Abstract
Genetic variation in a population can be summarized through principal component analysis (PCA) on genome-wide data. PCs derived from such analyses are valuable for genetic association studies, where they can correct for population stratification. We investigated how to capture the genetic population structure in a well-characterized sample from the Netherlands and in a worldwide data set and examined whether (1) removing long-range linkage disequilibrium (LD) regions and LD-based SNP pruning significantly improves correlations between PCs and geography and (2) whether genetic differentiation may have been influenced by migration and/or selection. In the Netherlands, three PCs showed significant correlations with geography, distinguishing between: (1) North and South; (2) East and West; and (3) the middle-band and the rest of the country. The third PC only emerged with minimized LD, which also significantly increased correlations with geography for the other two PCs. In addition to geography, the Dutch North–South PC showed correlations with genome-wide homozygosity (r=0.245), which may reflect a serial-founder effect due to northwards migration, and also with height (♂: r=0.142, ♀: r=0.153). The divergence between subpopulations identified by PCs is partly driven by selection pressures. The first three PCs showed significant signals for diversifying selection (545 SNPs - the majority within 184 genes). The strongest signal was observed between North and South for the functional SNP in HERC2 that determines human blue/brown eye color. Thus, this study demonstrates how to increase ancestry signals in a relatively homogeneous population and how those signals can reveal evolutionary history.
Keywords: PCA, linkage disequilibrium, population structure, migration, diversifying selection, Netherlands
Introduction
Population genetic studies are of great value for detecting population substructure and making inferences about human history regarding migrations, expansions, and human evolution.1 The genetic variation in a population can be summarized by uncorrelated principal components (PCs) through principal component analysis (PCA) on genome-wide data, usually with the explained variance monotonically decreasing with each PC. The PCs explaining most variation often show striking correlations with geography,2, 3, 4 a consequence of the decreasing genetic similarity as geographic distance increases. Such PCs are also of value in genetic association studies, where they are used to correct for allele frequency differences due to systematic ancestry differences, i.e., population stratification.5
When analyzing genome-wide genetic variants, one has to consider that some regions of the genome may be overrepresented in the PCs due to elevated levels of linkage disequilibrium (LD), diluting the genome-wide patterns that reflect ancestry differences. Very strong and/or long-range LD at a particular locus can even result in PCs that only reflect genetic variation at that specific locus.6, 7 Price et al7 therefore recommended the exclusion of long-range LD regions for PCAs, but advised against pruning for LD, as it did not significantly affect PCs in HapMap populations.6 We hypothesize that these LD artifacts may have larger confounding effects when carrying out a PCA in a single relatively small population, where ancestry differences are relatively small, than in a PCA that is run on a pooled data set of multiple populations with greater between-population differences. To test this hypothesis, we ran PCAs on different SNP sets with varying levels of LD on a large sample of Dutch individuals, and separately in a pooled dataset consisting of the populations from the 1000 Genomes Project8 covering five different continents (Europe, Africa, Asia, North-, and South America). Correlations between PCs and geography should be a good proxy for how well the PCs reflect ancestry differences. Correction for stratified phenotypes in association studies, such as height/stature, should be more effective in reducing false positives when using PCs that are a better reflection of one's ancestry.
The PCs showing the strongest ancestry signals are then used to further study the population substructure and genetic history of the Netherlands. The demographic history of this population is complex and still not completely understood. This is partly due to the highly variable Dutch geographic landscape. Large parts of the Netherlands were (and still are) well below sea level and our current landscape (apart from the urbanized areas) resembles the one of ∼1500 AD. Before that time, large parts of the Netherlands were still covered by sea (either permanently or under strong tidal influence) and uninhabitable; hence, population sizes were probably very low. The arrival of the first Romans (56 BC) also marked the beginning of waves of immigrants from various parts of Europe. Among the first were Batavians (from Germany), during later centuries followed by large groups of economic immigrants and religious refugees from throughout Europe (mainly Iberian, French, Belgian, German, British, and Scandinavian).9 It is estimated that the ancestors of ∼75% of what we currently call the ‘native' Dutch population (autochtonen in Dutch) have immigrated into the Netherlands during the past 20 centuries.9 Further genetic differentiation within Dutch subpopulations may have been induced by isolation due to geographic and/or social factors. Studies on marriage records of the 19th and early 20th century showed greater isolation within Southern provinces, and the North-West province of Friesland, while the urbanized West showed lower rates of homogamy.10, 11, 12 Religion also had a considerable role in maintaining Dutch (sub)populations during almost the entire second half of the last millennium, separating the Catholic South from the mostly Protestant North, but also maintaining substructures among the highly segregated Protestant groups. Strong religious assortment was detectable until well into the 20th century.13 With increasing secularization during the 1960s and 1970s, however, religious assortment started to decline.14
The first goal of this study is to explore the ability of a PCA to capture population differentiation in a relatively homogeneous population using different SNP sets varying in LD. PCs that represent ancestry differences then are to be employed to aid in investigating patterns of past human migration and the impact of selection on genetic variation in the geographically relatively small area of the Netherlands (41 543 km2; 16 039 mi2). Migration patterns were previously detected through correlations between distance from Addis Abbaba, Ethiopia, and genome-wide heterozygosity and LD.15, 16, 17, 18 To investigate the influence of adaptive selection pressures on the genetic differentiation within the Netherlands, the distribution of alleles will be compared between subpopulations identified by the PCs.
SUBJECTS AND METHODS
Participants
Subjects were registered at the Netherlands Twin Register (NTR, N=5509)19 or the Netherlands Study of Depression and Anxiety (NESDA, N=2038).20 Genotyping was performed on the Affymetrix Human Genome-Wide SNP 6.0 Array (Affymetrix, Santa Clara, CA, USA) according to the manufacturer's protocol.
Individuals with possible non-Dutch or non-European ancestry (N=258) were identified by projecting PCs from the 1000 Genomes individuals on the Dutch individuals, and with additional help of the birth country of their parents (see Supplementary Information).
Only unrelated individuals were analyzed. Unrelated individuals were chosen using GCTA,21 by excluding one of each pair of individuals with an estimated genetic relationship of >0.025 (ie, more related than third or fourth cousin), reducing the sample from 7547 to 4441 subjects.
The current living address was available for 4103 unrelated subjects (of which 1841 also had place of birth available). Adult height was available for 3714 unrelated subjects, self-reported eye color for 1581 unrelated subjects (coded as blue, intermediate or brown), and self-reported hair color for 1583 unrelated subjects (coded as blond, red, light brown, dark brown, or black).
PCA on three SNP sets
Three different SNP sets were created to run the PCAs on, varying in the amount of LD allowed: Panel 1: all SNPs that passed QC (499 849 SNPs); Panel 2: excluding 24 long-range LD regions identified by Price et al7 (487 672 SNPs); and Panel 3: an LD-pruned SNP set without long-range LD regions, where SNPs were pruned recursively in a sliding window (window size=50, number of SNPs to shift after each step=5) based on a variance inflation factor (VIF) of 2 (130 248 SNPs). See Supplementary Information for details on QC. PCAs were run with the EIGENSOFT package6 to compute 10 PCs for each of the three LD varying SNP sets using its default parameters.
Effects of LD on PCA
To determine which SNPs underlie the variation reflected by the PCs, δ (absolute allele frequency difference) was calculated for all SNPs between individuals with the highest and individuals with the lowest PC values (top and bottom 1000 for the Dutch dataset; top and bottom 250 for 1000 Genomes). To investigate the amount of LD that influenced a PC, an LD matrix of its top 500 SNPs (determined by δ) was calculated in Plink, after which all LD values (r2) were averaged (Table 1).
Table 1. The 95% confidence intervals (CI) of the mean r 2 values of the top 500 SNPs (determined by δ) for each PC for each data set.
| 1000 Genomes | The Netherlands | |||||
|---|---|---|---|---|---|---|
| Panel 1=all SNPs that passed QC | Panel 2=Panel 1 without the 24 long-range LD regions | Panel 3=Panel 2 with genome-wide LD-based SNP pruning | Panel 1=All SNPs that passed QC | Panel 2=Panel 1 without the 24 long-range LD regions | Panel 3=Panel 2 without genome-wide LD-based SNP pruning | |
| PC1 | 0.1550–0.1556 | 0.1534–0.1540 | 0.1536–0.1542 | 0.0669–0.0676 | 0.0044–0.0049a | 0.0037–0.0042a |
| PC2 | 0.1008–0.1012 | 0.1003–0.1008 | 0.1004–0.1008 | 0.0889–0.0900a | 0.0570–0.0586 | 0.0061–0.0066b |
| PC3 | 0.1639–0.1648 | 0.1680–0.1688 | 0.1759–0.1767 | 0.0930–0.0941 | 0.0465–0.0479b | 0.0102–0.0109c |
| PC4 | 0.1721–0.1729 | 0.1776–0.1784 | 0.1888–0.1896 | 0.0865–0.0880 | 0.0516–0.0529 | 0.0056–0.0061 |
| PC5 | 0.1699–0.1704 | 0.1661–0.1666 | 0.1496–0.1501 | 0.0995–0.1014 | 0.0662–0.0678 | 0.0037–0.0042 |
| PC6 | 0.2077–0.2086 | 0.1946–0.1955 | 0.1764–0.1772 | 0.0908–0.0926 | 0.0678–0.0694 | 0.0036–0.0040 |
| PC7 | 0.0315–0.0319 | 0.0310–0.0315 | 0.0460–0.0465 | 0.0691–0.0708 | 0.0547–0.0562 | 0.0049–0.0054 |
| PC8 | 0.0343–0.0349 | 0.0449–0.0454 | 0.0342–0.0348 | 0.0521–0.0536b | 0.0574–0.0590 | 0.0038–0.0042 |
| PC9 | 0.1011–0.1018 | 0.0824–0.0830 | 0.0852–0.0858 | 0.0626–0.0641 | 0.0558–0.0573 | 0.0045–0.0050 |
| PC10 | 0.0799–0.0806 | 0.0179–0.0186 | 0.0141–0.0145 | 0.0618–0.0633 | 0.0526–0.0541 | 0.0040–0.0044 |
The LD changes very little across panels for the 1000 Genomes dataset, as opposed to the Dutch datasets.
The PC with the highest correlation with the North–South gradient.
The PC with the highest correlation with the East–West gradient.
The PC that also showed a correlation with the East–West gradient, and separates individuals from the middle of the Netherlands from individuals from the rest of the country (illustrated in Figure 1d).
To test whether LD influences correlations of PCs with geography, we compared the correlations of PCs from the three panels with the latitude and longitude coordinates with the R package psych, which allows testing the difference between two dependent correlations sharing one variable (the geographic location in this case).22, 23
Traces of migration: F, haplotype block size, and Fst
F (genome-wide homozygosity) was calculated in Plink.24 Haplotype blocks were calculated per chromosome in Plink for different groups of individuals. This was done with pair-wise LD calculations for SNPs within 4000 kb (the size of the largest long-range LD region: the chromosome 8p23.1 inversion), using the largest SNP set (499 849 SNPs). The sizes of all autosomal haplotype blocks were then averaged. Fst was calculated as a measure for genetic differentiation between populations according to Weir and Cockerham25 by calculating it for every SNP and then averaging all Fst values to obtain a genome-wide point estimate of the genetic distance.
Selection pressures as a source of genetic differentiation
Selection pressures were identified in Bayescan 2.1.26 A comparison of several algorithms designed to achieve this goal through Fst outlier tests concluded that this software package had the lowest false negative and false positive rates.27 After computing Fst values for all 499 849 SNPs between the top 1000 and bottom 1000 individuals for three PCs reflecting ancestry, Fst coefficients are decomposed into a population-specific component (β), shared by all loci, and a locus-specific component (α), shared by both populations. If α differs significantly from 0, it is assumed that the locus was under-diversifying (α>0) or balancing (α<0) selection. Significance is based on FDR-corrected q-values (<0.05).
For a more detailed description of the methods, see Supplementary Information.
Results
Increasing genome-wide ancestry signals by reducing LD
PCAs were run on three SNP sets that differed in the amount of LD: Panel 1 (499 849 SNPs), Panel 2 (excluding 24 known long-range LD regions: 487 672 SNPs), and Panel 3 (24 long-range LD regions excluded, and LD pruning: 130 248 SNPs). PCAs were run for the 1000 Genomes data set (N=1014; no Dutch included), and for a data set consisting of Dutch individuals only (N=4441).
For the 1000 Genomes dataset, the three panels had almost identical components (see correlations in Supplementary Tables S2 and S3). The only PC reflecting a long-range LD region was PC10 from Panel 1 (the top 449 SNPs based on δ fall within the inversion on chromosome 8p23.1). PCs extracted from the Dutch data set showed large differences between the three Panels (see Supplementary Tables S4 and S5). In Panel 1, all top 10 PCs represent variation in long-range LD regions. For the first 9 PCs, the majority or all of the top 500 SNPs fall in the 24 long-range LD regions, and for PC10, 45% of the top 500 SNPs come from 1 of the 24 long-range LD regions. For Panel 2, the LD levels between the top 500 SNPs of the top 10 PCs are slightly lower, but still somewhat in the same range as for Panel 1 (except for PC1, the North–South PC; see Table 1), while in Panel 3, the LD levels are about 10-fold lower (Table 1), suggesting that these PCs are likely to represent more genome-wide patterns.
As long as current and past migration rates are not too high, correlations with geography should be a good proxy for how well the PCs reflect ancestry. For a subset of the current sample with place of birth as well as current living address available (N=1841), the mean distance between birthplace and current living address is 33.33 km (20.71 mi; see Supplementary Figure S1). To test whether the degree of LD influences the correlations of the PCs with geography significantly, correlations between PCs from the three SNP panels and the North–South/East–West gradient were compared. The results of these tests are shown in Table 2 (per SNP panel, only the correlation of the PC with the highest geographic correlation is shown and tested). The correlations with geography are significantly improved for Panel 2, and again after additional LD-based SNP pruning in Panel 3. Panel 3 is the only Panel where the North–South and East–West PCs show up as the first two PCs respectively. Panel 3 also shows an additional PC (PC3) with a significant correlation of 0.162 (P<0.001) with the East–West gradient (see Figure 1d).
Table 2. Comparison of correlations with geography and λ values in GWASs for height (N=3714) between PCs from the three SNP panels varying in LD.
| Correlations between PCs and North–South gradient (N=4103) | Correlations between PCs and East–West gradient (N=4103) | λ for GWAS on height including the North–South PC as a covariate | ||||
|---|---|---|---|---|---|---|
| Panel used for PCA | No. of SNPs for PCA | Pearson correlation | Difference test | Pearson correlation | Difference test | |
| Panel 1=all SNPs that passed QC | 499 849 | rPC2,↕=0.441 | — | rPC8,↔=0.219 | — | 1.03937 |
| Panel 2=Panel 1 without the 24 long-range LD regions | 487 672 | rPC1,↕=0.589 | P=2.0 × 10-59 (versus Panel 1) | rPC3,↔=0.270 | P=7.4 × 10-11 (versus Panel 1) | 1.03092 |
| Panel 3=Panel 2 with genome-wide LD-based SNP pruning | 130 248 | rPC1,↕=0.603 | P=2.8 × 10-5 (versus Panel 2) | rPC2,↔=0.378 | P=6.6 × 10-26 (versus Panel 2) | 1.02961 |
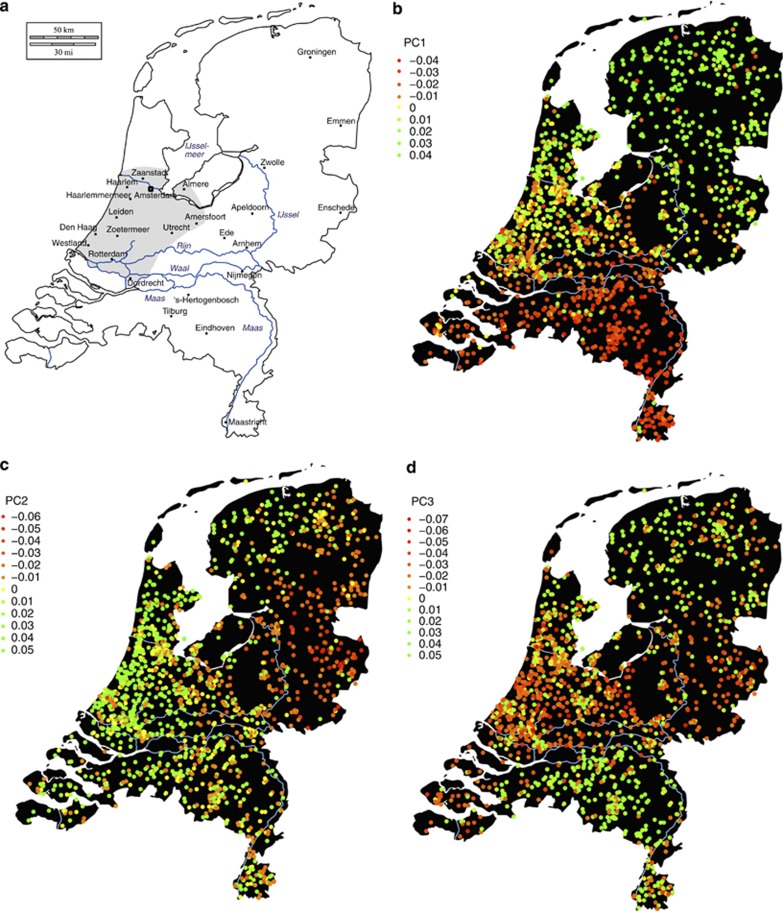
Figure 1.
The three PCs showing significant correlations with geography. (a): Map of the Netherlands, its major rivers, and its 26 largest municipalities (population size >100 000 as of April 2012). The gray area represents the highly urbanized Randstad area. (b), (c), and (d): The colors of the points indicate the mean value per postal code of PC1, PC2 and PC3 respectively from the LD-pruned SNP set without long-range LD regions (Panel 3). The plot is based on 4130 unrelated Dutch individuals spread out across 1635 postal codes of their current living address.
Population stratification of height
Height has been known as a stratifying variable across the world, even within relatively small areas, such as the Netherlands. Northern Dutch are taller on average than the Dutch from the Southern parts of the Netherlands.28 Also within Europe, height correlates with its North–South axis, with Northern Europeans being taller than Southern Europeans.29, 30 In our sample, however, height does not correlate very high with the North–South gradient of the current living address (males: r=0.036, P=0.232; females: r=0.050, P=0.020). The North–South PC, however, shows a higher and more significant correlation with height in both sexes (for Panel 3, the correlations are 0.142 for males and 0.153 for females, P-values <0.001). The fact that this PC does a better job of capturing the height differences between the subpopulations than their current living address, confirms that the PC is a better measure for ancestral origin than the geographical location, and that these height differences are indeed genetic.
As an illustration of the ability of the PCs to reflect the stratifying effects in the population, and the role of LD thereon, we conducted genome-wide association analyses for height in 3714 unrelated individuals, using the North–South PC from different SNP Panels to correct for population stratification within the Netherlands. When using only sex as a covariate in the GWAS on height, the lambda (λ) is 1.0543. Introducing the North–South PC from Panel 1, λ decreases to 1.0394, and continues to decline to 1.0296 as a result of excluding long-range LD regions and LD-based SNP pruning (see Table 2).
The first three PCs from Panel 3
The first three PCs of Panel 3 have the highest correlations with geography (the only ones with P<0.001), and the eigenvalues remain relatively constant in subsequent PCs (see Supplementary Figure S3). The following analyses will focus on these three PCs, which are plotted in Figure 1 and Supplementary Figures S4 and S5. The first PC, which we shall refer to as the North–South PC (Figure 1b), roughly differentiates the Southern provinces (Zeeland, Noord-Brabant, and Limburg) below the three major rivers (the Maas, the Waal, and the Rhine) from the Northern provinces, with the more urbanized West falling in between. The second PC, which we shall call the East–West PC (Figure 1c), mainly differentiates the Northeastern part of the Netherlands from the rest. The third PC, which we will call the middle-band PC (Figure 1d), separates the Northern and Southern provinces from the middle-band area of the Netherlands. There were 157 complete spouse pairs in the sample, for which we calculated the spouse correlations for each of the three PCs. The North–South PC has the highest and most significant spouse correlation (r=0.555, P<0.001). The East–West PC and the middle-band PC show nominally significant spouse correlations (r=0.164, P=0.040, and r=0.179, P=0.025 respectively).
Traces of migration in the Netherlands
The patterns of the first three PCs from Panel 3 resemble the expected patterns of the first three PCs of Novembre and Stephens31 (see Figure 1). Novembre and Stephens caution against drawing conclusions on migration events based on these patterns, because they resemble mathematical artifacts that may arise when PCA is conducted on spatial data where (genetic) similarity decreases with distance. The North–South PC, however, showed a moderate but significant correlation with F (inbreeding coefficient, a measure for genome-wide homozygosity) of 0.245 (P<0.001), indicating that the southern people are more heterozygous than the northern individuals (PC2 and PC3 did not show significant correlations with F). It was previously observed across populations that heterozygosity is negatively correlated with the distance from Addis Ababa, Ethiopia.16, 17 This phenomenon is consistent with a serial-founder effect, where populations expanded through successive migrations of smaller subsets of the populations out of the previous location, starting from a single origin in sub-Saharan Africa. This serial-founder effect also results in increased LD with increasing distance from Africa.15
Especially for PC1, the highly urbanized Randstad area shows an excess of intermediate PC values (see Figures 1a and b), which could be due to the admixture of Dutch subpopulations caused by high migration rates between rural areas and the urbanized West, as well as between the major cities in the West.32, 33 To investigate whether this could have influenced the correlation between PC1 and F, correlations with the North–South gradient and with F were calculated for PC1 for individuals from the 13 largest municipalities of Randstad area (i.e., with population >100 000), and individuals from the rest of the Netherlands separately (see paragraph The Randstad in the Supplementary Information, and Supplementary Tables S7 and S8). For individuals from the Randstad, the correlation with the North–South gradient is not significant and drops to around zero, while it increases for the rest of the country to 0.669. The correlation with F, however, is lower (r=0.170), but still very significant within the Randstad as well as for the rest of the country, where the correlation increases slightly (r=0.259). This indicates that the correlation between PC1 and homozygosity observed in the entire sample is not due to local admixture or inbreeding, making the serial-founder effect hypothesis a more plausible explanation.
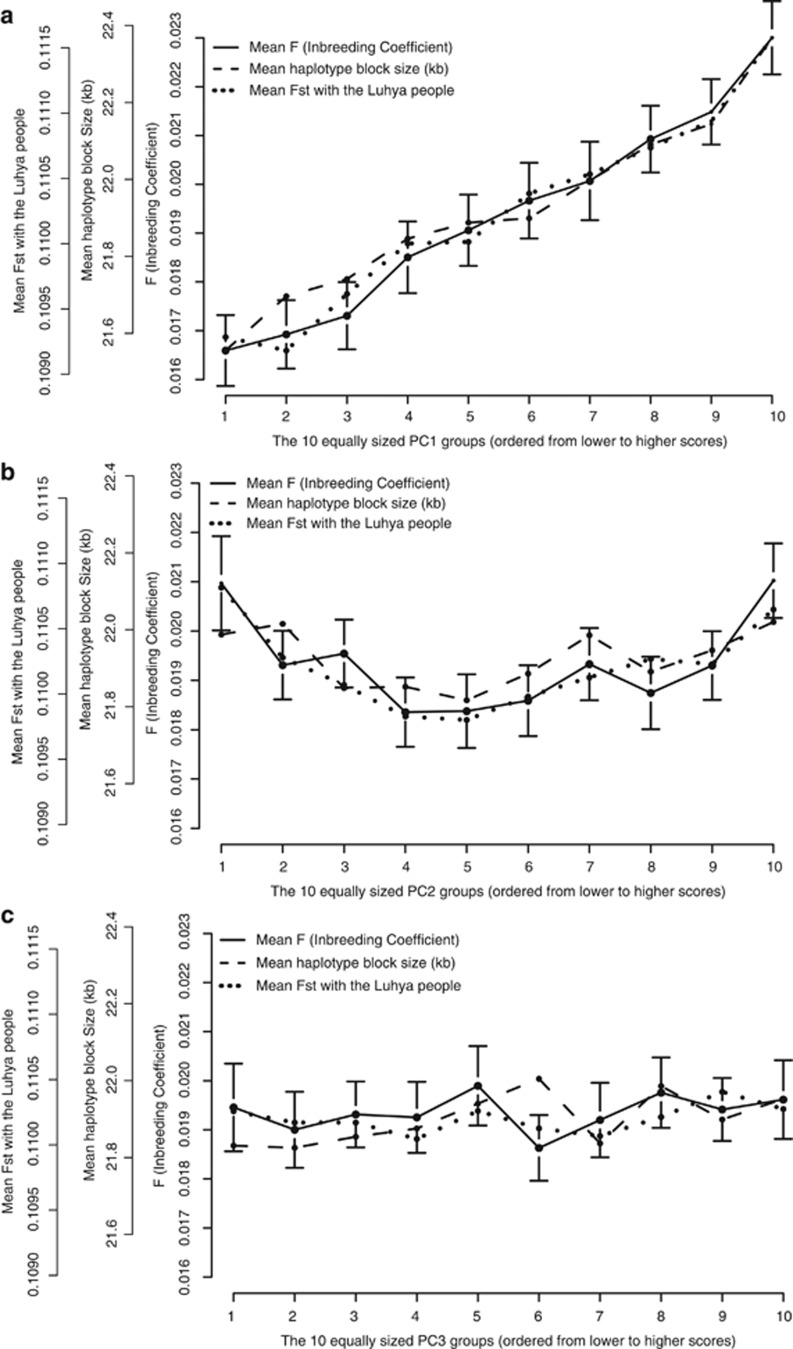
To further illustrate the relationship between the PCs and F, the Dutch subjects were ordered in an ascending order according to their PC value and divided into 10 equally sized groups (9 groups with N=444, and 1 group with N=445). For each group, the mean F was calculated, and plotted in Figure 2 with its 95% confidence interval. As an illustration, we calculated and plotted two related measures in the same Figure: genome-wide average haplotype block size, and the mean Fst with the Luhya people from 1000 Genomes (the 1000 Genomes population closest to Ethiopia). Figure 2 shows that all three measures show a similar linear increase as the North–South PC score increases, suggesting northwards migration. For PC2, the East–West PC, the homozygosity increases as one moves towards more positive as well as more negative values. This may be due to migration in multiple directions, but alternative explanations for this observation are still possible, such as local admixture and/or inbreeding. PC3 did not show significant differences between its 10 groups.
Figure 2.
F, haplotype block size and Fst with the Luhya people in relation to the PCs. For the first three PCs (a–c, respectively), the Dutch subjects were ordered in an ascending order according to their PC value (=PCs from Panel 3, the LD-pruned dataset without long-range LD regions) and divided into 10 equally sized groups (the first 9 groups with N=444, and group 10 with N=445). For each of the 10 groups, the mean F (inbreeding coefficient) is calculated and plotted with its 95% confidence interval. As an illustration of two related measures, the genome-wide average haplotype block size and the mean Fst with the Luhya people from 1000 Genomes, are plotted as well.
Selection pressures as a source of genetic differentiation
To investigate the extent of adaptive effects on the genetic differentiation within the Netherlands, Fst values were computed with Bayescan 2.126 for all 499 849 SNPs that passed QC. Fst values were computed between the top and bottom 1000 individuals for each PC depicted in Figure 1 (genome-wide mean Fst values: PC1=0.00059, PC2=0.00026, PC3=0.00021). Bayescan 2.1 then detected outliers with respect to Fst values using a Bayesian approach, allowing a distinction between divergence due to random drift and divergence that is more likely to be driven by selection pressures. After FDR correction, 273 SNPs reached significance for PC1, 172 SNPs were significant for PC2, and 100 SNPs for PC3 (q<0.05). All significant signals were in the direction of diversifying selection, which may be partly explained by the weak power to detect balancing selection in Fst outlier approaches.26, 27, 34 58.6% of the significant SNPs for PC1 fell within 88 genes, for PC2; 62.2% fell within 55 genes, and for PC3, 75% fell within 41 genes (as opposed to 51.4% of all 499 849 SNPs). These elevated proportions of genic SNPs among the outliers suggest that selection pressures on functional genetic variants had a role in the genetic differentiation between these Dutch subpopulations. Some of these genes have also been observed as highly differentiated within Europe, such as LCT (PC1), HERC2 (PC1), CADPS (PC1), IRF1 (PC1), SLC44A5 (PC1), R3HDM1 (PC1), ACOXL (PC3), and BTBD9 (PC3).35, 36, 37, 38, 39
The SNP with the highest Fst was observed in the North–South PC (PC1), falls within the HERC2 gene (rs8039195, Fst=0.0061, q=0), and has been strongly associated with hair- and eye color.40, 41, 42 In the current Dutch data set, this SNP was also highly predictive for both eye color and hair color when analyzed in a linear regression (eye color: P=3.59 × 10-133; hair color: P=1.65 × 10-22). As eye- and hair color are associated, we conducted an additional linear regression for rs8039195 on eye color with hair color as a covariate, and for hair color with eye color as a covariate. With covariates, the association was still highly significant for eye color (P=7.8 × 10-112), but not for hair color (P=0.218). The genotype frequencies of this SNP are also highly differentiated between 1000 Genomes populations, with the TT genotype having lower frequencies in populations with predominantly brown eyes, while in Northern European populations the genotype frequency can be as high as 93.5% in the Finnish, where blue eyes are much more prevalent (see Supplementary Table S6). To get a higher resolution of the Fst values within and around the HERC2 gene, Fst values were calculated for 3495 SNPs (chr15: 28 300 000 bp–28 600 000 bp) between the available 1000 Genomes Northern European populations (the British and Finnish) and Southern European populations (the Iberian and Toscan). Of the SNPs that were genotyped in the Dutch sample, rs8039195 had the highest Fst between these European populations. The highest Fst value of all 3495 available 1000 Genomes SNPs was observed for rs12913832, identified recently as the functional SNP for determining human blue-brown eye color.43, 44 This SNP is in high LD with rs8039195 (r2=0.394, D′=0.993), consistent with rs12913832 being responsible for the significant signal for diversifying selection in this population.
In addition, significant divergence was observed in all three PCs for a number of genes that have major roles in brain function, such as GRM7 (PC1; encodes a metabotropic glutamate receptor), GRIN2A (PC1; encodes a subunit for the NMDA receptor), BDNF (PC2; encodes the brain-derived neurotrophic factor), SLC6A4 (PC3; encodes the serotonin transporter), NRXN3 (PC3; encodes neurexin-3-alpha), AUTS2 (PC3; autism susceptibility candidate 2). When including genes from all three PCs that showed signals of selection pressures in the clustering algorithm of Ingenuity Pathway Analysis (IPA spring release 2012; Ingenuity Systems Inc., Redwood City, CA, USA), the top 11 biological functions are brain-related (P≤1.26 × 10-4; with a large degree of overlap in molecules between the functions), with the most significant being neurotransmission of nervous tissue with 11 molecules and P=2.2 × 10-6. To ensure this result is not due to a sampling bias (part of the sample consists of major depressive disorder (MDD) cases and controls), the Bayescan analysis was repeated, this time comparing MDD cases (N=966) with MDD controls (N=1522). This analysis showed no significant signals.
Other notable genes showing significant signals include FTO (PC1) and HCP5 (HLA Complex P5 gene; PC1 and PC2). A full list of significant SNPs, Bayescan statistics, and the genes they fall in is given in the supplementary file (supplementary_file.xls).
Discussion
In an effort to elucidate the genetic substructure in a well-characterized population that contributes to multiple GWAS efforts, PCAs were conducted followed by a variety of follow-up analyses. The main aims of this study were: (1) to determine which of the SNP sets (varying in the amount of LD) led to the best PCs in terms of reflecting ancestral origin, (2) using these PCs, to investigate patterns of past human migration, and (3) identifying genomic regions under selection pressures.
We first examined the effect of reducing LD on the ability of the PCs to capture the genome-wide patterns reflecting ancestry differences. In SNP panel 1, the PCA on the 1000 Genomes populations resulted in only 1 of the top 10 PCs (PC10) reflecting a long-range LD region, while in the Dutch data set, all top 10 PCs reflect these regions. Price et al6 showed that genome-wide LD-based SNP pruning did not lead to improved PCs in an analysis of HapMap data. This was confirmed in our analysis of the 1000 Genomes dataset. In the Dutch dataset however, LD-based SNP pruning did lead to improved PCs, as shown by: (1) a large decrease in LD in the top 500 SNPs of the top 10 PCs (Table 1); (2) significantly improved correlations of the PCs with geography (Table 2); (3) the emergence of a new PC among the top ten PCs (PC3, the middle-band PC) that correlates significantly with geography (Figure 1d). We thus conclude that both excluding long-range LD regions and LD pruning are necessary when studying a relatively small population, which may consist of overlapping subpopulations. Large GWAS efforts usually consist of meta-analyses of multiple cohorts consisting of relatively homogeneous populations, which often use PCs to account for population stratification. PCs extracted from SNP sets with less LD are better suited for this goal, as we show using height as an example.
The Dutch North–South component had the highest correlation with geography, and showed the strongest levels of differentiation based on genome-wide Fst values. The high spouse correlation for this PC (0.555) suggests that the North–South differentiation is at least to some extent still ongoing. In Europe, there also is a consistent and reproducible distinction between Northern and Southern populations.45, 46 When projected onto the Dutch individuals, the 1000 Genomes PC that differentiates between Northern and Southern European populations (1000 Genomes PC4 in Supplementary Figure S2) shows a high and significant correlation with the Dutch North–South PC (r=0.656, P<0.001, in unrelated Dutch individuals). The correlation with height is also in the same direction as in Europe (i.e., Northerners are taller than Southerners on average), and blue/brown eye color as well. The Dutch North–South PC also shows a decrease in heterozygosity and an increase in mean haplotype block size in Northern as compared to Southern Dutch individuals (Figure 2), which has been observed between Northern and Southern European populations as well,45 and is best explained by a serial-founder effect. This effect is in line with the European South–-North expansions expected to have occurred at least during Paleolithic, Mesolithic and Neolithic times.47, 48, 49, 50 This effect does not necessarily have to reflect an upward migration that took place within the Netherlands; it may also be that, more recently, Southern Europeans migrated more to the South of the Netherlands, while Northern Europeans migrated more to the Northern parts of the country, maintaining the North–South distribution within the country.
It seems that the genetic differentiation between the Dutch subpopulations led to some phenotypic differences as well, as can be seen for example in the significant correlation of height with the North–South PC. The divergence between these subpopulations is at least in part driven by diversifying selection pressures. The majority of SNPs with significant signals of selection pressures are within genes, of which several have been found to strongly differentiate within Europe as well.35, 36, 37, 38, 39
The highest Fst is observed for rs8039195 from the HERC2 gene. This signal is very likely coming from the neighboring SNP rs12913832 (the strongest blue-brown eye color determinant in humans).43, 44 It is not entirely clear yet why eye color was under such strong selection pressures. It has been proposed that European eye color may have been under frequency-dependant sexual selection,51 which is known to favor color polymorphisms and increase their diversity in many species. The strong signal this particular SNP shows is probably due to the large effect this SNP has on the trait under selection, increasing the selective pressure on this single polymorphism.
Genes involved in brain function are significantly overrepresented among the rest of the signals. Selection pressures on brain related genes in modern humans have been reported previously.52, 53 More research is needed on the exact variants under selection and their functional impact in order to hypothesize which of the wide range of brain functions may have been under selection and why.
Other notable genes include FTO (PC1) and HCP5 (PC1 and PC2). FTO has a role in metabolism, having a large enough effect on obesity to be consistently associated with it,54, 55 suggesting dietary-influenced selection pressures, such as those expected from the transition from hunter-gatherer to agricultural societies. Selection pressures on FTO and other genes involved in obesity have been observed before in other populations.56, 57 The lactase gene (LCT) is also a well-established target of dietary-influenced selection pressures, and also showed significant signals in PC1. HCP5 (HLA Complex P5 gene) from the MHC region (a long-range LD region that was excluded in the PCA) is one of two genes that appear in multiple PCs (PC1 and PC2), and has a role in the immune system. Strong divergence of several genes from the HLA complex has been observed within Europe,36, 37, 38 most likely due to high evolution rates in the highly polymorphic MHC region in order to maintain resistance to rapidly evolving pathogens.58, 59 Other immunity-related genes that showed significant signals of selection in this study as well as previous studies are: IRF1 (PC1), ACE (PC1), LRRC4C (PC2), PLCL1 (PC3), and HSPD1 (PC3).60
In interpreting these findings, one should consider the possibility of ascertainment bias in SNP selection for the microarray, which may have caused signals to be missed (especially for analyses on selection pressures). SNP selection of about half of the SNPs on this Affymetrix array however is random in order to provide sufficient genome-wide coverage, which may have decreased this bias.61
This is a unique population genetics study in terms of resolution, because of the large sample from a relatively small geographical area with detailed phenotypic information available for the majority of the subjects. Increasing signals for ancestry in this data set allowed for the investigation of traces left by migration and adaptation in the genome of a region where the subpopulations have relatively subtle genetic differences. Further research is needed to identify the functional variants in the genomic regions showing significant signals for diversifying selection pressures, as these are likely to influence traits that increased fitness and/or reproductive success. Our results also confirm the importance of considering stratification in association studies of complex traits designed to detect very small effects, even when analyzing smaller supposedly homogeneous populations. In computing PCs to correct for these subtle ancestry differences, the level of LD should be minimized.
Acknowledgments
Funding was obtained from the Netherlands Organization for Scientific Research (NWO: MagW/ZonMW grants 904-61-090, 985-10-002,904-61-193,480-04-004, 400-05-717, Addiction-31160008 Middelgroot-911-09-032, Spinozapremie 56-464-14192), Center for Medical Systems Biology (CSMB, NWO Genomics), NBIC/BioAssist/RK(2008.024), Biobanking and Biomolecular Resources Research Infrastructure (BBMRI – NL, 184.021.007), the VU University's Institute for Health and Care Research (EMGO+ ) and Neuroscience Campus Amsterdam (NCA), the European Science Foundation (ESF, EU/QLRT-2001-01254), the European Community's Seventh Framework Program (FP7/2007-2013), ENGAGE (HEALTH-F4-2007-201413); the European Science Council (ERC Advanced, 230374), Rutgers University Cell and DNA Repository (NIMH U24 MH068457-06), the Avera Institute for Human Genetics, Sioux Falls, South Dakota (USA) and the National Institutes of Health (NIH, R01D0042157-01A). Part of the genotyping and analyses were funded by the Genetic Association Information Network (GAIN) of the Foundation for the US National Institutes of Health, the (NIMH, MH081802) and by the Grand Opportunity grants 1RC2MH089951-01 and 1RC2 MH089995-01 from the NIMH. AA was supported by CSMB/NCA. Statistical analyses were carried out on the Genetic Cluster Computer (http://www.geneticcluster.org), which is financially supported by the Netherlands Scientific Organization (NWO 480-05-003), the Dutch Brain Foundation and the department of psychology and education of the VU University Amsterdam. We are also very grateful to Professor NG Martin for his feedback on the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Manni F. Interview with Luigi Luca Cavalli-Sforza: past research and directions for future investigations in human population genetics. Hum Biol. 2010;82:245–266. doi: 10.3378/027.082.0301. [DOI] [PubMed] [Google Scholar]
- Chen J, Zheng H, Bei JX, et al. Genetic structure of the Han Chinese population revealed by genome-wide SNP variation. Am J Hum Genet. 2009;85:775–785. doi: 10.1016/j.ajhg.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J, Johnson T, Bryc K, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zöllner S, Rosenberg NA. A quantitative comparison of the similarity between genes and geography in worldwide human populations. PLoS Genet. 2012;8:e1002886. doi: 10.1371/journal.pgen.1002886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nat Rev Genet. 2010;11:459–463. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Price AL, Weale ME, Patterson N, et al. Long-range LD can confound genome scans in admixed populations. Am J Hum Genet. 2008;83:132. doi: 10.1016/j.ajhg.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin RM, Altshuler DL, Abecasis GR, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalekamp JC. Bataven en Buitenlanders: 20 Eeuwen Immigratie in Nederland. Wind Publishers: Huizen, the Netherlands; 2009. pp. 15–40. [Google Scholar]
- Knippenberg H, Pater B. De Eenwording van Nederland: Schaalvergroting en Integratie Sinds 1800. SUN: Nijmegen, the Netherlands; 1997. pp. 169–205. [Google Scholar]
- Kok J.Vrijt daar je zijt': huwelijk en partnerkeuze in Zeeland tussen 1830 en 1950. K Mandemakers, O Hoogerhuis en A de Klerk (red), Over Zeeuwse mensen Demografische en sociale ontwikkelingen in Zeeland in de negentiende en begin twintigste eeuw Themanummer Zeeland19987131–143.
- van Poppel F. Verbreding van de horizon? Veranderingen in de geografische herkomst van huwelijkspartners. Acta Geograph Lovaniensia. 1994;34:79–88. [Google Scholar]
- Polman A.Geografische en confessionele invloeden bij de huwelijkskeuze in Nederland: Stenfert Kroese1951
- Hendrickx J, Lammers J, Ultee W. Religious assortative marriage in the Netherlands, 1938-1983. Rev Relig Res. 1991. pp. 123–145.
- Jakobsson M, Scholz SW, Scheet P, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Ramachandran S, Deshpande O, Roseman CC, Rosenberg NA, Feldman MW, Cavalli-Sforza LL. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci USA. 2005;102:15942. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton TJ, Absher D, Feldman MW, Myers RM, Rosenberg NA, Li JZ. Genomic patterns of homozygosity in worldwide human populations. Am J Hum Genet. 2012;91:275–292. doi: 10.1016/j.ajhg.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boomsma DI, De Geus EJC, Vink JM, et al. Netherlands Twin Register: from twins to twin families. Twin Res Hum Genet. 2006;9:849–857. doi: 10.1375/183242706779462426. [DOI] [PubMed] [Google Scholar]
- Penninx BWJH, BeekmanATF, Smit JH, et al. The Netherlands Study of Depression and Anxiety (NESDA): rationale, objectives and methods. Int J Method Psych. 2008;17:121–140. doi: 10.1002/mpr.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2010;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olkin I, Finn JD. Correlations redux. Psychol Bull. 1995;118:155. [Google Scholar]
- Steiger JH. Tests for comparing elements of a correlation matrix. Psychol Bull. 1980;87:245. [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS. Genetic Data Analysis II. Sunderland, MA, USA: Sinauer; 1996. [Google Scholar]
- Foll M, Gaggiotti O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics. 2008;180:977–993. doi: 10.1534/genetics.108.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narum SR, Hess JE. Comparison of FST outlier tests for SNP loci under selection. Mol Ecol Resour. 2011;11:184–194. doi: 10.1111/j.1755-0998.2011.02987.x. [DOI] [PubMed] [Google Scholar]
- CBS: Centraal Bureau voor de Statistiek Gezondheidskenmerken naar regio, 1995-19992012
- Allen HL, Estrada K, Lettre G, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchin MC, Chiang CWK, Palmer CD, Sankararaman S, Reich D, Hirschhorn JN. Evidence of widespread selection on standing variation in Europe at height-associated SNPs. Nat Genet. 2012;44:1015–1019. doi: 10.1038/ng.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J, Stephens M. Interpreting principal component analyses of spatial population genetic variation. Nat Genet. 2008;40:646–649. doi: 10.1038/ng.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesger C. Noord-Hollanders in beweging: economische ontwikkeling en binnenlandse migratie, ca. 1800-1930. CGM. 2003.
- Suurenbroek F. Binnenlandse migratie naar en uit Amsterdam (1870-1890) Centrum voor de Geschiedenis van Migranten. 2001.
- Beaumont MA, Balding DJ. Identifying adaptive genetic divergence among populations from genome scans. Mol Ecol. 2004;13:969–980. doi: 10.1111/j.1365-294x.2004.02125.x. [DOI] [PubMed] [Google Scholar]
- Chen H, Patterson N, Reich D. Population differentiation as a test for selective sweeps. Genome Res. 2010;20:393–402. doi: 10.1101/gr.100545.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath SC, Gut IG, Brennan P, et al. Investigation of the fine structure of European populations with applications to disease association studies. Eur J Hum Genet. 2008;16:1413–1429. doi: 10.1038/ejhg.2008.210. [DOI] [PubMed] [Google Scholar]
- McEvoy BP, Montgomery GW, McRae AF, et al. Geographical structure and differential natural selection among North European populations. Genome Res. 2009;19:804–814. doi: 10.1101/gr.083394.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskvina V, Smith M, Ivanov D, et al. Genetic differences between five European populations. Hum Hered. 2010;70:141–149. doi: 10.1159/000313854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson N, Macpherson JM, Tung JY, et al. Web-based, participant-driven studies yield novel genetic associations for common traits. PLoS Genet. 2010;6:e1000993. doi: 10.1371/journal.pgen.1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Kraft P, Nan H, et al. A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet. 2008;4:e1000074. doi: 10.1371/journal.pgen.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, Liu F, Janssens A, et al. Three genome-wide association studies and a linkage analysis identify HERC2 as a human iris color gene. Am J Hum Genet. 2008;82:411–423. doi: 10.1016/j.ajhg.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm RA, Duffy DL, Zhao ZZ, et al. A single SNP in an evolutionary conserved region within intron 86 of the HERC2 gene determines human blue-brown eye color. Am J Hum Genet. 2008;82:424–431. doi: 10.1016/j.ajhg.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser M, Kayser M, Palstra RJ. HERC2 rs12913832 modulates human pigmentation by attenuating chromatin-loop formation between a long-range enhancer and the OCA2 promoter. Genome Res. 2012;22:446–455. doi: 10.1101/gr.128652.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao O, Lu TT, Nothnagel M, et al. Correlation between genetic and geographic structure in Europe. Curr Biol. 2008;18:1241–1248. doi: 10.1016/j.cub.2008.07.049. [DOI] [PubMed] [Google Scholar]
- Seldin MF, Shigeta R, Villoslada P, et al. European population substructure: clustering of northern and southern populations. PLoS Genet. 2006;2:e143. doi: 10.1371/journal.pgen.0020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belle EMS, Landry PA, Barbujani G. Origins and evolution of the Europeans' genome: evidence from multiple microsatellite loci. Proc R Soc Lond B Biol Sci. 2006;273:1595–1602. doi: 10.1098/rspb.2006.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A. Princeton Univ Pr. New Jersey, USA; 1994. The History and Geography of Human Genes; pp. 255–299. [Google Scholar]
- Chikhi L, Nichols RA, Barbujani G, Beaumont MA. Y genetic data support the Neolithic demic diffusion model. Proc Natl Acad Sci USA. 2002;99:11008. doi: 10.1073/pnas.162158799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Bandelt HJ, Macaulay V, et al. A signal, from human mtDNA, of postglacial recolonization in Europe. Am J Hum Genet. 2001;69:844. doi: 10.1086/323485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost P. European hair and eye color: A case of frequency-dependent sexual selection. Evol Hum Behav. 2006;27:85–103. [Google Scholar]
- Mekel-Bobrov N, Gilbert SL, Evans PD, et al. Ongoing adaptive evolution of ASPM, a brain size determinant in Homo sapiens. Science. 2005;309:1720–1722. doi: 10.1126/science.1116815. [DOI] [PubMed] [Google Scholar]
- Evans PD, Gilbert SL, Mekel-Bobrov N, et al. Microcephalin, a gene regulating brain size, continues to evolve adaptively in humans. Science. 2005;309:1717–1720. doi: 10.1126/science.1113722. [DOI] [PubMed] [Google Scholar]
- Fawcett KA, Barroso I. The genetics of obesity: FTO leads the way. Trends Genet. 2010;26:266–274. doi: 10.1016/j.tig.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimentidis YC, Abrams M, Wang J, Fernandez JR, Allison DB. Natural selection at genomic regions associated with obesity and type-2 diabetes: East Asians and sub-Saharan Africans exhibit high levels of differentiation at type-2 diabetes regions. Hum Genet. 2011;129:407–418. doi: 10.1007/s00439-010-0935-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Corona E, Sikora M, et al. Type 2 diabetes risk alleles demonstrate extreme directional differentiation among human populations, compared to other diseases. PLoS Genet. 2012;8:e1002621. doi: 10.1371/journal.pgen.1002621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina T, Ota M, Shimizu S, et al. Rapid evolution of major histocompatibility complex class I genes in primates generates new disease alleles in humans via hitchhiking diversity. Genetics. 2006;173:1555–1570. doi: 10.1534/genetics.106.057034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton R, Wilming L, Rand V, et al. Gene map of the extended human MHC. Nat Rev Genet. 2004;5:889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- Barreiro LB, Quintana-Murci L. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat Rev Genet. 2009;11:17–30. doi: 10.1038/nrg2698. [DOI] [PubMed] [Google Scholar]
- Perkel J. SNP genotyping: six technologies that keyed a revolution. Nat Methods. 2008;5:447–454. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.