Abstract
Controlled twisting of individual, double-stranded DNA molecules provides a unique method to investigate the enzymes that alter DNA topology. Such twisting requires a single DNA molecule to be torsionally constrained. This constraint is achieved by anchoring the opposite ends of the DNA to two separate surfaces via multiple bonds. The traditional protocol for making such DNA involves a three-way ligation followed by gel purification, a laborious process that often leads to low yield both in the amount of DNA and the fraction of molecules that is torsionally constrained. We developed a simple ligation-free procedure for making torsionally constrained DNA via polymerase chain reaction (PCR). This PCR protocol used two ‘megaprimers’, 400-base-pair long double-stranded DNA that were labelled with either biotin or digoxigenin. We obtained a relatively high yield of gel-purified DNA (∼500 ng/100 µl of PCR reaction). The final construct in this PCR-based method contains only one labelled strand in contrast to the traditional construct in which both strands of the DNA are labelled. Nonetheless, we achieved a high yield (84%) of torsionally constrained DNA when measured using an optical-trap-based DNA-overstretching assay. This protocol significantly simplifies the application and adoption of torsionally constrained assays to a wide range of single-molecule systems.
INTRODUCTION
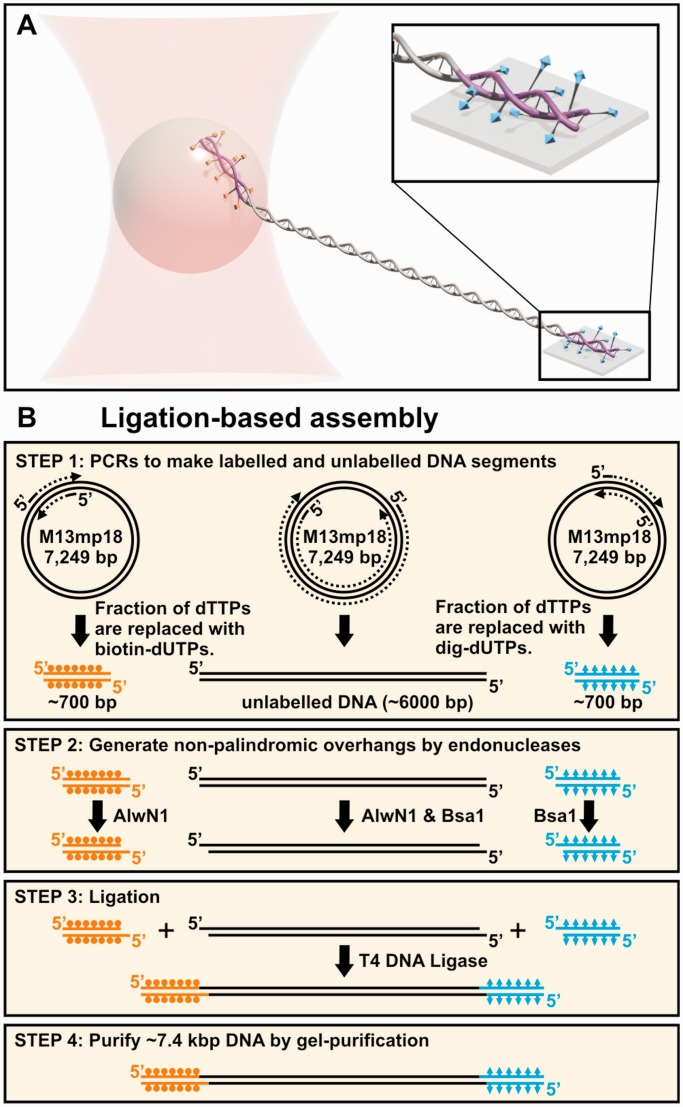
Fundamental processes in DNA metabolism can alter the topology of DNA (1). For instance, duplication of the parent molecule leads to two topologically intertwined daughter molecules (1). To overcome this and other issues in DNA topology, cells have a wide range of enzymes that manipulate the topological state of and the torsional stress within DNA. Traditionally, these enzymes, such as topoisomerase and gyrases, have been studied using a supercoiled plasmid-based assay (2). In the mid-1990s, the development of magnetic tweezers introduced the ability to twist a single DNA molecule (3). These studies initially focused on the polymer physics of supercoiled DNA (4) and, more recently, the mechanochemistry of topoisomerases and gyrases (5–11). Studies of torsionally controlled DNA are not limited to magnetic tweezers; optical traps are now being used to study the torsional properties of DNA (12–15) and its effect on DNA overstretching (16). All of these studies rely on torsionally constrained DNA that, in turn, require DNA that is nick free and anchored to two surfaces via multiple bonds (Figure 1A).
Figure 1.
Ligation-based assembly of DNA constructs for torsionally constrained assays. (A) Schematic of a single-molecule optical-trapping assay where the DNA is multiply labelled in both strands at both ends. Different tags [biotin (orange) and digoxigenin (cyan)] are incorporated into short DNA molecules (purple) that were ligated onto a central unlabelled DNA molecule. (B) An outline of the traditional method for making torsionally constrained DNA. In this method, three different DNA molecules are made by PCR, two of which are labelled with dig and biotin, respectively (Step 1). Restriction digest (Step 2) of all three DNAs facilitates the ligation necessary to assemble the final product (Step 3). Gel purification is necessary to achieve a uniform length product (Step 4).
The basic process for preparing the DNA for these assays has remained essentially unchanged since the invention of magnetic tweezers (Figure 1B), in contrast to the significant innovation in instrumentation and assays using torsionally constrained DNA (17–23). Unfortunately, this standard protocol is inefficient, involving ligating three molecules together to form the final construct. Specifically, the central DNA molecule is unlabelled, whereas the two end molecules are labelled with biotin and digoxigenin (dig), respectively. The labelled end strands are short [500–6000 base pair (bp)] relative to the central DNA molecule [5–48 kb (1.7–16 μm)] (3,24,25). Typically, the DNAs are made using polymerase chain reaction (PCR) with the extent of labelling for the end molecules controlled by the ratio of unlabelled dTTP to labelled dUTP (26). As a result, both strands of these DNA molecules are labelled. Next, to facilitate ligation, two pairs of non-palindromic overhangs are enzymatically introduced into these molecules. Nonetheless, the ligation reaction does not proceed to completion, yielding a mixture of products. Subsequent gel purification yields a uniform product but does not guarantee that both strands of the DNA have been ligated. Nicks in the DNA backbone that compromise the integrity of the substrate may still be present. Moreover, gel purification can introduce nicks into the DNA due to UV irradiation, even when illuminating with 365-nm light (27). As a result of these experimental difficulties, the concentration of the final product is typically low (0.1 ng/μl) (24), with a volume (50 μl). We note that, in our hands, this quantity of DNA is good for only a small number of our optical-trapping assays. Moreover, the percentage of molecules that are torsionally constrained is low, often going unreported. Thus, an improved method to simply and efficiently make torsionally constrained DNA is needed.
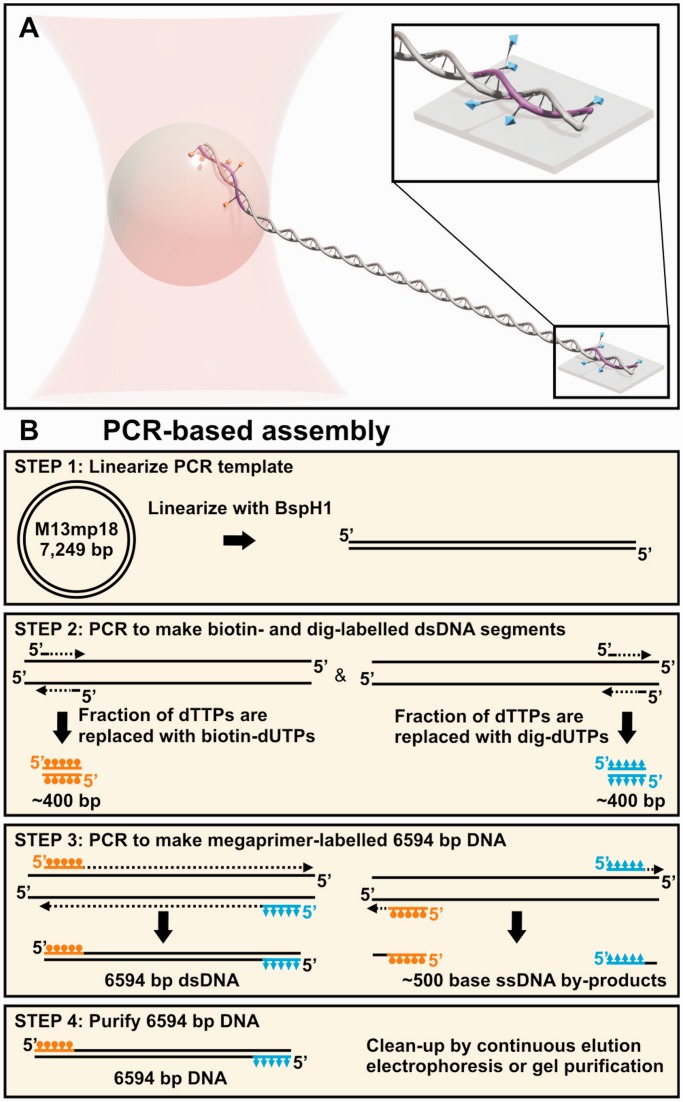
Although the basic steps of the standard method have remained constant, innovations in this process have improved the yield. Most notably, Seol et al. significantly improved the efficiency of the enzymatic digestion and ligation of labelled DNA (25). This method required carefully designing the sequences of the overhang so it did not contain any labels. With this improvement, the researchers increased the fraction of fully ligated molecules and, thereby, attained a 250-fmol yield of product (50 μl at 20 ng/μl). This work also led to a high fraction of torsionally constrained molecules (60%), but still required a long ligation step and gel purification for a uniform length product. Here, we present a simple ligation-free procedure for making torsionally constrained DNA via PCR. This PCR used two PCR-synthesized 400-bp-long double-stranded (ds) DNA ‘megaprimers’ that in our application were labelled with either biotin or digoxigenin. We obtained a relatively high yield of gel-purified DNA [∼120 fmol (500 ng/100 µl of reaction)] with substantially reduced time and labour. A potential concern with this ligation-free protocol is that each end of the DNA has only one labelled strand (Figure 2A). Nonetheless, we achieved a high percentage (∼84%) of torsionally constrained molecules when measured using an optical-trap-based DNA-overstretching assay.
Figure 2.
PCR-based assembly of DNA constructs for torsionally constrained assays. (A) Schematic of a single-molecule optical-trapping assay, where the DNA is multiply labelled on one strand at each end. Different tags [biotin (orange) and digoxigenin (cyan)] are incorporated into the DNA megaprimers (purple, 400-bp) that were used to amplify the target DNA during PCR. (B) An outline of our ligation-free method for making torsionally constrained DNA. Briefly, we linearized the template (Step 1) to suppress the amplification of off-target products. In Step 2, two sets of ∼25-bp ssDNA primers are used in PCR to make the two labelled 400-bp dsDNAs that becomes the megaprimers for the subsequent PCR (Step 3). Finally, the target DNA is gel-purified away from the 400-bp dsDNA megaprimers (Step 4).
MATERIALS AND METHODS
Our ligation-free method used megaprimer PCR (28) to efficiently incorporate different labels at the end of the desired DNA substrate (Figure 2B).
Generation of linearized template DNA
We made a linearized template by a restriction digest (Figure 2B, Step 1). M13mp18 RF vector [New England Biolabs (NEB), Ipswich, MA, USA] was enzymatically digested at a unique BspH1(NEB) site. The reaction was initially purified using the QiaQuick PCR Purification Kit (Qiagen, Valencia, CA, USA) followed by a gel extraction by Quantum Prep Freeze ‘N Squeeze DNA Gel Extraction Spin Columns (Bio-Rad, Hercules, CA, USA) to remove undigested product. Agarose gel extraction was followed by purification with the QiaQuick PCR Purification Kit (Qiagen, Valencia, CA, USA). The final product was eluted into 60 µl of elution buffer [EB: 10 mM Tris-Cl (pH 8.5)]. The resulting linear DNA was used as the template for subsequent PCR reactions.
Megaprimer design and amplification
For our optical-trapping assays, we wanted a relatively short substrate [∼2 μm long (6 kb)] for DNA overstretching studies with high spatiotemporal resolution (16). Prior work with magnetic tweezers has used ∼500–700-bp-long labelled regions to make torsionally constrained substrates (24,25,29). We chose to use shorter labelled regions to facilitate megaprimer-based PCR. To assist in designing primer sequences, we used PrimerQuest from Integrated DNA Technologies (IDT, Coralville, IA, USA), and all the primers were purchased from IDT. As shown in Supplementary Figure S1, the final design yielded a 6594-bp DNA molecule consisting of a 5743-bp central unlabelled section flanked by two ∼400-bp labelled regions. More specifically, two sets of ∼25-nucleotide-long primers were designed to amplifying the ∼400-bp-long megaprimers from the 1607–2024 and 509–912 regions of M13mp18 (Figure 2B, Step 2). We added a 5′-biotin and a 5′-dig label to the final construct using a terminally labelled forward and reverse primer (see Supplementary Table S1 in Supplementary Data for primer sequences and Supplementary Figure S1).
Amplification was carried out in 100 μl of reaction volume using the KOD Hot Start DNA Polymerase kit (Novagen, Darmstadt, Germany) and linearized M13mp18 as the template. We followed the manufacturer’s recommended reaction conditions and cycling parameters except that the extension time was increased by 10 s to allow for incorporation of labelled nucleotides (Supplementary Data). We incorporated labelled dUTPs by replacing dTTPs in the PCR reaction mix (26). Different labelling densities of the megaprimers were explored by changing the ratio of labelled dUTP to dTTP. Our biotin dUTP had a linker that was 4 carbons longer than the dig dUTP (biotin-16-dUTP and dig-11-dUTP, Roche, Indianapolis, IN, USA). The resulting DNA was purified using the QiaQuick PCR Purification Kit and eluted into 60 μl of EB. The DNA concentration was determined by optical density (OD260) measurements using a NanoDrop (Thermo Scientific, Wilmington, DE, USA) assuming the UV absorbance of the labels was negligible. DNA length was confirmed by gel analysis.
Generation of 2.2 -μm-long DNA using labelled megaprimers
We carried out megaprimer PCR (Figure 2B, Step 3) using the megaprimers described earlier in the text. The amplification conditions were based on the manufacturer’s recommended conditions except for using 10 nM of each primer and 36 ng (7.5 fmol) of template instead of the recommended 300 nM and 20 ng, respectively. The PCR product was initially purified using the QiaQuick PCR Purification Kit.
Purification of 2.2 -μm DNA away from megaprimers
The standard PCR clean-up kit removes only short single-stranded DNA (ss) primers (<40 nucleotides), while retaining longer pieces of dsDNA. We separated the desired product from the unincorporated 400-bp megaprimers using one of two different methods: conventional gel purification and continuous elution electrophoresis. The second method was investigated to see if the fraction of molecules that were torsionally constrained increased when exposure to UV light was avoided. Both gels were run under non-denaturating conditions.
For the continuous elution electrophoresis, we used a Mini-Prep Cell Model 491 (Bio-Rad, Hercules, CA, USA). The Mini-Prep Cell is designed to separate either proteins or nucleic acids through a gel run down a cylindrical tube. The separated product runs off the bottom of the gel into an elution chamber. This elution chamber traps the product with a dialysis membrane, and it is then drawn out by a peristaltic pump (Mini-Pump Variable Flow, VWR, Atlanta, GA, USA) and separated into fractions by a fraction collector (Spectra/Chrom CF-2, Spectrum Molecular Separations, Houston, TX, USA). We followed the Mini-Prep Cell instructions and Bio-Rad’s technical note 2203 (‘Preparative electrophoresis: purification of DNA fragments from 2 to 18 kb using the Model 491 Prep Cell’). Specifically, we first poured a 9-cm-long agarose separating gel (0.8%) on which a 1-cm-long agarose stacking gel (0.25%) was placed. Gels were prepared and run in Tris-borate-EDTA (pH 8.3) (45 mM Tris-borate, 1 mM EDTA). Typical separations times for the 6.6-kb DNA was 33 h. Fractions were collected in 30-min intervals with the flow rate set at 18.7 μl/min. The collected DNA was recovered by ethanol precipitation and resuspended in 20 μl of EB. The contents of each fraction were deduced via gel electrophoresis. The fractions containing the desired product were combined, and the final concentration was determined using OD260.
Conventional gel electrophoresis was done using a 0.5% agarose gel in Tris-acetate-EDTA. UV light exposure was limited to <30 s during gel cutting. Samples were recovered by Quantum Prep Freeze ‘N Squeeze DNA Gel Extraction Spin Columns (Bio-Rad), and further purified by QiaQuick PCR Purification Kit (Qiagen). The final product was eluted in 60 μl of EB.
Surface-coupled tethered-bead assay
We used a standard tethered-bead assay for stretching DNA with an optical trap (30). The tethered-bead assay was assembled by attaching the dig-labelled end of the DNA to an anti-dig-coated coverslip and the biotin-labelled end to a streptavidin-coated bead. Our flow cells were constructed from KOH-cleaned coverslips, double sticky tape, 5-min epoxy and microscope slides. The flow cell had an internal volume of ∼15 μl. We deposited anti-dig antibodies onto the KOH-cleaned coverslips by incubating the antibodies [50 μg/ml (Roche Diagnostics, Indianapolis, IN) in 100 mM phosphate buffer (pH 7.5)] in the flow cell for 1 h. Next, the flow cell was washed with ∼600 μl of wash buffer [WB: 20 mM Tris-Cl (pH 7.4), 50 mM NaCl, 0.3% Tween-20 and 3 mg/ml bovine serum albumin (BSA: Sigma, St. Louis, MO, USA)]. We further passivated the surface by incubating with 12 mg/ml BSA for 1 h. In parallel with this process, we prepared bead-DNA complexes by combining streptavidin-coated beads made from carboxyl-functionalized polystyrene beads (diameter = 780 nm, Life Technologies) with DNA at a 5:1 ratio, and incubating for 2 h in conjugation buffer [CB: 20 mM Tris-Cl (pH 7.4), 150 mM NaCl, 0.3% Tween-20 and 3 mg/ml BSA]. Next, we washed the flow cells with 400 μl of WB followed by 200 μl of CB and then introduced 30 μl of bead–DNA complex into the flow cell, and incubated for 1 h. A final wash with 600 μl of WB removed unbound beads. The trapping assay was conducted in 10 mM Tris-Cl (pH 7.4), 150 mM NaCl and 1 mM EDTA buffer at room temperature. Incubations were carried out at room temperature in humidity chambers. All the buffers used in the single-molecule assay were filtered with a 0.2 -μm filter. Additionally, the stock concentrations of BSA solution were also filtered.
Instrument and data acquisition
We used a highly stable optical trap capable of generating forces in excess of 120 pN. The details of this instrument and its calibration (31,32), as well as its use in overstretching assays (16), have been previously described. Briefly, we formed a stiff optical trap (∼0.7 pN/nm) by focusing a 1064-nm laser beam through an objective lens (NA = 1.4). We used a separate 810-nm laser to measure the bead position via back focal plane interferometry (33,34). The resulting data were digitized at 120 kHz with a 60-kHz anti-alias filter. Individual DNA molecules were stretched by holding the trap fixed, while moving the sample using a 3-axis, closed-loop piezo-electric stage (P-517.3 cd, Physik Instrumente, Auburn, MA, USA). The extension and force along the stretching axis were determined using the 2D geometry appropriate for a surface-coupled assay (30). Before overstretching the DNA, we selected for single DNA molecules based on the measured persistence length (p ≈ 45 nm) (35) and contour length (L ≈ 1.95–2.2 μm). This range of L accounted for variation in the number and location of anchor points. Formally, any L between 1.95 and 2.24 µm was possible, but we typically measured L between 2.0 and 2.1 µm. The force-extension records for determining the overstretching force (Fo) were measured by stretching to a predetermined extension at 5 μm/s. The stage was then moved backwards at the same velocity to its original location.
RESULTS
Torsionally constrained single-molecule assays require nick-free DNA constructs with multiple labels at each end. The traditional protocol for generating such DNA uses restriction digests and ligations (Figure 1B). In our present study, we developed a ligation-free PCR-based protocol for generating suitable DNA (Figure 2B). To do so, we used megaprimer PCR, which was originally developed for site-directed mutagenesis (28). Megaprimer PCR starts out with relatively long (∼400 bp) dsDNA molecules that will become the forward and reverse primers for the target DNA. During the high-temperature denaturation step of the PCR, the two different dsDNA molecules dissociate, generating the desired ssDNA primers along with two unused ssDNA molecules. To efficiently and cost-effectively incorporate multiple labels into these 400-bp megaprimers, we made them with PCR by partially replacing dTTP with dig- or bio-labelled dUTP (Figure 2B).
A key difference between the standard protocol and our ligation-free protocol is that the traditional method labels both strands of the DNA at each end, whereas our method labels only one strand (Figures 1A and 2A). Conceptually, one might expect that higher labelling densities would be required to achieve a similar level of torsional constraint when only one strand is labelled. Thus, we investigated the traditional labelling density (one labelled dUTP per nine unlabelled dTTP) (26) and a higher density. The percentage of torsionally constrained molecules is also affected by nicks in the DNA. Thus, we investigated two different methods for purifying the final product: standard gel electrophoresis and continuous gel elution, which avoids exposing the DNA to UV light. To demonstrate reproducibility, we used four separate replicates for this study, where by ‘replicate’ we mean that the DNA constructs were independently prepared from starting materials (Figure 2B, Steps 1–4).
Making labelled megaprimers by PCR
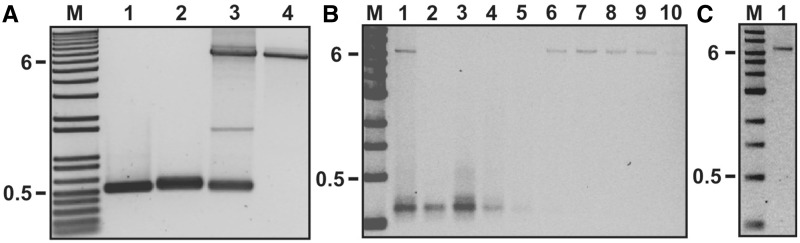
Using PCR, we made 400-bp-long labelled dsDNA megaprimers starting from two ∼25-nt-long primers and partially replacing dTTP with labelled dUTP (Figure 2B, Step 2). When 50% of the dTTP was replaced with biotin-16-dUTP, our megaprimer yield was 20 pmol (∼90 ng/μl at 60 μl) after PCR clean-up. Our yield for 25% replacement with dig-11-dUTP was comparable, but we had no measureable product at 50% replacement. Dig-11-dUTP modified nucleotides are known to adversely affect PCR efficiency more than biotin-16-dUTP (36). Gel analysis showed these amplifications yielded single bands (Figure 3A, Lane 1–2). As expected, the presence of the label led to gel shifts relative to an unlabelled DNA (Supplementary Figure S2). Decreasing the substitution of labelled dUTP for dTTP to 10% increased the yield of the megaprimer to ∼27 pmol (∼120 ng/μl at 60 μl) for both labels. We refer to these two sets of megaprimers as ‘high’ and ‘standard’ density megaprimers.
Figure 3.
Analysis of PCR products and purification technique using non-denaturating gel electrophoresis. (A) An agarose gel showing intermediate and final products. Lane M: DNA ladder (1 kb+, Invitrogen); Lane 1: Biotin-labelled megaprimer; Lane 2: Dig-labelled megaprimer; Lane 3: Pre-gel purified megaprimer PCR showing the desired product and unincorporated megaprimers; Lane 4: Final PCR product (6592 bp) after gel purification. (B) Fractions from continuous elution electrophoresis. Lane M: DNA ladder (1 kb, NEB); Lane 1: Mixture of PCR products (before separation); Lanes 2–5: Fraction 2, 3, 10, 15 (unreacted megaprimers); Lanes 6–10: Fraction 21, 22, 23, 25, 30 (6592-bp DNA). Fractions 21–30 were combined. (C) Gel analysis of the pooled product from the continuous elution electrophoresis. Lane M: DNA ladder (1 kb, NEB); Lane 1: Purified and pooled DNA.
PCR using labelled megaprimers
Our PCR using the high-density megaprimers yielded a strong band at the appropriate DNA length (Figure 3A, Lane 3). We also performed PCR using the standard density megaprimers. This PCR yielded the appropriate band and a band at a higher length. The higher molecular product did not interfere with our ability to gel purify the desired product (Supplementary Figure S3) and was most likely because of our template not being fully linearized.
Purification and yield of final product
We used two different methods for purifying the highly labelled product: standard gel electrophoresis and continuous gel elution. The standard gel electrophoresis in conjunction with Freeze ‘N Squeeze columns led to a single band (Figure 3A, Lane 4). The final yield of purified product was 120 fmol (500 ng/100 μl of PCR). Under our standard single-molecule assay conditions, this amount of DNA is sufficient for ∼300 slides with 40–50 tethered beads in an area of 100 × 100 μm2.
We also used continuous gel elution to purify the desired product from the megaprimer PCR (Figure 3B, Lane 1). Each ∼500 -μl fraction was concentrated to 20 μl during ethanol precipitation. Subsequent gel analysis used up half of the DNA (10 μl/fraction) and showed the target DNA band was successfully separated from the megaprimers (Figure 3B, Lanes 2–10). After purification and gel analysis, we recovered ∼30 fmol of DNA (∼140 ng/100 μl of reaction) (Figure 3C). This yield was 25–50% less than that from traditional gel purification using Freeze ‘N Squeeze columns. If necessary, this yield can be increased 2-fold, as the target DNA will elute in the same set of tubes and the gel analysis step can be omitted once the parameters of the continuous gel separation are fixed (voltage, volume per tube, gel density, flow rate and so forth).
Determination of torsionally constrained DNA
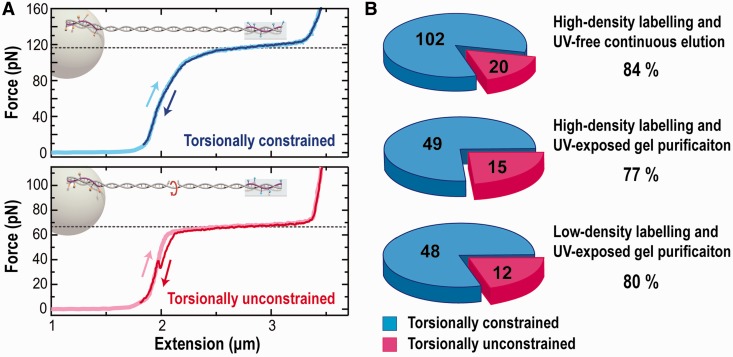
We used the overstretching force as a mechanical fingerprint to identify torsionally constrained DNA. For each replicate, we analysed at least 60 different individual DNA molecules. DNA molecules that overstretched at ∼110 pN were counted as torsionally constrained, whereas molecules that overstretched at 65 pN were designated as torsionally unconstrained (Figure 4A) (16,37). We note that a small fraction (∼18%) of these molecules showed an abrupt drop in Fo from 110 to 65 pN during the stretching–relaxation cycle (Supplementary Figure S4). These molecules were also counted as torsionally constrained (see Discussion for further details).
Figure 4.
Torsionally constrained DNA probed by measuring the overstretching force. (A) Representative force-extension curves of torsionally constrained (top) and unconstrained (bottom) DNA that exhibited overstretching forces at 110 and 65 pN, respectively. The molecules were first stretched (light colour) and then relaxed (dark colour), with hysteresis in the lower panel indicative of peeling from a nick or free end (16). (Inset) Cartoon of a torsionally constrained and an unconstrained DNA anchored to a surface via multiple bonds at each end. The bond colour indicates biotin (orange) and digoxigenin (cyan), respectively. (B) Pie charts showing the percentage of torsionally constrained DNA at three independent conditions: (top) DNA made with high-density megaprimers and purified with the continuous-elution method; (middle) DNA made with high-density megaprimers and purified by standard gel purification including UV irradiation; and (bottom) DNA made with standard density megaprimers and purified by standard gel purification.
Although we varied the degree of labelling and the purification method, we found that all four replicates tested had approximately the same high percentage (∼80%) of torsionally constrained DNA (Figure 4B). Each replicate was independent, including the separate amplification of the megaprimers done on separate days. Specifically, we compared three replicates made with highly labelled megaprimers but purified by two different methods. The two replicates purified with continuous gel elution were indistinguishable, with 84% of them torsionally constrained (n = 121). This percentage was slightly higher than DNA purified via standard gel electrophoresis (77%; n = 64). We next used megaprimers with the standard labelling density and performed traditional gel purification. This DNA continued to show excellent performance; 80% of the molecules were torsionally constrained (n = 60).
DISCUSSION
We have demonstrated that megaprimer-based PCR reaction provides a ligation-free method to create DNA for torsionally constrained single-molecule assays.
Simple, rapid and efficient production of torsionally constrained DNA
PCR is a simple and efficient way to assemble the desired product, a nick-free DNA that is multiply labelled with different labels at each end of the molecule. This protocol is rapid, eliminating several restriction digests and a three-way ligation. PCR-based assembly also avoids the significant issue of incomplete ligation. Moreover, megaprimer PCR produces enough DNA for a large number of single-molecule assays (∼300). A key scientific benefit of this large yield is that a single DNA preparation is sufficient for a whole set of experiments, improving their uniformity.
Low cost
We note that an ideal primer for our application would have a label once every helical turn (10 bp). Solid-state synthesis of DNA can yield 100–150-bp primers of this design, but such primers are expensive (∼$200 per label). A variation of this more expensive method has been successfully implemented by ligating on a short (62 bp) dsDNA segment containing six biotin tags (38). However, we did not pursue this option but rather focused on a more general approach that did not require costly primers for each new DNA construct. Fortunately, a high-density megaprimer was not necessary to achieve a high percentage of torsionally constrained DNA. By using the standard density megaprimers, we conserve the single most expensive material of the reaction, the labelled nucleotides (e.g. $350 for 50 nmol of biotin-labelled nucleotide). We also found that each PCR to make the labelled primers yielded enough megaprimers for several reactions. Thus, just $30 worth of labelled nucleotides is sufficient for ∼3000 single-molecule assays.
Linearizing the template improves yield
We found it critical to use a linear template to obtain clean product. In our megaprimer PCR reactions, the presence of even a small amount of circular plasmid led to an undesired side product, which decreased the yield of the desired target DNA. We note that when a linear DNA molecule is amplified from a circular template to be the template in a subsequent megaprimer PCR, then the original circular template is still present after standard PCR clean-up. This small amount of circular DNA led to an undesired product in subsequent megaprimer PCR when the template concentration was relatively high [75 pM (36 ng in 100 μl)], a template concentration that otherwise improved the yield of the megaprimer PCR. Thus, if the linearization of the circular template was incomplete, we gel purified the product.
Differences in purification protocols
Exposure to 365-nm UV light is known to cause nicks in DNA (27). We, therefore, explored a continuous gel elution method that avoided UV light (Figure 4B). Somewhat unexpectedly, this difference in the method of purification led to a minor (7%) change in the percentage of torsionally constrained DNA. The potential benefit of continuous gel elution should be reinvestigated if long (>15 kb) torsionally locked substrates are desired. The difference in sample handling (39) and the absence of UV light may maintain a high percentage of torsionally locked substrates for long DNA molecules.
Upper bound to percentage torsionally locked
Varying the density of labels in the megaprimers and the purification method led to minimal change in the percentage of torsionally constrained DNA. Our highest percentage of torsionally locked DNA (84%) was achieved when we used highly labelled megaprimers and continuous gel purification (Figure 4B). However, there was only a 4% decrease in the torsionally locked percentage when we used the standard density megaprimers and traditional gel purification that includes a brief UV exposure. Broadly speaking, this uniformly high level of torsionally constrained DNA is a major improvement. This result is ∼33% higher than the current state of the art (25). We attribute this difference to the absence of ligation in our protocol, because both studies used anti-dig-coated surfaces and streptavidin-coated beads. The uniformity of our results also suggests a common, unknown factor that limits the maximum percentage of torsionally constrained molecules. Two likely candidates are sample handling, which can introduce nicks (39), and the density of the surface-bound proteins responsible for binding the labelled DNA (Supplementary Figure S5). Future work is necessary to distinguish between these two potential mechanisms. Additionally, we certainly expect geometric constraints of the linker length and the 50-nm persistence length of dsDNA to prevent all of the incorporated labels from binding to the surface-bound proteins.
Overstretching assay applies atypically high forces
Torsionally constrained DNA is most often used in assays that apply low-to-moderate forces (0.1–20 pN) (3,5–11,13–15,17,19). In contrast, our overstretching assay applies a high force (>110 pN), with a small fraction (∼18%) of molecules showing an abrupt drop in F from 110 pN to 65 pN (Supplementary Figure S3). These force drops are consistent with a sudden decrease in the number of bonds at one end that compromise the torsional constraint. These abrupt drops should not affect the percentage of torsionally locked DNA in low-to-moderate force assays, as they occur only at high forces (>65 pN).
Force drops at high forces are expected given the strength of the dig–anti-dig (40) and biotin–streptavidin bond (41), which are ∼60 and ∼100 pN at a 10 nN/s loading rate, respectively. Interestingly, these force drops occur at an F above the average strength of a single dig–anti-dig bond and are generally not a saw-tooth-like series of small ruptures followed by an abrupt drop to 65 pN (Supplementary Figure S4). Such abrupt force drops suggest that the load is being shared across multiple bonds, rather than a single bond carrying the full load until it ruptures. Formally, force drops could also be due to the nicking of the DNA by laser-induced oxygen radicals. However, we think this was uncommon because for many molecules that lost torsional constraint, such constraint was recovered after the applied load was removed for a brief period. This recovery indicates that there were no nicks in the substrate.
Potential paths towards higher yield and longer DNA substrates
We have presented a substantially simplified method to make torsionally constrained DNA at high yield, both in terms of the amount of DNA and the percentage of torsionally locked DNA. These improvements save substantial time and labour and are sufficient for our overstretching studies and, we expect, most single-molecule assays. Nonetheless, there are still avenues for improvement for those that need a higher yield. We will briefly outline several such methods. First, different DNAPs are known to incorporate labelled nucleotides with different efficiencies (36). We, therefore, expect that one DNAP might be optimum for making labelled megaprimers, whereas a different DNAP would be best for using those megaprimers. Second, we are currently using a megaprimer concentration that is 30-fold lower than a standard primer concentration (0.3 µM). A higher template concentration helped compensate for this lower megaprimer concentration. As the yield of making the megaprimers increases, varying the template and megaprimer concentration becomes possible and should lead to improved yields. Finally, DNA yield should increase as the length of the megaprimer is decreased. We initially were worried that a 400-bp megaprimer labelled at high density on only one strand might not yield a high percentage of molecules that were torsionally locked. Not only did this DNA succeed, but DNA made with standard density megaprimers did as well. This independence of labelling density suggests shorter megaprimers would be a fruitful avenue of investigation.
Our ligation-free method relies on making the final product by PCR. We have demonstrated this technique with a 6.6-kb substrate. We expect PCR-based assembly to work without modification up to ∼10 kb after which there may be additional challenges. We note that commercial PCR kits (e.g. MasterAmp Extra-Long PCR Kit, Epicentre Biotechnologies, Madison, WI, USA) allow for 30–40 kb products with short unlabelled primers. Thus, there are prospects for making long torsionally constrained DNA, but such DNA molecules will require additional development effort.
CONCLUSION
Fundamental processes in DNA metabolism can alter the topology of DNA (1). Controlled twisting of individual DNA molecules provides a unique method to investigate the enzymes that alter the topology of DNA (4,18,42–46). Such twisting requires a single DNA molecule to be torsionally constrained. The traditional ligation-based method is time-consuming and often yields a small amount of DNA with a low percentage of the DNA that is torsionally constrained. Recent work has improved the yield through optimizing the DNA sequence at restriction sites (25). In this article, we present a ligation-free method for producing torsionally constrained DNA. The final product is assembled by megaprimer PCR. This PCR-based method produces a high yield and a high quality of DNA with less labour and time than the traditional ligation-based method. The high yield allows for ∼300 single-molecule assays per purification; the high percentage of torsionally constrained DNA (∼84%) demonstrates the high quality of the DNA. We expect the present protocol to significantly simplify the application and adoption of torsionally constrained assays to a wide range of single-molecule systems.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation [PHY-1125844]; and National Institute of Standards and Technology. Funding for open access charge: National Science Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Brad Baxley for help in figure production and Art Pardi for useful discussions. Mention of commercial products is for information only; it does not imply NIST’s recommendation or endorsement. TTP is a staff member of NIST’s quantum physics division.
REFERENCES
- 1.Wang JC. DNA topoisomerases. Annu. Rev. Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 2.Wang JC, Becherer K. Cloning of the gene topA encoding for DNA topoisomerase I and the physical mapping of the cysB-topA-trp region of Escherichia coli. Nucleic Acids Res. 1983;11:1773–1790. doi: 10.1093/nar/11.6.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strick TR, Allemand JF, Bensimon D, Bensimon A, Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 4.Strick TR, Allemand JF, Bensimon D, Croquette V. Stress-induced structural transitions in DNA and proteins. Annu. Rev. Biophys. Biomol. Struct. 2000;29:523–543. doi: 10.1146/annurev.biophys.29.1.523. [DOI] [PubMed] [Google Scholar]
- 5.Strick TR, Croquette V, Bensimon D. Single-molecule analysis of DNA uncoiling by a type II topoisomerase. Nature. 2000;404:901–904. doi: 10.1038/35009144. [DOI] [PubMed] [Google Scholar]
- 6.Koster DA, Croquette V, Dekker C, Shuman S, Dekker NH. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature. 2005;434:671–674. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- 7.Gore J, Bryant Z, Stone MD, Nollmann MN, Cozzarelli NR, Bustamante C. Mechanochemical analysis of DNA gyrase using rotor bead tracking. Nature. 2006;439:100–104. doi: 10.1038/nature04319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koster DA, Palle K, Bot ES, Bjornsti MA, Dekker NH. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature. 2007;448:213–217. doi: 10.1038/nature05938. [DOI] [PubMed] [Google Scholar]
- 9.Neuman KC, Charvin G, Bensimon D, Croquette V. Mechanisms of chiral discrimination by topoisomerase IV. Proc. Natl Acad. Sci. USA. 2009;106:6986–6991. doi: 10.1073/pnas.0900574106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basu A, Schoeffler AJ, Berger JM, Bryant Z. ATP binding controls distinct structural transitions of Escherichia coli DNA gyrase in complex with DNA. Nat. Struct. Mol. Biol. 2012;19:538–546. doi: 10.1038/nsmb.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seol Y, Zhang HL, Pommier Y, Neuman KC. A kinetic clutch governs religation by type IB topoisomerases and determines camptothecin sensitivity. Proc. Natl Acad. Sci. USA. 2012;109:16125–16130. doi: 10.1073/pnas.1206480109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.La Porta A, Wang MD. Optical torque wrench: angular trapping, rotation, and torque detection of quartz microparticles. Phys. Rev. Lett. 2004;92:190801. doi: 10.1103/PhysRevLett.92.190801. [DOI] [PubMed] [Google Scholar]
- 13.Forth S, Deufel C, Sheinin MY, Daniels B, Sethna JP, Wang MD. Abrupt buckling transition observed during the plectoneme formation of individual DNA molecules. Phys. Rev. Lett. 2008;100:148301. doi: 10.1103/PhysRevLett.100.148301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheinin MY, Forth S, Marko JF, Wang MD. Underwound DNA under tension: structure, elasticity, and sequence-dependent behaviors. Phys. Rev. Lett. 2011;107:108102. doi: 10.1103/PhysRevLett.107.108102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forth S, Deufel C, Patel SS, Wang MD. Direct measurements of torque during holliday junction migration. Biophys. J. 2011;101:L5–L7. doi: 10.1016/j.bpj.2011.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paik DH, Perkins TT. Overstretching DNA at 65 pN does not require peeling from free ends or nicks. J. Am. Chem. Soc. 2011;133:3219–3221. doi: 10.1021/ja108952v. [DOI] [PubMed] [Google Scholar]
- 17.Lipfert J, Wiggin M, Kerssemakers JW, Pedaci F, Dekker NH. Freely orbiting magnetic tweezers to directly monitor changes in the twist of nucleic acids. Nat. Commun. 2011;2:439. doi: 10.1038/ncomms1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bryant Z, Oberstrass FC, Basu A. Recent developments in single-molecule DNA mechanics. Curr. Opin. Struc. Biol. 2012;22:304–312. doi: 10.1016/j.sbi.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lipfert J, Kerssemakers JW, Jager T, Dekker NH. Magnetic torque tweezers: measuring torsional stiffness in DNA and RecA-DNA filaments. Nat. Methods. 2010;7:977–980. doi: 10.1038/nmeth.1520. [DOI] [PubMed] [Google Scholar]
- 20.Janssen XJA, Lipfert J, Jager T, Daudey R, Beekman J, Dekker NH. Electromagnetic torque tweezers: a versatile approach for measurement of single-molecule twist and torque. Nano Lett. 2012;12:3634–3639. doi: 10.1021/nl301330h. [DOI] [PubMed] [Google Scholar]
- 21.van Loenhout MT, de Grunt MV, Dekker C. Dynamics of DNA supercoils. Science. 2012;338:94–97. doi: 10.1126/science.1225810. [DOI] [PubMed] [Google Scholar]
- 22.Oberstrass FC, Fernandes LE, Bryant Z. Torque measurements reveal sequence-specific cooperative transitions in supercoiled DNA. Proc. Natl Acad. Sci. USA. 2012;109:6106–6111. doi: 10.1073/pnas.1113532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Vlaminck I, Henighan T, van Loenhout MT, Pfeiffer I, Huijts J, Kerssemakers JW, Katan AJ, van Langen-Suurling A, van der Drift E, Wyman C, et al. Highly Parallel Magnetic Tweezers by Targeted DNA Tethering. Nano Lett. 2011;11:5489–5493. doi: 10.1021/nl203299e. [DOI] [PubMed] [Google Scholar]
- 24.Lipfert J, Koster DA, Vilfan ID, Hage S, Dekker NH. Single-molecule magnetic tweezers studies of type IB topoisomerases. Methods Mol. Biol. 2009;582:71–89. doi: 10.1007/978-1-60761-340-4_7. [DOI] [PubMed] [Google Scholar]
- 25.Seol Y, Neuman KC. Magnetic tweezers for single-molecule manipulation. Methods Mol. Biol. 2011;783:265–293. doi: 10.1007/978-1-61779-282-3_15. [DOI] [PubMed] [Google Scholar]
- 26.Leger JF, Robert J, Bourdieu L, Chatenay D, Marko JF. RecA binding to a single double-stranded DNA molecule: a possible role of DNA conformational fluctuations. Proc. Natl Acad. Sci. USA. 1998;95:12295–12299. doi: 10.1073/pnas.95.21.12295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peak JG, Pilas B, Dudek EJ, Peak MJ. DNA breaks caused by monochromatic 365-nm ultraviolet-a radiation or hydrogen-peroxide and their repair in human epithelioid and xeroderma-pigmentosum cells. Photochem. Photobiol. 1991;54:197–203. doi: 10.1111/j.1751-1097.1991.tb02007.x. [DOI] [PubMed] [Google Scholar]
- 28.Sarkar G, Sommer SS. The “megaprimer” method of site-directed mutagenesis. Biotechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- 29.Leger JF, Romano G, Sarkar A, Robert J, Bourdieu L, Chatenay D, Marko JF. Structural transitions of a twisted and stretched DNA molecule. Phys. Rev. Lett. 1999;83:1066–1069. [Google Scholar]
- 30.Wang MD, Yin H, Landick R, Gelles J, Block SM. Stretching DNA with optical tweezers. Biophys. J. 1997;72:1335–1346. doi: 10.1016/S0006-3495(97)78780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carter AR, Seol Y, Perkins TT. Precision surface-coupled optical-trapping assays with 1 base-pair resolution. Biophys. J. 2009;96:2926–2934. doi: 10.1016/j.bpj.2008.12.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seol Y, Carpenter AE, Perkins TT. Gold nanoparticles: enhanced optical trapping and sensitivity coupled with significant heating. Opt. Lett. 2006;31:2429–2431. doi: 10.1364/ol.31.002429. [DOI] [PubMed] [Google Scholar]
- 33.Visscher K, Gross SP, Block SM. Construction of multiple-beam optical traps with nanometer-resolution position sensing. IEEE J. Sel. Top. Quant. Electron. 1996;2:1066–1076. [Google Scholar]
- 34.Gittes F, Schmidt CF. Interference model for back-focal-plane displacement detection in optical tweezers. Opt. Lett. 1998;23:7–9. doi: 10.1364/ol.23.000007. [DOI] [PubMed] [Google Scholar]
- 35.Seol Y, Li J, Nelson PC, Perkins TT, Betterton MD. Elasticity ofs DNA molecules: theory and experiment for contour lengths of 0.6-7 µm. Biophys. J. 2007;93:4360–4373. doi: 10.1529/biophysj.107.112995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson JP, Angerer B, Loeb LA. Incorporation of reporter-labeled nucleotides by DNA polymerases. Biotechniques. 2005;38:257–264. doi: 10.2144/05382RR02. [DOI] [PubMed] [Google Scholar]
- 37.van Mameren J, Gross P, Farge G, Hooijman P, Modesti M, Falkenberg M, Wuite GJ, Peterman EJ. Unraveling the structure of DNA during overstretching by using multicolor, single-molecule fluorescence imaging. Proc. Natl Acad. Sci. USA. 2009;106:18231–18236. doi: 10.1073/pnas.0904322106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deufel C, Forth S, Simmons CR, Dejgosha S, Wang MD. Nanofabricated quartz cylinders for angular trapping: DNA supercoiling torque detection. Nat. Methods. 2007;4:223–225. doi: 10.1038/nmeth1013. [DOI] [PubMed] [Google Scholar]
- 39.Adam RE, Zimm BH. Shear degradation of DNA. Nucleic Acids Res. 1977;4:1513–1537. doi: 10.1093/nar/4.5.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neuert G, Albrecht C, Pamir E, Gaub HE. Dynamic force spectroscopy of the digoxigenin-antibody complex. FEBS Lett. 2006;580:505–509. doi: 10.1016/j.febslet.2005.12.052. [DOI] [PubMed] [Google Scholar]
- 41.Merkel R, Nassoy P, Leung A, Ritchie K, Evans E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 1999;397:50–53. doi: 10.1038/16219. [DOI] [PubMed] [Google Scholar]
- 42.Strick T, Allemand J, Croquette V, Bensimon D. Twisting and stretching single DNA molecules. Prog. Biophys. Mol. Biol. 2000;74:115–140. doi: 10.1016/s0079-6107(00)00018-3. [DOI] [PubMed] [Google Scholar]
- 43.Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dulin D, Lipfert J, Moolman MC, Dekker NH. Studying genomic processes at the single-molecule level: introducing the tools and applications. Nat. Rev. Genet. 2013;14:9–22. doi: 10.1038/nrg3316. [DOI] [PubMed] [Google Scholar]
- 45.Neuman KC. Single-molecule measurements of DNA topology and topoisomerases. J. Biol. Chem. 2010;285:18967–18971. doi: 10.1074/jbc.R109.092437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Charvin G, Allemand JF, Strick TR, Bensimon D, Croquette V. Twisting DNA: single molecule studies. Contemp. Phys. 2004;45:383–403. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.