Abstract
DNA methylation is an important epigenetic regulation of gene transcription. Locus-specific DNA methylation can be used as biomarkers in various diseases including cancer. Many methods have been developed for genome-wide methylation analysis, but molecular diagnotics needs simple tools to determine methylation states at individual CpG sites in a gene fragment. In this report, we utilized the nanopore single-molecule sensor to investigate a base-pair specific metal ion/nucleic acids interaction, and explored its potential application in locus-specific DNA methylation analysis. We identified that divalent Mercury ion (Hg2+) can selectively bind a uracil-thymine mismatch (U-T) in a dsDNA. The Hg2+ binding creates a reversible interstrand lock, called MercuLock, which enhances the hybridization strength by two orders of magnitude. Such MercuLock cannot be formed in a 5-methylcytosine-thymine mismatch (mC-T). By nanopore detection of dsDNA stability, single bases of uracil and 5-methylcytosine can be distinguished. Since uracil is converted from cytosine by bisulfite treatment, cytosine and 5′-methylcytosine can be discriminated. We have demonstrated the methylation analysis of multiple CpGs in a p16 gene CpG island. This single-molecule assay may have potential in detection of epigenetic cancer biomarkers in biofluids, with an ultimate goal for early diagnosis of cancer.
The gene expression is not only controlled by the DNA sequence itself, but epigenome, the chemically modified DNAs and chromatin proteins1 that causes inherited alteration of gene expression without changing DNA sequences2,3,4,5. DNA methylation is one of the most commonly occurring epigenetic events in human genome6,7. It is a covalent addition of a methyl group to the cytosine ring by DNA methyltransferases8. Most DNA methylation occurs in CpG dinucleotides (5′-CG-3′)9,10, and over half of all the human genes have a CG rich stretch around promoters and/or the first exon regions, called CpG islands11,12. They are free of methylation in normal somatic cells1,2,11, but many CpG islands in cancer cells are aberrantly methylated13,14 to cause gene silencing13,15,16. Since abnormal DNA methylation in promoter CpG islands is a hall marker of all types of cancers and is chemically stable, it has emerged as a potential biomarker for cancer risk assessment, early detection, prognosis and therapeutic responses prediction13,17,18,19,20,21,22.
Many methods have been developed for DNA methylation detection, such as bisulfite sequencing23,24, CpG island microarray25, quantitative methylation–specific PCR (MSP)26,27 and mass spectrometry28. High-throughput microarrays and next generation sequencing are capable of analyzing genome-wide patterns of DNA methylation, and led to the discovery of many novel methylated genes in various types of tumors29. Other less-expensive and highly-sensitive methods, such as quantitative methylation-specific PCR (MethyLight) and combined bisulfite restriction analysis (COBRA) are useful in target validation or in a clinical diagnostic setting for detection of specific gene methylation in cancer and other diseases27,30. A cornerstone step in these assays is bisulfite treatment of DNA31 that introduces specific changes in the DNA strands. The changes depend on the methylation status of individual cytosine residues, yielding single nucleotide resolution information about the methylation status of a DNA segment. Recently, new techniques that integrate single-molecule and nanotechnology32,33 have emerged for base-specific determination of methylation status. Many of these reported methods, however, are not highly quantitative17,24,25,26,27,28,29,30,31,32,33,34. The detection employ expensive instrument, and the procedure is laborious, involving complex chemical labeling and amplification. These limit their applications in the clinical setting.
The nanopore technology provides a powerful single-molecule platform for the electric detection of nucleic acids35,36,37,38,39,40,41,42,43,44,45,46,47 at the single base level43,48,49,50,51,52. The nanopore has been developed for gene sequencing50,51,53,54,55 and the detection of gene damage56 and cancer-derived biomarkers57. Recently, both biological and synthetic nanopores have been proposed for DNA methylation detection58,59,60,61,62. In this report, we employed the protein nanopore to investigate a novel metal ion-bridged DNA interstrand lock, and explore its potential in locus-specific methylation detection. Metal ions are involved in almost all aspects of nucleic acid chemistry, playing a prominent role in maintaining nucleic acid structural integrity, determining RNA folding, working as catalytic co-factors of enzymes, and constructing biosensors and nanostructures (see reviews63,64,65). It has been known that divalent mercury ion (Hg2+) can specifically bind the thymine-thymine mismatched base pairs (T-T) for dsDNA stabilization66,67,68,69,70,71,72. This property has been applied in designing various DNA-based mercury sensors66,67,68,69,70,71,72,73,74. Recently, different groups have proposed the use of nanopore for mercury detection. The principle was identifying the translocation of a DNA hairpin that is stabilized by T-Hg-T bridged mismatches73,74. In this report, we initially proved that the nanopore can discriminate a single T-Hg-T bridged mismatch in a dsDNA (not the hairpin structure). More importantly, we uncovered that Hg2+ can not only bind the T-T mismatch, but also the uracil-thymine mismatch (U-T). The Hg2+ binding creates a reversible interstrand lock, called MercuLock, which enhances the hybridization strength by two orders of magnitude. Such MercuLock cannot be formed in a 5-methylcytosine-thymine mismatch (mC-T). Therefore we can use the nanopore to distinguish single bases between uracil and 5-methylcytosine in a sequence. As uracil is converted from unmethylated cytosine by bisulfite treatment, unmethylated cytosine and 5-methylcytosine in an original DNA can be discriminated.
Results
Stabilization of a single T-Hg-T interstrand MercuLock
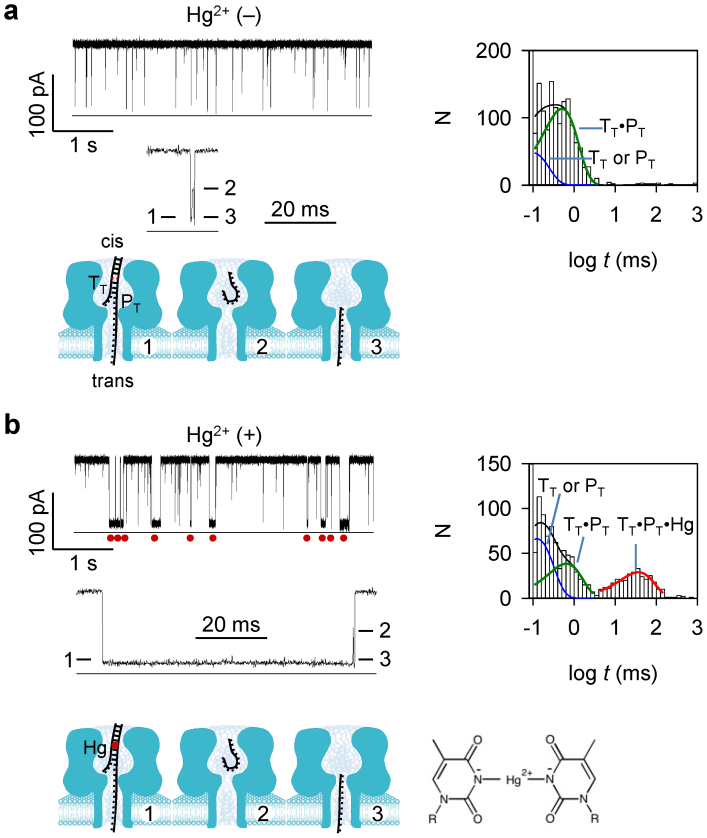
The 16-nt target DNA TT and its probe PT (1 μM/1 μM) was presented to the cis side of the nanopore (Table S1 for sequences). The TT·PT hybrid formed a T-T mismatch at T10. PT franked a poly (dC)30 tag at the 3′ end. As TT·PT was driven into the pore from cis entrance57, the tag threaded into the β-barrel, while the duplex domain was trapped in the nanocavity (Fig. 1a). The trapping of TT·PT generated a three-level conductance block (Fig. 1a). The block duration was 670 ± 140 μs (+130 mV). As studied earlier57, Level 1 of the block (IR/I = 10%) is for TT·PT unzipping; Level 2 (IR/I = 55%, ~0.23 ms) is for TT shortly residing in the nanocavity; and Level 3 (IR/I = 11%, ~0.12 ms) is for TT translocating through the β-barrel. In addition to the TT·PT blocks, Another type of short blocks with duration of 110 ± 20 μs should be attributed to the free TT or PT that translocate through the pore.
Figure 1. Detection of a single T-Hg-T MercuLock in the nanopore.
The mixture of target TT and probe PT were presented in cis solution. (a) and (b). Representative current traces, multi-level signature blocks, block duration histograms and corresponding diagram of molecular configurations, in the absence of Hg2+ (a) and in the presence of Hg2+ (b). The sequences of TT and PT are shown in Table S1. Traces were recorded at +130 mV (cis grounded) in 1 M KCl buffered with 10 mM Tris (pH 7.4). cis solution contained 1 μM TT and 1 μM PT. In b, 10 μM HgCl2 was presented in cis solution. Red dots under the trace in panel b mark the long block signatures for the TT·PT hybrid bound with a Hg2+ ion to the T-T mismatch. Values of block duration were given in Table S2. Red dot in the model in panel b represents the MercuLock formed in the DNA duplex.
When HgCl2 (10 μM) was added to cis solution, a new type of long three-level blocks appeared (Fig. 1b). They show similar Level 2 and Level 3 to the TT·PT signatures as in Fig. 1a. However, their Level 1 was prolonged over 50 folds, extending the entire block duration to 37 ± 6 ms. This type of blocks was not observed for other types of mismatches such as cytosine-thymine (C-T) at the same position in the DNA duplex, whether in the presence or in the absence of Hg2+ ions (Fig. S1). Furthermore, the block frequency continuously increased with increasing the Hg2+ concentration in a broad range from 1 nM to 10 μM (Fig. S2a), while the block duration was independent to the Hg2+ concentration (Fig. S2b). These observations suggest the formation of the TT·PT·Hg complex. We expected that Hg2+ binds to the T-T mismatch of the TT·PT duplex to form a T-Hg-T bridge-pair. This motif greatly stabilized the complex, resulting in a 50-fold prolonged unzipping time. Increasing the voltage across the pore can effectively shorten the unzipping time from 62 ± 7 ms at +100 mV to 28 ± 3 ms at +180 mV (Fig. S2c). In addition, the mass spectrometry (MS) result shows a main component for Hg2+ binding to the dsDNA containing a T-T mismatch (Fig. S3). The removal of two H+ ions from the Hg2+/dsDNA complex is consistent with the T-Hg-T structure suggested in the previous report75 (Fig. 1b). There were also minor peaks for Hg2+ binding with ssDNAs (Fig. S3). In the nanopore experiment, however, TT or PT alone only generated translocation blocks. It is uncertain whether Hg2+ binds to TT or PT in the nanopore detection, which is in different condition from the MS measurement (Fig. S3).
The equilibrium constant for the MercuLock can be evaluated by Kd = [TT·PT][Hg2+]/[TT·PT·Hg], where [TT·PT], [Hg2+] and [TT·PT·Hg] were concentrations of the three compounds. By comparing the block duration histograms in the absence (Fig. 1a) and in the presence of Hg2+ (Fig. 1b), we can evaluate the change in [TT·PT], which was assumed to be [TT·PT·Hg]. Thus Kd was calculated to be 2.9 μM. Furthermore, the ratio of the TT·PT·Hg and TT·PT block duration (τ+Hg/τ-Hg) allows evaluating the energy increase for unzipping the TT·PT·Hg complex upon Hg2+ binding, ΔG = RTln(τ+Hg/τ-Hg) = 8.1 k·mol−1. Therefore, the T-Hg-T bridge-pair functions as an interstrand lock, or MercuLock, that greatly stabilize dsDNA hybridization. The resulting nanopore signature for MercuLock can discriminate single T-T mismatches in a dsDNA.
Discrimination of uracil and 5′-methylcytosine with MercuLock
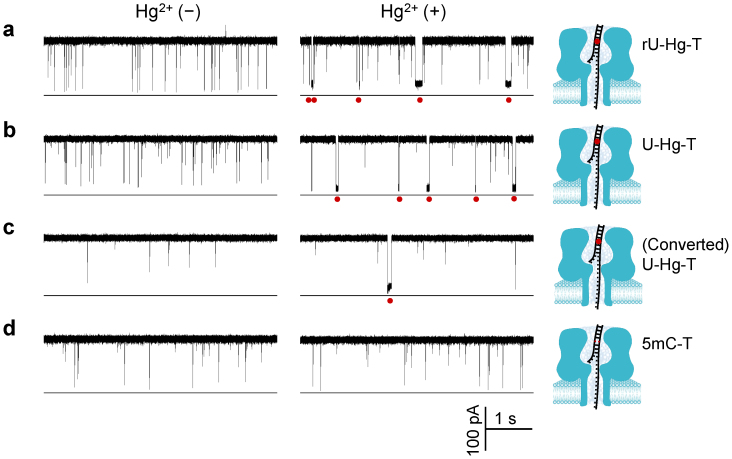
By utilizing the nanopore capability in single base-pair discrimination, we further examined whether the MercuLock can be formed with other types of mismatches. We focused on the uracil-thymine mismatch because RNAs use uracil instead of thymine for complementary pairing. The target TrU had one nucleotide difference from TT, with T10 substituted by a ribonucleoside uridine (rU) (Table S1). TrU can be hybridized with the same probe PT to form a rU-T mismatch. In the absence of Hg2+, the TrU·PT blocks were 820 ± 110 ms (Fig. 2a left trace). The addition of Hg2+ to cis solution generated distinct long blocks of 41 ± 6 ms (Fig. 2a right trace). This result is very similar to the T-T mismatch in the absence and in the presence of Hg2+ as in Fig. 1, suggesting that Hg2+ can bind the rU-T mismatch to form a stable rU-Hg-T MercuLock. We further tested another target TU, which has a deoxyuridine (U, Table S1) at the position T10. The TU·PT hybrid forms a U-T mismatch. We found that Hg2+ can also form MercuLock with the U-T mismatch (Fig. 2b). In the absence of Hg2+, we observed short blocks (1.0 ± 0.3 ms) for TU·PT (Fig.2b left trace), and in the presence of Hg2+ ions, we identified the characteristic long block (39 ± 5 ms) that acts as a signature for the TU·PT·Hg complex (Fig. 2b right panel). Overall, Hg2+ can also form a MercuLock with the uracil-thymine mismatch, which enhances the stability of the dsDNA by 40–50 times.
Figure 2. Discrimination of uracil and unmethylated cytosine with MercuLock.
(a) through (d) crrent trace showing signature blocks produced by various targe⃛probe hybrids TrU·PT (a), TU·PT (b), TC→U·PT (c) and TmC·PT (d) in the absence (left panel) and in the presence of Hg2+ (right panel). These hybrids contained a mismatch of uracil (uridine)-thymine (rU-T), uracil (deoxyuridine)-thymine mismatch (U-T), converted uracil-thymine (U-T), and 5-methyl cytosine-thymine (mC-T), respectively. TC→U was converted from target TC by bisulfite. Red dots under the traces marked the signature blocks for Hg2+ binding to the corresponding mismatches. Red dots in models represented the MercuLock formed in the DNA duplex. The sequences of targets TrU, TU, TC, TmC and probe PT were shown in Table S1. Traces were recorded at +130 mV in 1 M KCl solution buffered with 10 mM Tris (pH 7.4). cis solution contained 1 μM target DNAs and 1 μM PT, and 10 μM HgCl2 (right traces). The traces for TC·PT with and without Hg2+ were shown in Fig. S1. Values of block duration were given in Table S2.
It is common in methylation detection that the DNA will be pre-treated with bisulfite to convert cytosine into uracil. So we further examined whether the converted uracil can form MercuLock with thymine. The target TC, which has cytosine at the position 10, was treated by bisulfite; then the mixture of converted TC and the probe PT (not converted) was presented in cis solution. The current traces for converted TC→U·PT (Fig. 2c) are similar to TU·PT (Fig. 2b). The signature blocks for the TC→U·PT complex in the absence of Hg2+ was 1.3 ± 0.2 ms (Fig. 2c left trace). The TC→U·PT complex in the presence of Hg2+ generated a long signature block with duration of 31 ± 6 ms (Fig. 2c right panel). By comparison, Hg2+ did not bind the C-T mismatch in a TC·PT hybrid (Fig. S1). These findings confirm that cytosine has been converted to uracil and the MercuLocks is formed between the cytosine-converted uracil and thymine. The dsDNA stability can be enhanced over 20 folds upon Hg2+ binding. We also detected another target TmC that contained a 5′-methylcytosine in the same position. 5′methylcytosine cannot be converted by bisulfite treatment. In contrast to converted TC, the TmC·PT complex did not produce the long signature block, but we only observed short blocks either in the absence (1.7 ± 0.9 ms, Fig. 2d left trace) or in the presence (1.8 ± 0.4 ms, Fig. 2d right trace) of Hg2+, confirming that 5′-methylcytosine cannot form MercuLock with thymine. Overall, single bases of uracil and 5′-methyl cytosine can be discriminated by identifying the MercuLock formation in the nanopore. Since uracil is converted from unmethylated cytosine, in principle unmethylated cytosine can be distinguishable from 5′-methylcytosine in the original DNA sequence.
Multi-CpG methylation detection in a gene fragment
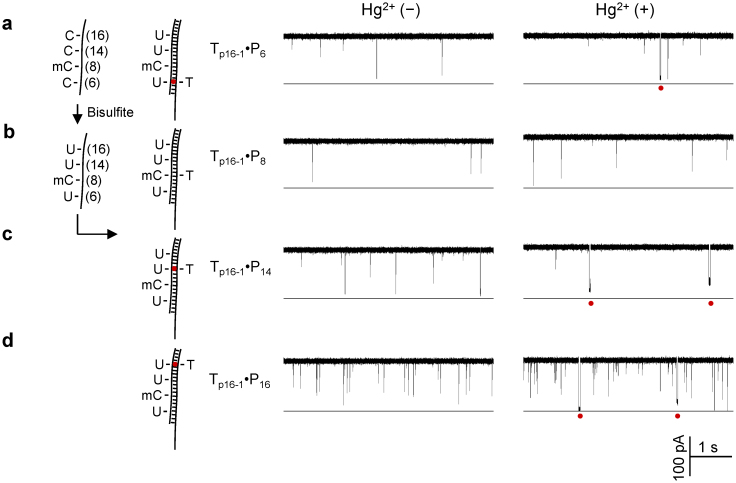
The p16 tumor suppressor gene (cyclin-dependent kinase inhibitor 2A, CDKN2A) performs an important role in regulating the cell cycle, and is a commonly studied target gene for cancer detection76,77,78,79. The methylation status in the p16 gene has been known to be related to the risk of developing a variety of cancers such as lung cancer and breast cancer76,77,78,79. Our target is a 22-nt fragment from the antisense chain of the p16 gene within CpG island 176 (Chromosome 9: 21,994,825-21,994,846, Fig. S4). This fragment includes 4 CpGs in positions 6, 8, 14 and 16 (Table S1). To target the bisulfite-converted sequence, we designed four probes, PC6, PC8, PC14 and PC16. Each probe employed a thymine to match one of CpG cytosines, and the four probes can detect all the four CpGs (6, 8, 14 and 16). There was a technical issue: the high GC content (70%) in this DNA fragment strengthens the target/probe hybridization, prolonging its dehybridization time for the DNA duplex containing an mC-T mismatch. This may affect the discrimination between the mC-T signatures and the U-Hg-T signatures. To solve this issue, we introduced three cytosines to each probe to form mismatches with the other three CpG cytosines of the target (Table S1), whether or not the target is converted. This design can significantly shortened the complex block duration in the absence of Hg2+, thus greatly enhanced the capability to discriminate MercuLock signatures.
The target Tp16-1 comprises a 5′-methylcytosine at C8, and cytosines at C6, C14 and C16. The bisulfite-treated target Tp16-1 was mixed with the four probes respectively. Their hybrids were detected in the nanopore individually. In a control experiment, Tp16-1 alone before and after conversion only generated spike-like rapid translocation blocks (Fig. S5). Fig. 3a–d shows the current traces for the four mixtures in the absence and in the presence of Hg2+. In the absence of Hg2+, we only observed short blocks for all four mixtures (2.2–2.6 ms, Fig. 3a–d left traces). The addition of Hg2+ ions produced long blocks for the mixtures of converted Tp16-1 and PC6 (11 ± 6 ms, Fig. 3a right trace), PC14 (36 ± 12 ms, Fig. 3c right trace) and PC16 (21 ± 8 ms, Fig. 3d right trace). The only sample that did not generate the long signature block in Hg2+ was the mixture with PC8. The distinct long blocks for PC6, PC14 and PC16 are consistent with cytosines at C6, C14 and C16, which have been converted to uracil to form the U-Hg-T MercuLock with the specific probe. In contrast, no long block signature observed in PC8 is in agreement with 5′-methylcytosine at C8 in Tp16-1, which cannot form the MercuLock.
Figure 3. Site-specific detection of DNA methylation with a MercuLock.
(a) through (d) were current traces for the bisufite-converted Tp16-1 hybridized with probes PC6 (a), PC8 (b), PC14 (c) and PC16 (d) in the absence of Hg2+ (left panel) and in the presence of Hg2+ (right panel ). The four probes were designed for detecting CpG cytosines at the positions C6, C8, C14 and C16. C8 was 5-methyl cytosine (mC) and remained unchanged after bisulfite treatment. The other three positions were unmethylated cytosine (C), and thus converted to uracil (U) by bisulfite treatment. Red dots under the traces mark the signature long blocks for Hg2+ ion binding to the U-T mismatches. Red dots in the models (left) marked the MercuLock in the DNA duplex.
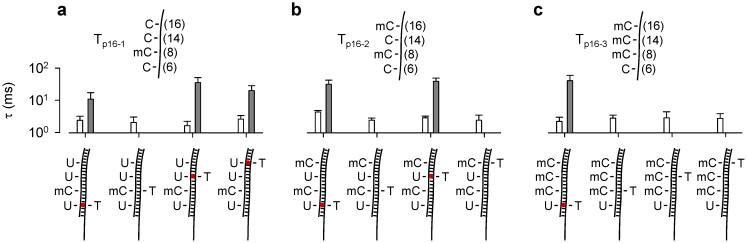
Finally we expanded to the targets carrying different numbers and distribution of 5 mC. Tp16-2 has two 5′-methylcytosines at C8 and C16, and Tp16-3 has three at C8, C14 and C16 positions. Both targets have cytosines at other CpG sites. Each converted target was mixed with the four probes (the same probes used for Tp16-1) respectively. Similar to Tp16-1 (Fig. 4a), the hybrids of Tp16-2 and Tp16-3 with each of the four probes only produced short blocks (2.1–3.7 ms) in the absence of Hg2+. For Tp16-2, the long block signatures can be observed with probes PC6 (32 ± 11 ms) and PC14 (40 ± 11 ms), and no such signature signals but only short blocks was observed with PC8 and PC16 in the presence of Hg2+ (Fig. 4b), verifying the formation of a U-Hg-T Merculock between converted Tp16-2·PC6 and Tp16-2·PC14, and no MercuLock is formed in mC-T mismatches in the Tp16-2·PC8 and Tp16-2·PC16 complexes. This result is consistent with the methylation distribution in Tp16-2: cytosine at C6 and C14, and 5-methylcytosine at C8 and C14. Similarly, the mixture of converted Tp16-3 with each of PC8, PC14 and PC16 cannot generate the long block signatures, and only short blocks (2.3–2.8 ms) was observed. The long block signatures were only observed with PC6 (42 ± 19 ms, Fig. 4c), thus verifying the methylation distribution in Tp16-3: cytosine at C6 and 5-methylcytosine at C8, C14 and C16.
Figure 4. Detection of DNA containing different numbers and distribution of methylated cytosines.
(a), (b) and (c) compared the duration of short and long signature blocks for targets Tp16-1 (a), Tp16-2 (b) and Tp16-3 (c) detected by four probes PC6, PC8, PC14 and PC16. The duration of signature blocks allowed determining the methylation status for each of four CpG cytosines. The DNA sequences of the three p16 fragments were given in Table S1. Duration values were given in Table S3. All traces were recorded at +130 mV in 1 M KCl and 10 mM Tris (pH 7.4).
Discussion
By using the nanopore sensor, we have uncovered a novel metal ion-nucleic acid interaction at the single base-pair level. The core of the finding is an Hg2+-bridged interstand lock that strongly and selectively stabilizes the uracil-thymine mismatch. The resulting significant difference in dsDNA stability leads to accurate single-base discrimination between uracil and thymine, and eventually the discrimination between cytosine and methylated cytosine. Comparing with current methylation analysis methodologies, this approach is label-free and does not require DNA amplification and sequencing. The single-molecule recognition of MercuLock formation is rapid and specific, therefore may have potential in methylation biomarker detection for diagnostics17,19. This laboratory finding-based method also faces challenges. Currently, each CpG site needs a specific probe and each nanopore measurement reads one CpG site. This detection mode is suitable for single locus DNA methylation detection, but is limited for genome-wide DNA methylation profiling, unless a high throughput nanopore platform36,80,81 is established. We are working on another nanopore strategy that can sequentially read several MercuLocks with one probe for multiple methylation analysis. The bisulfite pre-treatment may be incomplete although the conversion rate can reach at 99% with the current reagent82,83. In addition, the MercuLock formation is in equilibrium. There exists free U-T mismatch without Hg2+ labeling. These factors may not influence the recognition of MercuLock signatures in the nanopore, but may lower the accuracy for methylation quantification, i.e. determination of methylation percentage in the sample.
This work provides a powerful biophysical tool to explore metal ion-nucleic acids interactions in the living organisms and human. For example whether Hg2+ binding to T-T or U-T mismatched pair can compromise the DNA repair process in human, especially during tumorigenesis, needs further study. This work opens an avenue to the application of metal ion-nucleic acid interactions in rapid detection of single nucleotide alteration in gene sequence, such as pathological point mutations, single nucleotide polymorphism (SNPs) and DNA methylation in variety of disease states including cancer17,19.
Methods
DNAs samples
Oligonucleotides, including all targets and probes, were synthesized and HPLC-purified by Integrated DNA Technologies (Coralville, IA). They were dissolved in dd water to 1 mM and stored at −20°C as stocks. The target and probe DNAs were mixed at desire concentrations. The mixture was heated to 90°C for 5 minutes, then gradually cooled down to room temperature and stored at 4°C until use.
Nanopore electrical recording
The method for nanopore electrical recording has been described previously84. Briefly, we used 1,2-diphytanoyl-sn-glycerophosphatidylcholine (DPhPC, Avanti Polar Lipids) to form a lipid bilayer membrane over a ~150 μm orifice in the center of a 25-μm-thick Teflon film (Goodfellow) that partitioned between cis and trans recording solutions. The recording solutions on each side of the bilayer contained KCl at a desired concentration and were buffered with 10 mM Tris (pH 8.0). The α-hemolysin protein was added in the cis solution, from which the protein was inserted into the bilayer to form a nanopore. Target and probe DNAs and HgCl2 solutions were released to the cis solution. The voltage was given from trans solution and cis solution was grounded. In this configuration, a positive voltage can pull the negatively charged DNA through the pore from cis to trans. The ion current through the pore were recorded with an Axopatch 200B amplifier (Molecular Device Inc., Sunnyvale, CA), filtered with a built-in 4-pole low-pass Bessel Filter at 5 kHz, and acquired with Clampex 10 software (Molecular Device Inc.) through a Digidata 1440 A/D converter (Molecular Device Inc.) at a sampling rate of 20 kHz. The single-molecule events were analyzed using Clampfit 9.0 (Molecular Device Inc.), Excel (MicroSoft) and SigmaPlot (SPSS) software. In addition to the DNA duplex signature blocks (~10–100 ms), we also observed spike-like single-stranded DNA translocation events (~10–100 μs). These events were excluded from histogram construction and analysis. Data was presented as mean ± SD of at least three independent experiments. The nanopore measurements were conducted at 22 ± 2°C.
Bisulfite conversion
The bisulfite conversion for target DNAs was performed using the EZDNA Methylation-Gold Kit™ (ZYMO Research Corp.). Briefly, 10 μl of the target oligonucleotide sample (1 mM) were mixed with 10 μl water and 130 μl conversion reagent in a PCR tube. The PCR tube with the sample was placed in a thermal cycler, then heated at 98°C for 10 minutes and 64°C for 2.5 h. 600 μl M-binding buffer was added to a Zymo-spin IC™ column, then the sample was loaded into the column. After the conversion reaction, the column was centrifuged at 10,000 × g for 30 s, followed by washing with 100 μl wash buffer. After centrifuging for 30 s, 200 μl desulphonation buffer was loaded in the column and incubated at room temperature for 15–20 min. After incubation, the column was spun at 10,000 × g for 30 s, followed by washing twice with 200 μl wash buffer and spinning for 30 s. Purified olignucleotides were eluted with 10 μl elution buffer.
Author Contributions
I.K., Y.W. and C.R. designed the nanopore research, Y.F. and M.X.W. performed methylation analysis, L.Q.G. conceived the idea, all authors contributed to writing of the manuscript.
Supplementary Material
Supplementary Information - Kang et al
Acknowledgments
This investigation was partially supported by grants from the National Science Foundation 0546165 (L.Q.G.), the National Institutes of Health GM079613 (L.Q.G.) and the University of Missouri Intellectual Property Fast Track Initiative (A8881, M.X.W) and was conducted in a facility that was constructed with support from the Research Facilities Improvement Program Grant Number C06-RR-016489-01 from the National Centre for Research Resources, National Institutes of Health.
References
- Suzuki M. M. & Bird A. DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Gen. 9, 465–476 (2008). [DOI] [PubMed] [Google Scholar]
- Deaton A. M. & Bird A. CpG islands and the regulation of transcription. Genes Dev. 25, 1010–1022 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim P. S., Shannon M. F. & Hardy K. Epigenetic control of inducible gene expression in the immune system. Epigenomics 2, 775–795 (2010). [DOI] [PubMed] [Google Scholar]
- Jones P. A. & Baylin S. B. The fundamental role of epigenetic events in cancer. Nat. Rev. Gen. 3, 415–428 (2002). [DOI] [PubMed] [Google Scholar]
- Bannister A. J. & Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger G., Liang G., Aparicio A. & Jones P. A. Epigenetics in human disease and prospects for epigenetic therapy. Nature 429, 457–463 (2004). [DOI] [PubMed] [Google Scholar]
- Beck S. & Rakyan V. K. The methylome: approaches for global DNA methylation profiling. Trends Genet. 24, 231–237 (2008). [DOI] [PubMed] [Google Scholar]
- Bestor T. H. The DNA methyltransferases of mammals. Hum. Mol. Genet. 9, 2395–2402 (2000). [DOI] [PubMed] [Google Scholar]
- Bird A. The essentials of DNA methylation. Cell 70, 5–8 (1992). [DOI] [PubMed] [Google Scholar]
- Craig J. M. & Bickmore W. A. The distribution of CpG islands in mammalian chromosomes. NAT. GENET. 7, 376–382 (1994). [DOI] [PubMed] [Google Scholar]
- Illingworth R. S. & Bird A. P. CpG islands - ‘A rough guide'. FEBS Lett. 583, 1713–1720 (2009). [DOI] [PubMed] [Google Scholar]
- Bernstein B. E., Meissner A. & Lander E. S. The mammalian epigenome. Cell 128, 669–681 (2007). [DOI] [PubMed] [Google Scholar]
- Baylin S. B. & Jones P. A. A decade of exploring the cancer epigenome-biological and translational implications. Nat. Rev. Cancer 11, 726–734 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Molecular origins of cancer: Epigenetics in cancer. New Engl. J. Med. 358, 1148–1159+1096 (2008). [DOI] [PubMed] [Google Scholar]
- Basu R. & Zhang L. F. X chromosome inactivation: A silence that needs to be broken. Genesis 49, 821–834 (2011). [DOI] [PubMed] [Google Scholar]
- Baylin S. B. & Ohm J. E. Epigenetic gene silencing in cancer - A mechanism for early oncogenic pathway addiction? Nat. Rev. Cancer 6, 107–116 (2006). [DOI] [PubMed] [Google Scholar]
- Shi H., Wang M. X. & Caldwell C. W. CpG islands: Their potential as biomarkers for cancer. Expert Rev. Mol. Diagn. 7, 519–531 (2007). [DOI] [PubMed] [Google Scholar]
- Esteller M. Relevance of DNA methylation in the management of cancer. Lancet Oncol. 4, 351–358 (2003). [DOI] [PubMed] [Google Scholar]
- GrØnbæk K., Hother C. & Jones P. A. Epigenetic changes in cancer. APMIS 115, 1039–1059 (2007). [DOI] [PubMed] [Google Scholar]
- Holloway A. F. & Oakford P. C. Targeting epigenetic modifiers in cancer. Curr. Med. Chem. 14, 2540–2547 (2007). [DOI] [PubMed] [Google Scholar]
- Issa J. P. J. DNA methylation as a therapeutic target in cancer. Clin. Cancer Res. 13, 1634–1637 (2007). [DOI] [PubMed] [Google Scholar]
- Shivapurkar N. & Gazdar A. F. DNA methylation based biomarkers in non-invasive cancer screening. Curr. Mol. Med. 10, 123–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders J. & Paszkowski J. Bisulfite methylation profiling of large genomes. Epigenomics. 2, 209–220 (2010). [DOI] [PubMed] [Google Scholar]
- Harris R. A. et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat. Biotechnol. 28, 1097–1105 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adorján P. et al. Tumour class prediction and discovery by microarray-based DNA methylation analysis. Nucleic Acids Res 30, (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
-
Herman J. G., Graff J. R.,
 , Nelkin B. D. & Baylin S. B.
Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. PROC. NATL. ACAD. SCI. U. S. A.
93, 9821–9826 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
, Nelkin B. D. & Baylin S. B.
Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. PROC. NATL. ACAD. SCI. U. S. A.
93, 9821–9826 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar] - Eads C. A. et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res 28, (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrich M. et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. PROC. NATL. ACAD. SCI. U. S. A. 102, 15785–15790 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouse S. D., Nagarajan R. P. & Costello J. F. Genome-scale DNA methylation analysis. Epigenomics 2, 105–117 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eads C. A. & Laird P. W. Combined bisulfite restriction analysis (COBRA). Methods Mol. Biol. 200, 71–85 (2002). [DOI] [PubMed] [Google Scholar]
- Clark S. J., Statham A., Stirzaker C., Molloy P. L. & Frommer M. DNA methylation: Bisulphite modification and analysis. Nat. Protoc. 1, 2353–2364 (2006). [DOI] [PubMed] [Google Scholar]
- Song C. X. et al. Sensitive and specific single-molecule sequencing of 5-hydroxymethylcytosine. Nat. Methods 9, 75–77 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu R. et al. Nanomechanical recognition measurements of individual DNA molecules reveal epigenetic methylation patterns. Nat. Nanotechnol. 5, 788–791 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedeurwaerder S. et al. Evaluation of the Infinium Methylation 450 K technology. Epigenomics. 3, 771–784 (2011). [DOI] [PubMed] [Google Scholar]
- Bayley H. & Cremer P. S. Stochastic sensors inspired by biology. Nature 413, 226–230 (2001). [DOI] [PubMed] [Google Scholar]
- Bayley H. et al. Droplet interface bilayers. Mol. Biosyst. 4, 1191–1208 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L. Q. & Shim J. W. Single molecule sensing by nanopores and nanopore devices. Analyst 135, 441–451 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornblower B. et al. Single-molecule analysis of DNA-protein complexes using nanopores. Nat Methods 4, 315–317 (2007). [DOI] [PubMed] [Google Scholar]
- Howorka S. & Siwy Z. Nanopore analytics: Sensing of single molecules. Chemical Society Reviews 38, 2360–2384 (2009). [DOI] [PubMed] [Google Scholar]
- Movileanu L. Interrogating single proteins through nanopores: challenges and opportunities. Trends Biotechnol. 27, 333–341 (2009). [DOI] [PubMed] [Google Scholar]
- Ma L. & Cockroft S. L. Biological nanopores for single-molecule biophysics. Chembiochem 11, 25–34 (2010). [DOI] [PubMed] [Google Scholar]
- Majd S. et al. Applications of biological pores in nanomedicine, sensing, and nanoelectronics. Current Opinion in Biotechnology 21, 439–476 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olasagasti F. et al. Replication of individual DNA molecules under electronic control using a protein nanopore. Nat Nanotechnol. 5, 798–806 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan B. M. & Bashir R. Nanopore sensors for nucleic acid analysis. Nat Nanotechnol. 6, 615–624 (2011). [DOI] [PubMed] [Google Scholar]
- Wanunu M., Morrison W., Rabin Y., Grosberg A. Y. & Meller A. Electrostatic focusing of unlabelled DNA into nanoscale pores using a salt gradient. Nat. Nanotechnol. 5, 160–165 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanunu M. et al. Rapid electronic detection of probe-specific microRNAs using thin nanopore sensors. Nat. Nanotechnol. 5, 807–814 (2010). [DOI] [PubMed] [Google Scholar]
- Wendell D. et al. Translocation of double-stranded DNA through membrane-adapted phi29 motor protein nanopores. Nat Nanotechnol. 4, 765–772 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purnell R. F. & Schmidt J. J. Discrimination of single base substitutions in a DNA strand immobilized in a biological nanopore. ACS Nano 3, 2533–2538 (2009). [DOI] [PubMed] [Google Scholar]
- Chu J., Gonzalez-Lopez M., Cockroft S. L., Amorin M. & Ghadiri M. R. Real-time monitoring of DNA polymerase function and stepwise single-nucleotide DNA strand translocation through a protein nanopore. Angew. Chem. Int. Ed Engl. 49, 10106–10109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherf G. M. et al. Automated forward and reverse ratcheting of DNA in a nanopore at 5-A precision. Nat Biotechnol 30, 344–348 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrao E. A. et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat Biotechnol 30, 349–353 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J. et al. Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nanotechnol. 4, 265–270 (2009). [DOI] [PubMed] [Google Scholar]
- Hall A. R. et al. Hybrid pore formation by directed insertion of alpha-haemolysin into solid-state nanopores. Nat Nanotechnol. 5, 874–877 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branton D. et al. The potential and challenges of nanopore sequencing. Nature Biotechnology 26, 1146–1153 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasianowicz J. J., Brandin E., Branton D. & Deamer D. W. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. U. S. A. 93, 13770–13773 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- An N., Fleming A. M., White H. S. & Burrows C. J. Crown ether-electrolyte interactions permit nanopore detection of individual DNA abasic sites in single molecules. Proc Natl. Acad. Sci. U. S. A 109, 11504–11509 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zheng D., Tan Q., Wang M. X. & Gu L. Q. Nanopore-based detection of circulating microRNAs in lung cancer patients. Nat. Nanotechnol. 6, 668–674 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace E. V. B. et al. Identification of epigenetic DNA modifications with a protein nanopore. Chem. Commun. 46, 8195–8197 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrao E. A., Derrington I. M., Pavlenok M., Niederweis M. & Gundlach J. H. Nucleotide discrimination with DNA immobilized in the MspA nanopore. PLoS. One. 6, e25723 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J. et al. Detection and quantification of methylation in DNA using solid-state nanopores. Sci. Rep. 3, 1389; 10.1038/srep01389 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirsaidov U. et al. Nanoelectromechanics of methylated DNA in a synthetic nanopore. Biophys. J. 96, L32–L34 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanunu M. et al. Discrimination of methylcytosine from hydroxymethylcytosine in DNA molecules. J. Am. Chem. Soc. 133, 486–492 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clever G. H., Kaul C. & Carell T. DNA--metal base pairs. Angew. Chem Int. Ed Engl. 46, 6226–6236 (2007). [DOI] [PubMed] [Google Scholar]
- Interplay Between Metal ions and Nucleic Acids Sigel, A., Sigel, H. & Sigel, R. K. O. (eds.) (Springer, 2012). [Google Scholar]
- Muller J. Functional metal ions in nucleic acids. Metallomics. 2, 318–327 (2010). [DOI] [PubMed] [Google Scholar]
- Huang K. & Martí A. A. Recent trends in molecular beacon design and applications. Analytical and Bioanalytical Chemistry 402, 3091–3102 (2012). [DOI] [PubMed] [Google Scholar]
- Miyake Y. et al. Mercury II-mediated formation of thymine-Hg II-thymine base pairs in DNA duplexes. J. Am. Chem. Soc. 128, 2172–2173 (2006). [DOI] [PubMed] [Google Scholar]
- Nolan E. M. & Lippard S. J. Tools and tactics for the optical detection of mercuric ion. Chem. Rev. 108, 3443–3480 (2008). [DOI] [PubMed] [Google Scholar]
- Ono A. & Togashi H. Highly selective oligonucleotide-based sensor for mercury(II) in aqueous solutions. Angew. Chem. Int. Ed. 43, 4300–4302 (2004). [DOI] [PubMed] [Google Scholar]
- Ono A., Torigoe H., Tanaka Y. & Okamoto I. Binding of metal ions by pyrimidine base pairs in DNA duplexes. Chem. Soc. Rev. 40, 5855–5866 (2011). [DOI] [PubMed] [Google Scholar]
- Selid P. D., Xu H., Collins E. M., Face-Collins M. S. & Zhao J. X. Sensing mercury for biomedical and environmental monitoring. Sensors 9, 5446–5459 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y. et al. 15N- 15N J-coupling across Hg II: Direct observation of Hg II-mediated T-T base pairs in a DNA duplex. J. Am. Chem. Soc. 129, 244–245 (2007). [DOI] [PubMed] [Google Scholar]
- Wen S. et al. Highly sensitive and selective DNA-based detection of mercury(II) with alpha-hemolysin nanopore. J. Am. Chem. Soc. 133, 18312–18317 (2011). [DOI] [PubMed] [Google Scholar]
- Wang G., Zhao Q., Kang X. & Guan X. Probing mercury(II)-DNA interactions by nanopore stochastic sensing. J. Phys. Chem. B 117, 4763–4769 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake Y. et al. MercuryII-mediated formation of thymine-HgII-thymine base pairs in DNA duplexes. J Am. Chem Soc 128, 2172–2173 (2006). [DOI] [PubMed] [Google Scholar]
- Belinsky S. A. et al. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer Res. 66, 3338–3344 (2006). [DOI] [PubMed] [Google Scholar]
- Esteller M., Corn P. G., Baylin S. B. & Herman J. G. A gene hypermethylation profile of human cancer. Cancer Res. 61, 3225–3229 (2001). [PubMed] [Google Scholar]
- Marsit C. J. et al. Examination of a CpG island methylator phenotype and implications of methylation profiles in solid tumors. Cancer Res. 66, 10621–10629 (2006). [DOI] [PubMed] [Google Scholar]
- Wang Y. C., Hsu H. S., Chen T. P. & Chen J. T. Molecular diagnostic markers for lung cancer in sputum and plasma. Ann. N. Y. Acad. Sci. 1075, 179–184 (2006). [DOI] [PubMed] [Google Scholar]
- Baaken G., Sondermann M., Schlemmer C., Ruhe J. & Behrends J. C. Planar microelectrode-cavity array for high-resolution and parallel electrical recording of membrane ionic currents. Lab Chip 8, 938–944 (2008). [DOI] [PubMed] [Google Scholar]
- Baaken G., Ankri N., Schuler A. K., Ruhe J. & Behrends J. C. Nanopore-based single-molecule mass spectrometry on a lipid membrane microarray. ACS Nano 5, 8080–8088 (2011). [DOI] [PubMed] [Google Scholar]
- Krueger F., Kreck B., Franke A. & Andrews S. R. DNA methylome analysis using short bisulfite sequencing data. Nat. Methods 9, 145–151 (2012). [DOI] [PubMed] [Google Scholar]
- Sepulveda A. R. et al. CpG methylation analysis - Current status of clinical assays and potential applications in molecular diagnostics: A report of the association for molecular pathology. J. Mol. Diagn. 11, 266–278 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J. W., Tan Q. & Gu L. Q. Single-molecule detection of folding and unfolding of a single G-quadruplex aptamer in a nanopore nanocavity. Nucleic Acids Res 37, 972–982 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information - Kang et al