Abstract
RNA viruses, such as flaviviruses, are able to efficiently replicate and cap their RNA genomes in vertebrate and invertebrate cells. Flaviviruses use several specialized proteins to first make an uncapped negative strand copy of the viral genome that is used as a template for the synthesis of large numbers of capped genomic RNAs. Despite using relatively simple mechanisms to replicate their RNA genomes, there are significant gaps in our understanding of how flaviviruses switch between negative and positive strand RNA synthesis and how RNA capping is regulated. Recent work has begun to provide a conceptual framework for flavivirus RNA replication and capping and shown some surprising roles for genomic RNA during replication and pathogenesis.
Flaviviruses are the most prevalent mosquito-transmitted viral pathogens worldwide, and every year these viruses cause severe economic and human suffering. There are 35 known flaviviruses that cause human disease, and it has been estimated that approximately 2/3rd of the world population is at risk for infection by one or more of these pathogens. WNV has become endemic in the United States since 1999 and continues to cause significant problems with transplant recipients and other immunocompromised patients 1. Dengue viruses infect approximately 50 million individuals each year and are a leading cause of mortality in children in a number of Latin and Asian countries 2. Yellow fever virus is endemic in a number of African and South American countries, and causes 200,000 cases and 30,000 deaths in Africa even with effective vaccines available 3. There are currently no clinically useful antiviral drugs for the treatment of any flavivirus infection, and identification of novel points of intervention for drug development is an active area of research. Inhibiting flavivirus RNA genome replication is considered a potential approach to treating flavivirus infections, and in-depth understanding of the mechanisms that flaviviruses use to replicate their genomes is necessary for effective development of therapeutics and vaccines.
Flaviviruses are small, enveloped viruses with single-stranded RNA genomes of 11-12 Kb. The 5′ end of the positive strand genomic RNA possesses a N7 methylated (me7)-guanosine cap structure that directs viral polyprotein translation and protects the genome from 5′ exonuclease degradation 4,5. The 3′ end of the genome is non-polyadenylated and terminates in a stable stem-loop structure (3′ SL). The genome contains ~100 nucleotide 5′ and ~400-700 nucleotide 3′ untranslated regions (UTR) that contain RNA structures critical for RNA replication. Additional RNA structures are present in the ~3400 amino acid polyprotein coding region that are involved in cyclizing the positive strand genome during RNA replication. The flavivirus polyprotein encodes 11 mature viral proteins, three of which are involved in forming viral particles (Capsid (C), pre membrane (prM), and envelope (E)) and 8 non-structural proteins that are involved in RNA replication (NS1 (NS1′ in the JEV subgroup 6), NS2A, NS2B, NS3, NS4A, 2K, NS4B, NS5) 7.
Overview of the Flavivirus RNA Replication Cycle
Flavivirus particles enter cell via endocytosis, and the nucleocapsid is released from the virion via fusion of the viral membranes with the endosomal membrane 8. The viral genomic RNA is trafficked to the rough endoplasmic reticulum, where viral polyproteins are translated and co-translationally cleaved into mature proteins. The viral replication proteins induce membrane rearrangements that generate membranous compartments where RNA replication occurs. The positive strand genomic RNA is used as a template to produce low levels of uncapped negative sense RNA, which is used as a template for production of high levels of positive strand capped genomic RNA. A proportion of the newly synthesized positive strand RNAs are used for further protein translation to support virion production, some RNAs interact with and repress the RNAi and RNA decay pathways, and some interact with capsid proteins and are packaged into nascent virions. Virions mature through the trans-golgi system and are released into the extracellular milieu to spread virus infection throughout the host.
RNA Structures Involved in Negative Strand RNA Synthesis
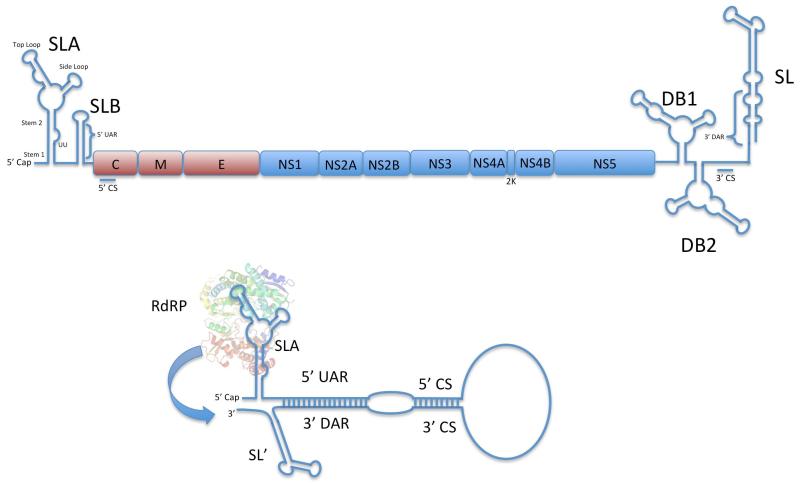
Several RNA structures are present in the coding and non-coding regions of flavivirus genomes that help direct RNA synthesis (Figure 1). The short 5′ untranslated region (UTR) contains several stem-loop structures critical for RNA synthesis and translation. A large stem-loop structure is present at the 5′ end of the 5′ UTR (Stem-Loop A (SLA)) that binds to NS5 and acts as a promoter for viral RNA synthesis 9,10. The core stem regions of SLA are conserved among the flaviviruses whereas the top and side loops are somewhat divergent, indicating the importance of the core stem regions. Along these lines, mutation of some but not all SLA regions that form the putative stem-loop significantly impair RNA replication. Mutations in SLA stems 1 and 2 reduced replication and gave rise to spontaneous revertants. Mutation of the UU bulge between stems 1 and 2 gave rise to a UA revertant, indicating that at least 1 U in that region is necessary for RNA replication. Deleting the terminal loop on the side loop blocked replication, whereas mutants with elongated side loop stems or sequence of the loop were viable. Internal base paring does not seem to be critical for stem 3, but disruption of base paring at the base of the top loop blocked replication and demonstrated a requirement for this loop region for efficient RNA replication.
Figure 1. Flavivirus Genomic RNA Structures.
A) Linear structure of a generic flavivirus RNA genome showing the positions of critical RNA structures and protein coding regions. B) Cyclization of Flavivirus positive strand genome promotes negative strand RNA synthesis. The 5′ end of the positive strand genomic RNA interacts with the 3′ end of the positive strand RNA via interactions between the CS and UAR/DAR regions. Hybridization between the 5′ UAR and 3′ DAR causes a re-organization of the 3′ SL structure, exposing the 3′ end of the positive strand RNA. The RdRP domain of NS5 binds to SLA on the 5′ end of the positive strand genome and utilizes the exposed 3′ end of the positive strand RNA as a template for negative strand RNA synthesis.
How does SLA contribute to RNA replication? At least part of this answer seems to be that SLA is involved in binding NS5. Filomatori et al used RNA mobility shift and in vitro RNA polymerization assays to demonstrate that the RNA dependent RNA polymerase (RdRP) domain of NS5 binds to SLA on the positive strand RNA to promote negative strand RNA synthesis 11. Interestingly, mutations in the top loop did not disrupt high affinity binding to NS5 but abolished RNA replication, suggesting that SLA has a functional role in RNA replication besides binding NS5. NS5 RdRP fingers domain mutants K456A and K457A were unable to initiate RNA synthesis in the absence of SLA while retaining the ability to bind to SLA, suggesting that this region is involved in SLA promoter dependent initiation 12. While these data do not definitively prove a functional interaction between the top loop and RdRP residues K456 and K457, they provide hints into how SLA may be interacting with the NS5 RdRP domain during negative strand synthesis.
The 3′ end of the positive strand genomic RNA is the starting point for negative strand RNA synthesis 13. The 3′ end of linear flavivirus genomes terminates in a stable stem-loop structure (3′ SL) where the conserved 3′ nucleotides are base paired at the base of the 3′ SL structure. It was originally thought that the 3′ SL structure was directly involved in negative strand synthesis, but several studies have indicated that the presence of the SL structure actually inhibits negative strand RNA synthesis 9,14. Base paring of the 3′ nucleotides presumably block RNA binding into the RdRP domain and not allow negative strand synthesis to occur. Recent work by the Brinton group has shown that specific hybridization between G7 and U75 in the 3′ SL is critical for the stability of the 3′ SL and mutation of these residues alter negative strand RNA synthesis and overall RNA replication 15. The 3′ SL structure is disrupted when the genomic RNA cyclizes via the hybridization of the 5′ and 3′ cyclization sequences and hybridization of 5′ UTR stem loop B 5′ Upstream of AUG (UAR) region and the 3′ Downstream of AUG (DAR) region at the 5′ end of 3′ SL 16,17 . Hybridization of the 5′-3′ cyclization sequences bring SLA and the 3′ end of the positive strand genome together and hybridization of the 5′UAR and 3′ DAR result in a conformational reorganization of SL into a more suitable single-stranded template for the RdRP (Figure 1). The close proximity of SLA to the single-stranded 3′ end positions in the cyclized genome positions NS5 to recognize the 3′ end as a template for negative strand RNA synthesis. The interaction between NS5 and SLA appears to be important as an RNA selectivity mechanism, and may allow SLA bound NS5 to recognize and use only positive strand genomes as negative strand RNA templates and avoid using cellular mRNAs also present on the rough endoplasmic reticulum as templates.
Positive Strand RNA Synthesis: A Puzzle with Missing Pieces
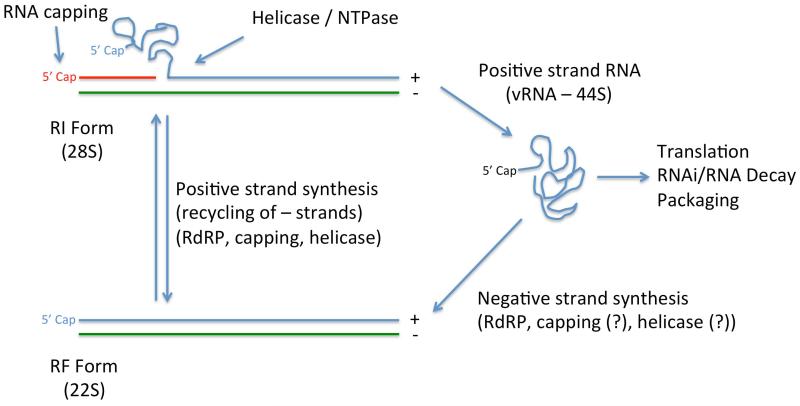
While a good deal of effort has been spent on understanding negative strand RNA synthesis, there is very little information about the molecular basis of positive strand synthesis during infection. The only in vitro model system for studying positive strand RNA replication uses cytoplasmic extracts from infected cells and monitors the production of the double-stranded 20S replicative form (RF), positive strand synthesizing 20-28S replicative intermediate form (RI), and completed 44S viral RNA (vRNA) 18. This approach provides a global view of flavivirus RNA replication, but without uncoupling negative strand and positive strand synthesis it is difficult to test specific hypotheses about positive strand RNA synthesis. We can deduce that positive strand synthesis is a much more complex process than negative strand RNA synthesis (Figure 2). Negative strand RNA synthesis seems to primarily require NS5 RdRP activity whereas positive strand RNA synthesis also incorporates 5′ RNA capping and RNA unwinding, requiring multiple additional enzymatic functions from NS3 (helicase/ATPase/RNA triphosphatase) and NS5 (guanylyltransferase/methyltransferase) that are described in more detail later in this article. The double-stranded RF RNA acts as the template for positive strand synthesis. Paradoxically, the positive strand 3′ SL RNA is thought to inhibit replication by forming double-stranded RNA and blocking RNA binding to the RdRP, and the minus strand 3′ end would also be double-stranded RF form which would be expected to block RNA binding by the RdRP. Because there is not a good in vitro system available to probe how positive strand replication occurs, most of our understanding of positive strand synthesis has necessarily been inferred from studies of the enzymatic activities of the NS3 and NS5 replication proteins and how they form the RNA replication complex.
Figure 2. Schematic of Flavivirus RNA Replication.
44S Positive strand viral RNAs (vRNA) are trafficked from incoming viral particles to the endoplasmic reticulum where viral polyproteins are translated. The newly synthesized NS5 RdRP generates a negative strand RNA (colored green) using the positive strand RNA as a template. It is unknown if RNA capping activity or RNA helicase activity occurs during negative strand RNA synthesis. The Replicative Form (RF) RNA is a duplex of negative and positive strand RNA thought to act as a template for additional capped positive strand RNA synthesis via the action of NS3 and NS5. The 28S Replicative Intermediate (RI) form is comprised of newly synthesized capped positive strand RNA (colored red) and displaced original capped positive strand RNA. The NS3 RNA triphosphatase and NS5 guanylyltransferase / methyltransferase enzymes generate a new RNA cap on the 5′ end of the nascent RNA strand. NS3 RNA helicase and NTPase activities are necessary for unwinding of dsRNA during positive strand RNA synthesis. Released positive strand RNAs can be used for additional protein translation, interference with RNAi or RNA decay pathways, packaging into viral particles, or generate additional RF forms.
The Flavivirus RNA Replication Complex: A Highly Integrated RNA Replication Machine
The nonstructural NS3 and NS5 proteins constitute the core flavivirus RNA replication complex. NS3 and NS5 possess all of the enzymatic functions necessary for replication of flavivirus genomes, while the remaining nonstructural proteins (NS1, NS2A, NS2B, NS4A, 2K, NS4B) are thought to provide support for replication and aid in the formation of the replication compartment 7. NS1 is located in the lumen of the endoplasmic reticulum, and NS2A, NS2B, NS4A, 2K, and NS2B are transmembrane proteins thought to reside within the viral replication compartment and surrounding membranes. The membrane associated proteins act to tether the NS3:NS5 complex to the membrane and NS1 helps organize the membrane-associated proteins and support the function of NS3 and NS5 during replication 19-23. Therefore, NS3 and NS5 represent the core replication complex and the other nonstructural proteins enhance or regulate NS3/NS5 function to aid in replication.
NS3 is a ~650 amino acid protein that possesses two distinct globular domains connected byã20 amino acid linker region. NS3 remains associated with the ER during replication and encodes several enzymatic functions critical for replication. NS3 encodes a N-terminal serine protease that cleaves the viral polyprotein in cis at several positions to form mature proteins 1,24. NS3 also encodes an ATP dependent C-terminal RNA helicase function that unwinds double-stranded RNA during replication 2,25,26 and is involved in positive strand RNA synthesis. Recent studies have indicated that the helicase domain can contribute to RNA annealing during replication which may contribute to cyclization of the positive strand RNA for negative strand RNA synthesis 3,27. The helicase domain also contains nucleotide triphosphatase (NTPase) activity used to power the helicase 4,5,28 and a RNA triphosphatase 6,29 activity that removes the γ–phosphate from newly synthesized positive strand RNAs to prepare the genome for RNA capping. The NS3 NTPase and RNA triphosphatase functions utilize the same Walker B motif to perform each reaction 7,30.
NS5 is a ~900 amino acid protein found at the C-terminus of the flavivirus polyprotein. NS5 possesses two distinct domains, a N-terminal methyltransferase/guanylyltransferase domain and a C-terminal RNA dependent RNA polymerase domain. The ~265 amino acid N-terminal “capping enzyme” was originally identified as a methyltransferase by the presence of a S-Adenosyl methionine binding motif homologous with the E. coli YdhB gene 8,31 and later empirically verified to possess 2′-O- 9,10,32 and guanine N7- 11,33 methyltransferase activities. In 2009, Issur et al demonstrated that the capping enzyme domain also possessed guanylyltransferase activity that was able to form a covalent protein:GMP adduct (protein guanylation) and transfer the GMP to di-phosphorylated RNA substrates to form the base cap structure 12,34. The C-terminal ~600 amino acids of NS5 encode a RNA dependent RNA polymerase responsible for synthesizing the negative and positive strand RNAs 13,35. A ~30 amino acid linker domain connects the capping enzyme and polymerase domain and has been implicated in association with NS3 helicase domain 9,14,36,37 and import of NS5 into the nucleus 15,38, although some crystallographic structures of the polymerase domain suggest that the flexible linker region lies within NS5 residues 260-270 and not 322-407 as previously thought 16,17,39,40.
NS3 and NS5 physically associate during replication to synergize their enzymatic activities and synthesize positive strand RNAs. The NS3 NTPase and RNA triphosphatase activities are higher in the presence of the NS5 RdRP domain than in isolation 18,41,42, and the NS5 capping enzyme guanylyltransferase activity is enhanced by the NS3 helicase domain 7,34. These findings suggest that NS3 and NS5 allosterically regulate each other’s function during RNA replication. It is currently unknown if NS3 helicase and NS5 RdRP activities are also allosterically regulated by their association with NS5 or NS3, respectively.
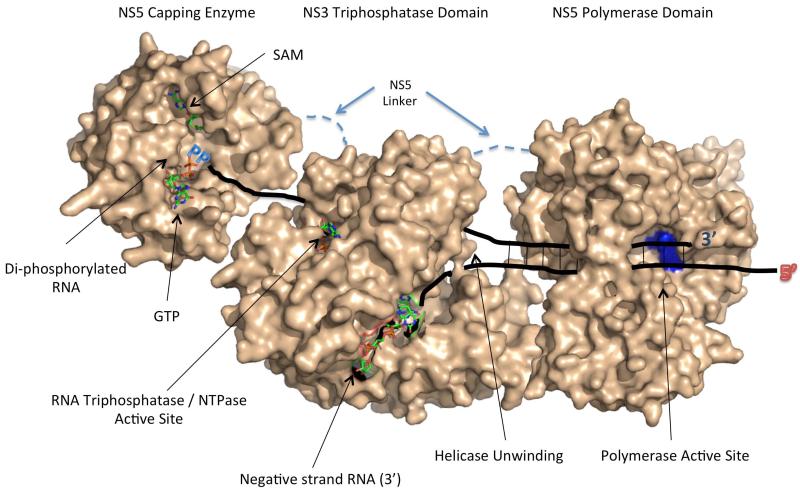
NS3 and NS5 work together to replicate genomes, but there is limited information about how these proteins physically associate or how allosteric regulation between the enzymes may occur during viral RNA replication. The development of a co-crystal structure between NS3 and NS5 would help illuminate how NS3 and NS5 interact, but at this time there is no such structure available. To help visualize what the replication complex structure may look like, we have developed a preliminary model for how NS3 and NS5 may physically interact (Figure 3). This model is based on the location of enzymatic active sites in each protein, limited physical interaction and compensatory mutation data, and the logical progression of RNA synthesis, RNA unwinding, and capping during flavivirus positive strand synthesis 19-23,34,36,37,41-46.
Figure 3. Model for NS3/NS5 Interaction Based on Known Structures and Enzymatic Active Sites.
In this model, the NS3 RNA triphosphatase / helicase domain interacts with NS5 via a flexible linker found between the capping enzyme and RdRP domains (dashed line). The NS3 protease domain is not included in this model for clarity. During positive strand RNA synthesis, the single-stranded negative strand RNA template enters the RdRP active site, and the polymerase catalyzes the elongation of a triphosphorylated positive strand RNA. The positive strand RNA is initially duplexed with the negative strand RNA, and this dsRNA is unwound by the helicase activity present in NS3. The positive strand RNA interacts with the RNA triphosphatase active site, which removes the γ-phosphate from the triphosphorylated RNA, resulting in a di-phosphorylated RNA substrate. The di-phosphorylated RNA is then fed into the NS5 capping enzyme where the guanylyltransferase function caps the RNA and the methyltransferase function methylates the RNA. The model was developed using the following PDB files (NS3 Helicase/RNA Triphosphatase domain (PDB codes: 2 JLR / 2JLU 69, NS5 capping enzyme (PDB Code: 3EVG 66, NS5 RdRP (PDB Code: 2J7U) 90).
Flavivirus RNA replication takes place in ER derived replication compartments
Flaviviruses rearrange intracellular membranes during replication, and several studies found that viral double-stranded RNA (dsRNA) were associated with rough endoplasmic membranes 47-49. Further ultrastructural studies have demonstrated that viral dsRNA and viral replication proteins NS1, NS2A, NS3, NS4A, and NS5 co-localized with ~100 nM vesicle-like structures termed vesicle packets 50-53. Biochemical studies demonstrated that viral RF and RI RNA forms were protected by membranes 54. Several studies indicated that genomic RNA in the vesicle packets are also protected from cellular antiviral responses during infection, including the RNAi and interferon responses, soon after infection 55,56. Recent 3D electron microscopy tomography studies have shed light onto the structure of the vesicle packets 57-59, which appear to be small spherical compartment structures contiguous with the endoplasmic reticulum membrane. Viral RNA is localized within these replication compartments, providing a protected environment for RNA replication to occur away from antagonistic cellular responses while retaining access to cytoplasmic resources (ribonucleotides, ions, ect.) through narrow pores. Data presented by Welsch et al. suggest that these replication compartments are in close proximity to sites of virion packaging, providing a mechanism to rapidly package genomic RNA into nascent virion particles while keeping cytoplasmic exposure to a minimum 57. Proliferation of the endoplasmic reticulum membrane appears to be involved in formation of these replication compartments, a process that appears to be at least partially controlled by the NS4A transmembrane protein 60,61. This process is likely linked with the observed perturbation of lipid homeostasis in infected cells 62 and virus induced alterations in lipid metabolism 63. The fatty acid synthetase enzyme has been observed in close proximity to replicating viral RNA and appears to be recruited to replication sites via interactions with NS3, providing lipids necessary for replication compartment formation 63. Inhibition of cholesterol biosynthesis has also been show to affect flavivirus RNA replication in replicon assays 64, although the role of cholesterol in replication compartment biosynthesis is still emerging.
RNA Capping in Flavivirus RNA Replication
The mature type cap 1 consists of a 5′-5′ linked me7 guanosine structure (me7-GpppN-me2). me7-GpppG-me2 capping of cellular mRNA molecules occurs co-transcriptionally in the nucleus by the action of four enzymes: RNA triphosphatase, RNA guanylyltransferase, guanine-7-methytransferase, and nucleoside 2′-O-methyltransferase. The 5′ end of the nascent RNA transcript (pppN) is hydrolyzed by the RNA triphosphatase to a di-phosphate end (ppN), which is then ligated with guanosine monophosphate (GMP) in a 5′-5′ linkage by the guanylyltransferase to form the base cap structure (GpppN). N7-methyltransferase transfers a methyl group from S-adenosyl methionine (SAM) to the guanine N7 position, resulting in me7-GpppN and S-adenosyl-L-homocysteine (SAH). This structure is known as cap 0. In a second enzymatic step, a methyl group is transferred from another AdoMet molecule to the 2′ hydroxyl position of the penultimate nucleotide by 2′-O-methyltransferase, generating the cap 1 structure (me7-GpppN-me2).
The cellular mRNA capping machinery is located in the nucleus, whereas flavivirus RNA replication occurs in ER-derived replication compartments in the cytoplasm. Because flaviviruses do not have access to the cellular capping machinery they must provide their own enzymes to produce capped RNA. Flavivirus genomic RNA is modified at the 5′ end of positive strand genomic RNA with a cap 1 structure (me7-GpppA-me2) generated by the virus encoded NS3 RNA triphosphatase 29, NS5 guanylyltransferase 34, NS5 2′-O-methyltransferase 32, and NS5 Guanine-N7-methyltransferase 65. X-ray crystal structures for each of these viral enzymes have been solved 26,32,65-67, providing a wealth of information about how these enzymes may function during RNA replication.
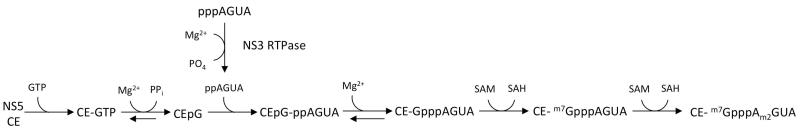
The order of RNA capping has not been completely defined for flaviviruses, but the canonical Ping-Pong mechanism for cap formation appears to be the most-likely scenario (Figure 4). The RNA triphosphatase is located in the NS3 helicase domain 68, and the RNA triphosphatase appears to overlap with the NTPase active site that powers the helicase 69,70. Newly synthesized negative and positive strand RNAs are triphosphorylated and would be appropriate substrates for the RNA triphosphatase. It is currently unknown if the negative strand RNA is modified by the RNA triphosphatase, but the observation that negative strand RNA is only found in the double-stranded form and not unwound by the NS3 helicase domain suggests that NS3 may not be a component of negative strand RNA synthesis. The γ–phosphate is removed from positive strand RNAs in a Mg2+ dependent reaction 30,68, resulting in a di-phosphorylated RNA. Di-phosphorylated RNA with a 5′ adenosine base is a substrate for the NS5 guanylytransferase within the capping enzyme domain 34. The capping enzyme binds GTP and forms a guanylated intermediate in a Mg2+ dependent manner through an as yet unknown mechanism, then transfers the GMP to the dephosphorylated RNA substrate. Studies on the methyltransferase function of the NS5 capping enzyme have taken place with capped RNAs 32,33,65,71, but it is unknown if the di-phosphorylated RNA is 2′O methylated prior to capping. Methylation does not appear to be required for the NS5 guanylyltransferase to cap the di-phosphorylated positive strand RNA, as an unmethylated in vitro generated RNA substrate was able to be capped by NS5 34.
Figure 4. Proposed Mechanism of Flavivirus RNA Capping.
NS3 RNA triphosphatase binds to and cleaves the γ-phosphate from newly synthesized positive strand RNAs, generating a di-phosphorylated RNA substrate. The NS5 capping enzyme (CE) binds GTP and forms the guanylated intermediate in a Mg2+ dependent reaction. The di-phosphorylated RNA substrate interacts with the guanylated NS5 protein, which transfers the GMP moiety to the di-phosphorylated RNA to form the base cap structure (GpppAGUAn). The base cap structure is first methylated at the guanine N7 position by the methyltransferase function within the capping enzyme, presumably by the action of a second NS5 capping enzyme protein. The cap 0 structure is then 2′O methylated to form the Cap 1 structure (m7GpppAm2GUAn). This model would allow the cap to be fully formed without repositioning.
Does phosphorylation control negative to positive strand RNA synthesis switching?
Early RNA replication is biased towards the production of uncapped negative strand template RNAs followed by capped positive strand RNAs later in infection. A major unanswered question about flavivirus replication is how the switch from negative strand RNA synthesis to positive strand RNA synthesis occurs. The majority of research on RNA synthesis during flavivirus infection has been focused on negative strand synthesis, but there are a few clues that may point to how this switch occurs. NS5 phosphorylation has been implicated in controlling the association of NS3 with NS5 during infection and retarget NS5 to the nucleus 43. Later work suggested that phosphorylation of the NS5 linker region by Casein Kinase II inhibited nuclear import of NS5 72, but it is still not known if Casein Kinase II phosphorylation directly plays a role in the association of NS5 and NS3. As mentioned above, there is little evidence that NS3 significantly functions in negative strand synthesis, and it is unclear if NS3 associates with NS5 during early RNA replication, but a reasonable hypothesis is that phosphorylation of the NS5 linker region early in infection may reduce NS3 association with NS5, effectively removing helicase and RNA triphosphatase activity from the replication complex. More recent data from the Striker group suggests that phosphorylation may inhibit NS5 methyltransferase activity. Phosphorylation of the yellow fever NS5 capping enzyme serine 56 was found to occur in transient transfection experiments, and phosphomimic experiments indicated that this phosphorylation event dramatically altered capping enzyme methyltransferase activity and RNA replication 73. Casein kinase 1 α was suggested to be the kinase responsible for serine 56 phosphorylation 74, although serine 56 is in the priming phosphorylation site of the consensus Casein kinase 1 α motif present in NS5 (S56XXS/T59) 75, so Casein kinase 1 α may not be the primary kinase utilized during replication. It is currently unknown of Casein kinase 1 α phosphorylation affects the RNA guanylyltransferase activity. If Casein kinase 1 α does affect NS5 capping enzyme guanylyltransferase activity, there are two potentially interesting outcomes. If phosphorylation down-regulates guanylyltransferase activity in a similar fashion to methyltransferase activity, then phosphorylation may effectively shut off RNA capping during replication. Alternatively phosphorylation may increase RNA guanylyltransferase activity and act as a switch between the enzymatic functions found within the capping enzyme domain. Either scenario would provide valuable insight into the regulation of RNA capping during replication. Protein Kinase G has also been implicated in phosphorylating the NS5 RdRP domain on residue threonine 449 in the RdRP fingers domain 76. Mutation of threonine 449 to histidine or glutamic acid but not serine in a dengue virus replicon aborted replication, indicating that the residue is critical for viral RNA replication. Pharmacologic treatment of dengue 2 infected cells with a Protein Kinase G inhibitor (8-Br-PET-cGMP) blocked viral replication. It is currently unclear what role threonine 449 phosphorylation has on RdRP function, and further studies need to be performed in this area. In each of the above cases it is unknown when these phosphorylation events occur during the RNA replication cycle, and a careful kinetic analysis of NS5 phosphorylation during infection needs to be performed to explicitly determine if phosphorylation is involved in temporal control of NS5 function and switching between negative and positive strand RNA synthesis.
RNA Capping, Methylation, and Structure Controls RNA Stability and Immune Evasion
RNA caps are critical to the function and stability of most translated RNAs. Capped mRNAs are specifically recognized by the translation initiation factor eIF4E in combination with eIF4A and eIF4G to form the eIF4F cap binding complex 77. Association of eIF4F with cap 1 structures is the first step in protein translation initiation and precedes ribosome 40S subunit recruitment. 5′ RNA cap structures block exonucleolytic cleavage of the RNA, increasing their lifespan and stability in cells 78. Additionally, cap 1 structures are used as an antiviral mechanism to discriminate self mRNAs from viral RNAs 79. Each of these properties demonstrates the importance of cap structures in cellular mRNA, and are especially important with viruses whose genomes are capped RNAs. The absence of a fully formed cap on many RNA virus genomes completely stops replication.
What happens to a flavivirus RNA when it is not capped? An obvious answer to this question is that the viral polyprotein would not be translated and the viral RNA would be degraded by the cellular RNA decay machinery. This, however, does not appear to be the whole story. Recent studies have demonstrated that a proportion of the flavivirus positive strand RNAs seem to be intentionally shunted into the RNA decay pathway to affect overall RNA stability and support RNA replication. In 2004 a short fragment of the Japanese Encephalitis virus positive strand RNA was identified by Northern blot that was originally hypothesized to be involved in RNA replication 80. Later work with Kunjin virus defined that the RNA fragment, now called the subgenomic flavivirus RNA (sfRNA), was an incomplete digestion product generated by the cellular 5′ exonuclease XRNI stalling on DB1 structure in the viral 3′ UTR 81. Similar results were demonstrated in yellow fever virus 82. Further work by Moon et al. demonstrated that sfRNA inactivates XRNI and results in increased cellular mRNA accumulation 83. XRN1 and several other P-body components have been found to be recruited to West Nile virus RNA replication sites, but interestingly RNA decapping enzymes such as DCP1 were not associated with RNA replication sites 84. XRN1 acts only on 5′ monophosphorylated RNA substrates, and if XRNI is degrading positive strand RNAs and being inactivated by the sfRNA then a logical hypothesis is that some proportion of positive strand RNAs are either uncapped during normal replication or are specifically targeted to the P-bodies for decapping and subsequent degradation. sfRNA deficient Kunjin viruses replicate to a similar extent as wild-type viruses in cell culture but display severely attenuated pathogenesis in mice 81. For a number of years there has been speculation that one of the flavivirus non-structural proteins interfered with the RNAi response, much like the B2 RNAi suppressor protein encoded by Flock House virus 85, but no such protein-based function has been found to date. Schnettler et al demonstrated that the sfRNA from West Nile virus was able to block siRNA and miRNA mediated gene suppression in mammalian and insect cells through an as yet described mechanism 86, providing an explanation for the partial resistance to RNAi observed during flavivirus infection. The ability of sfRNA to interfere with cellular RNA metabolism pathways suggest that the sfRNA does not play a direct role in RNA replication but is critical for pathogenesis in vivo, potentially by disturbing cellular gene expression during infection.
What role does RNA N7 and 2′O-methylation play during viral replication? N7-methylation of the RNA cap is used by the cellular cap binding protein eIF4E to recruit translation factors and ribosomes to the RNA 77. Without N7 cap methylation eIF4E cannot efficiently recognize the viral RNA and translation and replication is disrupted 33. West Nile virus mutants that selectively disrupt ribose 2′O methylation attenuated but did not completely block RNA replication in cell culture, indicating that 2′O methylation is not required for RNA replication 65. Interestingly, 2′O methylation defective viruses generated protective immunity against wild-type West Nile virus when used as a vaccine in mice. Michael Diamond and Volker Thiel elegantly demonstrated that cap 2′O methylation is used by cells as a molecular signature to discriminate self versus non-self RNAs, with 2′O methylation deficient West Nile virus RNAs being recognized by cellular IFIT proteins as non-self and stimulating robust interferon type I responses 79,87. These data show that RNA 2′O methylation acts as an immune evasion mechanism during flavivirus infection and potentially during other viral infections. More recently, Dong et al demonstrated that internal adenosine residues in the viral genome are 2′O methylated, and that this methylation affects viral translation, RNA replication, and potentially helps avoid the host immune response 88. Therefore, the RNA cap, RNA methylation, and RNA structures play a broader role in flavivirus biology than simply directing viral replication and polyprotein translation.
Conclusion and Perspectives
Our understanding of flavivirus replication has significantly advanced since the first demonstration in 1969 that flavivirus RNA was infectious 89. With the advent of modern molecular biology we have been able to build a conceptual model of how flavivirus genomes enter cells, generate large numbers of daughter genomes, and spread to naïve cells to propagate and cause disease.
However, there are still holes in our understanding of how flaviviruses replicate their genomes that need to be filled if we want to fully understand how these important pathogens replicate and cause disease. Studies of RNA synthesis during infection have focused almost exclusively on negative strand synthesis, and we still have only a very rudimentary understanding of the factors at play during positive strand RNA synthesis. The development of a manipulable in vitro positive strand replication system is required to dissect the molecular details of positive strand RNA synthesis. An in vitro positive strand replication system will likely need to include both full-length NS3 and NS5 and may require membranes and/or other cellular factors to proceed efficiently. The molecular regulation of negative-to-positive strand RNA switching is also an area where limited information exists. Replication events such as NS5 phosphorylation or formation of membranous replication compartments may trigger switching from negative to positive strand RNA synthesis, and further definition of how these events affect RNA replication is warranted. The composition of the ER-derived replication compartments and how RNA replication occurs within the compartments is also unclear. The volume within the replication compartment, which is roughly twice the outer diameter of a mature virion, appears to be large enough to accommodate one genome length RNA 57,58. The space constraints within the replication compartment suggest that there may only be enough space for one negative strand RNA in the replication compartment and that synthesis of positive strand RNAs may need to go to completion to before another round of RNA synthesis can occur. This scenario suggests that only one replication complex is active in each replication compartment and that only one positive strand RNA can be synthesized at a time, with the negative strand recycling within the replication compartment to provide a template for further rounds of positive strand RNA synthesis (Figure 5). This model correlates well with biochemical data presented by Chu and Westaway 18, although further definition of the replication compartment molecular organization is necessary. In summary, even though a good deal of information is now available about how flaviviruses replicate their genomes, regulation of genome replication is still an area with many unanswered questions.
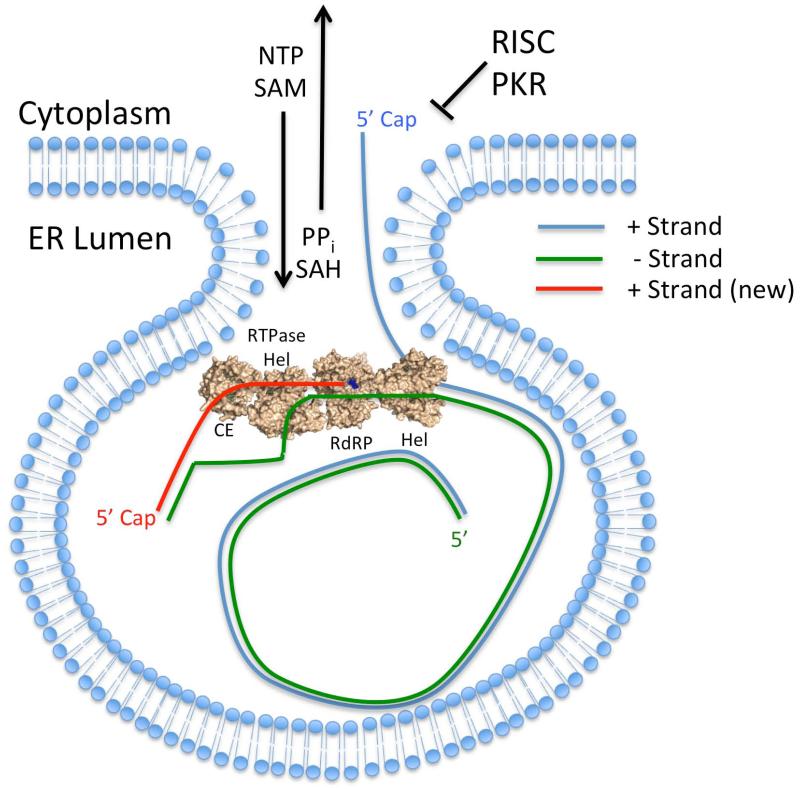
Figure 5. Potential Model for Positive Strand RNA Replication within the Replication Compartment.
Flavivirus RF RNA is entirely contained within replication compartments, which protects the negative strand RNA (colored green) from host antiviral factors such as RISC and PKR. dsRNA may initially interact with a separate NS3 helicase molecule (shown without the protease domain for clarity) that unwinds the RF form dsRNA and directs the original capped positive strand RNA out of the replication compartment for translation, interference with the miRNA and RNA decay pathways, and virion packaging. The 3′ end of unwound negative strand RNA enters the NS5 RdRP domain within the NS3:NS5 replication complex and results in the synthesis of a new capped positive strand RNAs (colored red) as described in Figure 2. The negative strand RNA likely forms a new dsRNA duplex with the nascent positive strand RNA to regenerate the RF form within the replication compartment, and the newly synthesized positive strand RNA would be released from the replication compartment during the next round of positive strand RNA synthesis.
Acknowledgements
We would like to thank Mr. Andy Gardner for assistance with this manuscript. This work was supported by a grant from the Rocky Mountain Regional Center of Excellence (NIH Grant U54 AI065357) to B.G.
Footnotes
No Conflict of Interest
Contributor Information
Bejan J. Saeedi, Department of Gastroenterology, University of Colorado Anschutz Medical Campus, Aurora, CO
Brian J. Geiss, Department of Microbiology, Immunology, and Pathology, Colorado State University, Fort Collins, CO; Department of Biochemistry and Molecular Biology, Colorado State University, Fort Collins, CO.
References
- 1.Pesko KN, Ebel GD. Infect Genet Evol. 2. Vol. 12. Elsevier B.V; Mar 1, 2012. West Nile virus population genetics and evolution; pp. 181–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guzman MG, Halstead SB, Artsob H, Buchy P, Farrar J, Gubler DJ, Hunsperger E, Kroeger A, Margolis HS, Martinez E, Nathan MB, Pelegrino JL, Simmons C, Yoksan S, Peeling RW. Nat Rev Microbiol. 12. Vol. 8. Nature Publishing Group; Dec 1, 2010. Dengue: a continuing global threat; pp. S7–S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellis BR, Barrett ADT. The enigma of yellow fever in East Africa. Rev. Med. Virol. 2008 Sep;18(5):331–46. doi: 10.1002/rmv.584. [DOI] [PubMed] [Google Scholar]
- 4.Cleaves GR, Dubin DT. Methylation status of intracellular dengue type 2 40 S RNA. Virology. 1979 Jul 15;96(1):159–65. doi: 10.1016/0042-6822(79)90181-8. [DOI] [PubMed] [Google Scholar]
- 5.Wengler G, Wengler G, Gross HJ. Studies on virus-specific nucleic acids synthesized in vertebrate and mosquito cells infected with flaviviruses. Virology. 1978 Sep 1;89(2):423–37. doi: 10.1016/0042-6822(78)90185-x. [DOI] [PubMed] [Google Scholar]
- 6.Mason PW. Maturation of Japanese encephalitis virus glycoproteins produced by infected mammalian and mosquito cells. Virology. 1989 Apr;169(2):354–64. doi: 10.1016/0042-6822(89)90161-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindenbach BD, Murray CL, Theil H-J, Rice CM. In: Field’s Virology: Flaviviridae. 6 ed. Knipe D, Howley P, editors. Lippincott WIlliams & Wilkins; 2013. pp. 712–46. [Google Scholar]
- 8.Smit JM, Moesker B, Rodenhuis-Zybert I, Wilschut J. Flavivirus Cell Entry and Membrane Fusion. Viruses. 2011 Feb 22;3(12):160–71. doi: 10.3390/v3020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filomatori CV. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 2006 Aug 15;20(16):2238–49. doi: 10.1101/gad.1444206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu L, Nomaguchi M, Padmanabhan R, Markoff L. Specific requirements for elements of the 5″ and 3″ terminal regions in flavivirus RNA synthesis and viral replication. Virology. 2008 Apr 25;374(1):170–85. doi: 10.1016/j.virol.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filomatori CV, Iglesias NG, Villordo SM, Alvarez DE, Gamarnik AV. RNA Sequences and Structures Required for the Recruitment and Activity of the Dengue Virus Polymerase. Journal of Biological Chemistry. 2011 Mar 4;286(9):6929–39. doi: 10.1074/jbc.M110.162289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iglesias NG, Filomatori CV, Gamarnik AV. The F1 Motif of Dengue Virus Polymerase NS5 Is Involved in Promoter-Dependent RNA Synthesis. J Virol. 2011 May 19;85(12):5745–56. doi: 10.1128/JVI.02343-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selisko B, Potisopon S, Agred R, Priet S, Varlet I, Thillier Y, Sallamand C, Debart F, Vasseur J, Canard B. Molecular Basis for Nucleotide Conservation at the Ends of the Dengue Virus Genome. PLoS Pathog. 2012 Sep 1;8(9):e1002912. doi: 10.1371/journal.ppat.1002912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.You S, Falgout B, Markoff L, Padmanabhan R. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5″- and 3-″terminal regions that influence RNA structure. J Biol Chem. 2001 May 11;276(19):15581–91. doi: 10.1074/jbc.M010923200. [DOI] [PubMed] [Google Scholar]
- 15.Davis WG, Basu M, Elrod EJ, Germann MW, Brinton MA. Identification of cis-Acting Nucleotides and a Structural Feature in West Nile Virus 3′-Terminus RNA That Facilitate Viral Minus Strand RNA Synthesis. J Virol. 2013 Jun 10;87(13):7622–36. doi: 10.1128/JVI.00212-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friebe P, Harris E. Interplay of RNA Elements in the Dengue Virus 5″ and 3″ Ends Required for Viral RNA Replication. J Virol. 2010 Jun 15;84(12):6103–18. doi: 10.1128/JVI.02042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friebe P, Peña J, Pohl MOF, Harris E. Virology. 2. Vol. 422. Elsevier Inc; Jan 20, 2012. Composition of the sequence downstream of the dengue virus 5′ cyclization sequence (dCS) affects viral RNA replication; pp. 346–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chu PW, Westaway EG. Replication strategy of Kunjin virus: evidence for recycling role of replicative form RNA as template in semiconservative and asymmetric replication. Virology. 1985 Jan 15;140(1):68–79. doi: 10.1016/0042-6822(85)90446-5. [DOI] [PubMed] [Google Scholar]
- 19.Falgout B, Pethel M, Zhang YM, Lai CJ. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J Virol. 1991 May;65(5):2467–75. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackenzie JM, Khromykh AA, Jones MK, Westaway EG. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology. 1998 Jun 5;245(2):203–15. doi: 10.1006/viro.1998.9156. [DOI] [PubMed] [Google Scholar]
- 21.Lindenbach BD, Rice CM. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J Virol. 1999 Jun;73(6):4611–21. doi: 10.1128/jvi.73.6.4611-4621.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erbel P, Schiering N, D’Arcy A, Renatus M, Kroemer M, Lim SP, Yin Z, Keller TH, Vasudevan SG, Hommel U. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat Struct Mol Biol. 2006 Apr 1;13(4):372–3. doi: 10.1038/nsmb1073. [DOI] [PubMed] [Google Scholar]
- 23.Miller S, Sparacio S, Bartenschlager R. Subcellular localization and membrane topology of the Dengue virus type 2 Non-structural protein 4B. J Biol Chem. 2006 Mar 31;281(13):8854–63. doi: 10.1074/jbc.M512697200. [DOI] [PubMed] [Google Scholar]
- 24.Chambers TJ, Weir RC, Grakoui A, McCourt DW, Bazan JF, Fletterick RJ, Rice CM. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proc Natl Acad Sci USA. 1990 Nov 1;87(22):8898–902. doi: 10.1073/pnas.87.22.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lain S, Riechmann JL, Martin MT, Garcia JA. Homologous potyvirus and flavivirus proteins belonging to a superfamily of helicase-like proteins. Gene. 1989 Oct;82(2):357–62. doi: 10.1016/0378-1119(89)90063-2. [DOI] [PubMed] [Google Scholar]
- 26.Wu J, Bera AK, Kuhn RJ, Smith JL. Structure of the Flavivirus Helicase: Implications for Catalytic Activity, Protein Interactions, and Proteolytic Processing. J Virol. 2005 Jul 28;79(16):10268–77. doi: 10.1128/JVI.79.16.10268-10277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gebhard LG, Kaufman SB, Gamarnik AV. Marcello A, editor. Novel ATP-Independent RNA Annealing Activity of the Dengue Virus NS3 Helicase. PLoS ONE. 2012 Apr 30;7(4):e36244. doi: 10.1371/journal.pone.0036244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warrener P, Tamura JK, Collett MS. RNA-stimulated NTPase activity associated with yellow fever virus NS3 protein expressed in bacteria. J Virol. 1993 Feb 1;67(2):989–96. doi: 10.1128/jvi.67.2.989-996.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wengler G. The NS 3 nonstructural protein of flaviviruses contains an RNA triphosphatase activity. Virology. 1993 Nov 1;197(1):265–73. doi: 10.1006/viro.1993.1587. [DOI] [PubMed] [Google Scholar]
- 30.Benarroch D, Selisko B, Locatelli G, Maga G, Romette J, Canard B. The RNA helicase, nucleotide 5″-triphosphatase, and RNA 5-″triphosphatase activities of Dengue virus protein NS3 are Mg-dependent and require a functional Walker B motif in the helicase catalytic core. Virology. 2004 Oct 25;328(2):208–18. doi: 10.1016/j.virol.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 31.Koonin EV. Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and lambda 2 protein of reovirus. J Gen Virol. 1993 Apr 1;74(Pt 4):733–40. doi: 10.1099/0022-1317-74-4-733. [DOI] [PubMed] [Google Scholar]
- 32.Egloff M-P, Benarroch D, Selisko B, Romette J-L, Canard B. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 2002 Jun 3;21(11):2757–68. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, Dong H, Deas TS, Zhou Y, Li H, Shi PY. West Nile virus 5″-cap structure is formed by sequential guanine N-7 and ribose 2-″O methylations by nonstructural protein 5. J Virol. 2006 Sep 1;80(17):8362–70. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Issur M, Geiss BJ, Bougie I, Picard-Jean F, Despins S, Mayette J, Hobdey SE, Bisaillon M. The flavivirus NS5 protein is a true RNA guanylyltransferase that catalyzes a two-step reaction to form the RNA cap structure. RNA. 2009 Dec;15(12):2340–50. doi: 10.1261/rna.1609709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grun JB, Brinton MA. Characterization of West Nile virus RNA-dependent RNA polymerase and cellular terminal adenylyl and uridylyl transferases in cell-free extracts. J Virol. 1986 Dec;60(3):1113–24. doi: 10.1128/jvi.60.3.1113-1124.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johansson M, Brooks AJ, Jans DA, Vasudevan SG. A small region of the dengue virus-encoded RNA-dependent RNA polymerase, NS5, confers interaction with both the nuclear transport receptor importin-beta and the viral helicase, NS3. J Gen Virol. 2001 Apr 1;82(Pt 4):735–45. doi: 10.1099/0022-1317-82-4-735. [DOI] [PubMed] [Google Scholar]
- 37.Brooks AJ. The Interdomain Region of Dengue NS5 Protein That Binds to the Viral Helicase NS3 Contains Independently Functional Importin beta 1 and Importin alpha /beta - Recognized Nuclear Localization Signals. Journal of Biological Chemistry. 2002 Sep 20;277(39):36399–407. doi: 10.1074/jbc.M204977200. [DOI] [PubMed] [Google Scholar]
- 38.Forwood JK, Brooks A, Briggs LJ, Xiao CY, Jans DA, Vasudevan SG. The 37-amino-acid interdomain of dengue virus NS5 protein contains a functional NLS and inhibitory CK2 site. Biochem Biophys Res Commun. 1999 Apr 21;257(3):731–7. doi: 10.1006/bbrc.1999.0370. [DOI] [PubMed] [Google Scholar]
- 39.Selisko B, Dutartre H, Guillemot J-C, Debarnot C, Benarroch D, Khromykh A, Despres P, Egloff M, Canard B. Comparative mechanistic studies of de novo RNA synthesis by flavivirus RNA-dependent RNA polymerases. Virology. 2006 Jul 20;351(1):145–58. doi: 10.1016/j.virol.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 40.Malet H, Egloff M-P, Selisko B, Butcher RE, Wright PJ, Roberts M, Gruez A, Sulzenbacher G, Vonrhein C, Bricogne G, Mackenzie JM, Khromykh AA, Davidons AD, Canard B. Crystal Structure of the RNA Polymerase Domain of the West Nile Virus Non-structural Protein 5. Journal of Biological Chemistry. 2007 Feb 2;282(14):10678–89. doi: 10.1074/jbc.M607273200. [DOI] [PubMed] [Google Scholar]
- 41.Cui T, Sugrue RJ, Xu Q, Lee AK, Chan YC, Fu J. Recombinant dengue virus type 1 NS3 protein exhibits specific viral RNA binding and NTPase activity regulated by the NS5 protein. Virology. 1998 Jul;246(2):409–17. doi: 10.1006/viro.1998.9213. [DOI] [PubMed] [Google Scholar]
- 42.Yon C, Teramoto T, Mueller N, Phelan J, Ganesh VK, Murthy KHM, Padmanabhan R. Modulation of the nucleoside triphosphatase/RNA helicase and 5′-RNA triphosphatase activities of Dengue virus type 2 nonstructural protein 3 (NS3) by interaction with NS5, the RNA-dependent RNA polymerase. J Biol Chem. 2005 Jul 22;280(29):27412–9. doi: 10.1074/jbc.M501393200. [DOI] [PubMed] [Google Scholar]
- 43.Kapoor M, Zhang L, Ramachandra M, Kusukawa J, Ebner KE, Padmanabhan R. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J Biol Chem. 1995 Aug 11;270(32):19100–6. doi: 10.1074/jbc.270.32.19100. [DOI] [PubMed] [Google Scholar]
- 44.Chen CJ, Kuo MD, Chien LJ, Hsu SL, Wang YM, Lin JH. RNA-protein interactions: involvement of NS3, NS5, and 3′ noncoding regions of Japanese encephalitis virus genomic RNA. J Virol. 1997 May 1;71(5):3466–73. doi: 10.1128/jvi.71.5.3466-3473.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vasudevan SG, Johansson M, Brooks AJ, Llewellyn LE, Jans DA. Characterisation of inter- and intra-molecular interactions of the dengue virus RNA dependent RNA polymerase as potential drug targets. Farmaco. 2001;56(1-2):33–6. doi: 10.1016/s0014-827x(01)01014-x. [DOI] [PubMed] [Google Scholar]
- 46.Kroschewski H, Lim SP, Butcher RE, Yap TL, Lescar J, Wright PJ, Vasudevan SG, Davidson AD. Mutagenesis of the dengue virus type 2 NS5 methyltransferase domain. J Biol Chem. 2008 Jul 11;283(28):19410–21. doi: 10.1074/jbc.M800613200. [DOI] [PubMed] [Google Scholar]
- 47.Stohlman SA, Wisseman CL, Eylar OR, Silverman DJ. Dengue virus-induced modifications of host cell membranes. J Virol. 1975 Oct;16(4):1017–26. doi: 10.1128/jvi.16.4.1017-1026.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ng ML, Pedersen JS, Toh BH, Westaway EG. Immunofluorescent sites in vero cells infected with the flavivirus Kunjin. Arch Virol. 1983;78(3-4):177–90. doi: 10.1007/BF01311313. [DOI] [PubMed] [Google Scholar]
- 49.Chu PW, Westaway EG. Molecular and ultrastructural analysis of heavy membrane fractions associated with the replication of Kunjin virus RNA. Arch Virol. 1992;125(1-4):177–91. doi: 10.1007/BF01309636. [DOI] [PubMed] [Google Scholar]
- 50.Mackenzie JM, Jones MK, Young PR. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology. 1996 Jun 1;220(1):232–40. doi: 10.1006/viro.1996.0307. [DOI] [PubMed] [Google Scholar]
- 51.Mackenzie JM, Kenney MT, Westaway EG. West Nile virus strain Kunjin NS5 polymerase is a phosphoprotein localized at the cytoplasmic site of viral RNA synthesis. Journal of General Virology. 2007 Apr 1;88(4):1163–8. doi: 10.1099/vir.0.82552-0. [DOI] [PubMed] [Google Scholar]
- 52.Westaway EG, Khromykh AA, Mackenzie JM. Nascent flavivirus RNA colocalized in situ with double-stranded RNA in stable replication complexes. Virology. 1999 May 25;258(1):108–17. doi: 10.1006/viro.1999.9683. [DOI] [PubMed] [Google Scholar]
- 53.Westaway EG, Mackenzie JM, Kenney MT, Jones MK, Khromykh AA. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J Virol. 1997 Sep;71(9):6650–61. doi: 10.1128/jvi.71.9.6650-6661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Uchil PD. Architecture of the Flaviviral Replication Complex: PROTEASE, NUCLEASE, AND DETERGENTS REVEAL ENCASEMENT WITHIN DOUBLE-LAYERED MEMBRANE COMPARTMENTS. Journal of Biological Chemistry. 2003 May 2;278(27):24388–98. doi: 10.1074/jbc.M301717200. [DOI] [PubMed] [Google Scholar]
- 55.Geiss BJ, Pierson TC, Diamond MS. Actively replicating West Nile virus is resistant to cytoplasmic delivery of siRNA. Virol J. 2005 Jun 28;2:53. doi: 10.1186/1743-422X-2-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoenen A, Liu W, Kochs G, Khromykh AA, Mackenzie JM. West Nile virus-induced cytoplasmic membrane structures provide partial protection against the interferon-induced antiviral MxA protein. Journal of General Virology. 2007 Nov 1;88(11):3013–7. doi: 10.1099/vir.0.83125-0. [DOI] [PubMed] [Google Scholar]
- 57.Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CKE, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. Composition and Three-Dimensional Architecture of the Dengue Virus Replication and Assembly Sites. Cell Host and Microbe. 2009 Jan 4;5(4):365–75. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gillespie LK, Hoenen A, Morgan G, Mackenzie JM. The Endoplasmic Reticulum Provides the Membrane Platform for Biogenesis of the Flavivirus Replication Complex. J Virol. 2010 Oct 15;84(20):10438–47. doi: 10.1128/JVI.00986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miorin L, Romero-Brey I, Maiuri P, Hoppe S, Krijnse-Locker J, Bartenschlager R, Marcello A. Three Dimensional Architecture of Tick-borne Encephalitis Virus Replication Sites and Trafficking of the Replicated RNA. J Virol. 2013 Apr 3; doi: 10.1128/JVI.03456-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller S, Kastner S, Krijnse-Locker J, Bühler S, Bartenschlager R. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J Biol Chem. 2007 Mar 23;282(12):8873–82. doi: 10.1074/jbc.M609919200. [DOI] [PubMed] [Google Scholar]
- 61.Roosendaal J, Westaway EG, Khromykh A, Mackenzie JM. Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. J Virol. 2006 May 1;80(9):4623–32. doi: 10.1128/JVI.80.9.4623-4632.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perera R, Riley C, Isaac G, Hopf-Jannasch AS, Moore RJ, Weitz KW, Pasa-Tolic L, Metz TO, Adamec J, Kuhn R. Dengue Virus Infection Perturbs Lipid Homeostasis in Infected Mosquito Cells. PLoS Pathog. 2012 Mar 22;8(3):e1002584. doi: 10.1371/journal.ppat.1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heaton NS, Perera R, Berger KL, Khadka S, LaCount DJ, Kuhn RJ, Randall G. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proceedings of the National Academy of Sciences. 2010 Oct 5;107(40):17345–50. doi: 10.1073/pnas.1010811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rothwell C, LeBreton A, Ng CY, Lim JYH, Liu W, Vasudevan S, Labow M, Gu F, Gaither LA. Virology. 1-2. Vol. 389. Elsevier Inc; Jun 20, 2009. Cholesterol biosynthesis modulation regulates dengue viral replication; pp. 8–19. [DOI] [PubMed] [Google Scholar]
- 65.Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard KA, Shi PY, Li H. Structure and function of flavivirus NS5 methyltransferase. J Virol. 2007 Apr 1;81(8):3891–903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Geiss BJ, Thompson AA, Andrews AJ, Sons RL, Gari HH, Keenan SM, Peersen OB. Analysis of flavivirus NS5 methyltransferase cap binding. J Mol Biol. 2009 Feb 6;385(5):1643–54. doi: 10.1016/j.jmb.2008.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu T, Sampath A, Chao A, Wen D, Nanao M, Chene P, Vasudevan SG, Lescar J. Structure of the Dengue virus helicase/nucleoside triphosphatase catalytic domain at a resolution of 2.4 A. J Virol. 2005 Aug 1;79(16):10278–88. doi: 10.1128/JVI.79.16.10278-10288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bartelma G, Padmanabhan R. Expression, purification, and characterization of the RNA 5′-triphosphatase activity of dengue virus type 2 nonstructural protein 3. Virology. 2002 Jul 20;299(1):122–32. doi: 10.1006/viro.2002.1504. [DOI] [PubMed] [Google Scholar]
- 69.Luo D, Xu T, Watson RP, Scherer-Becker D, Sampath A, Jahnke W, Yeong SS, Wang CH, Lim SP, Strongin A, Vasudevan SG, Lescar J. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 2008 Nov 13;27(23):3209–19. doi: 10.1038/emboj.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mastrangelo E, Bolognesi M, Milani M. Biochem Biophys Res Commun. 1. Vol. 417. Elsevier Inc; Jan 6, 2012. Flaviviral helicase: Insights into the mechanism of action of a motor protein; pp. 84–7. [DOI] [PubMed] [Google Scholar]
- 71.Dong H, Ren S, Li H, Shi PY. Separate molecules of West Nile virus methyltransferase can independently catalyze the N7 and 2′-O methylations of viral RNA cap. Virology. 2008 Jul 20;377(1):1–6. doi: 10.1016/j.virol.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Uchil PD, Kumar AVA, Satchidanandam V. Nuclear Localization of Flavivirus RNA Synthesis in Infected Cells. J Virol. 2006 Jun 1;80(11):5451–64. doi: 10.1128/JVI.01982-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bhattacharya D, Hoover S, Falk S, Weisblum B, Vestling M, Striker R. Phosphorylation of yellow fever virus NS5 alters methyltransferase activity. Virology. 2008 Oct 25;380(2):276–84. doi: 10.1016/j.virol.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bhattacharya D, Ansari IH, Striker R. The flaviviral methyltransferase is a substrate of Casein Kinase 1. Virus Res. 2009 Apr 1;141(1):101–4. doi: 10.1016/j.virusres.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Flotow H, Graves PR, Wang AQ, Fiol CJ, Roeske RW, Roach PJ. Phosphate groups as substrate determinants for casein kinase I action. J Biol Chem. 1990 Aug 25;265(24):14264–9. [PubMed] [Google Scholar]
- 76.Bhattacharya D, Mayuri, Best SM, Perera R, Kuhn RJ, Striker R. Protein Kinase G Phosphorylates Mosquito-Borne Flavivirus NS5. J Virol. 2009 Sep 15;83(18):9195–205. doi: 10.1128/JVI.00271-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gingras A-C, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–63. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 78.Coutts M, Krowczynska A, Brawerman G. Protection of mRNA against nucleases in cytoplasmic extracts of mouse sarcoma ascites cells. Biochim Biophys Acta. 1993 Apr 29;1173(1):49–56. doi: 10.1016/0167-4781(93)90241-5. [DOI] [PubMed] [Google Scholar]
- 79.Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, Siddell SG, Ludewig B, Thiel V. Nature Publishing Group. 2. Vol. 12. Nature Publishing Group; Jan 9, 2011. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5; pp. 137–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin KC, Chang HL, Chang RY. Accumulation of a 3′-Terminal Genome Fragment in Japanese Encephalitis Virus-Infected Mammalian and Mosquito Cells. J Virol. 2004 Apr 27;78(10):5133–8. doi: 10.1128/JVI.78.10.5133-5138.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Funk A, Truong K, Nagasaki T, Torres S, Floden N, Balmori-Melian E, Edmonds J, Dong H, Shi PY, Khromykh RNA Structures Required for Production of Subgenomic Flavivirus RNA. J Virol. 2010 Oct 7;84(21):11407–17. doi: 10.1128/JVI.01159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Silva PAGC, Pereira CF, Dalebout TJ, Spaan WJM, Bredenbeek PJ. An RNA Pseudoknot Is Required for Production of Yellow Fever Virus Subgenomic RNA by the Host Nuclease XRN1. J Virol. 2010 Nov 1;84(21):11395–406. doi: 10.1128/JVI.01047-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moon SL, Anderson JR, Kumagai Y, Wilusz CJ, Akira S, Khromykh AA, Wilusz J. A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. RNA. 2012 Oct 16;18(11):2029–40. doi: 10.1261/rna.034330.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chahar HS, Chen S, Manjunath N. Virology. 1. Vol. 436. Elsevier; Feb 5, 2013. P-body components LSM1, GW182, DDX3, DDX6 and XRN1 are recruited to WNV replication sites and positively regulate viral replication; pp. 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lu R, Maduro M, Li F, Li HW, Broitman-Maduro G, Li WX, Ding SW. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature. 2005 Aug 18;436(7053):1040–3. doi: 10.1038/nature03870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schnettler E, Sterken MG, Leung JY, Metz SW, Geertsema C, Goldbach RW, Vlak JM, Kohl A, Khromykh AA, Pijlman GP. Noncoding Flavivirus RNA Displays RNA Interference Suppressor Activity in Insect and Mammalian Cells. J Virol. 2012 Nov 19;86(24):13486–500. doi: 10.1128/JVI.01104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong H, Thiel V, Sen GC, Fensterl V, Klimstra WB, Pierson TC, Buller RM, Gale M, Shi PY, Diamond MS. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010 Nov 18;468(7322):452–6. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dong H, Chang DC, Hua MHC, Lim SP, Chionh YH, Hia F, Lee YH, Kukkaro P, Lok S, Dedon PC, Shi PY. Kuhn RJ, editor. 2′-O Methylation of Internal Adenosine by Flavivirus NS5 Methyltransferase. PLoS Pathog. 2012 Apr 5;8(4):e1002642. doi: 10.1371/journal.ppat.1002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Peleg J. Behaviour of infectious RNA from four different viruses in continuously subcultured Aedes aegypti mosquito embryo cells. Nature. 1969 Jan;221(5176):193–4. doi: 10.1038/221193a0. [DOI] [PubMed] [Google Scholar]
- 90.Yap TL, Xu T, Chen Y-L, Malet H, Egloff M-P, Canard B, Vasudevan SG, Lescar J. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J Virol. 2007 May 1;81(9):4753–65. doi: 10.1128/JVI.02283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]