Abstract
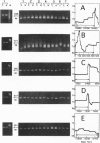
We have analyzed the level of intraindividual sequence variability (heteroplasmy) of mtDNA in human brain by denaturing gradient gel electrophoresis and sequencing. Single base substitutions, as well as insertions or deletions of single bases, were numerous in the noncoding control region (D-loop), and 35-45% of the molecules from a single tissue showed sequence differences. By contrast, heteroplasmy in coding regions was not detected. The lower level of heteroplasmy in the coding regions is indicative of selection against deleterious mutations. Similar levels of heteroplasmy were found in two brain regions from the same individual, while no heteroplasmy was detected in blood. Thus, heteroplasmy seems to be more frequent in nonmitotic tissues. We observed a 7.7-fold increase in the frequency of deletions/insertions and a 2.2-fold increase in the overall frequency of heteroplasmic mutations in two individuals aged 96 and 99, relative to an individual aged 28. Our results show that intraindividual sequence variability occurs at a high frequency in the noncoding regions of normal human brain and indicate that small insertions and deletions might accumulate with age at a lower rate than large rearrangements.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
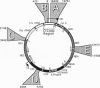
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Baumer A., Zhang C., Linnane A. W., Nagley P. Age-related human mtDNA deletions: a heterogeneous set of deletions arising at a single pair of directly repeated sequences. Am J Hum Genet. 1994 Apr;54(4):618–630. [PMC free article] [PubMed] [Google Scholar]
- Bendall K. E., Sykes B. C. Length heteroplasmy in the first hypervariable segment of the human mtDNA control region. Am J Hum Genet. 1995 Aug;57(2):248–256. [PMC free article] [PubMed] [Google Scholar]
- Bennett J. L., Clayton D. A. Efficient site-specific cleavage by RNase MRP requires interaction with two evolutionarily conserved mitochondrial RNA sequences. Mol Cell Biol. 1990 May;10(5):2191–2201. doi: 10.1128/mcb.10.5.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenteich A., Mitchell L. G., Merril C. R. A lifetime of retinal light exposure does not appear to increase mitochondrial mutations. Gene. 1991 Dec 15;108(2):305–309. doi: 10.1016/0378-1119(91)90451-g. [DOI] [PubMed] [Google Scholar]
- Boursot P., Yonekawa H., Bonhomme F. Heteroplasmy in mice with deletion of a large coding region of mitochondrial DNA. Mol Biol Evol. 1987 Jan;4(1):46–55. doi: 10.1093/oxfordjournals.molbev.a040421. [DOI] [PubMed] [Google Scholar]
- Cann R. L., Wilson A. C. Length mutations in human mitochondrial DNA. Genetics. 1983 Aug;104(4):699–711. doi: 10.1093/genetics/104.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavelier L., Jazin E. E., Eriksson I., Prince J., Båve U., Oreland L., Gyllensten U. Decreased cytochrome-c oxidase activity and lack of age-related accumulation of mitochondrial DNA deletions in the brains of schizophrenics. Genomics. 1995 Sep 1;29(1):217–224. doi: 10.1006/geno.1995.1234. [DOI] [PubMed] [Google Scholar]
- Corral-Debrinski M., Horton T., Lott M. T., Shoffner J. M., Beal M. F., Wallace D. C. Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nat Genet. 1992 Dec;2(4):324–329. doi: 10.1038/ng1292-324. [DOI] [PubMed] [Google Scholar]
- Cortopassi G. A., Arnheim N. Detection of a specific mitochondrial DNA deletion in tissues of older humans. Nucleic Acids Res. 1990 Dec 11;18(23):6927–6933. doi: 10.1093/nar/18.23.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortopassi G. A., Shibata D., Soong N. W., Arnheim N. A pattern of accumulation of a somatic deletion of mitochondrial DNA in aging human tissues. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7370–7374. doi: 10.1073/pnas.89.16.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Densmore L. D., Wright J. W., Brown W. M. Length variation and heteroplasmy are frequent in mitochondrial DNA from parthenogenetic and bisexual lizards (genus Cnemidophorus). Genetics. 1985 Aug;110(4):689–707. doi: 10.1093/genetics/110.4.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghivizzani S. C., Mackay S. L., Madsen C. S., Laipis P. J., Hauswirth W. W. Transcribed heteroplasmic repeated sequences in the porcine mitochondrial DNA D-loop region. J Mol Evol. 1993 Jul;37(1):36–37. doi: 10.1007/BF00170460. [DOI] [PubMed] [Google Scholar]
- Greenberg B. D., Newbold J. E., Sugino A. Intraspecific nucleotide sequence variability surrounding the origin of replication in human mitochondrial DNA. Gene. 1983 Jan-Feb;21(1-2):33–49. doi: 10.1016/0378-1119(83)90145-2. [DOI] [PubMed] [Google Scholar]
- Hale L. R., Singh R. S. Extensive variation and heteroplasmy in size of mitochondrial DNA among geographic populations of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8813–8817. doi: 10.1073/pnas.83.22.8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison R. G., Rand D. M., Wheeler W. C. Mitochondrial DNA size variation within individual crickets. Science. 1985 Jun 21;228(4706):1446–1448. doi: 10.1126/science.228.4706.1446. [DOI] [PubMed] [Google Scholar]
- Hauswirth W. W., Clayton D. A. Length heterogeneity of a conserved displacement-loop sequence in human mitochondrial DNA. Nucleic Acids Res. 1985 Nov 25;13(22):8093–8104. doi: 10.1093/nar/13.22.8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeh W. R., Blakley K. H., Brown W. M. Heteroplasmy suggests limited biparental inheritance of Mytilus mitochondrial DNA. Science. 1991 Mar 22;251(5000):1488–1490. doi: 10.1126/science.1672472. [DOI] [PubMed] [Google Scholar]
- Kim Y. L., Brown M. D., Wallace D. C. Single-strand conformation polymorphism analysis for the detection of point mutations in the mitochondrial DNA. Anal Biochem. 1995 Jan 20;224(2):608–611. doi: 10.1006/abio.1995.1096. [DOI] [PubMed] [Google Scholar]
- Kondo R., Horai S., Satta Y., Takahata N. Evolution of hominoid mitochondrial DNA with special reference to the silent substitution rate over the genome. J Mol Evol. 1993 Jun;36(6):517–531. doi: 10.1007/BF00556356. [DOI] [PubMed] [Google Scholar]
- Lee H. C., Pang C. Y., Hsu H. S., Wei Y. H. Ageing-associated tandem duplications in the D-loop of mitochondrial DNA of human muscle. FEBS Lett. 1994 Oct 31;354(1):79–83. doi: 10.1016/0014-5793(94)01063-3. [DOI] [PubMed] [Google Scholar]
- Lee H. C., Pang C. Y., Hsu H. S., Wei Y. H. Differential accumulations of 4,977 bp deletion in mitochondrial DNA of various tissues in human ageing. Biochim Biophys Acta. 1994 Apr 12;1226(1):37–43. doi: 10.1016/0925-4439(94)90056-6. [DOI] [PubMed] [Google Scholar]
- Monnat R. J., Jr, Loeb L. A. Nucleotide sequence preservation of human mitochondrial DNA. Proc Natl Acad Sci U S A. 1985 May;82(9):2895–2899. doi: 10.1073/pnas.82.9.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münscher C., Rieger T., Müller-Höcker J., Kadenbach B. The point mutation of mitochondrial DNA characteristic for MERRF disease is found also in healthy people of different ages. FEBS Lett. 1993 Feb 8;317(1-2):27–30. doi: 10.1016/0014-5793(93)81484-h. [DOI] [PubMed] [Google Scholar]
- Shigenaga M. K., Hagen T. M., Ames B. N. Oxidative damage and mitochondrial decay in aging. Proc Natl Acad Sci U S A. 1994 Nov 8;91(23):10771–10778. doi: 10.1073/pnas.91.23.10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soong N. W., Hinton D. R., Cortopassi G., Arnheim N. Mosaicism for a specific somatic mitochondrial DNA mutation in adult human brain. Nat Genet. 1992 Dec;2(4):318–323. doi: 10.1038/ng1292-318. [DOI] [PubMed] [Google Scholar]
- Stoneking M. Departure of human mitochondrial DNA variation from neutral expectations: an alternative explanation. J Mol Evol. 1990 Oct;31(4):343–350. doi: 10.1007/BF02101128. [DOI] [PubMed] [Google Scholar]
- Stoneking M., Hedgecock D., Higuchi R. G., Vigilant L., Erlich H. A. Population variation of human mtDNA control region sequences detected by enzymatic amplification and sequence-specific oligonucleotide probes. Am J Hum Genet. 1991 Feb;48(2):370–382. [PMC free article] [PubMed] [Google Scholar]
- Vigilant L., Stoneking M., Harpending H., Hawkes K., Wilson A. C. African populations and the evolution of human mitochondrial DNA. Science. 1991 Sep 27;253(5027):1503–1507. doi: 10.1126/science.1840702. [DOI] [PubMed] [Google Scholar]
- Walberg M. W., Clayton D. A. Sequence and properties of the human KB cell and mouse L cell D-loop regions of mitochondrial DNA. Nucleic Acids Res. 1981 Oct 24;9(20):5411–5421. doi: 10.1093/nar/9.20.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D. C. Diseases of the mitochondrial DNA. Annu Rev Biochem. 1992;61:1175–1212. doi: 10.1146/annurev.bi.61.070192.005523. [DOI] [PubMed] [Google Scholar]
- Wallace D. C. Mitochondrial DNA sequence variation in human evolution and disease. Proc Natl Acad Sci U S A. 1994 Sep 13;91(19):8739–8746. doi: 10.1073/pnas.91.19.8739. [DOI] [PMC free article] [PubMed] [Google Scholar]