Significance
Splicing factors are involved in the maturation of messenger RNAs that are translated into proteins. Select splicing factors have been implicated in RNA interference. However, the underlying mechanism is not clear. We found that in Drosophila the splicing factor SmD1 is indispensable for the production and function of small interfering RNAs, key components of RNAi, and that SmD1 interacts with both protein and RNA components of the RNAi machineries. Notably, defects in RNAi and in pre-mRNA splicing can be uncoupled, and the RNAi and splicing machineries are physically and functionally distinct. Our results suggest that Drosophila SmD1 plays a direct role in RNAi-mediated gene silencing independently of its pre-mRNA splicing activity.
Abstract
RNAi is an evolutionarily conserved gene regulatory process that operates in a wide variety of organisms. During RNAi, long double-stranded RNA precursors are processed by Dicer proteins into ∼21-nt siRNAs. Subsequently, siRNAs are incorporated into the RNA-induced silencing complexes (RISCs) that contain Argonaute-family proteins and guide RISC to target RNAs via complementary base pairing, leading to posttranscriptional gene silencing. Select pre-mRNA splicing factors have been implicated in RNAi in fission yeast, worms, and flies, but the underlying molecular mechanisms are not well understood. Here, we show that SmD1, a core component of the Drosophila small nuclear ribonucleoprotein particle implicated in splicing, is required for RNAi and antiviral immunity in cultured cells and in vivo. SmD1 interacts with both Dicer-2 and dsRNA precursors and is indispensable for optimal siRNA biogenesis. Depletion of SmD1 impairs the assembly and function of the small interfering RISC without significantly affecting the expression of major canonical siRNA pathway components. Moreover, SmD1 physically and functionally associates with components of the small interfering RISC, including Argonaute 2, both in flies and in humans. Notably, RNAi defects resulting from SmD1 silencing can be uncoupled from defects in pre-mRNA splicing, and the RNAi and splicing machineries are physically and functionally distinct entities. Our results suggest that Drosophila SmD1 plays a direct role in RNAi-mediated gene silencing independently of its pre-mRNA splicing activity and indicate that the dual roles of splicing factors in posttranscriptional gene regulation may be evolutionarily widespread.
RNAi is an ancient gene regulatory process (1). RNAi is triggered by ∼21- to 24-nt double-stranded siRNAs and microRNAs (miRNAs) (2–5), which are assembled into RNA-induced silencing complexes (RISCs). Once incorporated into the RISC, one strand of the siRNA or miRNA engages target mRNAs via complementary base pairing and modulates gene expression in a sequence-specific manner. RNAi generally represses gene expression posttranscriptionally by inhibiting translation and/or promoting destabilization of target mRNAs, or transcriptionally by modulating chromatin structure at the target loci. By regulating the expression of diverse target RNAs, RNAi plays a key role in numerous biological processes, including cancer, development, homeostasis, and antiviral defense.
In Drosophila melanogaster, siRNAs are generated from long dsRNA precursors that are either transcribed endogenously or introduced exogenously (6–10). Long dsRNAs are processed into duplex siRNAs by the ribonuclease III Dicer-2 (Dcr-2), which forms a stable complex with the cofactors loquacious (Loqs)-PD and R2D2 (11–14). The formation of mature RISC is a multistep process. First, the Dcr-2/R2D2 heterodimer gauges the thermodynamic stability of the ends of the siRNA duplex and associates with duplex siRNAs in a polarized pattern to form the RISC-loading complex (RLC) (15, 16). Several other proteins, including the nuclease Argonaute 2 (AGO2) and the chaperone proteins Hsc70 and Hsp90, join the RLC to form the pre-RISC (17, 18), leading to the loading of the siRNA duplex to AGO2. Following AGO2 loading, the duplex strand with the less stable 5′ terminus (the guide strand) is selectively retained, whereas the complementary (passenger) strand is cleaved by the slicer activity of AGO2, a process known as passenger-strand nicking (19–22). The siRNA duplex is subsequently unwound, and the sliced passenger strand is removed by the C3PO endoribonuclease complex (23), resulting in the formation of the mature RISC that contains the siRNA guide strand. Finally, target mRNAs are recruited to mature RISC by perfectly complementary base pairing with the siRNA guide strand and cleaved (sliced) by AGO2 (24, 25).
Recent studies have suggested that pre-mRNA splicing factors may be involved in RNAi. For example, mutations in genes encoding a subset of splicing factors impair RNAi-mediated heterochromatin formation and transcriptional silencing in Schizosaccharomyces pombe (26). In addition, genome-wide RNAi screens of Caenorhabditis elegans and cultured Drosophila cells, as well as a recent analysis of phylogenetic conservation of candidate RNAi factors, indicated that specific splicing factors are required for RNA-mediated gene silencing (27–30). Furthermore, SNR-3, the C. elegans homolog of SmD1, a core protein of the Drosophila small nuclear ribonucleoprotein particle (snRNP), associates with DCR-1 (31). Finally, Gemin3 and Gemin4, components of the Survival of Motor Neuron complex required for snRNP assembly, associate with AGO2 in human cell extract (32). These findings suggest that splicing factors may play dual roles in regulating gene expression; however, the molecular mechanisms underlying their roles in RNAi remain underexplored.
We previously identified a subset of splicing factors required for RNAi in Drosophila, including SmD1 (28). SmD1 and six additional Sm proteins (SmB, SmD2, SmD3, SmE, SmF, and SmG) form a heptameric ring structure that accommodates the U-rich snRNAs (33) and constitute core components of the snRNP essential for pre-mRNA splicing (34). In the current study, we investigated the molecular basis of the role of SmD1 in RNAi in Drosophila. We show that SmD1 impinges on multiple steps of the siRNA pathway, including siRNA biogenesis and small interfering RISC (siRISC) assembly/function. Moreover, SmD1 associates with canonical components of the siRNA biogenesis and effector machineries and is required for effective antiviral immunity. Finally, we provide evidence suggesting that SmD1 directly participates in RNAi independently of its pre-mRNA splicing functions.
Results
SmD1 Is Required for RNAi and Antiviral Immunity.
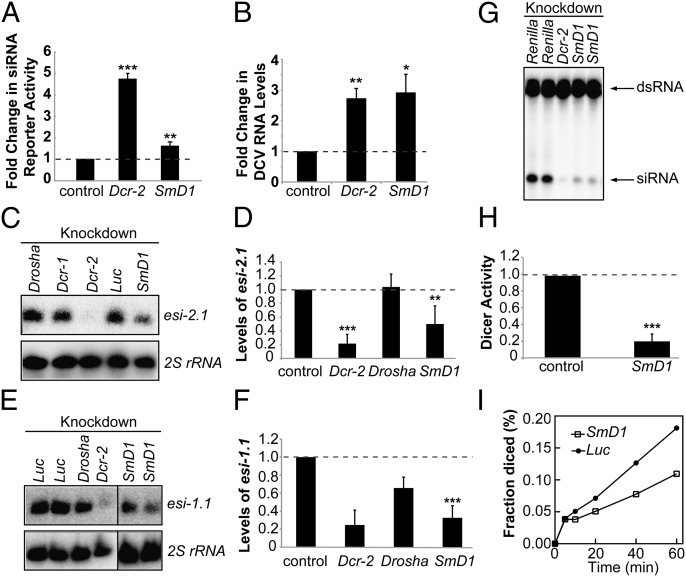
Previously we identified SmD1 as a candidate RNAi factor in Drosophila (28). To probe this further, we examined the effects of SmD1 and Dcr-2 silencing on RNAi in cultured Drosophila S2 cells expressing a luciferase sensor for the endogenous siRNA esi-2.1. We found that luciferase expression was derepressed in SmD1- or Dcr-2-depleted cells compared with control cells, indicating a defect in endogenous siRNA-mediated silencing of the reporter (Fig. 1A). In addition, results from an eye color-based RNAi assay suggest that SmD1 is required for effective RNAi in vivo (Fig. S1). RNAi plays a key role in antiviral immunity in Drosophila (35–37). To determine whether SmD1 is involved in this process, we treated S2 cells with SmD1 or Dcr-2 dsRNA and infected them with Drosophila C virus (DCV), a positive-strand RNA virus, and measured viral RNA levels in infected cells by RT-quantitative (q) PCR. We found that viral genomic RNA levels were higher in SmD1- or Dcr-2–depleted cells than in control cells (Fig. 1B). We also examined the effect of SmD1 knockdown in S2-NP cells persistently infected with Flock House virus (FHV). Analysis of deep-sequencing data revealed a significant increase in FHV genomic RNA levels in SmD1-depleted cells compared with control cells (Fig. S2). We conclude that SmD1 is required for RNAi and antiviral immunity.
Fig. 1.
Loss of SmD1 compromises RNAi and siRNA biogenesis. (A) S2 cells were transfected with Dcr-2, SmD1, or control (LacZ) dsRNA together with expression constructs for a Renilla luciferase reporter containing complementary binding sites for esi-2.1 and a firefly luciferase control reporter. Loss of RNAi was measured as derepression of the luciferase reporter activity (n ≥ 3). (B) S2 cells treated with various dsRNAs (below) were infected with DCV. DCV RNA levels were measured by RT-qPCR (n ≥ 3). (C) Northern blotting to measure levels of the endogenous siRNA esi-2.1 or 2S rRNA (loading control) in various dsRNA-treated S2 cells. (D) Quantification of esi-2.1 levels (n = 4) normalized against 2S rRNA levels and compared with controls. (E) Northern blotting to measure esi-1.1 and 2S rRNA levels in various knockdown cells. (F) Quantification of esi-1.1 levels. Dcr-2 and Drosha, n = 2; SmD1, n = 4. (G) Cytoplasmic lysates from S2 cells treated with various dsRNAs were incubated with radiolabeled dsRNA substrate to generate 21-nt siRNAs. RNAs were extracted and resolved by denaturing urea-PAGE. (H) The amount of siRNAs is quantified and normalized to controls (n = 3). (I) Kinetic analysis of Dcr-2 activity using lysates from control or SmD1 knockdown cells. Fraction of dsRNA substrate processed by Dcr-2 at various time points is shown. Data are shown as mean + SD in A, D, F, and H or mean + SEM in B. *P < 0.05, **P < 0.01, ***P < 0.001. Unless noted otherwise, dsRNAs against Renilla luciferase or firefly luciferase serve as controls.
SmD1 Interacts with Components of the siRNA Biogenesis Machinery and Is Indispensable for Optimal siRNA Biogenesis.
To determine whether SmD1 is required for siRNA biogenesis, we knocked down SmD1 in S2 cells and measured levels of esi-2.1 and esi-1.1 by Northern blotting. As controls, we also depleted Dcr-2 and key enzymes in miRNA biogenesis (Dcr-1 and Drosha). SmD1 knockdown reduced esi-2.1 and esi-1.1 levels by ∼50% and ∼70%, respectively, compared with control cells (Fig. 1 C–F). Because Dcr-2 is an essential RNase III in siRNA biogenesis, we asked whether loss of SmD1 affected Dcr-2 activity. S2 cells were treated with dsRNAs against SmD1 or Dcr-2, and the lysates were incubated with radiolabeled dsRNAs. Dcr-2 activity was measured by the production of 21-nt siRNAs. Notably, Dcr-2 activity was markedly reduced in cells depleted of SmD1 compared with the control cells at all time points examined (Fig. 1 G–I). Our results thus suggest a critical role for SmD1 in dsRNA processing.
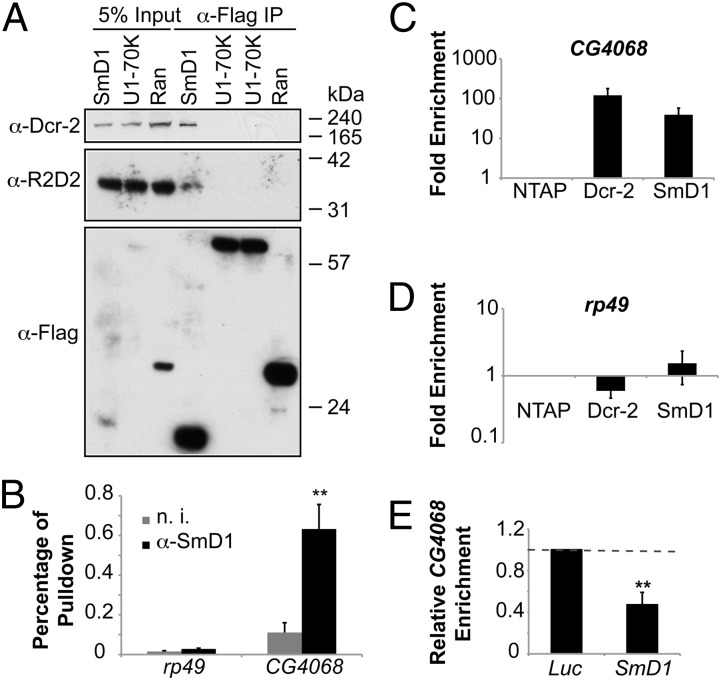
We next asked whether SmD1 physically associates with components of the siRNA biogenesis machinery. We found that endogenous Dcr-2 and R2D2 copurified with Flag-tagged SmD1 but not significantly with Ran or the U1 snRNP component U1-70K (Fig. 2A). To explore the possibility that SmD1 might modulate siRNA biogenesis by interacting with dsRNAs, thereby facilitating their recognition by Dcr-2, we performed RNA immunoprecipitation assays. This analysis revealed that CG4068 (the precursor of esi-2.1), but not the control mRNA rp49, was highly enriched in the SmD1 complex compared with the control sample (Fig. 2B). Similar results were obtained in immunopurified TAP-tagged SmD1 or Dcr-2 complexes (Fig. 2 C and D). Notably, the interaction between Dcr-2 and CG4068 was attenuated in cells depleted of SmD1 (Fig. 2E). In addition, analysis of RNA sequencing data revealed that compared with control cells SmD1-deficient cells accumulate higher levels of several structured transcripts that serve as precursors of endogenous siRNAs, indicating defects in Dcr-2–mediated processing of these RNAs into siRNAs (Table S1). Collectively, these data demonstrate that SmD1 associates with both components of the siRNA biogenesis machinery and dsRNA substrates, and that SmD1 is required for optimal Dcr-2–mediated processing of dsRNAs into siRNAs.
Fig. 2.
SmD1 interacts with components of the siRNA biogenesis machinery. (A) Lysates from S2 cells expressing Flag-tagged SmD1 or control proteins (U1-70K or Ran) were immunoprecipitated with anti-Flag antibody. Identical copies of input and immunoprecipitated material were subject to immunoblotting using various antibodies (left). (B) Total RNA was extracted from immunopurified SmD1 complexes and subject to RT-qPCR to measure levels of CG4068 (n = 4) or the control mRNA rp49. Samples recovered from a parallel immunoprecipitation using nonimmune serum (n. i.) serve as negative control. (C and D) Total RNA was extracted from immunopurified TAP-tagged Dcr-2 or SmD1 complexes and subject to RT-qPCR to measure levels of the esi-2.1 precursor transcript CG4068 (C) or the control mRNA rp49 (D). Fold enrichment (n = 3) compared with control samples (from cells expressing the TAP tag only) is shown on a log scale. (E) S2 cells stably expressing TAP–Dcr-2 were treated with control (Luc) or SmD1 dsRNAs. Relative enrichment of CG4068 in purified TAP–Dcr-2 complexes was measured by RT-qPCR (n = 3). Data are shown as mean + SD in E or mean + SEM in B, C, and D. **P < 0.01.
Loss of SmD1 Compromises siRISC Assembly and Function.
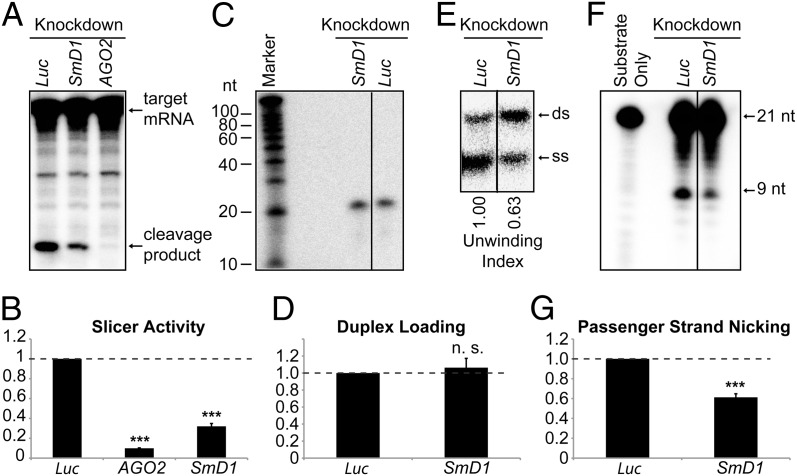
Having shown that SmD1 modulates siRNA biogenesis, we next asked whether SmD1 could influence siRNA function. To investigate this, we incubated cytoplasmic lysates from dsRNA-treated cells with a synthetic siRNA duplex. siRISC activity was assessed by monitoring the cleavage (slicing) of the cognate cap-labeled mRNA substrate. As expected, the slicer activity was abrogated in AGO2-depleted lysates. Importantly, SmD1-depleted lysates displayed significantly lower slicer activity than control lysates, suggesting a defect in siRISC assembly and/or function (Fig. 3 A and B). siRISC assembly involves multiple steps, including loading of the siRNA duplex into AGO2, nicking of the passenger strand, and duplex unwinding. To examine whether SmD1 is required for AGO2 loading, we immunopurified minimal RISC in the form of Flag–AGO2 immobilized onto agarose beads. Minimal RISC is incapable of assembling functional RISCs with siRNA duplexes unless supplemented with cell lysates containing essential RISC loading factors (17). Minimal RISC was incubated with an siRNA duplex radiolabeled on the guide strand and lysates from control or SmD1-depleted cells. Flag–AGO2-associated RNA was analyzed on a denaturing gel. Similar levels of the labeled siRNA guide strand were recovered in siRISCs formed with SmD1-depleted and control lysates, suggesting that SmD1 is not required for siRNA duplex loading into AGO2 (Fig. 3 C and D). In contrast, upon resolving AGO2-associated RNA by native PAGE to separate double-stranded from single-stranded (ss) siRNAs, we found that whereas the total amount of labeled guide strand recovered from the control and SmD1-depleted cells was similar, significantly lower levels of the ss guide strand were present in the siRISCs assembled with SmD1-depleted lysates than with control cell lysates (Fig. 3E). These results solidify the notion that SmD1 is dispensable for siRNA duplex loading but is required for duplex unwinding.
Fig. 3.
Loss of SmD1 abolishes siRISC assembly. (A) Cytoplasmic lysates from various knockdown cells (Top) were incubated with a synthetic siRNA duplex. The slicer activity of the resulting siRISC was measured as cleavage of the cap-radiolabeled target mRNA. (B) The slicer activity of the siRISC assembled in lysates from various knockdown cells (Bottom) was quantified (n ≥ 3). (C) Minimal RISC was immunopurified from Flag–AGO2 cell lysates using Flag-agarose beads. Purified minimal RISC was then incubated with lysates from control or SmD1-knockdown cells in the presence of an siRNA duplex radiolabeled on the guide strand. AGO2-bound RNAs were extracted from the beads and resolved by denaturing urea-PAGE. (D) siRNA duplex loading was quantified as the amount of radiolabeled siRNA guide strand recovered from the AGO2 siRISC assembled in lysates from various knockdown cells (n = 3). (E) siRNA duplex unwinding was measured as in C except that an excess of unlabeled guide siRNA strand was added to the reaction and AGO2-associated RNAs were resolved by native PAGE to separate ds-siRNAs and ss-siRNAs. The unwinding index was calculated and is shown at the bottom. A representative image from two independent experiments is shown. (F) Passenger-strand nicking was measured as described in C except that the siRNA duplex was radiolabeled on the passenger strand and an excess of 2′-O-methylated oligo complementary to the passenger strand was added. RNAs were extracted from the reaction mixture and resolved by denaturing urea-PAGE. (G) The passenger-strand nicking activity of lysates from various knockdown cells (Bottom) was quantified. Data are shown as mean + SEM in B, D, and G. ***P < 0.001; n.s., nonsignificant.
Because efficient duplex unwinding requires AGO2-mediated passenger-strand nicking, we examined whether SmD1 depletion affects this process. For this, minimal RISC was incubated with control or SmD1-knockdown cell lysates in the presence of an siRNA duplex radiolabeled on the passenger strand. RNA was isolated from the reaction mixture and resolved by denaturing PAGE. We found that RISCs formed with SmD1-depleted lysates produced ∼50% less cleaved passenger strand than RISCs formed with control lysates (Fig. 3 F and G). Taken together, these data demonstrate that SmD1 is required for passenger-strand nicking and duplex unwinding during siRISC assembly.
SmD1 Associates with Components of the siRISC.
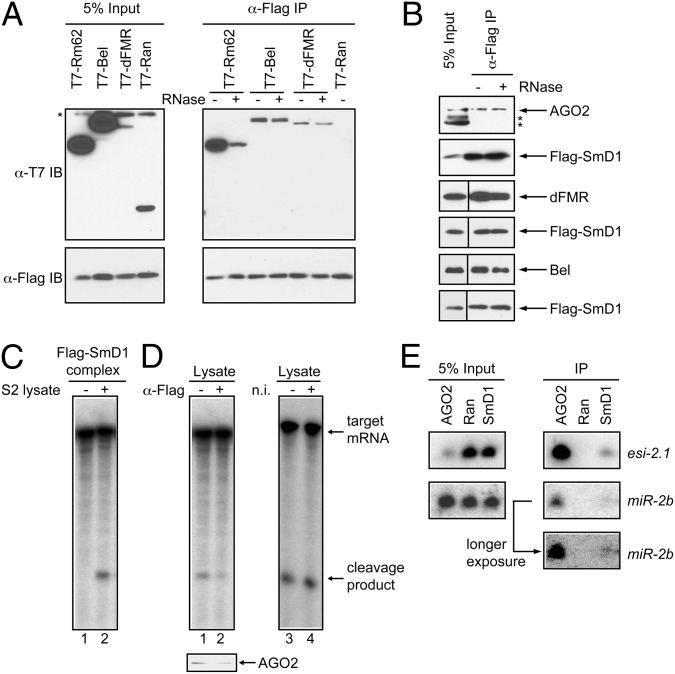
We next explored the possibility that SmD1 may associate with components of the siRISC machinery. To this end, we coexpressed in S2 cells Flag–SmD1 with T7-tagged Rm62, Bel, or dFMR, which associate with AGO2 and are required for efficient siRNA-mediated gene silencing (28, 38, 39). We found that T7-tagged Rm62, Bel, and dFMR, but not the control protein Ran, coimmunoprecipitated with Flag–SmD1 (Fig. 4A). We also detected endogenous AGO2, dFMR, and Bel in the Flag–SmD1 complex (Fig. 4B). Interestingly, the recovery of Rm62 was significantly reduced by RNase treatment, whereas that of AGO2, dFMR, and Bel was largely unaffected (Fig. 4 A and B). Notably, the interaction between SmD1 and AGO proteins seems to be evolutionarily conserved, because the mammalian orthologs also coimmunoprecipitated in an RNA-independent manner (Fig. S3).
Fig. 4.
SmD1 interacts with siRISC components. (A) Flag–SmD1 was coexpressed with various T7-tagged proteins (Top) in S2 cells. Cell lysates were immunoprecipitated with anti-Flag antibody. The isolated complexes were treated with or without RNase A then analyzed by immunoblotting with anti-Flag and anti-T7 antibodies. (B) Flag–SmD1 complexes were immunopurified from S2 cells, treated with or without RNase, and analyzed by immunoblotting with antibodies against AGO2, dFMR, Bel, or Flag. Asterisks in A and B indicate nonspecific bands. (C) Flag–SmD1 from lysates of stably transfected cells was immobilized onto anti-Flag agarose beads and then incubated with buffer or lysates from naïve S2 cells. RISC assembly was assessed by measuring slicer activity associated with the isolated beads (lanes 1 and 2). (D) Cell lysates were subject to slicer activity assays (Upper) before (lane 1) or after (lane 2) immunodepletion of Flag–SmD1 using anti-Flag agarose beads. AGO2 protein levels in aliquots of lysates were analyzed by immunoblotting (Lower). Control cell lysates were subject to a similar immunodepletion procedure using nonimmune (n. i.) serum (lanes 3 and 4). (E) Lysates from S2 cells expressing various Flag-tagged proteins (Top) were immunoprecipitated with anti-Flag antibody. Associated RNAs were extracted and analyzed by Northern blotting to detect esi-2.1 and miR-2b. Of note, expression of Flag–AGO2 reduced levels of total AGO2 (possibly owing to cosuppression) (45) and esi-2.1 in the input sample.
To establish functional relevance of the interactions between SmD1 and siRISC components, we immunopurified and incubated Flag–SmD1 complex with lysates from naïve S2 cells in the presence of a synthetic siRNA duplex. This assay revealed that the SmD1 complex could form functional siRISCs in vitro, reminiscent of the minimal RISC (Fig. 4C, lanes 1 and 2). Furthermore, upon removal of Flag–SmD1 by immunodepletion, the remaining lysates were found to be moderately depleted of AGO2 and showed a concomitant reduction in slicer activity (Fig. 4D, lanes 1 and 2). In contrast, slicer activity was unaffected in cell lysates treated with preimmune serum (Fig. 4D, lanes 3 and 4). These data thus show a functional interaction between SmD1 and AGO2. Next, we performed Northern blotting to determine whether small RNAs are present in the SmD1 complex. As expected, we detected high levels of esi-2.1 and moderate levels of miR-2b in complexes formed with Flag–AGO2, but not with the control Flag–Ran (Fig. 4E). Notably, low but clearly above-background levels of esi-2.1 and miR-2b were recovered in Flag–SmD1 complexes (Fig. 4E). It is unclear whether SmD1 interacts with esi-2.1 and miR-2b directly or through an intermediate such as AGO2. Taken together, these data demonstrate that SmD1 functionally associates with both protein and RNA components of the siRISC.
SmD1 Depletion Does Not Significantly Affect the Expression of Canonical siRNA Factors.
Because SmD1 is a core component of the spliceosome, the effects of SmD1 knockdown on RNAi could be due to aberrant expression of genes encoding canonical siRNA factors. To investigate this, we measured both protein and mRNA levels of several siRNA pathway components. With the exception of a reduction in Loqs-PD, however, we detected no significant changes in the steady-state levels of these factors (Fig. S4). To comprehensively identify changes in gene expression and mRNA splicing patterns upon loss of SmD1, we performed pair-end sequencing of RNA samples from S2 cells treated with SmD1 or control dsRNA. Overall, among the 16,882 expressed splice variants and 10,878 genes in our dataset, SmD1 silencing significantly affected the abundance of 3,187 (18.9%) splice variants from 2,442 (22.4%) genes, respectively (Dataset S1), consistent with the splicing role of SmD1. Genes involved in several functional categories were significantly enriched (Dataset S2). Assessing the relative abundance of the splice variants of several randomly selected transcripts by RT-PCR revealed a good concordance with the RNA sequencing (RNA-seq) data (Fig. S5 and Dataset S1). Of note, the RNA-seq data also confirmed that SmD1 depletion did not significantly affect the expression of most of the canonical RNAi genes, with a few exceptions (Table S2). A reduction in Loqs-PD levels could underlie the siRNA biogenesis defects upon SmD1 depletion; however, it is unlikely to contribute to the defects in siRISC assembly (Fig. 3), which does not require Loqs-PD. In addition, the mild reduction in levels of Hsp90/Hsc70 in SmD1-depleted cells is not sufficient to cause defects in siRNA duplex loading into AGO2 (Fig. 3 C and D). Furthermore, we detected a moderate increase, rather than a decrease, in levels of mRNAs encoding the C3PO components Translin and Trax. Collectively, these data show that loss of SmD1 does not significantly affect the expression of the major canonical siRNA factors and are consistent with the notion that SmD1 plays a direct role in RNAi.
Select Splicing Factors, But Not Splicing per se, Affect RNAi.
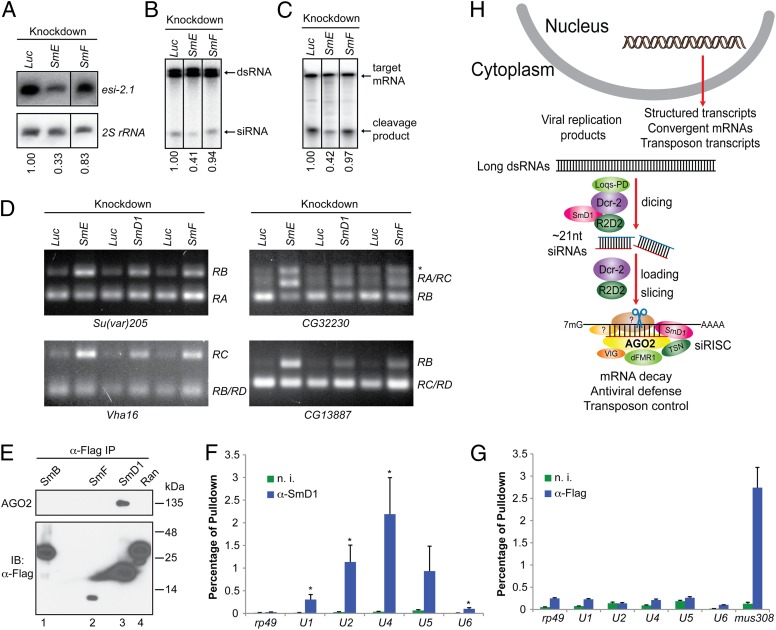
To determine whether a broader link exists between RNAi and splicing, we examined additional Sm proteins for their potential involvement in RNAi. We found that knockdown of SmE reproducibly reduced both dicing and slicing activities (Fig. 5 A–C). In contrast, we detected no defects in any aspect of RNAi in SmF-knockdown cells (Fig. 5 A–C), even though SmF-depleted cells displayed obvious alterations in the splicing pattern of the same set of mRNAs as in SmD1- or SmE-depleted cells (Fig. 5D and Fig. S5). In addition, SmF did not coimmunoprecipitate with endogenous AGO2, nor did SmB, another Sm protein (Fig. 5E). We could not test the remaining Sm proteins in this assay owing to poor expression of epitope-tagged proteins. Nonetheless, these data show that the RNAi defects elicited by SmD1 or SmE depletion can be uncoupled from pre-mRNA splicing defects and suggest that the spliceosome and siRISC are separate functional entities. To address this further, we immunopurified the siRISC and snRNP complexes and examined the presence of various snRNAs and mRNAs. As expected, snRNAs were highly enriched in the SmD1 complex (Fig. 5F). In contrast, similar to the negative control rp49 mRNA, snRNAs were largely absent in the AGO2 complex (Fig. 5G). However, the mus308 transcript, a validated target for esi-2.1 (6), was highly enriched in the AGO2 complex (Fig. 5G). Taken together, these data demonstrate that select splicing factors, but not splicing per se, influence RNAi and that the RNAi machinery and the spliceosome are physically and functionally distinct.
Fig. 5.
RNAi and splicing machineries are physically and functionally distinct. (A) S2 cells were treated with various dsRNAs. Levels of esi-2.1 and the control 2S rRNA were measured by Northern blotting. (B and C) Cytoplasmic lysates were assayed for (B) dsRNA processing by dicer assays and (C) mRNA cleavage by slicer assays. In A–C, relative levels of the indicated product are shown beneath the lanes. Representative images from two independent experiments are shown. (D) Splicing activity was assessed in various knockdown cells (Top) by examining splicing patterns of the indicated mRNAs (Bottom) by RT-PCR. The splice variants are shown on the right. The asterisk indicates an unannotated splice variant. (E) Lysates from S2 cells expressing various Flag-tagged proteins were immunoprecipitated using anti-Flag antibody. The immunoprecipitates were analyzed by immunoblotting with antibodies against AGO2 and Flag. (F and G) Total RNA was extracted from immunopurified SmD1 (F) or Flag–AGO2 complexes (G). Levels of various snRNAs or the control mRNAs mus308 and rp49 were measured by RT-qPCR. Results are the mean + SEM of n = 4; *P < 0.05. Samples recovered from a parallel immunoprecipitation using nonimmune serum (n. i.) serve as negative control. (H) A schematic depicting the proposed roles of SmD1 in the initiation (siRNA biogenesis) and effector (siRISC assembly and/or mRNA cleavage) steps of RNAi.
Discussion
In this study, we report that the core splicing factor SmD1 affects RNAi in cultured Drosophila cells and in vivo, and that SmD1 is essential for innate immunity against both acute (DCV) and persistent (FHV) viral infections. We found that SmD1 interacts with components of the siRNA biogenesis machinery and with siRNA precursor transcripts and is required for siRNA biogenesis. In addition, SmD1 associates with protein and RNA components of the siRISC and is indispensable for siRNA duplex unwinding and passenger-strand nicking during siRISC assembly. Furthermore, the role of SmD1 in RNAi seems to be independent of its pre-mRNA splicing activity. Our study thus demonstrates that SmD1 affects both initiation (siRNA biogenesis) and effector steps (siRISC assembly and function) of RNAi (Fig. 5H).
We detected Dcr-2 and R2D2 in immunopurified SmD1 complexes, indicating a direct or indirect interaction between SmD1 and components of the small RNA biogenesis machinery. CG4068, the precursor transcript for esi-2.1, was also enriched in the SmD1 complex, although it is unclear whether SmD1 binds CG4068 directly. Our results support two mutually nonexclusive models for the role of SmD1 in siRNA biogenesis. First, by interacting with both dsRNAs and Dcr-2, SmD1 could function as a molecular adaptor to facilitate the recognition of dsRNAs by Dcr-2. Our observation that SmD1 depletion compromised the interaction between Dcr-2 and CG4068 lends support to this model. Second, SmD1 could be required for the optimal catalytic activity of Dcr-2. Although a decrease in Loqs-PD could underlie the siRNA biogenesis defects elicited by SmD1 depletion, our findings that SmD1 coimmunoprecipitates with both Dcr-2 and dsRNAs are nonetheless consistent with the notion that SmD1 plays a direct role in siRNA biogenesis.
SmD1 is dispensable for AGO2 loading at the early stage of siRISC assembly but is required for the transition from pre-RISC to mature RISC, which involves AGO2-mediated nicking of the passenger strand and siRNA duplex unwinding. In addition, SmD1 associates with select siRISC components including AGO2, dFMR, Rm62, Bel, and small RNAs. Moreover, the SmD1 complex assembled into a functional siRISC in vitro (Fig. 4C). Future investigations will determine whether the association of SmD1 with these components occurs directly or via intermediates. Interestingly, binding between SmD1 and AGO2 was of sufficiently high affinity to cause moderate depletion of endogenous AGO2 and a concomitant reduction in siRISC activity (Fig. 4D). SmD1, therefore, seems to be an accessory factor required for siRISC maturation. It remains to be determined whether SmD1 affects product release in the dicing and slicing steps.
Purified recombinant SmD1 from Escherichia coli or baculovirus-infected cells did not restore the dicing and slicing activities in SmD1-knockdown lysates. It is possible that depletion of SmD1 leads to destabilization of select components of cognate complex(es). Thus, SmD1 complexes containing additional factors of appropriate stoichiometry may be necessary to achieve the rescue effect. Of note, combining SmD1-knockdown and SmD1-overexpressing cell lysates synergistically enhanced slicer activity (Fig. S6). Although the underlying cause could be that SmD1 modulates the expression of an unknown factor(s), the simplest explanation is that SmD1 plays a direct role in siRISC assembly and/or function.
SmD1 interacts with components of the RNAi machineries, which are devoid of snRNAs essential for pre-miRNA splicing. In addition, RNAi defects can be uncoupled from pre-mRNA splicing defects. Moreover, depletion of SmE, but not of SmF, phenocopies the RNAi defects elicited by the loss of SmD1. These findings imply that select splicing factors are capable of extraspliceosomal functions, analogous to the extraribosomal roles of certain ribosomal proteins (40), and strongly suggest that subcomplexes of splicing factors may facilitate optimal RNA-mediated gene silencing.
Together with other reports, our study reveals an evolutionarily conserved scheme in which splicing factors modulate RNAi in a manner independent of their pre-mRNA splicing activities (26–30). In an interesting reversal, recent studies report that small RNAs and components of the RNAi machinery modulate pre-mRNA splicing in flies and mammals (41, 42). Thus, the cross-talk between pre-mRNA splicing and small RNA-mediated gene silencing pathways may be considerably more extensive than previously appreciated.
Materials and Methods
Information regarding cell culture, dsRNA treatment, DNA constructs, antibodies, and fly strains can be found in Supporting Information.
Viral Infection.
S2 cells were subject to two rounds of dsRNA treatment. A suspension of Drosophila C virus in PBS was then added to the culture medium. Cells were harvested ∼6–8 h later and thoroughly washed with PBS. Total RNA was extracted with TRIzol (Invitrogen) and viral RNA was detected by RT-qPCR.
RNAi Assays.
Dicing, slicing, siRISC loading, duplex unwinding, and passenger-strand nicking assays were performed using cytoplasmic lysate from knockdown S2 cells, as previously described, with minor modifications (17, 43, 44). Supporting Information gives additional experimental procedures and information.
Supplementary Material
Acknowledgments
We thank Drs. Greg Hannon, Qinghua Liu, Mikiko Siomi, Haruhiko Siomi, Paul Lasko, Phil Zamore, and Richard Carthew for reagents and protocols, Gene Yeo for deep sequencing, and Rich Binari and Benjamin Czech for comments and discussions. This work was supported by start-up funds from the Sanford–Burnham Medical Research Institute (to R.Z.) and in part by National Institutes of Health grants (to N.P., I.J.M, A.S. and T.M.R.). N.P. is a Howard Hughes Medical Institute Investigator.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1315803110/-/DCSupplemental.
References
- 1.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101(1):25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 3.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 4.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 5.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286(5441):950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 6.Czech B, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453(7196):798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghildiyal M, et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320(5879):1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawamura Y, et al. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453(7196):793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- 9.Okamura K, et al. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453(7196):803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tuschl T, Zamore PD, Lehmann R, Bartel DP, Sharp PA. Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev. 1999;13(24):3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 12.Liu Q, et al. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301(5641):1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 13.Hartig JV, Esslinger S, Böttcher R, Saito K, Förstemann K. Endo-siRNAs depend on a new isoform of loquacious and target artificially introduced, high-copy sequences. EMBO J. 2009;28(19):2932–2944. doi: 10.1038/emboj.2009.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou R, et al. Processing of Drosophila endo-siRNAs depends on a specific Loquacious isoform. RNA. 2009;15(10):1886–1895. doi: 10.1261/rna.1611309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306(5700):1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 16.Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell. 2004;117(1):83–94. doi: 10.1016/s0092-8674(04)00258-2. [DOI] [PubMed] [Google Scholar]
- 17.Iwasaki S, et al. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol Cell. 2010;39(2):292–299. doi: 10.1016/j.molcel.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 18.Miyoshi T, Takeuchi A, Siomi H, Siomi MC. A direct role for Hsp90 in pre-RISC formation in Drosophila. Nat Struct Mol Biol. 2010;17(8):1024–1026. doi: 10.1038/nsmb.1875. [DOI] [PubMed] [Google Scholar]
- 19.Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19(23):2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123(4):607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 21.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123(4):621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 22.Leuschner PJ, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7(3):314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, et al. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science. 2009;325(5941):750–753. doi: 10.1126/science.1176325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwarz DS, Tomari Y, Zamore PD. The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Curr Biol. 2004;14(9):787–791. doi: 10.1016/j.cub.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305(5689):1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 26.Bayne EH, et al. Splicing factors facilitate RNAi-directed silencing in fission yeast. Science. 2008;322(5901):602–606. doi: 10.1126/science.1164029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tabach Y, et al. Identification of small RNA pathway genes using patterns of phylogenetic conservation and divergence. Nature. 2013;493(7434):694–698. doi: 10.1038/nature11779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou R, et al. Comparative analysis of argonaute-dependent small RNA pathways in Drosophila. Mol Cell. 2008;32(4):592–599. doi: 10.1016/j.molcel.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim JK, et al. Functional genomic analysis of RNA interference in C. elegans. Science. 2005;308(5725):1164–1167. doi: 10.1126/science.1109267. [DOI] [PubMed] [Google Scholar]
- 30.Parry DH, Xu J, Ruvkun G. A whole-genome RNAi Screen for C. elegans miRNA pathway genes. Curr Biol. 2007;17(23):2013–2022. doi: 10.1016/j.cub.2007.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duchaine TF, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124(2):343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 32.Mourelatos Z, et al. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16(6):720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kambach C, et al. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 1999;96(3):375–387. doi: 10.1016/s0092-8674(00)80550-4. [DOI] [PubMed] [Google Scholar]
- 34.Mount SM, Salz HK. Pre-messenger RNA processing factors in the Drosophila genome. J Cell Biol. 2000;150(2):F37–F44. doi: 10.1083/jcb.150.2.f37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang XH, et al. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312(5772):452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol. 2006;7(6):590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- 37.van Rij RP, et al. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20(21):2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16(19):2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ishizuka A, Siomi MC, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16(19):2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34(1):3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taliaferro JM, et al. Two new and distinct roles for Drosophila Argonaute-2 in the nucleus: Alternative pre-mRNA splicing and transcriptional repression. Genes Dev. 2013;27(4):378–389. doi: 10.1101/gad.210708.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ameyar-Zazoua M, et al. Argonaute proteins couple chromatin silencing to alternative splicing. Nat Struct Mol Biol. 2012;19(10):998–1004. doi: 10.1038/nsmb.2373. [DOI] [PubMed] [Google Scholar]
- 43.Nayak A, Andino R. Slicer activity in Drosophila melanogaster S2 extract. Methods Mol Biol. 2011;721:231–244. doi: 10.1007/978-1-61779-037-9_14. [DOI] [PubMed] [Google Scholar]
- 44.Yang B, Li H. Dicer assay in Drosophila S2 cell extract. Methods Mol Biol. 2011;721:215–229. doi: 10.1007/978-1-61779-037-9_13. [DOI] [PubMed] [Google Scholar]
- 45.Czech B, et al. Hierarchical rules for Argonaute loading in Drosophila. Mol Cell. 2009;36(3):445–456. doi: 10.1016/j.molcel.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.