Abstract
DNA-protein cross-links (DPCs) are ubiquitous, structurally diverse DNA lesions formed upon exposure to bis-electrophiles, transition metals, UV light, and reactive oxygen species. Because of their super-bulky, helix distorting nature, DPCs interfere with DNA replication, transcription, and repair, potentially contributing to mutagenesis and carcinogenesis. However, the biological implications of DPC lesions have not been fully elucidated due to the difficulty of generating site-specific DNA substrates representative of DPC lesions formed in vivo. In the present study, a novel approach involving post-synthetic reductive amination has been developed to prepare a range of hydrolytically stable lesions structurally mimicking the DPCs produced between the N7 position of guanine in DNA and basic lysine or arginine side chains of proteins and peptides.
Introduction
Exposure to common antitumor drugs, environmental toxins, transition metals, UV light, ionizing radiation, and free radical-generating systems can result in cellular proteins becoming covalently trapped on DNA.1 The resulting DNA-protein cross-links (DPCs) are unusually bulky, structurally diverse, and highly heterogeneous DNA lesions involving a wide range of proteins of varying size, hydrophobicity, and cellular functions.1–4 Our previous mass spectrometry-based studies have revealed that a wide range of proteins can become covalently bound to genomic DNA upon treatment of human cells with clinically relevant concentrations of chemotherapeutic drugs (cisplatin and mechlorethamine) and metabolically activated carcinogens such as 1,2,3,4-diepoxybutane.2,3,5 Some examples of the participating proteins include HSP 90, tubulins, DNA helicases, PCNA, Fen-1, KU 70, Ku 86, Ref-1, PARP, and DNA polymerase δ.2,3 MS/MS sequencing has shown that DNA-protein cross-linking is non-random, with specific amino acid side chains (cysteine, lysine, histidine, or arginine) participating in covalent conjugate formation to the N-7 position of guanine in DNA (Scheme 1).2,3,6 DPCs are also formed as a result UV irradiation, treatment with formaldehyde,7–9 and endogenous exposure to reactive oxygen species, lipid peroxidation products, and transition metals, and have been shown to accumulate in the brain and heart tissues with age.7,10–17 Recent studies with laboratory mice deficient in the Fanconi Anemia DNA repair pathway have implicated DPC formed by formaldehyde in the observed cellular toxicity.18,19
Scheme 1.
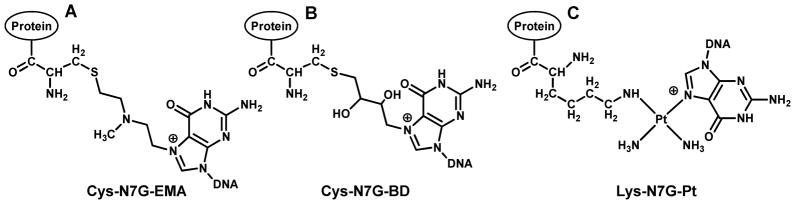
Structures of DNA-protein cross-links induced by bis-electrophiles: N-[2-(guan-7-yl)ethyl]alkylamine adducts formed by mechlorethamine (A); 4-(guan-7-yl)-2,3-butanediol adducts formed by 1,2,3,4-diepoxybutane (B), and 1,1-cis-diammine-2-(5-amino-5-carboxypentyl)amino-2-(2′-deoxyguanosine-7-yl)-platinum(II) adducts induced by cisplatin (C).
Due to their enormous size as compared to other DNA lesions, DPCs are believed to compromise genetic stability and cellular viability by interfering with normal DNA-protein interactions required for DNA replication, transcription, and repair.1 We have recently engineered protein monoepoxide agents that specifically induce chromosomal DPC.20 These DNA-reactive proteins induced significant levels of mutations and toxicity when introduced into human cells,20 probably because the resulting DPCs block DNA replication and transcription. However, relatively little is known about the influence of DPC adducts on DNA and RNA polymerases or their repair mechanisms in mammalian cells. Consequently, there is a pressing need to examine DNA replication and transcription in the presence of specific DPC lesions and to identify DNA repair mechanisms responsible for their removal from mammalian cells.
Any mechanistic investigations of the biological effects of DPC lesions in human cells require the availability of structurally defined DNA substrates containing DPC lesions at a specified site of DNA. However, the access to such DPC substrates has been limited due to the synthetic challenge of covalently linking two complex biomolecules (DNA and proteins) in a site-specific manner. Previously, model DPCs have been generated by covalently trapping various enzymes on their DNA substrates. For example, the Schiff base intermediate produced between T4-pdg glycosylase/AP lyase and apurinic/apyrimidinic site of DNA can be reduced to form a stable T4-pdg-DNA conjugate.21 A disulfide trapping strategy was used to attach N149C mutant of human 8-oxoguanine DNA glycosylase I (hOGG1) protein to a DNA duplex containing alkanethiol tether at the N4 position of cytosine.22 DNA methyltransferease has been cross-linked to C6 position of 5-aza-cytosine.23 More recently, a reductive amination strategy was used to generate DNA-protein/peptide conjugates by the reaction of the N2-guanine aldehyde functionality derived from acrolein-induced 3-(2′-deoxyrobo-1′-syl)-5,6,7,8-tetrahydro-8-hydroxypyrimido[1,2a]purin-10(3H)-one (γ-HOPdG) with proteins and peptides.24,25
The most common site of DNA involved in DPC formation following treatment with bis-electrophiles is the N-7 of guanine (Scheme 1). However, to our knowledge, no methods exist in the literature to generate N-7 guanine conjugated DPCs. One formidable obstacle in accomplishing this goal is that N-7 guanine alkylation destabilizes the β-glycosidic bond of the modified nucleoside, leading to spontaneous depurination. In the present study, we have developed a new methodology to create hydrolytically stable structural mimics of N-7 guanine conjugated DPCs by reductive amination reactions between the Lys and Arg side chains of proteins and acetaldehyde functionalities of the modified 7-deazaguanine residues within DNA. The resulting model DPCs are structurally analogous to N7 guanine adducts generated by antitumor nitrogen mustards, 1,2,3,4-diepoxybutane (Scheme 1) and chlorooxirane.26
Experimental Procedures
General
Synthetic oligonucleotides containing 7-deaza-7-(2,3-dihydroxypropan-1-yl)-2′-deoxy-guanosine (DHP-deaza-dG) and DHP-deaza-dG nucleoside were prepared as previously described.27 Fluorescein-dT phosphoramidite, protected 2′-deoxyribo-nucleoside-3′-phosphoramidites (dA-CE, Ac-dC-CE, dmf-dG-CE, dT-CE), Ac-dC-CPG ABI, dmf-dG-CPG ABI columns, and all other reagents required for automated DNA synthesis were purchased from Glen Research (Sterling, VA). Synthetic DNA oligonucleotides were synthesized by solid phase synthesis using an ABI 394 DNA synthesizer (Applied Biosystems, CA). All solvents and chemical reagents were obtained from commercial sources and used without further purification.
Preparation of Radiolabeled DNA Duplexes
Single stranded oligodeoxynucleotides 5′-G TCA CTG GTA DHP-deaza-dG CA AGC ATT G-3′ and 5′-C AGT GAC CAT CDHP-deaza-dGT TCG TAA C-3′ (2 nmol in 12 μL of water) were radiolabeled with γ-32P ATP using standard methods. Following heating at 65 °C for 10 min to inactivate the enzyme, excess γ-32P ATP was removed using Illustra microspin G25 columns (GE Healthcare, Pittsburgh, PA). To obtain double stranded DNA, 5′-32P-enlabeled oligomers were mixed with equimolar amounts of the complementary strands in 10 mM Tris buffer (pH 7) containing 50 mM NaCl and heated at 90 °C for 10 min, followed by gradual cooling overnight.
Reductive Amination to Generate DNA-Protein Cross-links
32P-endlabeled DNA duplexes (50 pmol in 8 μL water) were oxidized in the presence of 50 mM NaIO4 (5 μL) in 15 mM sodium phosphate buffer (pH 5.4, 5 μL) for 6 h at 4 °C in the dark to unmask the aldehyde moiety on DHP-deaza-dG (Scheme 2). Excess NaIO4 was quenched with 55 mM Na2SO3 (5 μL). Proteins and peptides of interest (0.5 – 2.5 nmol) were incubated with the aldehyde-containing DNA duplexes (50 pmol) in the presence of 25 mM NaCNBH3 at 37 °C overnight to generate stable DNA-protein cross-links. Aliquots of the reaction mixtures were withdrawn and resolved by 12% SDS-PAGE with or without proteinase K digestion (6 units, 48 h at 37 °C).
Scheme 2.
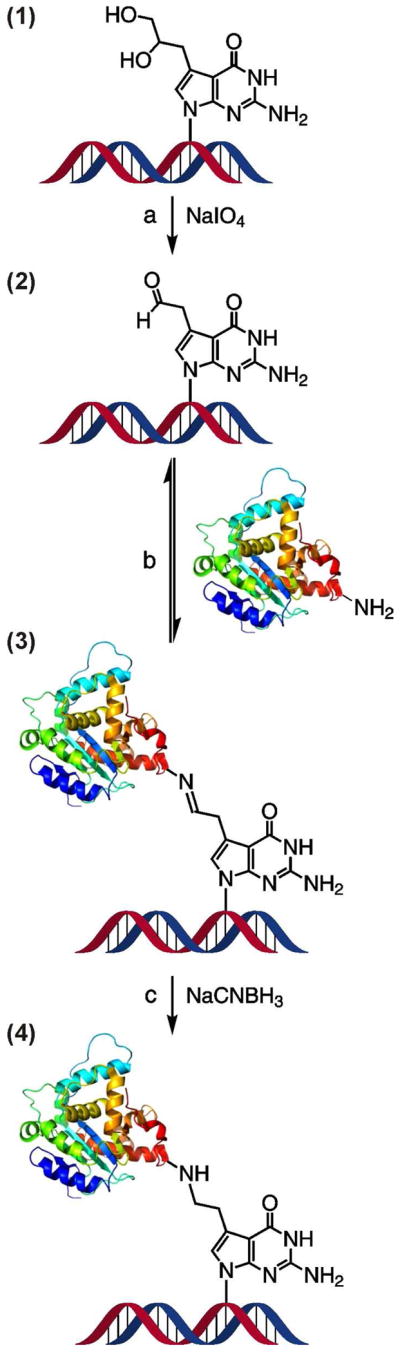
Synthesis of DPCs by a post-synthetic reductive amination strategy: (a) oxidative cleavage to unmask the reactive aldehyde moiety on the 7-deaza-7-(2,3-dihydroxypropyl)-guanine of the synthetic oligomer, (b) reaction of an amino group on the protein with the aldehyde on the DNA to form a Schiff base, (c) reduction of the imine to form a stable amine linkage.
Gel Electrophoretic Analysis of DNA-Protein Cross-links Generated by Reductive Amination
12% SDS-PAGE gel plates were pre-run at a constant voltage of 150 V for 30 min in 1× SDS running buffer. DPC reaction mixtures were dissolved in an equal volume of 0.1% TFA or 10% SDS (2 μL). The samples were reconstituted in SDS loading buffer and heated at 90 °C for 5 min prior to loading. The gels were run at a constant voltage of 150 V at ambient temperature. Radiolabeled DNA strands and DPCs were detected with a Storm 840 phosphorimager (Amersham Biosciences Corp., Piscataway, NJ) or a Typhoon FLA 7000 instrument (GE Healthcare, Pittsburgh, PA). Covalent DPCs were observed as slowly moving bands on the gel, and the reaction yields were calculated by volume analysis using Image Quant TL 8.0 (GE Healthcare, Pittsburgh, PA).
To visualize the proteins participating in DPC formation to aldehyde-containing DNA, NuPAGE Novex 12% Bis-Tris gels (Life Technologies, Grand Island, NY) was pre-run at a constant voltage of 100 V for 30 min in 1× NuPAGE MOPS SDS running buffer (Life Technologies, Grand Island, NY). The reaction mixtures obtained from DNA-protein cross-linking were dissolved in 10% SDS (2 μL) and reconstituted in NuPAGE LDS sample buffer (Life Technologies, Grand Island, NY). The samples were heated at 70 °C for 10 min prior to loading onto a gel. The gels were run at a constant voltage of 100 V at ambient temperature. The unreacted protein and DPC bands were visualized by staining with SimplyBlue SafeStain (Life Technologies, Grand Island, NY).
Sample Processing for Mass Spectrometry Analysis
DPCs were generated by reductive amination as described above using unlabeled DNA and proteins (15 to 20-fold excess) and either directly processed for mass spectrometric analysis or purified by PAGE as described below.
In direct processing experiments, the DNA component of DPCs was digested with PDE I (120 mU), PDE II (105 mU), DNase (35 U) and alkaline phosphatase (22 U) in 10 mM Tris-HCl/15 mM MgCl2 (pH 7) buffer containing at 37 °C overnight. The resulting protein-nucleoside conjugates were dried in vacuo, reconstituted in 100 mM NH4HCO3 (pH 7.9) (90 μL), and subjected to trypsin digestion using MS grade Trypsin Gold (Promega, Madison, WI) (2.5 μg) at 37 °C for 20 h. The digests were dried in vacuo, desalted using ZipTip with 0.6 μL C18 resin (Millipore, Billerica, MA), and the resulting peptides were reconstituted in 0.1% formic acid (10 μL) prior to MS analysis.
Alternatively, DNA-protein conjugates were first purified by 12% SDS-PAGE and stained with SimplyBlue SafeStain (Life Technologies, Grand Island, NY). DPC-containing gel bands were cut into slices and subjected to reduction using 300 mM DTT (10 μL) followed by alkylation with iodoacetamide (10 μL in 100 μL of 25 mM NH4HCO3, pH 7.9). Gel pieces were dehydrated with acetonitrile, dried under vacuum, reconstituted in 25 mM NH4HCO3 (pH 7.9) (75 μL) and incubated with PDE I (120 mU) at 37 °C overnight. Next, the samples were subjected to tryptic digestion and ZipTip desalting as described above, followed by MS analysis.
Characterization of DNA-Protein Cross-links by Mass Spectrometry
All HPLC-ESI+-MS/MS analyses were conducted with a Thermo Scientific LTQ Orbitrap Velos mass spectrometer interfaced with an Eksigent NanoLC-Ultra 2D HPLC system. Peptide mixtures (5 μL) were loaded onto a nano HPLC column (75 μm ID, 10 cm packed bed, 15 μm orifice) created by hand packing commercially purchased fused-silica emitters (New Objective, Woburn, MA) with Luna C18 5 μm separation media (Phenomenex, Torrance, CA). Liquid chromatography was carried out at an ambient temperature at a flow rate 0.3 μl/min using 0.1% formic acid (A) and acetonitrile (B). The solvent composition was changed linearly from 2% to 70% B over 60 min, then to 95% B over 1 min, kept at 95% B for further 5 min, and decreased to 2% B in 1 min. Finally, the flow rate was increased to 1 μl/min and kept at 2% B for an additional 7 min. Mass spectrometry was performed using the FTMS mass analyzer with a resolution of 60,000 ppm and with a scan range of m/z 300 – 2000. Peptide MS/MS spectra were collected using data-dependent scanning in which one full scan mass spectrum was followed by eight MS/MS spectra using an isolation width of 2.5 m/z, normalized CID collision energy of 35%, 1 repeat count, 20 s exclusion duration, with an exclusion mass width of ± 5 ppm.
Spectral data were analyzed using Thermo Proteome Discoverer 1.3 (Thermo Scientific, San Jose, CA) that linked raw data extraction, database searching, and probability scoring. The raw data were directly uploaded, without any format conversion, to search against the protein FASTA database. Search parameters included trypsin specificity and up to 2 missed cleavage sites. Protein N-terminus, lysine, or arginine residues were specified as possible modification sites by specifying the following dynamic modifications: (A) ethan-1,2-diyl cross-link to 7-deaza-7-ethan-1-yl-dG, +292.1172 Da (C13H16N4O4); (B) cross-link to 7-deaza-7-ethan-1-yl-guanine,+176.0698 Da (C8H8N4O); or (C) cross-link to 7-deaza-7-ethan-1-yl-dGMP, +372.0835 Da (C13H17N4O7P).
Synthesis and Characterization of 7-Deaza-7-(2-(N-acetyllysine)ethan-1-yl)-2′-deoxyguanosine and 7-deaza-7-(2-(N-acetylarginine)ethan-1-yl)-2′-deoxyguanosine Conjugates
Synthetic 7-deaza-7-(2,3-dihydroxypropan-1-yl)-2′-deoxyguanosine27 (10 nmol) was oxidized in the presence of 50 mM NaIO4 (4 μL) in 1 M sodium phosphate buffer (pH 5.4, 6 μl) for 6 h at 4 °C in the dark to generate 7-deaza-7-(formylmethan-1-yl)-2′-deoxyguanosine. Excess NaIO4 was quenched with 55 mM Na2SO3 (4 μl). N-acetyl protected Lys or Arg (4 μL of 5 mM solution) was added to the reaction mixture, followed by 0.5 M NaCNBH3 (4 μL) at 37 °C overnight to generate amino acid-nucleoside conjugates. The amino acid-nucleoside conjugates were isolated by HPLC on a Supelcosil-LC-18-DB (4.6 × 250 mm, 5 μm) column (Sigma Aldrich, Milwaukee, WI) using a gradient of 0.1% formic acid (A) and acetonitrile (B). The solvent composition was changed from 0 to 24% B over 24 min, then to 75% B over 6 min, and finally to 0% B over 2 min. HPLC fractions containing major components were collected, concentrated in vacuo and analyzed by capillary HPLC-ESI+-MSn on an Agilent 1100 capillary HPLC-ion trap mass spectrometer (Agilent Technologies, Inc., Wilmington, DE).
Synthesis and Characterization of Nucleoside-Peptide Conjugates
Synthetic nucleoside-peptide conjugates were prepared using 7-deaza-7-(2,3-dihydroxypropan-1-yl)-2′-deoxyguanosine and angiotensin I (DRVYIHPFHL) or substance P (RPKPQQFFGLMNH2) peptides by reductive amination procedure as described above. The reaction mixtures were dried in vacuo and desalted using ZipTip with 0.6 μL C18 resin (Millipore, Billerica, MA). The resulting mixtures were reconstituted in 0.1% formic acid (10 μL) and analyzed on a Thermo Scientific LTQ Orbitrap Velos mass spectrometer interfaced with an Eksigent NanoLC-Ultra 2D HPLC system as described below.
Desalted reaction mixtures (2 μL) were loaded onto a nano HPLC column (75 μm ID, 10 cm packed bed, 15 μm orifice) created by hand packing commercially purchased fused-silica emitters (New Objective, Woburn, MA) with Zorbax SB-C18 5 μm separation media (Phenomenex, Torrance, CA). Liquid chromatography was carried out using 0.1% formic acid (A) and acetonitrile (B) as solvents at an ambient temperature at an initial flow rate 1 μl/min at 2% B for 5.5 min. The flow rate was reduced to 0.3 μl/min in 30 s and the solvent composition was changed linearly from 2% to 50% B over 20 min, then to 95% B over 1 min, kept at 95% B for further 5 min, and decreased to 2% B in 1 min. Finally, the HPLC flow rate was increased to 1 μl/min and kept at 2% B for an additional 6 min. Mass spectrometry was performed using the FTMS mass analyzer with a resolution of 60,000 ppm and with a scan range of m/z 300 – 2000 in the full scan mode. MS2 spectra were collected using the iontrap with an isolation width of 2.5 m/z, normalized CID collision energy of 35%, while MS3 spectra were collected using the orbitrap with an isolation width of 2.5 m/z, and a normalized HCD collision energy of 35%.
Results
Experimental strategy for the generation of hydrolytically stable model DPC substrates
Since N7-guanine alkylation introduces a positive charge on the alkylated base, it destabilizes the N-glycosidic bond, leading to spontaneous depurination.28 Therefore, it is not practical to employ N7-guanine adducts in DNA replication and repair experiments. To avoid spontaneous degradation of our model DPC substrates, we have replaced the N-7 nitrogen of guanine with a carbon atom (7-deaza-G).27 To create a protein reactive group, the 2,3-dihydroxyprop-1-yl group was introduced at the same position (DHP-deaza-dG, compound 1 in Scheme 2). Treatment with periodate converts the diol group to the corresponding aldehyde (2 in Scheme 2), which then reacts with free amino groups of proteins (e.g., Lys or Arg side chains) to form a Schiff base (3 in Scheme 2). The latter can be quantitatively reduced with NaCNBH3 to produce a stable amine linkage (4 in Scheme 2). The aldehyde substrate (2 in Scheme 2) is a direct model for N7-(2-oxoethyl)-G, which is the major DNA adduct from exposure to chlorooxirane,26 and the resulting model cross-links are structurally analogous to DPCs formed by chlorooxirane and antitumor nitrogen mustards in cells (Scheme 3).
Scheme 3.
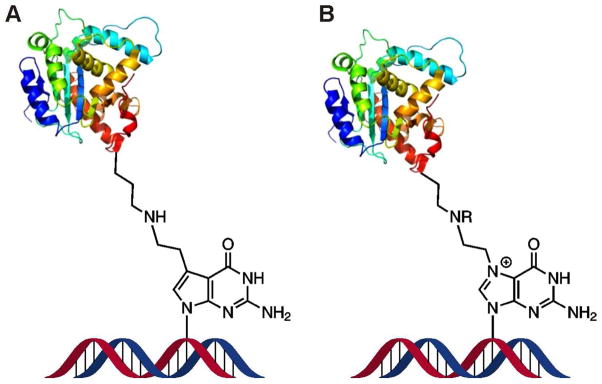
Structural similarity between the model DNA-protein cross-links obtained by the reductive amination of aldehyde containing DNA and proteins (A) and cellular DPCs formed in vivo upon reaction of antitumor nitrogen mustards with N7 guanine of DNA and proteins (B).2 Synthetic DPC substrates (A) are also a direct model for crosslinking of proteins to N7-(2-oxoethyl)-G, which is the major DNA adduct formed upon exposure to vinyl chloride.26
Characterization of DPC formation by denaturing gel electrophoresis
Our initial experiments were conducted with recombinant human AlkB protein, a DNA repair protein that contains multiple nucleophilic lysine and arginine residues and is known to bind to DNA.29,30 The formation of covalent AlkB-DNA conjugates was monitored by two independent methods. In the first approach, oligodeoxynucleotides containing the convertible nucleoside (DHP-deaza-dG) were radiolabeled with 32P-ATP. Following the cross-linking reaction, DPC formation was detected as the appearance of a new, low mobility band on denaturing PAGE (Figure 1). Alternatively, free proteins and DNA-protein conjugates were visualized by protein staining, and the presence of a cross-link was detected as a new protein band with reduced mobility (Figure 2).
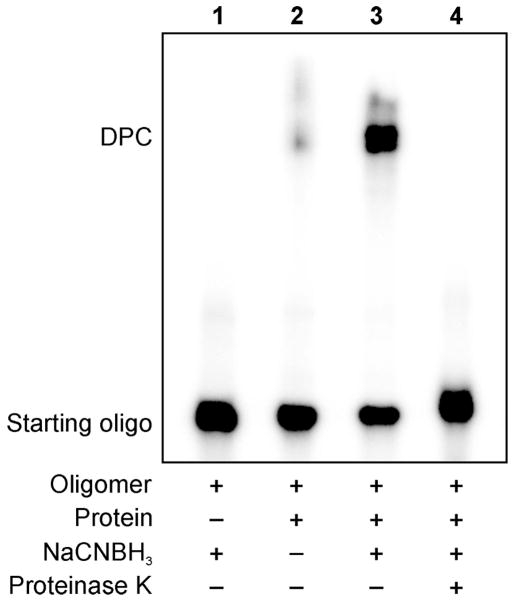
Figure 1.
Denaturing SDS-PAGE analysis of DNA-protein cross-links generated by reductive amination reaction (Scheme 2) between E. coli AlkB protein and aldehyde-containing DNA 20-mer, 5′-G TCA CTG GTA DHP-deaza-dG CA AGC ATT G-3′. DNA and DNA-protein conjugation products were visualized by 32P-end labeling. The formation of covalent DPC is revealed as a low mobility band on the gel. Lane 1: aldehyde containing oligonucleotide in the presence of a reducing agent (negative control); Lane 2: aldehyde containing oligonucleotide and AlkB protein in the absence of a reducing agent; Lane 3: reaction mixture of aldehyde containing oligonucleotide and the protein in the presence of a reducing agent; Lane 4: reaction mixture from lane 3 subjected to proteinase K digestion.
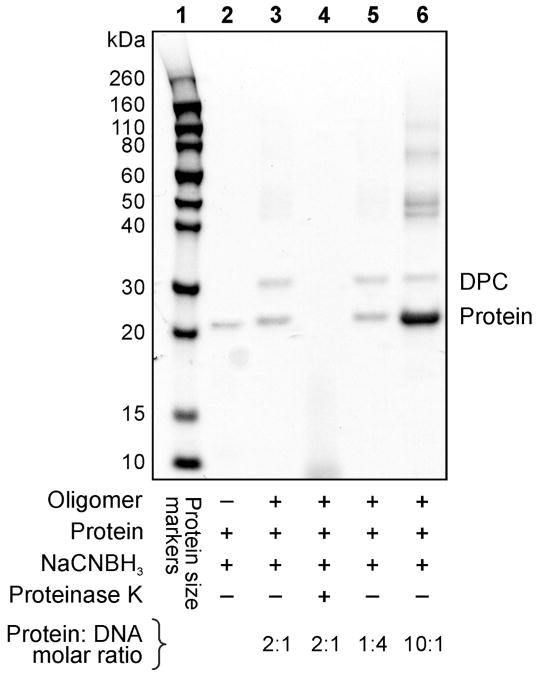
Figure 2.
Reductive amination-mediated cross-linking between aldehyde-containing DNA 20-mer (5′-G TCA CTG GTA DHP-deaza-dG CA AGC ATT G-3′) and E. coli AlkB protein visualized via SimplyBlue protein staining. Lane 1: protein size markers; Lane 2: AlkB protein (negative control); Lane 3: DPCs generated using 1:2 ratio of DNA to protein; Lane 4: proteinase K digested reaction mixture from lane 3; Lane 5: DPCs generated using 4:1 molar ratio of DNA to protein; Lane 6: DPCs generated using 1:10 ratio of DNA to protein. Multiple bands observed in the region > 40 kDa are due to protein aggregation.
As shown in Figure 1, the cross-linking reaction between DHP-deaza-dG containing DNA 18-mer and recombinant AlkB protein leads to the formation of covalent DPC conjugates as revealed by the appearance of a low mobility band on a denaturing polyacrylamide gel (Lane 3). This band is not observed in control experiments conducted in the absence of protein (Lane 1), and only trace amounts of conjugation are observed in the absence of the reducing agent (Lane 2). The DPC band disappears when the reaction mixture is subjected to proteinase K digestion, (Lane 4), confirming that it corresponds to covalent DNA-protein conjugates.
These results were further confirmed using protein staining to visualize the protein and the DPCs (Figure 2). Following reductive amination reaction between AlkB protein and DHP-deaza-dG containing 18-mer, a new band was observed with an increased molecular weight as compared to unreacted AlkB protein (22.9 kDa) (Lane 3). The size of the newly formed conjugate (~ 29 kDa) is consistent with the addition of 18-mer oligodeoxynucleotide (6.2 kDa) to the protein, and the cross-linking yield is dependent on DNA:protein ratio (Lanes 3, 5 and 6). In addition, several higher molecular weight bands (> 40 kDa) were observed due to the propensity of the GFP protein to oligomerize when present at a high concentration.
Effects of reaction conditions on DPC yields
The experimental conditions for each of the reaction steps (Scheme 2) were optimized by varying the reaction temperature, reaction time, and molar ratios. We found that the highest DPC yields are observed when the NaIO4-mediated oxidative cleavage (step a) is conducted at 4 °C, and the optimal temperature for reductive amination reaction (steps b and c) was 37 °C (Figure S1).
When the effect of reaction times on DPC yields was assessed, the best results were achieved when the time of periodate-mediated oxidative cleavage (step a) was limited to 2–6 h (Figure S2). This can be explained by a limited stability of aldehyde under oxidizing conditions. In contrast, the best yields of reductive amination reaction (steps b and c) were achieved at extended reaction times (12 to 24 h) (Figure S2).
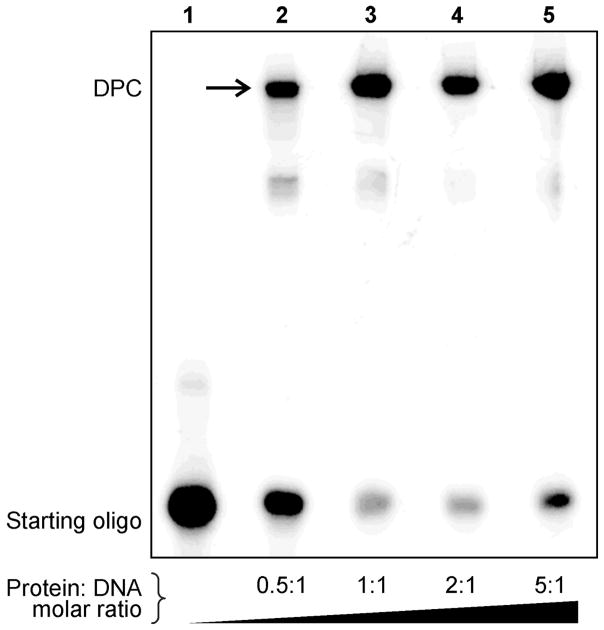
The influence of DNA:protein molar ratios on reaction yields was examined by keeping the concentration of one of the reagents (protein or DNA) constant while varying the equivalents of the other. When protein amounts were varied in respect to constant amounts of radiolabeled DNA, the highest DPC yields (~ 85%) were achieved when using an excess of the protein (Figure 3). When protein amounts were held constant and protein staining was employed to follow the reaction, increasing DNA concentrations similarly has led to increased DPC yields (Figure 2). These results suggest that the reversible Schiff base formation between the aldehyde functionality within DNA and the basic amino acid side chains of the protein can be driven forward towards product formation by employing a large molar excess of the other reagent. In a practical sense, generation of DNA substrates for replication and repair studies requires an excess of the protein in order to maximize the yields of DPC-containing DNA.
Figure 3.
Variation of DPC yields with increased protein:DNA molar ratio. 32P-endlabeled DNA strands containing site-specific DHP-deaza-dG were subjected oxidation to aldehyde and reductive amination in the presence of recombinant AlkB protein as described in Figure 1. The molar equivalents of AlkB protein to DNA were varied between 0.5:1 to 5:1, and DPCs were visualized by phosphorimaging. The production of covalent conjugates is increased in the presence excess of excess protein (Lanes 4–5).
Influence of protein identity on DPC formation
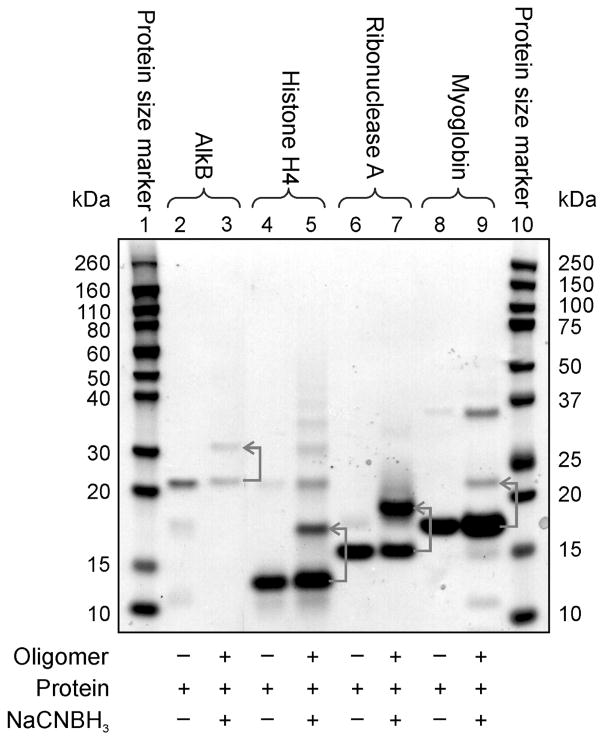
In theory, any protein containing lysine or arginine side chains can be cross-linked to DHP-deaza-dG containing DNA using the reductive amination strategy (Scheme 2). However, the reactivity and the accessibility of basic residues may vary depending on the protein identity. Therefore, the general applicability of our approach was examined using a range of proteins of different sizes and structures (Table 1 and Figure 4). We found that while some proteins (AlkB, NEIL1, Histone H4, GAPDH,) formed DPCs in a high yield (75 – 95%), significantly lower DPC yields (< 20%) were observed for others (trypsin, carboxypeptidase, myoglobin) (Table 1). In general, DNA binding proteins such as those involved in chromatin condensation and DNA repair produced DPCs in a higher yield than proteins that do not have an affinity for DNA. These results suggest that the formation of DPCs by reductive amination is facilitated by reversible DNA-protein interactions, which brings the two biomolecules into a close proximity to each other. Additional low mobility bands were observed for some proteins due to their dimerization (e.g. histone H4 and myoglobin, Figure 4).
Table 1.
Proteins and peptides used to generate DPCs using the reductive amination reactions with DHP-deaza-dG containing DNA.
| Protein/peptide | Molecular weight/kDa | No. of Lys per monomer | No. of Arg per monomer | % Yield of DPC |
|---|---|---|---|---|
| Histones | 11–15 | 14 | 9 | 99 |
| Substance P | 1.3 | 1 | 1 | 94 |
| Tat | 1.8 | 2 | 6 | 92 |
| Ribonuclease A | 14.7 | 8 | 4 | 89 |
| Histone H4 | 11.5 | 11 | 14 | 89 |
| GAPDH | 38 | 26 | 11 | 74 |
| Aprotinin | 6.5 | 4 | 7 | 73 |
| NEIL1 | 44.5 | 23 | 42 | 64 |
| Apomyoglobin | 17.3 | 19 | 2 | 63 |
| Insulin | 5.8 | 2 | 5 | 63 |
| AlkB | 22.9 | 8 | 13 | 59 |
| RNase | 14.7 | 8 | 4 | 56 |
| Hypertensin I | 0.9 | 0 | 1 | 43 |
| Pepsin | 34.6 | 11 | 4 | 25 |
| AGT | 21.9 | 12 | 6 | 20 |
| Myoglobin | 17.6 | 19 | 2 | 19 |
| T4 PNK | 132 | 29 | 19 | 18 |
| Carboxypeptidase A | 34 | 18 | 17 | 17 |
| Proteinase K | 28.9 | 14 | 17 | 9 |
| Trypsin | 23.3 | 8 | 3 | 4 |
| Pepstatin | 0.7 | 0 | 0 | 0 |
| Leupeptin | 0.5 | 0 | 1 | <1 |
Note: The reactions were carried out under optimized conditions using a 20-fold excess of protein/peptide with respect to the starting oligonucleotide.
Figure 4.
Influence of protein identity on DPC yield for reductive amination. Denaturing PAGE analysis of DPCs visualized via protein staining using SimplyBlue stain. Lane 1 and 10: protein size markers; Lanes 2–9: DPC formation between aldehyde-containing DNA and AlkB, histone H4, ribonuclease A, and myoglobin. Lanes 2, 4, 6, and 8 correspond to reactions conducted in the absence of DNA and NaCNBH3 (negative controls). The cross-linking reactions were conducted using the 2:1 protein:DNA molar ratio.
To gain insight into the identities of the amino acid residues participating in DPC formation, the cross-linking reactions were carried out with peptides of differing amino acid composition. High abundance DPC bands were observed for Tat and Substance P, which are rich in lysine and arginine residues (Figure S3, lanes 2 and 4) suggesting that amino groups of Lys and Arg may be involved in crosslinking to DNA. In contrast, no covalent conjugates were detected for pepstatin, which has no Lys or Arg residues (Figure S3, lane 5). A single DPC band observed for hypertensin I, which has only one Arg residue and no Lys (Figure S3, lane 3). Taken together, these results suggest that amino side chains of Lys and Arg within proteins can participate in reductive amination reactions with DHP-deaza-dG containing DNA.
Mass spectrometric characterization of DPCs
A mass spectrometry-based approach was employed to further characterize the structures of DNA-protein conjugates created by reductive amination and to identify the amino acids participating in reactions. The DNA components of DPCs to selected proteins (AlkB, RNase A, histone H4 and myoglobin) were digested with PDE I, PDE II, DNAse, and alkaline phosphatase. The resulting protein-nucleoside conjugates were cleaved with trypsin, and the peptides were analyzed by nano HPLC-ESI+-MS/MS on an Orbitrap Velos mass spectrometer. Tryptic peptides containing cross-links to deaza-dG were identified, and the cross-linking sites were determined by MS/MS sequencing (see an example in Figure 5). The mass spectral data were processed using Thermo Proteome Discoverer 1.3 (ThermoScientific, San Jose, CA) to identify the cross-linking sites. We found that for all four proteins examined, multiple lysine and arginine residues were engaged in the cross-linking reaction to aldehyde-containing DNA under reducing conditions (Tables 2, S1– S3, Figure S4).
Figure 5.
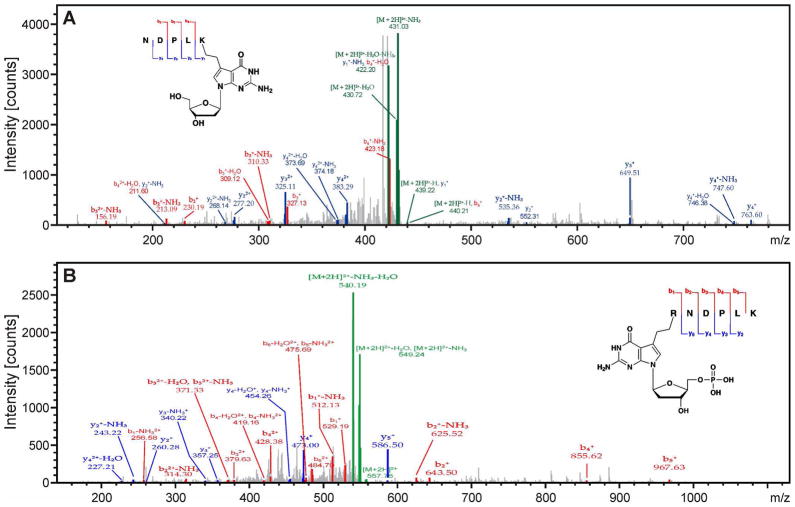
MS/MS spectra of AlkB tryptic peptides NDPLK* (m/z 439.7, doubly charged) and R*NDPLK (m/z 557.8, doubly charged), where K* and R* contain an ethyl cross-link to 7-deaza-2′-deoxyguanosine and 7-deaza-2′-deoxyguanosine 5′-monophosphate, respectively. Protein-DNA conjugates were generated by reductive amination reaction of recombinant AlkB protein with aldehyde-containing DNA 21-mer. The DNA component of the DPCs was digested with phosphodiesterases and alkaline phosphatase, and the resulting protein-nucleoside conjugate was subjected to tryptic digestion and peptide sequencing by nano HPLC-ESI+-MS/MS on Orbitrap Velos.
Table 2.
Sites of reductive amination-mediated crosslinking between DHP-deaza-dG containing DNA and recombinant AlkB protein as identified by nano HPLC-ESI+-MS/MS of tryptic digests.
| Amino acid positions | Amino acid sequence | Site of modification | Location within the protein |
|---|---|---|---|
| 25 – 35 | FAFNAAEQLIR | R35 | - |
| 122 – 127 | CVPGAK | K127 | DNA binding groove |
| 122 – 134 | CVPGAKLSLHQDK | K127 | DNA binding groove |
| 161 – 166 | RNDPLK | R161 | DNA binding groove |
| 162 – 166 | NDPLK | K166 | DNA binding groove |
| 162 – 167 | NDPLKR | K166 | DNA binding groove |
| 162 – 183 | RLLLEHGDVVVWGGESR | R167 | DNA binding groove |
| 194 – 204 | VGVHPLTTDCR | R204 | Active site |
| 194 – 210 | VGVHPLTTDCRYNLTFR | R204 | Active site |
| 194 – 210 | VGVHPLTTDCRYNLTFR | R210 | Active site |
| 205 – 215 | YNLTFRQAGKK | R210 | Active site |
HPLC-ESI+-MS/MS analysis of AlkB-DNA conjugates was repeated several times, yielding reproducible results. Peptide sequencing by HPLC-ESI+-MS/MS has revealed that two lysine residues (K127, K166) and five arginine residues of AlkB (R35, R161, R167, R204, R210) can participate in the AlkB-DNA cross-linking via reductive amination (Figure 6, Table 2). Examination of published crystal structures suggests that K134, R204, and R210 are located in the active site of AlkB, while K127, K166, and R167 reside in the DNA binding groove of the protein.29,30 These results suggest that specific AlkB-DNA binding facilitates covalent DPC formation. However, some of the residues participating in cross-linking (e.g. R35) are located outside of the DNA-binding domain. This can be explained by partial denaturation of the protein under the strongly reducing conditions used in our experiments.
Figure 6.
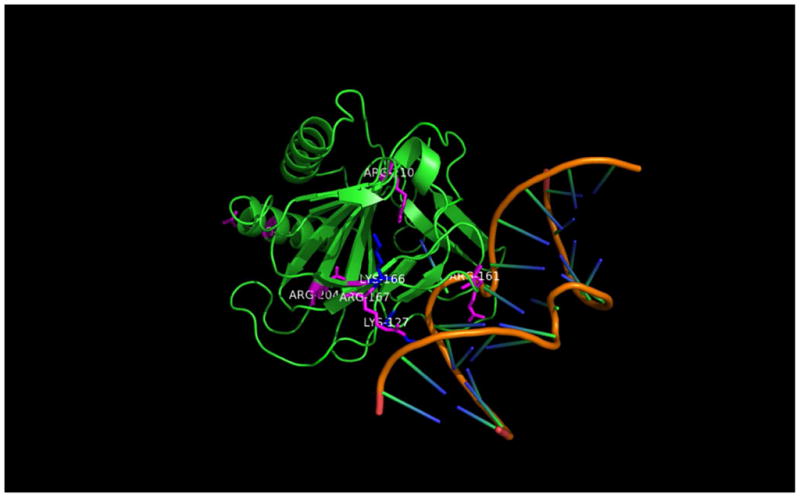
Crystal structure of AlkB protein bound to double stranded DNA showing the amino acids participating in DNA-protein crosslinking.
Synthesis and Structural Characterization of 7-Deaza-7-(2-(N-acetyllysine)ethan-1-yl)-2′-deoxyguanosine and 7-Deaza-7-(2-(N-acetylarginine)ethan-1-yl)-2′-deoxyguanosine Conjugates
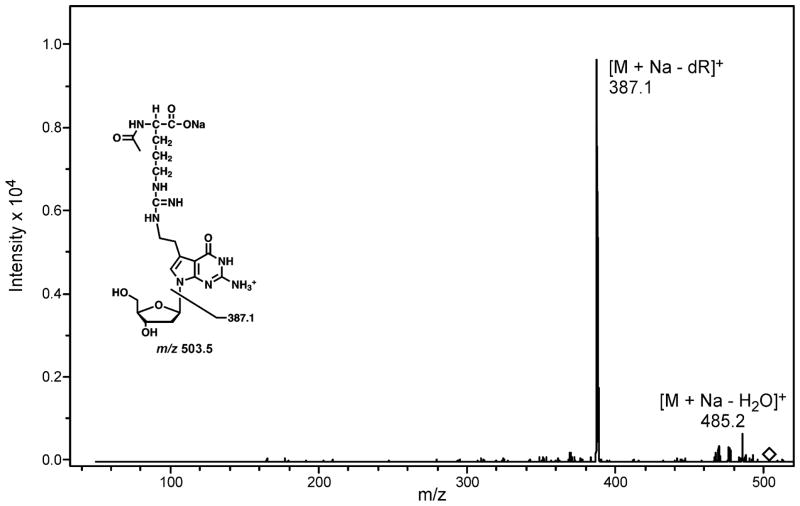
To confirm the exact chemical structure of the DNA-protein cross-links generated via reductive amination (Scheme 2), synthetic 7-deaza-7-(2,3-dihydroxypropan-1-yl)-2′-deoxyguanosine (DHP-deaza-dG) was allowed to react with N-acetyl protected Lys and Arg, and the resulting nucleoside-amino acid conjugates were isolated by HPLC and characterized by mass spectrometry. In the case of N-acetyl-Lys, the major conjugation product was observed at m/z 503.2, corresponding to the [M + Na]+ ions of 7-deaza-7-(2-(N-acetyl-lysine)ethan-1-yl)-2′-deoxyguanosine. MS/MS fragmentation pathway of this conjugate (Figure 7) was dominated by the product ions at m/z 387.1 and m/z 485.2, which correspond to the loss of deoxyribose, and a water molecule, respectively (Figure 7). Similar results were observed for N-acetyl-Arg. MS2 fragmentation of protonated 7-deaza-7-(2-(N-acetylarginine)ethan-1-yl)-2′-deoxyguanosine, [M+H]+, revealed the loss of water ([M+H-H2O]+), acetylamine ([M+H-MeCONH2]+) and deoxyribose ([M+H-dR]+) (Figure S5). Taken together, these results are consistent with the cross-linking mechanism shown in Scheme 2, e.g. Schiff base formation between the lysine amino side chain and the aldehyde functionality within oxidized DHP-deaza-dG, followed by imine reduction to generate a stable amino linkage.
Figure 7.
HPLC-ESI+-MS/MS spectrum of 7-deaza-7-(2-(N-acetyllysine)ethan-1-yl)-2′-deoxyguanosine.
Synthesis and Characterization of Nucleoside-Peptide Conjugates
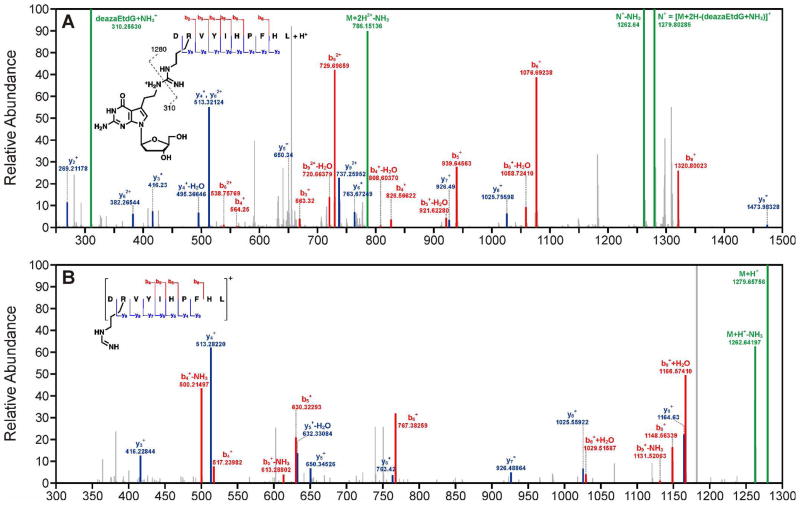
To further confirm the chemical structure and site of crosslinking of DPCs formed by this reductive amination strategy, nucleoside-peptide conjugates of 7-deaza-7-(2,3-dihydroxypropan-1-yl)-2′-deoxyguanosine (deaza-DHP-dG) to angiotensin I and substance P were characterized by mass spectrometry. MS2 spectrum of angiotensin I conjugate revealed that the site of crosslinking is side chain amino group of arginine (Figure 8A). The MS2 spectrum further revealed that 7-deaza-7-(1-aminoethan-2-yl)-2′deoxyguanosine is lost upon fragmentation of the doubly charged parent ion, [M+2H]+ = 794.9 m/z. The resulting singly charged daughter ion, [M+H]+ = 1279.8 m/z was further fragmented to observe additional b and y ions of the modified peptide that confirmed the site of crosslinking (Figure 8B).
Figure 8.
NanoLC-nanospary-MS2 (A) and MS3 (B) spectra of the peptide, Angiotensin I (DRVYIHPFHL) crosslinked to 7-deaza-7-(ethan-1-yl)-2′-deoxyguanosine.
Discussion
Dynamic DNA-protein interactions are crucial for many cellular functions including chromatin packaging,31 cell division, 32 DNA replication, gene expression,31,33 DNA damage response, and DNA repair.31,33 Proteins reversibly interact with DNA by a combination of electrostatic forces, hydrogen bonding, and stacking interactions, and their ability to dissociate from DNA is critical for their cellular functions. However, exposure to common antitumor drugs, environmental toxins, transition metals, UV light, ionizing radiation, and free radical-generating systems can result in proteins becoming covalently trapped on DNA.1,4 This generates super bulky, highly heterogeneous DNA-protein cross-links (DPCs) that can block DNA and RNA polymerases, causing toxicity and/or mutations in affected cells.2,3,10,17,34–37
Our previous mass spectrometry-based proteomics studies have revealed that covalent DNA-protein cross-links (DPCs) involving the N7 position of guanine are readily formed in human cells treated with clinically relevant concentrations of chemotherapeutic drugs (e.g., platinum compounds and nitrogen mustards)2,3 and metabolically activated carcinogens (e.g., 1,2,3,4-diepoxybutane).2,3 Additionally, covalent DPCs have been shown to accumulate in an age-dependent fashion in the brain and heart tissues, probably a result of exposure to endogenous reactive oxygen species, lipid peroxidation products, and transition metals.17 If not repaired, DPCs may contribute to the development of cancer, cardiovascular disease, and age-related neurodegeneration.16,17,37–41
Conflicting data exist in the literature regarding the mechanisms of cellular repair of DPC lesions. Reardon et al. examined the ability of reconstituted bacterial and mammalian excision nuclease systems to recognize the ring-open T4 pyrimidine DNA glycosylase-DNA cross-links.42 While the excision of DNA-protein conjugates was not detected, DPCs to short polypeptides were recognized and cleaved by mammalian protein extracts, leading to the hypothesis that DPCs are proteolytically degraded prior to their repair via the NER pathway.42 Similar conclusions were drawn by the Lloyd group when using a bacterial UvrABC system21 and by Baker et al. who examined model DPCs containing bacterial DNA methyltransferase-DPC (37 kDa) attached to the C-6 position of cytosine.23 Quievryn et al. observed reduced rates of repair of formaldehyde-induced DPCs in the presence of a protease inhibitor.43 In contrast, Nakano et al. reported that cytosolic ATP-dependent proteases are not involved in DPC removal.44,45 These authors proposed that homologous recombination repair is responsible for removing the majority of DPCs generated via oxanine, while only DPCs involving small proteins (< 12 kDa in bacteria and < 8–10 kDa in mammalian cells) are repaired by NER.44,45
Structural factors such as protein size, identity and lesion structures (e.g. major or minor groove of DNA) are also likely to affect the DPC lesion′s ability to be bypassed by DNA and RNA polymerases.1,46 For example, E. coli polymerase I and HIV-1 reverse transcriptase were completely blocked by cis-[diamminedichloridoplatinum (II)] (cisplatin) cross-link to histone H1.47 The A family human polymerase ν was blocked by DNA-peptide cross-links located in the minor groove via N2-dG.25 In contrast, chemically and structurally similar lesions located in the major groove of DNA via N6-dA were efficiently and accurately bypassed by both human polymerase ν and E. coli polymerase I.25,48
It is likely that the discrepancies between the mechanisms of DPC repair and bypass reported by different groups reflect structural differences between the model DPCs examined. Indeed, these previous studies have employed DNA-conjugates of diverse structure and size, including those where the protein was directly attached to ring open abasic sites in DNA.4,44,49 This underlines the need to reexamine the replication and repair of DPC-containing DNA using substrates resembling the lesions formed in cells. The most common site of DNA involved in DPC formation is the N7 of guanine.2,4,6 However, to our knowledge, no methods have been previously reported in the literature to generate N-7 guanine conjugated DPCs.
As mentioned in the introduction, currently available synthetic strategies to generate site-specific DPCs are limited to several main strategies. Lloyd et al.21 and Sancar et al.21 employed a semi-enzymatic approach to trap T4 pyrimidine dimer glycosylase/AP lyase (T4-pdg) on abasic sites of DNA in the presence of sodium borohydrate. A similar methodology has been used to attach oxoguanine glycosylase (Ogg) to DNA strands containing 8-oxo-dG. DNA methyltransferase (Dnmt) has been trapped on DNA containing 5-fluoro-5-methylcytosine.23 Another approach involves the use of oxanine (Ox) in DNA that spontaneously reacts with amino groups of proteins to give a pyrimidine ring-open structure,41 a strategy that is specific to nitric oxide-induced Oxa lesion, requires a large excess of the protein (425 to 3000-fold), as well as long incubation times (up to 48 h).41,45 Finally, Schiff base formation between acrolein-induced γ-HOPdG adducts and lysine residues of proteins and peptides can be stabilized in the presence of NaCNBH3.15,24,25 Either the N2 guanine or N6 adenine aldehyde functionality derived from acrolein-induced 3-(2′-deoxyribos-1′-yl)-5,6,7,8-tetrahydro-8-hydroxypyrimido [1,2a]purin-10(3H)-one (γ-HOPdG) can be reacted with proteins and peptides to produce a Schiff base, which was subsequently reduced to the corresponding amine with sodium cyanoborohydride.24,25 To our knowledge, no synthetic methodologies are available to generate N7-gianine DPCs such as those formed in vivo upon exposure to environmental carcinogens and antitumor agents.2,3
In the present study, a post-synthetic reductive amination strategy was employed to create hydrolytically stable structural mimics of N-7 guanine conjugated DPCs by reductive amination reactions between the Lys and Arg side chains of proteins and acetaldehyde functionalities of modified 7-deazaguanine residues of DNA. The main advantage of this approach is that it generates sequence specific DPC lesions structurally analogous to the lesions formed in vivo. Reductive amination methodology is highly versatile, as it can be used to generate DPCs to most proteins and peptides containing Lys and/or Arg residues (Table 1). The resulting structurally defined and hydrolytically stable DPCs can be used to study the biological fate of DPCs in vitro to better understand the effects of these lesions in cells. Furthermore, experimental methods are being developed in our laboratory to incorporate these substrates into plasmid DNA and study their repair in cells to identify the mechanisms responsible for the removal of these lesions in vivo.
Model DPC lesions generated in this work resemble the adducts induced by antitumor nitrogen mustards (Scheme 3).2,6 We have previously identified 39 proteins that form covalent DPC in human fibrosarcoma (HT1080) cells treated with mechlorethamine.50 However, it is not known to what extent DPC formation contributes to toxicity of nitrogen mustards in cancer cells. The availability of hydrolytically stable model DPCs substrates will, for the first time, enable structural and biological evaluation of these super-bulky lesions. Based on our recent studies with DNA-reactive protein reagents that specifically induce DPCs in cells,20 we hypothesize that spontaneous and xenobiotic-induced DPCs, if not repaired, compromise the efficiency and the accuracy of DNA replication and are responsible for a major portion of the toxicity and mutagenicity induced by bis-alkylating agents, UV light, reactive oxygen species, and γ-radiation.
The model DPC substrates created by reductive amination (Scheme 2) are site-specific in respect to DNA, but may involve multiple possible cross-linking sites within the protein (Figure 6, Tables 2, S1–S3). This limited specificity in respect to the protein side chains should not affect the ability to employ these model conjugates in biological studies since the “real” DPC lesions formed in cells are also heterogenous in nature. However, it may not be practical to use this approach to generate DNA-protein conjugates for structural studies by NMR or X-ray crystallography, especially in the case of proteins that contain multiple basic residues available for reaction with DNA. Other types of conjugations that employ bioorthogonal reactive groups in each biomolecule (protein and DNA) may be more appropriate for this purpose. We are currently exploring the use of copper-catalyzed [3+2] Huisgen cycloaddition (click reaction) between azide-functionalized proteins51 and alkyne-containing DNA to generate site specific DPC conjugates.
Supplementary Material
Acknowledgments
We thank Prof. Chuan He (University of Chicago) for providing recombinant AlkB protein and Robert Carlson (Masonic Cancer Center, University of Minnesota) for preparing the figures for this manuscript. This research was supported by grants from the National Cancer Institute (CA100670 (NT) and CA165911 (OS)) and a brainstorm award from the University of Minnesota Masonic Cancer Center (NT).
List of Abbreviations
- DHP-deaza-dG
7-deaza-7-(2,3-dihydroxypropan-1-yl)-2′-deoxy-guanosine
- DTT
dithiothreitol
- DPCs
DNA-protein cross-links
- MALDI-TOF-MS
matrix assisted laser desorption ionization time of flight mass spectrometry
- PDE I
phosphodiesterase I (snake venom exonuclease)
- PDE II
phosphodiesterase II (bovine spleen exonuclease)
- RP-HPLC-ESI+-MS/MS
reversed phase high performance liquid chromatography – electrospray ionization tandem mass spectrometry
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
Footnotes
Supporting Information Available: Supplement includes tables showing the sites of reductive amination-mediated cross-linking between DHP-deaza-dG containing DNA and recombinant histone H4, myoglobin, and RNase I; figures illustrating the influence of reaction temperature and time on cross-linking yields, SDS-PAGE analysis of DPCs prepared using various peptides, and crystal structures of myoglobin and ribonuclease A indicating the cross-linking sites, HPLC-ESI+-MS/MS analysis of 7-deaza-7-(2-(N-acetylarginine)ethan-1-yl)-2′-deoxyguanosine. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Barker S, Weinfeld M, Murray D. DNA-protein crosslinks: their induction, repair, and biological consequences. Mutat Res. 2005;589:111–135. doi: 10.1016/j.mrrev.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Loeber RL, Michaelson-Richie ED, Codreanu SG, Liebler DC, Campbell CR, Tretyakova NY. Proteomic analysis of DNA-protein cross-linking by antitumor nitrogen mustards. Chem Res Toxicol. 2009;22:1151–1162. doi: 10.1021/tx900078y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michaelson-Richie ED, Loeber RL, Codreanu SG, Ming X, Liebler DC, Campbell C, Tretyakova NY. DNA-Protein Cross-Linking by 1,2,3,4-Diepoxybutane. J Proteome Res. 2010;9:4356–4367. doi: 10.1021/pr1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ide H, Shoulkamy MI, Nakano T, Miyamoto-Matsubara M, Salem AMH. Repair and biochemical effects of DNA-protein crosslinks. Mutat Res. 2011;711:113–122. doi: 10.1016/j.mrfmmm.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Gherezghiher TB, Ming X, Villalta PW, Campbell C, Tretyakova NY. 1,2,3,4-Diepoxybutane-induced DNA-protein cross-linking in human fibrosarcoma (HT1080) cells. J Proteome Res. 2013;12:2151–2164. doi: 10.1021/pr3011974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loeber R, Michaelson E, Fang Q, Campbell C, Pegg AE, Tretyakova N. Cross-linking of the DNA repair protein O6-alkylguanine DNA alkyltransferase to DNA in the presence of antitumor nitrogen mustards. Chem Res Toxicol. 2008;21:787–795. doi: 10.1021/tx7004508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao W, Pen G, Yang X. DNA-protein crosslinks induced by formaldehyde and its repair process. Int J Environ Pollut. 2009;37:299–308. [Google Scholar]
- 8.Lu K, Collins LB, Ru H, Bermudez E, Swenberg J. Distribution of DNA adducts caused by inhaled formaldehyde is consistent with induction of nasal carcinoma but not leukemia. Toxicol Sci. 2010;116:441–451. doi: 10.1093/toxsci/kfq061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu K, Ye W, Zhou L, Collins LB, Chen X, Gold A, Ball LM, Swenberg J. Structural characterization of formaldehyde-induced cross-links between amino acids and deoxynucleosides and their oligomers. J Am Chem Soc. 2010;132:3388–3399. doi: 10.1021/ja908282f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa M, Zhitkovich A, Toniolo P. DNA-protein cross-links in welders: molecular implications. Cancer Res. 1993;53:460–463. [PubMed] [Google Scholar]
- 11.Lei YX, Zhang Q, Zhuang ZX. Study on DNA-protein crosslinks induced by chromate and nickel compounds in vivo with 125I-postlabelling assay. Mutat Res. 1995;329:197–203. doi: 10.1016/0027-5107(95)00047-m. [DOI] [PubMed] [Google Scholar]
- 12.Xie J, Fan R, Meng Z. Protein oxidation and DNA-protein crosslink induced by sulfur dioxide in lungs, livers, and hearts from mice. Inhal Toxicol. 2007;19:759–765. doi: 10.1080/08958370701399885. [DOI] [PubMed] [Google Scholar]
- 13.Johansen ME, Muller JG, Xu X, Burrows CJ. Oxidatively induced DNA-protein cross-linking between single-stranded binding protein and oligodeoxynucleotides containing 8-oxo-7,8-dihydro-2′-deoxyguanosine. Biochemistry. 2005;44:5660–5671. doi: 10.1021/bi047580n. [DOI] [PubMed] [Google Scholar]
- 14.Xu X, Muller JG, Ye Y, Burrows CJ. DNA-protein cross-links between guanine and lysine depend on the mechanism of oxidation for formation of C5 vs C8 guanosine adducts. J Am Chem Soc. 2008;130:703–709. doi: 10.1021/ja077102a. [DOI] [PubMed] [Google Scholar]
- 15.Minko IG, Kozekov ID, Kozekova A, Harris TM, Rizzo CJ, Lloyd RS. Mutagenic potential of DNA-peptide crosslinks mediated by acrolein-derived DNA adducts. Mutat Res. 2008;637:161–172. doi: 10.1016/j.mrfmmm.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin LJ. DNA damage and repair: relevance to mechanisms of neurodegeneration. J Neuropathol Exp Neurol. 2008;67:377–387. doi: 10.1097/NEN.0b013e31816ff780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zahn RK, Zahn-Daimler G, Ax S, Hosokawa M, Takeda T. Assessment of DNA-protein crosslinks in the course of aging in two mouse strains by use of a modified alkaline filter elution applied to whole tissue samples. Mech Ageing Dev. 1999;108:99–112. doi: 10.1016/s0047-6374(98)00151-1. [DOI] [PubMed] [Google Scholar]
- 18.Garaycoechea JI, Crossan GP, Langevin F, Daly M, Arends MJ, Patel KJ. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 2012;489:571–575. doi: 10.1038/nature11368. [DOI] [PubMed] [Google Scholar]
- 19.Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011;475:53–58. doi: 10.1038/nature10192. [DOI] [PubMed] [Google Scholar]
- 20.Tretyakova NY, Michaelson-Richie ED, Gherezghiher TB, Kurtz J, Ming X, Wickramaratne S, Campion M, Kanugula S, Pegg AE, Campbell C. DNA-reactive protein monoepoxides induce cell death and mutagenesis in mammalian cells. Biochemistry. 2013;52:3171–3181. doi: 10.1021/bi400273m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minko IG, Zou Y, Lloyd RS. Incision of DNA-protein crosslinks by UvrABC nuclease suggests a potential repair pathway involving nucleotide excision repair. Proc Natl Acad Sci U S A. 2002;99:1905–1909. doi: 10.1073/pnas.042700399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banerjee A, Yang W, Karplus M, Verdine GL. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005;434:612–618. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 23.Baker DJ, Wuenschell G, Xia L, Termini J, Bates SE, Riggs AD, O’Connor TR. Nucleotide excision repair eliminates unique DNA-protein cross-links from mammalian cells. J Biol Chem. 2007;282:22592–22604. doi: 10.1074/jbc.M702856200. [DOI] [PubMed] [Google Scholar]
- 24.VanderVeen L, Harris TM, Jen-Jacobson L, Marnett LJ. Formation of DNA-protein cross-links between gamma-hydroxypropanodeoxyguanosine and EcoRI. Chem Res Toxicol. 2008;21:1733–1738. doi: 10.1021/tx800092g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamanaka K, Minko IG, Takata Ki, Kolbanovskiy A, Kozekov ID, Wood RD, Rizzo CJ, Lloyd RS. Novel Enzymatic Function of DNA Polymerase ν in Translesion DNA Synthesis Past Major Groove DNA-Peptide and DNA-DNA Cross-Links. Chem Res Toxicol. 2010:689–695. doi: 10.1021/tx900449u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fedtke N, Boucheron JA, Walker VE, Swenberg JA. Vinyl chloride-induced DNA adducts. II: Formation and persistence of 7-(2′-oxoethyl)guanine and N2,3-ethenoguanine in rat tissue DNA. Carcinogenesis. 1990;11:1287–1292. doi: 10.1093/carcin/11.8.1287. [DOI] [PubMed] [Google Scholar]
- 27.Angelov T, Guainazzi A, Schãrer OD. Generation of DNA interstrand cross-links by post-synthetic reductive amination. Org Lett. 2009;11:661–664. doi: 10.1021/ol802719a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gates KS, Nooner T, Dutta S. Biologically relevant chemical reactions of N7-alkylguanine residues in DNA. Chem Res Toxicol. 2004;17:839–856. doi: 10.1021/tx049965c. [DOI] [PubMed] [Google Scholar]
- 29.Westbye MP, Feyzi E, Aas PA, Vagbo CB, Talstad VA, Kavli B, Hagen L, Sundheim O, Akbari M, Liabakk N, Slupphaug G, Otterlei M, Krokan HE. Human AlkB homolog 1 is a mitochondrial protein that demethylates 3-methylcytosine in DNA and RNA. J Biol Chem. 2008;283:25046–25056. doi: 10.1074/jbc.M803776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang CG, Yi C, Duguid EM, Sullivan CT, Jian X, Rice PA, He C. Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature. 2008;452:961–965. doi: 10.1038/nature06889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 32.Bednar J, Horowitz RA, Grigoyev SA, Carruthers LM, Hansen JC, Koster AJ, Woodcock CL. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci U S A. 1998;95:14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ritzefeld M, Sewald N. Real-time analysis of specific protein-DNA interactions with surface plasmon resonance. J Amino Acids. 2012;2012:1–19. doi: 10.1155/2012/816032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaham J, Bomstein Y, Meltzer A, Kaufman Z, Palma E, Ribak J. DNA--protein crosslinks, a biomarker of exposure to formaldehyde--in vitro and in vivo studies. Carcinogenesis. 1996;17:121–125. doi: 10.1093/carcin/17.1.121. [DOI] [PubMed] [Google Scholar]
- 35.Tsapakos MJ, Hampton TH, Wetterhahn KE. Chromium(VI)-induced DNA lesions and chromium distribution in rat kidney, liver, and lung. Cancer Res. 1983;43:5662–5667. [PubMed] [Google Scholar]
- 36.Cupo DY, Wetterhahn KE. Binding of Chromium to Chromatin and DNA from Liver and Kidney of Rats Treated with Sodium Dichromate and Chromium (III) Chloride in vivo. Cancer Res. 1985;45:1146–1151. [PubMed] [Google Scholar]
- 37.Reddy VP, Zhu X, Perry G, Smith MA. Oxidative stress in diabetes and Alzheimer’s disease. J Alzheimers Dis. 2009;16:763–774. doi: 10.3233/JAD-2009-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Flora S, Izzotti A, Randerath K, Randerath E, Bartsch H, Nair J, Balansky R, van Schooten F, Degan P, Fronza G, Walsh D, Lewtas J. DNA adducts and chronic degenerative disease. Pathogenetic relevance and implications in preventive medicine. Mutat Res. 1996;366:197–238. [PubMed] [Google Scholar]
- 39.Bandyopadhyay U, Das D, Banerjee RK. Reactive oxygen species: oxidative damage and pathogenesis. Curr Sci. 1999;77:658–666. [Google Scholar]
- 40.Lovell MA, Markesbery WR. Oxidative DNA damage in mild cognitive impairment and late-stage Alzheimer’s disease. Nucleic Acids Res. 2007;35:7497–7504. doi: 10.1093/nar/gkm821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakano T, Terato H, Asagoshi K, Masaoka A, Mukuta M, Ohyama Y, Suzuki T, Makino K, Ide H. DNA-protein cross-link formation mediated by oxanine. A novel genotoxic mechanism of nitric oxide-induced DNA damage. J Biol Chem. 2003;278:25264–25272. doi: 10.1074/jbc.M212847200. [DOI] [PubMed] [Google Scholar]
- 42.Reardon JT, Cheng Y, Sancar A. Repair of DNA-Protein Cross-links in Mammalian Cells. Cell Cycle. 2006;5:1366–1370. doi: 10.4161/cc.5.13.2892. [DOI] [PubMed] [Google Scholar]
- 43.Quievryn G, Zhitkovich A. Loss of DNA-protein crosslinks from formaldehyde-exposed cells occurs through spontaneous hydrolysis and an active repair process linked to proteosome function. Carcinogenesis. 2000;21:1573–1580. [PubMed] [Google Scholar]
- 44.Nakano T, Katafuchi A, Matsubara M, Terato H, Tsuboi T, Masuda T, Tatsumoto T, Pack SP, Makino K, Croteau DL, Van Houten B, Iijima K, Tauchi H, Ide H. Homologous recombination but not nucleotide excision repair plays a pivotal role in tolerance of DNA-protein cross-links in mammalian cells. J Biol Chem. 2009;284:27065–27076. doi: 10.1074/jbc.M109.019174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakano T, Morishita S, Katafuchi A, Matsubara M, Horikawa Y, Terato H, Salem AMH, Izumi S, Pack SP, Makino K, Ide H. Nucleotide excision repair and homologous recombination systems commit differentially to the repair of DNA-protein crosslinks. Mol Cell. 2007;28:147–158. doi: 10.1016/j.molcel.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 46.Oleinick NL, Chiu SM, Ramakrishnan N, Xue LY. The formation, identification, and significance of DNA-protein cross-links in mammalian cells. Br J Cancer. 1987;8:135–140. [PMC free article] [PubMed] [Google Scholar]
- 47.Chvalova K, Brabec V, Kasparkova J. Mechanism of the formation of DNA-protein cross-links by antitumor cisplatin. Nucleic Acids Res. 2007;35:1812–1821. doi: 10.1093/nar/gkm032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamanaka K, Minko IG, Finkel SE, Goodman MF, Lloyd RS. Role of high-fidelity Escherichia coli DNA polymerase I in replication bypass of a deoxyadenosine DNA-peptide cross-link. J Bacteriol. 2011;193:3815–3821. doi: 10.1128/JB.01550-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Minko IG, Kurtz AJ, Croteau DL, Van Houten B, Harris TM, Lloyd RS. Initiation of repair of DNA-polypeptide cross-links by the UvrABC nuclease. Biochemistry. 2005;44:3000–3009. doi: 10.1021/bi0478805. [DOI] [PubMed] [Google Scholar]
- 50.Michaelson-Richie ED, Ming X, Codreanu SG, Loeber RL, Liebler DC, Campbell C, Tretyakova NY. Mechlorethamine-induced DNA-protein cross-linking in human fibrosarcoma (HT1080) cells. J Proteome Res. 2011;10:2785–2796. doi: 10.1021/pr200042u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duckworth BP, Chen Y, Wollack JW, Sham Y, Mueller JD, Taton TA, Distefano MD. A universal method for the preparation of covalent protein-DNA conjugates for use in creating protein nanostructures. Angew Chem Int Ed Engl. 2007;46:8819–8822. doi: 10.1002/anie.200701942. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.