Abstract
Cellular compartmentalization into organelles serves to separate biological processes within the environment of a single cell. While some metabolic reactions are specific to a single organelle, others occur in more than one cellular compartment. Specific targeting of proteins to compartments inside of eukaryotic cells is mediated by defined sequence motifs. To achieve multiple targeting to different compartments cells use a variety of strategies. Here, we focus on mechanisms leading to dual targeting of peroxisomal proteins. In many instances, isoforms of peroxisomal proteins with distinct intracellular localization are encoded by separate genes. But also single genes can give rise to differentially localized proteins. Different isoforms can be generated by use of alternative transcriptional start sites, by differential splicing or ribosomal read-through of stop codons. In all these cases different peptide variants are produced, of which only one carries a peroxisomal targeting signal. Alternatively, peroxisomal proteins contain additional signals that compete for intracellular targeting. Dual localization of proteins residing in both the cytoplasm and in peroxisomes may also result from use of inefficient targeting signals. The recent observation that some bona fide cytoplasmic enzymes were also found in peroxisomes indicates that dual targeting of proteins to both the cytoplasm and the peroxisome might be more widespread. Although current knowledge of proteins exhibiting only partial peroxisomal targeting is far from being complete, we speculate that the metabolic capacity of peroxisomes might be larger than previously assumed.
Keywords: peroxisomes, protein import, alternative splicing, ribosomal read-through, glycolysis
Introduction
Peroxisomes are near-ubiquitous eukaryotic organelles that have been first described as microbodies in murine kidney-cells (Rhodin, 1954). Later, these organelles were shown to contain enzymes involved in the turnover of hydrogen peroxide (H2O2), which gave rise to the term peroxisomes (deDuve and Bauduin, 1966). One of the major sources of H2O2 is β-oxidation of fatty acids that occurs in peroxisomes throughout the eukaryotic kingdoms (Poirier et al., 2006). Beside these common tasks of fatty acid degradation and peroxide detoxification, peroxisomes exhibit a wide variety of other metabolic functions (Nyathi and Baker, 2006; van der Klei et al., 2006; Wanders and Waterham, 2006). A high degree of functional and metabolic specialization in different organisms even led to specific naming of peroxisomal subtypes. In plants and fungi, glyoxysomes harbor enzymes of the glyoxylate cycle, which is required for the anabolic use of acetyl-CoA as carbon source (Breidenbach and Beevers, 1967; Zimmermann and Neupert, 1980; Kionka and Kunau, 1985). In trypanosomes, the majority of glycolytic enzymes reside in specialized peroxisomes called glycosomes (Opperdoes and Borst, 1977). Filamentous ascomycetes contain “Woronin bodies” that play a mechanical role and seal septal pores (Jedd and Chua, 2000). In spite of their obvious functional and metabolic diversity all types of peroxisomes share a highly conserved import system for their matrix proteins (Gabaldon, 2010). Import is mediated by peroxisomal targeting sequences (PTS), that reside either at the C-terminus (PTS1) or at the N-terminus (PTS2) of proteins (Rucktäschel et al., 2011). C-terminal PTS1 motifs consist of about 12 amino acids that contain at the very end a characteristic tripeptide derived from the prototype sequence SKL (Gould et al., 1987, 1989; Brocard and Hartig, 2006). The commonly used consensus motif for C-terminal tripeptides is (S/A/C)-(K/R/H)-(L/M), while some studies suggest a more degenerated consensus (Lametschwandtner et al., 1998; Reumann et al., 2007). A few proteins contain internal motifs acting as PTS (Peterson et al., 1997; Klein et al., 2002; Gunkel et al., 2004; Oshima et al., 2008; Galland et al., 2010). PTS1 containing proteins are recognized by the cytoplasmic receptor Pex5 and are imported into peroxisomes in their fully folded, oligomeric and even cofactor bound form (Brocard et al., 1994; Glover et al., 1994; McNew and Goodman, 1994). A minority of proteins contains an N-terminal PTS2-motif, which is recognized by the soluble receptor Pex7 (Swinkels et al., 1991; Marzioch et al., 1994; Rucktäschel et al., 2011). PTS2-motifs exhibit the consensus sequence (R/K)(L/V/I)-X5-(H/Q)(L/A) (Petriv et al., 2004). Interestingly, some species completely lack the PTS2 import pathway (Motley et al., 2000; Gonzalez et al., 2011). Some proteins have been described that lack any detectable PTS-motifs but are nevertheless found in peroxisomes. A quite unusual way to achieve peroxisomal import of proteins without PTS is “piggy-backing”, since proteins can also be imported as oligomers (Glover et al., 1994; McNew and Goodman, 1994; Yang et al., 2001). It has been demonstrated that peroxisomal import of the copper containing superoxide dismutase (SOD) is mediated via interaction with a chaperone that harbors a PTS1 (Islinger et al., 2009).
For several peroxisomal proteins dually targeted isoforms have been described. These isoforms execute the same or a similar function in at least one other place (for an overview see Table 1). In general, cells can use various mechanisms to achieve dual or multiple targeting of proteins. One of the best systems studied are mitochondrial proteins some of which occur also in other cellular compartments (for review Yogev and Pines, 2011). Here, we address the diversity of mechanisms to mediate dual targeting of peroxisomal proteins and distinguish five different strategies (see Figure 1): (1) gene duplication, (2) generation of alternative transcripts from single genes, (3) leaky scanning of start and stop codons, (4) competition between multiple targeting signals (5) partial peroxisomal localization of proteins, since the targeting signal is either modified or inefficient.
Table 1.
Overview on peroxisomal proteins with dual localization.
| Enzyme | Organism | Mechanism for dual localizationa | Localization | References |
|---|---|---|---|---|
| Malate dehydrogenase | S. cerevisiae | Gene duplication | Cyt, Mito, Pex | McAlister-Henn and Thompson, 1987; Minard and McAlister-Henn, 1991; Steffan and McAlister-Henn, 1992 |
| NADPH-dependent isocitrate dehydrogenase | S. cerevisiae | Gene duplication | Cyt, Mito, Pex | Haselbeck and McAlister-Henn, 1991; Henke et al., 1998; Loftus et al., 1994; van Roermund et al., 1998 |
| Citrate synthase | S. cerevisiae | Gene duplication | Mito, Pex | Kim et al., 1986; Lewin et al., 1990; Rosenkrantz et al., 1986 |
| NADPH-dependent isocitrate dehydrogenase | A. nidulans | Alternative transcription initiation sites | Cyt, Mito, Pex | Szewczyk et al., 2001 |
| Malate dehydrogenase | Y. lipolytica | Differential splicing | Cyt, Pex | Kabran et al., 2012 |
| 6-phosphogluconate dehydrogenase | C. albicans | Differential splicing | Cyt, Pex | Strijbis et al., 2012 |
| Hydroxypyruvate reductase | Cucurbita sp. (cv. Kurokawa Amakuri Nankin) | Differential splicing | Cyt, Pex | Hayashi et al., 1996 |
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | U. maydis | Differential splicing | Cyt, Pex | Freitag et al., 2012 |
| Phosphoglycerate kinase (PGK) | A. nidulans | Differential splicing | Cyt, Pex | Freitag et al., 2012 |
| Phosphoglycerate kinase (PGK) | U. maydis | Ribosomal read-through | Cyt, Pex | Freitag et al., 2012 |
| Triose phosphate isomerase (TPI) | U. maydis | Ribosomal read-through | Cyt, Pex | Freitag et al., 2012 |
| Iron-containing superoxide dismutase | L. polyedrum | Alternative start codons | Plas/Mito, Pex | Bodyl and Mackiewicz, 2007 |
| 70-kDa heat shock protein | Citrullus vulgaris | Alternative start codons | Pex, Plas | Wimmer et al., 1997 |
| 3-Hydroxy-3-methylglutaryl coenzyme A lyase | H. sapiens (liver cells) | Multiple targeting signals | Mito, Pex | Ashmarina et al., 1999 |
| Type II NAD(P)H dehydrogenase | A. thaliana | Multiple targeting signals | Mito, Pex | Carrie et al., 2008; Xu et al., 2013 |
| Catalase A | S. cerevisiae | Multiple targeting signals | Mito, Pex | Petrova et al., 2004 |
| NAD+-dependent glycerol 3-phosphat dehydrogenase | S. cerevisiae | Protein modification (Phosphorylation) | Pex, Cyt/Nuc | Jung et al., 2010 |
| Glucose-6-phosphat dehydrogenase | A. thaliana | Redox dependent heterodimerization | Pex, Plas | Meyer et al., 2011 |
| Alanine-glyoxylate aminotransferase | H. sapiens | Protein folding | Cyt, Pex | Fodor et al., 2012 |
| Epoxide hydrolase | H. sapiens (liver cells, kidney cells) | Level of expression, quarternary structure | Cyt, Pex | Arand et al., 1991; Enayetallah et al., 2006; Luo et al., 2008 |
Abbreviations: Cyt, cytosol; Mito, mitochondrion; Nuc, nucleus; Pex, peroxisome; Plas, plastid.
Figure 1.
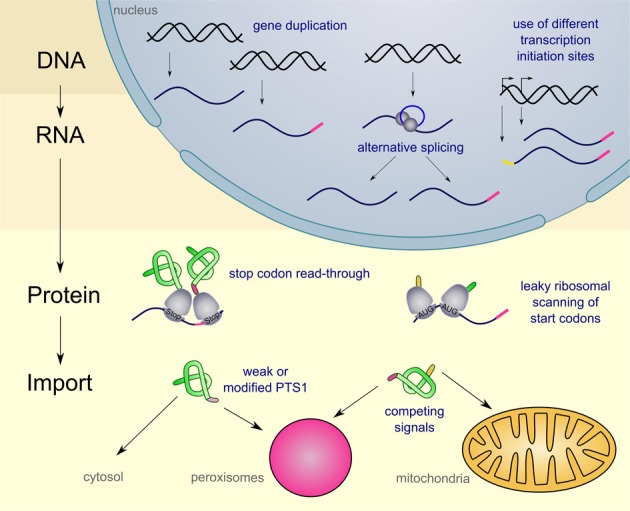
Schematic overview on the diversity of mechanism leading to dual targeting of peroxisomal proteins. Peroxisomal targeting signals are indicated in pink, while mitochondrial signal sequences are indicated in yellow. For further explanations see text.
Dual targeting of peroxisomal proteins by gene duplication.
Enzymes that are part of shuttle systems to maintain homeostasis between organelles and the surrounding cytoplasm usually display dual targeting. E.g. redox homeostasis is reached by exchange of reduced small intermediates with their oxidized counterparts. In mitochondria, recycling of NAD+/NADH is achieved via a malate/aspartate shuttle system (Bakker et al., 2001). In peroxisomes, regeneration of NAD+ during β-oxidation also depends on a related malate shuttle since the peroxisomal membrane is permeable for small molecules but impermeable for NADH (van Roermund et al., 1995; Antonenkov et al., 2004).
In Saccharomyces cerevisiae three genes encoding malate dehydrogenases have been identified. These code for distinct isoforms, which localize in the cytoplasm, mitochondria and peroxisomes, respectively (McAlister-Henn and Thompson, 1987; Minard and McAlister-Henn, 1991; Steffan and McAlister-Henn, 1992). The different isoenzymes not only carry different targeting signals, but also fulfill distinct functions and show specific regulation. All three malate dehydrogenases are involved in maintaining redox homeostasis. In addition, mitochondrial Mdh1 participates in the tricarboxylic acid cycle, cytosolic Mdh2 is required for gluconeogenesis and peroxisomal Mdh3 is an essential component of the glyoxylate cycle (McAlister-Henn and Thompson, 1987; Minard and McAlister-Henn, 1991; Steffan and McAlister-Henn, 1992)
NADP+/NADPH homeostasis in peroxisomes and mitochondria is reached via an alternative shuttle that involves the exchange of isocitrate with alpha-ketoglutarate. This reaction is catalyzed by NADPH dependent isocitrate dehydrogenase (Idp). Similar to malate dehydrogenase, discrete genes (idp1, idp2 and idp3) encode the mitochondrial, cytosolic and peroxisomal isoforms of Idp in S. cerevisiae (Haselbeck and McAlister-Henn, 1991; Loftus et al., 1994; Henke et al., 1998; van Roermund et al., 1998). Also, the mitochondrial (Cit1) and peroxisomal isoforms (Cit2) of citrate synthase are encoded by different genes in S. cerevisiae (Kim et al., 1986; Rosenkrantz et al., 1986; Lewin et al., 1990).
In yeast this type of multiple localization might be preferred since this species has undergone a large scale genome duplication during its evolution (Kellis et al., 2004). This allowed to attribute different cellular functions and localizations to these paralogous genes. In addition encoding isozymes by separate genes facilitates differential control and regulation not only on the transcriptional level, but also at the level of enzyme kinetics and allosteric control. It has been shown that in S. cerevisiae, which only contains a very limited number of introns, protein composition is nearly exclusively regulated by differential transcription (Goffeau et al., 1996).
In contrast, other eukaryotes make extensive use of post-transcriptional processes such as alternative splicing to adapt the proteome to environmental changes (Nilsen and Graveley, 2010).
Dual localization resulting from different transcripts derived from a single gene
In contrast to budding yeast, where gene duplication is commonly used for dual targeting of isozymes, other species often use single genes to produce dually targeted proteins. In Aspergillus nidulans synthesis of cytoplasmic, mitochondrial and peroxisomal isoforms of NADP-dependent Idp results from alternative use of transcription initiation sites of the idpA gene (Szewczyk et al., 2001). The longer transcript encodes a protein which contains both an N-terminal mitochondrial targeting sequence (MTS) and a C-terminal PTS1. Downstream transcription initiation leads to a shorter idpA transcript coding for a protein without the MTS (Szewczyk et al., 2001). While the longer form is preferentially located in mitochondria, the shorter form lacking the MTS is targeted both to peroxisomes and the cytosol. The functional dominance of the N-terminal mitochondrial over the C-terminal peroxisomal targeting signal is most likely due to commitment to mitochondrial import occurring co-translationally before the C-terminus is synthesized (Danpure, 1997).
Differential splicing is another mechanism to generate compartment-specific isoforms from single genes (Yogev and Pines, 2011). In the yeast Yarrowia lipolytica the cytoplasmic and peroxisomal isoforms of malate dehydrogenase are generated from alternatively spliced transcripts that differ in their intron size by only four nucleotides. The resulting proteins are highly similar but only one of the Mdh isoenzymes carries a functional PTS1 (Kabran et al., 2012). A related mechanism has been reported for dual targeting of 6-phosphogluconate dehydrogenase (Gnd1) in Candida albicans. In this human pathogenic fungus, alternative splicing of gnd1 transcripts leads to expression of a PTS2 containing isoform (Strijbis et al., 2012). Gnd1 is an essential enzyme of the oxidative branch of the pentose phosphate pathway. This pathway is used to generate NADPH and predominantly resides in the cytosol but has also been found in peroxisomes (Antonenkov, 1989; Corpas et al., 1998; Frederiks and Vreeling-Sindelarova, 2001; Boren et al., 2006; Reumann et al., 2007). Two other enzymes of this pathway, the glucose-6-phosphate dehydrogenase Zwf1 and the 6-phosphogluconolactonase Sol3, have been observed in peroxisomes in C. albicans (Strijbis et al., 2012). Differential splicing allows for regulation of dual targeting. In pumpkin leaves the ratio of peroxisomal and cytosolic isoforms of hydroxypyruvate reductase is achieved by light dependent differentially splicing (Hayashi et al., 1996; Mano et al., 1999, 2000).
An unexpected case of dual targeting by alternative splicing was recently described for fungal enzymes involved in glycolysis. This metabolic pathway is considered to be cytoplasmic and glycolytic proteins such as glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and phosphoglycerate kinase (PGK) often serve as cytoplasmic marker proteins in cell biology. A notable exception are trypanosomes, which have transferred the cytoplasmic glycolytic pathway into peroxisome-derived glycosomes (Opperdoes and Borst, 1977). This is considered as an adaptation to the unique lifestyle of these parasites in the bloodstream of vertebrates (Michels et al., 2006). In the basidiomycetous fungus Ustilago maydis a C-terminal extended peroxisomal isoform of GAPDH is expressed from an alternatively spliced transcript (Freitag et al., 2012). Inspection of other fungal species revealed that dual targeting of glycolytic enzymes is widespread. In the ascomycetous fungus Aspergillus nidulans the peroxisomal isoform of PGK but not of GAPDH is generated by differential splicing (Freitag et al., 2012).
Dual localization via “leaky” start and stop codons
Further bioinformatic analysis of fungal genes coding for glycolytic enzymes revealed a novel molecular mechanism for dual targeting. Peroxisomal targeting of GAPDH, PGK and triose phosphate isomerase (TPI) is reached by ribosomal read-through of stop codons resulting in a fraction of C-terminally extended proteins ending with a PTS1 (Freitag et al., 2012). Stop codon read-through has been described for retroviral systems where it is used to enlarge protein diversity, but was also observed for cellular transcripts (Bertram et al., 2001; Jungreis et al., 2011). The efficiency of ribosomes to recognize stop codons is affected by sequence context and RNA secondary structure and might be subject of control (Bertram et al., 2001). Isoforms generated by ribosomal read-through correspond to a single transcript and escape detection by transcriptomics. Therefore, this novel mode of dual targeting is difficult to observe.
A related way to produce different isoforms from a single transcript is the use of alternative start codons. The iron-containing SOD of the dinoflagellate Lingulodinium polyedrum catalyzes dismutation of superoxide radicals to hydrogen peroxide and oxygen as the first line of defense against reactive oxygen species (ROS) (Bodyl and Mackiewicz, 2007; McCord and Fridovich, 1969). Translation initiation at the first start codon results in an SOD, which contains both an N-terminal targeting signal for plastids and mitochondria and a C-terminal PTS1. This form was suggested to reside in plastids and in mitochondria. Efficient peroxisomal targeting of SOD appears to depend on leaky ribosomal scanning and initiation at a downstream in-frame start codon resulting in an isoform lacking the N-terminal signal sequence (Bodyl and Mackiewicz, 2007). In watermelon cotyledons the mRNA molecules of a 70-kDa heat shock protein contain two in frame start codons. Translational initiation at the first start codon leads to a longer isoform, which carries a N-terminal presequence mediating plastid import, while the shorter isoform localizes to peroxisomes due to a PTS2 (Wimmer et al., 1997).
Dual targeting of proteins with multiple targeting signals
Although it is commonly assumed that N-terminal signal sequences are dominant over PTS1 (Danpure, 1997), several examples are known where substantial peroxisomal targeting occurs even in the presence of an N-terminal MTS. 3-Hydroxy-3-methylglutaryl coenzyme A lyase (HL) catalyzes the conversion of β-hydroxy-β –methylglutaryl-CoA to acetoacetate, which is important during sterol biosynthesis in mitochondria. In human liver cells HL shows dual localization in mitochondria and peroxisomes. Peroxisomal HL still contains the N-terminal mitochondrial signal sequence, suggesting that dual localization of HL results from an intricate balance between mitochondrial and peroxisomal uptake (Ashmarina et al., 1999).
Another protein with competing signals is type II NAD(P)H dehydrogenase. This enzyme is typically located at the inner mitochondrial membrane but is also found in chloroplasts or peroxisomes (Xu et al., 2013). Three of the seven Arabidopsis thaliana genes encoding type II NAD(P)H dehydrogenases (ND) give rise to proteins which are dually targeted both to mitochondria and peroxisomes. These proteins carry an additional C-terminal signal for peroxisomal targeting. Intracellular distribution of the ND proteins with competing signals was shown to depend on the affinity of their signal sequences for their respective receptors/chaperones (Carrie et al., 2008).
If proteins contain competing targeting signals, localization studies with fluorescent proteins may result in ambiguous results. A number of A. thaliana acyl-activating enzymes localize either to peroxisomes or to other compartments depending on whether the fluorescent reporter protein was fused at the N- or the C-terminus (Hooks et al., 2012). Therefore it is still unclear whether these proteins occur outside of peroxisomes also in the natural situation.
Competition between the two targeting signals may also be affected by environmental factors. Catalase A (Cta1) of S. cerevisiae contains in addition to a non-canonical mitochondrial targeting signal, two peroxisomal targeting signals, an internal signal and a C-terminal PTS1 (Petrova et al., 2004). Both PTSs were shown to be sufficient to target Cta1 to peroxisomes (Kragler et al., 1993). The distribution of Cta1 between peroxisomes and mitochondria is influenced by growth conditions. In the presence of nutrients enhancing H2O2 formation, like oleic acid, catalase A is predominantly targeted to peroxisomes. In contrast, cultivation of yeast in raffinose leads to increased mitochondrial localization of Cta1. However, the molecular base for this differential targeting is still obscure (Petrova et al., 2004).
Proteins carrying regulated or inefficient PTS
In all examples discussed above, multiple targeting signals are involved in dual localization residing either concomitantly in a single polypeptide or in different isoforms. In the case of proteins that occur both in peroxisomes and the cytoplasm alternative mechanisms may operate. Dual localization can also result from modified or weak PTS1 signals leading to inefficient import into peroxisomes. Partial peroxisomal localization is difficult to visualize with fluorescent marker proteins, since cytoplasmic fluorescence usually prevents detection of the peroxisomal localization. Therefore this type of dual targeting is likely to be missed in microscopic studies. For the similar case of partial mitochondrial targeting a lacZ-complementation assay has been successfully applied to verify dual targeting (Ben-Menachem et al., 2011). Photobleaching of the cytosolic fraction can also be used to visualize partial peroxisomal localization (Buch et al., 2009).
In the NAD+-dependent glycerol 3-phosphate dehydrogenase (Gpd1) of S. cerevisiae, protein modification via phosphorylation is used to interfere with peroxisomal targeting (Jung et al., 2010). Gpd1 catalyzes the conversion of dihydroxyacetone phosphate (DHAP) to glycerol 3-phosphate (G3P) to cope with osmotic stress (Merkel et al., 1982; Chen et al., 1987). Gpd1 harbors an N-terminal PTS2, however, the subcellular distribution of Gpd1 depends on environmental factors. Upon osmotic cell stress, Gpd1 is relocated to both the cytosol and the nucleus. This altered localization is triggered by phosphorylation of two serine residues close to the PTS2, thus impairing peroxisomal import (Jung et al., 2010). In A. thaliana, peroxisomal import of glucose-6-phosphate dehydrogenase (G6PD1) is triggered by redox signaling and results in relocalization of G6PD1 from chloroplasts to peroxisomes. In this case, formation of a disulfide bridge allows recognition of an internal PTS (Meyer et al., 2011).
A weak/non-canonical PTS1 motif has recently been shown to be critical for proper folding of a PTS1 bearing protein due to prolonged duration of cytosolic localization prior to transfer into peroxisomes (Williams et al., 2012). Especially the import of proteins bearing non-canonical PTS1 motifs may depend on correct protein folding. Even minor misfolding may result in cytosolic localization as was demonstrated for the alanine-glyoxylate aminotransferase (AGT) of humans (Fodor et al., 2012). AGT is known to exhibit a variable distribution in mitochondria and/or peroxisomes in a variety of mammalian species (Danpure, 1997). Similarly, the non-canonical PTS1 motif of human epoxide hydrolase triggers peroxisomal import as a function of concentration and quaternary structure of the protein (Arand et al., 1991; Enayetallah et al., 2006; Luo et al., 2008).
Recent comprehensive studies of the peroxisomal proteome revealed additional proteins that have been previously annotated as cytosolic. Especially in plant peroxisomes, a variety of proteins with unconventional PTS1-motifs has been identified (Reumann et al., 2007; Reumann, 2011). Some of these proteins turned out to reside exclusively in peroxisomes, while others localize in the cytoplasm as determined by microscopy (Reumann et al., 2009). But this does not prove that these proteins are cytosolic since a minor fraction may reside in peroxisomes. At least for one of these proteins, a glutathione reductase, carrying a quite unusual PTS1 (-TNL), partial targeting to peroxisomes was demonstrated (Kataya and Reumann, 2010).
In fungi, proteome studies confirmed the partial peroxisomal localization of glycolytic enzymes and revealed a further candidate, fructose-bisphosphate aldolase (FBA) (Kiel et al., 2009; Managadze et al., 2010). Partial peroxisomal targeting of FBA is presumably mediated by a conserved C-terminal non-canonical PTS1-like motif (Kiel et al., 2009; Freitag et al., 2012). In U. maydis, this motif is able to trigger complete peroxisomal import if fused as a dodecamer to GFP, while a full-length GFP-FBA fusion protein results in cytoplasmic fluorescence. This suggests that partial peroxisomal import requires additional features of the protein that interfere with recognition of the unconventional PTS1. The combination of bioinformatic and experimental strategies revealed a heterogeneity of functional PTS1 motifs both in plants and fungi (Reumann, 2011; Freitag et al., 2012). Taken together these data indicate that partial peroxisomal targeting may occur more often than previously assumed. It has even been suggested that all cytosolic proteins may be found in any organelle at least in tiny amounts probably due to mistargeting. This hypothesis was proposed to explain the transfer of whole metabolic pathways from one compartment to another during evolution e.g. that of glycolysis in trypanosomes (Martin, 2010).
Concluding remarks
The large variety of mechanisms leading to dual targeting of peroxisomal proteins (summarized in Figure 1) suggests that the metabolic capacity of peroxisomes might have been underestimated in the past. This idea is supported by the recent discovery of several hitherto unrecognized peroxisomal metabolic pathways. These include glycolysis in fungal peroxisomes, biotin synthesis in plants and fungi, as well as biosynthesis of secondary metabolites such as siderophores and antibiotics (Bartoszewska et al., 2011; Magliano et al., 2011; Tanabe et al., 2011; Freitag et al., 2012; Grundlinger et al., 2013). Especially for mammals, knowledge of the role of peroxisomes appears to be far from being complete (Schrader and Fahimi, 2008; Islinger et al., 2012). We envision that also in mammals the metabolic capacity of peroxisomes may be of greater variability with dual targeting playing a growing role.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Julia Ast received a fellowship from the Marburg University Research Academy (MARA). Julia Ast and Alina C. Stiebler were supported by the German Science Foundation (DFG) Graduate School GRK 1216 and Johannes Freitag was supported by the SYNMIKRO Center for Synthetic Microbiology, Marburg. Michael Bölker received a grant from the German Science Foundation (DFG) Research Group 1334.
References
- Antonenkov V. D. (1989). Dehydrogenases of the pentose phosphate pathway in rat liver peroxisomes. Eur. J. Biochem. 183, 75–82 10.1111/j.1432-1033.1989.tb14898.x [DOI] [PubMed] [Google Scholar]
- Antonenkov V. D., Sormunen R. T., Hiltunen J. K. (2004). The rat liver peroxisomal membrane forms a permeability barrier for cofactors but not for small metabolites in vitro. J. Cell. Sci. 117, 5633–5642 10.1242/jcs.01485 [DOI] [PubMed] [Google Scholar]
- Arand M., Knehr M., Thomas H., Zeller H. D., Oesch F. (1991). An impaired peroxisomal targeting sequence leading to an unusual bicompartmental distribution of cytosolic epoxide hydrolase. FEBS Lett. 294, 19–22 10.1016/0014-5793(91)81333-4 [DOI] [PubMed] [Google Scholar]
- Ashmarina L. I., Pshezhetsky A. V., Branda S. S., Isaya G., Mitchell G. A. (1999). 3-Hydroxy-3-methylglutaryl coenzyme A lyase: targeting and processing in peroxisomes and mitochondria. J. Lipid Res. 40, 70–75 [PubMed] [Google Scholar]
- Bakker B. M., Overkamp K. M., van Maris A. J., Kotter P., Luttik M. A., van Dijken J. P., et al. (2001). Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25, 15–37 10.1111/j.1574-6976.2001.tb00570.x [DOI] [PubMed] [Google Scholar]
- Bartoszewska M., Opalinski L., Veenhuis M., van der Klei I. J. (2011). The significance of peroxisomes in secondary metabolite biosynthesis in filamentous fungi. Biotechnol. Lett. 33, 1921–1931 10.1007/s10529-011-0664-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Menachem R., Tal M., Shadur T., Pines O. (2011). A third of the yeast mitochondrial proteome is dual localized: a question of evolution. Proteomics 11, 4468–4476 10.1002/pmic.201100199 [DOI] [PubMed] [Google Scholar]
- Bertram G., Innes S., Minella O., Richardson J., Stansfield I. (2001). Endless possibilities: translation termination and stop codon recognition. Microbiology 147, 255–269 [DOI] [PubMed] [Google Scholar]
- Bodyl A., Mackiewicz P. (2007). Analysis of the targeting sequences of an iron-containing superoxide dismutase (SOD) of the dinoflagellate Lingulodinium polyedrum suggests function in multiple cellular compartments. Arch. Microbiol. 187, 281–296 10.1007/s00203-006-0194-5 [DOI] [PubMed] [Google Scholar]
- Boren J., Ramos-Montoya A., Bosch K. S., Vreeling H., Jonker A., Centelles J. J., et al. (2006). In situ localization of transketolase activity in epithelial cells of different rat tissues and subcellularly in liver parenchymal cells. J. Histochem. Cytochem. 54, 191–199 10.1369/jhc.5A6745.2005 [DOI] [PubMed] [Google Scholar]
- Breidenbach R. W., Beevers H. (1967). Association of the glyoxylate cycle enzymes in a novel subcellular particle from castor bean endosperm. Biochem. Biophys. Res. Commun. 27, 462–469 10.1016/S0006-291X(67)80007-X [DOI] [PubMed] [Google Scholar]
- Brocard C., Hartig A. (2006). Peroxisome targeting signal 1: is it really a simple tripeptide? Biochim. Biophys. Acta 1763, 1565–1573 10.1016/j.bbamcr.2006.08.022 [DOI] [PubMed] [Google Scholar]
- Brocard C., Kragler F., Simon M. M., Schuster T., Hartig A. (1994). The tetratricopeptide repeat-domain of the PAS10 protein of Saccharomyces cerevisiae is essential for binding the peroxisomal targeting signal-SKL. Biochem. Biophys. Res. Commun. 204, 1016–1022 10.1006/bbrc.1994.2564 [DOI] [PubMed] [Google Scholar]
- Buch C., Hunt M. C., Alexson S. E., Hallberg E. (2009). Localization of peroxisomal matrix proteins by photobleaching. Biochem. Biophys. Res. Commun. 388, 355–359 10.1016/j.bbrc.2009.08.013 [DOI] [PubMed] [Google Scholar]
- Carrie C., Murcha M. W., Kuehn K., Duncan O., Barthet M., Smith P. M., et al. (2008). Type II NAD(P)H dehydrogenases are targeted to mitochondria and chloroplasts or peroxisomes in Arabidopsis thaliana. FEBS Lett. 582, 3073–3079 10.1016/j.febslet.2008.07.061 [DOI] [PubMed] [Google Scholar]
- Chen S. M., Trumbore M. W., Osinchak J. E., Merkel J. R. (1987). Improved purification and some molecular and kinetic properties of sn-glycerol-3-phosphate dehydrogenase from Saccharomyces cerevisiae. Prep. Biochem. 17, 435–446 10.1080/00327488708062506 [DOI] [PubMed] [Google Scholar]
- Corpas F. J., Barroso J. B., Sandalio L. M., Distefano S., Palma J. M., Lupianez J. A., et al. (1998). A dehydrogenase-mediated recycling system of NADPH in plant peroxisomes. Biochem. J. 330(Pt 2), 777–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danpure C. J. (1997). Variable peroxisomal and mitochondrial targeting of alanine: glyoxylate aminotransferase in mammalian evolution and disease. Bioessays 19, 317–326 10.1002/bies.950190409 [DOI] [PubMed] [Google Scholar]
- deDuve C., Bauduin P. (1966). Peroxisomes (Microbodies and related particles). Physiol. Rev. 46, 323–357 [DOI] [PubMed] [Google Scholar]
- Enayetallah A. E., French R. A., Barber M., Grant D. F. (2006). Cell-specific subcellular localization of soluble epoxide hydrolase in human tissues. J. Histochem. Cytochem. 54, 329–335 10.1369/jhc.5A6808.2005 [DOI] [PubMed] [Google Scholar]
- Fodor K., Wolf J., Erdmann R., Schliebs W., Wilmanns M. (2012). Molecular requirements for peroxisomal targeting of alanine-glyoxylate aminotransferase as an essential determinant in primary hyperoxaluria type 1. PLoS Biol. 10:e1001309 10.1371/journal.pbio.1001309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederiks W. M., Vreeling-Sindelarova H. (2001). Localization of glucose-6-phosphate dehydrogenase activity on ribosomes of granular endoplasmic reticulum, in peroxisomes and peripheral cytoplasm of rat liver parenchymal cells. Histochem. J. 33, 345–353 10.1023/A:1012427224822 [DOI] [PubMed] [Google Scholar]
- Freitag J., Ast J., Bolker M. (2012). Cryptic peroxisomal targeting via alternative splicing and stop codon read-through in fungi. Nature 485, 522–525 10.1038/nature11051 [DOI] [PubMed] [Google Scholar]
- Gabaldon T. (2010). Peroxisome diversity and evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 765–773 10.1098/rstb.2009.0240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galland N., de Walque S., Voncken F. G., Verlinde C. L., Michels P. A. (2010). An internal sequence targets Trypanosoma brucei triosephosphate isomerase to glycosomes. Mol. Biochem. Parasitol. 171, 45–49 10.1016/j.molbiopara.2010.01.002 [DOI] [PubMed] [Google Scholar]
- Glover J. R., Andrews D. W., Rachubinski R. A. (1994). Saccharomyces cerevisiae peroxisomal thiolase is imported as a dimer. Proc. Natl. Acad. Sci. U.S.A. 91, 10541–10545 10.1073/pnas.91.22.10541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A., Barrell B. G., Bussey H., Davis R. W., Dujon B., Feldmann H., et al. (1996). Life with 6000 genes. Science 274, 546, 563–547. [DOI] [PubMed] [Google Scholar]
- Gonzalez N. H., Felsner G., Schramm F. D., Klingl A., Maier U. G., Bolte K. (2011). A single peroxisomal targeting signal mediates matrix protein import in diatoms. PLoS ONE 6:e25316 10.1371/journal.pone.0025316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. G., Keller G. A., Subramani S. (1987). Identification of a peroxisomal targeting signal at the carboxy terminus of firefly luciferase. J. Cell Biol. 105, 2923–2931 10.1083/jcb.105.6.2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. J., Keller G. A., Hosken N., Wilkinson J., Subramani S. (1989). A conserved tripeptide sorts proteins to peroxisomes. J. Cell Biol. 108, 1657–1664 10.1083/jcb.108.5.1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundlinger M., Yasmin S., Lechner B. E., Geley S., Schrettl M., Hynes M., et al. (2013). Fungal siderophore biosynthesis is partially localized in peroxisomes. Mol. Microbiol. 88, 862–875 10.1111/mmi.12225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunkel K., van Dijk R., Veenhuis M., van der Klei I. J. (2004). Routing of Hansenula polymorpha alcohol oxidase: an alternative peroxisomal protein-sorting machinery. Mol. Biol. Cell 15, 1347–1355 10.1091/mbc.E03-04-0258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haselbeck R. J., McAlister-Henn L. (1991). Isolation, nucleotide sequence, and disruption of the Saccharomyces cerevisiae gene encoding mitochondrial NADP(H)-specific isocitrate dehydrogenase. J. Biol. Chem. 266, 2339–2345 [PubMed] [Google Scholar]
- Hayashi M., Tsugeki R., Kondo M., Mori H., Nishimura M. (1996). Pumpkin hydroxypyruvate reductases with and without a putative C-terminal signal for targeting to microbodies may be produced by alternative splicing. Plant Mol. Biol. 30, 183–189 10.1007/BF00017813 [DOI] [PubMed] [Google Scholar]
- Henke B., Girzalsky W., Berteaux-Lecellier V., Erdmann R. (1998). IDP3 encodes a peroxisomal NADP-dependent isocitrate dehydrogenase required for the beta-oxidation of unsaturated fatty acids. J. Biol. Chem. 273, 3702–3711 10.1074/jbc.273.6.3702 [DOI] [PubMed] [Google Scholar]
- Hooks K. B., Turner J. E., Graham I. A., Runions J., Hooks M. A. (2012). GFP-tagging of Arabidopsis acyl-activating enzymes raises the issue of peroxisome-chloroplast import competition versus dual localization. J. Plant Physiol. 169, 1631–1638 10.1016/j.jplph.2012.05.026 [DOI] [PubMed] [Google Scholar]
- Islinger M., Grille S., Fahimi H. D., Schrader M. (2012). The peroxisome: an update on mysteries. Histochem. Cell Biol. 137, 547–574 10.1007/s00418-012-0941-4 [DOI] [PubMed] [Google Scholar]
- Islinger M., Li K. W., Seitz J., Volkl A., Luers G. H. (2009). Hitchhiking of Cu/Zn superoxide dismutase to peroxisomes–evidence for a natural piggyback import mechanism in mammals. Traffic 10, 1711–1721 10.1111/j.1600-0854.2009.00966.x [DOI] [PubMed] [Google Scholar]
- Jedd G., Chua N. H. (2000). A new self-assembled peroxisomal vesicle required for efficient resealing of the plasma membrane. Nat. Cell Biol. 2, 226–231 10.1038/35008652 [DOI] [PubMed] [Google Scholar]
- Jung S., Marelli M., Rachubinski R. A., Goodlett D. R., Aitchison J. D. (2010). Dynamic changes in the subcellular distribution of Gpd1p in response to cell stress. J. Biol. Chem. 285, 6739–6749 10.1074/jbc.M109.058552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungreis I., Lin M. F., Spokony R., Chan C. S., Negre N., Victorsen A., et al. (2011). Evidence of abundant stop codon readthrough in Drosophila and other metazoa. Genome Res. 21, 2096–2113 10.1101/gr.119974.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabran P., Rossignol T., Gaillardin C., Nicaud J. M., Neuveglise C. (2012). Alternative splicing regulates targeting of malate dehydrogenase in Yarrowia lipolytica. DNA Res. 19, 231–244 10.1093/dnares/dss007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataya A. R., Reumann S. (2010). Arabidopsis glutathione reductase 1 is dually targeted to peroxisomes and the cytosol. Plant Signal. Behav. 5, 171–175 10.4161/psb.5.2.10527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M., Birren B. W., Lander E. S. (2004). Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428, 617–624 10.1038/nature02424 [DOI] [PubMed] [Google Scholar]
- Kiel J. A., van den Berg M. A., Fusetti F., Poolman B., Bovenberg R. A., Veenhuis M., et al. (2009). Matching the proteome to the genome: the microbody of penicillin-producing Penicillium chrysogenum cells. Funct. Integr. Genomics 9, 167–184 10.1007/s10142-009-0110-6 [DOI] [PubMed] [Google Scholar]
- Kim K. S., Rosenkrantz M. S., Guarente L. (1986). Saccharomyces cerevisiae contains two functional citrate synthase genes. Mol. Cell. Biol. 6, 1936–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kionka C., Kunau W. H. (1985). Inducible beta-oxidation pathway in Neurospora crassa. J. Bacteriol. 161, 153–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A. T., van den Berg M., Bottger G., Tabak H. F., Distel B. (2002). Saccharomyces cerevisiae acyl-CoA oxidase follows a novel, non-PTS1, import pathway into peroxisomes that is dependent on Pex5p. J. Biol. Chem. 277, 25011–25019 10.1074/jbc.M203254200 [DOI] [PubMed] [Google Scholar]
- Kragler F., Langeder A., Raupachova J., Binder M., Hartig A. (1993). Two independent peroxisomal targeting signals in catalase A of Saccharomyces cerevisiae. J. Cell Biol. 120, 665–673 10.1083/jcb.120.3.665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lametschwandtner G., Brocard C., Fransen M., Van Veldhoven P., Berger J., Hartig A. (1998). The difference in recognition of terminal tripeptides as peroxisomal targeting signal 1 between yeast and human is due to different affinities of their receptor Pex5p to the cognate signal and to residues adjacent to it. J. Biol. Chem. 273, 33635–33643 10.1074/jbc.273.50.33635 [DOI] [PubMed] [Google Scholar]
- Lewin A. S., Hines V., Small G. M. (1990). Citrate synthase encoded by the CIT2 gene of Saccharomyces cerevisiae is peroxisomal. Mol. Cell. Biol. 10, 1399–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus T. M., Hall L. V., Anderson S. L., McAlister-Henn L. (1994). Isolation, characterization, and disruption of the yeast gene encoding cytosolic NADP-specific isocitrate dehydrogenase. Biochemistry 33, 9661–9667 10.1021/bi00198a035 [DOI] [PubMed] [Google Scholar]
- Luo B., Norris C., Bolstad E. S., Knecht D. A., Grant D. F. (2008). Protein quaternary structure and expression levels contribute to peroxisomal-targeting-sequence-1-mediated peroxisomal import of human soluble epoxide hydrolase. J. Mol. Biol. 380, 31–41 10.1016/j.jmb.2008.04.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magliano P., Flipphi M., Arpat B. A., Delessert S., Poirier Y. (2011). Contributions of the peroxisome and beta-oxidation cycle to biotin synthesis in fungi. J. Biol. Chem. 286, 42133–42140 10.1074/jbc.M111.279687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Managadze D., Wurtz C., Wiese S., Meyer H. E., Niehaus G., Erdmann R., et al. (2010). A proteomic approach towards the identification of the matrix protein content of the two types of microbodies in Neurospora crassa. Proteomics 10, 3222–3234 10.1002/pmic.201000095 [DOI] [PubMed] [Google Scholar]
- Mano S., Hayashi M., Nishimura M. (1999). Light regulates alternative splicing of hydroxypyruvate reductase in pumpkin. Plant J. 17, 309–320 10.1046/j.1365-313X.1999.00378.x [DOI] [PubMed] [Google Scholar]
- Mano S., Hayashi M., Nishimura M. (2000). A leaf-peroxisomal protein, hydroxypyruvate reductase, is produced by light-regulated alternative splicing. Cell Biochem. Biophys. 32(Spring), 147–154 10.1385/CBB:32:1-3:147 [DOI] [PubMed] [Google Scholar]
- Martin W. (2010). Evolutionary origins of metabolic compartmentalization in eukaryotes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 847–855 10.1098/rstb.2009.0252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzioch M., Erdmann R., Veenhuis M., Kunau W. H. (1994). PAS7 encodes a novel yeast member of the WD-40 protein family essential for import of 3-oxoacyl-CoA thiolase, a PTS2-containing protein, into peroxisomes. EMBO J. 13, 4908–4918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlister-Henn L., Thompson L. M. (1987). Isolation and expression of the gene encoding yeast mitochondrial malate dehydrogenase. J. Bacteriol. 169, 5157–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord J. M., Fridovich I. (1969). Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 244, 6049–6055 [PubMed] [Google Scholar]
- McNew J. A., Goodman J. M. (1994). An oligomeric protein is imported into peroxisomes in vivo. J. Cell Biol. 127, 1245–1257 10.1083/jcb.127.5.1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkel J. R., Straume M., Sajer S. A., Hopfer R. L. (1982). Purification and some properties of sn-glycerol-3-phosphate dehydrogenase from Saccharomyces cerevisiae. Anal. Biochem. 122, 180–185 10.1016/0003-2697(82)90268-8 [DOI] [PubMed] [Google Scholar]
- Meyer T., Holscher C., Schwoppe C., von Schaewen A. (2011). Alternative targeting of Arabidopsis plastidic glucose-6-phosphate dehydrogenase G6PD1 involves cysteine-dependent interaction with G6PD4 in the cytosol. Plant J. 66, 745–758 10.1111/j.1365-313X.2011.04535.x [DOI] [PubMed] [Google Scholar]
- Michels P. A., Bringaud F., Herman M., Hannaert V. (2006). Metabolic functions of glycosomes in trypanosomatids. Biochim. Biophys. Acta 1763, 1463–1477 10.1016/j.bbamcr.2006.08.019 [DOI] [PubMed] [Google Scholar]
- Minard K. I., McAlister-Henn L. (1991). Isolation, nucleotide sequence analysis, and disruption of the MDH2 gene from Saccharomyces cerevisiae: evidence for three isozymes of yeast malate dehydrogenase. Mol. Cell. Biol. 11, 370–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley A. M., Hettema E. H., Ketting R., Plasterk R., Tabak H. F. (2000). Caenorhabditis elegans has a single pathway to target matrix proteins to peroxisomes. EMBO Rep. 1, 40–46 10.1093/embo-reports/kvd010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen T. W., Graveley B. R. (2010). Expansion of the eukaryotic proteome by alternative splicing. Nature 463, 457–463 10.1038/nature08909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyathi Y., Baker A. (2006). Plant peroxisomes as a source of signalling molecules. Biochim. Biophys. Acta 1763, 1478–1495 10.1016/j.bbamcr.2006.08.031 [DOI] [PubMed] [Google Scholar]
- Opperdoes F. R., Borst P. (1977). Localization of nine glycolytic enzymes in a microbody-like organelle in Trypanosoma brucei: the glycosome. FEBS Lett. 80, 360–364 10.1016/0014-5793(77)80476-6 [DOI] [PubMed] [Google Scholar]
- Oshima Y., Kamigaki A., Nakamori C., Mano S., Hayashi M., Nishimura M., et al. (2008). Plant catalase is imported into peroxisomes by Pex5p but is distinct from typical PTS1 import. Plant Cell Physiol. 49, 671–677 10.1093/pcp/pcn038 [DOI] [PubMed] [Google Scholar]
- Peterson G. C., Sommer J. M., Klosterman S., Wang C. C., Parsons M. (1997). Trypanosoma brucei: identification of an internal region of phosphoglycerate kinase required for targeting to glycosomal microbodies. Exp. Parasitol. 85, 16–23 10.1006/expr.1996.4114 [DOI] [PubMed] [Google Scholar]
- Petriv O. I., Tang L., Titorenko V. I., Rachubinski R. A. (2004). A new definition for the consensus sequence of the peroxisome targeting signal type 2. J. Mol. Biol. 341, 119–134 10.1016/j.jmb.2004.05.064 [DOI] [PubMed] [Google Scholar]
- Petrova V. Y., Drescher D., Kujumdzieva A. V., Schmitt M. J. (2004). Dual targeting of yeast catalase A to peroxisomes and mitochondria. Biochem. J. 380, 393–400 10.1042/BJ20040042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y., Antonenkov V. D., Glumoff T., Hiltunen J. K. (2006). Peroxisomal beta-oxidation–a metabolic pathway with multiple functions. Biochim. Biophys. Acta 1763, 1413–1426 10.1016/j.bbamcr.2006.08.034 [DOI] [PubMed] [Google Scholar]
- Reumann S. (2011). Toward a definition of the complete proteome of plant peroxisomes: where experimental proteomics must be complemented by bioinformatics. Proteomics 11, 1764–1779 10.1002/pmic.201000681 [DOI] [PubMed] [Google Scholar]
- Reumann S., Babujee L., Ma C., Wienkoop S., Siemsen T., Antonicelli G. E., et al. (2007). Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19, 3170–3193 10.1105/tpc.107.050989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S., Quan S., Aung K., Yang P., Manandhar-Shrestha K., Holbrook D., et al. (2009). In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant Physiol. 150, 125–143 10.1104/pp.109.137703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodin J. (1954). Correlation of Ultrastructural Organization and Function in Normal and Experimentally Changed Proximal Convoluted Tubule Cells of the Mouse Kidney, an Electron Microscopic Study. Thesis, Karolinska Institute, Stockholm. [Google Scholar]
- Rosenkrantz M., Alam T., Kim K. S., Clark B. J., Srere P. A., Guarente L. P. (1986). Mitochondrial and nonmitochondrial citrate synthases in Saccharomyces cerevisiae are encoded by distinct homologous genes. Mol. Cell. Biol. 6, 4509–4515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rucktäschel R., Girzalsky W., Erdmann R. (2011). Protein import machineries of peroxisomes. Biochim. Biophys. Acta 1808, 892–900 10.1016/j.bbamem.2010.07.020 [DOI] [PubMed] [Google Scholar]
- Schrader M., Fahimi H. D. (2008). The peroxisome: still a mysterious organelle. Histochem. Cell Biol. 129, 421–440 10.1007/s00418-008-0396-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan J. S., McAlister-Henn L. (1992). Isolation and characterization of the yeast gene encoding the MDH3 isozyme of malate dehydrogenase. J. Biol. Chem. 267, 24708–24715 [PubMed] [Google Scholar]
- Strijbis K., van den Burg J., Visser W. F., van den Berg M., Distel B. (2012). Alternative splicing directs dual localization of Candida albicans 6-phosphogluconate dehydrogenase to cytosol and peroxisomes. FEMS Yeast Res. 12, 61–68 10.1111/j.1567-1364.2011.00761.x [DOI] [PubMed] [Google Scholar]
- Swinkels B. W., Gould S. J., Bodnar A. G., Rachubinski R. A., Subramani S. (1991). A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 10, 3255–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk E., Andrianopoulos A., Davis M. A., Hynes M. J. (2001). A single gene produces mitochondrial, cytoplasmic, and peroxisomal NADP-dependent isocitrate dehydrogenase in Aspergillus nidulans. J. Biol. Chem. 276, 37722–37729 10.1074/jbc.M105645200 [DOI] [PubMed] [Google Scholar]
- Tanabe Y., Maruyama J., Yamaoka S., Yahagi D., Matsuo I., Tsutsumi N., et al. (2011). Peroxisomes are involved in biotin biosynthesis in Aspergillus and Arabidopsis. J. Biol. Chem. 286, 30455–30461 10.1074/jbc.M111.247338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Klei I. J., Yurimoto H., Sakai Y., Veenhuis M. (2006). The significance of peroxisomes in methanol metabolism in methylotrophic yeast. Biochim. Biophys. Acta 1763, 1453–1462 10.1016/j.bbamcr.2006.07.016 [DOI] [PubMed] [Google Scholar]
- van Roermund C. W., Hettema E. H., Kal A. J., van den Berg M., Tabak H. F., Wanders R. J. (1998). Peroxisomal beta-oxidation of polyunsaturated fatty acids in Saccharomyces cerevisiae: isocitrate dehydrogenase provides NADPH for reduction of double bonds at even positions. EMBO J. 17, 677–687 10.1093/emboj/17.3.677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roermund C. W., van den Berg M., Wanders R. J. (1995). Localization of peroxisomal 3-oxoacyl-CoA thiolase in particles of varied density in rat liver: implications for peroxisome biogenesis. Biochim. Biophys. Acta 1245, 348–358 10.1016/0304-4165(95)00099-2 [DOI] [PubMed] [Google Scholar]
- Wanders R. J., Waterham H. R. (2006). Biochemistry of mammalian peroxisomes revisited. Annu. Rev. Biochem. 75, 295–332 10.1146/annurev.biochem.74.082803.133329 [DOI] [PubMed] [Google Scholar]
- Williams C., Bener Aksam E., Gunkel K., Veenhuis M., van der Klei I. J. (2012). The relevance of the non-canonical PTS1 of peroxisomal catalase. Biochim. Biophys. Acta 1823, 1133–1141 10.1016/j.bbamcr.2012.04.006 [DOI] [PubMed] [Google Scholar]
- Wimmer B., Lottspeich F., van der Klei I., Veenhuis M., Gietl C. (1997). The glyoxysomal and plastid molecular chaperones (70-kDa heat shock protein) of watermelon cotyledons are encoded by a single gene. Proc. Natl. Acad. Sci. U.S.A. 94, 13624–13629 10.1073/pnas.94.25.13624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Law S. R., Murcha M. W., Whelan J., Carrie C. (2013). The dual targeting ability of type II NAD(P)H dehydrogenases arose early in land plant evolution. BMC Plant Biol. 13:100 10.1186/1471-2229-13-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Purdue P. E., Lazarow P. B. (2001). Eci1p uses a PTS1 to enter peroxisomes: either its own or that of a partner, Dci1p. Eur. J. Cell Biol. 80, 126–138 10.1078/0171-9335-00144 [DOI] [PubMed] [Google Scholar]
- Yogev O., Pines O. (2011). Dual targeting of mitochondrial proteins: mechanism, regulation and function. Biochim. Biophys. Acta 1808, 1012–1020 10.1016/j.bbamem.2010.07.004 [DOI] [PubMed] [Google Scholar]
- Zimmermann R., Neupert W. (1980). Biogenesis of glyoxysomes. Synthesis and intracellular transfer of isocitrate lyase. Eur. J. Biochem. 112, 225–233 10.1111/j.1432-1033.1980.tb07198.x [DOI] [PubMed] [Google Scholar]


