Background: MicroRNAs have critical roles in T cell development under normal and stress conditions.
Results: Increasing miR-185 levels attenuate T cell development via the targeting of Mzb1, Nfatc3, and Camk4.
Conclusion: Elevations in miR-185 impair thymopoiesis at several developmental stages.
Significance: miR-185 levels are reduced in 22q11.2 deletion syndrome patients and the identification of its gene targets is clinically informative.
Keywords: Calcium Intracellular Release, Genetic Diseases, Immunodeficiency, MicroRNA, T Cell Biology, Transcription Target Genes, Transgenic Mice, 22q11.2 Deletion Syndrome
Abstract
miR-185 is a microRNA (miR) that targets Bruton's tyrosine kinase in B cells, with reductions in miR-185 linked to B cell autoantibody production. In hippocampal neurons, miR-185 targets both sarcoplasmic/endoplasmic reticulum calcium ATPase 2 and a novel Golgi inhibitor. This miR is haploinsufficient in 90–95% of individuals with chromosome 22q11.2 deletion syndrome, patients who can present with immune, cardiac, and parathyroid problems, learning disorders, and a high incidence of schizophrenia in adults. The reduced levels of miR-185 in neurons cause presynaptic neurotransmitter release. Many of the 22q11.2 deletion syndrome patients have a thymic hypoplasia, which results in a peripheral T cell lymphopenia and unusual T helper cell skewing. The molecular targets of miR-185 in thymocytes are unknown. Using an miR-185 T cell transgenic approach, increasing levels of miR-185 attenuated T cell development at the T cell receptor β (TCRβ) selection checkpoint and during positive selection. This caused a peripheral T cell lymphopenia. Mzb1, Nfatc3, and Camk4 were identified as novel miR-185 targets. Elevations in miR-185 enhanced TCR-dependent intracellular calcium levels, whereas a knockdown of miR-185 diminished these calcium responses. These effects concur with reductions in Mzb1, an endoplasmic reticulum calcium regulator. Consistent with their haploinsufficiency of miR-185, Mzb1 levels were elevated in thymocyte extracts from several 22q11.2 deletion syndrome patients. Our findings indicate that miR-185 regulates T cell development through its targeting of several mRNAs including Mzb1.
Introduction
MicroRNAs (miRs)2 are small noncoding RNAs (20–24 nucleotides in length) that regulate gene expression by causing mRNA degradation and/or translational inhibition (1, 2). Functional roles of the miRs in the immune system are being partly elucidated with loss- and gain-of-function approaches. For example, conditional knock-out lines of Dicer, an RNase III enzyme critical for miR biogenesis, in thymic epithelial cells causes a premature thymic involution, in part via the diminished expression of miR-29 (3). The elimination of Dicer at the DN3 stage (CD4−CD8−CD44−CD25+) of thymopoiesis reduces thymic cellularity (4). For pro-B cells, this knock-out impairs their transition to pre-B cells, whereas its loss in mature CD19+ B cells causes B cell autoantibody production (5, 6). This was linked to the loss of miR-185 and the up-regulation of its target, Bruton's tyrosine kinase (Btk) (5). Gain-of-function approaches have uncovered roles for miR-146 and miR-155 in regulating IL-2 production in T cell and increasing NK cell numbers and cytokine production, respectively (7, 8).
miR-185 is a stress-responsive miR expressed in the thymus (9). It is encoded on human chromosome 22q11.2 and is haploinsufficient in 22q11.2/DiGeorge syndrome patients (10). Such patients can have a thymic hypoplasia, hypoparathyroidism, cardiac anomalies, and/or learning disabilities (11). Some patients have an increased frequency of autoimmune disorders and T helper cell alterations (12, 13). One-third of 22q11.2 deletion syndrome patients will develop schizophrenia as adults (14). Individuals with a duplication of chromosome 22q11.2 (trisomy 22q11.2) can have similar clinical presentations, suggesting that both reductions and elevations in miR-185 are clinically relevant (15, 16). Mouse models of this syndrome have shown that the haploinsufficiency of miR-185 elevates the expression of one of its neuronal targets, SERCA2 (Sarcoplasmic/endoplasmic reticulum calcium ATPase 2) (17). High levels of SERCA2 enhance presynaptic neurotransmitter release in hippocampal neurons (17). This contributes to an age-dependent cognitive impairment. A second target increased in neurons is 2310044H10Rik, a Golgi-associated inhibitor (18). Although many miR-185 targets have been identified, it remains unknown whether there are additional targets in the immune system.
We developed transgenic mice with increasing levels of miR-185 expressed in thymocytes and peripheral T cells. This attenuated both pre-TCR and positive selection, with an ensuing peripheral T cell lymphopenia. Several novel targets of miR-185 were identified, including marginal zone B and B1 cell-specific protein (Mzb1), nuclear factor of activated T cells, cytoplasmic, calcineurin-dependent 3 (Nfatc3), and calcium/calmodulin-dependent protein kinase IV (Camk4). miR-185 transgenic thymocytes had higher levels of calcium influx upon TCR stimulation. A knockdown of miR-185 resulted in reduced TCR-driven calcium responses, with corresponding increases in Mzb1. The protein levels of Mzb1 were also elevated in thymocytes from many 22q11.2 deletion syndrome samples. Taken together, our findings indicate that alterations in the expression of miR-185 can affect T cell development and activation by controlling the expression of several novel mRNA targets including Mzb1.
EXPERIMENTAL PROCEDURES
Mouse Lines
The miR-185 transgenic (Tg) lines were generated by the University of Texas Southwestern Medical Center Transgenic Core facility. The VA-hCD2 transgenic cassette containing 600 bp of genomic DNA with miR-185 was injected into C57BL/6 fertilized eggs. miR-185 expression was confirmed by Southern blotting, Northern blotting, and PCR. OTII/miR-185 double Tg mice were obtained from crosses between the OTII Tg line and miR-185 Tg-35. All mouse procedures were carried out in accordance with the Institutional Animal Care and Use Committee at the University of Texas Southwestern Medical Center (IACUC number 2010-0053). Animal use adheres to applicable requirements such as the Animal Welfare Act, the Guide for the Care and Use of Laboratory Animals, and the United States Government Principals regarding the care and use of animals. Unless otherwise indicated, mice 5–7 weeks of age were used in all experiments.
Human Thymus Tissues
Human thymus was obtained from patients undergoing corrective heart surgery at Children's Medical Center in Dallas, TX from 2012 to 2013. Informed consent was obtained for all patients and control subjects (Institutional Review Board number 072010-003).
Cell Isolation, Culture, and Flow Cytometry
Lymphocytes were prepared as described previously (19). Antibodies used were from BD Biosciences unless otherwise indicated. CD44−CD25+ (CD4−CD8−TCRγδ−NK1.1−B220−CD11b−CD11c−) DN3 thymocytes were sorted with a Mo-Flo high speed cell sorter (Cytomation). Intracellular TCRβ staining was performed using Cytoperm/Cytofix kit (BD Biosciences). Quantification of apoptosis was determined by staining with annexin V antibody. Thymocytes from OTII Tg and OTII/miR-185 Tg-35 mice were isolated and incubated (2 × 106 cells/ml; 24-well plate) with 10 μm SIINFEKL or OVA class II peptides for 20 h at 37 °C. Live percentages of DP thymocytes were determined by gating on annexin V− 7-aminoactinomycin D− cells. Natural regulatory T cells were stained using the mouse regulatory T cell staining kit (eBioscience). Total CD4+ T cells (5 × 105 cells/ml), purified from the lymph nodes using magnetic beads (BD Biosciences), were stimulated with anti-CD3ϵ (3 mg/ml; 145.2C11) and anti-CD28 (3 mg/ml; 37.51). After a 48-h culture period, IL-2 levels were measured using the mouse IL-2 ELISA MAX kit (BioLegend).
RNA Analysis
Northern blotting was performed as described previously (9, 20). For real-time PCR, RNA was isolated using the Ambion RNAqueous micro kit (Invitrogen) and treated with DNase. cDNA was made from 100 ng of RNA using the High Capacity cDNA reverse transcription kit (Applied Biosystems). The real-time PCR steps were performed with 5 ng of cDNA, 2× Maxima SYBR Green quantitative PCR master mix, and 100 nm ROX (5′-carboxy-X-rhodamine) passive reference dye (Thermo Scientific). The real-time cycling parameters were as follows: 1 cycle at 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s on an ABI 7300 series PCR machine. Relative expression was calculated by the comparative threshold method (ΔΔCT).
Microarray Analyses
RNA was isolated from DN3 cells that were sorted from normal C57BL/6 mice or the miR-185 Tg-6 lines (n = 6 mice pooled). cDNA synthesis and hybridization onto Illumina SingleColor MouseWG-6_V2_0_R1 platform were performed by the University of Texas Southwestern (UTSW) Genomics and Microarray Core Facility. GenomeStudio Data Analysis software version 1.8.0 was used. Using GeneSpring GX 11 version 4.0 (Agilent Technologies), quantile normalization of all samples was performed to obtain a gene-level expression file. Next, unpaired Student's t test was performed to identify significantly (p < 0.05) deregulated genes among the wild type and miR-185 Tg samples.
MicroRNA Target Validation
miR-185 (∼600-bp genomic DNA) was cloned into pCDNA3.1 (Invitrogen). The 3′-untranslated regions (3′-UTRs) of target genes (Btk, Mzb1, Mcm10, Camk4, Hmga1, Nfatc3, Igf1r, and Dusp4) were amplified by PCR and ligated into the firefly luciferase reporter construct (pMIR-REPORT, Invitrogen). Reporter constructs and a β-galactosidase vector were co-transfected into COS-1 cells (1 × 105 cells/ml; 24-well plate) along with either pCDNA3.1 or pCDNA3.1-miR-185, using the FuGENE 6 transfection reagent (Roche Applied Science). Cells were processed using the luciferase assay kit (Promega). Relative luciferase activity was calculated by normalizing the firefly luciferase to the β-galactosidase. The murine Mzb1 coding sequences (CDS) were cloned into the pEF1/myc-His B plasmid (Invitrogen). Transfections were done in HEK293T cells (1 × 105 cells/ml; 24-well plate) using the X-tremeGENE 9 DNA kit (Roche Applied Science). Mutations in the 3′-UTR and CDS of Mzb1 were introduced with QuikChange site-directed mutagenesis kit (Stratagene).
MicroRNA Knockdown and Immunoblotting
Jurkat T cells (2–2.5 × 105 cells/ml) were transfected with either miR control inhibitor (microRNA Hairpin Inhibitor Negative Control 1, cel-miR-67) or miR-185 inhibitor (10–40 nm, miRIDIAN, Thermo Scientific) using PepMute siRNA transfection reagent (SignaGen). Immunoblotting was performed as described previously with the following antibodies: Mzb1 (11454-1-AP, Proteintech), NFATc3 (SC-8321, Santa Cruz Biotechnology), β-actin (4967, Cell Signaling), GFP (632380, Clontech), Myc epitope (2272, Cell Signaling), anti-rabbit HRP-conjugated secondary antibody, and anti-mouse IgG HRP-conjugated secondary antibody (19). Expression levels were quantified using the ImageJ software (version 1.46r). X-ray films, developed using chemiluminescence, were scanned with the Canon CanoScan 8800r. They were saved as TIFF images with a resolution of 300 dpi. These files were converted to 8-bit grayscale images using ImageJ software. Band quantifications were performed according to the vendor's instructions. For the Western blots, multiple exposures were obtained.
Measurement of Intracellular Calcium Responses
Thymocytes (1 × 107 cells/ml) were loaded with Fluo-3-AM (4 μm, Invitrogen) in 1× Hanks' balanced salt solution (Cellgro) and incubated at 37 °C for 30 min. Base-line fluorescence was monitored for 45 s at 37 °C before adding biotinylated anti-CD3ϵ and anti-CD4 (1 μg/ml, BioLegend). The fluorescence intensity was measured for 45 s followed by streptavidin (2 μg/ml) and monitoring for an additional 330 s. Jurkat T cells were stimulated with the anti-clonotypic antibody, C305.2. Maximal calcium responses were determined by adding ionomycin (1 μm), which was quenched by the addition of MnCl2 (1 mm). In certain experiments, the SERCA pump inhibitor thapsigargin (1 μm, Invitrogen) and ionomycin (2 μm) were added onto Fluo-3 AM-loaded thymocytes that had been washed and resuspended in calcium-free medium. All sample acquisition was performed at 37 °C.
Statistical Analyses
GraphPad Prism Software was used to calculate mean values, S.D., and S.E. and to perform statistical analyses. The statistical significance was designated with asterisks (*, p < 0.05, **, p < 0.01, and ***, p < 0.001), and p more than 0.05 was considered as nonsignificant.
RESULTS
Elevations in miR-185 Attenuate T Cell Development
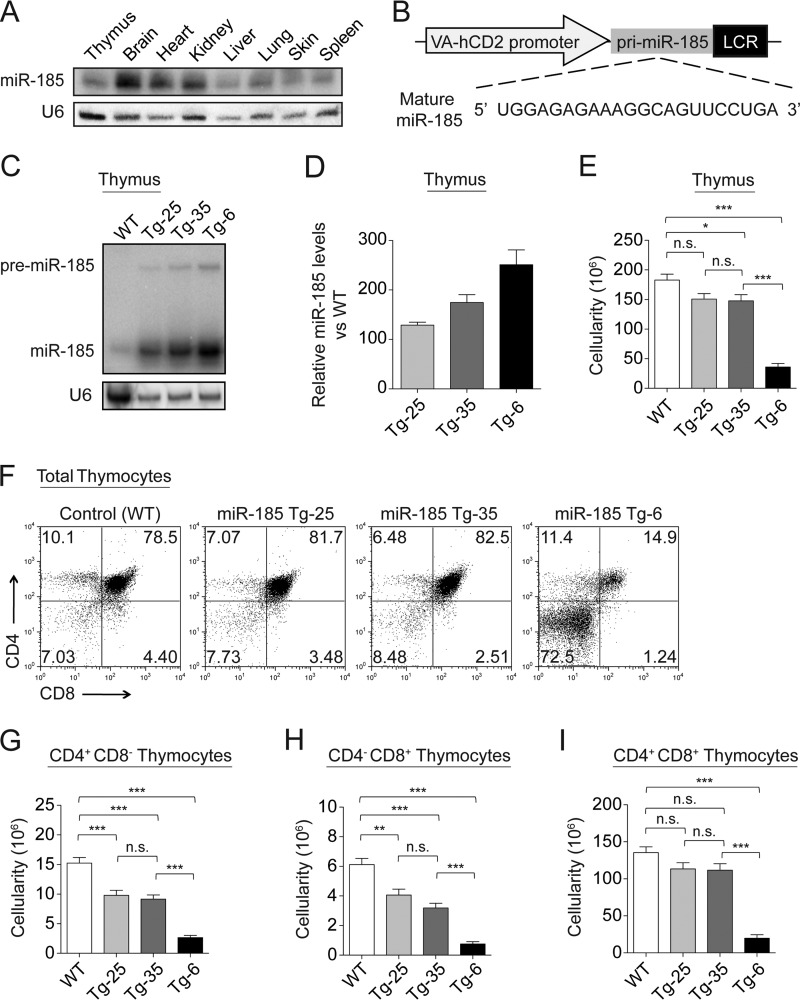
miR-185 is highly conserved and expressed in most tissues including the thymus, brain, heart, kidney, liver, lung, skin, and spleen (Fig. 1A). It is expressed in immature thymocytes, mature CD4+CD8− and CD4−CD8+ T cells, and B cells (5, 9, 21, 22). To determine whether miR-185 levels impact T cell functions, we utilized a gain-of-function transgenic approach (23) in which the murine primary miR-185 (pri-miR-185) was overexpressed in thymocytes and peripheral T cells (Fig. 1B). Three transgenic lines, designated as miR-185 Tg-25, miR-185 Tg-35, and miR-185 Tg-6, were selected based on their increasing levels of miR-185 expression. miR-185 was overexpressed 130-, 175-, and 250-fold in Tg-25, Tg-35, and Tg-6 lines, respectively, when compared with nontransgenic littermates (Fig. 1, C and D). Overall thymic cellularity was slightly reduced in Tg-25 and Tg-35 lines, whereas a severe thymic hypoplasia was noted in the Tg-6 line (Fig. 1E). Increasing levels of miR-185 caused a statistically significant decrease in the percentage and number of CD4+CD8− and CD4−CD8+ SP cells when compared with control WT mice, suggesting an impairment at the DP stage (Fig. 1, F–H). The highest overexpressing line, miR-185 Tg-6, had statistically significant reductions in both the DP and the SP subsets, reflected as a dramatic loss in overall thymic cellularity (Fig. 1, E–I). Elevated DN percentages in the Tg-6 line further indicated an attenuated early thymopoiesis (Fig. 1F). The Tg-6 line had no defined cortical region, an absent cortico-medullary junction, and a pronounced stromal component in the medulla (data not shown).
FIGURE 1.
Elevations in miR-185 impair T cell development. A, miR-185 expression in various tissues assessed by Northern blotting. U6 probe was used as the endogenous control. B, VA-hCD2 transgenic cassette. Primary miR-185 (pri-miR-185) was cloned under control of the human CD2 promoter, which enables mature miR-185 expression in T cells. C, a representative Northern blot demonstrating the expression levels of miR-185 in the thymus of the control and transgenic lines. D, relative overexpression levels of miR-185 in different transgenic lines were determined by Northern blotting. The wild type control was set as 1. Bars show the mean -fold changes ± S.E. normalized to the U6 levels from two independent experiments. E, total thymus cellularity in the control and miR-185 Tg mice. F, total thymocytes from control and miR-185 Tg mice were stained for CD4 and CD8 and analyzed by FACS. G–I, bar graphs show absolute cell numbers of the CD4+CD8− thymocytes (G), CD4−CD8+ thymocytes (H), and CD4+CD8+ thymocytes (I) in the control and miR-185 Tg mice. Data are of the mean ± S.E. from WT (n = 50), Tg-25 (n = 20), Tg-35 (n = 40), and Tg-6 (n = 70) mice (n.s. = nonsignificant, *, p < 0.05, **, p < 0.01, ***, p < 0.001; one-way ANOVA followed by Tukey's post hoc test).
Thymopoiesis in miR-185 Transgenic Lines Is Affected at Two Development Checkpoints
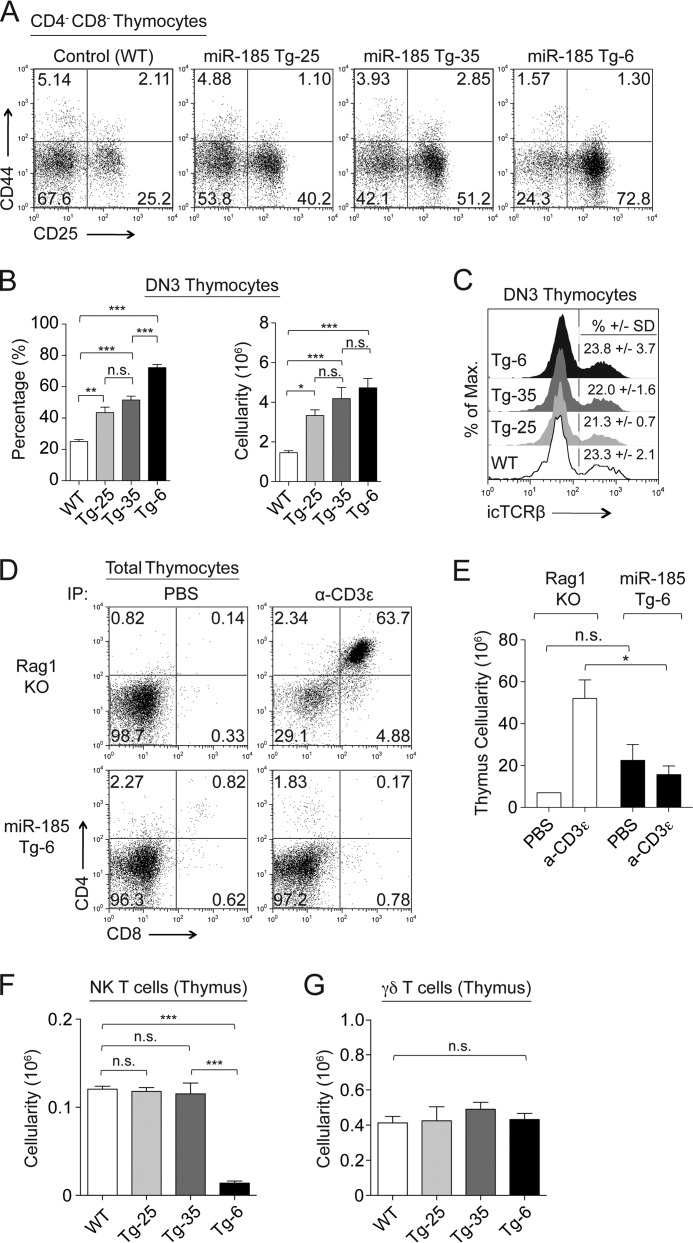
The attenuated T cell development in the miR-185 transgenic mice appeared to be during pre-TCR and TCR selection stages. To assess whether there was a defect at the pre-TCR checkpoint, CD4−CD8− (DN) thymocytes were profiled for CD44 and CD25 cell surface expression, which marks four DN subsets, including those thymocytes at pre-TCR selection stage (DN3). Increasing miR-185 levels matched the severity of the block at the DN3 (CD44−CD25+) stage, with the percentage of DN3 cells increasing from 25% in controls to 40, 51, and 73% in the miR-185 Tg-25, Tg-35, and Tg-6 lines, respectively (Fig. 2A). This was statistically significant for all the Tg lines when compared with the wild type controls, and between Tg-6 versus Tg-35 and Tg-25 (Fig. 2B). No change in intracellular TCRβ expression was detected in the DN3 subsets (Fig. 2C). To induce TCR signals, we next injected the mice intraperitoneally with anti-CD3ϵ. This normally causes a differentiation of DN3 thymocytes to the DP stage in Rag1-deficient mice, which is coupled with an increased thymic cellularity. The DN3 thymocytes in the miR-185 Tg-6 mice were unable to progress to the DP stage (Fig. 2, D and E). Surface expression levels of CD5 on DN3 thymocytes were normal in Tg-25 and Tg-35 lines, but slightly elevated in the Tg-6 line, an indication of normal pre-TCR engagement and signal strength (data not shown). With regard to other cell populations, the percentage and number of NK T cells were similar in the miR-185 Tg lines, except for the Tg-6 line that had very low NK T cell numbers (Fig. 2F). γδ T cell numbers in all Tg lines were similar to the control, whereas their percentages were slightly increased (Fig. 2G).
FIGURE 2.
Increasing levels of miR-185 attenuate T cell development at pre-TCRβ selection checkpoint. A, surface expression of CD25 and CD44 gated on CD4−CD8− (B220−, NK1.1−, TCRγδ−, CD11b−, and CD11c−) thymocytes of the control and miR-185 Tg mice. B, percentages (left) and absolute cell numbers (right) of DN3 (CD25+CD44−) thymocytes are shown as the mean ± S.E. using at least six mice per group (n.s. = nonsignificant, *, p < 0.05, **, p < 0.01, ***, p < 0.001; one-way ANOVA followed by Tukey's post hoc test). C, histograms show expression levels of intracellular TCRβ (icTCRβ) in DN3 thymocytes. The mean percentages ± S.D. values of intracellular TCRβ expression were shown for the miR-185 Tg and control mice (n > 2 mice per group). D, total thymocytes from PBS or anti-CD3ϵ treated Rag1−/− and miR-185 Tg-6 mice were stained for CD4 and CD8 and analyzed by FACS at 5 days after intraperitoneal (IP) injection. E, total thymus cellularity of anti-CD3ϵ and PBS injected Rag1−/− and miR-185 Tg-6 mice (n = 3 mice per group; n.s. = nonsignificant, *, p < 0.05, **, p < 0.01, ***, p < 0.001; two-way ANOVA followed by Bonferroni's post hoc test). F and G, bar graphs represent the mean ± S.E. cell numbers of NK T cells (F) and γδ T cells (G) in the thymus of the control and miR-185 Tg mice using at least three mice per group (n.s. = nonsignificant, *, p < 0.05, **, p < 0.01, ***, p < 0.001; one-way ANOVA followed by Tukey's post hoc test).
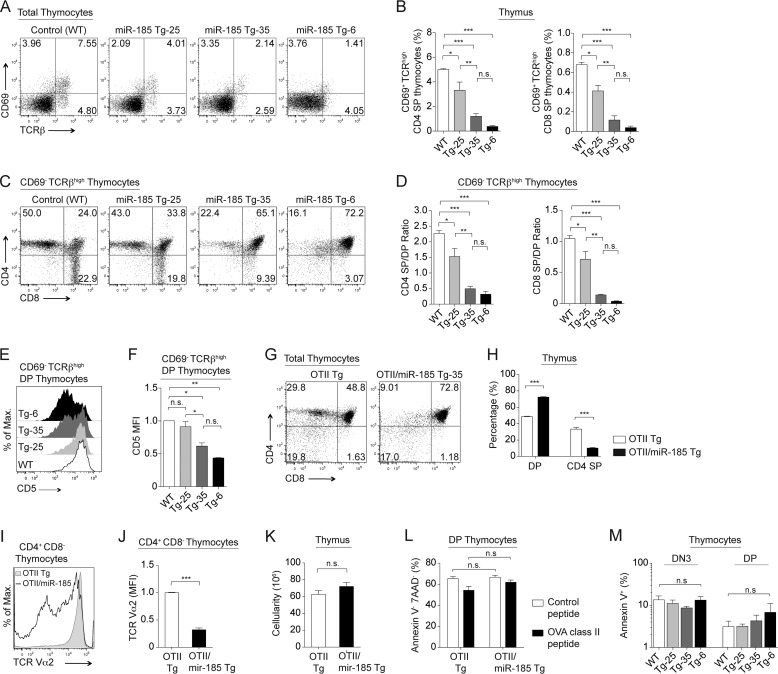
The reduced SP thymocytes in all the miR-185 Tg lines suggested an impairment of positive selection. Consistent with this, the percentages of CD69+TCRβhigh total thymocytes, including CD69+TCRβhigh CD4 and CD8 SP thymocytes, were reduced in a statistically significant manner (Fig. 3, A and B). The reduction in positive selection was consistent with an increased percentage of DP thymocytes that were CD69−TCRβhigh (Fig. 3, C and D). Differences between the closely matched Tg-25 and Tg-35 lines were also of statistical significance when the reductions in CD4+CD8− and CD4−CD8+ subsets were compared (Fig. 3, C and D). Negligible numbers of mature CD69−TCRβhigh SP thymocytes were found in the miR-185 Tg-6 line (Fig. 3, C and D). Moreover, CD5 expression on CD69−TCRβhigh DP thymocytes was lower in miR-185 Tg lines, supporting impaired positive selection (Fig. 3, E and F). Attenuated positive selection was further established by comparing the number of OTII-specific TCR transgenic T cells developing in the miR-185 Tg-35 lines. Their numbers were significantly reduced in the OTII/miR-185 Tg-35 double Tg lines when compared with the OTII Tg parental line (Fig. 3, G and H), and those residual SP cells lost the expression of the transgenic TCRα subunit (Fig. 3, I and J). Severe loss of CD4 SP thymocytes in the OTII/miR-185 Tg-35 line could reflect enhanced negative selection. However, the total number of thymocytes was equivalent in both the OTII and the OTII/miR-185 Tg-35 lines (Fig. 3K). Furthermore, in vitro treatment of thymocytes from the OTII/miR-185 Tg-35 line with an OVA class II peptide induced DP cell death, indicating that negative selection was intact and similar (Fig. 3L). The impairment at the DN3 and DP stages was not due to increased cell death because annexin V+ percentages were similar in the miR-185 Tg lines (Fig. 3M). Taken together, these findings demonstrate that increases in miR-185 reduced the effectiveness of TCRβ and positive selection.
FIGURE 3.
Increasing levels of miR-185 attenuate T cell development at TCR-positive selection checkpoint. A, flow cytometric analysis of CD69 and TCRβ expression on total thymocytes from the control and miR-185 Tg mice. B, percentages of CD69+TCRβhigh CD4 SP and CD69+TCRβhigh CD8 SP thymocytes are shown. C, plots represent CD4 by CD8 profiles of CD69−TCRβhigh thymocytes. D, cellularity ratios of the CD69−TCRβhigh CD4 SP (left) and the CD69−TCRβhigh CD8 SP (right) to the CD69−TCRβhigh DP thymocytes were established for the results shown in C. E, histogram shows CD5 expression on CD69−TCRβhigh DP thymocytes. F, relative mean fluorescence intensity (MFI) levels of CD5 in CD69−TCRβhigh DP thymocytes. A–F, each bar is the mean ± S.E. of three independent experiments (n.s. = nonsignificant, *, p < 0.05, **, p < 0.01, ***, p < 0.001; one-way ANOVA followed by Tukey's post hoc test). G, flow cytometric analysis of CD4 and CD8 expression on total thymocytes from OTII Tg and OTII/miR-185 Tg-35 mice. H, average percentages of DP and CD4 SP thymocytes are shown. I, surface expression of TCR Vα2 gated on CD4+CD8− thymocytes from OTII Tg (dark gray) and OTII/miR-185 Tg-35 mice (black line). J, relative mean fluorescence intensity (MFI) levels of TCR Vα2 on CD4+CD8− thymocytes. K, total thymus cellularity of OTII Tg and OTII/miR-185 Tg mice was represented. G–K, data are of at least six mice per group. Bar graphs represent the mean ± S.E. values (*, p < 0.05, **, p < 0.01, ***, p < 0.001; two-tailed unpaired Student's t test). L, live (annexin V− 7-aminoactinomycin D− (7AAD−)) percentages of DP thymocytes upon in vitro treatment of OTII Tg and OTII/miR-185 Tg thymocytes with SIINFEKL peptide as a negative control and OVA class II peptide. Each bar is the mean ± S.E. of three mice per group (n.s. = nonsignificant, *, p < 0.05, **, p < 0.01, ***, p < 0.001; two-way ANOVA followed by Bonferroni's post hoc test). M, graph shows the mean ± S.E. percentages of annexin V+ DN3 and DP thymocytes from control (white), Tg-25 (light gray), Tg-35 (dark gray), and Tg-6 (black) mice (n = 3 mice per group) (n.s. = nonsignificant, *, p < 0.05, **, p < 0.01, ***, p < 0.001; one-way ANOVA followed by Tukey's post hoc test).
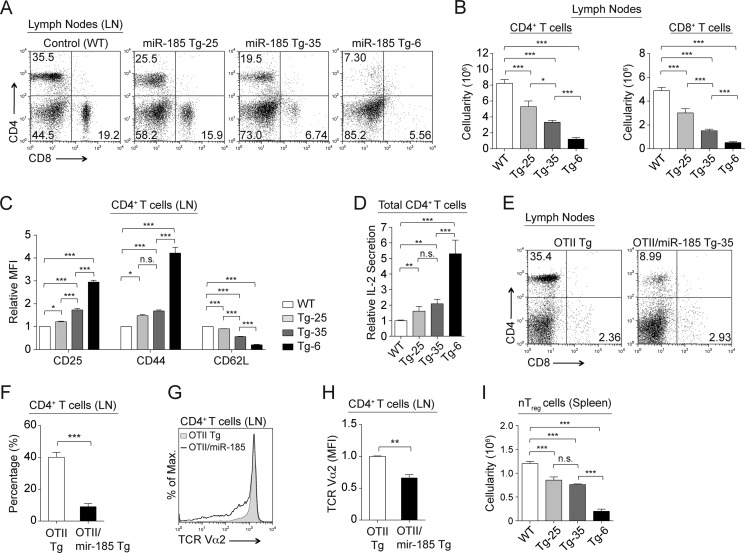
The miR-185 Transgenic Lines Have a Peripheral T Cell Lymphopenia
An miR-185 dose-dependent reduction in the percentage and number of mature peripheral CD4+CD8− and CD4−CD8+ T cells occurred in all three miR-185 Tg lines and was most pronounced in Tg-6 (Fig. 4, A and B). Peripheral T cells from all the miR-185 Tg lines displayed spontaneous hyperactivated phenotype with increased CD25 and CD44 expression and decreased CD62L expression (Fig. 4C). Consistent with this activated phenotype, total CD4+ T cells had a statistically significant increase in IL-2 production (Fig. 4D). Total CD4+ T cells from the miR-185 Tg-6 line were unable to proliferate, whereas CD4+ T cells from the miR-185 Tg-25 and Tg-35 lines had a slightly diminished proliferative response, evident only after 48 h but not 72 h of stimulation, likely due to decreased percentages of naive T cells in miR-185 Tg mice (data not shown). In vitro TCR stimulation with anti-CD3ϵ/CD28 led to an increase in apoptosis of total CD4+ T cells, the severity of which matched increasing miR-185 levels (data not shown). A significant reduction was noted both in numbers and in TCR density of mature peripheral CD4+CD8− T cells from the OTII/miR-185 Tg-35 mice (Fig. 4, E–H). Moreover, the number of natural T regulatory (Foxp3+ CD25+ CD4+) cells was also reduced gradually in the spleen of all miR-185 Tg lines (Fig. 4I).
FIGURE 4.
Elevated levels of miR-185 cause a peripheral T cell lymphopenia. A, flow cytometric analysis of CD4+ and CD8+ T cells in the lymph nodes from normal and miR-185 Tg mice. B, bar graphs show absolute numbers of CD4+ and CD8+ T cells in the lymph nodes in the control and miR-185 Tg mice. Data are of the mean ± S.E. from WT (n = 40), Tg-25 (n = 16), Tg-35 (n = 46), and Tg-6 (n = 59) mice (*, p < 0.05, **, p < 0.01, ***, p < 0.001; one-way ANOVA followed by Tukey's post hoc test). C, relative mean fluorescence intensity (MFI) levels ± S.E. of CD25, CD44, and CD62L markers on CD4+ T cells in the lymph nodes of WT (white), Tg-25 (light gray), Tg-35 (dark gray), and Tg-6 (black) mice using at least five mice per group (n.s. = nonsignificant, *, p < 0.05, **, p < 0.01, ***, p < 0.001; one-way ANOVA followed by Tukey's post hoc test). D, graph represents relative IL-2 secretion from anti-CD3ϵ/CD28 stimulated total CD4+ T cells in miR-185 Tg lines and the control (WT), set to 1. Each bar is the mean ± S.E. of at least five independent experiments (n.s. = nonsignificant, *, p < 0.05, **, p < 0.01, ***, p < 0.001; Two-tailed unpaired Student's t test). E, lymphocytes from the lymph nodes of OTII Tg and OTII/miR-185 Tg mice were stained for CD4 and CD8 and analyzed by FACS. F, average percentages of CD4+CD8− T cells in the lymph nodes. G, surface expression of TCR Vα2 gated on CD4+CD8− T cells from the lymph nodes of OTII Tg (dark gray) and OTII/miR-185 Tg mice (black line). H, relative mean fluorescence intensity (MFI) levels of TCR Vα2 on CD4+CD8− lymphocytes. E–H, data are of at least six mice per group. Bar graphs represent the mean ± S.E. values (*, p < 0.05, **, p < 0.01, ***, p < 0.001; two-tailed unpaired Student's t test). I, graph shows absolute numbers of natural regulatory T (nTreg) cells in the spleen of the control and miR-185 Tg mice. Data are of the mean ± S.E. from at least three mice per group (n.s. = nonsignificant, *, p < 0.05, **, p < 0.01, ***, p < 0.001; one-way ANOVA followed by Tukey's post hoc test).
miR-185 Targets a Number of Genes Implicated in Thymopoiesis, Including the Endoplasmic Reticulum Calcium Regulator, Mzb1
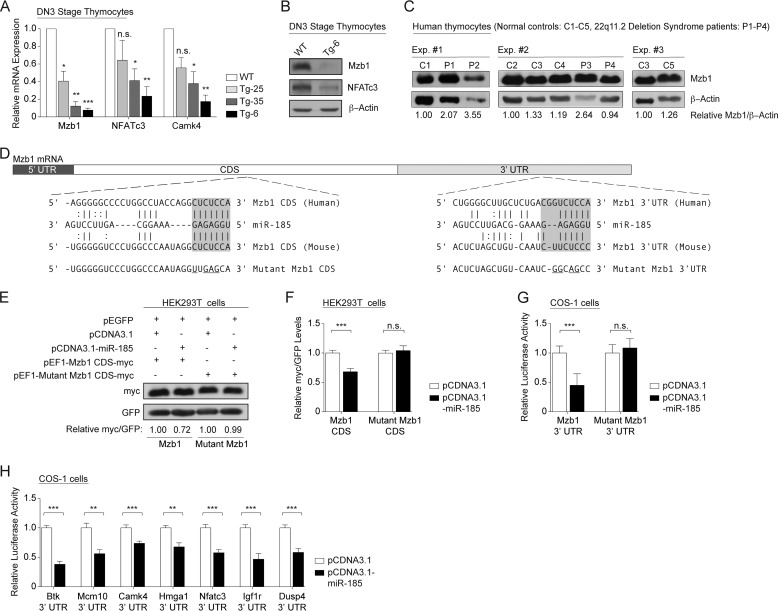
The miR-185 effects on T cell development suggested that genes coupled to pre-TCR- and TCR-driven selection were targeted by this miR. Gene expression comparisons were performed on DN3 sorted thymocytes from wild type mice and pooled miR-185 Tg-6 mice. 234 genes were down- and 317 were up-regulated more than 1.5-fold in the DN3 thymocytes of miR-185 Tg-6 mice when compared with normal controls (p < 0.05) (supplemental Tables S1 and S2). An miR target prediction database (miRWalk) parsed the down-regulated genes to those containing putative miR-185 binding sites on their 3′-UTRs and/or CDS (24). The top 25 candidates are listed (Table 1). Quantitative RT-PCR with gene-specific probes for Mzb1 (also known as 2010001M09Rik, PACAP, or pERp1), Nfatc3, and Camk4 revealed a direct, and statistically significant, miR-185 dose-dependent decrease in the expression of each target (Fig. 5A). Protein expression comparisons confirmed a substantial loss of Mzb1 and NFATc3 in sorted DN3 cells (Fig. 5B). Patients with the 22q11.2 deletion are haploinsufficient in miR-185 (10). To determine whether the levels of Mzb1 were altered in their thymocytes, immunoblotting was performed using protein extracts prepared from five independently prepared normal controls and four individuals with confirmed deletions on 22q11.2. Mzb1 was up-regulated >2-fold in 3 of the 4 patient samples (Fig. 5C).
TABLE 1.
The top 25 down-regulated genes with predicted miR-185 binding sites on their 3′-UTR and/or coding sequences
This table provides -fold changes of the down-regulated genes in miR-185 Tg DN3 thymocytes as compared with the wild type control. miRWalk target prediction database was used to identify putative miR-185 target genes.
| Gene symbol | Fold | Description |
|---|---|---|
| Mzb1 | −4.84 | Marginal zone B and B1-cell specific protein |
| Hk2 | −4.19 | Hexokinase II |
| Hoxa7 | −3.81 | Homeo box A7 |
| Tpst1 | −3.79 | Protein-tyrosine sulfotransferase 1 |
| Mcm10 | −3.26 | Minichromosome maintenance deficient 10 |
| Camk4 | −3.24 | Calcium/calmodulin-dependent protein kinase IV |
| Stab1 | −3.20 | Stabilin 1 |
| E130012A19Rik | −3.10 | RIKEN cDNA E130012A19 gene |
| 2310044H10Rik | −3.06 | RIKEN cDNA 2310044H10 gene, MIRTA22 |
| Abcf2 | −2.94 | ATP-binding cassette, subfamilyF (GCN20), member 2 |
| Gja1 | −2.75 | Gap junction membrane channel protein α 1 |
| Gga2 | −2.73 | Golgi-associated, gamma adaptin ear-containing, ARF-binding protein 2 |
| Nsf | −2.59 | N-Ethylmaleimide sensitive fusion protein |
| Ccdc53 | −2.52 | Coiled-coil domain containing 53 |
| Mns1 | −2.41 | Meiosis-specific nuclear structural protein 1 |
| Ncl | −2.40 | Nucleolin |
| Hmga1 | −2.38 | High mobility group AT-hook 1 |
| Shmt1 | −2.36 | Serine hydroxymethyltransferase 1 |
| Mcm5 | −2.26 | Minichromosome maintenance deficient 5 |
| Igf2bp3 | −2.22 | Insulin-like growth factor 2 mRNA binding protein 3 |
| Nfatc3 | −2.21 | Nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 |
| Rrm1 | −2.19 | Ribonucleotide reductase M1 |
| Suz12 | −2.18 | Suppressor of zeste 12 homolog (Drosophila) |
| Igf1r | −2.14 | Insulin-like growth factor I receptor |
| Rcl1 | −2.13 | RNA terminal phosphate cyclase-like 1 |
FIGURE 5.
miR-185 targets a number of genes in developing thymocytes. A, relative mRNA levels of Mzb1, Nfatc3, and Camk4 in DN3 thymocytes, normalized to the endogenous Gapdh levels, were determined by real-time quantitative PCR. WT values were set to 1. Data shown are of the mean ± S.E. of at least three independent experiments performed in triplicates. Bars are representative of WT (white), Tg-25 (light gray), Tg-35 (dark gray), and Tg-6 (black) mice. (n.s. = nonsignificant, *, p < 0.05, **, p < 0.01, ***, p < 0.001; versus the threshold set as 1; one sample Student's t test) B, immunoblot analysis of Mzb1 and NFATc3 expression in miR-185 Tg-6 DN3 thymocytes when compared with the wild type control. β-Actin was used as the endogenous control. C, Mzb1 protein expression levels in human thymocytes obtained from five normal individuals (C1–C5) and four patients with 22q11.2 deletion syndrome (P1–P4). β-Actin was used as the endogenous control. Band intensities of Mzb1 and β-actin were measured using the ImageJ software. The Mzb1/β-actin ratio was calculated by dividing the band intensity of Mzb1 to that of the β-actin for each sample. Relative Mzb1 levels were then determined for each experiment (Exp #1–Exp. #3.) indicated as a group. This was done by normalizing the Mzb1/β-actin ratio for each sample relative to the first control sample. The first control sample was set as 1 in each of three independent experiments. D, the Mzb1 CDS and 3′-UTR each contain one putative miR-185 binding site. The diagram shows conserved miR-185 base pairing with human and murine Mzb1 mRNA. Mutated Mzb1 sequences are underlined. E, miR-185 directly targets Mzb1 CDS. A representative blot was shown from HEK293T cells transfected with the plasmid (pEF1) containing either wild type Mzb1 CDS-Myc or mutant Mzb1 CDS-Myc fusion, along with the empty vector (white) or pCDNA3.1/miR-185 (black). A GFP-expressing plasmid (pEGFP) was used as the transfection control. Band intensities of Myc and GFP were calculated for each lane using the ImageJ software. Relative Myc levels in the wild type Mzb1 or mutant Mzb1 transfectants were determined by normalizing the Myc/GFP ratio of pCDNA3.1/miR-185 to that of the pCDNA3.1 control, which was set as 1. F, graph shows the mean ± S.E. of relative Myc/GFP levels from four independent experiments performed in at least duplicates (n.s. = nonsignificant, *, p < 0.05, **, p < 0.01, ***, p < 0.001; two-tailed unpaired Student's t test). G, the Mzb1 3′-UTR is a direct target of miR-185. H, validation of additional miR-185 targets. G–H, luciferase activity was normalized to the β-galactosidase of COS-1 cells transfected with the luciferase plasmids containing the indicated 3′-UTR, along with either the empty vector or pCDNA3.1-miR-185. Normalized luciferase activity of the pCDNA3.1/miR-185 (black) transfectant was determined relative to that of the empty pCDNA3.1 vector (white), which was set as 1. Btk 3′-UTR, a previously validated target of miR-185, was used as a positive control. Data shown are of the mean ± S.E. from four independent experiments performed in at least triplicates (*, p < 0.05, **, p < 0.01, ***, p < 0.001; two-tailed unpaired Student's t test).
Prediction software suggests that there are two miR-185 target sites in Mzb1 mRNA, one actually in the coding sequence (Fig. 5D). In transfection assays with just the CDS of Mzb1, miR-185 reduced its expression when compared with the vector control in a statistically significant manner (Fig. 5, E and F). Mutating the target sequence in Mzb1 prevented its down-regulation (Fig. 5, E and F). To confirm targeting of the 3′-UTR of Mzb1, luciferase reporter assays were performed. Relative to the control, the luciferase activity of Mzb1 3′-UTR was decreased more than 2-fold (Fig. 5G). Mutations of the target sequence within the Mzb1 3′-UTR restored control luciferase levels (Fig. 5G). A number of additional targets identified in the microarray (Mcm10, Camk4, Hmga1, Nfatc3, Igf1r, and Dusp4) were validated as novel miR-185 targets in luciferase reporter assays, with the Btk 3′-UTR included as a positive control (Fig. 5H).
miR-185 Levels Affect TCR-driven Intracellular Calcium Responses
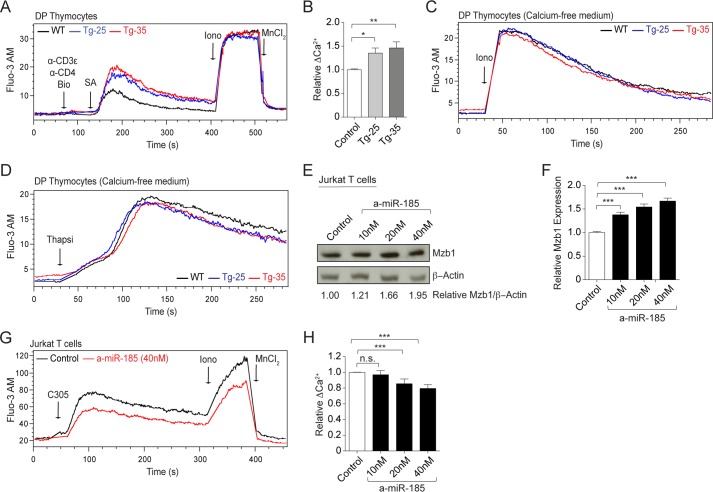
One of the miR-185 targets, Mzb1, is an endoplasmic reticulum-associated protein that regulates B cell receptor-driven calcium responses (25). We examined the changes TCR-driven intracellular Ca2+ responses in DP thymocytes from the miR-185 Tg lines. DP thymocytes from normal mice increased intracellular Ca2+ responses 30–40 s after TCR/CD4 cross-linking (Fig. 6A). The magnitude of the TCR-induced Ca2+ response was significantly higher in the Tg-25 and Tg-35 lines (Fig. 6, A and B). The addition of the calcium ionophore, ionomycin, revealed an identical capacity of all groups of thymocytes to internalize Ca2+, indicating that the differences in the cells were TCR signaling-dependent (Fig. 6C). Thymocytes with varying levels of miR-185 exhibited similar Ca2+ responses following thapsigargin addition (Fig. 6D). These experiments indicated that all the Tg lines had similar endoplasmic reticulum Ca2+ stores as wild type mice. In a loss-of-function approach, chemically modified miR inhibitors (antagomirs) were used to reduce miR-185 activity in Jurkat T cells. Inhibiting miR-185 increased Mzb1 protein expression in a dose-dependent and statistically significant manner when compared with the control inhibitor (cel-miR-67) (Fig. 6, E and F). The knockdown of miR-185, which resulted in higher Mzb1 levels, reduced TCR-mediated intracellular calcium levels in Jurkat T cells (Fig. 6, G and H). In summary, miR-185 directly affects TCR-triggered calcium responses in developing thymocytes and Jurkat T cells.
FIGURE 6.
miR-185 controls TCR-stimulated intracellular calcium responses. A, intracellular calcium flux was analyzed by flow cytometry in DP thymocytes from the WT (black line), Tg-25 (blue line), and Tg-35 (red line) mice. DP thymocytes were gated by size. Black arrows indicate the time points for each treatment. Fluo-3 AM-loaded thymocytes were treated with biotinylated (Bio) anti-CD3ϵ and anti-CD4 followed by streptavidin (SA), ionomycin (Iono), and MnCl2. B, graph shows relative changes in TCR-triggered peak Ca2+-influx over the base line. Each bar represents the mean ± S.E. of six independent experiments (*, p < 0.05, **, p < 0.01, ***, p < 0.001; two-tailed unpaired Student's t test). C and D, Fluo-3 AM-loaded thymocytes were treated with ionomycin (Iono) (C) and thapsigargin (Thapsi) (D) in the absence of extracellular calcium. DP thymocytes were gated electronically. Experiments were repeated two times, and representative plots are shown from the WT (black line), Tg-25 (blue line), and Tg-35 (red line) mice. E, representative immunoblot shows Mzb1 expression in Jurkat T cells transfected with varying concentrations of miR-185 inhibitor (a-miR-185) when compared with the control (miR negative control inhibitor). β-Actin was used as the endogenous control. Band intensities of Mzb1 and β-actin were measured using the ImageJ software. The relative amounts of Mzb1 protein were shown as normalized to the control inhibitor, which was set as 1. This was done by dividing the Mzb1/β-actin ratio of each sample to that of the control sample. F, graph represents the mean ± S.E. of relative Mzb1 levels in Jurkat T cells transfected with the control and miR-185 inhibitor from five independent experiments, performed in at least duplicates (*, p < 0.05, **, p < 0.01, ***, p < 0.001; two-tailed unpaired Student's t test). G, intracellular calcium responses were analyzed by flow cytometry over time in Jurkat T cells following transfection with a-miR-185 (red line) and the control inhibitor (black line). Fluo-3 AM-loaded Jurkat T cells were treated with the mAb C305.2 (C305) (anti-TCRβ), ionomycin (Iono), and MnCl2. Black arrows indicate the time points for each treatment. H, graph shows relative changes in TCR-triggered peak Ca2+ influx over the base line. Each bar represents the mean ± S.E. of four independent experiments (n.s. = nonsignificant, *, p < 0.05, **, p < 0.01, ***, p < 0.001; two-tailed unpaired Student's t test).
DISCUSSION
Gain- and loss-of-function approaches were used to characterize the function role of miR-185 in T cells and identify its mRNA targets. A transgene-driven overexpression of miR-185 caused a developmental impairment in thymopoiesis.
The most down-regulated target, Mzb1, was first identified as a novel gene induced during B to plasma cell differentiation, regulating proper assembly and secretion of mature IgM (26, 27). It is highly expressed in marginal zone B cells and regulates intracellular Ca2+ flux upon B cell receptor stimulation (25). Our findings indicate that Mzb1 is also highly expressed in DN3 thymocytes and is present in Jurkat T cells, consistent with prior Northern blotting results (26–28). miR-185 targets two highly conserved sites in Mzb1, one in the CDS. This likely contributes to the reduced efficiency of thymopoiesis as pre-TCR- and αβ TCR-driven intracellular calcium responses are likely too high to support expansion or positive selection, respectively. Reductions in NFATc3 and CAMK4 also affect these pathways. In fact, the targeted elimination of Nfatc3 causes a very similar development block at the pre-TCR selection stage and during positive selection, with a resulting peripheral T cell lymphopenia (29, 30).
Our data raise important questions as to whether miR-185 affects T and B cell functions in humans. Patients with 22q11.2 deletion syndrome, haploinsufficient for miR-185, have an increased prevalence of autoimmune disorders and B cell defects (10). We suggest that reductions in miR-185 affect the expression of both Btk and Mzb1 in B cells, enhancing autoantibody production (5, 25). These patients have abnormal T helper cell skewing (31). Further experiments are needed to elucidate the contribution of miR-185 in both thymocytes and thymic epithelial/mesenchymal cells. Mouse models of 22q11.2 deletion syndrome confirm a reduction of miR-185, with an age-dependent reduction in other miRs (17). A distinct miR-185 target in neurons is the calcium regulator, SERCA2 (17). Because SERCA2 is expressed at very low levels in thymocytes, the principal targets of miR-185 in thymocytes are likely distinct. In fact, Mcm10, Hmga1, Igf1r, and Dusp4 were additional targets. miR-185 can target Six1, RhoA, and Cdc42, genes involved in controlling cell cycle progression in various cancer cells (32–34). Interestingly, although these were not identified in our thymocyte screen, they may play a role in the peripheral T cells (35). It will be important to assess the consequences of both the haploinsufficiency and the trisomy of miR-185 on the novel targets reported herein as Nfatc3 and Camk4 are expressed in many tissues and organs affected by the deletions on chromosome 22q11.2.
In summary, our findings implicate a role of miR-185 in the T cell development through its targeting of genes with validated importance during thymopoiesis.
Acknowledgments
We thank Angela Mobley for assistance with flow cytometry. In addition, we thank Ashley Hoover for helpful comments. We sincerely appreciate the suggestions provided by Rhonda Bassel-Duby and other members of the Eric Olson laboratory in the Department of Molecular Biology at UT Southwestern Medical Center. Dr. Jen Liou (Department of Physiology) provided insights into calcium responses. We thank the transgenic and knock-out facility for generation of the mice. We greatly appreciate Dr. Ellen V. Rothenberg (Division of Biology at California Institute of Technology, Pasadena, CA) for critically reviewing the manuscript. We also thank Dr. Shaheen Khan, Dr. Navin Fatema Chowdhury, and Dr. Igor Dozmorov for assistance with the analysis of microarray data.
This work was supported in part by National Institutes of Health Grants R21 AI083827-01 (to N. S. C. v. O.). This work was also supported by Children's Medical Center Foundation (to N. S. C. v. O. and M. T. d. l. M.) and the Jeffrey Modell Foundation (to M. T. d. l. M.).

This article contains supplemental Tables S1 and S2.
Data reported in this paper were submitted to GEO database under accession number GSE49057.
- miR
- microRNA
- CDS
- coding sequence
- DN
- CD4−CD8−
- DP
- CD4+CD8+
- CD4 SP
- CD4+CD8−
- CD8 SP
- CD4−CD8+
- Btk
- Bruton's tyrosine kinase
- SERCA2
- sarcoplasmic/endoplasmic reticulum calcium ATPase 2
- Mzb1
- marginal zone B and B1-cell specific protein
- NFATc3
- nuclear factor of activated T cells, cytoplasmic, calcineurin-dependent 3
- CAMK4
- calcium/calmodulin-dependent protein kinase IV
- Mcm10
- minichromosome maintenance deficient 10
- Hmga1
- high mobility group AT-hook 1
- Igf1r
- insulin-like growth factor I receptor
- Dusp4
- dual specificity phosphatase 4
- NK
- natural killer
- Tg
- transgenic
- ANOVA
- analysis of variance
- TCR
- T cell receptor
- OVA
- ovalbumin
- OTII
- TCR transgenic mice specific for ovalbumin peptide with I-Ab.
REFERENCES
- 1. Bartel D. P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 2. Kloosterman W. P., Plasterk R. H. (2006) The diverse functions of microRNAs in animal development and disease. Dev. Cell 11, 441–450 [DOI] [PubMed] [Google Scholar]
- 3. Papadopoulou A. S., Dooley J., Linterman M. A., Pierson W., Ucar O., Kyewski B., Zuklys S., Hollander G. A., Matthys P., Gray D. H., De Strooper B., Liston A. (2012) The thymic epithelial microRNA network elevates the threshold for infection-associated thymic involution via miR-29a mediated suppression of the IFN-α receptor. Nat. Immunol. 13, 181–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cobb B. S., Nesterova T. B., Thompson E., Hertweck A., O'Connor E., Godwin J., Wilson C. B., Brockdorff N., Fisher A. G., Smale S. T., Merkenschlager M. (2005) T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J. Exp. Med. 201, 1367–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Belver L., de Yébenes V. G., Ramiro A. R. (2010) MicroRNAs prevent the generation of autoreactive antibodies. Immunity 33, 713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Koralov S. B., Muljo S. A., Galler G. R., Krek A., Chakraborty T., Kanellopoulou C., Jensen K., Cobb B. S., Merkenschlager M., Rajewsky N., Rajewsky K. (2008) Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell 132, 860–874 [DOI] [PubMed] [Google Scholar]
- 7. Curtale G., Citarella F., Carissimi C., Goldoni M., Carucci N., Fulci V., Franceschini D., Meloni F., Barnaba V., Macino G. (2010) An emerging player in the adaptive immune response: microRNA-146a is a modulator of IL-2 expression and activation-induced cell death in T lymphocytes. Blood 115, 265–273 [DOI] [PubMed] [Google Scholar]
- 8. Trotta R., Chen L., Costinean S., Josyula S., Mundy-Bosse B. L., Ciarlariello D., Mao C., Briercheck E. L., McConnell K. K., Mishra A., Yu L., Croce C. M., Caligiuri M. A. (2013) Overexpression of miR-155 causes expansion, arrest in terminal differentiation and functional activation of mouse natural killer cells. Blood 121, 3126–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Belkaya S., Silge R. L., Hoover A. R., Medeiros J. J., Eitson J. L., Becker A. M., de la Morena M. T., Bassel-Duby R. S., van Oers N. S. (2011) Dynamic modulation of thymic microRNAs in response to stress. PLoS One 6, e27580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de la Morena M. T., Eitson J. L., Dozmorov I. M., Belkaya S., Hoover A. R., Anguiano E., Pascual M. V., van Oers N. S. (2013) Signature microRNA expression patterns identified in humans with 22q11.2 deletion/DiGeorge syndrome. Clin. Immunol. 147, 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kobrynski L. J., Sullivan K. E. (2007) Velocardiofacial syndrome, DiGeorge syndrome: the chromosome 22q11.2 deletion syndromes. Lancet 370, 1443–1452 [DOI] [PubMed] [Google Scholar]
- 12. Piliero L. M., Sanford A. N., McDonald-McGinn D. M., Zackai E. H., Sullivan K. E. (2004) T-cell homeostasis in humans with thymic hypoplasia due to chromosome 22q11.2 deletion syndrome. Blood 103, 1020–1025 [DOI] [PubMed] [Google Scholar]
- 13. Kanaya Y., Ohga S., Ikeda K., Furuno K., Ohno T., Takada H., Kinukawa N., Hara T. (2006) Maturational alterations of peripheral T cell subsets and cytokine gene expression in 22q11.2 deletion syndrome. Clin. Exp. Immunol. 144, 85–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Karayiorgou M., Simon T. J., Gogos J. A. (2010) 22q11.2 microdeletions: linking DNA structural variation to brain dysfunction and schizophrenia. Nat. Rev. Neurosci. 11, 402–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ensenauer R. E., Adeyinka A., Flynn H. C., Michels V. V., Lindor N. M., Dawson D. B., Thorland E. C., Lorentz C. P., Goldstein J. L., McDonald M. T., Smith W. E., Simon-Fayard E., Alexander A. A., Kulharya A. S., Ketterling R. P., Clark R. D., Jalal S. M. (2003) Microduplication 22q11.2, an emerging syndrome: clinical, cytogenetic, and molecular analysis of thirteen patients. Am. J. Hum. Genet. 73, 1027–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yobb T. M., Somerville M. J., Willatt L., Firth H. V., Harrison K., MacKenzie J., Gallo N., Morrow B. E., Shaffer L. G., Babcock M., Chernos J., Bernier F., Sprysak K., Christiansen J., Haase S., Elyas B., Lilley M., Bamforth S., McDermid H. E. (2005) Microduplication and triplication of 22q11.2: a highly variable syndrome. Am. J. Hum. Genet. 76, 865–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Earls L. R., Fricke R. G., Yu J., Berry R. B., Baldwin L. T., Zakharenko S. S. (2012) Age-dependent microrna control of synaptic plasticity in 22q11 deletion syndrome and schizophrenia. J. Neurosci. 32, 14132–14144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu B., Hsu P. K., Stark K. L., Karayiorgou M., Gogos J. A. (2013) Derepression of a neuronal inhibitor due to miRNA dysregulation in a schizophrenia-related microdeletion. Cell 152, 262–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Becker A. M., Blevins J. S., Tomson F. L., Eitson J. L., Medeiros J. J., Yarovinsky F., Norgard M. V., van Oers N. S. (2010) Invariant NKT cell development requires a full complement of functional CD3 ζ immunoreceptor tyrosine-based activation motifs. J. Immunol. 184, 6822–6832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pall G. S., Codony-Servat C., Byrne J., Ritchie L., Hamilton A. (2007) Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA, and piRNA by northern blot. Nucleic Acids Res. 35, e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kirigin F. F., Lindstedt K., Sellars M., Ciofani M., Low S. L., Jones L., Bell F., Pauli F., Bonneau R., Myers R. M., Littman D. R., Chong M. M. (2012) Dynamic microRNA gene transcription and processing during T cell development. J. Immunol. 188, 3257–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kuchen S., Resch W., Yamane A., Kuo N., Li Z., Chakraborty T., Wei L., Laurence A., Yasuda T., Peng S., Hu-Li J., Lu K., Dubois W., Kitamura Y., Charles N., Sun H. W., Muljo S., Schwartzberg P. L., Paul W. E., O'Shea J., Rajewsky K., Casellas R. (2010) Regulation of microRNA expression and abundance during lymphopoiesis. Immunity 32, 828–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhumabekov T., Corbella P., Tolaini M., Kioussis D. (1995) Improved version of a human CD2 minigene based vector for T cell-specific expression in transgenic mice. J. Immunol. Methods 185, 133–140 [DOI] [PubMed] [Google Scholar]
- 24. Dweep H., Sticht C., Pandey P., Gretz N. (2011) miRWalk – database: Prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 44, 839–847 [DOI] [PubMed] [Google Scholar]
- 25. Flach H., Rosenbaum M., Duchniewicz M., Kim S., Zhang S. L., Cahalan M. D., Mittler G., Grosschedl R. (2010) Mzb1 protein regulates calcium homeostasis, antibody secretion, and integrin activation in innate-like B cells. Immunity 33, 723–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van Anken E., Pena F., Hafkemeijer N., Christis C., Romijn E. P., Grauschopf U., Oorschot V. M., Pertel T., Engels S., Ora A., Lástun V., Glockshuber R., Klumperman J., Heck A. J., Luban J., Braakman I. (2009) Efficient IgM assembly and secretion require the plasma cell induced endoplasmic reticulum protein pERp1. Proc. Natl. Acad. Sci. U.S.A. 106, 17019–17024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shimizu Y., Meunier L., Hendershot L. M. (2009) pERp1 is significantly up-regulated during plasma cell differentiation and contributes to the oxidative folding of immunoglobulin. Proc. Natl. Acad. Sci. U.S.A. 106, 17013–17018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hoffman B. G., Williams K. L., Tien A. H., Lu V., de Algara T. R., Ting J. P., Helgason C. D. (2006) Identification of novel genes and transcription factors involved in spleen, thymus, and immunological development and function. Genes Immun. 7, 101–112 [DOI] [PubMed] [Google Scholar]
- 29. Oukka M., Ho I. C., de la Brousse F. C., Hoey T., Grusby M. J., Glimcher L. H. (1998) The transcription factor NFAT4 is involved in the generation and survival of T cells. Immunity 9, 295–304 [DOI] [PubMed] [Google Scholar]
- 30. Raman V., Blaeser F., Ho N., Engle D. L., Williams C. B., Chatila T. A. (2001) Requirement for Ca2+/calmodulin-dependent kinase type IV/Gr in setting the thymocyte selection threshold. J. Immunol. 167, 6270–6278 [DOI] [PubMed] [Google Scholar]
- 31. Zemble R., Luning Prak E., McDonald K., McDonald-McGinn D., Zackai E., Sullivan K. (2010) Secondary immunologic consequences in chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome). Clin. Immunol. 136, 409–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Imam J. S., Buddavarapu K., Lee-Chang J. S., Ganapathy S., Camosy C., Chen Y., Rao M. K. (2010) MicroRNA-185 suppresses tumor growth and progression by targeting the Six1 oncogene in human cancers. Oncogene 29, 4971–4979 [DOI] [PubMed] [Google Scholar]
- 33. Liu M., Lang N., Chen X., Tang Q., Liu S., Huang J., Zheng Y., Bi F. (2011) miR-185 targets RhoA and Cdc42 expression and inhibits the proliferation potential of human colorectal cells. Cancer Lett 301, 151–160 [DOI] [PubMed] [Google Scholar]
- 34. Takahashi Y., Forrest A. R., Maeno E., Hashimoto T., Daub C. O., Yasuda J. (2009) MiR-107 and MiR-185 can induce cell cycle arrest in human non small cell lung cancer cell lines. PLoS One 4, e6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P. P. (2011) A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]