Abstract
Nucleotide excision repair (NER) is critical for maintaining genome integrity. How chromatin dynamics are regulated to facilitate this process in chromatin is still under exploration. We show here that a histone H2A variant, Htz1 (H2A.Z), in nucleosomes has a positive function in promoting efficient NER in yeast. Htz1 inherently enhances the occupancy of the histone acetyltransferase Gcn5 on chromatin to promote histone H3 acetylation after UV irradiation. Consequently, this results in an increased binding of a NER protein, Rad14, to damaged DNA. Cells without Htz1 show increased UV sensitivity and defective removal of UV-induced DNA damage in the Htz1-bearing nucleosomes at the repressed MFA2 promoter, but not in the HMRa locus where Htz1 is normally absent. Thus, the effect of Htz1 on NER is specifically relevant to its presence in chromatin within a damaged region. The chromatin accessibility to micrococcal nuclease in the MFA2 promoter is unaffected by HTZ1 deletion. Acetylation on previously identified lysines of Htz1 plays little role in NER or cell survival after UV. In summary, we have identified a novel aspect of chromatin that regulates efficient NER, and we provide a model for how Htz1 influences NER in Htz1 nucleosomes.
INTRODUCTION
The eukaryotic genome is packaged with histones and non-histone proteins to form chromatin. This characteristic genome organization in eukaryotes makes chromatin the primary platform for almost all DNA-based events, including replication, transcription and DNA repair (1–3). Hence, efforts have long been made to understand how chromatin impinges on genome organization and to decipher the epigenetic information encoded within chromatin. In addition to post-translational histone modifications (4) and ATP-dependent chromatin remodelling (5), incorporation of histone variants into nucleosomes (6,7) is another means by which cells regulate chromatin dynamics in response to internal and environmental cues.
The Saccharomyces cerevisiae Htz1 protein is a histone H2A variant that belongs to the highly conserved family of H2A.Z proteins (8). Sequence comparison indicates that H2A.Z proteins across species share more sequence identity with each other than with canonical histone H2A from the same species, suggesting that H2A.Z may have unique functions that are different from canonical histone H2A (9). Htz1 is incorporated into the nucleosomes in the form of a Htz1–H2B dimer by the ATP-dependent SWR complex to replace H2A–H2B, and the incorporation is independent of DNA replication (10–13).
The vast majority of S. cerevisiae promoters have a stereotypical chromatin architecture, characterized by two well-positioned nucleosomes flanking a nucleosome-depleted region (14–16). Genome-wide studies have shown that Htz1 is significantly enriched at the promoter-proximal nucleosomes of genes and in the euchromatin/heterochromatin boundaries (17–21). Acetylation at four lysines in the N-terminus is required for many of the functions of Htz1, including normal chromosome segregation (22) and maintenance of heterochromatin boundaries (23). At Htz1K14, the most abundantly acetylated site, acetylation is enriched at actively transcribed genes (24), and Htz1 acetylation is required for the efficient activation of GAL1 (25) and oleate-induced genes (26). Reflecting these multiple roles, htz1 alleles lacking the N-terminal acetylation sites have genetic interactions with genes involved in silencing, chromosome transmission, and transcription (27). Htz1 has also been linked to genome stability (28,29). The absence of Htz1 affects cell cycle progression and causes lethality or sickness in combination with checkpoint mutations (30). These results, together with the fact that htz1Δ cells are sensitive to some DNA-damaging reagents, suggest a role for Htz1 in maintaining genome integrity (12,22,23). Htz1 occupancy is dynamic near DNA strand breaks during double strand break repair (31–33), and Htz1 is necessary for strand resection and checkpoint activation at persistent DNA double-strand breaks (33). Other than this, the function of Htz1 in DNA repair remains largely unclear.
Nucleotide excision repair (NER) is a major and highly conserved repair pathway that cells use to remove a broad range of DNA damage, including cyclobutane pyrimidine dimers (CPDs) induced by UV irradiation (34). Compared with the vast efforts that have been made to comprehend the regulation of chromatin dynamics during transcription activation, studies on how NER operates in the chromatin environment are generally modest; therefore, our knowledge is limited. Early studies concentrated on the physical hindrance of nucleosomes on NER and revealed that lesions in the linker DNA and towards the ends of positioned nucleosomes are repaired faster than those in the centre of the nucleosomes (35–38). Meanwhile, a hypothetical model of ‘access-repair-restore’ was proposed, in an effort to rationalize the occurrence of chromatin remodelling during NER (39). More recently, direct evidence of roles for histone modifications and chromatin remodelling in NER has started to emerge. We previously showed that histone H3 acetylation by Gcn5 is activated in the repressed MFA2 promoter after UV irradiation in S. cerevisiae (40,41). This UV-induced histone H3 hyperacetylation is necessary for the efficient repair of UV-induced CPDs in this locus. Rad16, a SWI/SNF-like NER protein responsible for the repair of DNA damage in non-transcribed DNA, governs the occupancy of Gcn5 on chromatin to regulate histone H3 acetylation after DNA damage (42,43). Others also showed that Dot1-controlled constitutive H3K79 methylation is required for the repair of CPDs in the RPB2 and HML loci (44,45). In addition to histone modifications, chromatin remodelling by the SWI/SNF (46) and INO80 (47) ATP-dependent complexes was also found to be involved in the removal of CPDs from the transcriptionally silent HML locus by NER. Subunits of these chromatin remodelling complexes interact with Rad4-Rad23 in a UV dependent manner (46,47). The INO80 complex does not remodel the chromatin in the early stages after DNA damage. Instead, it is required for the restoration of chromatin after DNA damage is removed (47).
Here we examined how Htz1 contributes to the repair of UV-induced CPDs in the Htz1-containing nucleosomes in the MFA2 promoter. Our data show that in wild-type cells, Htz1 is present in the two nucleosomes in the MFA2 promoter, and Htz1 in these nucleosomes has a positive function for the efficient repair of UV-induced CPDs by NER. Deletion of HTZ1 results in a significant reduction in the occupancy of Gcn5 on these two nucleosomes. This leads to a reduced response in histone H3 acetylation after UV. Consequently, the binding of Rad14 protein to damaged DNA is compromised, and the removal of CPDs is defective at MFA2 in the absence of Htz1. However, deletion of HTZ1 has no effect on histone H3 acetylation, the binding of Rad14 to damaged DNA and the repair of CPDs in the silent HMRa1 locus where Htz1 is absent in wild-type cells bearing a functional HTZ1 gene. This indicates a function for Htz1 in promoting NER in the Htz1-containing nucleosomes by regulating the occupancy of Gcn5 on chromatin, the UV-induced histone H3 acetylation and the binding of Rad14 to damaged DNA in the nucleosomes. Our results reveal a novel regulation of chromatin dynamics to ensure efficient repair of DNA damage.
MATERIALS AND METHODS
Yeast strains, growth, UV treatment, survival and repair
Yeast strains and plasmids are listed in Table 1. Yeast strains were grown in yeast complete medium, Yeast Extract-Peptone-Dextrose (YPD) at 30°C. To tag GCN5 and RAD14 with the Myc epitope, the plasmid p3747, a kind gift from Dr Richard A. Young, was used as a template to generate polymerase chain reaction (PCR) products containing the Myc epitope coding sequence and URA3 selectable marker flanked by homologous regions designed to recombine at the 3′ end of the GCN5 and RAD14, respectively. The PCR products were transformed into the BY4742, and its derivative htz1Δ mutant strain. Clones were selected for growth on uracil-minus plates. Insertion of the epitope coding sequence was confirmed by PCR and sequencing, and the expression of the epitope-tagged protein was confirmed by western blotting using an anti-Myc antibody. To generate pCM302, the HTZ1 sequence was amplified, and cloned into the HindIII–SalI sites of the yeast centromeric CEN/ARS, URA3 vector pRS416 (48). Point mutations were introduced into the HTZ1 coding sequence using the QuikChange II Site-Directed Mutagenesis Kit (Stratagene) to generate pCM544 and pCM566. Mutations were verified by sequencing. For strains harbouring these plasmids, cells were grown in minimal medium without uracil. For UV survival experiments, cells at exponential phase (2–4 × 107 cells/ml) from overnight culture were collected and resuspended in distilled water. Appropriate amount of cells were then spread onto YPD plates and subjected to various doses of UV treatment. Plates were kept in the dark at 30°C for 3–4 days before colonies were scored. For repair and post-UV chromatin immunoprecipitation (ChIP) experiments, cells at exponential phase (2–4 × 107 cells/ml) were resuspended to a final concentration of 2 × 107 cells/ml in pre-chilled phosphate buffered saline. The cell suspension was then either mock treated (without UV) or irradiated with 254 nm UV light at a dose of 100 J/m2. Mock-treated and irradiated cells were collected and resuspended in fresh YPD to allow for the indicated repair times in the dark at 30°C with shaking.
Table 1.
Yeast strains
| Strains | Genotype | Source |
|---|---|---|
| WT, BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Euroscarf |
| htz1Δ | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 htz1:: KanMX4 | Euroscarf |
| swr1Δ | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 swr1:: KanMX4 | Euroscarf |
| yaf9Δ | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 yaf9:: KanMX4 | Euroscarf |
| bdf1Δ | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 bdf1:: KanMX4 | Euroscarf |
| gcn5Δ | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 gcn5:: KanMX4 | Euroscarf |
| WT, HTZ1-HA | MATα HHT1-HHF1Δ(HHT2-HHF2) leu2–3,112 ura3-52 lys2Δ201 HTZ1-HA | Zhang et al., 2005 |
| WT, BY4742, RAD14-Myc | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 RAD14::13Myc-URA3 | This study |
| htz1Δ, RAD14-Myc | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 RAD14::13Myc-URA3 htz1:: NAT1 | This study |
| WT, BY4742, GCN5-Myc | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 GCN5::18Myc-URA3 | This study |
| htz1Δ, GCN5-Myc | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 htz1Δ::kanMX4 GCN5::18Myc-URA3 | This study |
| WT, W303 | MATα ade2–1 leu2–3, 112 his3–1 ura3–52 trpl-100can1–100 HTZ1–3Flag-KanMX4 | Babiarz et al., 2006 |
| W303, htz1K3,8,10,14R, | MATα ade2–1 leu2–3, 112 his3–1 ura3–52 trpl-100can1–100 htz1K3, 8, 10, 14R-3Flag-KanMX4 | Babiarz et al., 2006 |
| pCM302 | CEN6-ARS4 URA3 HTZ1 | This study |
| pCM544 | CEN6-ARS4 URA3 htz1K3,8,10,14R | This study |
| pCM566 | CEN6-ARS4 URA3 htz1K3,8,10,14Q | This study |
Gel assay to analyse CPD repair in the overall genome
About 10 µg of DNA from each sample was treated with 5 µl of the CPD-specific endonuclease, obtained from Micrococcus luteus, for 1 h at 37°C, and then subjected to agarose gel electrophoresis (1.5%) under denaturing conditions (50 mM of NaOH, 1mM of EDTA in 1× Tris-acetate-EDTA [TAE] running buffer). The gel was washed, neutralized and stained with 1% ethidium bromide. The number of CPDs per kb DNA was calculated as described by Sutherland and Shih (49) and Bespalov et al. (50).
Chromatin immunoprecipitation
ChIP was performed as described before (40). In brief, 50 ml of cells (2 × 107 cells/ml) were cross-linked with 1.4 ml formaldehyde (37%) at room temperature for 20 min with shaking. Glycine (5.5 ml, 2.5 M) was added to stop cross-linking. Cells were collected, washed and then transferred to 2 ml Eppendorf tubes in 0.5 ml of ChIP buffer (50 mM HEPES-KOH, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.25% sodium dodecyl sulphate (SDS), 5 µl of protease inhibitor cocktail [Sigma UK, Cat No. P8215]). After addition of 0.5 ml of glass beads (425∼600 μm, Sigma), samples were vortexed at 4°C for 10 min. The cell lysate was collected for sonication. Sonication was carried out by using a Bioruptor (Diagenode) at 4°C, with power on ‘high’, 30 s on and 30 s off for 10 cycles to yield fragmented DNA of ∼300 bp in length. For experiments involving mononucleosomes, 300 μl of cell lysate in ChIP buffer after bead beating was supplemented with MgCl2 and CaCl2 to final concentrations of 20 and 5 mM, respectively, and then treated with 300 U of micrococcal nuclease (MNase; Nuclease S7, Roche) for 10 min at 37°C. The reaction was terminated by adding 1/50 volume of 0.5 M EDTA. The soluble MNase-fragmented chromatin was further homogenized by a brief sonication. Size analysis of purified DNA from samples after the treatment confirmed that only mononucleosomes are generated. For immunoprecipitation, 100 μl of sheared chromatin solution was incubated for 2 h at room temperature in a total volume of 1 ml in ChIP buffer containing specific antibodies. The amounts of antibodies used were 5 μg of anti-Htz1 (Abcam, ab4626), 2 μg of anti-HA (Upstate, 05–904), 5 μl of anti-histone H3 (Upstate, 17–10 046), 3 μg of anti-acetyl histone H3 (Upstate, 06–599; this antibody was raised against acetylated H3 at K9/K14) and 5 μg of anti-Myc (9B11, Cell Signalling technology, #2276). Fifty microlitres of protein G beads Sepharose (GE Healthcare Life Sciences) was added to the samples, and the mixture was incubated for another hour with shaking. The beads were washed, and DNA was eluted from the beads. After the cross-linking was reversed, DNA was finally purified using a Qiagen PCR purification kit. Quantitative PCR was performed in real time by using the iQ SYBR Green Supermix (Bio-Rad) in the Bio-Rad iCycler. Reactions were performed in triplicate for each sample, and melting curves were executed to ensure a single PCR product. For samples involving UV treatment, DNA damage was removed before PCR by using a PreCR DNA repair kit (New England Biolab). For mononucleosome ChIP, primers were designed to amplify the sequences within individual nucleosomes in the nucleosomes −2 and −1 in the MFA2 promoter, and the nucleosomes 4 and 5 in the HMRa1 locus. Otherwise, primers were designed to amplify sequences covering the above two nucleosomes in the MFA2 and the HMRa1 locus, respectively. Primer sequences are available on request.
Analysis of CPDs at nucleotide resolution
This was carried out as previously described in detail (51). Repair analysis focused on both strands of the HaeIII fragment (−517 to +83) in the MFA2 promoter and the top strand of the RsaI-BglII fragment (+61 to +476) in the HMRa1 locus. The primers used were for detection of CPDs in the bottom strand of the MFA2 promoter, 5′biotin-GATAGCTTTTTTCCCTCATCTATTTTCTCGGAAAACTTGGTG3′; for detection of CPDs in the top strand of the MFA2 promoter, 5′biotin-GATAGCTTTTTTCCCTTGATTATATAGATTGTCTTTCTTTTCAGAGGAT3′; for detection of CPDs in the top strand of HMRa1, 5′biotin-GTAAGCTTTTTTTCATACGTTTATTTATGAACTACAAATTGT3′. Autoradiographs were obtained after scanning gels with a Typhoon Trio (GE Healthcare Life Sciences), and the signal for each band was quantified using ImageQuant 5.0 software. The repair rate was presented as the time needed to repair 50% of the initial damage (T50%). Bands close to each other in the gel and having the same repair rate were treated as one group. The data plotted in the graphs represent the average from at least three independent experiments. The statistical analysis to compare the repair rate between different strains was carried out using the student’s t-test.
MNase sensitivity assay
Chromatin extraction and MNase treatment were performed as described previously (52). In brief, 400 ml of cells in 5 ml of lysis solution (1 M sorbitol, 5 mM β-mercaptoethanol) was treated with Zymolyase-20T (ICN Biochemicals) at a concentration of 25 mg per 1 g of wet cells for 30 min at 30°C. The spheroplasts were collected and lysed in 7 ml Ficoll solution (18% w/v Ficoll, 20 mM KH2PO4, pH 6.8, 1 mM MgCl2, 0.25 mM EGTA and 0.25 mM EDTA) by one stroke through a syringe. The nuclear pellets were collected, washed and resuspended in 1 ml of digestion buffer (15 mM Tris–HCl, pH 7.4, 75 mM NaCl, 3 mM MgCl2, 1.5 mM CaCl2, 1 mM β-mercaptoethanol). Two hundred fifty microlitres aliquots were treated with 0, 5, 15, 30 and 50 U of MNase (Nuclease S7, Roche) for 10 min at 37°C. The reaction was terminated by addition of 30 μl of stop solution to final concentrations of 1% SDS and 5 mM EDTA, respectively. DNA was purified, and digested with HaeIII restriction enzyme. The detailed labelling procedures were as described previously (52). The primers used were the same as in the section of ‘analysis of CPDs at nucleotide resolution’ (see earlier section).
Quantitative western blotting
Fifty millilitres of cells (2 × 107 cells/ml) were lysed by beating with glass beads (425∼600 μm, Sigma) in 0.5 ml of dialysis buffer (20 mM HEPES-KOH, 10 mM MgSO4, 10 mM EGTA, 20% glycerol and 5 mM dithiothreitol, 5 μl of protease inhibitor cocktail [Sigma UK, Cat No. P8215]). The supernatant was collected for western blotting. Proteins were separated by 6% polyacrylamide–SDS gel electrophoresis. After transferring to the nitrocellulose membrane, the blots were probed with anti-Myc (9B11, Cell signalling, 2276) and anti-beta Actin (Abcam, ab8224) antibodies before with Cy5 conjugated anti-mouse IgG secondary antibodies (GE Healthcare Life Science, PA45009). Membranes were scanned via a Typhoon Trio (GE Healthcare) and images were quantified by ImageQuant software.
Slot blot assay to analyse CPD levels
One hundred nanograms of DNA was denatured by adding NaOH to a final concentration of 0.4 M, and then transferred to a GeneScreen Plus nylon-based membrane via a slot-blot transfer apparatus (Bio-Dot Microfilitration Apparatus, Bio-Rad). The membrane was then probed with an antibody specific for CPDs (Anti-Thymine Dimer Clone KTM53, Kamiya Biomedical Company, Seattle). The detection was achieved via an ECF detection system (ECF western blotting reagent pack, GE Healthcare Life Sciences). The image was scanned with the Typhoon Trio (GE Healthcare Life Sciences), and quantified by ImageQuant 5.0 software.
RESULTS
Cells lacking the HTZ1 and SWR1 genes show increased UV sensitivity and slower removal of CPDs from the overall genome
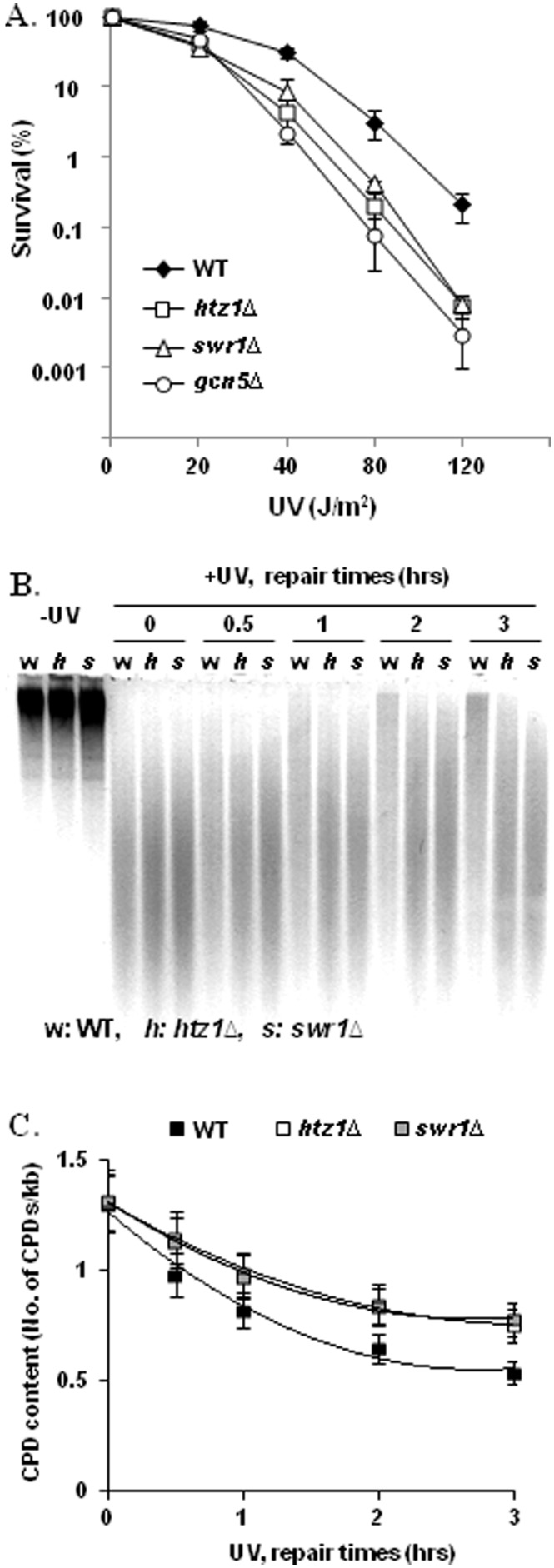
Htz1 is incorporated into the nucleosomes by the SWR complex of which Swr1 is the catalytic subunit (10–12). Both of the htz1Δ and swr1Δ cells are sensitive to the DNA-damaging agent methyl methanesulfonate, which induces DNA strand breaks (12,22). We first tested whether deletion of HTZ1 or SWR1 will result in increased sensitivity to UV irradiation. We also included the gcn5Δ mutant for comparison. The gcn5Δ mutant is moderately sensitive to UV, and Gcn5 controls UV-induced histone H3 acetylation in the MFA2 promoter, which is required for efficient repair of CPDs via NER (40,53). As shown in Figure 1A, both the htz1Δ and swr1Δ cells are indeed more sensitive to UV than the wild type. This prompted us to further investigate whether the increased UV sensitivity in the htz1Δ and swr1Δ mutant cells is related to less-efficient repair of UV-induced CPDs via NER. We used a gel-based assay to assess the repair of CPDs via NER in the total genomic DNA. Genomic DNA from samples taken before and at various times after UV irradiation (100 J/m2) was treated with a CPD-specific endonuclease and subjected to agarose gel electrophoresis under denaturing conditions. Recognition and then cleavage of damaged DNA at CPD sites by the enzyme generates fragmented DNA. The repair of CPDs in DNA by NER restores DNA to longer fragments that run more slowly in the gel. Figure 1B shows a representative gel examining the restoration of DNA towards longer fragments due to NER during a 3 h repair period; it indicates that the restoration of high molecular weight DNA occurs more slowly in the htz1Δ and swr1Δ mutants, compared with that in the wild type. The gel was also densitometrically scanned, and the CPD content in each sample was calculated based on a method described by Sutherland and Shih (49) and Bespalov et al. (50). The quantitative results (Figure 1C) show that the htz1Δ and swr1Δ mutants have more CPDs in their genome than the wild type does after 3 h repair time, indicating deletion of HTZ1 or SWR1 results in a defect in the repair of UV-induced CPDs from a considerable portion of the genome.
Figure 1.
Htz1 is required for cell survival after UV and global CPD repair in the overall genome. (A) The UV sensitivity of wild-type cells and of cells lacking the HTZ1, SWR1 or GCN5 genes. Data are the average of at least three independent experiments ± SD. (B) Repair of CPDs from the genome. DNA from samples taken before and after UV (100 J/m2) was treated with a CPD-specific endonuclease. The digestion products were then separated by gel electrophoresis under denaturing conditions and visualized with EtBr. Here, 0, 0.5, 1, 2 and 3 indicate repair times (hours). (C) CPD content in genomic DNA. The gel in (B) was scanned using a Typhoon Trio (GE Healthcare Life Sciences), and the median migrating distances of the DNA smear were calculated using ImageQuant 5.0. CPD content (CPDs/kb) was calculated as described by Sutherland and Shih (49) and Bespalov et al. (50). The data were fitted to a second-order polynomial. Values represent the mean ± SD of three independent experiments.
Acetylation of Htz1 at K3, 8, 10, 14 does not contribute to UV survival and CPD repair
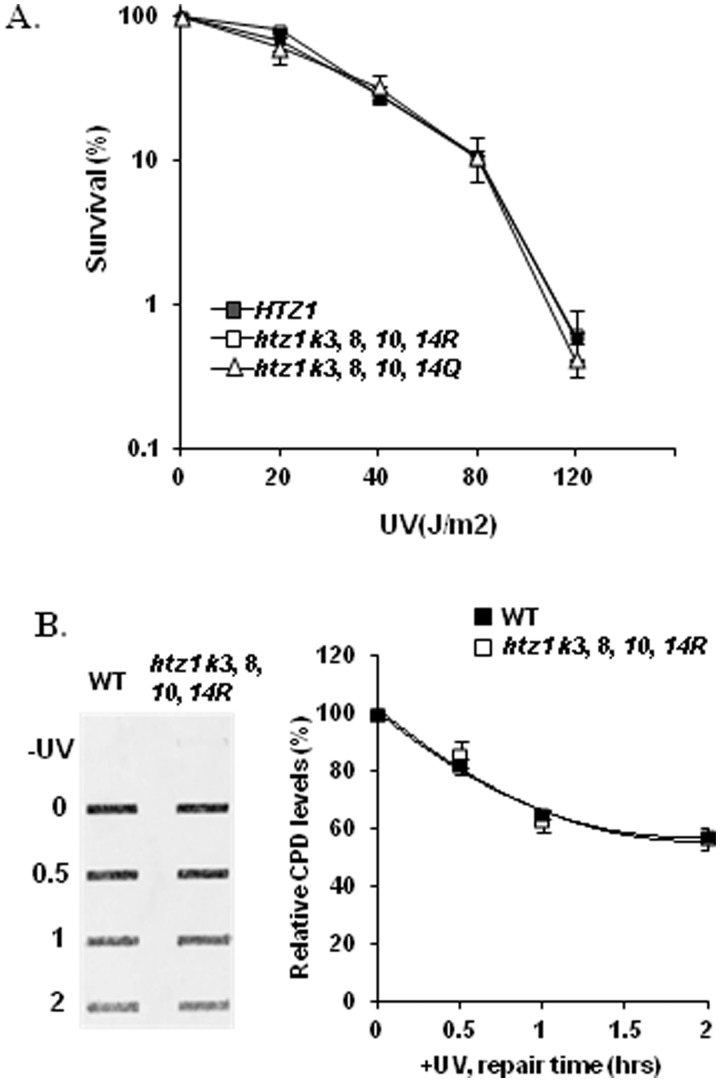
Both Htz1 and Gcn5 are important for cell survival after UV irradiation. As removal of Gcn5 also affects the acetylation of Htz1 (23,24), one explanation could be that acetylation of Htz1 is important for cell survival and CPD repair after UV irradiation. To test this, we generated yeast centromeric plasmids carrying wild-type HTZ1; htz1 K3, 8, 10, 14R; or htz1 K3, 8, 10, 14Q alleles, and expressed them in the htz1Δ strain. In the htz1 K3, 8, 10, 14R plasmid, the four acetylatable lysines are replaced by arginines (R), which mimic the unacetylated state, whereas in the htz1 K3, 8, 10, 14Q plasmid, the four lysines were mutated to glutamines (Q), which mimic acetylated lysines. As indicated in Figure 2A, strains with these three plasmids showed the same sensitivity to UV. We also measured the content of CPDs in genomic DNA over a period of 2 h after UV treatment in cells harbouring wild-type HTZ1 and htz1 K3, 8, 10, 14R plasmids by a slot-blot assay with antibodies against CPDs. The result indicates that there is no difference in CPD content between cells with wild-type HTZ1 and the mutated htz1 K3, 8, 10, 14R (Figure 2B). To ensure that the phenomenon we observed with these strains is not due to plasmid expression, we tested cell survival and CPD repair from the overall genome after UV irradiation in a different wild type (W303) and its isogenic mutant in which all the four lysines of the endogenous Htz1 were mutated to arginines (R). These two strains again showed the same survival rate and repair of CPDs after UV irradiation (Supplementary Figure S1). Therefore, we conclude that acetylation at these sites does not contribute to CPD repair by NER and cell survival after UV irradiation.
Figure 2.
Htz1 acetylation at sites K3, 8, 10, 14 is not required for UV survival and CPD repair. (A) The sensitivity to UV irradiation of strains with wild-type HTZ1 or htz1 alleles in which the 4 N-terminal acetylation sites have been substituted by arginine or glutamine residues. Data are the average of at least three independent experiments ± SD. (B) CPD levels in the genome were detected by the slot-blot assay. DNA from samples taken after UV (100 J/m2) was probed with CPD-specific antibodies. Indicated on the left side of the image are the repair times in hours. CPD levels are presented relative to the amount in samples taken immediately after UV irradiation. Data were the average of at least three independent experiments ± SD, and fitted to a second-order polynomial.
The absence of Htz1 reduces NER at the repressed MFA2 promoter, but not at the HMRa1 locus
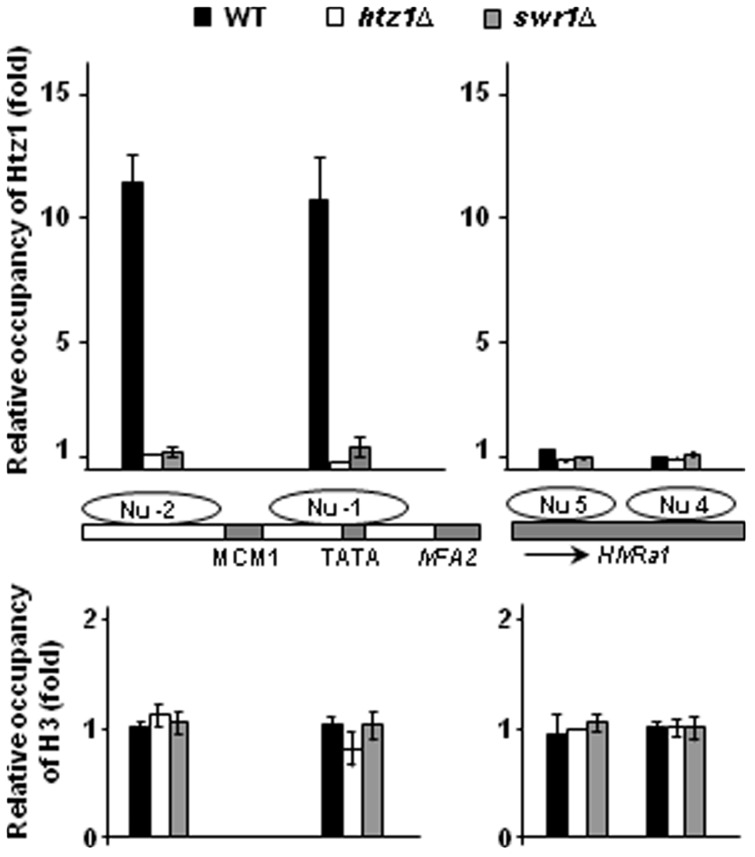
The data thus far show that Htz1, but not its acetylation, is required for NER. The results of the global repair assay in Figure 1B and C could either indicate a partial defect in the capability of the whole NER machinery or a defect in the repair of UV-induced CPDs after UV irradiation in certain regions, e.g. the Htz1 bearing nucleosomes, of the genome. Therefore, we further investigated the repair of CPDs at specific genetic loci that are either enriched or depleted for Htz1. To identify and select these, we examined available genome-wide data of Htz1 occupancy (20), and we chose two loci for further investigation: the MFA2 promoter (nucleosome -2 and -1) (52) and the inner sequence (nucleosome 4 and 5) in the HMRa1 locus (54). Both MFA2 and HMRa1 are transcriptionally inactive in our strains (α mating type), and have well-positioned nucleosomes. Mononucleosome ChIP using anti-Htz1 antibodies confirmed that Htz1 occupies the two nucleosomes in the repressed MFA2 promoter, but it is absent from the HMRa1 locus in wild-type cells bearing a functional Htz1 gene (Figure 3). As expected, deletion of SWR1 reduces the presence of Htz1 in the repressed MFA2 promoter to almost the level of that in the htz1Δ mutant cells. The occupancy of histone H3 remains the same in MFA2 and HMRa1, and deletion of HTZ1 and SWR1 does not influence this.
Figure 3.
Htz1 occupies the MFA2 promoter, but not the HMRa1 inner sequence, in a Swr1-dependent manner. Mononucleosome ChIP analysis of Htz1 and H3 occupancy was performed using anti-Htz1 and anti-H3 antibodies. Gene structures are illustrated in the middle of the graph. Shaded areas are binding sites for regulatory factors and gene bodies. Htz1 occupancy is presented as the relative levels to that in the htz1Δ mutant, and H3 levels were normalized against that at Nu 5 in the HMRa1 locus in the htz1Δ mutant. Data are the average of at least three independent experiments ± SD.
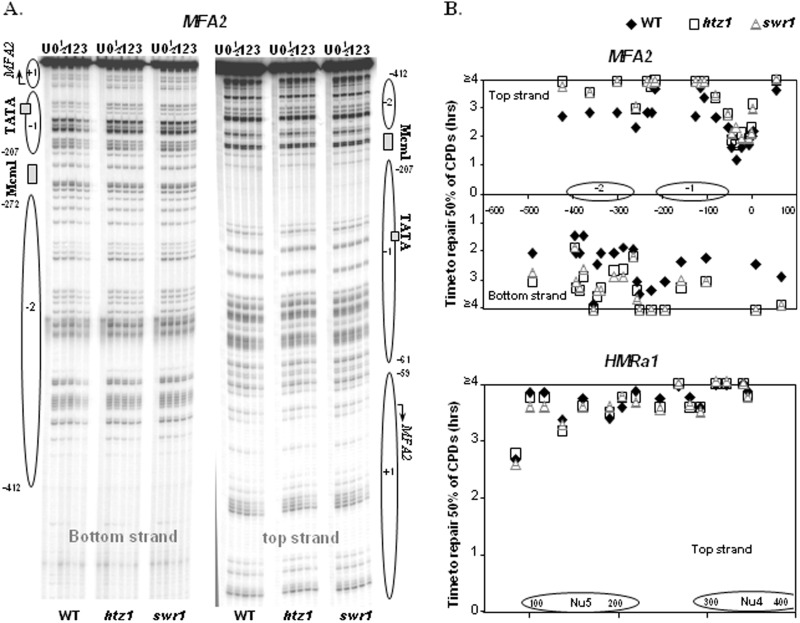
We next investigated how deletion of HTZ1 and SWR1 affects the repair of CPDs after UV irradiation in the MFA2 and the HMRa1 loci. Typical gels for the high resolution CPD repair analysis in the MFA2 promoter are shown in Figure 4A. The gels for CPD repair in the HMRa1 locus are shown in Supplementary Figure S2. Repair efficiency is quantitatively presented in Figure 4B as the time needed to remove 50% of initial CPDs (T50%) at specific sites. In the MFA2 promoter, the repair of CPDs is significantly reduced at almost all sites across the two original Htz1-bearing nucleosome region, including the linker, in the htz1Δ and swr1Δ mutant cells compared with that in the wild-type cells (t-test, WT versus htz1Δ, P = 2.39 × 10−5, WT versus swr1Δ, P = 8.44 × 10−5). There is no evidence that the repair difference only occurs in certain regions within the two nucleosomes. Meanwhile, the repair of CPDs in these two nucleosomes in the htz1Δ and swr1Δ mutant cells are similar (t-test, P = 0.71). However, in the HMRa1 locus where we analysed the repair in one strand, the repair of CPDs in the original non-Htz1 nucleosomes is equally efficient in all of these three strains (t-test, WT versus htz1Δ, P = 0.31; WT versus swr1Δ, P = 0.33; htz1Δ versus swr1Δ, P = 0.54). This clearly indicates that deletion of HTZ1 or SWR1 influences the repair of CPDs in the Htz1-containing nucleosomes in the MFA2 promoter, but not in the non-Htz1 nucleosomes in the HMRa1 locus. This cannot be attributed to a down-regulation of NER because repair of CPDs at HMRa1 remains unchanged in htz1Δ cells. Therefore, Htz1 in the nucleosomes in wild-type cells has a direct effect on NER at the MFA2 promoter.
Figure 4.
Deletion of HTZ1 or SWR1 reduces the repair of CPDs in the MFA2 promoter. (A) Gels depicting CPDs in the top and bottom strands of a HaeIII restriction fragment (−516 to +83) in the MFA2 promoter after a UV dose of 100 J/m2. Lane U is DNA from mock-irradiated cells, while 0, 1/2, 1, 2 and 3 are DNA from irradiated cells after 0, 1/2, 1, 2 and 3 h repair, respectively. Alongside the gels are symbols representing nucleosome positions at MFA2. Nucleotide positions are allocated in relation to the MFA2 start codon. (B) Time to remove 50% of the initial CPDs (T50%) at given sites. T50% of a single CPD or a group of CPDs with a similar repair rate was calculated (T50% < 3 h) or extrapolated (T50% > 3 h). The T50% of slowly repaired CPDs (≥4 h) was shown at the same level (≥4 h) on the graph. Data are the average of three to five independent experiments.
Depletion of HTZ1 does not change the MNase sensitivity of DNA in chromatin in the MFA2 promoter
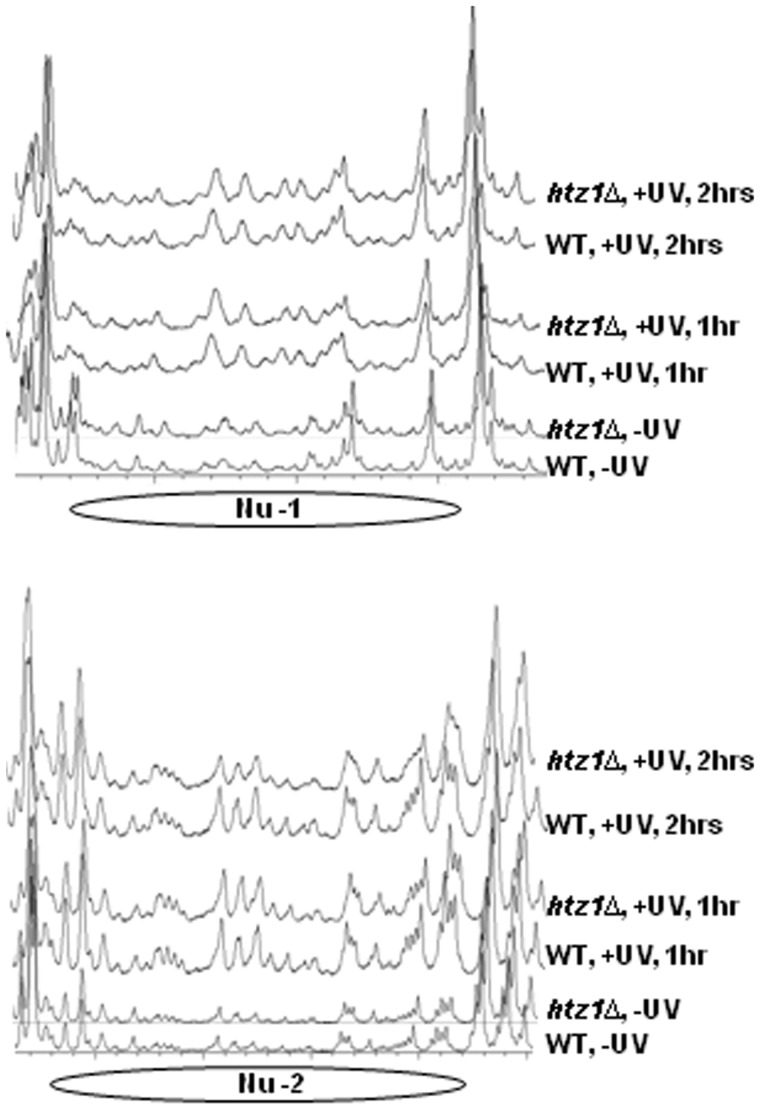
Incorporation of Htz1 into the nucleosomes could potentially change their stability, and also their susceptibility to chromatin remodelling when required. Both the default chromatin structure and chromatin remodelling after UV have a significant influence on CPD repair in the MFA2 promoter (40,43). To address whether deletion of HTZ1 results in any changes in chromatin accessibility in the MFA2 promoter, and whether the deletion influences chromatin remodelling at this locus after UV treatment, we used a high-resolution nucleosome mapping approach (52) to examine the susceptibility of DNA in chromatin in the MFA2 promoter to MNase digestion in wild-type and the htz1Δ cells. This method is capable of assessing the sensitivity to MNase digestion of every site in a DNA sequence of interest in chromatin, and it has single nucleotide resolution. All gels are presented in Supplementary Figure S3. We scanned these gels to produce graphs of MNase sensitivity at individual cutting sites within the chromatin, and presented them in Figure 5. These graphs reveal that MNase digestion in the MFA2 promoter is almost identical between chromatin from wild-type cells and the htz1Δ cells; firstly, wild-type and the htz1Δ cells have the same nucleosome positioning patterns in the MFA2 promoter; secondly, DNA within the two original Htz1 nucleosomes in the MFA2 promoter is similarly sensitive to MNase in these two strains under normal growth condition; thirdly, DNA within the two nucleosomes in these two strains has similar degrees of increase in MNase sensitivity after UV irradiation (Figure 5). We observed the same extra peaks and peak heights within the nucleosomes in UV 1 h and UV 2 h samples irrespective of the presence of Htz1. Thus, we conclude that deletion of HTZ1 does not measurably change the accessibility of the nucleosomal DNA in the MFA2 promoter, nor does it influence the kinetics of chromatin becoming more sensitive to MNase due to chromatin remodelling in this region after UV treatment.
Figure 5.
Deletion of HTZ1 does not change the relative sensitivity of the nucleosomal DNA to MNase in the MFA2 promoter. The graphs were obtained after scanning the lanes with 5 U of MNase in gels for the top and bottom strands of MFA2 as shown in Supplementary Figure S3. The peaks indicate MNase-sensitive sites, which increase in size and frequency after UV treatment but are not different between wild-type and the htz1Δ cells. This is a representative of at least three independent experiments.
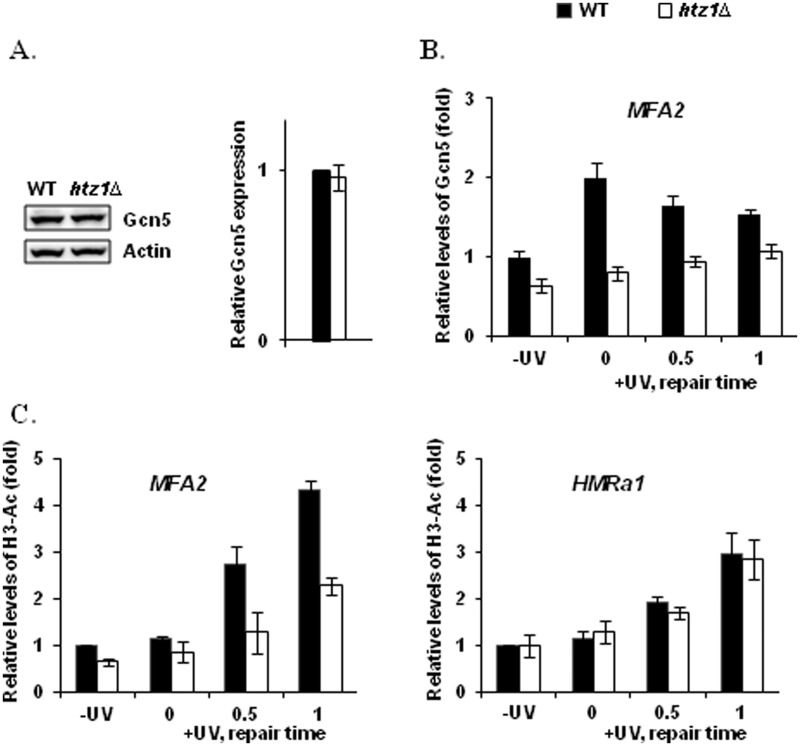
Htz1 promotes the occupancy of Gcn5 and histone H3 hyperacetylation in Htz1 nucleosomes in the MFA2 promoter after UV irradiation
Our data on MNase sensitivity and repair indicate that deletion of HTZ1 reduces the efficiency in repair of CPDs in the Htz1-containing nucleosomes in the MFA2 promoter without causing major detectable changes in chromatin accessibility both before and after UV treatment. This is similar to what we observed with the gcn5Δ strain that also exhibits reduced NER at the repressed MFA2 gene (40). In our previous reports (43) we showed that Rad7 and Rad16 proteins mediate the increased occupancy of the histone acetyltransferase Gcn5 on the nucleosomes at the MFA2 promoter after UV irradiation, and this leads to histone hyperacetylation at H3K9/K14 which is required for efficient repair of UV-induced CPDs (40,43). Hence, we now focused our investigation on how Htz1 influences the occupancy of Gcn5 on chromatin and histone H3 acetylation by performing ChIP assays. We first measured the total expression level of Gcn5 protein in wild-type and the htz1Δ mutant by quantitative western blot. Our data indicated that the expression levels of Gcn5 remain the same in these two strains (Figure 6A). For ChIP assays, to ensure efficient amplification by PCR of DNA with UV-induced damage, we included a repair procedure in which DNA damage in all immunoprecipitated DNA was removed by a mixture of DNA repair enzymes (see ‘Materials and Methods’ section) before PCR. The levels of Gcn5 occupancy at the two nucleosomes in the MFA2 promoter were normalized against those in the HMRa1 locus. This reflects the binding of Gcn5 at MFA2 in relation to that at HMRa1. The histone H3 acetylation levels at MFA2 and HMRa1 were normalized against the H3 levels at these loci, respectively. At MFA2, we observed an increase in the occupancy of Gcn5 in the wild-type cells after UV treatment as we did previously [Figure 6B (43)]. Deletion of HTZ1 results in a reduction in the occupancy of Gcn5 at this locus both before and after UV treatment when compared with that at the HMRa1 locus. Although the levels of Gcn5 occupancy are slightly increased in the htz1Δ cells after UV treatment, they are still significantly lower than that seen in wild type (Figure 6B). In relation to this, the htz1Δ mutant cells exhibit a lower constitutive level of histone H3K9/K14 acetylation (0.65 ± 0.07 in htz1Δ versus 1 in WT) in the promoter of MFA2 before UV irradiation than in the wild-type cells (Figure 6C). More importantly, the increase in the levels of H3K9/K14 acetylation after UV irradiation in the htz1Δ mutant cells is significantly less than that in the wild-type cells (e.g. 2.28 ± 0.17 fold increase in htz1Δ versus 4.34 ± 0.22 fold increase in WT, 2 h after UV). In the HMRa1 locus where Htz1 is normally absent, an increase in histone H3K9/K14 acetylation after UV treatment was also observed. Deletion of HTZ1 has no influence on the extent of the increase (Figure 6C). To conclude, our data indicate that histone H3 acetylation at K9/K14 is activated at both the MFA2 and HMRa1 loci after UV, and specifically Htz1 in the nucleosomes of the MFA2 promoter promotes the occupancy of the histone acetyltransferase Gcn5 on chromatin and histone H3K9/K14 acetylation on these nucleosomes.
Figure 6.
The occupancy of Gcn5 and induction of H3 acetylation at the MFA2 promoter is partially dependent on Htz1. (A) Gcn5 expression in wild-type and the htz1Δ mutant. Gcn5 was detected with anti-Myc antibodies and Cy5 conjugated anti-mouse IgG secondary antibodies in GCN5-myc strains. The expression levels were normalized against those of Actin. (B) ChIP analysis of the occupancy of Gcn5 was performed with anti-Myc antibodies in GCN5-myc strains. The levels of Gcn5 binding at MFA2 was normalized against those at the HMRa1 locus, and then presented as the fold change relative to the mock irradiated sample in wild type. (C) ChIP analysis of Histone H3 acetylation (H3-Ac) was performed using Ac-H3 (K9, K14) antibodies. The H3 acetylation levels were first normalized against the H3 levels, and then presented as the fold change relative to the mock irradiated sample in wild type. −UV: mock irradiated samples; 0: cells received 100 J/m2 of UV without repair; 0.5 h repair or 1 h repair: cells were irradiated with UV and then were allowed to repair in YPD for the number of hours indicated. Data are the average of at least three independent experiments ± SD.
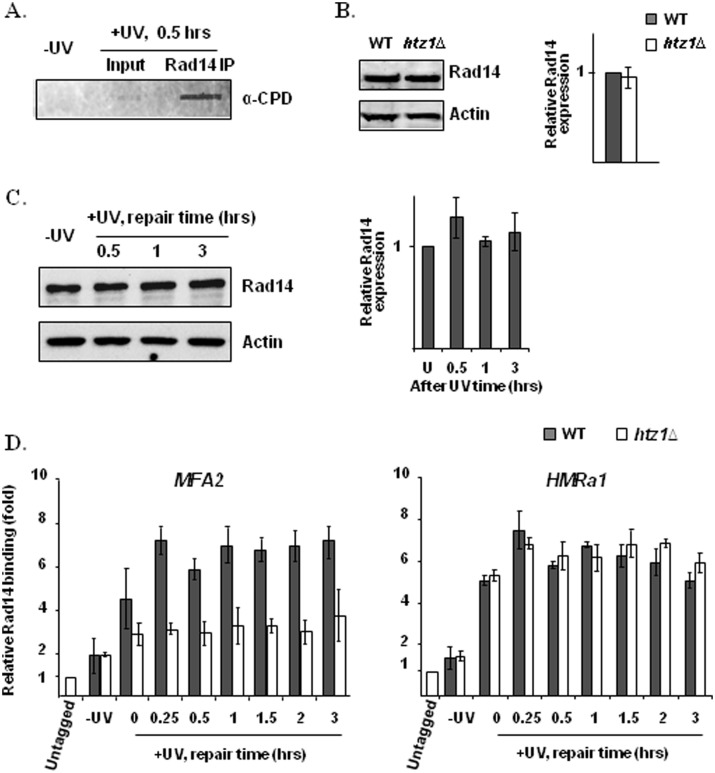
Htz1 promotes the binding of Rad14 to damaged DNA at the MFA2 promoter after UV
The presence of Htz1 in the nucleosomes in the MFA2 promoter helps to promote the increased occupancy of Gcn5 and histone H3K9/K14 acetylation in this region after UV treatment. These events are linked to a more efficient NER. We hypothesized that Htz1-associated histone H3 acetylation status is critical for the binding of core NER factors to damaged DNA in nucleosomes, and this binding decides the rate of lesion removal. To test this, we investigated how deletion of HTZ1 influences the binding of a key NER factor, Rad14, to damaged DNA in the Htz1 nucleosomes at MFA2 after UV. Rad14 is believed to act as a DNA damage recognition factor in the early steps of NER (55). After UV treatment, Rad14 bound DNA showed enriched CPDs (Figure 7A), confirming that Rad14 is preferentially associated with damaged DNA after UV irradiation. Quantitative western blotting showed that deletion of HTZ1 does not change the expression level of Rad14 (Figure 7B), and the expression of Rad14 is unaffected by UV treatment (Figure 7C). However, in ChIP assays in which the immunoprecipitated DNA was again treated with the DNA repair enzyme kit before PCR, we observed a significant reduction in Rad14 binding to the original Htz1 nucleosomes at the MFA2 promoter in the htz1Δ cells compared with that in the wild-type cells, while the binding of Rad14 at the HMRa1 locus was not affected (Figure 7D). This result suggests a positive function for Htz1 in Htz1-containing nucleosomes to promote the binding of Rad14 to damaged DNA in chromatin after UV irradiation. We also noticed that Rad14 binds to both the MFA2 promoter and the HMRa1 inner sequence even before UV treatment (Figure 7D, comparing −UV samples with untagged samples), but this binding is intensified after UV irradiation. The increased binding of Rad14 to damaged DNA occurs so fast that we observed a significant increase in Rad14 binding to both loci immediately after UV irradiation (Figure 7D, 0 repair time). We did not observe any reduction of Rad14 binding to both loci over the repair time up to 3 h. At the UV dose used, there is still DNA damage that needs to be repaired after 3 h, and therefore the continued enhanced binding of Rad14 to DNA is expected.
Figure 7.
Rad14 binding to damaged DNA after UV treatment is enhanced by Htz1. Rad14 immunoprecipitation was performed with anti-Myc antibodies in RAD14-myc strains. Quantitative expression of Rad14 was determined with anti-Myc antibodies and Cy5 conjugated anti-mouse IgG secondary antibodies in RAD14-myc strains. (A) CPD content in DNA (10 ng) from UV mock treated, UV treated and Rad14 immunoprecipitated samples, as detected by the slot-blot assay. The amount of DNA was measured by a Nanodrop. (B) Rad14 expression in wild-type and the htz1Δ cells. (C) Rad14 expression in wild-type cells before and after UV irradiation. The quantitative levels of Rad14 expression in (B) and (C) were normalized against those of Actin. (D) The binding of Rad14 to the MFA2 promoter and the HMRa1 inner sequence. In (D), the level of Rad14 binding is present as the fold change relative to those from the untagged strain. Quantitative data are the average of at least three independent experiments ± SD.
DISCUSSION
Emerging evidence suggests that efficient repair of DNA damage in the chromatin environment requires histone modifications and chromatin remodelling (56,57). Our previous studies, using the S. cerevisiae MFA2 gene as an example, revealed that histone H3 acetylation and chromatin remodelling are activated after UV irradiation, and both events are required for NER to efficiently process UV-induced CPDs in chromatin (40). The UV-stimulated H3 acetylation is achieved via increased occupancy of Gcn5 at MFA2, which is regulated by the global genome NER (GG-NER) complex Rad7/Rad16/Abf1 (42,43). These data have shed considerable insight in how GG-NER operates on repressed chromatin, and they have led us to propose a model to explain how chromatin dynamics are regulated to permit NER (43). In the absence of UV histone H3 acetylation is maintained at a basal level, and chromatin remains in a closed structure at the promoter of MFA2. After UV irradiation, Rad7 and Rad16 promote the occupancy of Gcn5 at this promoter, resulting in elevated levels of histone H3 acetylation. This promotes an open chromatin structure at MFA2. The GG–NER complex has DNA translocase activity, but, unlike some SWI/SNF superfamily complexes, it is unable to slide nucleosomes (58). This is critical, as it prevents transcription initiation sites being exposed to transcription initiation factors during the process of repair. In this way, the GG–NER complex can promote specific changes in chromatin structure required for efficient GG-NER, while preventing unregulated gene transcription at the same time.
New features in chromatin structure and their implications in the relevant biological events have been rapidly identified. H2A.Z (Htz1 in S. cerevisiae) nucleosomes are found in many organisms, and these nucleosomes have particular functions that cannot be replaced by canonical H2A nucleosomes (59). Here, we found that deletion of HTZ1 has a significant influence on the repair of UV-induced CPDs. By performing the same analysis in the Htz1-containing nucleosomes at the MFA2 promoter and the non-Htz1-containing nucleosomes in the HMRa1 locus, our experiments revealed that this defect is directly related to the absence of Htz1 in the nucleosomes. This implicates intrinsic properties of Htz1 nucleosomes in facilitating more efficient NER in these nucleosomes in wild-type cells. The positive functions of Htz1 nucleosomes influence NER in two ways. First, Htz1 promotes the occupancy of the histone acetyltransferase Gcn5 and therefore the UV-induced histone H3 acetylation in the Htz1-containing nucleosomes at MFA2. Second, Htz1 promotes the efficient binding of Rad14 to damaged DNA in the Htz1-bearing nucleosomes, thereby promoting NER after UV irradiation. These events are not mutually exclusive, and it is likely that the former enables the latter. This study has revealed an additional layer of information in chromatin at the MFA2 promoter that cells use to regulate chromatin dynamics to ensure efficient DNA damage repair after UV irradiation.
Htz1 promotes NER in the Htz1-containing nucleosomes by either inherently influencing nucleosome stability or by its specific residues serving as modification sites for the binding of other molecules. Acetylation of Htz1 at one or more sites at lysines 3, 8, 10 and 14 is important for multiple functions of Htz1 (22–25,27). However, our data indicate that acetylation at these sites plays no roles in enabling more efficient NER and in influencing cell survival after UV. Others have also reported that point mutations at these sites do not recapitulate the sensitivity to the DNA-damaging agent methyl methanesulfonate that one sees with the htz1Δ mutant (22,23). This indicates that when cells encounter DNA-damaging agents, Htz1 has more functions in maintaining genome stability than those related to its N-terminal acetylation.
Incorporation of Htz1 into the nucleosomes to replace canonical histone H2A could possibly affect the nucleosome stability, higher order chromatin structure and their susceptibility to chromatin remodelling. However, available data failed to reach an exclusive conclusion regarding these issues. Various in vitro and in vivo studies gave contradictory results, with evidence being reported that the H2A.Z nucleosome is either more (18,60–62) or less stable (20,63,64) than the canonical H2A nucleosome. Our high-resolution MNase assay, which is capable of assessing the sensitivity of many sites in chromatin to MNase in vivo revealed that deletion of HTZ1 has no effect on the accessibility of nucleosomal DNA to MNase in the original Htz1 nucleosomes at the repressed MFA2 promoter. This is consistent with the available studies regarding the effect of Htz1 on chromatin structure (18,65). In addition, our assay also revealed that, with or without Htz1, these nucleosomes at MFA2 all become more sensitive to MNase and to similar extents in response to UV irradiation. This suggests that using this assay Htz1 has no influence on chromatin remodelling in this region after UV irradiation. Hence the positive function of Htz1 in NER cannot be simply explained by the general stability of the nucleosomes.
On the other hand, our data indicate that these Htz1 nucleosomes display a unique feature in that Htz1 promotes the binding of Gcn5 to chromatin and hence histone H3 hyperacetylation in these nucleosomes after UV irradiation, and this acetylation is required for efficient NER (40). For NER to process DNA damage in repressive chromatin, for example, in the repressed MFA2 promoter, NER proteins need to get access to DNA damage in chromatin. This may be facilitated by histone H3 acetylation and chromatin remodelling. Our Rad14 binding data showed that deletion of HTZ1 reduces the binding of Rad14 to damaged DNA by 50% after UV in the original Htz1 nucleosomes in the MFA2 promoter, but not in the HMRa1 locus where Htz1 is absent in wild-type cells with a functional HTZ1 gene. This is an indication of a unique function of Htz1 in the nucleosomes in promoting the binding of an important repair protein, Rad14, to damaged DNA in the nucleosomes after UV. It is directly reflected in the reduction in NER at MFA2 in the absence of Htz1. Although Rad14 is a damaged DNA-binding protein (55) and there is no evidence so far indicating Rad14 interacts with histones, the presence of Htz1 may produce a favourable environment after UV, for example, through helping achieve an optimal level of histone H3 acetylation, to facilitate the enhanced binding of Rad14 to damaged DNA. Recently, two studies revealed a similar phenomenon in mammalian cells in processes other than NER (66,67). That is, H2A.Z promotes histone H4 acetylation and in combination with H4 acetylation, Htz1 promotes the recruitment of downstream effector proteins to chromatin. These events occur during transcription activation (66) and DNA double strand break repair (67). Taking all these data together, it suggests that this function of Htz1 (H2A.Z) may be conserved from yeast to humans, and that it has roles in different processes.
Our data revealed an intrinsic property of Htz1 in regulating chromatin modifications to facilitate NER, and this function is confined to Htz1 containing nucleosomes. Htz1 is only present in a portion of the nucleosomes in the yeast genome (20,21). CPDs in the canonical H2A nucleosomes are also repaired after UV. We provided data for one of those regions, namely, the HMRa1 inner sequence, to indicate this. The question as to whether Htz1-containing nucleosomes mark regions for faster repair of CPDs by NER genome-wide can only be answered by entire genome studies using either ChIP-on-chip or ChIP seq approaches, and on which we are currently embarking.
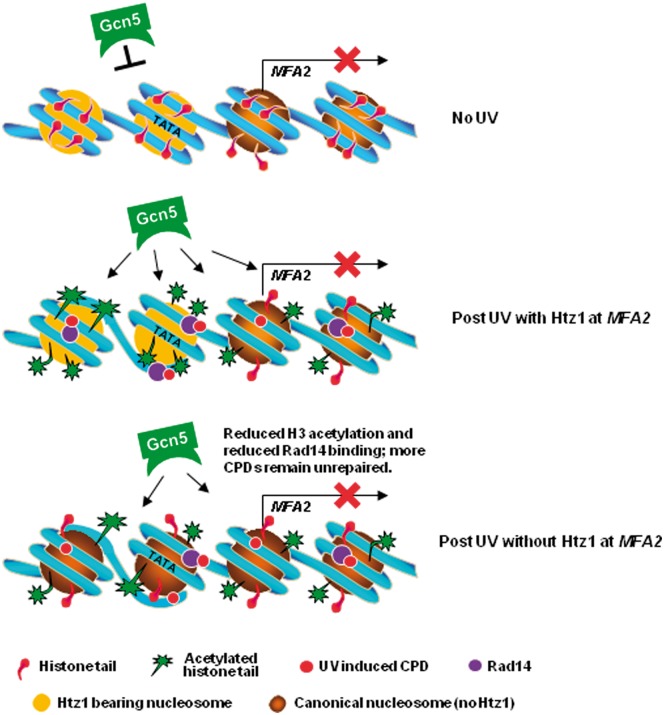
This study, in addition to our previous ones (40,42,43), demonstrates that multiple elements at the nucleosome level in chromatin decide the dynamics of chromatin after UV irradiation at a single genomic location, namely MFA2. They serve to facilitate efficient removal of DNA damage by NER. Based on these data, we extend our model (43) on how chromatin is regulated for efficient repair of UV-induced DNA damage, and summarize it in Figure 8 to include the novel role for Htz1 described in this article. After UV irradiation, Rad7 and Rad16 regulate the occupancy of Gcn5 at the MFA2 promoter to acetylate histone H3. Htz1 in the nucleosomes in this region promotes and maintains increased binding of Gcn5 to the nucleosomes, thus resulting in increased acetylation levels on histone H3. This is critical for the required setting of a specific chromatin structure for efficient NER. The increased levels of H3 acetylation enhance the recruitment of Rad14, and possibly other NER factors to the damage sites. This leads to more efficient lesion removal. Without Htz1 in the nucleosomes the higher acetylation levels on histone H3 cannot be achieved due to a less binding of Gcn5 to these nucleosomes, which results in reduced binding of Rad14 and, perhaps, other repair proteins to damaged DNA. Thereby, the lesion removal is compromised. This function of Htz1 cannot be replaced by canonical histone H2A because NER is compromised in both the htz1Δ strain and in the swr1Δ strain, which cannot deposit Htz1 to replace H2A/H2B in these nucleosomes.
Figure 8.
Model for chromatin remodelling at MFA2 during NER. Top panel. In the absence of UV histone H3 tails remain unacetylated and chromatin remains repressive. Middle Panel. After UV irradiation, the increased occupancy of Gcn5 that is mediated via the Rad7/Rad16 GG-NER complex is promoted by the presence of Htz1. This helps achieve enhanced acetylation levels on histone H3, which are critical for the specific chromatin setting required for optimal NER, so ensuring maximal binding of repair proteins, e.g. Rad14, to damaged DNA. This leads to efficient lesion removal. Lower Panel. Without Htz1 in these nucleosomes, the occupancy of Gcn5 and the acetylation levels on histone H3 are reduced. Therefore, the maximal binding of repair proteins to damaged DNA cannot be achieved and more CPDs remain unrepaired.
NER is an essential DNA repair process that is required for genome stability in many organisms and that its absence in humans is linked to the cancer-prone condition xeroderma pigmentosum. Despite knowing the details of how NER operates on naked DNA in vitro (34) we are still unravelling the nuances of how it operates in the context of chromatin. Here we have demonstrated that its functionality within the yeast genome can be enhanced by the presence of the histone variant Htz1. This is the first evidence that a histone variant impinges on NER and links this histone variant to another important epigenetic event that governs NER efficiency, namely histone H3 acetylation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
UK Medical Research Council Programme Award (to R.W.); Career Development Fellowship from the Wellcome Trust (to C.B.M.). Funding for open access charge: UK Medical Research Council Programme Award.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Bradley R. Cairns, Jasper Rine, James E. Haber and Richard A. Young for providing yeast strains and plasmids, and our colleagues for their intellectual inspiration and technical assistance.
REFERENCES
- 1.Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116:259–272. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 2.Li G, Reinberg D. Chromatin higher-order structures and gene regulation. Curr. Opin. Genet. Dev. 2011;21:175–186. doi: 10.1016/j.gde.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luger K, Dechassa ML, Tremethick DJ. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat. Rev. Mol. Cell. Biol. 2012;13:436–447. doi: 10.1038/nrm3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Cairns BR. Chromatin remodeling complexes: strength in diversity, precision through specialization. Curr. Opin. Genet. Dev. 2005;15:185–190. doi: 10.1016/j.gde.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Henikoff S, Furuyama T, Ahmad K. Histone variants, nucleosome assembly and epigenetic inheritance. Trends Genet. 2004;20:320–326. doi: 10.1016/j.tig.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 7.Talbert PB, Henikoff S. Histone variants–ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell. Biol. 2010;11:264–275. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 8.Jackson JD, Falciano VT, Gorovsky MA. A likely histone H2A.F/Z variant in Saccharomyces cerevisiae. Trends Biochem. Sci. 1996;21:466–467. doi: 10.1016/s0968-0004(96)20028-3. [DOI] [PubMed] [Google Scholar]
- 9.Zlatanova J, Thakar A. H2A.Z: view from the top. Structure. 2008;16:166–179. doi: 10.1016/j.str.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 10.Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 12.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 13.Luk E, Ranjan A, Fitzgerald PC, Mizuguchi G, Huang Y, Wei D, Wu C. Stepwise histone replacement by SWR1 requires dual activation with histone H2A.Z and canonical nucleosome. Cell. 2010;143:725–736. doi: 10.1016/j.cell.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 15.Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 16.Cairns BR. The logic of chromatin architecture and remodeling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 17.Guillemette B, Bataille AR, Gevry N, Adam M, Blanchette M, Robert F, Gaudreau L. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li B, Pattenden SG, Lee D, Gutierrez J, Chen J, Seidel C, Gerton J, Workman JL. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl Acad. Sci. USA. 2005;102:18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5' ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 22.Keogh MC, Mennella TA, Sawa C, Berthelet S, Krogan NJ, Wolek A, Podolny V, Carpenter LR, Greenblatt JF, Baetz K, et al. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes. Dev. 2006;20:660–665. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Babiarz JE, Halley JE, Rine J. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes. Dev. 2006;20:700–710. doi: 10.1101/gad.1386306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Millar CB, Xu F, Zhang K, Grunstein M. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes. Dev. 2006;20:711–722. doi: 10.1101/gad.1395506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halley JE, Kaplan T, Wang AY, Kobor MS, Rine J. Roles for H2A.Z and its acetylation in GAL1 transcription and gene induction, but not GAL1-transcriptional memory. PLoS Biol. 2010;8:e1000401. doi: 10.1371/journal.pbio.1000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wan Y, Saleem RA, Ratushny AV, Roda O, Smith JJ, Lin CH, Chiang JH, Aitchison JD. Role of the histone variant H2A.Z/Htz1p in TBP recruitment, chromatin dynamics, and regulated expression of oleate-responsive genes. Mol. Cell. Biol. 2009;29:2346–2358. doi: 10.1128/MCB.01233-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mehta M, Braberg H, Wang S, Lozsa A, Shales M, Solache A, Krogan NJ, Keogh MC. Individual lysine acetylations on the N terminus of Saccharomyces cerevisiae H2A.Z are highly but not differentially regulated. J. Biol. Chem. 2010;285:39855–39865. doi: 10.1074/jbc.M110.185967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 29.Bandyopadhyay S, Mehta M, Kuo D, Sung MK, Chuang R, Jaehnig EJ, Bodenmiller B, Licon K, Copeland W, Shales M, et al. Rewiring of genetic networks in response to DNA damage. Science. 2010;330:1385–1389. doi: 10.1126/science.1195618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dhillon N, Oki M, Szyjka SJ, Aparicio OM, Kamakaka RT. H2A.Z functions to regulate progression through the cell cycle. Mol. Cell. Biol. 2006;26:489–501. doi: 10.1128/MCB.26.2.489-501.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papamichos-Chronakis M, Krebs JE, Peterson CL. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes. Dev. 2006;20:2437–2449. doi: 10.1101/gad.1440206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Attikum H, Fritsch O, Gasser SM. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 2007;26:4113–4125. doi: 10.1038/sj.emboj.7601835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalocsay M, Hiller NJ, Jentsch S. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol. Cell. 2009;33:335–343. doi: 10.1016/j.molcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 34.Reed SH, Waters R. DNA repair. In: Cooper DN, editor. Nature Encyclopaedia of the Human Genome. London: Nature Publishing Group; 2003. pp. 148–154. [Google Scholar]
- 35.Wellinger RE, Thoma F. Nucleosome structure and positioning modulate nucleotide excision repair in the non-transcribed strand of an active gene. EMBO J. 1997;16:5046–5056. doi: 10.1093/emboj/16.16.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li S, Smerdon MJ. Nucleosome structure and repair of N-methylpurines in the GAL1–10 genes of Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:44651–44659. doi: 10.1074/jbc.M206623200. [DOI] [PubMed] [Google Scholar]
- 37.Powell NG, Ferreiro J, Karabetsou N, Mellor J, Waters R. Transcription, nucleosome positioning and protein binding modulate nucleotide excision repair of the Saccharomyces cerevisiae MET17 promoter. DNA Repair (Amst) 2003;2:375–386. doi: 10.1016/s1568-7864(02)00239-2. [DOI] [PubMed] [Google Scholar]
- 38.Ferreiro JA, Powell NG, Karabetsou N, Kent NA, Mellor J, Waters R. Cbf1p modulates chromatin structure, transcription and repair at the Saccharomyces cerevisiae MET16 locus. Nucleic Acids Res. 2004;32:1617–1626. doi: 10.1093/nar/gkh324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Green CM, Almouzni G. When repair meets chromatin. First in series on chromatin dynamics. EMBO Rep. 2002;3:28–33. doi: 10.1093/embo-reports/kvf005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu Y, Teng Y, Liu H, Reed SH, Waters R. UV irradiation stimulates histone acetylation and chromatin remodeling at a repressed yeast locus. Proc. Natl Acad. Sci. USA. 2005;102:8650–8655. doi: 10.1073/pnas.0501458102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu Y, Waters R. Histone acetylation, chromatin remodeling and nucleotide excision repair: hint from the study on MFA2 in Saccharomyces cerevisiae. Cell Cycle. 2005;4:1043–1045. doi: 10.4161/cc.4.8.1928. [DOI] [PubMed] [Google Scholar]
- 42.Teng Y, Liu H, Gill HW, Yu Y, Waters R, Reed SH. Saccharomyces cerevisiae Rad16 mediates ultraviolet-dependent histone H3 acetylation required for efficient global genome nucleotide-excision repair. EMBO Rep. 2008;9:97–102. doi: 10.1038/sj.embor.7401112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu S, Teng Y, Waters R, Reed SH. How chromatin is remodelled during DNA repair of UV induced DNA damage in Saccharomyces cerevisiae. PLoS Genet. 2011;7:e1002124. doi: 10.1371/journal.pgen.1002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaudhuri S, Wyrick JJ, Smerdon MJ. Histone H3 Lys79 methylation is required for efficient nucleotide excision repair in a silenced locus of Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:1690–1700. doi: 10.1093/nar/gkp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tatum D, Li S. Evidence that the histone methyltransferase Dot1 mediates global genomic repair by methylating histone H3 on lysine 79. J. Biol. Chem. 2011;286:17530–17535. doi: 10.1074/jbc.M111.241570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gong F, Fahy D, Smerdon MJ. Rad4-Rad23 interaction with SWI/SNF links ATP-dependent chromatin remodeling with nucleotide excision repair. Nat. Struct. Mol. Biol. 2006;13:902–907. doi: 10.1038/nsmb1152. [DOI] [PubMed] [Google Scholar]
- 47.Sarkar S, Kiely R, McHugh PJ. The Ino80 chromatin-remodeling complex restores chromatin structure during UV DNA damage repair. J. Cell Biol. 2010;191:1061–1068. doi: 10.1083/jcb.201006178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sutherland BM, Shih AG. Quantitation of pyrimidine dimer contents of non-radioactive deoxyribonucleic acid by electrophoresis in alkaline agarose gels. Biochemistry. 1983;22:745–749. doi: 10.1021/bi00273a006. [DOI] [PubMed] [Google Scholar]
- 50.Bespalov VA, Conconi A, Zhang X, Fahy D, Smerdon MJ. Improved method for measuring the ensemble average of strand breaks in genomic DNA. Environ. Mol. Mutagen. 2001;38:166–174. doi: 10.1002/em.1068. [DOI] [PubMed] [Google Scholar]
- 51.Teng Y, Li S, Waters R, Reed SH. Excision repair at the level of the nucleotide in the Saccharomyces cerevisiae MFA2 gene: mapping of where enhanced repair in the transcribed strand begins or ends and identification of only a partial rad16 requisite for repairing upstream control sequences. J. Mol. Biol. 1997;267:324–337. doi: 10.1006/jmbi.1996.0908. [DOI] [PubMed] [Google Scholar]
- 52.Teng Y, Yu S, Waters R. The mapping of nucleosomes and regulatory protein binding sites at the Saccharomyces cerevisiae MFA2 gene: a high resolution approach. Nucleic Acids Res. 2001;29:E64. doi: 10.1093/nar/29.13.e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Teng Y, Yu Y, Waters R. The Saccharomyces cerevisiae histone acetyltransferase Gcn5 has a role in the photoreactivation and nucleotide excision repair of UV-induced cyclobutane pyrimidine dimers in the MFA2 gene. J. Mol. Biol. 2002;316:489–499. doi: 10.1006/jmbi.2001.5383. [DOI] [PubMed] [Google Scholar]
- 54.Ravindra A, Weiss K, Simpson RT. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating-type locus HMRa. Mol. Cell. Biol. 1999;19:7944–7950. doi: 10.1128/mcb.19.12.7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guzder SN, Sung P, Prakash L, Prakash S. Yeast DNA-repair gene RAD14 encodes a zinc metalloprotein with affinity for ultraviolet-damaged DNA. Proc. Natl Acad. Sci. USA. 1993;90:5433–5437. doi: 10.1073/pnas.90.12.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Czaja W, Mao P, Smerdon MJ. The emerging roles of ATP-dependent chromatin remodeling enzymes in nucleotide excision repair. Int. J. Mol. Sci. 2012;13:11954–11973. doi: 10.3390/ijms130911954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waters R, Evans K, Bennett M, Yu S, Reed S. Nucleotide excision repair in cellular chromatin: studies with yeast from nucleotide to gene to genome. Int. J. Mol. Sci. 2012;13:11141–11164. doi: 10.3390/ijms130911141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu S, Smirnova JB, Friedberg EC, Stillman B, Akiyama M, Owen-Hughes T, Waters R, Reed SH. ABF1-binding sites promote efficient global genome nucleotide excision repair. J. Biol. Chem. 2009;284:966–973. doi: 10.1074/jbc.M806830200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jackson JD, Gorovsky MA. Histone H2A.Z has a conserved function that is distinct from that of the major H2A sequence variants. Nucleic Acids Res. 2000;28:3811–3816. doi: 10.1093/nar/28.19.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park YJ, Dyer PN, Tremethick DJ, Luger K. A new fluorescence resonance energy transfer approach demonstrates that the histone variant H2AZ stabilizes the histone octamer within the nucleosome. J. Biol. Chem. 2004;279:24274–24282. doi: 10.1074/jbc.M313152200. [DOI] [PubMed] [Google Scholar]
- 61.Thambirajah AA, Dryhurst D, Ishibashi T, Li A, Maffey AH, Ausio J. H2A.Z stabilizes chromatin in a way that is dependent on core histone acetylation. J. Biol. Chem. 2006;281:20036–20044. doi: 10.1074/jbc.M601975200. [DOI] [PubMed] [Google Scholar]
- 62.Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat. Struct. Biol. 2000;7:1121–1124. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- 64.Placek BJ, Harrison LN, Villers BM, Gloss LM. The H2A.Z/H2B dimer is unstable compared to the dimer containing the major H2A isoform. Protein Sci. 2005;14:514–522. doi: 10.1110/ps.041026405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morillo-Huesca M, Clemente-Ruiz M, Andújar E, Prado F. The SWR1 histone replacement complex causes genetic instability and genome-wide transcription misregulation in the absence of H2A.Z. PLoS One. 2010;5:e12143. doi: 10.1371/journal.pone.0012143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Draker R, Ng MK, Sarcinella E, Ignatchenko V, Kislinger T, Cheung P. A Combination of H2A.Z and H4 acetylation recruits Brd2 to chromatin during transcriptional activation. PLoS Genet. 2012;8:e1003047. doi: 10.1371/journal.pgen.1003047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol. Cell. 2012;48:723–733. doi: 10.1016/j.molcel.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.