Abstract
The prevailing demographic model for Drosophila melanogaster suggests that the colonization of North America occurred very recently from a subset of European flies that rapidly expanded across the continent. This model implies a sudden population growth and range expansion consistent with very low or no population subdivision. As flies adapt to new environments, local adaptation events may be expected. In order to describe demographic and selective events during North American colonization, we have generated a dataset of 35 individual whole genome sequences from inbred lines of D. melanogaster from a west coast US population (Winters, California, USA) and compared them with a public genome dataset from Raleigh (Raleigh, North Carolina, USA). We analyzed nuclear and mitochondrial genomes and describe levels of variation and divergence within and between these two North American D. melanogaster populations. Both populations exhibit negative values of Tajima’s D across the genome, a common signature of demographic expansion. We also detected a low but significant level of genome-wide differentiation between the two populations, as well as multiple allele surfing events, which can be the result of gene drift in local subpopulations on the edge of an expansion wave. In contrast to this genome-wide pattern, we uncovered a 50 kilobases segment in chromosome arm 3L that showed all the hallmarks of a soft selective sweep in both populations. A comparison of allele frequencies within this divergent region among six populations from three continents allowed us to cluster these populations in two differentiated groups, providing evidence for the action of natural selection on a global scale.
Keywords: whole genome, demographic expansion, population differentiation, positive selection, soft selective sweep, global pattern
Introduction
It is generally accepted that D. melanogaster originated in equatorial Africa from a D. melanogaster-D. simulans ancestor (Lachaise et al. 1988; Stephan & Li 2007). Li & Stephan (2006) determined that a demographic and range expansion occurred about 60,000 years ago. Colonization of Eurasia took place after the last Pleistocene glaciation about 10–15 thousand years ago (David & Capy 1988). Due to the small size of the founder populations, this colonization event was associated with a severe bottleneck (Li & Stephan 2006). The colonization required adaptation to more temperate and cold climates leading to the fixation of a large number of beneficial mutations. Thus, the overall pattern of genetic variation among European D. melanogaster populations can be explained by a combination of demographic and selective processes (Li & Stephan 2006).
Colonization of the Americas appears to have taken place in two steps. The first step occurred a few hundred years ago with the introduction of flies from Tropical Africa to Tropical America, likely following the trade of slaves (David & Capy 1988). The second step occurred as late as the mid 19th century and involved the colonization of North America from European D. melanogaster populations (David & Capy 1988). D. melanogaster was first described in New York in 1875, and subsequently found in many other parts of the continent (Keller 2007), most likely as a result of a rapid demographic expansion. Similar to the Eurasian colonization, the colonization of North America possibly involved a population bottleneck. Since North America was colonized by a subset of European flies, which in turn derived from the ancestral African pool, we would expect low genetic variation among North American D. melanogaster populations. Contrary to this expectation, Caracristi & Schlötterer (2003) found high levels of polymorphism among North American flies. Notably, they observed substantial divergence between European and North American populations and a greater proportion of shared alleles between African and eastern North American flies than between African and European samples. These authors suggested that this could be the result of an admixture between Caribbean and North American flies with the Caribbean populations as a source of African alleles. More recently, Duchen et al. (2013) revisited the demographic origin of the North American populations using an Approximate Bayesian Computation approach, and found that admixture between Africa and Europe most likely generated the North American populations, with an estimated proportion of African ancestry of 15%.
Contradictory results exist regarding genetic structure among North America D. melanogaster populations. For example, Kreitman & Aguadé (1986) and Coyne & Milstead (1987) found high levels of gene flow based on RFLP of the Adh locus and mark-recapture experiments respectively. Conversely, allozyme studies of Johnson & Schaffer (1973) and Singh & Long (1992), as well as RFLP analyses of the Pgd locus by Begun & Aquadro (1994) and a chromosomal inversion survey of Mettler et al. (1977), showed genetic differentiation among North American flies. All these types of molecular markers are now suspected to be affected by natural selection, and hence any demographic signal may be masked by selection. In an attempt to remove the effects of natural selection, Caracristi & Schlötterer (2003) conducted a study of 48 microsatellite loci and found significant differentiation between East coast and West coast North American populations. Yet, a large-scale effort is needed in order to understand the relative contribution of demography and selection in shaping the patterns of polymorphism and population subdivision among North American D. melanogaster.
To resolve this issue, we have focused on whole-genome data, which are particularly useful in understanding to which extent demography and selection have shaped genetic variation within and between populations. Demographic processes affect the entire genome, whereas natural selection acts on specific loci. Genome-wide analysis of genetic polymorphism should help to distinguish between demographic and selective forces, and identify those genes that are involved in local adaptation (Biswas & Akey 2006; Turner et al. 2010; Yi et al. 2010). However, it is worth noting that a recent series of papers have challenged this view, suggesting a pervasive role of natural selection in shaping the polymorphism patterns of the genome of certain species, like D. melanogaster (Hahn 2008; Wright & Andolfatto 2008; Sella et al. 2009).
To our knowledge, only six published studies have analyzed whole genome sequences in Drosophila species from a population genomics perspective to date. Begun et al. (2007) sequenced seven lines of D. simulans and one of D. yakuba and compared them with the reference sequence for D. melanogaster. They selected these fly lines to capture variation in ancestral geographic regions, recent cosmopolitan populations, and the three highly diverged mitochondrial haplotypes described for this species. Sackton et al. (2009) used high- throughput sequencing to generate a low coverage dataset of nine D. melanogaster lines. This pilot project tested the accuracy of population genetic inferences using shallow sequencing depth. Although the authors sequenced flies from two different regions, North America and Africa, they did not perform a population comparison due to the limitations of their datasets. More recently, Mackay et al. (2012) conducted a large population genomic and phenotypic analysis in a panel of 168 D. melanogaster inbred lines, and performed genome-wide association studies to identify SNPs that are likely affecting the phenotypes. A population comparison was not possible in this study as all flies were sampled in a single location. Langley et al. (2012) obtained whole genome sequences of a number of inbred genotypes from two different populations, and performed an exhaustive analysis of polymorphism, divergence and linkage disequilibrium across the euchromatic portion of the genome. Finally, Kolaczkowski et al. (2011) and Fabian et al. (2012) used a pooled-sequencing approach to conduct an outlier scan between populations along latitudinal clines, in Australia and in the east coast of North America, respectively. In both cases, the authors found several genomic regions that might have been differentiated due to environment-specific selection.
Here, we report a whole-genome resequencing effort for 35 D. melanogaster genotypes originally sampled from an organic orchard in Winters, CA (Yang & Nuzhdin 2003). We describe genome-wide levels of polymorphism in this set of fly genotypes from Winters and in a recently published set of genomes from Raleigh, NC (Mackay et al. 2012). Using this dataset, we conduct several population genomic analyses with the following objectives: (i) Test the hypothesis of a recent population expansion, as implied by the prevailing demographic model of colonization of North America (David & Capy 1988); (ii) Estimate the level of genetic differentiation between these two populations (Winters and Raleigh), and test the hypothesis of population subdivision among North American D. melanogaster; (iii) Look for signatures of positive selection across the genome; (iv) Compare allele frequencies at candidate regions among different populations from all over the world in an attempt to identify common patterns of variation and to get a better understanding of how selection might be acting on such genome regions.
Materials and Methods
Fly lines, library construction and sequencing
D. melanogaster natural genotypes were collected from an orchard in Winters, California in 1998 (Yang & Nuzhdin 2003) and were made isogenic by at least 40 generations of full-sibling inbreeding. Flies were reared on standard medium at 25°C with a 12 h light: 12 h dark cycle. The names of these lines are: w23, w26, w33, w34, w35, w36, w37, w38, w40, w43, w47, w49, w50, w52, w54, w55, w56, w59, w60, w62, w63, w64, w66, w67, w68, w69, w74, w76, w79, w80, w82, w84, w86, w87, w114. DNA was extracted from whole-body female flies using Qiagen’s DNeasy Blood and Tissue Kit (Qiagen) and sheared to a fragment length of ~300 bp using the Covaris S2 (Covaris). Subsequent library preparation was done according to standard Illumina protocols. Libraries were sequenced on an Illumina Genome Analyzer IIx (Illumina) in 76 bp and 108 bp single end format runs. The fastq files containing the sequencing reads have been deposited in the NCBI Sequence Read Archive (SRA) database under the Accession Number SRP009033.3.
We also extracted the DNA of 23 isofemale D. melanogaster lines from 12 locations in the southeast United States and Caribbean islands. These lines were collected in the summers of 2004 and 2005 (Yukilevich & True 2008) and were maintained on standard medium with a 12h light: 12h dark cycle. We designed a pair of primers in order to amplify a fragment of the coding sequence of the gene Obst-F (FlyBase ID: FBgn0036947), with a length of 537 bp. The primers were Obst-F-F: TCACTATGGAGCCTACTTCC, and Obst-F-R: TATTATCACTTTTGGAAGC. PCR products were run in a 1.2% agarose gel, from which we excised the corresponding band. The gel band was subsequently purified using Zymoclean Gel DNA Recovery Kit (Zymo Research) and submitted for sequencing (Laragen: Sequencing and Genotyping, Culver City, CA) with the primer Obst-F-F.
We retrieved Illumina high-throughput sequencing data from the Sequence Read Archive (SRA) database for a subset of 33 D. melanogaster genotypes included in the DGRP panel (Mackay et al. 2012; http://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?study=SRP000694) and in the Drosopila Population Genomics Project (www.dpgp.org). These lines are: RAL-208, RAL-301, RAL-303, RAL-304, RAL-313, RAL-324, RAL-335, RAL-357, RAL-358, RAL-362, RAL-365, RAL-375, RAL-379, RAL-380, RAL-399, RAL-427, RAL-437, RAL-486, RAL-517, RAL-555, RAL-639, RAL-705, RAL-707, RAL-712, RAL-714, RAL-730, RAL-732, RAL-765, RAL-774, RAL-786, RAL-799, RAL-820, RAL-852. We restricted our analysis to this subset of 33 Raleigh lines for two reasons: (i) to have a similar sample size in both populations and (ii) because for most of these lines, sequencing data was available from both sources (DGRP and DPGP). We combined the sequencing data from both sources.
Allele counts per position for two genome regions (3L: 18,000,000–19,000,000 and 3L: 20,190,000–20,240,000) were obtained for four other populations: (1) Povoa de Varzim, North Portugal (http://www.popoolation.at/pgt), (2) New Jersey (USA) (Remolina et al. 2012), and two Australian locations from (3) Queensland and (4) Tasmania (Kolaczkowski et al. 2011).
Mapping and SNP calling
We trimmed all the reads based on quality using the SolexaQA package with default parameters (Cox et al. 2010), and discarded those reads that were shorter than 25 bp after trimming. Then we employed Bowtie 2 (ver. beta 5) to map all the reads to the FlyBase reference genome ver. 5.41, using the default “very-sensitive” and “–N=1” parameters (Salzberg & Langmead 2012). After mapping the reads, we used GATK (dePristo et al. 2011) to perform a local realignment step around indels, and then the Picard Tools package (http://picard.sourceforge.net) to mark all PCR and optical duplicates.
All the previous steps were separately done for each genotype. We then used the Unified Genotyper included in the GATK package, setting all parameters to recommended default values, to simultaneously call SNPs in all samples. Even though all fly lines included in this study have been inbred for many generations, there might still be polymorphic positions within individual lines due to residual heterozygosity and new mutations. Heterozygotic positions, and nucleotide positions with no coverage at any given genotype were set to “N”, and not included in the analysis.
Identity by descent
A potential problem that can arise when sampling multiple individuals from the same location is that some of the collected genotypes may share a certain proportion of their genomes due to kinship. Therefore, the amount of genetic polymorphism that is estimated from that sample is not reflecting the actual level of genetic diversity in the population. To avoid such an effect, we performed pairwise comparisons of all genotypes within each population using a sliding-windows approach. For every pair of genotypes, we compared SNPs in windows of 1Mb and shifted the window every 100 Kb. Genomic regions with an identity of 95% of higher were considered as identical by descent (IBD). Such IBD regions were subsequently masked in the genotype with lower coverage for downstream analysis.
Nuclear genome diversity pattern
In order to describe the level of genetic polymorphism in the two populations, Raleigh and Winters, we estimated the common summary statistics π, which is the average number of pairwise nucleotide differences per site (Tajima 1983), and θ (Watterson 1975), the population mutation parameter, which is an unbiased estimator of the number of segregating sites. We calculated the Tajima’s D statistic (Tajima 1989) to scan the genome for signatures of selection and/or demographic events. This test is based on the site frequency spectrum, and it is sensitive to either selection or demographic changes. In the absence of selection, Tajima’s D test yields negative values in the event of a population expansion. These three statistics were calculated both per site and using a sliding windows approach (non-overlapping windows of 100 Kb). We estimated an average value of π, θ and Tajima’s D for each of the five major chromosome arms (X, 2L, 2R, 3L, and 3R) in each population. We divided the data into different categories (CDS, including synonymous and non-synonymous, exon, 5′ UTR, 3′ UTR, intron, and intergenic) and estimated all the previously mentioned statistics for each category. For the estimation of these population parameters, we requested at least 75% of valid calls at any given site in order to be included in the analysis (i.e. 25 valid calls in the Raleigh sample, and 27 in the Winters). Once a site passes this threshold, all valid bases are used for the calculations. In order to account for missing data in each site, the sample size of included sites was adjusted with the number of valid bases. Genome sites that did not pass the threshold were not included in the analysis. Chromosome and category estimates were done averaging over the total number of included sites. All these calculations of population parameters were done using custom Python scripts.
Population differentiation at the nuclear genome
To estimate the level of genetic differentiation between Raleigh and Winters, we used the θ statistic as described in Weir & Cockerham (1984; equation on page 1363). We applied a multiple alleles correction for two populations that have recently descended from a non-inbred ancestral population (see appendix in Weir & Cockerham 1984), since this appears to be the case for North American D. melanogaster populations (David & Capy 1988). Because this statistic is analogous to Wright’s FST (Wright 1951), we denote it here as θST to avoid confusion with the population mutational parameter described above. The calculation was done by genome site.
To empirically test whether the two populations were significantly more differentiated than expected under the null model of panmixia, we performed a permutation analysis. We set up the null distribution by combining all allele counts at every site, randomly reassigning population labels, and re-computing θST. From this null distribution we annotated the θST value that corresponded to the 99% quantile (i.e. the value above which we find 1% of all values) and repeated this process 1,000 times for each chromosome. Finally, we compared the actual 99% cut-off θST value with that expected under panmixia. These calculations were done using Python custom scripts.
Because data for New Jersey, Portugal, Queensland and Tasmania were based on pooled sequences, we normalized allele counts prior to calculate pairwise θST. To normalize, we estimated allele frequencies per position for all six populations, multiplied the frequency by 100, and used these normalized allele counts for θST calculations.
Detection of selection
Demographic processes can promote allele frequency differences between populations, via genetic drift, at random positions across the genome. Conversely, an aggregation of highly differentiated positions in a relatively short genome region may be an indicator of the action of natural selection (Lewontin & Krakauer 1973). Non-synonymous changes are more likely to be affected by selection, since they directly affect the amino acid sequence of the proteins. In order to detect traces of local adaptation events in the Raleigh and the Winters populations, we plotted the θST values for all non-synonymous polymorphic positions along each chromosome and searched for aggregations in the top 0.1% quantile.
For practical purposes, and in order to be conservative, we arbitrarily considered as candidate outliers those regions of length equal to or smaller than 50 Kb, containing three or more non-synonymous positions above the top 0.1% quantile of the chromosome in which they are located. Among the candidate regions identified, we focused our analysis on the region with the highest number of non-synonymous positions in the top quantile.
To confirm that the most differentiated genome region we observed (see Results) is a significant outlier we performed a permutation test according to the following procedure: we randomly sampled a region of the same chromosome containing an equal number of non-synonymous positions as our candiate outlier region, and calculated the mean θST value. We repeated this sampling process 100,000 times, recorded all the θST values, and created a null distribution. Finally, we compared the actual observed θST value of the candidate region with that null distribution.
We also investigated whether that significant outlier genome region could simply be the result of demographic events rather than selection using coalescent simulations. As detailed in the Introduction, the prevailing demographic model for the colonization of North America by D. melanogaster implies that a subset of European flies first arrived to the east coast of North America, and then expanded throughout the continent (David & Capy 1988; Keller 2007). This model, however, does not take into account the admixture between African and North American flies, as suggested by by Caracristi & Schlötterer (2003) and Duchen et al. (2013). Using the program ms (Hudson 2002), we simulated an autosome-linked region of the same length as our top candidate outlier, in a population of 35 chromosomes, evolving without selection for 1,280 generations. We assumed 10 generations per year (a common assumption for D. melanogaser natural populations) and 128 years after the colonization, which is the time that has passed between the first report of D. melanogaser in North America (Keller 2007) and the year the Winters genotypes were collected (Yang & Nuzhdin 2003). We used a mutation rate of 1.45 × 10−9 per site per generation (Li & Stephan 2006). The population-scaled recombination rate (ρ) was estimated with the program LDhat v.2.2 (McVean et al. 2004). The demographic model we simulated consisted of an initial effective population size N2 (the European source population), a post-bottleneck North American founder population with size N1, and a current North American population of size N0, after 1,280 generations of exponential growth. We calculated N2 to be 1.43 × 106 for autosomal-linked loci, which is the estimated current effective population size for the X chromosome in the European population (Li & Stephan 2006) multiplied by 4/3 to account for the difference in effective size between chromosome X and autosomes. We assumed the ratio N2/N0 to be 1.5, which is the ratio between the θ estimate for non-coding X-linked loci for the current European population (Li & Stephan 2006) and the average of our estimates of θ for intergenic and intronic sites on the chromosome X in the Winters population. To model the strength of the bottleneck and the growth rate after the colonization, we assumed a set of different ratios N1/N0: 0.1, 0.01, 0.001, 0.0001, and 0.00001. Using these parameters we ran 106 simulations for every value of the N1/N0 ratio, and compared the actul polymorphism values of the outlier region with the simulated values.
Mitochondrial DNA analysis
We assembled entire mitochondrial genome sequences for all individuals analyzed, visually inspected the aligned sequences with the program SeaView ver. 4 (Gouy et al. 2010) and filtered the dataset removing those gene sequences with no or very low variability and regions with no coverage in any of the flies. In order to describe the level of genetic variation of these mitochondrial sequences, we estimated haplotypic diversity (Hd) and nucleotide diversity (π) values using the program Arlequin ver. 3.5 (Excoffier & Lischer 2010).
Using the same software, we looked for traces of a demographic expansion event. We performed the Tajima’s D test and a mismatch analysis for each population separately. For the mismatch analysis, Arlequin applies a Sum of Squares Deviations approach (SSD test) to compare the observed frequency of pairwise sequence differences (mismatch distribution) to the expected number of sequence differences under a sudden expansion model. The statistical significance of these tests was assessed by 1,000 coalescent simulations.
Both Tajima’s D and SSD tests are sensitive to selection and demography. Under selective neutrality, a significant negative value for Tajima’s D or a very low value of SSD, may suggest a scenario of demographic expansion. Besides, due to the small size and the lack of recombination in the mitochondrial genome, all genes share the same genealogical history, thus it is possible that a selection event acting on one locus will affect the entire molecule (Ballard & Rand 2005), leading to a misinterpretation of demographic and/or selective patterns. In order to check whether the mitochondrial sequences are under selection, we conducted the McDonald-Kreitman test (McDonald & Kreitman 1991) with the DnaSP software using D. simulans as an outgroup.
Finally, we estimated the amount of genetic differentiation between the Winters and the Raleigh populations using the FST statistic as implemented in Arlequin.
Results
Nuclear genome diversity pattern
We have obtained whole-genome sequences of 35 isogenic D. melanogaster strains originally collected in Winters, CA (Yang & Nuzhdin 2003), using a next-generation sequencing technology (Illumina GAIIx). The mean sequencing depth was 4.7X, and on average 87% of the euchromatic genome was covered. Table 1 shows the mean estimates of π, θ and Tajima’s D for all chromosome arms and the X chromosome, for this set of flies and for a subset of 33 fly lines of the DGRP (Mackay et al. 2012). Supplementary table 1 shows the values of these polymorphism indices per chromosome and site category. There was no statistical evidence for a difference in distribution of π and θ estimates for autosomes (Mann-Whitney U, p = 0.2508 for both statistics) or the X chromosome (Mann-Whitney U, p = 0.3306 for π and p = 0.5361 for θ) between Raleigh and Winters populations. As expected, synonymous and non-synonymous positions showed the highest and the lowest level of polymorphism respectively, and coding regions were less variable than non-coding in both populations. A sliding windows analysis (non-overlapping windows of 100 Kb; Supplementary figure 1) showed that these statistics were generally uniform across chromosomes, being lower near the centromere and the telomeres. Overall, our estimates are consistent with previously reported polymorphism values for the Raleigh set of genotypes (Mackay et al. 2012; Langley et al. 2012). In addition, π estimates for synonymous and non-synonymous sites are very similar to those reported by Langley et al. (2012).
Table 1.
Mean π, θ and Tajima’s D values for all autosomal chromosome arms and chromosome X in each population.
| Chromosome | Raleigh | Winters | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| π | θ | Tajima’s D | π | θ | Tajima’s D | |
| 2L | 0.00647 | 0.00672 | −0.15040 | 0.00521 | 0.00557 | −0.25552 |
| 2R | 0.00584 | 0.00610 | −0.16712 | 0.00491 | 0.00524 | −0.24330 |
| 3L | 0.00584 | 0.00610 | −0.16543 | 0.00472 | 0.00513 | −0.31176 |
| 3R | 0.00491 | 0.00522 | −0.24044 | 0.00412 | 0.00446 | −0.30037 |
| X | 0.00393 | 0.00398 | −0.05646 | 0.00357 | 0.00371 | −0.15004 |
For Raleigh and Winters samples, π values were lower than θ for autosomes and chromosome X resulting in genome-wide negative Tajima’s D values (Table 1). Demographic processes affect the entire genome, but selection is thought to only affect specific loci. Therefore, this result seems to be consistent with a demographic expansion pattern for both populations. On the other hand, Tajima’s D values were lower in the Winters sample, which might be an indicator of a more recent expansion in this population.
Population differentiation at the nuclear genome
Table 2 shows the results of the θST analysis. Genome-wide average differentiation level between Winters and Raleigh samples was low (θST =0.036). The permutation test yielded an expected 99% cut-off θST of around 0.21 for all chromosomes under the null hypothesis of panmixia. The actual percentage of positions with a θST value above the cut-off was, for all chromosomes, more than double the expected under panmixia. This result indicates a statistically significant amount of genetic divergence between Raleigh and Winters. Notably, we did not find any fixed difference between the two populations (i.e. a position with a θST value of 1) along the genome.
Table 2.
Results of the θST analysis between Winters and Raleigh populations per chromosome. “Positions” indicate the number of positions analyzed after discarding those with less than 15 genotypes with high-quality base calls per population. “Mean” refers to the average θST value across the chromosome. “Panmixia” is the 99% cut-off θST value expected under panmixia, obtained through 1,000 simulations. “% Above” indicates the actual percentage of positions with a θST above the cut-off value.
| Chromosome | Positions | Mean | Panmixia | % Above |
|---|---|---|---|---|
| 2L | 21,151,117 | 0.038 | 0.207 | 2.6 |
| 2R | 19,378,120 | 0.035 | 0.207 | 2.3 |
| 3L | 22,442,999 | 0.035 | 0.208 | 2.3 |
| 3R | 26,666,954 | 0.037 | 0.210 | 2.6 |
| X | 20,404,483 | 0.034 | 0.212 | 2.1 |
When a population is expanding its geographic range, usually small groups of pioneer individuals advance and found local subpopulations. Because the effective size of these local subpopulations is generally low, allele frequencies may change with respect to the source population due to genetic drift. This demographic effect is called allele surfing, and it is typically found at the edges of range expansion waves (Edmonds et al. 2004). We have found a number of highly differentiated positions larger than expected under the null hypothesis of panmixia that are randomly distributed across the genome. This polymorphism pattern supports a model of multiple allele surfing events during a range expansion process.
Detection of outliers
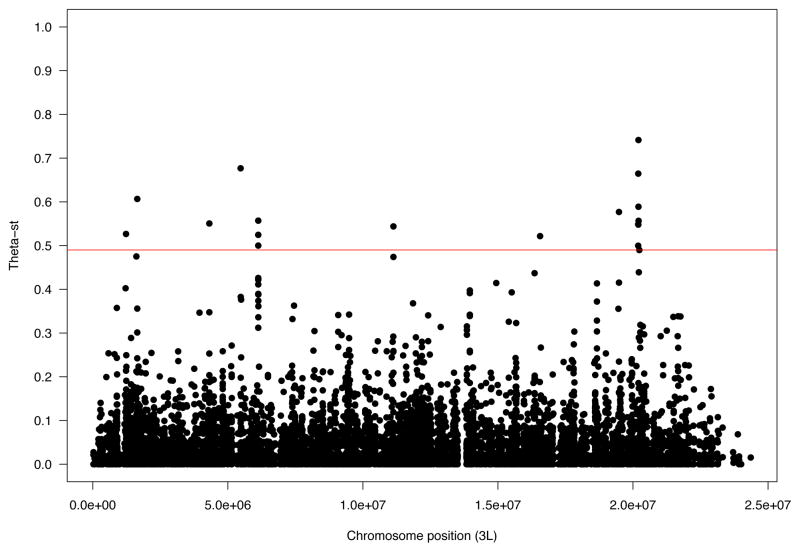
We plotted the θST values for all non-synonymous polymorphic positions along each chromosome and searched for aggregations in the top 0.1% quantile (Figure 1 and Supplementary figure 2). We identified seven relatively short genome segments (≤50 Kb) containing three or more non-synonoymous changes above the 0.1% θST threshold (Supplementary table 2). Among these segments, the highest number of differentiated non-synonymous sites was located in a 50 Kb region of chromosome 3L, between positions 20,190,000 and 20,240,000.
Figure 1.
Plot of θST values for all non-synonymous positions across chromosome 3L. The red line indicates the 0.1% quantile.
Following the permutation approach described in Materials and Methods, we generated a null distribution of estimated θST values for chromosome regions containing the same total number of non-synonymous sites as the candidate region in chromosome 3L. The observed θST value for this region falls outside the distribution, providing statistical evidence that this candidate region is a significant outlier.
The mean θST across this 50 Kb divergent region in chromosome 3L was 0.17, roughly five times the genome average. The percentage of positions in this region with a θST value above the expected under panmixia was 28.8%, more than ten times higher than the percentage for the entire chromosome (Table 2). A third of the non-synonymous sites within this region showed a θST value above the 0.1% threshold for chromosome 3L. These highly differentiated non-synonymous mutations were located in only five of the fifteen protein-coding genes located in this region (Supplementary table 3). Interestingly, these five genes are all annotated as structural constituents of the peritrophic membrane according to Flybase. Polymorphism levels in this region are reduced with respect to the mean genome value in both populations (Raleigh: π=0.00169, θ=0.00205; Winters: π=0.00122, θ=0.00151). We performed coalescent simulations under the standard neutral model, assuming the prevailing demographic scenario for the colonization of North America by D. melanogaster, as detailed in Materials and Methods. The observed polymorphism values are below the lower 0.001% quantile value from the simulations (for all N1/N0 ratios tested), indicating that demography alone is not sufficient to explain the low polymorphism levels observed. It is worth noting that our demographic model, does not take into account the admixture between African and North American flies, suggested by Caracristi & Schlötterer (2003) and Duchen et al. (2013). Therefore, the simulated polymorphism value of the North American founder population (N1) is likely lower than the actual value, making our test more conservative. Taken together, these results provide evidence for the action of natural selection on this genome region.
A hard sweep selection episode is expected to cause a dramatic reduction in polymorphism in the surrounding area coupled with increased LD levels at both sides, but not across the selected site (Pennings & Hermisson 2006). In contrast, in soft sweep events, in which the beneficial mutation is already present in the population, the decrease in polymorphism is usually weaker (Hermisson & Pennings 2005). Also, LD is expected to extend throughout the region in the soft sweep case (Pennings & Hermisson 2006). In order to understand the type of selective sweep the divergent region of chromosome 3L is undergoing, we estimated linkage disequilibrium across the region for each population separately. Linkage disequilibrium between pairs of polymorphic positions was calculated using the statistics D, D′, and R, assesing their significance with a Fisher exact test. All these tests were done with the DnaSP software (Librado & Rozas 2009). The results are given in Supplementary table 4. We found statistically significant LD between variable sites situated in both ends of the region (i.e. spanning the entire region) in both populations. In general, this pattern is consistent with a soft selective sweep event affecting the highly differentiated region we found in chromosome 3L. The fact that all alleles are present in both populations indicates that the sweep started from standing variation. On the other hand, alternate alleles at the divergent non-synonymous sites are at high frequency in both populations. This indicates that either the favored alleles in one population are deleterious in the other, or that opposite alleles are positively selected in different populations.
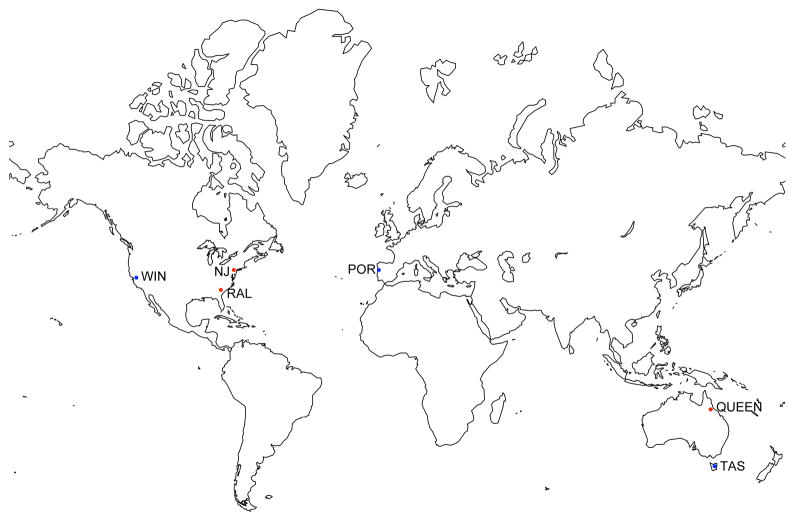
In order to identify worldwide patterns of allele frequencies distribution that could help us to understand how selection may be affecting this divergent genomic region, we compared allele frequencies among D. melanogaster populations from different geographic areas. We calculated pairwise θST between six populations for the 50 Kb differentiated region (3L: 20,190,000–20,240,000) (Table 3). The six populations fell into two groups (Figure 2): Winters, Portugal, and Tasmania versus Raleigh, New Jersey, and Queensland. Mean θST between Winters, Portugal, and Tasmania was 0.06, and between Raleigh, New Jersey, and Queensland was 0.123. However mean θST between the two groups of populations was 0.498. As a comparison, we estimated pairwise θST for another region in chromosome 3L (positions 18,000,000–19,000,000) that contains 2.6% of the positions above the panmixia cut-off. Mean θST between Winters, Portugal, and Tasmania was 0.1. Mean θST between Raleigh, New Jersey, and Queensland was 0.143. And θST between these two groups of populations was 0.123. According to these results, there seems to be a global pattern of differential selection, with opposite alleles selected in different groups of populations for the chromosome region 3L: 20,190,000–20,240,000.
Table 3.
Pairwise θST values between Winters, Portugal, Tasmania, Raleigh, New Jersey and Queensland populations for the highly differentiated region in chromosome 3L (20,190,000–20,240,000) (below diagonal) and another region of the same chromosome not suspected to be under selection (above diagonal) (18,000,000–19,000,000).
| Population | Winters | Portugal | Tasmania | Raleigh | New Jersey | Queensland |
|---|---|---|---|---|---|---|
| Winters | - | 0.08 | 0.09 | 0.04 | 0.15 | 0.10 |
| Portugal | 0.05 | - | 0.13 | 0.08 | 0.18 | 0.13 |
| Tasmania | 0.05 | 0.08 | - | 0.13 | 0.19 | 0.13 |
| Raleigh | 0.42 | 0.35 | 0.47 | - | 0.15 | 0.09 |
| New Jersey | 0.49 | 0.40 | 0.53 | 0.06 | - | 0.19 |
| Queensland | 0.63 | 0.54 | 0.65 | 0.14 | - |
Figure 2.
Map showing the six populations analyzed in this paper for the highly differentiated region at chromosome 3L: 20,190,000–20,240,000. WIN: Winters (CA, USA); RAL: Raleigh (CA, USA); NJ: New Jersey (NJ, USA); POR: Povoa de Varzim (Portugal); QUEEN: Queensland (Australia); TAS: Tasmania (Australia). Red and blue dots indicate populations grouping together.
The question arises, what can account for this global pattern of differential selection between these two groups of populations? Tasmania and Queensland are situated at the ends of a well-studied latitudinal cline (Hoffman & Weeks, 2007), ranging from temperate to tropical areas, and Kolaczkowski et al. (2011) found high differentiation for some non-synonymous positions within the same chromosome region. Therefore, a potential explanation might be the difference in latitude between the two groups of populations. Winters and the Portuguese populations are situated at close latitudes (38°30′N and 41°22′N, respectively) in temperate regions of the northen hemisphere, and the Tasmanian flies were collected at two locations within the same latitude range in the southern hemisphere (41.2°S to 42.7°S). The Queensland flies were collected from tropical latitudes in the southern hemisphere (15.4°S and 16.9° S). However, even though Raleigh and New Jersey are situated at more temperate latitudes (35°46′N and ~40°N respectively), these populations grouped with Queensland. Interestingly, Caracristi & Schlötterer (2003) suggested the existence of an admixture zone between Caribbean and east coast North American flies, proposing the Caribbean populations as a source of African alleles (Yukilevich et al. 2010). Duchen et al. (2013) tested several demographic models using Approximate Bayesian Computation, and found strong statistical support for the admixture hypothesis, suggesting that such admixture between European and African D. melanogaster likely generated the North American populations. A scenario of introgression of tropical alleles into Raleigh and New Jersey from Caribbean locations would explain the clustering pattern we have observed. To further test this hypothesis, we amplified and sequenced a fragment of 537 bp of Obst-F gene in the Caribbean and southeast US fly lines described in Materials and Methods. This gene is located within the 3L divergent region (Supplementary table 3). We aligned these sequences with the homologous sequences in Raleigh and Winters flies and calculated pairwise FST based on haplotype frequencies using Arlequin (Excoffier & Lische 2010). All pairwise comparisons involving the Winters population were statistically significant whereas none of the others were (Supplementary table 5). These results provide additional support for the existence of an admixture zone in eastern North America, as proposed by Caracristi & Schlötterer (2003), and explain the presence of tropical alleles in temperate populations (Raleigh and New Jersey).
Mitochondrial DNA analysis
From the alignment of all mitochondrial genomes, we obtained a final dataset of 4,976 bp, which included the genes ATP8, ATP6, COIII, COII, COI, and Cytb. Table 4 contains a summary of the population genetic parameters and statistics. Diversity values (both Hd and π) were much higher in the Raleigh than in the Winters population. The McDonald-Kreitman test did not show a significant deviation from the neutral model for this dataset. Therefore, the results of the Tajima’s D test and the mismatch distribution analysis can be interpreted from a demographic perspective, as we cannot reject neutrality. Tajima’s D test yielded statistically significant negative values for both populations. The SSD statistic (for the mismatch distribution) showed very low estimates, and the null hypothesis of population expansion cannot be rejected. Altogether, these results support a pattern of demographic expansion for both populations. On the other hand, both tests yielded lower values for the Winters sample. This result, combined with lower values of polymorphism in Winters, suggest that the expansion started more recently in this population.
Table 4.
Intra- and interpopulation analysis of the mitochondrial dataset for Winters and Raleigh populations. The dataset includes the genes ATP8, ATP6, COIII, COII, COI and Cytb (4,976 bp in total). Hd is the haplotypic diversity; π is nucleotide diversity; D is the Tajima’s D test; SSD stands for Sum of Squared Differences; McD-K is the McDonald and Kreitman test.
| Hd | π | D | SSD | McD-K | FST | |
|---|---|---|---|---|---|---|
| Winters | 0.49580 | 0.00015 | −2.35870(p<0.01) | 0.0016(p=0.65) | 0.622 (p=0.29) | 0.135 (p<0.01) |
| Raleigh | 0.90731 | 0.00085 | −2.30857(p=0.00) | 0.0064(p=0.52) |
Regarding population structure, based on mtDNA haplotype frequency differences, we obtained a FST value of 0.135 (p-value < 0.05) indicating a significant level of differentiation between the two populations.
Discussion
Genome-wide levels of polymorphism in North American D. melanogaster populations
Our estimates of π and θ for the subset of 35 DGRP genotypes from Raleigh are very similar to those obtained by Mackay et al. (2012) and Langley et al. (2012), based on 168 and 37 genotypes respectively. This agreement with previously published works serves as a validation of our results, and confirms those previous estimates.
Sackton et al. (2009) reported θ values for a pooled sample of six Raleigh lines, some of which have been used in this paper, in Mackay et al. (2012), and in Langley et al. (2012). Their estimates were lower than our values and those in other studies, and they specifically compare with Hutter et al. (2007). Sackton et al. (2009) suggest that this could be due to unaccounted sequencing errors in Hutter et al. (2007), an actual difference in polymorphism level between populations, or an overly conservative correction in their own estimates. Polymorphism estimates in Hutter et al. (2007), Mackay et al. (2012), Langley et al. (2012), and in the present study are very similar suggesting that indeed Sackton et al. (2009) may have used a too conservative approach.
Can we still detect a signal of demographic expansion in the genome of D. melanogaster?
The prevailing demographic model for D. melanogaster suggests that the colonization of North America took place very recently with Europe as the source of the founder flies (David & Capy 1988). This model implies a rapid demographic growth involving both population and range expansion from eastern to western North America.
In the present study, we have found support for a demographic expansion scenario in both populations, Raleigh and Winters. Our results also suggest that this expansion probably started more recently in the western population (Winters). This result is supported by both nuclear and mitochondrial genome datasets. We have also found a pattern of polymorphism consistent with multiple allele surfing events, suggesting a range expansion process in the two populations. Altogether, our results provide support for the prevailing demographic scenario for D. melanogaster (David & Capy 1988). Under this scenario, the Winters flies would be at the front of a demographic and range expansion wave from eastern to western North America after a single colonization event from Europe.
Interestingly, several recent papers have suggested that polymorphism patterns in the genome of D. melanogaster, and other species with very large effective population sizes, may be affected by pervasive natural selection (Hahn 2008; Wright & Andolfatto 2008; Sella et al. 2009). In fact, there is experimental evidence that a large proportion of genomic sites might be functional in D. melanogaster (The modENCODE Consortium, 2010), and therefore potential targets of selection. Even synonymous sites, which have been traditionally thought to be selectively neutral, seem to be under selection (Wright & Andolfatto 2008; Zeng & Charlesworth 2010). If true, this would make it very challenging to distinguish between the effects of selection and demography in shaping genetic variation patterns. Indeed, current statistical methods are unable to distinguish between demography and selection (Li et al. 2012). Therefore, even though there is non-molecular evidence suggesting a very recent colonization of North America (Keller 2007), the demographic expansion hypothesis needs to be further revisited once adequate statistical methods are developed.
Genome-wide pattern of population differentiation in North American D. melanogaster
Different studies published to date have yielded contradictory results regarding population structure in North America. Some have suggested a lack of structure (Kreitman & Aguadé 1986; Coyne & Milstead 1987), whereas others observed population subdivision (Johnson & Schaffer 1973; Singh & Long 1992; Begun & Aquadro 1994; Mettler et al. 1977; Caracristi & Schlötterer 2003). Particularly, Caracristi & Schlötterer (2003) found significant differentiation between a population from northern California (Groth Winery, Napa Valley) and three populations from the eastern US, but no differentiation among the latter. Fabian et al. (2012) reported very similar levels of genome-wide differentiation to those in Caracristi & Schlötterer (2003), but between three populations along the east coast (Maine, Pennsylvania, and Florida).
We have found a statistically significant level of genetic differentiation between the sample from the west coast (Winters, California) and the sample from the eastern region of North America (Raleigh, North Carolina) with both nuclear and mitochondrial genome datasets. The amount of divergence between populations found in the present study (θST = 0.036) is very similar to that reported in Caracristi & Schlötterer (2003) and Fabian et al. (2012).
Based on a demographic model of recent colonization and rapid spread over North America (David & Capy 1988), Caracristi & Schlötterer (2003) suggested that this pattern of differentiation could be accounted for by local episodes of genetic drift. Consistent with this hypothesis, the low but significant level of genetic divergence found in the present paper between the Winters and the Raleigh populations may be explained by the accumulation of multiple allele surfing events that occured as the species expanded its range after the colonization of North America.
Evidence for selection
In our genome-wide comparison of allele frequencies between Winters and Raleigh populations, we have found a highly differentiated 50 Kb long region in chromosome 3L, between positions 20,190,000 and 20,240,000. This region contains a very large number of divergent non-synonymous mutations concentrated in only five genes. The polymorphism level is reduced in this chromosome segment with respect to the genome average in both populations, and there is significant linkage disequilibrium spanning across the entire region. Using coalescent simulations under the neutral model, we have shown that the reduced polymorphism levels observed in this region cannot be explained by demography alone. These results provide strong evidence that this region of chromosome 3L is affected by selection, and it is likely undergoing a soft selective sweep (Hermisson & Pennings 2005; Pennings & Hermisson 2006).
A global pattern of selection
In order to obtain a better insight on how selection may be acting on this genome region, we compared allele frequencies among six populations from all over the world in an attempt to identify common patterns of variation. These populations clearly clustered in two differentiated groups: Winters, Portugal and Tasmania in one group, and New Jersey, Raleigh and Queensland in the other group. The level of divergence between groups was much higher than within groups, indicating that natural selection is acting in opposite directions in both groups of populations. Two hypotheses can explain this pattern of allele frequencies distribution. First, Caracristi & Schlötterer (2003) proposed the existence of an admixture zone in the east coast of North America with introgression from tropical flies from the Caribbean into temperate populations of North America. Duchen et al. (2013), and our results provide additional support for this hypothesis, which would explain the presence of tropical alleles in New Jersey and Raleigh. Therefore, the allele frequencies distribution we observe may be the result of a tropical-temperate differentiation with opposite alleles positively selected at different latitudes. A caveat to this hypothesis, however, is the implication that introgression has to be stronger than selection in order to maintain tropical alleles in temperate populations at high frequency.
A second explanation that may account for the global distribution of allele frequencies we have found could involve the Mediterranean climate as the selective agent. Mediterranean-climate regions are generally found between 31 and 40 degrees latitude north and south of the equator, on the western side of continents (Ritter 2006). Winters, Portugal and Tasmania are situated in areas with Mediterranean climate, whereas New Jersey, Raleigh and Queensland are not. This hypothesis is not exclusive with the admixture and introgression scenario suggested by Caracristi & Schlötterer (2003), Duchen et al. (2013), and our data.
A larger sampling effort, including populations from tropical and temperate areas with Mediterranean and non-Mediterranean climate will be needed in order to uncover the causes of the global pattern of selection we have found for the region in chromosome 3L.
Mechanism of selection
Without a better characterization of the environmental differences between the populations and a deeper analysis of the genotype-phenotype connection for the selected alleles, one can only speculate about the mechanism of selection acting on them. However, there are some interesting aspects of the chromosome region under selection that may provide useful insights. The five genes showing highly divergent frequencies at non-synoymous positions present the same biological function. They are consituents of the peritrophic matrix, which is a protein barrier secreted in the midgut of the flies that protects against pathogens and toxins entering with the food (Lehane 1997). Chandler et al. (2011) showed that diet plays a major role in shaping the Drosophila bacterial microbiome, and suggest that the flies exercise some level of control over the bacteria that inhabits its digestive tract. A possible mechanism for the flies to exercise this control over their microbiome might be through changes on the proteins that form the peritrophic matrix. Therefore, selection for different alleles in different latitudinal/climatic areas, would lead to differences in the microbiome composition. A comparison of the diet and gut microbiome composition between D. melanogaster flies from tropical and temperate regions and/or from Mediterranean versus non-Mediterranean areas would be needed to test this idea.
Supplementary Material
Supplementary figure 1. Plots of a) π, b) θ and c) Tajima’s D, across the genome based on sliding windows analysis, with non-overlapping windows of 100 Kb. The orange line represents the Winters population, and the blue line is the estimate for Raleigh.
Supplementary figure 2. Plots of θST between Winters and Raleigh populations for non-synonymous positions for all chromosome arms except 3L (Figure 2). Red lines represent the 0.1% quantile.
Supplementary table 1. Mean estimates of π, θ and Tajima’s D for all chromosome arms and the X chromosome for all site categories.
Supplementary table 2. List of highly differentiated genomic regions spanning less than 50 Kb.
Supplementary table 3. List of genes with highly divergent non-synonymous changes between Raleigh and Winters. “NSYN” indicates the total number of non-synonymous changes and θST is the mean θST value across the non-synonymous positions.
Supplementary table 4. Linkage disequlibrium analysis for the highly differentiated region in chromosome 3L (20,190,000–20,240,000) between Winters and Raleigh.
Supplementary table 5. Pairwise FST analysis for the gene Obst-F (FBgn0036947) between Winters, Raleigh, and two sets of samples from the Southeastern US (SEUS) and several Caribbean locations.
Acknowledgments
Funding
We are grateful to NIH for supporting this research through the following grants: P50 HG002790, NIH MH091561 and GM076643.
We would like to thank the USC (University of Southern California) undergraduate students Oliver Gantz, Alexander Lofthus and Srna Vlaho for their invaluable help with fly and molecular work, Fabrizio Ghiselli (University of Bologna) for his useful comments and suggestions, Matt Salomon (USC) for his comments and his help with the mapping and SNP calling procedure, and Bryan Kolaczkowski (University of Florida) and Peter Chang (USC) for providing polymorphism data from the two Australian populations and from New Jersey respectively.
Footnotes
- Illumina fastq files containing original reads: NCBI SRA SRP009033.3
- Obst-F sequences for Southeast and Caribbean isofemale lines: Genbank accession numbers JN885138-JN885158.
- COI sequences for Southeast and Caribbean isofemale lines: Genbank accession numbers JN885159-JN885180.
Sequence alignments and SNP data: Dryad doi:10.5061/dryad.kt062.
Literature Cited
- Ballard JW, Rand DM. The population biology of the mitochondrial DNA and its phylogenetic implications. Annual Review of Ecology, Evolution, and Systematics. 2005;36:621–642. [Google Scholar]
- Begun D, Aquadro CF. Evolutionary inferences from DNA variation at the 6-phosphogluconate dehydrogenase locus in natural populations of Drosophila: selection and geographic differentiation. Genetics. 1994;136:155–171. doi: 10.1093/genetics/136.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry A, Kreitman M. Molecular analysis of an allozyme cline: alcohol dehydrogenase in Drosophila melanogaster at the east coast of North America. Genetics. 1993;134:869–893. doi: 10.1093/genetics/134.3.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Akey JM. Genomics insights into positive selection. Trends in genetics. 2006;22(8):437–446. doi: 10.1016/j.tig.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Black WC, Baer CF, Antolin MF, DuTeau NM. Population genomics: genome-wide sampling of insect populations. Annual Review of Entomology. 2001;46:441–469. doi: 10.1146/annurev.ento.46.1.441. [DOI] [PubMed] [Google Scholar]
- Burke MK, Dunham JP, Shahrestani P, Thornton KR, Rose MR, Long AD. Genome-wide analysis of a long-term evolution experiment with Drosophila. Nature. 2010;467:587–592. doi: 10.1038/nature09352. [DOI] [PubMed] [Google Scholar]
- Caracristi G, Schlötterer C. Genetic differentiation between American and European Drosophila melanogaster populations could be attributed to admixture of African alleles. Molecular Biology and Evolution. 2003;20(5):792–799. doi: 10.1093/molbev/msg091. [DOI] [PubMed] [Google Scholar]
- Chandler JA, Lang JM, Bhatnagar S, Eisen JA, Kopp A. Bacterial communities of diverse Drosophila species: ecological context of a host-microbe model system. PLoS Genetics. 2011;7(9):e1002272. doi: 10.1371/journal.pgen.1002272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MP, Peterson DA, Biggs PJ. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics. 2010;11:485. doi: 10.1186/1471-2105-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Milstead B. Long-distance migration of Drosophila: dispersal of D. melanogaster alleles from a Maryland orchard. The American Naturalist. 1987;130:170–82. [Google Scholar]
- David JR, Capy P. Genetic variation of Drosophila melanogaster natural populations. Trends in Genetics. 1988;4(4):106–111. doi: 10.1016/0168-9525(88)90098-4. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchen P, Živković D, Hutter S, Stephan W, Laurent S. Demographic inference reveals African and European admixture in the North American Drosophila melanogaster population. Genetics. 2013;193:291–301. doi: 10.1534/genetics.112.145912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds CA, Lillie AS, Cavalli-Sforza L. Mutations arising in the wave front of an expanding population. Proceedings of the Natural Academy of Sciences USA. 2004;101(4):975–979. doi: 10.1073/pnas.0308064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Lischer HL. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. Department of Genome Sciences, University of Washington; Seattle: 2005. [Google Scholar]
- Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution. 2010;27(2):221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Hahn MW. Toward a selection theory of molecular evolution. Evolution. 2008;62(2):255–265. doi: 10.1111/j.1558-5646.2007.00308.x. [DOI] [PubMed] [Google Scholar]
- Hermisson J, Pennings PS. Soft sweeps: molecular population genetics of adaptation from standing variation. Genetics. 2005;169:2335–2352. doi: 10.1534/genetics.104.036947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Kliman M. Population genetics and phylogenetics of DNA sequence variation at multiple loci within the Drosophila melanogaster species complex. Molecular Biology and Evolution. 1993;10(4):804–822. doi: 10.1093/oxfordjournals.molbev.a040044. [DOI] [PubMed] [Google Scholar]
- Hoffman AA, Weeks AR. Climatic selection on genes and traits after a 100 year-old invasion: a critical look at the temperate-tropical clines in Drosophila melanogaster from eastern Australia. Genetica. 2007;129:133–147. doi: 10.1007/s10709-006-9010-z. [DOI] [PubMed] [Google Scholar]
- Hudson RR. Generating samples under a Wright-Fisher neutral model. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- Hutter S, Li H, Beisswanger S, De Lorenzo D, Stephan W. Distinctly different sex ratios in African and Europen populations of Drosophila melanogaster inferred from chromosomewide single nucleotide polymorphism data. Genetics. 2007;177:469–480. doi: 10.1534/genetics.107.074922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson FM, Schaffer E. Isozyme variability in species of the genus Drosophila. VII. Genotype- environment relationships in populations of D. melanogaster from the eastern United States. Biochemical Genetics. 1973;10:149–163. doi: 10.1007/BF00485762. [DOI] [PubMed] [Google Scholar]
- Jordan KW, Carbone MA, Yamamoto A, Morgan TJ, Mackay TFC. Quantitative genomics of locomotor behavior in Drosophila melanogaster. Genome Biology. 2007;8:R172. doi: 10.1186/gb-2007-8-8-r172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A. Drosophila melanogaster’s history as a human comensal. Current Biology. 2007;17:R77–R81. doi: 10.1016/j.cub.2006.12.031. [DOI] [PubMed] [Google Scholar]
- Kim SY, Lohmueller KE, Albrechtsen A, Li Y, Korneliussen T, Tian G, Grarup N, Jiang T, Andersen G, Witte D, Jorgensen T, Hansen T, Pedersen O, Wang J, Nielsen R. Estimation of allele-frequency and association mapping using next generartion sequencing data. BMC Bioinformatics. 2011;12:231. doi: 10.1186/1471-2105-12-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. The Neutral Theory of Molecular Evolution. Cambridge University Press; New York: 1983. [Google Scholar]
- Kolaczkowski B, Kern AD, Holloway AK, Begun DJ. Genomic differentiation between temperate and tropical Australian populations of Drosophila melanogaster. Genetics. 2011;187:245–260. doi: 10.1534/genetics.110.123059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitman M, Aguadé M. Genetic uniformity in two populations of Drosophila melanogaster as revealed by filter hybridization of four-nucleotide-recognizing restriction enzyme digests. Proceedings of the Natural Academy of Sciences USA. 1986;83:3562–3566. doi: 10.1073/pnas.83.10.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaise D, Cariou M-L, David JR, Lemeunier F, Tsacas L, Ashburner M. Historical biogeography of the Drosophila melanogaster species subgroup. Evolutionary Biology. 1988;22:159–225. [Google Scholar]
- Langley CH, Stevens K, Cardeno C, Lee YC, Schrider DR, Pool JE, Langley SA, Suarez C, Corbett-Detig RB, Kolaczkowski B, Fang S, Nista PM, Holloway AK, Kern AD, Dewey CN, Song YS, Hahn MW, Begun DJ. Genomic variation in natural populations of D. melanogaster. Genetics. 2012;192:533–598. doi: 10.1534/genetics.112.142018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehane MJ. Peritrophic matrix structure and function. Annual Review of entomology. 1997;42:525–550. doi: 10.1146/annurev.ento.42.1.525. [DOI] [PubMed] [Google Scholar]
- Lewontin RC, Krakauer J. Distribution of gene frequency as a test of the theory of selective neutrality of polymorphisms. Genetics. 1973;74:175–195. doi: 10.1093/genetics/74.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long A, Singh R. Molecules versus morphology: the detection of selection acting on morphological characters along a cline in Drosophila melanogaster. Heredity. 1995;74:569–589. doi: 10.1038/hdy.1995.81. [DOI] [PubMed] [Google Scholar]
- Li J, Li H, Jakobsson M, Li S, Sjödin, Lascoux M. Joint analysis of demography and selection in population genetics: where do we stand and where we could go? Molecular Ecology. 2012;21:28–44. doi: 10.1111/j.1365-294X.2011.05308.x. [DOI] [PubMed] [Google Scholar]
- Li H, Stephan W. Inferring the demographic history and rate of adaptive substitution in Drosophila. PloS Genetics. 2006;2(10):e166. doi: 10.1371/journal.pgen.0020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lynch M. Estimation of nucleotide diversity, disequilibrium coefficients, and mutation rates from high-coverage genome-sequencing projects. Molecular Biology and Evolution. 2008;25:2421–2431. doi: 10.1093/molbev/msn185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC, Richards S, Stone EA, et al. The Drosophila melanogaster Genetic Reference Panel. Nature. 2012;482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- McVean GAT, Myers SR, Hunt S, Deloukas P, et al. The fine-scale structure of recombination rate variation in the human genome. Science. 2004;304:581–584. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- Mettler LE, Voelker RA, Mukai T. Inversion clines in populations of Drosophila melanogaster. Genetics. 1977;87:169–176. doi: 10.1093/genetics/87.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R. Molecular signatures of natural selection. Annual Review of Genetics. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- Pandey RV, Kofler R, Orozco-terWengel P, Nolte V, Schlötterer C. PoPoolation DB: a user-friendly web-based database for the retrieval of natural polymorphisms in Drosophila. BMC Genetics. 2011;12:27. doi: 10.1186/1471-2156-12-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennings PS, Hermisson J. Soft sweeps III: the signature of positive selection from recurrent mutation. PLoS Genetics. 2006;2(12):e186. doi: 10.1371/journal.pgen.0020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Pickrell JK, Graham C. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Current Biology. 2010;20(4):208–215. doi: 10.1016/j.cub.2009.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski M, Coop G, Wall JD. The signature of positive selection on standing variation. Evolution. 2005;59(11):2312–2323. [PubMed] [Google Scholar]
- Remolina SC, Chang PL, Leips J, Nuzhdin SV, Hughes KA. Genomic basis of aging and life-history evolution in Drosophila melanogaster. Evolution. 2012 doi: 10.1111/j.1558-5646.2012.01710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds J, Weir BS, Cockerham CC. Estimation of the coancestry coefficient: Basis for a short-term genetic distance. Genetics. 1983;105:767–779. doi: 10.1093/genetics/105.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter ME. The physical environment: an introduction to physical geography. Visited April. 2006;24:2013. http://www.uwsp.edu/geo/faculty/ritter/geog101/textbook/title_page.html. [Google Scholar]
- Sackton TB, Kulathinal RJ, Bergman CM, Quinlan AR, Dopman EB, Carneiro M, Marth GT, Hartl DL, Clark AG. Population genomic inferences from sparse high-throughput sequencing of two populations of Drosophila melanogaster. Genome Biology and Evolution. 2009;1:449–465. doi: 10.1093/gbe/evp048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg SL, Langmead B. Fast gapped-read alignment with Bowtie2. Nature Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer L, Hennessy M, Peixoto A, Rosato E, Parkinson H, Costa R, Kyriacou C. Natural variation in a Drosophila clock gene and temperature compensation. Science. 1997;278:2117–2120. doi: 10.1126/science.278.5346.2117. [DOI] [PubMed] [Google Scholar]
- Sella G, Petrov DA, Przeworski M, Andolfatto P. Pervasive natural selection in the Drosophila genome? PloS Genetics. 2009;5 (6):e1000495. doi: 10.1371/journal.pgen.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RS, Long A. Geographic variation in Drosophila: from molecules to morphology and back. Trends in Ecology and Evolution. 1992;7:340–345. doi: 10.1016/0169-5347(92)90127-W. [DOI] [PubMed] [Google Scholar]
- Stephan W, Li H. The recent demographic and adaptive history of Drosophila melanogaster. Heredity. 2007;98:65–68. doi: 10.1038/sj.hdy.6800901. [DOI] [PubMed] [Google Scholar]
- Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutationhypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The modENCODE Consortium. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TL, Bourne EC, Von Wettberg EJ, Hu TT, Nuzhdin SV. Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nature Genetics. 2010;42:260–263. doi: 10.1038/ng.515. [DOI] [PubMed] [Google Scholar]
- Watterson GA. On the number of segregating sites in genetical models without recombination. Theoretical Population Biology. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38(6):1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Wright SI, Andolfatto P. The impact of natural selection on the genome: emerging patterns in Drosophila and Arabidopsis. The Annual Review of Ecology, Evolution, and Systematics. 2008;39:193–213. [Google Scholar]
- Yang HP, Nuzhdin SV. Fitness costs of Doc expression are insufficient to stabilize its copy number in Drosophila melanogaster. Molecular Biology and Evolution. 2003;20:800–804. doi: 10.1093/molbev/msg087. [DOI] [PubMed] [Google Scholar]
- Yi X, Lian Y, Huerta-Sanchez E, et al. Sequencing of 50 human exomes reveals adapatation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukilevich R, True JR. Incipient sexual isolation among cosmopolitan Drosophila melanogaster populations. Evolution. 2008;62(8):2112–2121. doi: 10.1111/j.1558-5646.2008.00427.x. [DOI] [PubMed] [Google Scholar]
- Yukilevich R, Turner TL, Aoki F, Nuzhdin SV, True JR. Patterns and processes of genome-wide divergence between North American and African Drosophila melanogaster. Genetics. 2010;186:219–239. doi: 10.1534/genetics.110.117366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng K, Charlesworth B. Studying patterns of recent evolution at synonymous sites and intronic sites in Drosophila melanogaster. Journal of Molecular Evolution. 2010;70:116–128. doi: 10.1007/s00239-009-9314-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1. Plots of a) π, b) θ and c) Tajima’s D, across the genome based on sliding windows analysis, with non-overlapping windows of 100 Kb. The orange line represents the Winters population, and the blue line is the estimate for Raleigh.
Supplementary figure 2. Plots of θST between Winters and Raleigh populations for non-synonymous positions for all chromosome arms except 3L (Figure 2). Red lines represent the 0.1% quantile.
Supplementary table 1. Mean estimates of π, θ and Tajima’s D for all chromosome arms and the X chromosome for all site categories.
Supplementary table 2. List of highly differentiated genomic regions spanning less than 50 Kb.
Supplementary table 3. List of genes with highly divergent non-synonymous changes between Raleigh and Winters. “NSYN” indicates the total number of non-synonymous changes and θST is the mean θST value across the non-synonymous positions.
Supplementary table 4. Linkage disequlibrium analysis for the highly differentiated region in chromosome 3L (20,190,000–20,240,000) between Winters and Raleigh.
Supplementary table 5. Pairwise FST analysis for the gene Obst-F (FBgn0036947) between Winters, Raleigh, and two sets of samples from the Southeastern US (SEUS) and several Caribbean locations.