertrophy by targeting c-Jun. Nat Med. 2012;18(11):1643–1650. doi: 10.1038/nm.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dominy JE, Jr, Lee Y, Jedrychowski MP, et al. The deacetylase Sirt6 activates the acetyltransferase GCN5 and suppresses hepatic gluconeogenesis. Mol Cell. 2012;48(6):900–913. doi: 10.1016/j.molcel.2012.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sebastian C, Zwaans BM, Silberman DM, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012;151(6):1185–1199. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mostoslavsky R, Chua KF, Lombard DB, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124(2):315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 62.Kanfi Y, Naiman S, Amir G, et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483(7388):218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 63.Grob A, Roussel P, Wright JE, et al. Involvement of SIRT7 in resumption of rDNA transcription at the exit from mitosis. J Cell Sci. 2009;122(Pt 4):489–498. doi: 10.1242/jcs.042382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006;20(9):1075–1080. doi: 10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vakhrusheva O, Smolka C, Gajawada P, et al. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res. 2008;102(6):703–710. doi: 10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]
- 66.Roth M, Chen WY. Sorting out functions of sirtuins in cancer. Oncogene. 2013. [Epub] [DOI] [PMC free article] [PubMed]
- 67.Hiraike H, Wada-Hiraike O, Nakagawa S, et al. Expression of DBC1 is associated with nuclear grade and HER2 expression in breast cancer. Exp Ther Med. 2011;2(6):1105–1109. doi: 10.3892/etm.2011.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee H, Kim KR, Noh SJ, et al. Expression of DBC1 and SIRT1 is associated with poor prognosis for breast carcinoma. Hum Pathol. 2011;42(2):204–213. doi: 10.1016/j.humpath.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 69.Wu M, Wei W, Xiao X, et al. Expression of SIRT1 is associated with lymph node metastasis and poor prognosis in both operable triple-negative and non-triple-negative breast cancer. Med Oncol. 2012;29(5):3240–3249. doi: 10.1007/s12032-012-0260-6. [DOI] [PubMed] [Google Scholar]
- 70.Chen WY, Wang DH, Yen RC, et al. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123(3):437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 71.Eades G, Yao Y, Yang M, et al. miR-200a regulates SIRT1 expression and epithelial to mesenchymal transition (EMT)-like transformation in mammary epithelial cells. J Biol Chem. 2011;286(29):25992–26002. doi: 10.1074/jbc.M111.229401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Holloway KR, Barbieri A, Malyarchuk S, et al. SIRT1 positively regulates breast cancer associated human aromatase(CYP19A1) expression. Mol Endocrinol. 2013;27(3):480–490. doi: 10.1210/me.2012-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Y, Zhang M, Dong H, et al. Deacetylation of cortactin by SIRT1 promotes cell migration. Oncogene. 2009;28(3):445–460. doi: 10.1038/onc.2008.388. [DOI] [PubMed] [Google Scholar]
- 74.Choi HK, Cho KB, Phuong NT, et al. SIRT1-Mediated FoxO1 Deacetylation Is Essential for Multidrug Resistance-Associated Protein 2 Expression in Tamoxifen-Resistant Breast Cancer Cells. Mol Pharm. 2013 doi: 10.1021/mp400287p. [Epub] [DOI] [PubMed] [Google Scholar]
- 75.Suzuki K, Hayashi R, Ichikawa T, et al. SRT1720, a SIRT1 activator, promotes tumor cell migration, and lung metastasis of breast cancer in mice. Oncol Rep. 2012;27(6):1726–1732. doi: 10.3892/or.2012.1750. [DOI] [PubMed] [Google Scholar]
- 76.Wang F, Chan CH, Chen K, Guan X, Lin HK, Tong Q. Deacetylation of FOXO3 by SIRT1 or SIRT2 leads to Skp2-mediated FOXO3 ubiquitination and degradation. Oncogene. 2012;31(12):1546–1557. doi: 10.1038/onc.2011.347. [DOI] [PubMed] [Google Scholar]
- 77.Huffman DM, Grizzle WE, Bamman MM, et al. SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res. 2007;67(14):6612–6618. doi: 10.1158/0008-5472.CAN-07-0085. [DOI] [PubMed] [Google Scholar]
- 78.Kojima K, Ohhashi R, Fujita Y, et al. A role for SIRT1 in cell growth and chemoresistance in prostate cancer PC3 and DU145 cells. Biochem Biophys Res Commun. 2008;373(3):423–428. doi: 10.1016/j.bbrc.2008.06.045. [DOI] [PubMed] [Google Scholar]
- 79.Kang H, Suh JY, Jung YS, Jung JW, Kim MK, Chung JH. Peptide switch is essential for Sirt1 deacetylase activity. Mol Cell. 2011;44(2):203–213. doi: 10.1016/j.molcel.2011.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang B, Hasan MK, Alvarado E, Yuan H, Wu H, Chen WY. NAMPT overexpression in prostate cancer and its contribution to tumor cell survival and stress response. Oncogene. 2011;30(8):907–921. doi: 10.1038/onc.2010.468. [DOI] [PubMed] [Google Scholar]
- 81.Lovaas JD, Zhu L, Chiao CY, Byles V, Faller DV, Dai Y. SIRT1 enhances matrix metalloproteinase-2 expression and tumor cell invasion in prostate cancer cells. Prostate. 2013;73(5):522–530. doi: 10.1002/pros.22592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Byles V, Zhu L, Lovaas JD, et al. SIRT1 induces EMT by cooperating with EMT transcription factors and enhances prostate cancer cell migration and metastasis. Oncogene. 2012;31(43):4619–4629. doi: 10.1038/onc.2011.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nakane K, Fujita Y, Terazawa R, et al. Inhibition of cortactin and SIRT1 expression attenuates migration and invasion of prostate cancer DU145 cells. Int J Urol. 2012;19(1):71–79. doi: 10.1111/j.1442-2042.2011.02888.x. [DOI] [PubMed] [Google Scholar]
- 84.Herranz D, Maraver A, Canamero M, et al. SIRT1 promotes thyroid carcinogenesis driven by PTEN deficiency. Oncogene. 2013;32(34):4052–4056. doi: 10.1038/onc.2012.407. [DOI] [PubMed] [Google Scholar]
- 85.Noh SJ, Baek HA, Park HS, et al. Expression of SIRT1 and cortactin is associated with progression of non-small cell lung cancer. Pathol Res Pract. 2013;209(6):365–370. doi: 10.1016/j.prp.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 86.Tseng RC, Lee CC, Hsu HS, Tzao C, Wang YC. Distinct HIC1-SIRT1-p53 loop deregulation in lung squamous carcinoma and adenocarcinoma patients. Neoplasia. 2009;11(8):763–770. doi: 10.1593/neo.09470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xie M, Liu M, He CS. SIRT1 regulates endothelial Notch signaling in lung cancer. PLoS One. 2012;7(9):e45331. doi: 10.1371/journal.pone.0045331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu G, Su L, Hao X, et al. Salermide up-regulates death receptor 5 expression through the ATF4-ATF3-CHOP axis and leads to apoptosis in human cancer cells. J Cell Mol Med. 2012;16(7):1618–1628. doi: 10.1111/j.1582-4934.2011.01401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Menssen A, Hydbring P, Kapelle K, et al. The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop. Proc Natl Acad Sci U S A. 2012;109(4):E187–E196. doi: 10.1073/pnas.1105304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kriegl L, Vieth M, Kirchner T, Menssen A. Up-regulation of c-MYC and SIRT1 expression correlates with malignant transformation in the serrated route to colorectal cancer. Oncotarget. 2012;3(10):1182–1193. doi: 10.18632/oncotarget.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nosho K, Shima K, Irahara N, et al. SIRT1 histone deacetylase expression is associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Mod Pathol. 2009;22(7):922–932. doi: 10.1038/modpathol.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Akao Y, Noguchi S, Iio A, Kojima K, Takagi T, Naoe T. Dysregulation of microRNA-34a expression causes drug-resistance to 5-FU in human colon cancer DLD-1 cells. Cancer Lett. 2011;300(2):197–204. doi: 10.1016/j.canlet.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 93.Feng AN, Zhang LH, Fan XS, et al. Expression of SIRT1 in gastric cardiac cancer and its clinicopathologic significance. Int J Surg Pathol. 2011;19(6):743–750. doi: 10.1177/1066896911412181. [DOI] [PubMed] [Google Scholar]
- 94.Cha EJ, Noh SJ, Kwon KS, et al. Expression of DBC1 and SIRT1 is associated with poor prognosis of gastric carcinoma. Clin Cancer Res. 2009;15(13):4453–4459. doi: 10.1158/1078-0432.CCR-08-3329. [DOI] [PubMed] [Google Scholar]
- 95.Zhu H, Xia L, Zhang Y, et al. Activating transcription factor 4 confers a multidrug resistance phenotype to gastric cancer cells through transactivation of SIRT1 expression. PLoS One. 2012;7(2):e31431. doi: 10.1371/journal.pone.0031431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen HC, Jeng YM, Yuan RH, Hsu HC, Chen YL. SIRT1 promotes tumorigenesis and resistance to chemotherapy in hepatocellular carcinoma and its expression predicts poor prognosis. Ann Surg Oncol. 2012;19(6):2011–2019. doi: 10.1245/s10434-011-2159-4. [DOI] [PubMed] [Google Scholar]
- 97.Choi HN, Bae JS, Jamiyandorj U, et al. Expression and role of SIRT1 in hepatocellular carcinoma. Oncol Rep. 2011;26(2):503–510. doi: 10.3892/or.2011.1301. [DOI] [PubMed] [Google Scholar]
- 98.Portmann S, Fahrner R, Lechleiter A, et al. Antitumor effect of SIRT1 inhibition in human HCC tumor models in vitro and in vivo. Mol Cancer Ther. 2013;12(4):499–508. doi: 10.1158/1535-7163.MCT-12-0700. [DOI] [PubMed] [Google Scholar]
- 99.Chen J, Zhang B, Wong N, et al. Sirtuin 1 is upregulated in a subset of hepatocellular carcinomas where it is essential for telomere maintenance and tumor cell growth. Cancer Res. 2011;71(12):4138–4149. doi: 10.1158/0008-5472.CAN-10-4274. [DOI] [PubMed] [Google Scholar]
- 100.Wang H, Liu H, Chen K, et al. SIRT1 promotes tumorigenesis of hepatocellular carcinoma through PI3K/PTEN/AKT signaling. Oncol Rep. 2012;28(1):311–318. doi: 10.3892/or.2012.1788. [DOI] [PubMed] [Google Scholar]
- 101.Jang KY, Noh SJ, Lehwald N, et al. SIRT1 and c-Myc promote liver tumor cell survival and predict poor survival of human hepatocellular carcinomas. PLoS One. 2012;7(9):e45119. doi: 10.1371/journal.pone.0045119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lou W, Chen Q, Ma L, et al. Oncolytic adenovirus co-expressing miRNA-34a and IL-24 induces superior antitumor activity in experimental tumor model. J Mol Med (Berl) 2013;91(6):715–725. doi: 10.1007/s00109-012-0985-x. [DOI] [PubMed] [Google Scholar]
- 103.Bae HJ, Noh JH, Kim JK, et al. MicroRNA-29c functions as a tumor suppressor by direct targeting oncogenic SIRT1 in hepatocellular carcinoma. Oncogene. 2013. [Epub] [DOI] [PubMed]
- 104.Zhao G, Cui J, Zhang JG, et al. SIRT1 RNAi knockdown induces apoptosis and senescence, inhibits invasion and enhances chemosensitivity in pancreatic cancer cells. Gene Ther. 2011;18(9):920–928. doi: 10.1038/gt.2011.81. [DOI] [PubMed] [Google Scholar]
- 105.Zhang JG, Zhao G, Qin Q, et al. Nicotinamide prohibits proliferation and enhances chemosensitivity of pancreatic cancer cells through deregulating SIRT1 and Ras/Akt pathways. Pancreatology. 2013;13(2):140–146. doi: 10.1016/j.pan.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 106.Wauters E, Sanchez-Arevalo Lobo VJ, Pinho AV, et al. Sirtuin-1 regulates acinar-to-ductal metaplasia and supports cancer cell viability in pancreatic cancer. Cancer Res. 2013;73(7):2357–2367. doi: 10.1158/0008-5472.CAN-12-3359. [DOI] [PubMed] [Google Scholar]
- 107.Xu J, Zhu W, Xu W, et al. Up-regulation of MBD1 promotes pancreatic cancer cell epithelial-mesenchymal transition and invasion by epigenetic down-regulation of E-cadherin. Curr Mol Med. 2013;13(3):387–400. [PubMed] [Google Scholar]
- 108.Jang KY, Kim KS, Hwang SH, et al. Expression and prognostic significance of SIRT1 in ovarian epithelial tumours. Pathology. 2009;41(4):366–371. doi: 10.1080/00313020902884451. [DOI] [PubMed] [Google Scholar]
- 109.Benayoun BA, Georges AB, L’Hote D, et al. Transcription factor FOXL2 protects granulosa cells from stress and delays cell cycle:role of its regulation by the SIRT1 deacetylase. Hum Mol Genet. 2011;20(9):1673–1686. doi: 10.1093/hmg/ddr042. [DOI] [PubMed] [Google Scholar]
- 110.Allison SJ, Jiang M, Milner J. Oncogenic viral protein HPV E7 upregulates the SIRT1 longevity protein in human cervical cancer cells. Aging (Albany NY) 2009;1(3):316–327. doi: 10.18632/aging.100028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Marshall GM, Liu PY, Gherardi S, et al. SIRT1 promotes N-Myc oncogenesis through a positive feedback loop involving the effects of MKP3 and ERK on N-Myc protein stability. PLoS Genet. 2011;7(6):e1002135. doi: 10.1371/journal.pgen.1002135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kim MJ, Ahn K, Park SH, et al. SIRT1 regulates tyrosine hydroxylase expression and differentiation of neuroblastoma cells via FOXO3a. FEBS Lett. 2009;583(7):1183–1188. doi: 10.1016/j.febslet.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 113.Chang CJ, Hsu CC, Yung MC, et al. Enhanced radiosensitivity and radiation-induced apoptosis in glioma CD133-positive cells by knockdown of SirT1 expression. Biochem Biophys Res Commun. 2009;380(2):236–242. doi: 10.1016/j.bbrc.2009.01.040. [DOI] [PubMed] [Google Scholar]
- 114.Dixit D, Sharma V, Ghosh S, Mehta VS, Sen E. Inhibition of Casein kinase-2 induces p53-dependent cell cycle arrest and sensitizes glioblastoma cells to tumor necrosis factor(TNFalpha)-induced apoptosis through SIRT1 inhibition. Cell Death Dis. 2012;3:e271. doi: 10.1038/cddis.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ma JX, Li H, Chen XM, et al. Expression patterns and potential roles of SIRT1 in human medulloblastoma cells in vivo and in vitro. Neuropathology. 2013;33(1):7–16. doi: 10.1111/j.1440-1789.2012.01318.x. [DOI] [PubMed] [Google Scholar]
- 116.Kozako T, Aikawa A, Shoji T, et al. High expression of the longevity gene product SIRT1 and apoptosis induction by sirtinol in adult T-cell leukemia cells. Int J Cancer. 2012;131(9):2044–2055. doi: 10.1002/ijc.27481. [DOI] [PubMed] [Google Scholar]
- 117.Jang KY, Hwang SH, Kwon KS, et al. SIRT1 expression is associated with poor prognosis of diffuse large B-cell lymphoma. Am J Surg Pathol. 2008;32(10):1523–1531. doi: 10.1097/PAS.0b013e31816b6478. [DOI] [PubMed] [Google Scholar]
- 118.Bradbury CA, Khanim FL, Hayden R, et al. Histone deacetylases in acute myeloid leukaemia show a distinctive pattern of expression that changes selectively in response to deacetylase inhibitors. Leukemia. 2005;19(10):1751–1759. doi: 10.1038/sj.leu.2403910. [DOI] [PubMed] [Google Scholar]
- 119.Heltweg B, Gatbonton T, Schuler AD, et al. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 2006;66(8):4368–4377. doi: 10.1158/0008-5472.CAN-05-3617. [DOI] [PubMed] [Google Scholar]
- 120.Cea M, Soncini D, Fruscione F, et al. Synergistic interactions between HDAC and sirtuin inhibitors in human leukemia cells. PLoS One. 2011;6(7):e22739. doi: 10.1371/journal.pone.0022739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li L, Wang L, Li L, et al. Activation of p53 by SIRT1 inhibition enhances elimination of CML leukemia stem cells in combination with imatinib. Cancer Cell. 2012;21(2):266–281. doi: 10.1016/j.ccr.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yuan H, Wang Z, Li L, et al. Activation of stress response gene SIRT1 by BCR-ABL promotes leukemogenesis. Blood. 2012;119(8):1904–1914. doi: 10.1182/blood-2011-06-361691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang Z, Yuan H, Roth M, Stark JM, Bhatia R, Chen WY. SIRT1 deacetylase promotes acquisition of genetic mutations for drug resistance in CML cells. Oncogene. 2013;32(5):589–598. doi: 10.1038/onc.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dickson BC, Riddle ND, Brooks JS, Pasha TL, Zhang PJ. Sirtuin 1 (SIRT1): a potential immunohistochemical marker and therapeutic target in soft tissue neoplasms with myoid differentiation. Hum Pathol. 2013;44(6):1125–1130. doi: 10.1016/j.humpath.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 125.Firestein R, Blander G, Michan S, et al. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS One. 2008;3(4):e2020. doi: 10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang RH, Sengupta K, Li C, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14(4):312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Noguchi A, Li X, Kubota A, et al. SIRT1 expression is associated with good prognosis for head and neck squamous cell carcinoma patients. Oral Surg Oral Med Oral Pathol Oral Radiol. 2013;115(3):385–392. doi: 10.1016/j.oooo.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 128.Kabra N, Li Z, Chen L, et al. SirT1 is an inhibitor of proliferation and tumor formation in colon cancer. J Biol Chem. 2009;284(27):18210–18217. doi: 10.1074/jbc.M109.000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jang SH, Min KW, Paik SS, Jang KS. Loss of SIRT1 histone deacetylase expression associates with tumour progression in colorectal adenocarcinoma. J Clin Pathol. 2012;65(8):735–739. doi: 10.1136/jclinpath-2012-200685. [DOI] [PubMed] [Google Scholar]
- 130.Lim JH, Lee YM, Chun YS, Chen J, Kim JE, Park JW. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1alpha. Mol Cell. 2010;38(6):864–878. doi: 10.1016/j.molcel.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 131.Simic P, Williams EO, Bell EL, Gong JJ, Bonkowski M, Guarente L. SIRT1 suppresses the epithelial-to-mesenchymal transition in cancer metastasis and organ fibrosis. Cell Rep. 2013;3(4):1175–1186. doi: 10.1016/j.celrep.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yuan J, Minter-Dykhouse K, Lou Z. A c-Myc-SIRT1 feedback loop regulates cell growth and transformation. J Cell Biol. 2009;185(2):203–211. doi: 10.1083/jcb.200809167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cho IR, Koh SS, Malilas W, et al. SIRT1 inhibits proliferation of pancreatic cancer cells expressing pancreatic adenocarcinoma up-regulated factor (PAUF), a novel oncogene, by suppression of beta-catenin. Biochem Biophys Res Commun. 2012;423(2):270–275. doi: 10.1016/j.bbrc.2012.05.107. [DOI] [PubMed] [Google Scholar]
- 134.Srisuttee R, Koh SS, Kim SJ, et al. Hepatitis B virus X (HBX) protein upregulates beta-catenin in a human hepatic cell line by sequestering SIRT1 deacetylase. Oncol Rep. 2012;28(1):276–282. doi: 10.3892/or.2012.1798. [DOI] [PubMed] [Google Scholar]
- 135.Srisuttee R, Koh SS, Malilas W, et al. SIRT1 sensitizes hepatocellular carcinoma cells expressing hepatitis B virus X protein to oxidative stress-induced apoptosis. Biochem Biophys Res Commun. 2012;429(1–2):45–50. doi: 10.1016/j.bbrc.2012.10.102. [DOI] [PubMed] [Google Scholar]
- 136.Hiratsuka M, Inoue T, Toda T, et al. Proteomics-based identification of differentially expressed genes in human gliomas:down-regulation of SIRT2 gene. Biochem Biophys Res Commun. 2003;309(3):558–566. doi: 10.1016/j.bbrc.2003.08.029. [DOI] [PubMed] [Google Scholar]
- 137.Peters CJ, Rees JR, Hardwick RH, et al. Oesophageal Cancer Clinical and Molecular Stratification (OCCAMS) Study Group A 4-gene signature predicts survival of patients with resected adenocarcinoma of the esophagus, junction, and gastric cardia. Gastroenterology. 2010;139(6):1995–2004. doi: 10.1053/j.gastro.2010.05.080. e15. [DOI] [PubMed] [Google Scholar]
- 138.He X, Nie H, Hong Y, Sheng C, Xia W, Ying W. SIRT2 activity is required for the survival of C6 glioma cells. Biochem Biophys Res Commun. 2012;417(1):468–472. doi: 10.1016/j.bbrc.2011.11.141. [DOI] [PubMed] [Google Scholar]
- 139.Li Y, Matsumori H, Nakayama Y, et al. SIRT2 down-regulation in HeLa can induce p53 accumulation via p38 MAPK activation-dependent p300 decrease, eventually leading to apoptosis. Genes Cells. 2011;16(1):34–45. doi: 10.1111/j.1365-2443.2010.01460.x. [DOI] [PubMed] [Google Scholar]
- 140.Zuo Q, Wu W, Li X, Zhao L, Chen W. HDAC6 and SIRT2 promote bladder cancer cell migration and invasion by targeting cortactin. Oncol Rep. 2012;27(3):819–824. doi: 10.3892/or.2011.1553. [DOI] [PubMed] [Google Scholar]
- 141.Liu PY, Xu N, Malyukova A, et al. The histone deacetylase SIRT2 stabilizes Myc oncoproteins. Cell Death Differ. 2013;20(3):503–514. doi: 10.1038/cdd.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Dan L, Klimenkova O, Klimiankou M, et al. The role of sirtuin 2 activation by nicotinamide phosphoribosyltransferase in the aberrant proliferation and survival of myeloid leukemia cells. Haematologica. 2012;97(4):551–559. doi: 10.3324/haematol.2011.055236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lai CC, Lin PM, Lin SF, et al. Altered expression of SIRT gene family in head and neck squamous cell carcinoma. Tumour Biol. 2013;34(3):1847–1854. doi: 10.1007/s13277-013-0726-y. [DOI] [PubMed] [Google Scholar]
- 144.Zhang CZ, Liu L, Cai M, et al. Low SIRT3 expression correlates with poor differentiation and unfavorable prognosis in primary hepatocellular carcinoma. PLoS One. 2012;7(12):e51703. doi: 10.1371/journal.pone.0051703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhang YY, Zhou LM. Sirt3 inhibits hepatocellular carcinoma cell growth through reducing Mdm2-mediated p53 degradation. Biochem Biophys Res Commun. 2012;423(1):26–31. doi: 10.1016/j.bbrc.2012.05.053. [DOI] [PubMed] [Google Scholar]
- 146.Someya S, Yu W, Hallows WC, et al. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143(5):802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yu W, Dittenhafer-Reed KE, Denu JM. SIRT3 protein deacetylates isocitrate dehydrogenase 2 (IDH2) and regulates mitochondrial redox status. J Biol Chem. 2012;287(17):14078–14086. doi: 10.1074/jbc.M112.355206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bell EL, Emerling BM, Ricoult SJ, Guarente L. SirT3 suppresses hypoxia inducible factor 1alpha and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011;30(26):2986–2996. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Inuzuka H, Gao D, Finley LW, et al. Acetylation-dependent regulation of Skp2 function. Cell. 2012;150(1):179–193. doi: 10.1016/j.cell.2012.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ashraf N, Zino S, Macintyre A, et al. Altered sirtuin expression is associated with node-positive breast cancer. Br J Cancer. 2006;95(8):1056–1061. doi: 10.1038/sj.bjc.6603384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Alhazzazi TY, Kamarajan P, Joo N, et al. Sirtuin-3 (SIRT3), a novel potential therapeutic target for oral cancer. Cancer. 2011;117(8):1670–1678. doi: 10.1002/cncr.25676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kamarajan P, Alhazzazi TY, Danciu T, D’silva NJ, Verdin E, Kapila YL. Receptor-interacting protein (RIP) and Sirtuin-3 (SIRT3) are on opposite sides of anoikis and tumorigenesis. Cancer. 2012;118(23):5800–5810. doi: 10.1002/cncr.27655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li S, Banck M, Mujtaba S, Zhou MM, Sugrue MM, Walsh MJ. p53-induced growth arrest is regulated by the mitochondrial SirT3 deacetylase. PLoS One. 2010;5(5):e10486. doi: 10.1371/journal.pone.0010486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Csibi A, Fendt SM, Li C, et al. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell. 2013;153(4):840–854. doi: 10.1016/j.cell.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Marquardt JU, Fischer K, Baus K, et al. SIRT6 dependent genetic and epigenetic alterations are associated with poor clinical outcome in HCC patients. Hepatology. 2013;58(3):1054–1064. doi: 10.1002/hep.26413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yang B, Zwaans BM, Eckersdorff M, Lombard DB. The sirtuin SIRT6 deacetylates H3K56Ac in vivo to promote genomic stability. Cell Cycle. 2009;8(16):2662–2663. doi: 10.4161/cc.8.16.9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459(7243):113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Min L, Ji Y, Bakiri L, et al. Liver cancer initiation is controlled by AP-1 through SIRT6-dependent inhibition of survivin. Nat Cell Biol. 2012;14(11):1203–1211. doi: 10.1038/ncb2590. [DOI] [PubMed] [Google Scholar]
- 159.Van Meter M, Mao Z, Gorbunova V, Seluanov A. SIRT6 overexpression induces massive apoptosis in cancer cells but not in normal cells. Cell Cycle. 2011;10(18):3153–3158. doi: 10.4161/cc.10.18.17435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang JC, Kafeel MI, Avezbakiyev B, et al. Histone deacetylase in chronic lymphocytic leukemia. Oncology. 2011;81(5–6):325–329. doi: 10.1159/000334577. [DOI] [PubMed] [Google Scholar]
- 161.Khongkow M, Olmos Y, Gong C, et al. SIRT6 modulates paclitaxel and epirubicin resistance and survival in breast cancer. Carcinogenesis. 2013;34(7):1476–1486. doi: 10.1093/carcin/bgt098. [DOI] [PubMed] [Google Scholar]
- 162.Bauer I, Grozio A, Lasiglie D, et al. The NAD+-dependent histone deacetylase SIRT6 promotes cytokine production and migration in pancreatic cancer cells by regulating Ca2+ responses. J Biol Chem. 2012;287(49):40924–40937. doi: 10.1074/jbc.M112.405837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.De Nigris F, Cerutti J, Morelli C, et al. Isolation of a SIR-like gene, SIR-T8, that is overexpressed in thyroid carcinoma cell lines and tissues. Br J Cancer. 2002;87(12):1479. doi: 10.1038/sj.bjc.6600636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kim JK, Noh JH, Jung KH, et al. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology. 2013;57(3):1055–1067. doi: 10.1002/hep.26101. [DOI] [PubMed] [Google Scholar]
- 165.Vakhrusheva O, Braeuer D, Liu Z, Braun T, Bober E. Sirt7-dependent inhibition of cell growth and proliferation might be instrumental to mediate tissue integrity during aging. J Physiol Pharmacol. 2008;59(Suppl 9):201–212. [PubMed] [Google Scholar]
- 166.Hubbi ME, Hu H, Kshitiz NF, Gilkes DM, Semenza GL. Sirtuin-7 inhibits the activity of hypoxia-inducible factors. J Biol Chem. 2013;288(29):20768–20775. doi: 10.1074/jbc.M113.476903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang Z, Chen W. Emerging roles of SIRT1 in cancer drug resistance. Genes and Cancer. 2013;4(3–4):82–90. doi: 10.1177/1947601912473826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lavu S, Boss O, Elliott PJ, Lambert PD. Sirtuins – novel therapeutic targets to treat age-associated diseases. Nat Rev Drug Discov. 2008;7(10):841–853. doi: 10.1038/nrd2665. [DOI] [PubMed] [Google Scholar]
- 169.Cen Y. Sirtuins inhibitors:the approach to affinity and selectivity. Biochim Biophys Acta. 2010;1804(8):1635–1644. doi: 10.1016/j.bbapap.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 170.Bruzzone S, Parenti MD, Grozio A, et al. Rejuvenating sirtuins: the rise of a new family of cancer drug targets. Curr Pharm Des. 2013;19(4):614–623. doi: 10.2174/138161213804581954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Mahajan SS, Leko V, Simon JA, Bedalov A. Sirtuin modulators. Handb Exp Pharmacol. 2011;206:241–255. doi: 10.1007/978-3-642-21631-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lain S, Hollick JJ, Campbell J, et al. Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator. Cancer Cell. 2008;13(5):454–463. doi: 10.1016/j.ccr.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Amengual JE, Clark-Garvey S, Kalac M, et al. Sirtuin and pan-class I/II deacetylase (DAC) inhibition is synergistic in preclinical models and clinical studies of lymphoma. Blood. 2013;120:2725. doi: 10.1182/blood-2013-02-485441. [DOI] [PubMed] [Google Scholar]
- 174.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families:a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11(5):384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 175.Finnin MS, Donigian JR, Pavletich NP. Structure of the histone deacetylase SIRT2. Nat Struct Biol. 2001;8(7):621–625. doi: 10.1038/89668. [DOI] [PubMed] [Google Scholar]
- 176.Jin L, Wei W, Jiang Y, et al. Crystal structures of human SIRT3 displaying substrate-induced conformational changes. J Biol Chem. 2009;284(36):24394–24405. doi: 10.1074/jbc.M109.014928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Schuetz A, Min J, Antoshenko T, et al. Structural basis of inhibition of the human NAD+-dependent deacetylase SIRT5 by suramin. Structure. 2007;15(3):377–389. doi: 10.1016/j.str.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 178.Pan PW, Feldman JL, Devries MK, Dong A, Edwards AM, Denu JM. Structure and biochemical functions of SIRT6. J Biol Chem. 2011;286(16):14575–14587. doi: 10.1074/jbc.M111.218990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zhao X, Allison D, Condon B, et al. The 2.5 Å crystal structure of the SIRT1 catalytic domain bound to nicotinamide adenine dinucleotide (NAD+) and an indole (EX527 analogue) reveals a novel mechanism of histone deacetylase inhibition. J Med Chem. 2013;56(3):963–969. doi: 10.1021/jm301431y. [DOI] [PubMed] [Google Scholar]
- 180.Disch JS, Evindar G, Chiu CH, et al. Discovery of thieno[3,2-d] pyrimidine-6-carboxamides as potent inhibitors of SIRT1, SIRT2, and SIRT3. J Med Chem. 2013;56(9):3666–3679. doi: 10.1021/jm400204k. [DOI] [PubMed] [Google Scholar]
- 181.Pan M, Yuan H, Brent M, Ding EC, Marmorstein R. SIRT1 contains N- and C-terminal regions that potentiate deacetylase activity. J Biol Chem. 2012;287(4):2468–2476. doi: 10.1074/jbc.M111.285031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hubbard BP, Gomes AP, Dai H, et al. Evidence for a common mechanism of SIRT1 regulation by allosteric activators. Science. 2013;339(6124):1216–1219. doi: 10.1126/science.1231097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103(27):10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Sundaresan NR, Pillai VB, Wolfgeher D, et al. The deacetylase SIRT1 promotes membrane localization and activation of Akt and PDK1 during tumorigenesis and cardiac hypertrophy. Sci Signal. 2011;4(182):ra46. doi: 10.1126/scisignal.2001465. [DOI] [PubMed] [Google Scholar]
- 185.Fu M, Liu M, Sauve AA, et al. Hormonal control of androgen receptor function through SIRT1. Mol Cell Biol. 2006;26(21):8122–8135. doi: 10.1128/MCB.00289-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yamamori T, DeRicco J, Naqvi A, et al. SIRT1 deacetylates APE1 and regulates cellular base excision repair. Nucleic Acids Res. 2010;38(3):832–845. doi: 10.1093/nar/gkp1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lee IH, Cao L, Mostoslavsky R, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008;105(9):3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Nakahata Y, Kaluzova M, Grimaldi B, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134(2):329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wu X, Kong X, Chen D, et al. SIRT1 links CIITA deacetylation to MHC II activation. Nucleic Acids Res. 2011;39(22):9549–9558. doi: 10.1093/nar/gkr651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Mao B, Zhao G, Lv X, et al. Sirt1 deacetylates c-Myc and promotes c-Myc/Max association. Int J Biochem Cell Biol. 2011;43(11):1573–1581. doi: 10.1016/j.biocel.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 191.Jeong H, Cohen DE, Cui L, et al. Sirt1 mediates neuroprotection from mutant huntingtin by activation of the TORC1 and CREB transcriptional pathway. Nat Med. 2011;18(1):159–165. doi: 10.1038/nm.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Liu Y, Dentin R, Chen D, et al. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456(7219):269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Peng L, Yuan Z, Ling H, et al. SIRT1 deacetylates the DNA methyltransferase 1 (DNMT1) protein and alters its activities. Mol Cell Biol. 2011;31(23):4720–4734. doi: 10.1128/MCB.06147-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Pradhan AK, Kuila N, Singh S, Chakraborty S. EVI1 up-regulates the stress responsive gene SIRT1 which triggers deacetylation and degradation of EVI1. Biochim Biophys Acta. 2011;1809(4–6):269–275. doi: 10.1016/j.bbagrm.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 195.Mattagajasingh I, Kim CS, Naqvi A, et al. SIRT1 promotes endothelium-dependent vascular relaxation by activating endothelial nitric oxide synthase. Proc Natl Acad Sci U S A. 2007;104(37):14855–14860. doi: 10.1073/pnas.0704329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Yu EJ, Kim SH, Heo K, Ou CY, Stallcup MR, Kim JH. Reciprocal roles of DBC1 and SIRT1 in regulating estrogen receptor alpha activity and co-activator synergy. Nucleic Acids Res. 2011;39(16):6932–6943. doi: 10.1093/nar/gkr347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Daitoku H, Hatta M, Matsuzaki H, et al. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci U S A. 2004;101(27):10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Brunet A, Sweeney LB, Sturgill JF, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303(5666):2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 199.Motta MC, Divecha N, Lemieux M, et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116(4):551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 200.van Loosdregt J, Vercoulen Y, Guichelaar T, et al. Regulation of Treg functionality by acetylation-mediated Foxp3 protein stabilization. Blood. 2010;115(5):965–974. doi: 10.1182/blood-2009-02-207118. [DOI] [PubMed] [Google Scholar]
- 201.Kemper JK, Xiao Z, Ponugoti B, et al. FXR acetylation is normally dynamically regulated by p300 and SIRT1 but constitutively elevated in metabolic disease states. Cell Metab. 2009;10(5):392–404. doi: 10.1016/j.cmet.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Dioum EM, Chen R, Alexander MS, et al. Regulation of hypoxia-inducible factor 2alpha signaling by the stress-responsive deacetylase sirtuin 1. Science. 2009;324(5932):1289–1293. doi: 10.1126/science.1169956. [DOI] [PubMed] [Google Scholar]
- 203.Westerheide SD, Anckar J, Stevens SM, Jr, Sistonen L, Morimoto RI. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009;323(5917):1063–1066. doi: 10.1126/science.1165946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Jeong J, Juhn K, Lee H, et al. SIRT1 promotes DNA repair activity and deacetylation of Ku70. Exp Mol Med. 2007;39(1):8–13. doi: 10.1038/emm.2007.2. [DOI] [PubMed] [Google Scholar]
- 205.Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol Cell. 2007;28(1):91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 206.Zocchi L, Sassone-Corsi P. SIRT1-mediated deacetylation of MeCP2 contributes to BDNF expression. Epigenetics. 2012;7(7):695–700. doi: 10.4161/epi.20733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Fulco M, Schiltz RL, Iezzi S, et al. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12(1):51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 208.Yuan Z, Zhang X, Sengupta N, Lane WS, Seto E. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol Cell. 2007;27(1):149–162. doi: 10.1016/j.molcel.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Yeung F, Hoberg JE, Ramsey CS, et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23(12):2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Libert S, Pointer K, Bell EL, et al. SIRT1 activates MAO-A in the brain to mediate anxiety and exploratory drive. Cell. 2011;147(7):1459–1472. doi: 10.1016/j.cell.2011.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Zhou Y, Schmitz KM, Mayer C, Yuan X, Akhtar A, Grummt I. Reversible acetylation of the chromatin remodelling complex NoRC is required for non-coding RNA-dependent silencing. Nat Cell Biol. 2009;11(8):1010–1016. doi: 10.1038/ncb1914. [DOI] [PubMed] [Google Scholar]
- 212.Guarani V, Deflorian G, Franco CA, et al. Acetylation-dependent regulation of endothelial Notch signalling by the SIRT1 deacetylase. Nature. 2011;473(7346):234–238. doi: 10.1038/nature09917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Bouras T, Fu M, Sauve AA, et al. SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. J Biol Chem. 2005;280(11):10264–10276. doi: 10.1074/jbc.M408748200. [DOI] [PubMed] [Google Scholar]
- 214.Luo J, Nikolaev AY, Imai S, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107(2):137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 215.Vaziri H, Dessain SK, Ng Eaton E, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107(2):149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 216.Rajamohan SB, Pillai VB, Gupta M, et al. SIRT1 promotes cell survival under stress by deacetylation-dependent deactivation of poly(ADP-ribose) polymerase 1. Mol Cell Biol. 2009;29(15):4116–4129. doi: 10.1128/MCB.00121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Asher G, Gatfield D, Stratmann M, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134(2):317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 218.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434(7029):113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 219.Akieda-Asai S, Zaima N, Ikegami K, et al. SIRT1 Regulates Thyroid-Stimulating Hormone Release by Enhancing PIP5Kgamma Activity through Deacetylation of Specific Lysine Residues in Mammals. PLoS One. 2010;5(7):e11755. doi: 10.1371/journal.pone.0011755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ikenoue T, Inoki K, Zhao B, Guan KL. PTEN acetylation modulates its interaction with PDZ domain. Cancer Res. 2008;68(17):6908–6912. doi: 10.1158/0008-5472.CAN-08-1107. [DOI] [PubMed] [Google Scholar]
- 221.Donmez G, Wang D, Cohen DE, Guarente L. SIRT1 suppresses beta-amyloid production by activating the alpha-secretase gene ADAM10. Cell. 2010;142(2):320–332. doi: 10.1016/j.cell.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 222.Wong S, Weber JD. Deacetylation of the retinoblastoma tumour suppressor protein by SIRT1. Biochem J. 2007;407(3):451–460. doi: 10.1042/BJ20070151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Kume S, Haneda M, Kanasaki K, et al. SIRT1 inhibits transforming growth factor beta-induced apoptosis in glomerular mesangial cells via Smad7 deacetylation. J Biol Chem. 2007;282(1):151–158. doi: 10.1074/jbc.M605904200. [DOI] [PubMed] [Google Scholar]
- 224.Ponugoti B, Kim DH, Xiao Z, et al. SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. J Biol Chem. 2010;285(44):33959–33970. doi: 10.1074/jbc.M110.122978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Nie Y, Erion DM, Yuan Z, et al. STAT3 inhibition of gluconeogenesis is downregulated by SirT1. Nat Cell Biol. 2009;11(4):492–500. doi: 10.1038/ncb1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Wang RH, Zheng Y, Kim HS, et al. Interplay among BRCA1, SIRT1, and Survivin during BRCA1-associated tumorigenesis. Mol Cell. 2008;32(1):11–20. doi: 10.1016/j.molcel.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450(7168):440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- 228.Pagans S, Pedal A, North BJ, et al. SIRT1 regulates HIV transcription via Tat deacetylation. PLoS Biol. 2005;3(2):e41. doi: 10.1371/journal.pbio.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Peng L, Ling H, Yuan Z, et al. SIRT1 negatively regulates the activities, functions, and protein levels of hMOF and TIP60. Mol Cell Biol. 2012;32(14):2823–2836. doi: 10.1128/MCB.00496-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Li K, Casta A, Wang R, et al. Regulation of WRN protein cellular localization and enzymatic activities by SIRT1-mediated deacetylation. J Biol Chem. 2008;283(12):7590–7598. doi: 10.1074/jbc.M709707200. [DOI] [PubMed] [Google Scholar]
- 231.Wang FM, Chen YJ, Ouyang HJ. Regulation of unfolded protein response modulator XBP1s by acetylation and deacetylation. Biochem J. 2011;433(1):245–252. doi: 10.1042/BJ20101293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Fan W, Luo J. SIRT1 regulates UV-induced DNA repair through deacetylating XPA. Mol Cell. 2010;39(2):247–258. doi: 10.1016/j.molcel.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 233.Ming M, Shea CR, Guo X, et al. Regulation of global genome nucleotide excision repair by SIRT1 through xeroderma pigmentosum C. Proc Natl Acad Sci U S A. 2010;107(52):22623–22628. doi: 10.1073/pnas.1010377108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Suzuki K, Koike T. Mammalian Sir2-related protein (SIRT) 2-mediated modulation of resistance to axonal degeneration in slow Wallerian degeneration mice:a crucial role of tubulin deacetylation. Neuroscience. 2007;147(3):599–612. doi: 10.1016/j.neuroscience.2007.04.059. [DOI] [PubMed] [Google Scholar]
- 235.Jing E, Gesta S, Kahn CR. SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab. 2007;6(2):105–114. doi: 10.1016/j.cmet.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Snider NT, Leonard JM, Kwan R, Griggs NW, Rui L, Omary MB. Glucose and SIRT2 reciprocally mediate the regulation of keratin 8 by lysine acetylation. J Cell Biol. 2013;200(3):241–247. doi: 10.1083/jcb.201209028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Rothgiesser KM, Erener S, Waibel S, Luscher B, Hottiger MO. SIRT2 regulates NF-kappaB dependent gene expression through deacetylation of p65 Lys310. J Cell Sci. 2010;123(Pt 24):4251–4258. doi: 10.1242/jcs.073783. [DOI] [PubMed] [Google Scholar]
- 238.Black JC, Mosley A, Kitada T, Washburn M, Carey M. The SIRT2 deacetylase regulates autoacetylation of p300. Mol Cell. 2008;32(3):449–455. doi: 10.1016/j.molcel.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Beirowski B, Gustin J, Armour SM, et al. Sir-two-homolog 2 (Sirt2) modulates peripheral myelination through polarity protein Par-3/atypical protein kinase C (aPKC) signaling. Proc Natl Acad Sci U S A. 2011;108(43):E952–E961. doi: 10.1073/pnas.1104969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Jiang W, Wang S, Xiao M, et al. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol Cell. 2011;43(1):33–44. doi: 10.1016/j.molcel.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Serrano L, Martinez-Redondo P, Marazuela-Duque A, et al. The tumor suppressor SirT2 regulates cell cycle progression and genome stability by modulating the mitotic deposition of H4K20 methylation. Genes Dev. 2013;27(6):639–653. doi: 10.1101/gad.211342.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Narayan N, Lee IH, Borenstein R, et al. The NAD-dependent deacetylase SIRT2 is required for programmed necrosis. Nature. 2012;492(7428):199–204. doi: 10.1038/nature11700. [DOI] [PubMed] [Google Scholar]
- 243.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A. 2006;103(27):10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Shulga N, Wilson-Smith R, Pastorino JG. Sirtuin-3 deacetylation of cyclophilin D induces dissociation of hexokinase II from the mitochondria. J Cell Sci. 2010;123(Pt 6):894–902. doi: 10.1242/jcs.061846. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 245.Hafner AV, Dai J, Gomes AP, et al. Regulation of the mPTP by SIRT3-mediated deacetylation of CypD at lysine 166 suppresses age-related cardiac hypertrophy. Aging (Albany NY) 2010;2(12):914–923. doi: 10.18632/aging.100252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Tseng AH, Shieh SS, Wang DL. SIRT3 deacetylates FOXO3 to protect mitochondria against oxidative damage. Free Radic Biol Med. 2013;63C:222–234. doi: 10.1016/j.freeradbiomed.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 247.Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol. 2008;382(3):790–801. doi: 10.1016/j.jmb.2008.07.048. [DOI] [PubMed] [Google Scholar]
- 248.Sundaresan NR, Samant SA, Pillai VB, Rajamohan SB, Gupta MP. SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. Mol Cell Biol. 2008;28(20):6384–6401. doi: 10.1128/MCB.00426-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Hirschey MD, Shimazu T, Goetzman E, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464(7285):121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Pillai VB, Sundaresan NR, Kim G, et al. Exogenous NAD blocks cardiac hypertrophic response via activation of the SIRT3-LKB1-AMP-activated kinase pathway. J Biol Chem. 2010;285(5):3133–3144. doi: 10.1074/jbc.M109.077271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Shimazu T, Hirschey MD, Hua L, et al. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 2010;12(6):654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Tao R, Coleman MC, Pennington JD, et al. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell. 2010;40(6):893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Yang Y, Cimen H, Han MJ, et al. NAD+-dependent deacetylase SIRT3 regulates mitochondrial protein synthesis by deacetylation of the ribosomal protein MRPL10. J Biol Chem. 2010;285(10):7417–7429. doi: 10.1074/jbc.M109.053421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Ahn BH, Kim HS, Song S, et al. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci U S A. 2008;105(38):14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Hallows WC, Yu W, Smith BC, et al. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell. 2011;41(2):139–149. doi: 10.1016/j.molcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Cimen H, Han MJ, Yang Y, Tong Q, Koc H, Koc EC. Regulation of succinate dehydrogenase activity by SIRT3 in mammalian mitochondria. Biochemistry. 2010;49(2):304–311. doi: 10.1021/bi901627u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Finley LW, Haas W, Desquiret-Dumas V, et al. Succinate dehydrogenase is a direct target of sirtuin 3 deacetylase activity. PLoS One. 2011;6(8):e23295. doi: 10.1371/journal.pone.0023295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Laurent G, German NJ, Saha AK, et al. SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase. Mol Cell. 2013;50(5):686–698. doi: 10.1016/j.molcel.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Nakagawa T, Guarente L. Sirtuins at a glance. J Cell Sci. 2011;124(Pt 6):833–838. doi: 10.1242/jcs.081067. [DOI] [PMC free article] [PubMed] [Google Scholar]