Abstract
Bromo-adjacent homology (BAH) domains are commonly found in chromatin-associated proteins and fall into two classes; Remodels the Structure of Chromatin (RSC)-like or Sir3-like. Although Sir3-like BAH domains bind nucleosomes, the binding partners of RSC-like BAH domains are currently unknown. The Rsc2 subunit of the RSC chromatin remodeling complex contains an RSC-like BAH domain and, like the Sir3-like BAH domains, we find Rsc2 BAH also interacts with nucleosomes. However, unlike Sir3-like BAH domains, we find that Rsc2 BAH can bind to recombinant purified H3 in vitro, suggesting that the mechanism of nucleosome binding is not conserved. To gain insight into the Rsc2 BAH domain, we determined its crystal structure at 2.4 Å resolution. We find that it differs substantially from Sir3-like BAH domains and lacks the motifs in these domains known to be critical for making contacts with histones. We then go on to identify a novel motif in Rsc2 BAH that is critical for efficient H3 binding in vitro and show that mutation of this motif results in defective Rsc2 function in vivo. Moreover, we find this interaction is conserved across Rsc2-related proteins. These data uncover a binding target of the Rsc2 family of BAH domains and identify a novel motif that mediates this interaction.
INTRODUCTION
Chromatin remodeling complexes alter the interactions between DNA and histone proteins to regulate aspects of DNA metabolism, such as transcription, replication and repair. In budding yeast, Remodels the Structure of Chromatin (RSC) is one such complex, which is highly abundant and comprises 17 subunits. (1). It is known to be important for transcriptional regulation and can act as both an activator and a repressor. In addition, it is involved in multiple DNA repair pathways (2–4), in establishment of cohesion (5) and in kinetochore regulation (6).
The homologous complex in higher eukaryotes is called PBAF (or SWI/SNF-B) (7) and has been implicated in DNA damage responses in addition to its known roles in transcriptional regulation (8–11), therefore suggesting a conserved functional role.
In yeast, RSC exists as two isoforms, containing either the Rsc1 or Rsc2 subunit (12). These proteins share a similar domain architecture, containing two bromodomains followed by a bromo-adjacent homology (BAH) domain. Interestingly, in PBAF, there is a single polypeptide termed BAF180 (or Polybromo) that appears to be a fusion of yeast Rsc1, Rsc2 and Rsc4 and contains six sequential bromodomains followed by two BAH domains (7,13). The function of the BAH domains in these proteins is currently not known.
BAH domains are found in a number of chromatin-associated proteins or protein complexes (13,14) and can be subdivided into two classes by amino acid sequence analysis (14,15). The first, which we refer to as ‘RSC-like’, includes BAH domains from Rsc1 and Rsc2 and their higher eukaryotic homologue BAF180. This family includes a diverse range of chromatin-associated proteins, such as Ash1 homologues and DNA methyltransferases. The second, ‘Sir3-like’ class is more restricted and only includes Orc1 homologues and the budding yeast protein Sir3, which arose from a gene duplication of Orc1 in this species (16).
Transcriptional silencing in budding yeast exists at three locations—telomeres, mating type switching donor cassettes (HML/R) and ribosomal DNA (rDNA) repeats (17). Both Orc1 and Sir3 are important for mediating transcriptional silencing at telomeres and HML/R, and the BAH domain of each protein is critical for this function (18).
Yeast Orc1 BAH interacts directly with the silent information regulator Sir1 (19), whereas in higher eukaryotes, Orc1 interacts with the heterochromatin-associated protein HP1 (20). In addition to these interactions, Orc1 also binds directly to nucleosomes (21,22), and in higher eukaryotes, this is mediated by an interaction with the tail of histone H4 dimethylated at lysine 20 (23). The Sir3 BAH domain is also able to bind to nucleosomes (24,25) via the Loss of Ribosomal Silencing (LRS) region of the nucleosome, which includes sequences from both H3 and H4 (26).
To date, no binding partners or molecular functions have been identified for the RSC-like class of BAH domains. Notably, neither the Sir1-interacting region of Orc1 (H-domain) nor the nucleosome-interacting regions of Orc1 or Sir3 are conserved at the amino acid level in the RSC family of BAH domains, making predictions about function or binding partners particularly difficult.
Here, we identify a role for Rsc2 in rDNA silencing, prompting us to ask whether its BAH domain plays a comparable role to those of Sir3 and Orc1. We found that the Rsc2 BAH domain is able to interact with chromatin both in vivo and in vitro. However, Rsc2 BAH specifically interacts with histone H3, suggesting that the mechanism of binding to chromatin is distinct from both Sir3 and Orc1. In support of this, the crystal structure of Rsc2 BAH, determined at 2.4 Å, reveals major differences to these other BAH domains, especially in the regions of their structure that mediate nucleosome interactions. Using targeted mutagenesis based on our structural information, we identified residues in Rsc2 that are important for H3 binding in vitro and for RSC function in vivo. Interestingly, amino acid sequence conservation within this region of the BAH domain suggests that a subset of RSC-like BAH domains are also likely to share this H3-binding mechanism. In support of this, we find that the BAH domains from Rsc1 and BAF180 are also able to bind H3 but not the other core histones in vitro. These data identify the first binding partner of the RSC-like family of BAH domains and provide insights into the function of Rsc2 in the RSC remodeling complex.
MATERIALS AND METHODS
Strains and plasmids
Information on yeast strains and plasmids is available in the Supplementary Information.
Silencing assays
Serial dilutions of logarithmically growing cultures were plated onto synthetic complete (SC) or media lacking leucine (−LEU) for plasmid selection and the same media lacking uracil (SC/−URA or −LEU/−URA) as a measure of silencing activity. Colonies were imaged or counted following 3–5 days incubation at 30°C. Survival was calculated from colonies on media lacking URA relative to those on URA-containing media. The data shown are the mean of at least three independent experiments ±1 SD.
Chromatin immunoprecipitation assays
Full-length Rsc2-myc was immunoprecipitated from YNK179-191 (rsc2). BAH-CT1 was immunoprecipitated from rsc2-BY4741 containing the indicated BAH-CT1-myc overexpression plasmid. Chromatin immunoprecipitation (ChIP) assays were performed as (27). Following qPCR quantification, data were normalized to input, and fold-enrichment was calculated relative to the untagged control. Data shown are the mean enrichment of at least three independent experiments ±1 SD. Data calculated as percentage of input are provided in Supplementary Table S1.
Expression and purification of recombinant proteins
Expression and purification of recombinant His or GST-tagged BAH-CT1, BAHRsc1 and BAH1BAF180 proteins were carried out by standard chromatographic methods using Talon (TaKaRa Bio) or Glutathione Sepharose 4 Fast Flow (GE Healthcare) affinity resins and a HiLoad Superdex 75 or 200 size exclusion column (GE Healthcare). For full details, see Supplementary Information.
Nucleosome pull-down assays
Native mononucleosomes were prepared from BY4741 essentially as (28). Either glutathione sepharose or Ni-agarose beads were equilibrated in wash buffer [15 mM HEPES (pH 7.6), 110 mM NaCl, 1 mM DTT] and incubated with 2 µg of GST-BAH-CT1 or His-BAH-CT1. In all, 180 µl of wash buffer and 20 µl of nucleosomes or nucleosome storage buffer [15 mM HEPES (pH 7.5), 400 mM NaCl, 1 mM DTT, 0.15 mM spermine, 0.5 mM spermidine, 10% glycerol] were added. Reactions were incubated overnight at 4°C and washed three times. Bound protein was eluted by boiling and analyzed by western blotting using an anti-H2B (29) or anti-H4 antibodies (Abcam).
Mononucleosome gel shift assays
Mononucleosomes were assembled in vitro onto a radiolabeled DNA fragment and gel purified as (30) using histones purified from HEK293 cells. Purified recombinant His-BAH-CT1 protein (0.24, 0.48, 0.72, 0.96 or 1.2 µg) was incubated for 1 h at room temperature with 3 µl of mononucleosomes [final 10 µl of reaction contained 9.5 mM Tris–HCl (pH 7.5), 0.9 mM HEPES (pH 7.5), 47.7 mM KCl, 11.5 mM NaCl, 0.05 mM EDTA, 0.02 mM TCEP, 9.5% glycerol, 0.4 µg/ml BSA]. Samples were run on 4.5%/0.4x TBE acrylamide gels at 120 V at 4°C.
DNA gel shift assays
Reactions contained 12.5 fmol of 25 or 49 bp annealed oligonucleotides or a 167 bp PCR fragment. DNA was end-labeled using 32P-γATP and PNK then passed over a G-50 spin column (GE Healthcare). Binding reactions contained 15 mM HEPES (pH 7.6), 5% glycerol, 2 mM MgCl2, 150 mM NaCl and 18 µg His-BAH-CT1 or 800 ng MBP-Ies6 (positive control) in a 10 µl of reaction volume. Reactions were incubated for 1 h at RT and resolved on 6% acrylamide/0.5× TBE gel.
GST pull-down assays
GST beads or beads bound to the indicated GST–BAH protein construct were equilibrated with binding buffer [15 mM HEPES (pH 7.5), 400 mM NaCl, 110 mM KCl, 5% glycerol, 0.5% NP40) or [50 mM Tris (pH 7.5), 1 M NaCl, 1% NP40]. Reactions using 100 µg of calf thymus histones (Sigma) were incubated at 4°C for 1 h. Beads were washed with 3× 1 ml of buffer. Bound protein was eluted by boiling and analyzed by western blotting with anti-H2A (31), anti-H2B (29), anti-H3 (Abcam) or anti-H4 (Abcam).
For recombinant histone pull-downs, GST beads or beads bound to the indicated GST-BAH protein construct were equilibrated with the binding buffer above supplemented with 1 mg/ml BSA and 2 M urea. Binding reactions using 10 µg of recombinant human H3 (NEB) were incubated at 4°C for 1 h, before washing, elution and analysis as aforementioned.
Co-immunoprecipitations
Extracts with sheared chromatin were prepared from wt or H3 A75V mutant H3-containing strains, each harboring either a myc-tagged Rsc2 expression plasmid or an empty vector control in IP buffer [50 mM HEPES (pH 7.5), 10 mM MgOAc, 5 mM EGTA, 0.1 mM EDTA, 150 mM KCl, 0.2% NP40, 5 mM β-mercaptoethanol, 200 µg/ml PMSF, 2 mM leupeptin, 2 mg/ml pepstatin A and 1 mg/ml aprotinin]. Lysates were incubated for 40 min at 4°C with anti-myc (9E10; Sigma). Protein G Dynabeads (Invitrogen) were added, and samples were incubated for a further 40 min at 4°C before 3 × 1 ml of washes with IP buffer. Bound protein was eluted from beads by boiling and analysed by western blotting using anti-myc (Sigma) or anti-H2A (31).
Crystallization data
Crystallization trials, data collection, phasing and refinement were performed as described in Supplementary Information. Statistics for the data collection, details of the model and Ramachandran and Molprobity statistics are given in Table 1.
Table 1.
Data collection and refinement statistics
| Data collectiona | |
| Space group | P 21 |
| Cell dimensions | |
| a, b, c (Å) | 64.09, 64.07, 136.84 |
| α, β, γ (°) | 90.00, 95.47, 90.00 |
| Resolution (Å) | 45.00–2.40 (2.53–2.40)b |
| Rmerge | 0.142 (0.424) |
| Mean I/σ I | 5.8 (2.7) |
| Completeness (%) | 98.7 (99.0) |
| Redundancy | 3.5 (3.6) |
| Wilson B (Å2) | 25.5 |
| Refinement | |
| Resolution (Å) | 44.50–2.40 |
| No. reflections | 42819 |
| Rwork/Rfree | 0.22/0.29 |
| No. atoms | |
| Protein | |
| Chain A | 1893 |
| Chain B | 1889 |
| Chain C | 1893 |
| Chain D | 1913 |
| Ligand/ion | 8 (Cl−) |
| 6 (glycerol) | |
40 ( ) ) |
|
| Water | 541 |
| B-factors (average) | |
| Protein | |
| Chain A | 34.13 |
| Chain B | 32.85 |
| Chain C | 17.34 |
| Chain D | 16.63 |
| Ligand/ion | 36.24 (Cl−) |
| 27.88 (glycerol) | |
39.11 (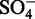 ) ) |
|
| Water | 26.35 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 0.772 |
| Model Qualityc | |
| MolProbity score | 1.89 (96th percentile) |
| All atom clashscore | 13.40 (87th percentile) |
| Rotamer outliers | 10/815 |
| Ramachandran outliers | 0/927 |
| Ramachandran favoured | 898/927 (96.9%) |
aData were collected from a single crystal.
bValues in parentheses are for highest-resolution shell.
cDetermined by MolProbity v3.15.
Survival assays
The 5-fold serial dilutions of logarithmically growing cultures were spotted onto Yeast extract, Peptone, Adenine, Dextrose (YPAD) plates containing the indicated amounts of hydroxyurea (HU) or dimethyl sulfoxide (DMSO). Plates were incubated for 2–3 days at 30°C.
RESULTS
Rsc2 is important for rDNA silencing
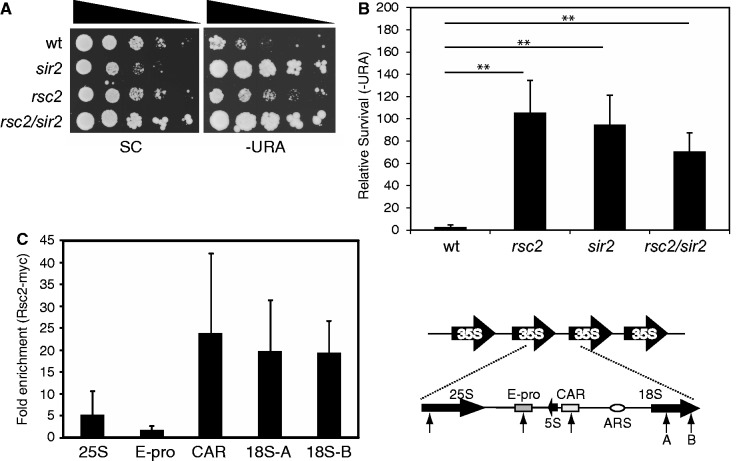
To investigate the potential contribution of the Rsc2 protein to transcriptional silencing at the rDNA repeats, we created an rsc2 deletion in a strain carrying a URA3 reporter gene in this region (32). In a wild-type strain, survival on media lacking URA is low due to transcriptional silencing of URA3. In contrast, the rsc2 mutant strain had much higher survival, indicating a defect in silencing (Figure 1A). The rsc2 strain has a slow growth phenotype, resulting in generally smaller colony sizes of the rsc2 strain compared with wild-type or the sir2 strain, which makes comparisons between the strains in this assay difficult. Therefore, we also quantified the relative survival on media lacking URA to give a more robust indication of the survival defect (Figure 1B).
Figure 1.
Rsc2 is important for mediating silencing of rDNA. (A) Silencing was monitored in strains containing a URA3 reporter gene in the rDNA (32) by assaying survival on media lacking URA relative to survival on SC medium. (B) Survival of silencing reporter strains as in (A) on media lacking URA was quantitated, and the data are represented as the mean ± 1 SD of at least three independent experiments. Statistical analysis was performed using an unpaired t-test. Asterisks indicates P ≤ 0.01. (C) Chromatin IP analysis of Rsc2-myc at various locations across the rDNA repeat. Data shown are the mean enrichment of at least three independent experiments ± 1 SD. The right hand panel shows a schematic of the 9.1 kb rDNA repeats with an expanded view of a single repeat. The 25S, 5S and 18S coding sequences are indicated with solid arrows. The relative positions of the E-pro promoter region, cohesin-associated region and autonomously replicating sequence (ARS) are indicated. Locations of primer pairs are indicated with arrows.
Sir2 is a histone deacetylase that functions in transcriptional silencing at all three heterochromatic loci (HML/R, rDNA and telomeres). Although Sir2 is a common factor, it functions in concert with distinct protein complexes at the rDNA and at telomeres (17). At the rDNA, Sir2 is a part of the RENT complex that comprises Sir2, Net1 and Cdc14, whereas at telomeres and HML/R, Sir2 works in concert with Sir3 and Sir4 (17). The silencing defect in the rsc2 mutant strain was similar to that seen in a sir2 mutant (Figure 1A and B), prompting us to examine the phenotype of an rsc2/sir2 double mutant. We found the defect in the double mutant strain is neither worse nor significantly different to that of the sir2 single mutant (Figure 1B), suggesting that rsc2 may function in the same pathway as sir2 in mediating rDNA silencing.
The effect of RSC on rDNA silencing could be due to direct action at the rDNA or indirect effects of transcriptional misregulation in the absence of Rsc2. We therefore investigated whether Rsc2 was associated with chromatin in the rDNA repeats. To do this, we created a C-terminally 13 Myc-tagged full-length Rsc2 construct under the control of its own promoter (Figure 2A). We determined that the presence of the Myc tag has no detectable effect on Rsc2 activity in vivo (data not shown) and used this to perform ChIP assays. Using primer pairs at various sites within the rDNA repeats, we found that Rsc2 is associated with multiple locations across the rDNA repeat (Figure 1C; right panel), with substantial enrichment at the cohesion-associated region and the 18S coding region (Figure 1C; left panel).
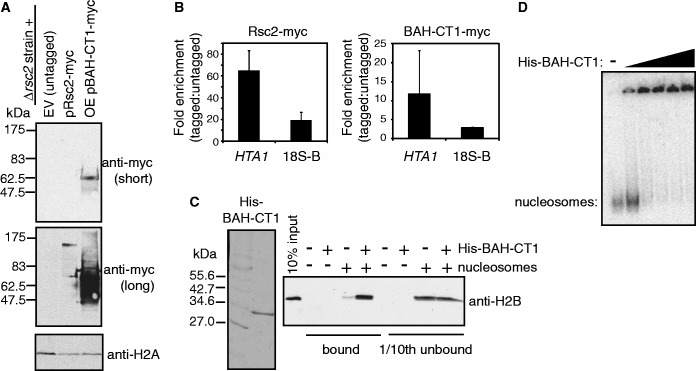
Figure 2.
The Rsc2 BAH-CT1 domain directly interacts with chromatin. (A) Western blot analysis of rsc2 null strains carrying an empty vector (lane 1; EV), a plasmid with myc-tagged full-length RSC2 under the control of its own promoter (lane 2; pRsc2-myc) and an overexpression construct of myc-tagged BAH-CT1 under the control of the GAPDH promoter (lane 3; OE pBAH-CT1-myc). Westerns were analyzed with anti-myc (top two panels) or anti-H2A (bottom panel) as a loading control. (B) Full-length Rsc2 and Rsc2 BAH-CT1 are associated with chromatin in vivo. ChIP assays examining enrichment of full-length Myc-tagged Rsc2 (top panel) or Myc-tagged overexpressed BAH-CT1 (bottom panel) relative to the untagged control at the HTA1 promoter or the 18S region of the rDNA (18S-B on Figure 1C). Data shown are the mean enrichment of at least three independent experiments ± 1 SD. (C) Coomassie stained gel of recombinant His-tagged BAH-CT1 (left panel). Western blot analysis of pull-down assays with nickel agarose using anti-H2B (right panel). Lane 1 shows 10% input of yeast mononucleosomes used in pull-down assays. The total bound fraction and 10% of the unbound fraction of each pull-down reaction were analysed for the presence of H2B by western blotting. (D) Gel shift analysis using mononucleosomes assembled onto radiolabeled DNA (lane 1) with increasing concentrations of recombinant His-tagged BAH-CT1.
The LRS region of the nucleosome, which is contacted by the Sir3 BAH domain, is important for transcriptional silencing at all three heterochromatic loci. However, as described above Sir3 functions only at the telomeres and HML/R loci. This raises the possibility that a different BAH domain might contact the LRS region of the nucleosome at the rDNA. Given our finding that Rsc2 is important for rDNA silencing and is physically associated with rDNA repeats, we set out to investigate its BAH domain in more detail.
The BAH domain of Rsc2 directly interacts with chromatin
We began by investigating whether the isolated Rsc2 BAH domain, like full-length Rsc2, was capable of binding to chromatin in vivo. The RSC complex contains several other protein subunits with direct chromatin binding activity [e.g. (33,34)]. Additionally, Rsc2 itself has two bromodomains and an AT hook upstream of the BAH domain that may also contribute to chromatin binding in vivo. Therefore, to investigate whether the BAH domain is able to independently interact with chromatin in vivo, we created a Myc-tagged Rsc2 yeast expression construct lacking the potential chromatin binding bromodomains and AT hook motif (Figures 2A and 4A). This construct also lacked the C-terminal region (CT2) required for assembly of Rsc2 into the RSC complex (12) and corresponded to the BAH domain and the highly conserved C-terminal region 1 (BAH-CT1) used in the structural analyses described later in the text (Figure 4A).
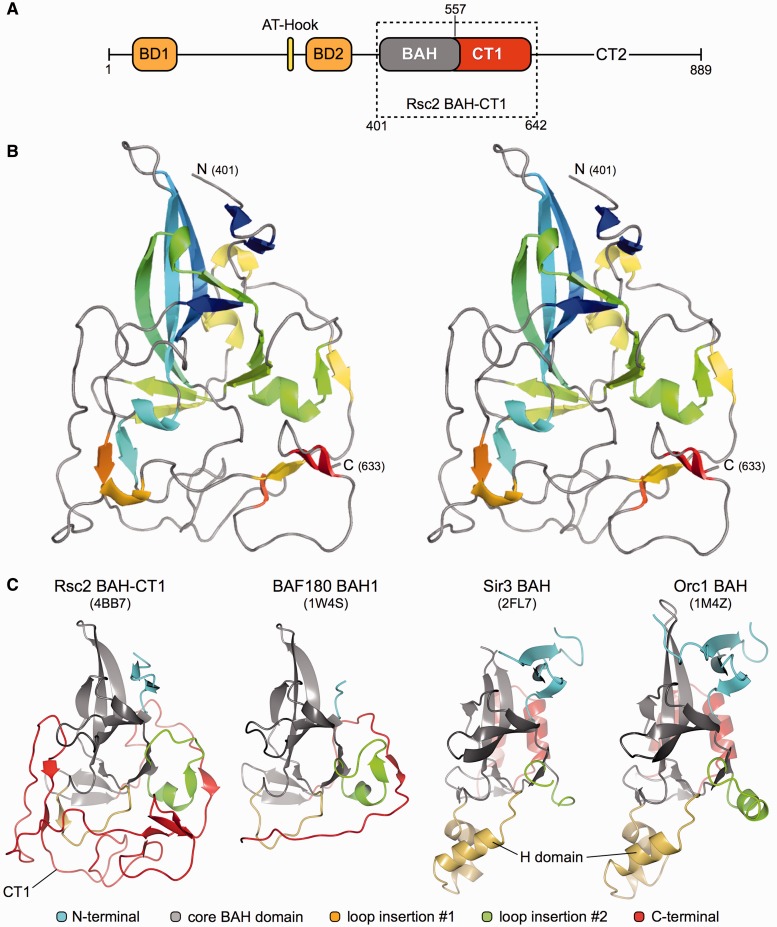
Figure 4.
Structure of the Rsc2 BAH-CT1. (A) Schematic representation of the functional domains of Saccharomyces cerevisiae Rsc2 where BD, BAH and CT indicate bromodomain, BAH domain and C-terminal conserved regions, respectively. The boxed region indicates the amino acid boundaries of the RSC2-BAH-CT1 expression construct used in this study. (B) Stereo-pair secondary structure cartoon of RSC2-BAH-CT1, colored blue to red from the visible N-terminus at residue 401 to the C-terminus at residue 633. (C) Side-by-side comparison of the BAH domain structures of Rsc2 BAH-CT1 (this study), BAF180, Sir3 and Orc1. The core canonical BAH domain fold is colored gray in each case. N- and C-terminal additions/extensions to the fold are colored blue and red, respectively, with loop insertions at two points colored in green and yellow. PDB accession codes are shown in parentheses.
In addition to testing occupancy in the rDNA, we used the HTA1 promoter as a positive control, as Rsc2 was previously found to be associated with this region (35). We found that both full-length Rsc2 and the Rsc2 BAH-CT1 construct were enriched at both genomic loci (Figure 2B), showing that the BAH-CT1 protein is independently capable of binding to chromatin in vivo. The lower level of BAH-CT1 enrichment in chromatin, despite much higher expression levels compared with full-length Rsc2, is consistent with the idea that other domains and components of RSC contribute to the overall affinity and/or specificity of the intact RSC complex. We also find that overexpression of BAH-CT1 has no detectable influence on the phenotype of either wt or rsc2 mutant yeast expressing this construct (data not shown), indicating that it does not interfere with RSC function in vivo.
To investigate whether this was a direct interaction, we isolated native mononucleosomes from yeast and tested these in a pull-down assay using purified recombinant His-tagged BAH-CT1 protein (Figure 2C, left panel) and nickel-agarose beads. Although there was a low level of non-specific histone binding to the beads alone, the signal was substantially enriched when BAH-CT1 was present (Figure 2C, right panel), which strongly suggests that BAH-CT1 is able to directly interact with nucleosomes. To further substantiate this, we also investigated BAH-CT1 nucleosome binding in vitro with mononucleosomes in gel shift assays. These mononucleosomes were prepared using native human histones assembled onto a radiolabelled, 247 bp fragment of DNA (30). In this assay, we also detected an interaction (Figure 2D), supporting the conclusion that the Rsc2 BAH domain binds directly to nucleosomes.
The BAH domain of Rsc2 interacts with histone H3
To determine which component(s) of the nucleosome were responsible for the interaction detected with the BAH-CT1 protein, we individually tested DNA and histones. First, we used the His-BAH-CT1 protein in gel shift assays with multiple different DNA substrates, including the 247 bp substrate used to reconstitute mononucleosomes used in the gel shift assays aforementioned. We could not detect any interaction with free DNA under a variety of binding conditions (Figure 3A and data not shown).
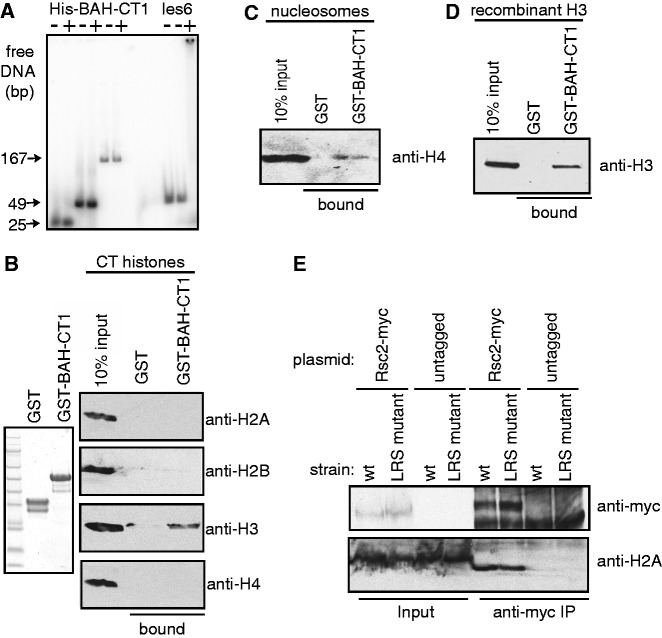
Figure 3.
The BAH-CT1 domain of Rsc2 interacts with histone H3. (A) Gel shift analysis using radiolabeled DNA with recombinant His-tagged BAH-CT1 or recombinant MBP-tagged Ies6 as a positive control. (B) Coomassie-stained gel of recombinant GST or GST-BAH-CT1 (left panel) used in pull-down assays with native calf thymus histones. Bound proteins were analyzed by western blotting with antibodies against each of the four core histones (right panel). (C) Pull-down assay using GST or GST-BAH-CT1 and native yeast mononucleosomes analyzed with anti-H4 antibody. (D) Pull-down assay using GST or GST-BAH-CT1 and recombinant purified histone H3, analyzed with antibodies against histone H3. (E) Co-immunoprecipitation of chromatin with wt Rsc2 is unaffected by LRS mutation A75V of histone H3. Co-IP was performed from the indicated strains containing either myc-tagged Rsc2 expression plasmid (Rsc2-myc) or empty vector with anti-myc antibody. Input and bound proteins were analyzed with anti-myc (top panel) or anti-H2A (bottom panel).
We next tested interactions between BAH-CT1 and histones. To do this, we created a GST-BAH-CT1 construct to circumvent non-specific binding problems with free histones (Figure 3B, left panel). Using a mixture containing all four core histones isolated from calf thymus, we found that histone H3, but no other core histones, interacted specifically with BAH-CT1 in pull-down assays (Figure 3B, right panel). Under these conditions, H3 and H4 can potentially interact, raising the possibility that when BAH-CT1 binds H3, it displaces H4. To investigate this, we repeated the pull-down assay using native mononucleosomes purified from yeast and probed for the presence of H4. We found that H4 was pulled-down by BAH-CT1 when in the context of the nucleosome (Figure 3C), indicating that BAH-CT1 binding to H3 does not disrupt H3–H4 interaction. Taken together with the nucleosome binding assays in Figure 2, these data suggest that the nucleosome structure is not grossly perturbed by BAH-CT1 binding.
We then performed pull-down assays using purified recombinant histone H3 and also detected an interaction with the BAH-CT1 protein (Figure 3D), demonstrating that the interaction is direct. Because the H3 used in these experiments was recombinant protein purified from Escherichia coli, this result also demonstrates that covalent modifications of histone H3 are not required for the interaction with BAH-CT1.
The LRS region of the nucleosome, which is bound by Sir3 at telomeres and HML/R, is made up of H3 and H4 surfaces, and the BAH domain of Sir3 makes extensive contacts with all four core histones (26). Therefore, our data suggest that the mechanism of chromatin binding by Rsc2 BAH is distinct from that of the Sir3 BAH domain. To further investigate this, we tested whether a mutation in the LRS region of the nucleosome, which impairs the ability of the Sir3 protein to bind (24), would also impair the ability of Rsc2 to bind. We immunoprecipitated Rsc2 from extracts with sheared chromatin prepared from wt or LRS mutant (H3 A75V) yeast and found no difference in associated histones (Figure 3E). This result suggests that LRS mutations that have previously been shown to disrupt Sir3 binding do not disrupt Rsc2 binding, consistent with the idea that the Rsc2 BAH domain is binding to chromatin in a mechanistically distinct manner from the Sir3 BAH domain.
Structure of the Rsc2 BAH domain
To gain insights into the mechanism of chromatin binding, we set out to investigate the structure of the Rsc2 BAH domain. Through a combination of a priori knowledge, using our previously determined crystal structure of the first (proximal) BAH domain from BAF180 (PDB: 1W4S), multiple sequence alignment and the Phyre protein-fold recognition server (http://www.sbg.bio.ic.ac.uk/∼phyre), we were able to predict the likely domain boundaries of the Rsc2 BAH domain. We transformed E. coli with different constructs exploring these limits, purified the expressed recombinant proteins by standard chromatographic procedures (see ‘Materials and Methods’ section) and then put them into crystallization trials. Diffracting crystals were only obtained for a construct encoding amino acids 401-642 (BAH-CT1), encompassing the BAH domain and the highly conserved C-terminal 1 (CT1) region of Rsc2 (Figure 4A).
The structure was solved by molecular replacement (using the BAF180-BAH1 domain as a search model) and refined at a resolution of 2.4 Å (PDB: 4BB7; see Table 1). The protein crystallized in spacegroup P21, with four molecules comprising the asymmetric unit (see ‘Materials and Methods’ section). Readily interpretable electron density spanned amino acids Asp401 to Thr633 (Figure 4B) for each molecule.
The N-terminal part of BAH-CT1 (Asp 401 to Lys 524) essentially conforms to the definition of a canonical BAH domain [described in (14,15)], which comprises a distorted β-barrel core with additional β-strand and 310 helical elements. In contrast, the CT1 region (Pro 557 to Ile 632, colored red in Figure 4C) follows an extended ‘meander’, containing only a few secondary structure elements, which wraps around and packs back against the core BAH domain fold (Figure 4C and Supplementary Figure S1). Interestingly, amino acids Lys 524 through Asn 617 also form an unusual helical super-structure, containing several β-turn motifs.
BAH domains and their interactions with histones
Comparison of the BAH domains from Rsc2, BAF180, Sir3 and Orc1 (Figure 4C) clearly highlights the structural diversity possible in this type of domain, and therefore the necessary sub-categorization into either RSC-like or Sir3-like classes. In each case, the fold of the core is strongly conserved between each protein, but then extensively elaborated, via both N and C-terminal additions and large loop insertions at two positions within the fold (Figure 4C). For Sir3 and Orc1, this includes the inserted H-domain that is required for interaction with their respective protein partners, and the C-terminal helix-turn-helix motif required for specific nucleosome contacts—with both of these structural motifs notably absent from both Rsc2 BAH-CT1 and BAF180 BAH1.
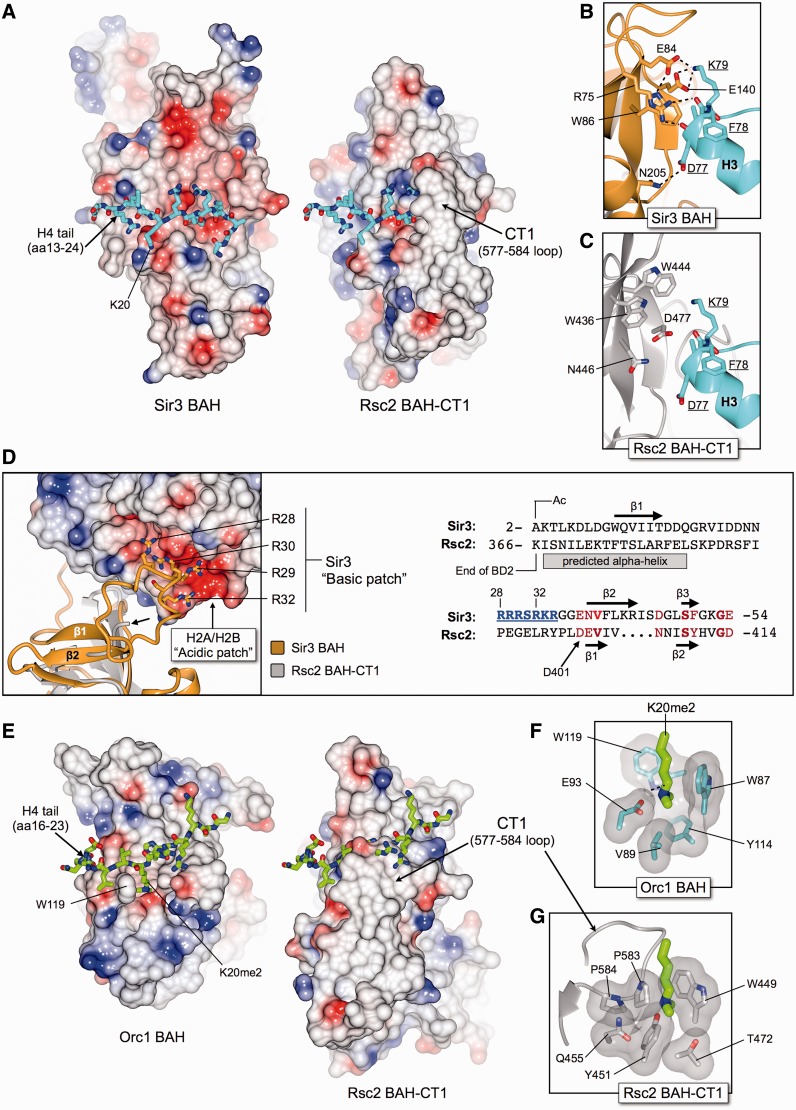
The recent X-ray crystal structures of the yeast Sir3 BAH domain in complex with a nucleosome core particle [PDB: 3TU4, (26)] and of the mouse Orc1 BAH domain in complex with an H4K20me2 peptide [PDB: 4DOW, (23)] allows a detailed examination of their respective histone interfaces and a direct comparison with the structurally equivalent regions of Rsc2 BAH-CT1 (Figure 5).
Figure 5.
The Rsc2 BAH-CT1 domain differs from both Sir3 and Orc1 BAH domains in regions critical for making histone-specific contacts, indicating that the mechanism of H3 binding in Rsc2 is distinct from these proteins. (A) Molecular surface representations of Sir3 BAH (left) and Rsc2 BAH-CT1 (right) coloured by electrostatic potential. The tail of histone H4 (amino acids 13-24) bound to Sir3 is shown in stick representation, with carbon atoms colored in cyan. The equivalent path for the histone tail in Rsc2 was determined by superposing the two structures. The overall charge distribution on the surface of Rsc2 is different to Sir3, being much less acidic and more hydrophobic in nature and therefore unlikely to bind to the histone H4 tail. Furthermore, the presence of the CT1-loop (amino acids 577–584) also prevents an interaction by blocking the expected binding path. (B) Molecular details of the Sir3-BAH interaction with helix α1 and loop L1 of histone H3, including several residues comprising the LRS region of the nucleosome core particle. Potential hydrogen bonds are indicated by black dotted lines. (C) Molecular details for the equivalent region of Rsc2 BAH-CT1 as shown in (B), highlighting that the identities of the amino acids involved in H3 binding are not conserved between the two BAH domains. (D) The N-terminus of Sir3 contains a ‘basic patch’ (amino acids 28–32), which interacts with a complementarily charged ‘acidic patch’ formed between histones H2A and H2B on the surface of the nucleosome core particle, as shown by the molecular cartoon and electrostatic surface (left). Sir3 BAH is shown in gold, and superposed Rsc2 BAH-CT1 in gray. The visible N-terminus (Asp401) of Rsc2 is indicated by the short arrow. Amino acid sequence alignment and secondary structure prediction (right) indicates that a similar basic patch is not present in Rsc2, and that the protein could not interact with the acidic patch in the same manner as Sir3. BD2 = bromodomain 2. (E) As for (A), but showing the interaction between Orc1 and the tail of histone H4 (amino acids 16–23) dimethylated at lysine 20. (F) Molecular details of the Orc1 BAH H4K20me2 binding pocket. (G) Molecular details for the equivalent region of Rsc2 BAH-CT1, as shown in (F), highlighting that the identities of the amino acids involved in K20me2 binding are not conserved between the two BAH domains, and do not form a methyl-lysine binding pocket.
Sir3 makes multiple interactions with the protein surface of the nucleosome. First, an extended interface is made with the tail of histone H4 (amino acids 13–24), which binds to a complementarily charged channel traversing one face of the BAH domain (Figure 5A, left). Residues Lys16, and His18 of the histone tail also sit within a small charged pocket, making a number of specific contacts with the side chains of Sir3. Calculation of surface electrostatic potential and superposing of structures (CCP4MG; McNicholas et al. Acta Cryst. 2011) highlights that (i) the equivalent face of Rsc2 BAH-CT1 (Figure 5A, right) does not have the same overall charge distribution as that of Sir3, being substantially less negatively charged and more hydrophobic in nature, and therefore unlikely to be able to bind to the predominantly basic histone H4 tail; (ii) the pocket accommodating the side-chains Lys16 and His18 of histone H4 is not present in Rsc2; and (iii) the trajectory of the bound histone H4 tail is blocked in Rsc2, by the loop comprising amino acids 577–584 of the CT1 region (CT1-loop).
Second, Sir3 makes specific hydrogen bonds to histone H3, in particular with the end of helix α1 and loop L1, of which five residues comprise part of the LRS region of the nucleosome, namely, Gln76, Asp77, Phe78, Lys79 and Thr80 (Figure 5B). In Sir3, the guanidinium head group of Arg75 makes hydrogen bonds to the backbone carbonyls of both Asp77 and Phe78. The equivalent side chain of Rsc2 is Trp436, which would be both functionally and structurally unable to make the same contacts (Figure 5C). In Rsc2, Trp444 and Asp477 are respectively in the equivalent position to Glu84 and Glu140 of Sir3, which interact with the side-chain of Lys79 from histone H3. Furthermore, the overall position of the H3 α1 helix and the Lys79 side chain are stabilized by van der Waals interactions with Trp86—which is equivalent to Asn446 in Rsc2. In each case, the conformationally equivalent residues of Rsc2 would not be able to recapitulate the observed Sir3/histone H3 interaction.
Third, at its N-terminus, Sir3 contains a ‘basic patch’ motif (Figure 5D, left) comprised of residues Arg28, Arg29, Arg30, Arg32, Lys33 and Arg34. In the Sir3/nucleosome structure, this motif lies in close proximity to the complementarily charged acidic patch, found on the combined H2A/H2B histone surface. The Rsc2 BAH-CT1 construct used for crystallization studies starts at amino acid Asp401 and therefore does not directly contain information about the structurally equivalent region. However, amino acid sequence alignment and secondary structure predictions [PsiPred, (36)] indicate that Rsc2 does not contain such a basic patch and could therefore not interact with H2A/H2B in the same manner as Sir3 (Figure 5D, right).
The interaction of the BAH domain from Orc1 with the histone H4 tail (dimethylated on Lys20) is also structurally distinct from that observed for Sir3 (23). The histone tail (amino acids 16–23) sits instead across a predominantly hydrophobic face of Orc1, with the methylated lysine bound into a small pocket, gated on one side by the side chain of Trp119 (Figure 5E, left). The CT1-loop, which packs back up against the core BAH domain fold in Rsc2, would again sterically prevent the tail of histone H4 from binding in a similar manner (Figure 5E, right). Furthermore, residues that form the H4K20me2 binding pocket in Orc1 (Trp87, Val89, Glu93, Tyr114 and Trp119) are not conserved in Rsc2 (Figure 5F and G) and do not form a methyl-lysine binding pocket. Moreover, there is no equivalent to Trp119, which forms an essential part of this pocket—instead two prolines (583 and 584) from the distal CT1-loop pack against the side-chain of Tyr451 that, in combination with its hydroxyl group, fill and block any potential binding pocket in Rsc2.
Our comparisons of the Rsc2 BAH-CT1 structure with both Sir3 and Orc1 in complex with nucleosomes/histone tails strongly indicate that the observed interaction of Rsc2 with histone H3 must occur via a unique and distinct interface, using an alternative set of surface amino acid residues.
Identification of an H3 binding interface on the Rsc2 BAH domain
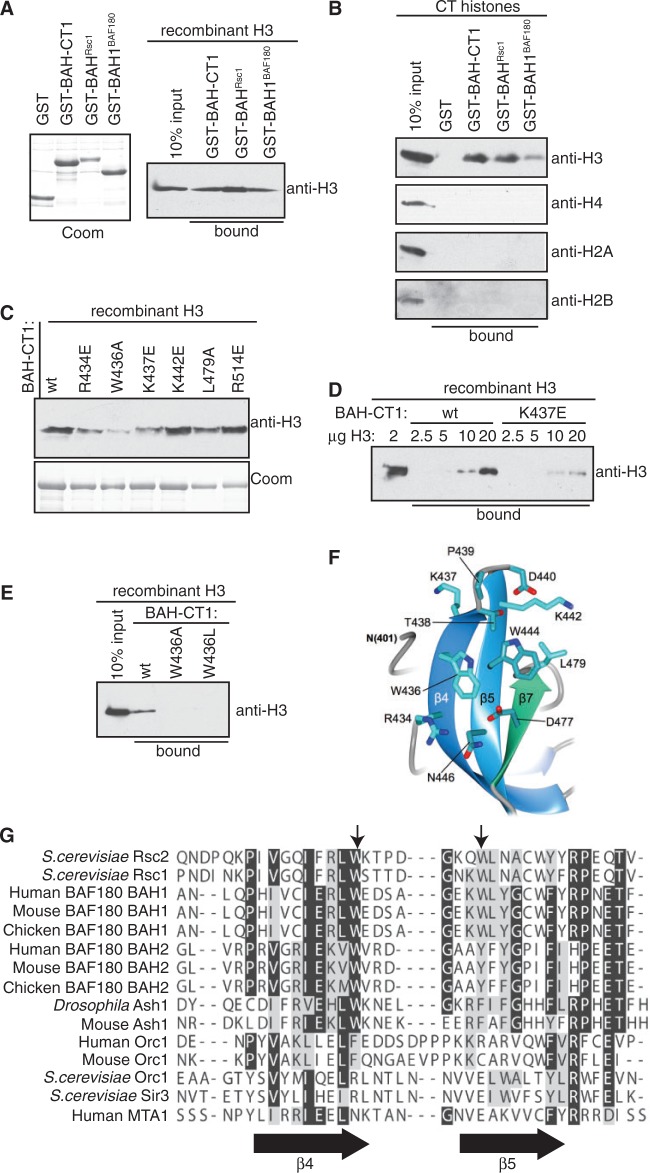
We previously determined that the related Rsc1 and Rsc2 proteins have both redundant and individual functions in mediating DNA damage responses (37). In addition, we found that replacing the BAH domain of Rsc2 with that of Rsc1 has no effect on survival after DNA damage but changes the remodeling activity of Rsc2 at a DNA DSB in vivo (37), suggesting that the BAH domains themselves have both overlapping and distinct functions. We therefore set out to test whether the Rsc1 BAH-CT1 domain was also capable of binding to histone H3. When pull-down assays were performed using recombinant purified GST-BAHRsc1, an interaction with H3 was detected (Figure 6A). We then tested the proximal BAH domain (BAH1) of human BAF180 using purified recombinant GST-BAH1BAF180, and this protein was also capable of interacting with H3 in pull-down assays (Figure 6A). To determine whether selective binding to H3 is a feature of this family of BAH domains, we examined the specificity of binding of BAHRsc1 and BAH1BAF180 from a mixture of all four histones prepared from calf thymus. All three BAH domains interacted with H3, but not with any of the other core histones (Figure 6B). These results suggest that the ability to bind to histone H3 is a conserved feature of the BAH domains of the Rsc2 homologues. In addition, because the BAF180 construct did not contain the downstream CT1 region, these data suggest that the interface with H3 is contained within the BAH domain itself.
Figure 6.
A conserved motif on Rsc2 BAH mediates the interaction with H3. (A) Left panel: Coomassie stained gel of GST, GST-BAH domain from Rsc1 (GST-BAHRsc1) and GST-BAH1 from BAF180 (GST-BAH1BAF180). Right panel: GST pull-down assay using Rsc2 BAH-CT1 (GST-BAH-CT1), GST-BAHRsc1 or GST-BAH1BAF180 and recombinant histone H3. Bound protein was analyzed by western blotting using anti-H3. (B) Rsc2 GST-BAH-CT1, GST-BAHRsc1 and GST-BAH1BAF180 proteins were assayed for the ability to specifically interact with histone H3 from a mixture of calf thymus core histones in pull-down assays. Bound protein was analyzed by western blotting using antibodies specific for each of the four core histones. (C) A panel of mutant Rsc2 GST-BAH-CT1 proteins was assayed for the ability to interact with histone H3 in pull-down assays. Bound protein was analyzed by western blotting using anti-H3 antibody. Coomassie stained gel of the GST-BAH-CT1 proteins used in the pull-down assays (bottom panel). (D) GST-BAH-CT1 or GST-BAH-CT1-K437E was used in pull-down assays containing the indicated amount of recombinant H3 and bound protein was analysed by western blotting using anti-H3. (E) Molecular details of the β4–β5 connecting loop of Rsc2 BAH-CT1 showing the positions of Trp436, Lys437 and Trp444. (F) GST pull-down assay using wt, W436A and W436L GST-BAH-CT1 constructs. Bound protein was analyzed by western blotting using anti-H3. (G) Sequence alignment of the region of the BAH domain encompassing β4, β5 (indicated by arrows below the alignment) and the connecting loop. The positions of Trp436 and Trp444 in Rsc2 are indicated by arrows above the alignment.
To further define the nature of the interaction between Rsc2 BAH-CT1 and H3, we selected a number of residues on the surface of the molecule for mutagenesis. These were guided by our crystal structure and selected based on solvent accessibility with a focus on those residues conserved between the BAH domains of Rsc2, Rsc1 and BAF180. The mutant GST-BAH-CT1 constructs were tested in pull-down assays with H3 as aforementioned, and we found that several of these were no longer able to bind to H3 as well as the wild-type protein (Figure 6C).
Of the mutations tested, W436A and K437E reproducibly showed reduced binding to histone H3 in pull-down assays (Figure 6D and E and data not shown). Interestingly, both Trp436 and Lys437 are both located at one end of beta-strand 4 (β4), close to the loop connecting β4 to β5. Both residues are solvent accessible, but face in opposite directions. Trp436 is only partially packed against Trp444 at its C-beta position, which in turn makes van der Waals contacts with the side chain of Leu479 (Figure 6F). We were concerned that mutation of a large hydrophobic residue might drastically destabilize the Rsc2 BAH-CT1 protein. We therefore decided to make an alternative mutation replacing the tryptophan with a LEU—W436L—intended to partially recapitulate the observed hydrophobic stacking with Trp444. Notably, this mutation also showed a histone H3 binding defect (Figure 6E). We then tested each mutant in a thermal denaturation experiment to assess the overall stability/fold of each protein. As might be expected, the Lys437 charge reversal mutant had little effect, as indicated by a similar temperature midpoint (Tm), for the transition from folded to unfolded protein, as the wild-type protein (45.6 and 46.4°C, respectively). Of the two Trp436 mutants, W436A had a somewhat lower Tm (41.3°C) than W436L (43.0°C), indicating that there was, at least, some perturbation of the fold (Supplementary Figure S2). However, as each recombinant protein produced a single, cooperative, sigmoidal-shaped denaturation curve; was readily expressed in, then purified from E. coli with no apparent chaperone contaminant (i.e. GroEL); eluted as a single monomeric peak, at the same volume, from a size exclusion chromatography column; and could be concentrated to >5 mg/ml, we surmise that the apparent reduction in Tm is most likely due to localized changes in amino acid side chain packing, rather than a global misfolding of the protein.
We examined the amino acid sequence of this area from BAH domains of a number of other proteins and find that the pair of hydrophobic residues flanking the β4–β5 connecting loop is conserved among Rsc2 homologues as well as a subset of RSC-like BAH domains. In contrast, they are not found in the Orc1 and Sir3 proteins (Figure 6G).
Mutations in the H3 binding interface of the BAH domain result in impaired Rsc2 activity in vivo
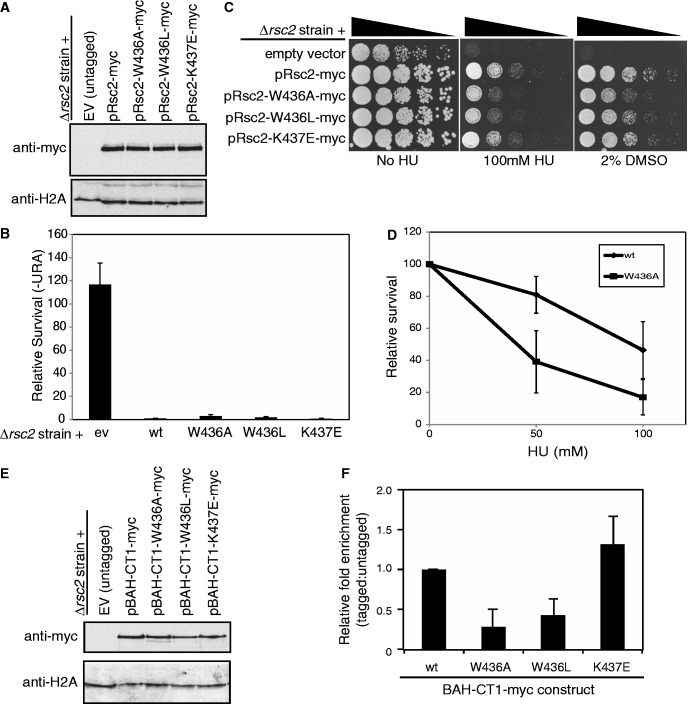
To determine whether mutation of the residues that impaired H3 binding in vitro had any consequence to RSC activity in vivo, we introduced the W436A, W436L and K437E mutations into a full-length, myc-tagged Rsc2 expression construct. These were introduced into an rsc2 null strain (alongside a wt control), and expression was analyzed by western blotting. We found all three mutant proteins were stably expressed in vivo (Figure 7A).
Figure 7.
Mutation of the conserved motif in the Rsc2 BAH domain that is important for H3 binding in vitro disrupts some Rsc2 functions in vivo. (A) Western blot analysis of whole cell lysates prepared from rsc2 null strains carrying empty vector or expression constructs with full-length Myc-tagged Rsc2 (wt, W436A, W436L or K437E mutant) using anti-myc (top panel) or anti-H2A (bottom panel) as a loading control. (B) Silencing was monitored as in Figure 1B using the rsc2 null strain carrying either empty vector, wt Rsc2 or mutant Rsc2 constructs, and the data are represented as the mean ± 1 SD of at least three independent experiments. (C) The W436A and W436L mutant Rsc2 proteins are unable to fully complement the hypersensitivity of an rsc2 null strain. Serial dilutions of mid-log cultures were plated onto media containing the indicated drug and incubated for 2–3 days at 30°C before imaging. (D) Survival of the W436A mutant strain compared with wt Rsc2. Three independent cultures of each strain were grown and plated onto media containing the indicated amount of HU. Survival was calculated relative to media lacking HU and the data are shown ± 1 SD. (E) Western blot analysis of whole cell lysates prepared from rsc2 null strains carrying either empty vector or overexpression BAH-CT1-myc constructs as indicated using anti-myc (top panel) or anti-H2A (bottom panel) as a loading control. (F) W436A and W436L mutations in Rsc2 BAH-CT1 impair chromatin association in vivo. ChIP assays examining enrichment of Myc-tagged overexpressed BAH-CT1 relative to the untagged control at the 18S region of the rDNA. Data shown are the mean enrichment of at least three independent experiments ± 1 SD.
We then investigated the ability of these mutant constructs to support rDNA silencing. Perhaps surprisingly, all three mutants were able to rescue the silencing defect of a rsc2 null strain to apparently wild-type levels (Figure 7B). We note that the W436A mutant strain appears to have a mild silencing defect when compared with wt, but this is not statistically significant. We considered several possible reasons for this finding. First, BAH-CT1 proteins bearing these mutations were still able to weakly bind H3 in vitro, and it is possible that the effect of such mutations in vivo is too small to disrupt activity, particularly when there are multiple other chromatin binding domains still present in the RSC complex. In this case, we would predict that all of Rsc2’s functions would be intact in the BAH mutant-containing strains. Second, it is possible that the H3 binding activity of the BAH domain does not contribute to the function of Rsc2 in mediating silencing at the rDNA but is important for other functions of Rsc2. We therefore set out to test the mutants in other cellular assays.
To determine whether these mutations had any effect on the role of Rsc2 in mediating DNA damage responses, we tested the ability of the mutant Rsc2-containing strains to survive in the presence of DNA damage. We found that mutation of Trp436 is unable to fully complement the hypersensitivity of an rsc2 null strain to the DNA damaging agent HU (Figure 7C). Because the phenotypes of the mutant strains in this assay were subtle, we quantitated survival in the W436A mutant strain and found there is a significant reduction in survival in the presence of HU when compared with wt (Figure 7D).
RSC also functions in transcriptional regulation, and one set of genes that is misregulated in strains lacking RSC activity is related to cell wall biogenesis (38). Consistent with this, we find that rsc2 null strains are hypersensitive to DMSO (Figure 7C). When tested in the presence of DMSO, we found that there was a small but reproducible decrease in survival, particularly of the W436A mutant strain, when compared with wt (Figure 7C). Collectively, these data suggest that mutation of this region of the BAH domain impacts on at least a subset of Rsc2 functions in vivo.
Based on our biochemical data, we predicted that the observed phenotypes in the mutant strains were due to decreased ability of the mutant BAH domains to bind to H3. To test this, we performed ChIP assays. However, when we did this using full-length Rsc2 constructs, we did not find a decrease in chromatin enrichment in the mutants compared with wt (data not shown). One possible explanation for this is that the effect of BAH domain mutations on chromatin binding may be small when they are present within the full-length protein, which, when incorporated into the RSC complex, will be associated with multiple chromatin binding domains. To circumvent this issue, we created the mutations in the BAH-CT1 overexpression construct to allow us to assess the ability of these mutant proteins to bind to chromatin in vivo when examined in isolation. The three mutant BAH-CT1 domains were expressed at similar levels to wt (Figure 7E). When ChIP assays were performed, we found the chromatin enrichment of the K437E mutant construct was not significantly different from wt (Figure 7F), suggesting the residual H3 binding activity we detect in vitro (Figure 6D) is sufficient for chromatin binding in vivo. In contrast, and consistent with the greater effect of these mutations on H3 binding in vitro, the enrichment of the two tryptophan mutants (W436A and W436L) in chromatin was substantially reduced compared with wt (Figure 7F). To examine whether this loss of binding was specific to the rDNA locus, chromatin enrichment of the mutant BAH-CT1 domains was analyzed at a further two loci; the promoter regions of the HTA1 and HTZ1 genes. The mutant proteins displayed largely the same profile of chromatin association at these loci as at the rDNA locus, suggesting that their defect in chromatin binding is global (Supplementary Figure S3).
DISCUSSION
In this work, we identified a role for the Rsc2 subunit of RSC in transcriptional silencing at the rDNA, which prompted us to focus on the BAH domain of this subunit. These studies uncovered a direct interaction between the Rsc2 BAH domain and histone H3 and led to the identification of an interaction interface that is conserved in a subset of Rsc-like BAH domains. These data give us a number of insights into the role of Rsc2 and its BAH domain in the context of RSC activity in cells.
The mutations in the BAH domain that impact on H3 binding in vitro result in relatively subtle phenotypes in vivo. As the RSC complex has multiple subunits that are known or predicted to make contacts with chromatin, such as the bromodomains of Rsc2, Rsc4 and Sth1, it is reasonable to speculate that impairment of any single domain is not likely to abolish chromatin binding or activity of the complex. In support of this, we found that the mutant proteins were not defective in chromatin binding when tested in the context of the full-length protein in vivo, but they only showed differences from wt when tested in the context of the BAH-CT1 construct. Perhaps more surprisingly, we found that the mutations did not appear to have an impact on silencing (although we note that there may be a weak effect of the W436A mutation), but they did affect other Rsc2-dependent functions. This shows that the BAH domain is not specifically mediating RSC-dependent rDNA silencing, but it is instead playing a more general role.
We found that the BAH domains of Rsc2 homologues are also able to bind H3 in vitro, and the presence of a tryptophan at the end of beta strand 4 (β4) is conserved in these proteins. In addition, they have a second large hydrophobic residue present at the other end of the β4–β5 connecting loop, analogous to Rsc2 Trp444, which makes hydrophobic contacts with Rsc2 Trp436 (Figure 6F and G). Unsurprisingly, in Sir3 and Orc1 BAH domains, which use distinct nucleosome-binding surfaces, neither of these residues is conserved nor are they conserved across the entire RSC-like class of BAH domains (e.g. Human MTA1, see Figure 6G). Interestingly, however, there are a number of other proteins, such as Ash1 (Figure 6G), within the RSC-like class of BAH domains that, like Rsc2 and its homologues, have a conserved tryptophan in β4 and a large hydrophobic residue in β5. Based on our findings, we speculate that these proteins may also interact in a similar manner with H3.
Despite the fact that the BAH domains from both Rsc2 and Rsc1 can bind H3 in vitro, we found that replacing the BAH domain of Rsc2 with that of Rsc1 alters its specificity of remodeling (37), suggesting that they are not functionally equivalent in vivo. One possibility is that the modification pattern of H3 may be differentially affecting interactions with the two BAH domains. However, it is also possible that there are other functions or interaction partners of the BAH domains that are not interchangeable.
Genes encoding several subunits of PBAF, including BAF180, are commonly mutated or misregulated in cancer and have been shown to function as tumor suppressor genes (39,40). Mutations in BAF180 were found in lung cancer, breast cancer and renal carcinoma cells (39,41), and protein expression levels were altered in head and neck squamous cell carcinoma (HNSCC) (42). The loss of BAF180 activity results in compromised p53 activity (41,43). Interestingly, a squamous cell lung cancer sample in the Catalogue of Somatic Mutations in Cancer database possesses a W948L missense mutation within the first BAH domain of BAF180 (the mutation data were obtained from the Sanger Institute Catalogue Of Somatic Mutations In Cancer web site, http://www.sanger.ac.uk/cosmic) (44). This substitution is the equivalent of W436L of Rsc2, which we found to be defective in H3 binding. The vast majority of the mutations identified in the renal cell carcinoma study resulted in truncated proteins, but 9 missense and 2 in-frame deletions were identified, two of which were in the second BAH domain (39). Based on our data, we would predict that the 6 amino acid deletion in BAH2 (M1209-E1214, DMFYKKE) in BAF180 (corresponding to L463-E468, DLFYKNEof Rsc2) would lead to a highly destabilized or unfolded protein, due to the removal of several residues forming part of a hydrophobic core. The second BAH2 mutation identified was D1285G, which corresponds to D540 in Rsc2. Interestingly, this mutation would not be expected to affect the fold but would alter the overall localized negative charge in this region.
The identification of the BAH domain of the RSC-like class of BAH domains as a histone H3 binding module will aid studies of the RSC/PBAF chromatin remodeling complexes and lead to a greater understanding of the molecular mechanism underpinning their cellular activities.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [45–58].
FUNDING
Cancer Research UK CEA [C7905/A10269 to A.L.C. and J.A.D.]; Cancer Research UK Programme [C302/A8265 to A.W.O. and L.H.P.]. Funding for open access charge: Cancer Research UK.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank members of the Downs laboratory for helpful discussions, and Peter Brownlee additionally for help with strain construction.
REFERENCES
- 1.Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 2.Kent NA, Chambers AL, Downs JA. Dual Chromatin Remodeling Roles for RSC during DNA Double Strand Break Induction and Repair at the Yeast MAT Locus. J. Biol. Chem. 2007;282:27693–27701. doi: 10.1074/jbc.M704707200. [DOI] [PubMed] [Google Scholar]
- 3.Shim EY, Hong SJ, Oum JH, Yanez Y, Zhang Y, Lee SE. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol. Cel.l Biol. 2007;27:1602–1613. doi: 10.1128/MCB.01956-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faucher D, Wellinger RJ. Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS Genet. 2010;6:pii: e1001082. doi: 10.1371/journal.pgen.1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gartenberg M. Heterochromatin and the cohesion of sister chromatids. Chromosome Res. 2009;17:229–238. doi: 10.1007/s10577-008-9012-z. [DOI] [PubMed] [Google Scholar]
- 6.Desai P, Guha N, Galdieri L, Hadi S, Vancura A. Plc1p is required for proper chromatin structure and activity of the kinetochore in Saccharomyces cerevisiae by facilitating recruitment of the RSC complex. Mol. Genet. Genomics. 2009;281:511–523. doi: 10.1007/s00438-009-0427-9. [DOI] [PubMed] [Google Scholar]
- 7.Mohrmann L, Verrijzer CP. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim. Biophys. Acta. 2005;1681:59–73. doi: 10.1016/j.bbaexp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Park JH, Park EJ, Hur SK, Kim S, Kwon J. Mammalian SWI/SNF chromatin remodeling complexes are required to prevent apoptosis after DNA damage. DNA Repair (Amst) 2008;8:29–39. doi: 10.1016/j.dnarep.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 9.Lee HS, Park JH, Kim SJ, Kwon SJ, Kwon J. A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 2010;29:1434–1445. doi: 10.1038/emboj.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng G, Yim EK, Dai H, Jackson AP, Burgt I, Pan MR, Hu R, Li K, Lin SY. BRIT1/MCPH1 links chromatin remodelling to DNA damage response. Nat. Cell Biol. 2009;11:865–872. doi: 10.1038/ncb1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bochar DA, Wang L, Beniya H, Kinev A, Xue Y, Lane WS, Wang W, Kashanchi F, Shiekhattar R. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell. 2000;102:257–265. doi: 10.1016/s0092-8674(00)00030-1. [DOI] [PubMed] [Google Scholar]
- 12.Cairns BR, Schlichter A, Erdjument-Bromage H, Tempst P, Kornberg RD, Winston F. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol. Cell. 1999;4:715–723. doi: 10.1016/s1097-2765(00)80382-2. [DOI] [PubMed] [Google Scholar]
- 13.Goodwin GH, Nicolas RH. The BAH domain, polybromo and the RSC chromatin remodelling complex. Gene. 2001;268:1–7. doi: 10.1016/s0378-1119(01)00428-0. [DOI] [PubMed] [Google Scholar]
- 14.Callebaut I, Courvalin JC, Mornon JP. The BAH (bromo-adjacent homology) domain: a link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 1999;446:189–193. doi: 10.1016/s0014-5793(99)00132-5. [DOI] [PubMed] [Google Scholar]
- 15.Oliver AW, Jones SA, Roe SM, Matthews S, Goodwin GH, Pearl LH. Crystal structure of the proximal BAH domain of the polybromo protein. Biochem J. 2005;389:657–664. doi: 10.1042/BJ20050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hickman MA, Rusche LN. Transcriptional silencing functions of the yeast protein Orc1/Sir3 subfunctionalized after gene duplication. Proc. Natl Acad. Sci. USA. 2010;107:19384–19389. doi: 10.1073/pnas.1006436107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gartenberg MR. The Sir proteins of Saccharomyces cerevisiae: mediators of transcriptional silencing and much more. Curr. Opin. Microbiol. 2000;3:132–137. doi: 10.1016/s1369-5274(00)00064-3. [DOI] [PubMed] [Google Scholar]
- 18.Norris A, Boeke JD. Silent information regulator 3: the Goldilocks of the silencing complex. Genes Dev. 2010;24:115–122. doi: 10.1101/gad.1865510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu HC, Stillman B, Xu RM. Structural basis for origin recognition complex 1 protein-silence information regulator 1 protein interaction in epigenetic silencing. Proc. Natl Acad. Sci. USA. 2005;102:8519–8524. doi: 10.1073/pnas.0502946102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duncker BP, Chesnokov IN, McConkey BJ. The origin recognition complex protein family. Genome Biol. 2009;10:214. doi: 10.1186/gb-2009-10-3-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noguchi K, Vassilev A, Ghosh S, Yates JL, DePamphilis ML. The BAH domain facilitates the ability of human Orc1 protein to activate replication origins in vivo. EMBO J. 2006;25:5372–5382. doi: 10.1038/sj.emboj.7601396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller P, Park S, Shor E, Huebert DJ, Warren CL, Ansari AZ, Weinreich M, Eaton ML, MacAlpine DM, Fox CA. The conserved bromo-adjacent homology domain of yeast Orc1 functions in the selection of DNA replication origins within chromatin. Genes Dev. 2010;24:1418–1433. doi: 10.1101/gad.1906410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuo AJ, Song J, Cheung P, Ishibe-Murakami S, Yamazoe S, Chen JK, Patel DJ, Gozani O. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature. 2012;484:115–119. doi: 10.1038/nature10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Norris A, Bianchet MA, Boeke JD. Compensatory interactions between Sir3p and the nucleosomal LRS surface imply their direct interaction. PLoS Genet. 2008;4:e1000301. doi: 10.1371/journal.pgen.1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Onishi M, Liou GG, Buchberger JR, Walz T, Moazed D. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol. Cell. 2007;28:1015–1028. doi: 10.1016/j.molcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 26.Armache KJ, Garlick JD, Canzio D, Narlikar GJ, Kingston RE. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 A resolution. Science. 2011;334:977–982. doi: 10.1126/science.1210915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 28.Lacoste N, Utley RT, Hunter J, Poirier GG, Cote J. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J. Biol. Chem. 2002;277:30421–30424. doi: 10.1074/jbc.C200366200. [DOI] [PubMed] [Google Scholar]
- 29.Foster ER, Downs JA. Methylation of H3 K4 and K79 is not strictly dependent on H2B K123 ubiquitylation. J. Cell. Biol. 2009;184:631–638. doi: 10.1083/jcb.200812088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langst G, Bonte EJ, Corona DFV, Becker P. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999;97:843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- 31.Downs JA, Lowndes NF, Jackson SP. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature. 2000;408:1001–1004. doi: 10.1038/35050000. [DOI] [PubMed] [Google Scholar]
- 32.Huang J, Brito IL, Villen J, Gygi SP, Amon A, Moazed D. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev. 2006;20:2887–2901. doi: 10.1101/gad.1472706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kasten M, Szerlong H, Erdjument-Bromage H, Tempst P, Werner M, Cairns BR. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng HH, Robert F, Young RA, Struhl K. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev. 2002;16:806–819. doi: 10.1101/gad.978902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 37.Chambers AL, Brownlee PM, Durley SC, Beacham T, Kent NA, Downs JA. The two different isoforms of the RSC chromatin remodeling complex play distinct roles in DNA damage responses. PLoS One. 2012;7:e32016. doi: 10.1371/journal.pone.0032016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Angus-Hill ML, Schlichter A, Roberts D, Erdjument-Bromage H, Tempst P, Cairns BR. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol. Cell. 2001;7:741–751. doi: 10.1016/s1097-2765(01)00219-2. [DOI] [PubMed] [Google Scholar]
- 39.Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weissman B, Knudsen KE. Hijacking the chromatin remodeling machinery: impact of SWI/SNF perturbations in cancer. Cancer Res. 2009;69:8223–8230. doi: 10.1158/0008-5472.CAN-09-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia W, Nagase S, Montia AG, Kalachikov SM, Keniry M, Su T, Memeo L, Hibshoosh H, Parsons R. BAF180 is a critical regulator of p21 induction and a tumor suppressor mutated in breast cancer. Cancer Res. 2008;68:1667–1674. doi: 10.1158/0008-5472.CAN-07-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ralhan R, Desouza LV, Matta A, Chandra Tripathi S, Ghanny S, Datta Gupta S, Bahadur S, Siu KW. Discovery and verification of head-and-neck cancer biomarkers by differential protein expression analysis using iTRAQ labeling, multidimensional liquid chromatography, and tandem mass spectrometry. Mol. Cell. Proteomics. 2008;7:1162–1173. doi: 10.1074/mcp.M700500-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burrows AE, Smogorzewska A, Elledge SJ. Polybromo-associated BRG1-associated factor components BRD7 and BAF180 are critical regulators of p53 required for induction of replicative senescence. Proc. Natl Acad. Sci. USA. 2010;107:14280–14285. doi: 10.1073/pnas.1009559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bamford S, Dawson E, Forbes S, Clements J, Pettett R, Dogan A, Flanagan A, Teague J, Futreal PA, Stratton MR, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br. J. Cancer. 2004;91:355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanny JC, Kirkpatrick DS, Gerber SA, Gygi SP, Moazed D. Budding yeast silencing complexes and regulation of Sir2 activity by protein-protein interactions. Mol. Cell. Biol. 2004;24:6931–6946. doi: 10.1128/MCB.24.16.6931-6946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeMase D, Zeng L, Cera C, Fasullo M. The Saccharomyces cerevisiae PDS1 and RAD9 checkpoint genes control different DNA double-strand break repair pathways. DNA Repair (Amst) 2005;4:59–69. doi: 10.1016/j.dnarep.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 47.Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 48.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992;vol. 26 [Google Scholar]
- 49.CCP4. The CCP4 suite: programs for protein crystallography. Acta. Crystallogr. D. Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 50.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta. Crystallogr. D. Biol. Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta. Crystallogr. D. Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 52.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of coot. Acta. Crystallogr. D. Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta. Crystallogr. D. Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davis IW, Murray LW, Richardson JS, Richardson DC. MolProbity: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ericsson UB, Hallberg BM, Detitta GT, Dekker N, Nordlund P. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Analyt. Biochem. 2006;357:289–298. doi: 10.1016/j.ab.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 56.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Livingstone CD, Barton GJ. Protein sequence alignments: a strategy for the hierarchical analysis of residue conservation. Comput. Appl. Biosci. 1993;9:745–756. doi: 10.1093/bioinformatics/9.6.745. [DOI] [PubMed] [Google Scholar]
- 58.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.