Significance
Most studies in neuroscience focus on average, or typical, behavior; however, much less is known about how neurobiological differences between individuals contribute to their cognitive and emotional characteristics. In this study, we use outbred rats to investigate if CREB, a key protein required for memory formation, participates in the natural emergence of memory-related individual differences. We show that CREB is activated at lower levels in rats with trait-like deficits in emotional memory. Thus, even in genetically diverse populations, naturally occurring differences in the function of specific proteins may contribute to defining common behavioral traits.
Keywords: individual differences, fear conditioning, novelty
Abstract
Much of what is known about the neurobiology of learning and memory comes from studies of the average behavior. In contrast, intersubject differences that emerge within groups are difficult to study systematically and are often excluded from scientific discussion. Nevertheless, population-wide variability is a virtually universal feature of both complex traits, such as intelligence, and hardwired responses, such as defensive behaviors. Here, we use outbred rats to investigate if cAMP response element-binding protein (CREB), a transcription factor that has long been known in experimental settings to be crucial for associative plasticity, participates in natural memory phenotypes. Using a combination of behavioral, biochemical, and viral techniques, we show that a subset of rats with trait-like deficits in aversive memory have basally reduced CREB activity in the lateral amygdala but can be induced to perform at average levels by directly or indirectly enhancing pretraining CREB phosphorylation. These data suggest that endogenous CREB activity in the amygdala may set a critical threshold for plasticity during memory formation.
The neurobiology of long-term memory (LTM) has been studied at length through the lens of experiments designed to induce changes, on average, in the majority of individuals. In contrast, considerably less research has focused attention on the biology associated with behavioral outcomes that deviate from the norm. LTM formation is an essential and highly conserved cellular process, yet it is also variable: both inbred and outbred rodent strains are widely observed to differ substantially in their capacities to learn and retain associations (1, 2). In experimental settings, these differences appear more often as a hindrance than as a subject of research; however, understanding the mechanisms that give rise to outliers may provide key insights into the biological constraints on, and requirements for, normal mammalian cognition.
Outbred Sprague–Dawley rats individually differ on numerous cognitive measures (3), including LTM strength, which appears to be normally distributed (4, 5). Such differences likely represent a product of both heritable genetic traits and experience-dependent epigenetic and developmental programming. These factors have previously been shown to contribute to phenotypes such as novelty seeking (6), impulsivity (7), aggressiveness (8), anxiety, resilience (9), or susceptibility to stress (10). However, little is known about how basal expression of plasticity-related proteins (PRPs) relates to stable phenotypes revealed during normal learning.
During Pavlovian threat (fear) conditioning, an aversive unconditioned stimulus (US, i.e., footshock) and a neutral conditioned stimulus (CS, i.e., tone) concurrently activate sensory pathways that synapse onto common lateral amygdala (LA) neurons, leading to an increase in CS value (11). This process activates numerous molecular cascades that regulate gene expression required for memory consolidation (12). The transcription factor, cAMP response element binding protein (CREB), is widely viewed as a core component of this machinery, important for the integration of convergent intra- and extracellular signals (13, 14). The transcriptional regulation of plasticity-related genes is particularly associated with phosphorylation of CREB on ser133, which correlates with both physiological and behavioral measures of plasticity (15–17). Moreover, functional CREB augmentation can enhance associative plasticity, network excitability (18, 19), and the likelihood of neuronal participation in memories (20–22), whereas genetic and pharmacological attenuation of CREB function lead to neuroplasticity impairments (23–27).
Despite CREB’s established importance for synaptic plasticity, its endogenous contributions to memory are not well characterized. Here, we investigate the link between basal CREB phosphorylation and natural phenotypic variability in nonpathological fear memory among genetically heterogeneous rodents. In particular, we focus on biochemical differences that emerge in rats with extreme behavioral traits associated with aversive memory. These data support the idea that amygdala CREB activity may regulate individual differences in a threshold for plasticity that can be shifted by exposure to stimulating environmental experience.
Results
Distribution of Phenotypes in Outbred Rats.
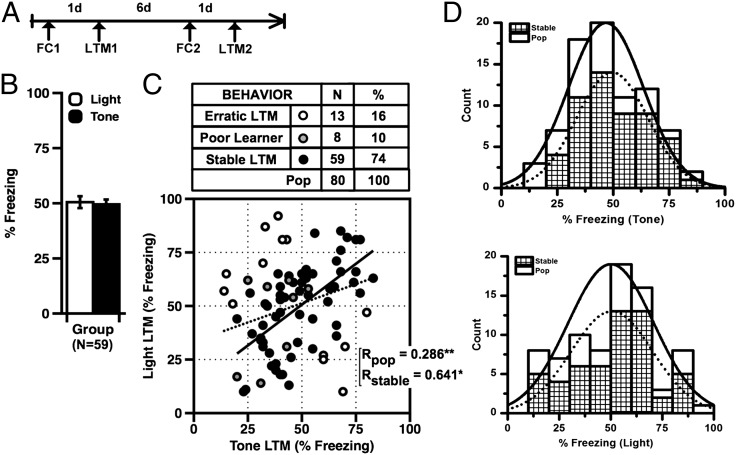
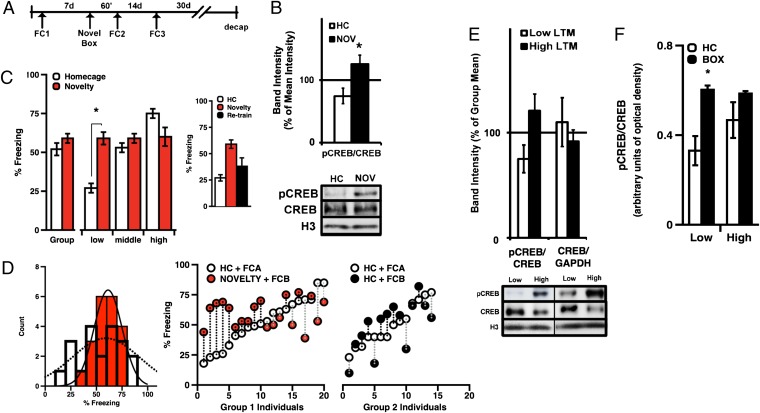
Freezing elicited by an aversive CS is a highly reproducible measure in rodents and known to correlate tightly with physiological and neural processes associated with aversive learning (28). Although individual differences in freezing are generally observed in rodent groups, it is unclear how state-dependent factors (i.e., stress, fatigue, etc.) that do not reflect traits influence the distribution of behaviors. Here, we initially asked what percentage of individual rats show stable, or trait-like, CS-elicited freezing when trained using two different moderate threat (fear)-conditioning protocols, given on separate occasions (Fig. 1A). All rats were trained twice in separate contexts with six 20-s presentations of either a 5-kHz tone or a 0.2-Hz flashing light (CS), each coterminating with a mild 0.6-mA footshock (US). As a population, rats showed typical acquisition of stimulus-elicited freezing, with no significant effects of stimulus type, CS order, or session sequence (Fig. 1B). Tone and light LTM for both the original population (n = 80) and the stable subset (n = 59) were significantly correlated (Fig. 1C). In addition, neither LTM test distribution significantly deviated from expected normal values, and both data sets correlated well with values drawn from an ideal cumulative distribution (Fig. S1, Left and Right). To identify rats with trait-like (i.e., predictable) behavioral performance, we defined criteria to differentiate stable from erratic performers (Materials and Methods). Sixteen percent of rats fit our definition of erratic and were excluded from biochemistry. In addition, eight additional rats were excluded for poor initial learning (mean <40%) to minimize interaction between deficits in acquisition and memory (Fig. 1C). Thus, rats identified as phenotypically stable were defined as individuals from the original population that learned the task and showed predictable performance on two LTM measures (Fig. 1D, Upper, tone; Lower, light). Analysis of each distribution confirmed that both stable and parent populations met criteria for normality [Kolmogorov–Smirnov (K–S) test].
Fig. 1.
(A) Schematic of procedure for assessing phenotypes. (B) Mean tone and light-elicited freezing (LTM) did not significantly differ (tone vs. light LTM: paired t tests, P > 0.05). (C) Tone and light LTM were significantly correlated for both the original population (pop) and the stable subset (pop: R = 0.286, P = 0.001; stable: R = 0.620, P < 0.001). (D) Frequency histograms for tone (Upper) and light (Lower), showing original (n = 80), overlaid with stable (n = 59) population measures. Both distributions met criteria for normality (K–S, df = 80; light, P = 0.3837; tone, P = 0.5130). Error bars indicate ± SEM.
Basal Differences in Amygdala CREB Activation.
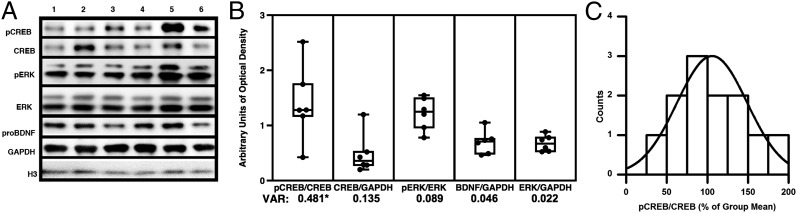
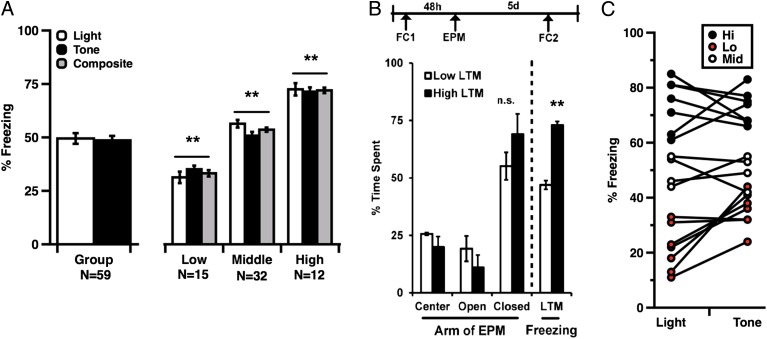
Although numerous inbred mouse strains have been genetically and biochemically differentiated (29, 30), relatively little is known about baseline variability in PRP expression among healthy, genetically unrelated individuals. Here we used Western blots to compare levels of PRPs in LA tissue from randomly selected naive rats (Fig. 2A). We focused specifically on proteins known to be translated and/or activated in response to memory-related cellular signals, including phospho-CREB-Ser133 (pCREB), probrain derived neurotrophic factor (proBDNF), c-Fos, and phospho-extracellular signal-regulated kinase (pERK). Quantification of the bands of interest revealed significant differences in protein-to-protein variances among these targets across individuals in a cohort (Fig. 2B). Levels of total CREB varied significantly more than levels of total ERK, and relative measures of pCREB (pCREB/CREB) varied significantly more than other biochemical ratios. Based on this observation, we asked if the distribution of pCREB/CREB fit a Gaussian distribution and satisfied the K-S criteria for normality. Western blot analysis of pCREB in naive rats (n = 12) indicated that population-wide measures were statistically consistent with standard values drawn from a normal distribution (Fig. 2C) although it is important to note that, compared with behavioral groups, protein distribution analyses involved smaller samples; thus, interpretations of these interindividual data may be limited. We also asked if variability in CREB correlates with trait-like differences in amygdala-dependent emotional memory. To obtain baseline estimates of protein levels, rats identified as behaviorally stable (Fig. 1C) were separated into three phenotypic categories: high (H-LTM), middle (M-LTM), and low (L-LTM) (Fig. 3A). Groups were assigned using a composite LTM score derived from two light + two tone trials, and all three groups were significantly different on both tone and light LTM measures (Fig. 3A).
Fig. 2.
(A) Blot and quantified protein expression from LA tissue extracted from six naive rats. (B) A series of representative Western blots depicting proteins probed in tissue from six individual rats. Quantification of protein levels from randomly selected naive rats revealed significantly greater variance in expression of phospho-S133-CREB/CREB with respect to phospho-ERK/ERK (F-test, pCREB/CREB vs. BDNF/GAPDH, F = 10.558, P = 0.011; pCREB/CREB vs. pERK/ERK, F = 5.405, P = 0.044 and total CREB/GAPDH vs. ERK/GAPDH F = 6.197, P = 0.033). (C) Histogram depicting frequency distribution for pCREB/CREB in naive rats. Quantified band intensities were consistent with expected normal values (K–S test of normality, n = 12; K–S, P = 0.950). Error bars indicate ± SEM.
Fig. 3.
(A) Group means, tone, light, and composite LTM scores from phenotypically stable rats. All phenotypic groups differed significantly from each other (ANOVA, phenotype: F(1,56) = 187.75; P < 0.001; post hoc: Low vs. Middle: t (23) = 7.89, P < 0.001; Low vs. High: t (23) = 10.69, P < 0.001; High vs. Middle, t = 4.152: P < 0.001, Bonferroni). (B) Stable differences in memory expression did not significantly correlate with anxiety on the elevated plus maze (H-LTM vs. L--LTM, closed arm time, t test, P > 0.05). (C) H-LTM and L-LTM rats selected for biochemical analysis showed stable performance on tone and light memory tests. Error bars indicate ± SEM; **P < 0.001.
Interestingly, despite stable differences in fear LTM, none of the phenotypic groups differed significantly in the elevated plus maze, which tests anxiety-like behavior (Fig. 3B). Thus, phenotypes analyzed in the current study are unlikely to reflect a general difference in arousal or aversion. We also asked if H- and L-LTM rats differed in expression of hippocampus (HPC)-dependent contextual memory because the HPC is strongly implicated in spatial and temporal aspects of memory consolidation. Rats expressing high (n = 5) and low (n = 5) cue-elicited aversive memory (hippocampus-independent) were therefore tested for context fear (Fig. S2); however, these groups did not significantly differ.
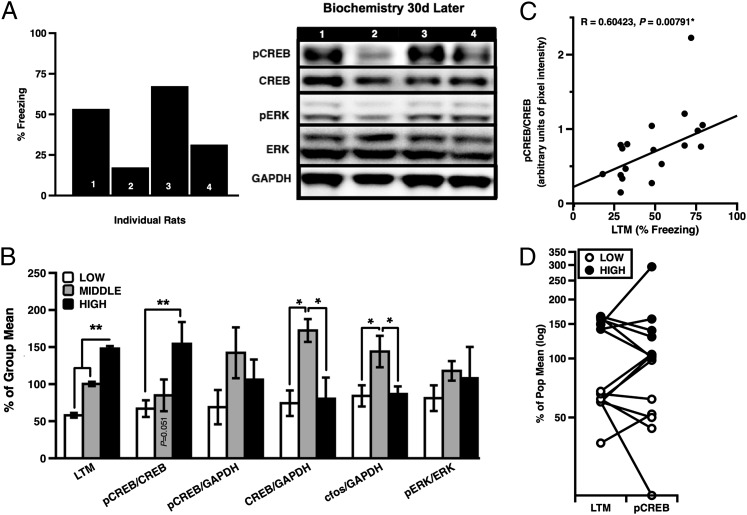
To relate baseline biochemistry with behavior, a subset of stable H-LTM (n = 6) and L-LTM (n = 8) rats were killed 30 d after their last memory test, and LA tissue was extracted for Western blots (Fig. 3C). In addition, tissue was extracted from a small group of M-LTM rats (n = 4) for general comparison. Blots from previously trained rats were probed for pCREB, CREB, and pERK. Results of these analyses revealed significantly higher baseline levels of pCREB/CREB in LA protein from H-LTM rats, relative to both L-LTM and M-LTM rats (Fig. 4A). Unexpectedly, we also observed higher levels of total amygdala CREB and c-fos in M-LTM performers compared with H- and L-LTM rats (Fig. 4B). In addition, levels of pCREB/CREB were significantly correlated with LTM strength across phenotypes (Fig. 4C), whereas pERK/ERK did not significantly distinguish any group. Although data from this population subset were relatively limited, the observed difference in total CREB among rats near the center of the distribution highlights the potential significance of relative, as opposed to absolute, levels of pCREB with respect to CREB.
Fig. 4.
(A) A series of Western blots assessing pCREB, CREB, pERK, ERK, and GAPDH from individual rats; (B) Group biochemical measures in LA tissue from L-LTM and H-LTM rats 30 d after memory test (ANOVA, phenotype: F(2,15) = 5.325, P = 0.018, post hoc: High vs. Low, P = 0.016, Tukey HSD; CREB: F(2,15) = 4.793, P = 0.025; c-fos F(2,14) = 5.323, P = 0.019; post hoc (CREB): Middle vs. Low, P = 0.026; Middle vs. High, P = 0.049, Tukey HSD; post hoc (c-fos): Middle vs. Low, P = 0.020; Middle vs. High, P = 0.043, Tukey HSD). (C) Basal pCREB/CREB significantly correlated with memory (n = 18, R = 0.604, P = 0.007). (D) LTM performance was a relatively good predictor of pCREB/CREB levels. Error bars indicate ± SEM; *P < 0.05; **P < 0.001.
Using tissue from the same H- and L-LTM rats, we also measured pCREB/CREB levels in the HPC, another brain area known to be important for learning and memory. We observed a noteworthy, although nonsignificant, trend toward inverse correlation (Pearson’s R = −0.641, P = 0.087) between pCREB/CREB ratios in HPC and cue-elicited freezing (Fig. S3).
LTM Priming with Spatial Novelty.
CREB has previously been shown to influence neuronal excitability; therefore, we asked if increasing CREB in rats with poor behavioral performance would lower their threshold for memory consolidation, allowing them to perform at average levels (31). To test the idea that transient enhancement of CREB activity might selectively reduce the threshold for consolidation in L-LTM rats, we used a procedure for novelty “priming,” similar to that previously shown to facilitate consolidation of weak learning (32). Past studies have found that exposure to spatial novelty shortly before subthreshold training or stimulation can facilitate CREB-dependent gene expression, natural memory formation, and artificially induced plasticity (32–34). Here, we asked whether novel box exposure before training could similarly enable memory formation in rats with trait-like impairments in memory retention after standard training conditions (Fig. 5A).
Fig. 5.
(A) Protocol used for novelty experiments. (B) Relative to total CREB, novelty increased LA pCREB 1 h later [n = 5/4; t test, t (7) = −3.028, P = 0.019]. (C) Novelty enhanced L-LTM performance when training occurred 1 h [t test, t (5) = 5.55, P = 0.003], but not 2 wk, later (t test, planned comparison: L-LTM in s1 vs. s3, P > 0.05). (D) Pretraining novelty significantly reduced variance in LTM scores (n = 20; F = 3.20, P = 0.015). (E) Thirty days after LTM test, L-LTM rats expressed lower baseline pCREB than H-LTM rats [t test, t (9) = 2.14, P = 0.061]. (F) L-LTM (n = 5) but not H-LTM (n = 6) rats express significantly higher pCREB 1 h after novelty (ANOVA, novelty: F(2,19) = 9.165, P = 0.007; interaction: F(1,19) = 4.916, P = 0.039, post hoc: L-LTM + novelty (n = 5): t(4.6) = 4.014, P = 0.012). Representative Western blots depict independent measures of CREB, pCREB, and histone H3 protein levels. Error bars indicate ±SEM; *P < 0.05.
We first confirmed that novelty exposure was sufficient to induce a general increase in LA pCREB 1 h later. Compared with naive rats (n = 5), rats exposed to novelty for 15 min (n = 4) expressed significantly higher levels of LA pCREB 1 h later (Fig. 5B). We then repeated the same dual-training procedure; however, this time, rats received a 15-min novel box exposure 1 h before session 2 (s2) of training. Thus, phenotypes were assigned based on memory of the unprimed training session 1 (s1), with the top and bottom 30% of individuals populating the L-LTM and H-LTM groups, respectively.
In LTM test s1 (before priming), all groups significantly differed in behavior (Fig. 5C, ANOVA, F(2,17) = 56.475, P < 0.0001); however, in LTM test s2, all a priori phenotypic differences between subgroups were abolished (ANOVA, P > 0.05). Interestingly, despite the effect of novelty on population variance, it did not significantly affect the group mean. Rather, L-LTM, but not M- or H-LTM, were enhanced by pretraining novelty exposure; thus, M-LTM rats remained unchanged as a result of priming, whereas H-LTM rats showed a nonsignificant trend toward diminished freezing (Fig. 5C). Taken together, we found that brief exposure to spatial novelty normalized the previously significant difference between phenotypes and significantly reduced the overall variance of the group (Fig. 5D). These effects were not due to cross-training interactions, as rats in an unprimed control population showed no significant phenotype difference or shift in population variance between s1 and s2 (Fig. 5D, Right, n = 14, F-test, s1 vs. s2: F = 1.158, P > 0.05).
In addition, we asked if the effect of novelty on weak memory was association-specific or if rats performing poorly in s1 would also perform poorly after retraining. To address this question, rats were subjected to a third session (s3) of unprimed training 2 wk later (CS = white noise). Consistent with the idea of memory-specific priming, L-LTM rats, which expressed improved memory in s2, returned to poor performance in s3. (Fig. 5C, Inset). Moreover, compared with s2, the s3 population variance was significantly greater and did not differ from the population variance of behavior in s1 (F-test, s1 vs. s3: F = 1.20, P > 0.05). To biochemically corroborate this finding, we also collected protein samples from L-LTM and H-LTM individuals 30 d after training in s3. As in our initial observation, ratios of pCREB/CREB in L-LTM (n = 5) rats remained generally lower than in H-LTM (n = 6) rats, despite falling just short of statistical significance (Fig. 5E, P = 0.061). Thus, L-LTM rats expressed improved memory when exposed to spatial novelty 1 h before conditioning, yet they reemerged as poor performers after retraining 2 wk later and showed a trend toward lower LA pCREB/CREB ratios at 30 d.
As a follow-up question, we also asked if L- and H-LTM rats express acute differences in pCREB/CREB 1 h after novel box exposure coinciding with the time of training (Fig. 5 B–D). We therefore analyzed LA protein collected from separate groups of rats killed 1 h (rather than 30 d) after novelty (Fig. 5F). Quantification of blots from 1-h and 30-d samples run in parallel suggest that box-exposed L-LTM rats had significantly higher levels of pCREB/CREB 1 h later compared with L-LTM rats killed under baseline conditions. In contrast, H-LTM rats did not show a significant box-induced increase in pCREB/CREB at 1 h relative to baseline (30 d) H-LTM counterparts. Thus, novelty appeared to exert an acute affect on pCREB expression, which more dramatically affected L-LTM rats (Fig. 5F).
Low LTM Expressers Are Selectively Augmented by CREBY134F.
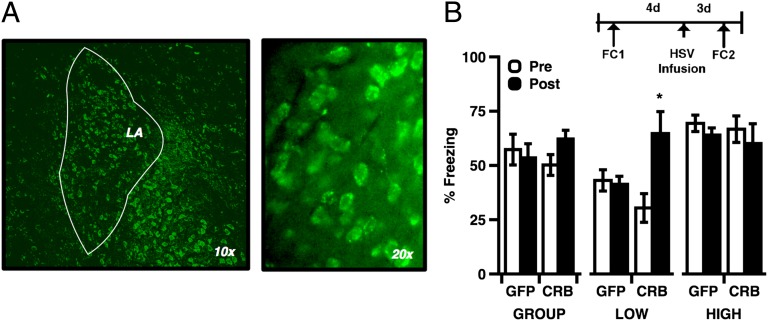
To test the direct effect of enhanced CREB function in L-LTM rats, our final experiment used intra-LA microinfusions of an HSV viral vector to express constitutively active CREBY134F in rats with strong LTM phenotypes. Published research has shown that N terminus fusion of CREB with GFP does not compromise CREB activity (35), and studies using similar methods have reported increased CREB activity and CRE-mediated transcription (36, 37). We hypothesized that, like box exposure, direct viral enhancement of CREB function before learning might be sufficient improve memory in L-LTM rats. Four days before viral infusion, rats were trained, tested, and assigned to either group 1 (L-LTM, lowest 30%) or group 2 (M/H-LTM, upper 60%). Following this preinfusion test of LTM, rats from both groups received intra-LA infusions of either HSV-GFP (GFP, n = 13) or HSV-GFP-CREBY134F (CREBY134F, n = 11, Fig. 6A). Two days after viral infection, all rats received a second round of training, and LTM was tested 24 h later. Comparison of pre- vs. postinfusion LTM revealed a significant main effect of phenotype but no main effect of CREBY134F (P > 0.05). We did, however, observe a significant interaction between viral construct and phenotype, such that postinfusion memory was enhanced only among L-LTM rats receiving CREBY134F (Fig. 6B). Moreover, preinfusion behavioral differences between phenotypic groups were abolished for memory acquired after CREBY134F expression. In contrast, performance among L-LTM controls infused with HSV-GFP remained significantly lower than M/H-LTM rats when tested for memory acquired after HSV infusion.
Fig. 6.
(A) GFP fluorescence from HSV-infected neurons. Cell counts estimated from samples of HSV-infected tissue indicate that 11.7% of basal/lateral amygdala cells were GFP(+) relative to DAPI. (B) Viral expression of CREBY134F selectively enhanced L-LTM memory (ANOVA, phenotype: F(2,19) = 15.604, P < 0.001; interaction: (session) F(1,20) = 32.556, P < 0.001; (construct) F(1,20) = 4.599, P = 0.044; post hoc, Low vs. Middle/High, Pre- vs. Postinfusion: t (9) = −3.788, P = 0.004). L-LTM behavior was not affected by HSV-GFP infusion (post hoc, Low vs. Middle/High, Pre- vs. Postinfusion: t (11) = −4.326, P = 0.001). Error bars indicate ± SEM; *P < 0.05.
Discussion
Individuals with outlier behavioral phenotypes represent an important, but under-studied subset of normal populations that could provide critical insights into our understanding of normal cognition as well as behavioral pathology. Such individual differences are likely the result of genetic, epigenetic, and developmental events that influence both internal homeostatic properties and external stimulus-driven responses (38). However, most relevant studies to date have focused either on genetically amplified traits or on acute responses to experience-dependent activity. In contrast, few studies have analyzed stable differences in basal biochemical properties with respect to natural (noninbred) behavioral traits. Here we show that CREB, which has long been implicated in basic mechanisms for neuronal plasticity and excitability (18), may also function as an endogenous regulator of innate cognitive and emotional characteristics. Our biochemical data further suggest that relative ratios of endogenous pCREB/CREB may better predict behavior than either pCREB or CREB levels alone. Moreover, these data raise questions about how average-performing individuals differ from those on the extremes in terms of behavioral flexibility, biochemical profile, and systems-level circuit dynamics. Future investigation will be needed to determine if endogenous variability in pCREB/CREB ratios also corresponds to differentially distributed pools of protein within individual amygdala neurons.
The current findings also raise questions about how CREB and other downstream plasticity-related proteins, in conjunction with neuromodulators such as dopamine, coordinate network interactions across brain areas. The tendency for hippocampal and amygdala pCREB/CREB to relate oppositely to cued behavioral responses suggests that competitive or homeostatic mechanisms may be important for balancing plasticity at interconnected sites. This observation also mirrors patterns reported in other circuits and is consistent with the theory that feedback between anatomically distinct neural circuits is essential for healthy cognition. Given that threat-related pathologies are often marked by excessive cue responsiveness coupled with impaired context discrimination, further investigation of hippocampal–amygdalar biochemical relationships would be informative.
Finally, we find that, although generally stable across time, individual plasticity can be transiently facilitated either with exposure to spatial novelty or with direct augmentation of amygdala CREB function by viral overexpression. Together, these data indicate that the canonical involvement of CREB in plasticity-related transcription (39) extends to the control of natural memory efficacy and that common downstream protein-signaling dynamics may contribute to core behavioral attributes, even among genetically distinct individuals.
Materials and Methods
Subjects.
All procedures were conducted in compliance with the National Institutes of Health Guide for the Care and Use of Experimental Animals and the New York University Animal Care and Use Committee. Male Sprague–Dawley rats (250–300 g, Hilltop Lab Animals) were individually housed on a 12h/12h light/dark cycle with food and water ad libitum for the duration of experiments.
Behavior.
Rats were habituated, trained, and tested in yoked sound-insulated conditioning chambers and recorded with infrared digital cameras (Coulbourn Instruments, Graphic State 2). Sequential (counterbalanced) tone and light conditioning sessions were given 1 wk apart. In each session, rats were habituated to the training box for 30 min the day before training. Training consisted of 290 s of acclimation followed by six CS presentations (either a 5-kHz, 80-dB tone or a 0.2-Hz flashing light) coterminating with a 0.5-s footshock US (0.6 mA; mean intertrial interval = 120 s). The LTM test entailed two CS presentations in a novel context, given 1 d after training. Rat identification numbers were coded, and freezing was scored manually by a blind rater. In experiments with novelty, rats received 15-min in a novel box 1 h before training. For postnovelty biochemistry, rats received a 15-min novelty exposure 1 h before decapitation.
Phenotype/Exclusion Criteria.
For the initial biochemistry and population-wide distribution, rats were omitted from the “stable” subset used for biochemistry if (i) learning was impaired, as indicated by a mean acquisition score of <40% across trials 2–6 of tone and light sessions (10-trial mean) or (ii) mean tone and light LTM measures were “erratic,” or deviated more than one SD from the mean difference for |light – tone|. After applying these exclusions, rats were subdivided into low, middle, and high phenotypic categories according to their composite LTM score, defined as the mean of all light and tone test trials. Rats were considered L- or H-LTM if their composite LTM scores were >1*SD from the mean (large population analysis) or were in the top/bottom 30% of the group (smaller behavior experiments). Both standards identified characteristically similar individuals.
Viral Constructs.
CREBY134F and the control gene, LacZ, were fused with a GFP reporter under control of the constitutive IE 4/5 HSV promoter. Genes of interest were cloned into the HSV amplicon (HSV-PrpUC) and packaged using a replication-defective helper virus (with 5dl1.2 deletion) as previously described (40). Virus was purified on a sucrose gradient, pelleted, and resuspended in 10% sucrose.
Viral Infusions.
Rats were assigned to groups based on light LTM scores. Two days later, rats received intra-LA infusions of HSV-GFP or HSV-GFP-CREBY134F. Three days postinfection (7 d posttraining), rats were trained in a different context and tested the next day. Following completion of behavior, rats were perfused and brains were cut at 40 μm and mounted to confirm injector placements and GFP expression (ProLong-Gold Antifade with DAPI, Invitrogen). Rats were excluded either if the injector missed the LA or if GFP was absent. Images were acquired by epifluorescence and coded to conceal experimental groups. Regions of interest were drawn around the amygdala in ImageJ, and DAPI was used to guide anatomical boundaries. Infection rates were calculated from manual cell counts of GFP/DAPI.
Western Blotting.
Rats were deeply anesthetized with 25% chloral hydrate (wt/vol) and decapitated for fresh LA tissue extraction. Briefly, freshly removed brains were horizontally bisected at the level of the rhinal fissure, and LA tissue was removed in whole-nucleus form using a 1-mm curette (Fig. S4). Protein homogenates were prepared by sonication in ice-cold buffer [50 mM Tris⋅HCl, pH 7.5; 150 KCl; 1 DTT; 1 EDTA; protease and phosphatase inhibitor mixtures (Sigma)]. Protein concentrations were determined by bicinchoninic acid assay protein assay (Pierce). Samples were prepared for SDS/PAGE on 4–12% (wt/vol) acrylamide gel (Invitrogen) and transferred to PVDF. Blots were blocked in 0.2% iBlock and incubated with anti–phopho-(ser133)-CREB IgG, phospho-42/44-ERK, and/or total ERK p42/p44 (Cell Signaling). Twenty-four hours later, blots were treated with an HRP-conjugated secondary IgG (Promega), and protein bands were detected by chemiluminescence (ECL-Plus; GE Healthcare) on a KODAK 4000MM imager. Blots were stripped overnight [25 mM glycine, 2% (wt/vol) SDS, pH 2.0] and reprobed with total CREB IgG and/or GAPDH and histone H3 (Cell Signaling). Bands were visualized on a Kodak imager and quantified using GelEval software (FrogDance).
Statistics.
Differences were significant if P < 0.05. For data analyzed by ANOVA, a Fisher least significant difference post hoc test was used for significant main effects, and a Bonferroni post hoc correction was used for significant interactions in the absence of main effects (or when more than three comparisons were involved). For population data, normality was verified with the K–S test, and variances were compared by F-test. For Western blots, local background was subtracted from the sum pixel value of each band, and differences were normalized to a measure of total protein on the same blot (GAPDH or histone H3). Final values were calculated as a mean of two to four replications/sample, and statistical outliers were defined as samples >2*SD from the mean on repeated runs. One H-LTM tissue sample with very high pCREB band intensity was excluded using these criteria.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants F31 MH083472 to K.K.C.; NS034007 and NS047384 to E.K.; and MH046516 and MH038774 to J.E.L.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304665110/-/DCSupplemental.
References
- 1. Camp MC, et al. (2012) Genetic strain differences in learned fear inhibition associated with variation in neuroendocrine, autonomic, and amygdala dendritic phenotypes. Neuropsychopharmacology 37(6):1534–1547. [DOI] [PMC free article] [PubMed]
- 2.Yilmazer-Hanke DM. Morphological correlates of emotional and cognitive behaviour: Insights from studies on inbred and outbred rodent strains and their crosses. Behav Pharmacol. 2008;19(5-6):403–434. doi: 10.1097/FBP.0b013e32830dc0de. [DOI] [PubMed] [Google Scholar]
- 3.Curé M, Rolinat JP. Behavioral heterogeneity in Sprague-Dawley rats. Physiol Behav. 1992;51(4):771–774. doi: 10.1016/0031-9384(92)90114-h. [DOI] [PubMed] [Google Scholar]
- 4.Bush DE, Sotres-Bayon F, LeDoux JE. Individual differences in fear: Isolating fear reactivity and fear recovery phenotypes. J Trauma Stress. 2007;20(4):413–422. doi: 10.1002/jts.20261. [DOI] [PubMed] [Google Scholar]
- 5. Matzel LD, et al. (2003) Individual differences in the expression of a “general” learning ability in mice. J Neurosci 23(16):6423–6433. [DOI] [PMC free article] [PubMed]
- 6.Duclot F, Hollis F, Darcy MJ, Kabbaj M. Individual differences in novelty-seeking behavior in rats as a model for psychosocial stress-related mood disorders. Physiol Behav. 2011;104(2):296–305. doi: 10.1016/j.physbeh.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalley JW, et al. Nucleus accumbens D2/3 receptors predict trait impulsivity and cocaine reinforcement. Science. 2007;315(5816):1267–1270. doi: 10.1126/science.1137073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lindberg J, et al. (2005) Selection for tameness has changed brain gene expression in silver foxes. Curr Biol 15(22):R915–R916. [DOI] [PubMed]
- 9.Krishnan V, et al. Molecular adaptations underlying susceptibility and resistance to social defeat in brain reward regions. Cell. 2007;131(2):391–404. doi: 10.1016/j.cell.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 10. Schmidt MV, et al. (2010) Individual stress vulnerability is predicted by short-term memory and AMPA receptor subunit ratio in the hippocampus. J Neurosci 30(50):16949–16958. [DOI] [PMC free article] [PubMed]
- 11.Maren S. Synaptic mechanisms of associative memory in the amygdala. Neuron. 2005;47(6):783–786. doi: 10.1016/j.neuron.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 12.Frankland PW, et al. Consolidation of CS and US representations in associative fear conditioning. Hippocampus. 2004;14(5):557–569. doi: 10.1002/hipo.10208. [DOI] [PubMed] [Google Scholar]
- 13.Kida S, et al. CREB required for the stability of new and reactivated fear memories. Nat Neurosci. 2002;5(4):348–355. doi: 10.1038/nn819. [DOI] [PubMed] [Google Scholar]
- 14. Josselyn SA, et al. (2001) Long-term memory is facilitated by cAMP response element-binding protein overexpression in the amygdala. J Neurosci 21(7):2404–2412. [DOI] [PMC free article] [PubMed]
- 15.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2(8):599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 16.Bito H, Deisseroth K, Tsien RW. CREB phosphorylation and dephosphorylation: a Ca(2+)- and stimulus duration-dependent switch for hippocampal gene expression. Cell. 1996;87(7):1203–1214. doi: 10.1016/s0092-8674(00)81816-4. [DOI] [PubMed] [Google Scholar]
- 17.Montminy M. Transcriptional regulation by cyclic AMP. Annu Rev Biochem. 1997;66:807–822. doi: 10.1146/annurev.biochem.66.1.807. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Y, et al. CREB regulates excitability and the allocation of memory to subsets of neurons in the amygdala. Nat Neurosci. 2009;12(11):1438–1443. doi: 10.1038/nn.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Won J, Silva AJ. Molecular and cellular mechanisms of memory allocation in neuronetworks. Neurobiol Learn Mem. 2008;89(3):285–292. doi: 10.1016/j.nlm.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Suzuki A, et al. (2011) Upregulation of CREB-mediated transcription enhances both short- and long-term memory. J Neurosci 31(24):8786–8802. [DOI] [PMC free article] [PubMed]
- 21.Weeber EJ, Sweatt JD. Molecular neurobiology of human cognition. Neuron. 2002;33(6):845–848. doi: 10.1016/s0896-6273(02)00634-7. [DOI] [PubMed] [Google Scholar]
- 22.Middei S, et al. CREB selectively controls learning-induced structural remodeling of neurons. Learn Mem. 2012;19(8):330–336. doi: 10.1101/lm.025817.112. [DOI] [PubMed] [Google Scholar]
- 23. Chen G, Zou X, Watanabe H, van Deursen JM, Shen J (2010) CREB binding protein is required for both short-term and long-term memory formation. J Neurosci 30(39):13066–13077. [DOI] [PMC free article] [PubMed]
- 24.Josselyn SA, Kida S, Silva AJ. Inducible repression of CREB function disrupts amygdala-dependent memory. Neurobiol Learn Mem. 2004;82(2):159–163. doi: 10.1016/j.nlm.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 25.Gass P, et al. Deficits in memory tasks of mice with CREB mutations depend on gene dosage. Learn Mem. 1998;5(4–5):274–288. [PMC free article] [PubMed] [Google Scholar]
- 26.Bourtchuladze R, et al. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell. 1994;79(1):59–68. doi: 10.1016/0092-8674(94)90400-6. [DOI] [PubMed] [Google Scholar]
- 27.Pittenger C, et al. Reversible inhibition of CREB/ATF transcription factors in region CA1 of the dorsal hippocampus disrupts hippocampus-dependent spatial memory. Neuron. 2002;34(3):447–462. doi: 10.1016/s0896-6273(02)00684-0. [DOI] [PubMed] [Google Scholar]
- 28.Iwata J, LeDoux JE. Dissociation of associative and nonassociative concomitants of classical fear conditioning in the freely behaving rat. Behav Neurosci. 1988;102(1):66–76. doi: 10.1037//0735-7044.102.1.66. [DOI] [PubMed] [Google Scholar]
- 29.Sung JY, et al. Learning strategy selection in the water maze and hippocampal CREB phosphorylation differ in two inbred strains of mice. Learn Mem. 2008;15(4):183–188. doi: 10.1101/lm.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pollak DD, et al. Strain-dependent regulation of plasticity-related proteins in the mouse hippocampus. Behav Brain Res. 2005;165(2):240–246. doi: 10.1016/j.bbr.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 31.Viosca J, Lopez de Armentia M, Jancic D, Barco A. Enhanced CREB-dependent gene expression increases the excitability of neurons in the basal amygdala and primes the consolidation of contextual and cued fear memory. Learn Mem. 2009;16(3):193–197. doi: 10.1101/lm.1254209. [DOI] [PubMed] [Google Scholar]
- 32. Moncada D, Viola H (2007) Induction of long-term memory by exposure to novelty requires protein synthesis: Evidence for a behavioral tagging. J Neurosci 27(28):7476–7481. [DOI] [PMC free article] [PubMed]
- 33.Moncada D, Viola H. Phosphorylation state of CREB in the rat hippocampus: A molecular switch between spatial novelty and spatial familiarity? Neurobiol Learn Mem. 2006;86(1):9–18. doi: 10.1016/j.nlm.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 34.Li S, Cullen WK, Anwyl R, Rowan MJ. Dopamine-dependent facilitation of LTP induction in hippocampal CA1 by exposure to spatial novelty. Nat Neurosci. 2003;6(5):526–531. doi: 10.1038/nn1049. [DOI] [PubMed] [Google Scholar]
- 35.Chao JR, et al. Characterization of the mouse adenylyl cyclase type VIII gene promoter: Regulation by cAMP and CREB. Eur J Neurosci. 2002;16(7):1284–1294. doi: 10.1046/j.1460-9568.2002.02186.x. [DOI] [PubMed] [Google Scholar]
- 36.Barrot M, et al. CREB activity in the nucleus accumbens shell controls gating of behavioral responses to emotional stimuli. Proc Natl Acad Sci USA. 2002;99(17):11435–11440. doi: 10.1073/pnas.172091899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Olson VG, et al. (2005) Regulation of drug reward by cAMP response element-binding protein: Evidence for two functionally distinct subregions of the ventral tegmental area. J Neurosci 25(23):5553–5562. [DOI] [PMC free article] [PubMed]
- 38.Antoniou K, et al. Individual responses to novelty are associated with differences in behavioral and neurochemical profiles. Behav Brain Res. 2008;187(2):462–472. doi: 10.1016/j.bbr.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 39.Xia M, et al. Identification of compounds that potentiate CREB signaling as possible enhancers of long-term memory. Proc Natl Acad Sci USA. 2009;106(7):2412–2417. doi: 10.1073/pnas.0813020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carlezon WA, Jr, et al. Regulation of cocaine reward by CREB. Science. 1998;282(5397):2272–2275. doi: 10.1126/science.282.5397.2272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.