Abstract
The differentiation of CD8 T cells in response to acute infection results in the acquisition of hallmark phenotypic effector functions, however the epigenetic mechanisms that program this differentiation process on a genome-wide scale are largely unknown. Here we report the DNA methylomes of antigen-specific naïve and day 8 effector CD8 T cells following acute LCMV infection. During effector CD8 T cell differentiation, DNA methylation was remodeled such that changes in DNA methylation at gene promoter regions negatively correlated with gene expression. Importantly, differentially methylated regions (DMRs2) were enriched at cis-elements, including enhancers active in naïve T cells. DMRs were associated with cell type-specific transcription factor binding sites, and these transcription factors clustered into modules that define networks targeted by epigenetic regulation and control of effector CD8 T cell function. Changes in the DNA methylation profile following CD8 T cell activation, revealed numerous cellular processes, cis-elements, and transcription factor networks targeted by DNA methylation. Together, the results demonstrated that DNA methylation remodeling accompanies the acquisition of the CD8 T cell effector phenotype and repression of the naïve cell state. These data therefore provide the framework for an epigenetic mechanism that is required for effector CD8 T cell differentiation and adaptive immune responses.
Introduction
In response to acute infection naïve CD8 T cells differentiate into effector cells capable of killing infected cells and clearing the infection. Effector CD8 T cell function is characterized by the induction of a specific transcriptional program that drives rapid proliferation, expression of key cytokines and effector proteins necessary for cell killing, and the capacity to migrate into infected tissue (1–4). Upon antigen clearance, 90% of effector cells undergo apoptosis while the remaining cells complete their differentiation into a pool of memory CD8 T cells (5). A number of the critical transcription factors that drive this differentiation program, such as Blimp-1 (Prdm1), Tbet (Tbx21), and Eomesodermin (Eomes) have been identified and characterized (2, 6, 7). However, little is known about the epigenetic programs that enforce the induced gene expression changes and permit faithful inheritance of effector function during the proliferative phase of infection.
Epigenetic mechanisms ensure the maintenance and inheritance of gene expression programs through cell division and include DNA methylation and histone modifications (8, 9). Mammalian DNA methylation primarily involves the methylation of CpG dinucleotides and is associated with a repressed epigenetic state when found in gene promoters (8, 10–12). DNA methylation is maintained or deposited de novo by one of three DNA methyltransferases (DNMT1, DNMT3A, and DNMT3B). Methylated CpG DNA is recognized/interpreted by a family of methyl-CpG binding proteins (10, 13, 14). DNA methylation readers and writers are vital components of the adaptive immune response. Deletion of the maintenance methyltransferase DNMT1 during T cell development resulted in normal lineage formation but led to homeostatic defects and the inability to silence lineage specific genes in CD4 T cell differentiation (15). Similarly, deletion of the de novo methyltransferase DNMT3A in CD4 T cells did not affect lineage specification but permitted ectopic cytokine expression and increased lineage plasticity (16). Interestingly, conditional deletion of DNMT1 at the time of CD8 T cell activation resulted in a diminished effector pool, fewer memory CD8 T cells, and reduced ability to clear antigen (17). In contrast, deletion of the DNA methylation reader MBD2 had no effect on proliferation but inhibited the formation of a functional memory compartment following viral challenge (18). These studies demonstrate the importance of DNA methylation in maintaining phenotypic programs during the rapid proliferation of effector CD8 T cells and indicate that correct interpretation of methylated sequences is functionally important for the adaptive immune response.
The dynamic nature of DNA methylation in hematopoietic cell types has been cataloged during early differentiation of the myeloid and lymphoid lineages (19), mouse erythropoiesis (20), and between human regulatory T cells (Tregs) and naïve CD4 cells (21). However, none of these studies profiled a clonal or antigen-specific population of cells. A recent study in CD4 T cells demonstrated that developmental T cell receptor (TCR) specific signaling can establish a preexisting methylation profile such that only CD4 T cells that upregulated Foxp3 and exhibited a specific methylation epitype were able to differentiate into the Treg lineage (22). These data suggest epigenetic heterogeneity in the naïve CD4 T cell pool and highlight the importance and value of profiling the linear differentiation of cells that possess a single TCR.
Dynamic DNA methylation in CD8 T cells has previously been studied only at the single gene level. The effector cytokine Ifng and inhibitory receptor Pdcd1 genes are methylated and silenced in naïve CD8 T cells, demethylated and expressed at the effector stage, and remethylated when expression is silenced in memory CD8 T cells (23, 24). Methylation changes at the Pdcd1 locus occurred in key regulatory regions (23, 25). Changes in DNA methylation at cell-type specific enhancer regions for Ifng have been described during CD4 T cell lineage specification but only at the promoter in CD8 T cells (24, 26). These studies show that DNA methylation is dynamically regulated during differentiation of CD8 T cells and suggests that it is targeted to cis-regulatory elements. Following differentiation cues in ES cells, DNA-binding factors direct sequence-specific modifications to the epigenome, influencing local epigenetic states, including DNA methylation (27). Therefore, mapping the sites of differential DNA methylation during differentiation could identify novel regulatory elements and the sequences bound by factors that drive epigenetic remodeling. Moreover, such maps provide a foundation for how CD8 T cell differentiation programs are imprinted within the genome.
Infection of C57Bl/6 mice with the lymphocytic choriomeningitis virus (LCMV) Armstrong strain results in an acute infection that is cleared typically within 8 days. At this time point, antigen-specific CD8 T cell numbers have peaked and the cells have fully differentiated into effector cells. To derive an understanding of the epigenetic mechanisms controlling this differentiation, we mapped genome-wide DNA methylation in naïve and day 8 (D8) effector CD8 T cells following LCMV Armstrong infection. DNA methylation was globally remodeled during effector cell differentiation and promoter DNA methylation changes inversely correlated with gene expression. We examined active enhancers and observed that an overall gain in methylation occurred at these regions in D8 effector CD8 T cells, suggesting that an epigenetic silencing mechanism is functioning at these sites during their differentiation. Importantly, we found that differentially methylated regions (DMRs) were enriched for transcription factor motifs that coordinated effector CD8 T cell differentiation and function in response to external stimuli. Together, these data identify novel regulatory regions and transcription factor networks, which suggest that DNA methylation is an important and dynamic epigenetic mechanism contributing to the formation and/or maintenance of the effector CD8 T cell state.
Materials & Methods
Isolation of antigen-specific CD8 T cells
LCMV-specific splenic CD8 T cells were obtained from naïve transgenic P14 mice that have a transgenic TCR recognizing the H-2 Db GP33-41 epitope of LCMV (28). Chimeric mice were generated by intravenous adoptive transfer of 105 congenically labeled Thy1.1+ naïve LCMV-specific CD8 T cells into Thy1.2+ C57BL/6 recipients. 24 hours after adoptive transfer, chimeric mice were infected with 2×105 pfu of LCMV Armstrong. LCMV-specific effector CD8 T cells were obtained from the spleen 8 days post-infection (D8 effectors). CD8 T cells were purified by FACS using fluorescently labeled CD90.1 (Thy1.1) and CD8 antibodies as previously described (1, 5, 18, 24). Naïve P14 cells were FACS purified and used as antigen-specific naïve CD8 T cells. All animal experiments were approved by the Emory University Institutional Animal Care and Use Committee.
Methyl-DNA Immunoprecipitation sequencing (MeDIP-seq)
MeDIP-seq was performed as described previously (29, 30). Briefly, 2 μg of pooled genomic DNA from three independent isolations of naïve and D8 effector CD8 T cells, and input control were sonicated to an average size of 300 bp, end repaired, A-tailed, and sequencing adaptors ligated according to the manufacturer’s recommended protocol (Illumina, Inc). Following agarose gel size selection, naïve and D8 effector CD8 T cell samples were immunoprecipitated with 1 μg of an anti-5-methylcytosine antibody (Eurogentec) at 4°C over night. Methylated DNA was purified and 1/3 of the sample amplified for 12 cycles by PCR along with the input fraction to generate sequencing libraries. Libraries were quantitated by Agilent BioAnalyzer, and each library was sequenced using a single-end, 50bp protocol on a single lane of a HiSeq2000 instrument at the Southern California Genotyping Consortium. All sequencing data is freely available via the GEO database (http://www.ncbi.nlm.nih.gov/geo/) accession number GSE44638.
MeDIP-seq Analysis
Raw sequencing reads were mapped to the mouse genome (mm9) using Bowtie (31) and only those uniquely mappable and non-redundant reads were used in subsequent analyses. Manipulation, annotation, and analysis of sequence data were performed using HOMER software (32) and custom R and PERL scripts, which are available upon request. To identify DMRs, we used the MEDIPS package that makes use of a sliding window approach (33). Briefly, we extended each read to 300 bp such that they represent the average fragment size of immunoprecipitated DNA as determined by the Agilent BioAnalyzer trace. We compared the average enrichment measured as the mean reads per million (rpm) in 10 bp bins, for 200 bp sliding windows, and incremented the window 100 bp at a time. Bin, window, and increment parameters that produced the fewest number of DMRs not containing a CpG were chosen after comparison of a range of values. Regions where naïve and D8 effector CD8 T cell samples did not contain enrichment greater than the 90th percentile of the input (rpm) were eliminated due to lack of enrichment. Regions that contained input, naïve, or D8 effector enrichment above the 99.999 percentile were eliminated as sequencing artifacts. P-values calculated by MEDIPS were corrected for multiple hypotheses testing using the Benjamini-Yekutieli correction. A false discovery rate (FDR) of less than 0.01 was considered significant. Significance of DMR enrichment at promoters and enhancers was determined by permutation testing. The locations of the promoters or enhancers were shuffled 1,000 times and the number of DMRs that overlapped the permuted set of promoters or enhancers recalculated and compared that to the actual number of overlaps. The P-value was equal to the number of times the permuted data had a larger overlap than the actual data, divided by the permutations.
Bisulfite sequencing
10 ng of genomic DNA was treated with sodium bisulfite to convert unmethylated cytosines to uracils with the EpiTech Bisulfite Kit according to the manufacturer’s protocol (Qiagen, Inc). Bisulfite converted DNA was amplified by PCR and the amplicons cloned using Topo TA vectors (Invitrogen). Positive colonies were purified and sequenced (Beckman Coulter Genomics). Bisulfite sequences were aligned to their respective in silico bisulfite converted genomic sequences using the Bioconductor Biostrings R package and custom R scripts, which are available upon request. Only sequences that contained data for all CpGs and with a bisulfite conversion rate of greater than 95% (as determined by conversion of non-CpG cytosines to uracil and subsequent amplification as thymine) were included in the analysis. Significance was determined by a one sided Fisher’s exact test. All bisulfite PCR primers are listed in Supplemental Table 1.
Gene Expression and Histone Data Analysis
Differential gene expression between naïve and D8 effector CD8 T cells has been described previously (4). Microarray data were downloaded from the GEO database, accession number GSE9650, and genes that changed greater than 1.5 fold with an FDR less than 0.05 were identified by Significance Analysis of Microarrays (34). Histone modification profiles present in the thymus were identified by ChIP-seq as part of the ENCODE project (35). All ENCODE data sets used in this study are listed in Supplemental Table 1. Raw ChIP-seq reads were mapped to the mouse genome (mm9) using Bowtie (31) and significantly enriched peaks relative to input control identified by HOMER software (32) using the “histone” setting. Overlapping and unique peaks were determined using Bedtools (36).
Results
DNA methylation of naïve and D8 effector P14 CD8 T cells
DNA methylation at Ifng and Pdcd1 gene promoters in CD8 T cells changed during their differentiation and negatively correlated with transcription of these genes (23, 24), suggesting that other genes or perhaps many genes within CD8 T cells use DNA methylation to mediate control of gene expression. To investigate the global role for DNA methylation in CD8 T cells during an acute viral infection, the LCMV Armstrong strain was used as an infection model. P14 transgenic mice harbor a knock-in T-cell receptor specific for the H2-Db-restricted GP33-41 epitope of LCMV (28). The use of P14 transgenic mice ensure that TCR affinity for peptide-MHC-I molecules is equal among clonal P14 CD8 T cells during development and an immune response (22). Adoptive transfer of naïve P14 CD8 T cells into wild-type mice, followed by infection with LCMV Armstrong leads to a robust adaptive immune response and viral clearance (1, 5, 37). Thus, naïve splenic (CD44lowCD8+) P14 CD8 T cells were purified and adoptively transferred into wild-type mice, which were subsequently infected with LCMV Armstrong. Eight days post-infection, at the peak of the adaptive immune response (1), antigen-specific P14 effector CD8 T cells (D8 effector) were isolated based on expression of the congenic marker Thy1.1 (Figure 1A). Naïve and D8 effector CD8 T cells were purified in triplicate and were found to exhibit surface expression of key phenotypic markers typical of their respective developmental stage (Supplemental Figure 1)(1, 4, 37).
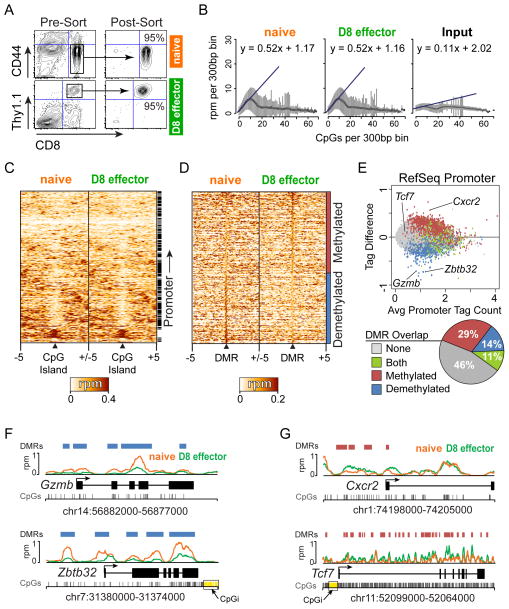
Figure 1. DNA methylation is globally dynamic during effector CD8 T cell differentiation in response to LCMV Armstrong infection.
(A) Representative plots of naïve CD8 T cells (CD8+CD44low) isolated by FACS from P14 transgenic mice and adoptively transferred into wt hosts and infected with LCMV Armstrong. Eight days post-infection, antigen-specific effector CD8 T cells (D8 effectors) (CD8+Thy1.1+) were purified by FACS. (B) The density of CpGs in 300bp windows across the genome was calculated and MeDIP-seq data in each window was plotted as reads per million (rpm). MeDIP-seq signal was consistently enriched at low-density CpG regions in both naïve and D8 effectors. No enrichment was observed for Input control. (C) Heatmap showing MeDIP-seq read density surrounding all 16,206 murine CpG islands. Each row represents 5 kb surrounding one CpG island with read density normalized to reads per million (rpm). Rows are hierarchically clustered and the right annotation bar denotes CpG islands that overlap a promoter. (D) Heatmap of MeDIP-seq read density at 646,673 differentially methylated regions (DMRs) identified between naïve and D8 effector CD8 T cells is shown. 296,007 DMRs became demethylated and 350,666 became methylated greater than 1.5-fold during D8 effector differentiation. Each row represents 5 kb surrounding one DMR with read density normalized to rpm. Rows are sorted by decreasing change in methyl-DNA enrichment. (E) Scatter plot of methylation difference (D8 effector – naïve) versus average MeDIP-seq tag densities at all 25,387 murine RefSeq promoters is shown. Promoter regions were defined as the upstream 2000 bp and downstream 500 bp from the transcription start site of each gene. RefSeq promoters that overlapped a DMR are color-coded and the percentage of total DMRs that overlap promoters is portrayed in a pie chart. (F) The genes for the CD8 T cell effector protease Gzmb and the transcription factor Zbtb32 demonstrating significant demethylation changes are plotted depicting the location of the DMRs (blue), CpGs (grey), CpG islands (CpGi – yellow), gene structure, and read density for naïve (Orange) and D8 effectors (Green). (G) The chemokine receptor Cxcr2 and T cell developmental transcription factor Tcf7 are significantly methylated in D8 effectors. Each gene is plotted as above showing methylated DMRs (red).
DNA from naïve and D8 effector samples was isolated and the respective samples pooled and subjected to methyl-DNA immunoprecipitation (MeDIP) using a 5-methylcytosine specific antibody. The precipitated DNA and input control were analyzed by deep sequencing. Sequencing was performed to a depth that generated roughly ten-fold coverage for half the mouse genomic CpGs (29, 30, 38). Analysis of the MeDIP-seq data indicated a robust linear enrichment at low-density CpG loci that was highly similar between both naïve and D8 effectors and greater than that observed at CpG dense regions or the input control (Figure 1B). This is consistent with previous observations that areas of low CpG density are typically methylated compared to those regions of high CpG density, such as CpG islands, which remain unmethylated (27, 33, 38, 39). Analysis of all mouse CpG islands indicated that the majority are unmethylated in naïve and D8 effectors (Figure 1C). The small percentage of heavily methylated CpG islands were primarily located outside of promoter regions, which has been noted previously in humans and mice (40, 41).
Global DNA methylation dynamics during differentiation of naïve to effector CD8 T cells
To identify DMRs between naïve and D8 effectors, we applied a genome-wide sliding window approach to identify methyl-enrichment changes greater than 1.5-fold with a false discovery rate (FDR) less than 0.01 that were enriched above background (33). This approach identified 296,007 demethylated and 350,666 methylated DMRs in D8 effectors compared to naïve CD8 T cells (Figure 1D). The changes in promoter methylation for all 25,387 murine RefSeq promoters were analyzed as promoter-proximal sequences represent focal control points for transcriptional regulation. RefSeq promoters were annotated for the presence of a DMR, and their change in methylation was plotted with respect to the average methylation enrichment in naïve and D8 effectors (Figure 1E). This revealed that 54% of RefSeq promoters contained at least one DMR (29% methylated, 14% demethylated, and 11% both). The overlap of both methylated and demethylated DMRs with promoters was greater than that expected by chance (P < 0.001). Promoters that contained a DMR displayed varying levels of methylation change but overall trended in the same direction as the DMR they overlapped. The 11% of promoters that included both methylated and demethylated DMRs displayed the largest variation in methylation changes. The presence of both DMRs suggested that these promoters might contain both negative and positive regulatory elements that become activated or repressed epigenetically.
DMRs occurred at biologically relevant gene promoters that are necessary for effector CD8 T cell function. For example, genes with demethylated promoter DMRs included Gzmb and Zbtb32 (Figure 1F). The Gzmb gene encodes the serine protease Granzyme B, which is important for effector CD8 T cell cytotoxic function (42) and in D8 effectors compared to naïve cells was strongly demethylated in the gene body and almost completely lacked a methyl-DNA signal at its promoter region. Zbtb32, a transcription factor induced in activated lymphocytes (43, 44), demonstrated significant demethylation at the transcription start site (TSS), gene body, and upstream elements. Cxcr2 and Tcf7 significantly gained methylation in D8 effectors compared to naïve cells (Figure 1G). Cxcr2 encodes a chemokine receptor implicated in cellular senescence (45) and contained several methylated DMRs that were concentrated at the promoter region. The transcription factor Tcf7, which is essential for lymphocyte development and differentiation (46), was heavily methylated in the gene body, but did not contain a DMR in the promoter due to the presence of a CpG island, which is likely constitutively unmethylated (Figure 1E). In hematopoietic development, sequences immediately proximal to CpG islands, termed CpG island shores, were shown to exhibit highly dynamic DNA methylation levels compared to CpG islands (19). Similarly, we identified DMRs at CpG island shores at both Zbtb32 and Tcf7. Overall, the MeDIP-seq data suggested that DNA methylation was globally reprogrammed and possibly targeted to specific regulatory regions that may be important for controlling the expression of genes central to the adaptive immune response and CD8 T cell function.
Bisulfite sequencing validates MeDIP-seq DMRs
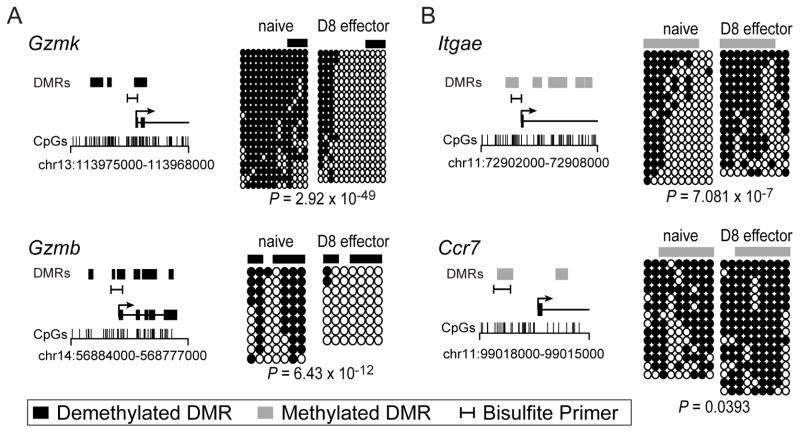
To verify the MeDIP-seq data and provide a single-CpG resolution map of select regions, we validated a number of DMRs by clonal bisulfite sequencing (cBS) from two to three independent isolations of naïve and D8 effector CD8 T cells. DMRs in biologically relevant and statistically representative TSSs were chosen for validation. As indicated by the MeDIP-seq and cBS, genes encoding the serine proteases important for CD8 T cell cytotoxic activity, Gzmb and Gzmk contained promoter DMRs that became almost completely demethylated in D8 effectors (Figure 2A). cBS of the integrin Itgae gene demonstrated increased methylation directly at the TSS while the chemokine receptor Ccr7 showed significant gains in methylation upstream of the TSS (Figure 2B). Together, these data validated the methylation differences identified by MeDIP-seq and provide a single-CpG resolution of the dynamics at promoter regions for genes important in the immune response.
Figure 2. Bisulfite sequencing validated MeDIP-seq methylation changes.
A representative set of methylated and demethylated DMRs were validated by bisulfite sequencing. Two loci that contained demethylated DMRs (A) Gzmk and Gzmb, and two loci (B) Itgae and Ccr7 that exhibited methylated DMRs were chosen. For each gene the location of bisulfite primers, DMRs, and CpGs are indicated in relation to the transcription start site. Each line of bisulfite sequencing data represents an individual clone with black circles denoting methylated CpGs and open circles representing unmethylated CpGs. The location of the DMR analyzed is diagramed above each bisulfite data set. All loci were validated from two to three independent samples of naïve and D8 effector time points and demonstrated statistically significant changes according to a one-sided Fisher’s exact test.
DNA methylation inversely correlates with gene expression
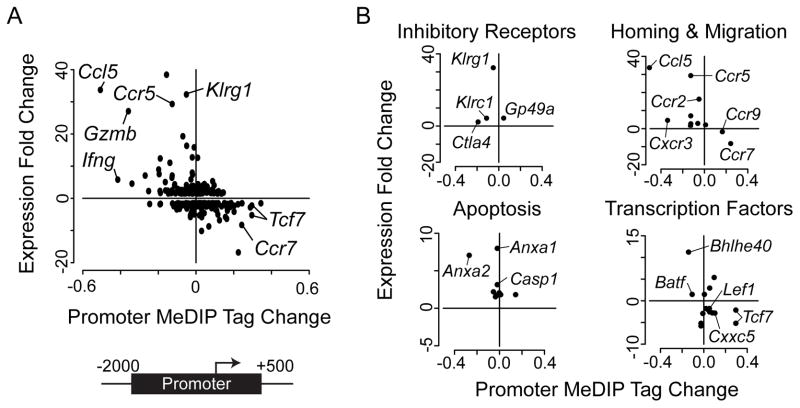
DNA methylation is a repressive epigenetic modification and has previously been shown to negatively influence transcription (12, 14). The CD8 T cell response to LCMV results in global transcriptional remodeling during differentiation with hundreds of genes differentially expressed in D8 effectors compared to the naïve state (4). To investigate the relationship between DNA methylation and gene expression in CD8 T cells, we correlated the change in promoter methylation in D8 effectors with known transcriptional changes in this system. DNA methylation demonstrated a significant negative correlation with gene expression (Spearman’s rank correlation ρ = −0.41, P < 0.001)(Figure 3A). Genes that increased in expression largely were demethylated at their promoters (top left quadrant) while genes that became transcriptionally silenced gained methylation in D8 effectors (bottom right quadrant). For example, the chemokine receptor Ccr7 was significantly methylated and repressed following CD8 T cell differentiation, a change we validated by cBS (Figure 2B). The Ifng promoter was one of the strongest demethylated and induced genes, a result that supported previous observations in CD8 and CD4 T cells (24, 26). Interestingly, the chemokine receptor Ccr5 and its ligand Ccl5 were both robustly upregulated and demethylated.
Figure 3. Promoter DNA methylation inversely correlates with gene expression.
(A) Scatter plot showing the mean change in promoter methyl-DNA enrichment (rpm) relative to gene expression fold-change for all genes differentially expressed between naïve and D8 effector CD8 T cells is presented. Promoter regions were defined as the upstream 2000 bp and downstream 500 bp from the transcription start site (TSS) of each gene. Gene expression was negatively correlated with promoter methylation (Spearman’s rank correlation ρ = −0.41, P < 0.001). (B) Scatter plots of methyl-DNA enrichment difference and gene expression fold change for four functional groups of genes important to effector CD8 T cell function.
The above results suggest that functional families of genes may be coordinately regulated by DNA methylation. To determine if this was the case, all differentially expressed genes between naïve and D8 effectors were grouped according to functional classes as previously described (4), and the association of promoter methylation change with gene expression was plotted. DNA methylation at promoters inversely correlated with gene expression within functional groups, suggesting that these genes may be regulated by coordinated epigenetic mechanisms (Figure 3B). For example, three inhibitory receptor genes Klrg1, Klrc1, and Ctla4 were upregulated and demethylated in D8 effectors. Genes important for homing and migration displayed the most significant trend as the majority of induced genes were demethylated. Ccr7 and Ccr9, two cytokine receptor genes important for naïve T cell development and homeostasis were both down regulated and methylated. Additionally, genes involved in apoptosis, such as Casp1, and the calcium-signaling proteins Anxa1 and Anxa2 were upregulated and demethylated and may play a role in promoting cell death of effector CD8 T cells at later stages of the process.
Transcription factors represent essential components of T cell effector function that dynamically integrate TCR signals and external stimuli to drive cellular differentiation. The Wnt pathway is necessary for T cell development and homeostasis and is repressed at the effector stage (46–48). Three members of the Wnt signaling pathway Tcf7, Lef1, and Cxxc5 were down regulated and methylated. Interestingly, Batf, which modulates energy metabolism and mediates T cell differentiation and exhaustion (49, 50) was upregulated and demethylated along with Bhlhe40, a factor involved in cellular differentiation and circadian rhythm (51–53). DNA methylation did not completely correlate with gene expression in all instances. For example, the inhibitory receptor Gp49a was upregulated but gained DNA methylation in its promoter (Figure 3B, top left). These changes could be due to the methylation of inhibitory sequences or a temporal delay in the acquisition of repressive epigenetic marks and effects on mRNA levels. Nevertheless, the majority of genes demonstrated a significant inverse correlation between changes in promoter DNA methylation and gene expression levels. The relationship of DNA methylation and gene expression implicates a key role for epigenetics in establishing the D8 effector transcriptional program.
Thymic enhancers gain DNA methylation in effector CD8 T cells
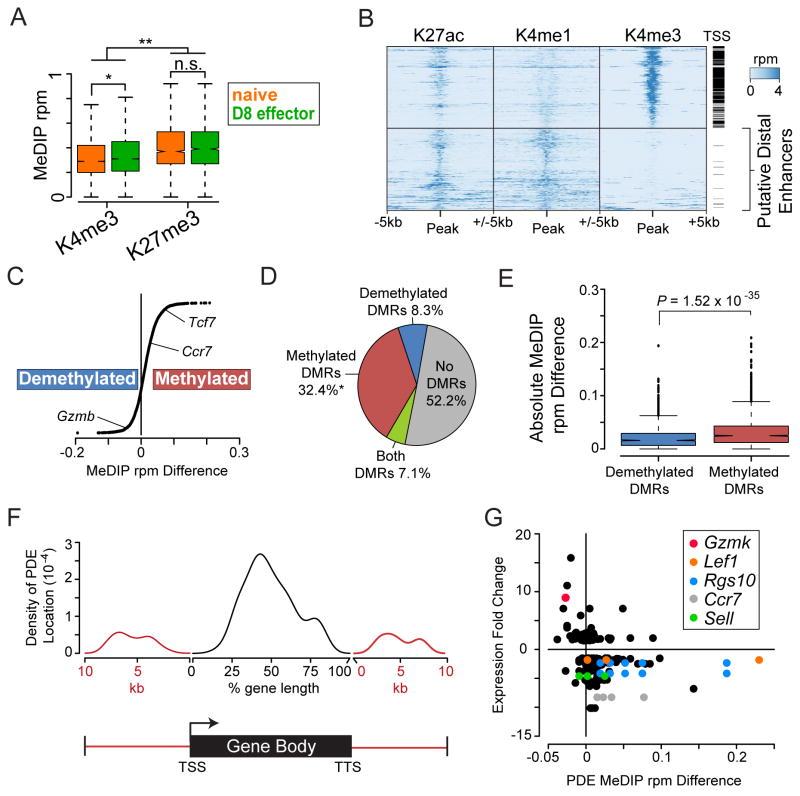
The Encyclopedia of DNA elements (ENCODE) project previously mapped histone modifications in whole thymus, an organ that harbors the developing T cell lineage (Supplemental Table 1)(35, 54). We hypothesized that histone patterns in the thymus would provide insight into loci important for T cell development and/or maintenance of a naïve T cell. The histone H3 lysine 4 trimethylation modification (H3K4me3) marks the TSS of genes and is positively associated with transcription (55) and negatively associated with DNA methylation (56, 57). Conversely, H3K27me3 is a repressive epigenetic modification and has been mechanistically linked to DNA methylation at a number of loci (58). We characterized the average MeDIP-seq signal at regions of H3K4me3 or H3K27me3 in the thymus. DNA methylation was significantly enriched at loci containing repressive histone marks in both naïve and D8 effectors as compared to regions that contain H3K4me3 (Figure 4A). Additionally, D8 effectors contained significantly more DNA methylation at regions of H3K4me3 than naïve, consistent with observations that more TSS gained DNA methylation in D8 effectors (Figure 1E). DNA methylation at regions enriched for H3K27me3 in the thymus were globally unchanged, suggesting these regions are permanently repressed once they acquire H3K27me3.
Figure 4. Enhancers active in the thymus become methylated in D8 effector CD8 T cells.
(A) Histone maps generated from the thymus were downloaded from the ENCODE project (35). Regions containing the repressive histone modification H3K27me3 were significantly enriched for DNA methylation compared to regions with the active histone mark H3K4me3 (n.s. = not significant, * = P < 10−17, ** = P < 10−32 Student’s t-test). (B) Heatmaps of H3K4me3, H3K4me1, and H3K27ac read density (rpm) at 19,365 regions in the adult mouse thymus. Each region is annotated for proximity (within 1 kb) to a TSS with a black bar. Proximal promoter elements (top) contain H3K4me3 whereas putative distal enhancers (PDEs) (bottom) lack H3K4me3 and are marked by high levels of H3K4me1 and H3K27ac. Rows for both proximal promoter elements and PDEs were hierarchically clustered separately across all three histone modifications. (C) Methylation changes at all 5,471 active thymic PDEs showed a trend towards becoming methylated in D8 effectors. Methylation change is calculated as average MeDIP rpm in D8 effectors minus naïve for each PDE. Gene annotations were predicted based on mapping each element to the closest TSS. (D) PDEs primarily overlap methylated DMRs. The overlap of DMRs and PDEs was computed and percentages indicated in a pie chart. Asterisk indicates significant overlap of methylated DMRs with PDE regions greater than random chance (P < 0.001). (E) Box plot of absolute methylation difference in PDEs that overlap methylated and demethylated DMRs. PDEs that overlapped methylated DMRs had significantly greater changes in methylation than those that overlapped demethylated DMRs. Significance was calculated by Student’s t-test. (F) Density plot summarizing the distribution of PDEs located within the gene body or surrounding 10kb of genes differentially expressed between naïve and D8 effector CD8 T cells. Location of PDEs within gene bodies was calculated as the distance from the TSS and position normalized to a percentage of the gene body length. TSS, transcription start site; TTS, transcription termination sequence. (G) Scatter plot displaying the mean change in PDE DNA methylation relative to gene expression fold-change for all genes differentially expressed and annotated with a PDE. Select PDEs that map to CD8 relevant genes are highlighted in the figure key.
Transcriptional enhancers provide critical control points for cell type-specific gene expression and have been shown to undergo epigenetic changes, including DNA methylation, during cellular differentiation (27, 57, 59). For example, during embryonic stem (ES) cell differentiation, DNA methylation was found to be more dynamic at distal regulatory regions outside of core promoters (57), suggesting that similar events may also occur in differentiating T cells. The presence of both H3K4me1 and H3K27 acetylated (H3K27ac) histone modifications in the absence of H3K4me3 separates active distal enhancers from promoter-proximal TSS elements (60). Putative distal enhancers in thymic cells were identified by mapping regions enriched for both H3K4me1 and H3K27ac modifications. The regions were then stratified by the presence or absence of H3K4me3. This approach established 5,670 putative distal enhancers (PDEs), 99% of which were at least 2 kb away from the nearest TSS (Figure 4B). To investigate the role for DNA methylation at thymic enhancers, we analyzed the average change in DNA methylation from naïve to D8 effectors. Although PDEs became both methylated and demethylated, there was a trend towards more enhancers gaining DNA methylation in D8 effectors (Figure 4C). PDEs methylated in D8 effectors were identified around genes that became repressed, such as Tcf7 and Ccr7. In addition, genes that became activated in D8 effectors were associated with PDEs that became demethylated such as Gzmb. We annotated the PDEs for overlap with DMRs and found that 36.3% of the PDEs active in the thymus overlapped with a methylated DMR while only 8% overlapped with a demethylated DMR (Figure 4D). The overlap of methylated DMRs was more than expected by random chance (P < 0.001), whereas there was not a significant enrichment of demethylated DMRs in thymic enhancers. Additionally, the change in MeDIP-seq signal was significantly greater at PDEs that overlapped a methylated DMR than a demethylated DMR (Figure 4E). These data indicate that enhancers active in the thymus robustly gain DNA methylation in D8 effectors compared to naïve cells.
To assess the relationship between the DNA methylation status of the PDEs and the expression of the genes that were activated or repressed in this system, PDE’s were mapped to gene bodies and to the sequences 10 kb upstream of the TSS or downstream of the transcription termination sequence (TTS). 164 PDEs were identified within the 10 kb limit for 76 differentially expressed genes. PDEs were enriched inside the gene body and were depleted around transcription start and termination sites (Figure 4F). Upstream and downstream PDEs demonstrated a biphasic distribution, suggesting that both short (<5kb) and long (>5kb) range enhancers exist for these genes. Changes in PDE DNA methylation were next correlated with changes in gene expression (Figure 4G). Inverse correlations between PDE DNA methylation and gene expression were observed for many of the genes with several genes having multiple PDEs that changed in a similar manner (Lef1, RGs10, CCR7, and Sel1). The annotation of these genes with PDEs has potentially identified an additional layer of epigenetic regulation outside of promoter regions, and suggest that DNA methylation may play an important role in regulating enhancer activity during effector CD8 T cell differentiation.
DMRs are enriched for functional transcription factor motifs
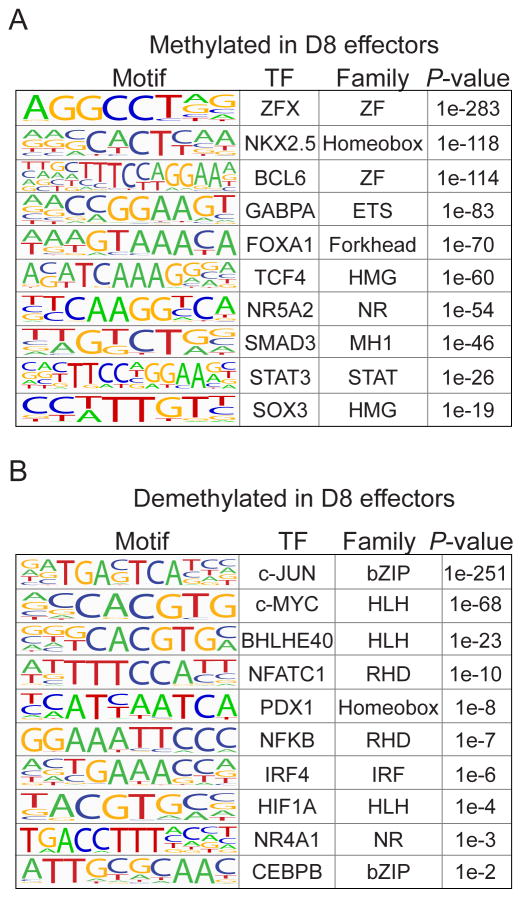
We demonstrated that DNA methylation dynamically changed at both distal enhancers and promoters for genes differentially expressed in CD8 T cells in response to LCMV infection. These data predicted that DMRs contain cis-elements and could be bound by transcription factors important in CD8 T cell function and that demethylated and methylated DMRs may have unique functions. Using consensus DNA-binding sequences termed “logos”, which were generated from published ChIP-seq experiments, we searched for transcription factor motifs that were significantly enriched in the methylated DMRs versus the demethylated DMRs, and vice versa. DMRs methylated in D8 effectors contained motifs for developmental transcription factors such as the ETS (GABPA), High Mobility Group (SOX3 and TCF4), Forkhead (FOXA1), and Zinc Finger (ZFX and BCL6) families (Figure 5A and Supplemental Table 2). Methylation of these sites may be a repressive epigenetic mechanism to silence these transcriptional networks that are active in the developing or naïve T cell. In contrast, DMRs demethylated in D8 effectors were highly enriched for transcription factor families known to play roles in the effector response, such as the bZIP (c-JUN and CEBPB), Rel Homology Domain (NFATc1 and NFKB), and IRF (IRF4) families (Figure 5B and Supplemental Table 2). Loss of methylation at these DMRs may lead to an accessible and open chromatin environment that facilitates binding of these factors and promote effector differentiation and function in response to external stimuli. DNA methylation has previously been reported to prevent DNA binding for a number of proteins (61–65). Separate from correlating with a repressive local chromatin environment, DNA methylation may physically restrict DNA binding for those factors that contain a CpG in their recognition motif, providing multiple epigenetic mechanisms to regulate transcription factor activity.
Figure 5. DMRs are enriched for TF binding sites important for naïve and effector function.
Transcription factor motifs significantly enriched in either (A) methylated DMRs or (B) demethylated DMRs were identified. The binding motif, transcription factor, and DNA binding domain family are indicated for the most significant site for each motif. Enriched motifs in methylated DMRs are mainly developmental transcription factors that may be important for naïve T cell maturation or homeostasis. Demethylated DMRs were enriched for motifs of transcription factors that signal from the TCR and respond to external stimuli. (ZF, Zinc Finger; ETS, E-Twenty Six; HMG, High-Mobility Group; NR, Nuclear Receptor; MH1, MAD Homology 1; STAT, Signal Transducer and Activator of Transcription; bZIP, basic leucine Zipper; HLH, Helix-Loop-Helix; RHD, Rel Homology Domain; IRF, Interferon Response Factor).
Transcription Factor Modules in D8 effector function
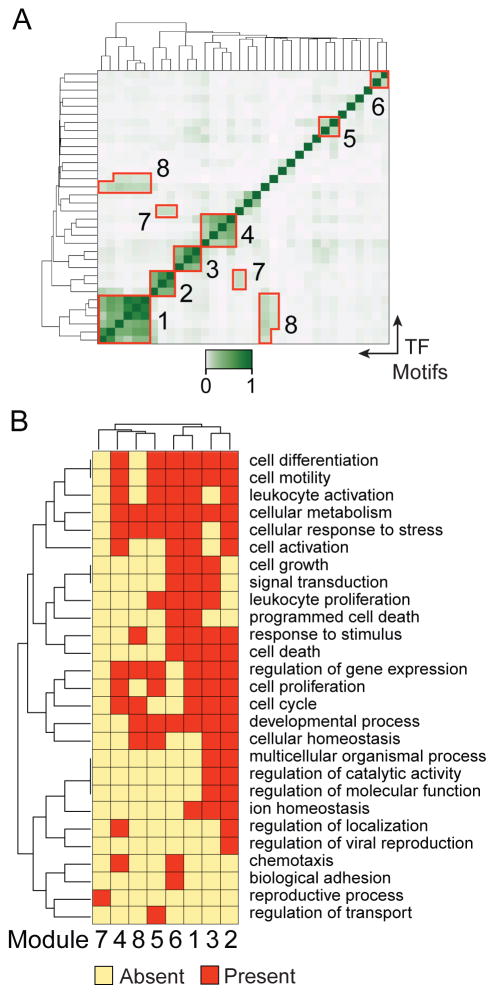
Transcription factors cooperatively bind cis-elements to regulate transcription in multifactor modules thereby integrating multiple signaling pathways to regulate a biological process (32, 66, 67). To identify transcription factor modules that cooperate to control effector CD8 T cell function, we identified motifs that co-occurred within the same DMRs, by correlating the number of motifs that occur in each DMR between any two transcription factors. Transcription factor motif co-occurrence, measured by Spearman’s rank correlation ρ, was used to hierarchically cluster transcription factors, revealing modules commonly bound at the same DMRs (Figure 6A). This analysis identified eight modules enriched in the demethylated DMRs (Figure 6A and Table I). Six of the eight modules correlated along the axis and contained motifs for the calcineurin pathway factor NFAT, AP-1 transcription factors c-JUN and JUND, nuclear receptors, homeobox factors, and helix-loop-helix DNA binding proteins. Additionally, GATA3 interacted with the AP-1 factors in Module 3, and HIF1A and NRF1 were predicted to interact with the helix-loop-helix factors in Module 8. Transcription factor motifs enriched in methylated DMRs also cluster into five modules designated as A-E (Supplemental Figure 2A). These modules included factors belonging to the ETS and STAT family of transcription factors (Supplemental Figure 2B).
Figure 6. Transcription factor modules cooperate to promote effector CD8 T cell function.
(A) Transcription factor co-occurrence at demethylated DMRs was hierarchically clustered to identify factors that potentially cooperate in modules at the same DMR. Co-occurrence is calculated by Spearman’s rank correlation of the number of transcription factor binding sites in each DMR between any two of the 35 significant transcription factor motifs. Eight modules used in subsequent analyses are indicated in red outlines. (B) DMRs containing each module were mapped to the nearest gene and gene ontology analysis was performed to identify biological processes each module affected. Significant GO terms enriched in any of the eight modules were clustered and shared and unique processes depicted in a heatmap. Each row represents the presence or absence of the indicated GO term and each column is a transcription factor module.
Table I.
Transcription factor modules present in demethylated DMRs
| Module | Transcription Factor Motifs |
|---|---|
|
| |
| 1 | ATF3, USF1, BHLHE40, c-MYC, MAX |
| 2 | CRE (cAMP responsive element), JUND, c-JUN |
| 3 | NFE2, c-JUN, AP-1 |
| 4 | NANOG, HOXB4, PDX1 |
| 5 | NFAT-AP-1, NFATC1 |
| 6 | NR4A1, RXR |
| 7 | GATA3, JUND, c-JUN |
| 8 | HIF1A, NRF1, ATF3, USF1, BHLHE40, c-MYC, MAX |
To determine the biological function of each module, the significantly enriched Gene Ontology (GO) terms present in the genes regulated by each module were identified. GO terms were clustered to identify shared processes between transcription factors modules (Figure 6B). For the modules identified in the methylated DMRs, no significant GO terms could be found to assign specific functions to those modules. In contrast, with the exception of module 7, all of the modules in demethylated DMRs shared broad overlapping functions with roles in processes, such as cellular differentiation, metabolism, and cell motility, and were analyzed further. Six modules were enriched for the regulation of gene expression ontology, suggesting that they play a role integrating signals that fine-tune downstream transcriptional networks. Module 6, comprised of nuclear receptors, was enriched for known functions in apoptosis (68), but interestingly a putative novel role in chemotaxis was uncovered. Module 1 was involved in diverse cellular processes such as differentiation, leukocyte activation, programmed cell death, and leukocyte proliferation. Module 1 integrated signals from the cyclic-AMP pathway through ATF3 and E-box binding factors MYC, MAX, USF1, and BHLHE40. While roles in the immune response have been described for the other factors in module 1 (69–72), no functional role has previously been described for BHLHE40 in CD8 T cells. Interestingly, Modules 2 and 3 included the AP-1 factors, but uniquely Module 2 contained CREB and Module 3 NFE2. Distinct functions for Module 2 consisted of leukocyte and cell activation while unique processes for Module 3 included cell growth, leukocyte proliferation, and signal transduction. These results suggest separate roles for the CREB-AP1 module in promoting T cell activation while the MAP kinase-AP1 pathways are predicted to regulate T cell expansion and proliferation.
Discussion
The differentiation from naïve to effector CD8 T cells following acute infection is a critical step in the adaptive immune response. The gene expression changes that occur following differentiation cues have been studied (2, 4), however little is known about how the epigenome is reprogrammed to permit the acquisition of new cellular functions. Here, we report the genome-wide DNA methylation profile of naïve and D8 effector CD8 T cells using MeDIP-seq. Consistent with the scale of epigenetic changes previously reported during ES cell differentiation (57), we find extensive global reprogramming of DNA methylation during effector CD8 T cell differentiation. Gene expression and promoter DNA methylation were negatively correlated, demonstrating that previous findings at the Ifng (24) and Pdcd1 (23) loci defined a broad epigenetic mechanism for gene regulation in CD8 T cells. Methylated and demethylated DMRs were enriched in 54% of murine RefSeq promoters, which greatly exceeds the known transcriptional reprogramming events during CD8 T cell differentiation. Changes in DNA methylation may result in the fine-tuning of gene expression that is not detectable by microarray technology, a shift to a new set of cis-elements that drive/inhibit gene expression in D8 effectors and not naïve cells, and/or define chromatin structural changes that serve to maintain the D8 effector cell fate program. In this vein, we identified 76 genes with expression changes that could be annotated to one or more PDEs whose DNA methylation status changed between naive and D8 effector CD8 T cells. Thus, the correlation of gene expression, promoter DNA methylation, and PDE DNA methylation, implies that the epigenome is actively reprogrammed as CD8 T cells differentiate to facilitate the emergence of new gene expression patterns and effector CD8 T cell function.
The finding that some DMRs were enriched for functional transcription factor motifs provided an additional layer of epigenetic regulation for DNA methylation in CD8 T cell differentiation. Overall, consensus motifs that were found to be in demethylated DMRs in D8 effectors were correlated with transcription factors that contribute to the effector function, such as NFATc1 and c-JUN. Conversely, motifs in DMRs that were in methylated in D8 effectors were mostly associated with transcription factors that function in developmental/differentiation pathways. Thus, the changes in DNA methylation appear to coincide with a cell fate program that has shifted from the ability to differentiate cells to one that is focused on effector outcome and function.
The unique transcription factor motifs enriched in demethylated and methylated DMRs indicated that changes in DNA methylation might restrict the accessibility to DNA for transcription factors. In fact, the physical binding of c-JUN, JUND (65), c-MYC (64), CREB/ATF (62), CTCF (61), and ETS1 (63) to DNA has previously been demonstrated to be methylation sensitive. Interestingly, these factors are ubiquitously expressed during CD8 T cell differentiation (2, 4). Therefore, as CD8 T cells differentiate into effector cells and the DNA methylation landscape is remodeled, so too are the accessible binding sites for these factors. This would predict that a precise order of events occurs for inactive (methylated) elements in naïve CD8 T cells as they transition into an active state (demethylated) in D8 effectors. For example, in CD8 T cells, the Pdcd1 (PD-1) locus undergoes such a process. In naïve CD8 T cells, Pdcd1 is extensively methylated and silent (23, 25). Pdcd1 is induced following the binding of NFATc1 to its cis-element (25), suggesting that NFATc1, which contains no CpG in its DNA-binding site, can access the Pdcd1 DNA and initiate expression of Pdcd1. Upon NFATc1 binding, the chromatin structure of the region adopts an active conformation (i.e., histone H3 and H4 acetylation) and during the initial stages of the effector response, the region loses its DNA methylation (23, 25). Likewise, many of the enriched transcription factor motifs do not contain a CpG in their binding motif. Therefore, there may be a temporal sequence of transcription factor binding events that first initiates an open chromatin conformation and demethylation of the DNA. This would be followed by the binding of DNA methylation-sensitive transcription factors, and new gene expression patterns.
Transcription factor networks integrate external signaling pathways to modulate gene expression and initiate cellular differentiation. Originally termed enhanceosomes (66), cis-acting regulatory elements frequently contain multiple transcription factor binding sites that form a module to integrate signaling pathways, a process that has been best studied in ES cells (67) and between macrophages and B cells (32). Our DNA sequence centric approach differs from previous transcription-focused module analyses in CD8 T cells that analyzed gene expression correlations (2). Although our prediction of DNA-binding motifs relied on high quality ChIP-seq data that is not available for every transcription factor, we identified transcription factor modules of both known and novel function in effector CD8 T cells. For example, module 5 consisted of the known NFAT-AP-1 interaction (73). In addition, this approach allowed us to dissect out two roles for the AP-1 family factors in modulating calcium and MAP kinase signaling pathways. Many modules participated in overlapping functions, suggesting a complex integration of signaling networks may be required to maintain effector function during clonal expansion. Moreover, our results predict broader roles in CD8 T cell function for a nuclear receptor module consisting of NR4A1 and RXR. The nuclear receptors may regulate integral CD8 T cell processes, such as activation, proliferation, metabolism, and chemotaxis. These findings highlight the complex interaction of signaling pathways that is required to maintain a highly dynamic and metabolically active cell population.
Our experiments revealed that both gain and loss of DNA methylation occurred during CD8 T cell differentiation. The acquisition of new DNA methylation implies that the de novo DNA methyltransferases, DNMT3A and 3B, were active during this process. Indeed, gene expression analyses for the DNMTs showed that during effector differentiation all components of the methylation machinery were expressed during this process (2, 4, 23). DNMT3A and 3B can be recruited to DNA by interactions with the histone methyltransferase G9a (74) or via interaction with DNMT3L (56). DNMT3A-DNMT3L complex binds chromatin containing unmethylated histone H3K4 (H3K4me0)(56). Active TSSs and PDEs are surrounded by nucleosomes that are H3K4me3 and H3K4me1 modified respectively, which implies that there is a stepwise process that must first remove the H3K4 methylation before the region can become a strong substrate for de novo methylation. The H3K4me1/2 histone demethylase LSD1 inactivates enhancers during ES cell differentiation (75) and may be active in this system to prepare these sites for de novo methylation. Additionally, the transcriptional repressor E2F6 can recruit DNMT3B directly to DNA (76), suggesting alternative chromatin-independent mechanisms exist for the acquisition of novel DNA methylation patterns.
The loss of DNA methylation in D8 effectors could have occurred by two general processes: passive and active. Following activation, CD8 T cells expand exponentially, allowing simple failure of DNMT1 to remethylate CpGs as a possible mechanism. Passive demethylation can also occur by the recruitment of factors that block DNMT1 binding, such as GATA3 (77). Evidence for active demethylation also exists as demethylation of the Il-2 promoter in CD4 T cells and the Ifng promoter in memory CD8 T cells was observed during experimental conditions of cell cycle arrest, suggesting an active process at these sites (24, 78). The TET family of enzymes can hydroxylate 5-methylcytosine to begin the process of demethylation that utilizes the base-excision repair pathway and culminates in an unmodified cytosine (79–81). The recent discovery and characterization of these enzymes provide credence to the notion that active DNA demethylation can occur. While NANOG can recruit TET1 and TET2 to promoters during induced pluripotent stem cell (iPS) reprogramming (82), no targeting mechanism has been identified for locus-specific active demethylation in CD8 T cells. A cellular demand for rapid gene expression kinetics may require one method over another, but both active and passive mechanisms are likely occurring at the demethylated DMRs in CD8 T cells.
The data presented here revealed the global acquisition of novel DNA methylation patterns in effector CD8 T cells that facilitated the expression of the effector phenotype while repressing the naïve transcriptional program. Importantly, the sites of DNA methylation remodeling identified both trans- and cis-factors important for naïve and effector CD8 T cell identity and function. These data have therefore drawn a blueprint for the foundation of one of the epigenetic programs associated with CD8 T cell differentiation. Further understanding of the mechanisms that write, erase, and interpret this blueprint will ultimately be important for manipulating the epigenome of these cells for the creation of novel vaccines and therapeutic treatments involving CD8 T cells.
Supplementary Material
Acknowledgments
We would like to thank members of the Boss and Ahmed laboratories for helpful comments and critiques during this work, as well as the Southern California Genotyping Consortium for sequencing and the Emory University Flow Core for cell isolation.
Footnotes
This work was supported by PO1AI 080192-05 and U19 AI05726-08 to JMB and RA; and by an American Cancer Society postdoctoral fellowship PF-09-134-01-MPC to BAY.
Abbreviations: cBS, clonal bisulfite sequencing; ChIP-seq, chromatin immunoprecipitation-sequencing; DMR, differentially methylated regions; ENCODE, encyclopedia of DNA elements; FDR, false discovery rate; GO, Gene Ontology; H3K4me, histone H3 lysine 4 methyl; LCMV, lymphocytic choriomeningitis virus; meDIP, methyl DNA immune precipitation; PDE, putative distal enhancers; rpm, reads per million; TSS, transcription start sites;
References
- 1.Blattman JN, Antia R, Sourdive DJ, Wang X, Kaech SM, Murali-Krishna K, Altman JD, Ahmed R. Estimating the precursor frequency of naive antigen-specific CD8 T cells. The Journal of experimental medicine. 2002;195:657–664. doi: 10.1084/jem.20001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doering TA, Crawford A, Angelosanto JM, Paley MA, Ziegler CG, Wherry EJ. Network Analysis Reveals Centrally Connected Genes and Pathways Involved in CD8(+) T Cell Exhaustion versus Memory. Immunity. 2012;37:1130–1144. doi: 10.1016/j.immuni.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parish IA, Kaech SM. Diversity in CD8(+) T cell differentiation. Current opinion in immunology. 2009;21:291–297. doi: 10.1016/j.coi.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, Subramaniam S, Blattman JN, Barber DL, Ahmed R. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 5.Murali-Krishna K, Altman JD, Suresh M, Sourdive DJ, Zajac AJ, Miller JD, Slansky J, Ahmed R. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity. 1998;8:177–187. doi: 10.1016/s1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- 6.Kallies A, Xin A, Belz GT, Nutt SL. Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity. 2009;31:283–295. doi: 10.1016/j.immuni.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 7.Intlekofer AM, Banerjee A, Takemoto N, Gordon SM, Dejong CS, Shin H, Hunter CA, Wherry EJ, Lindsten T, Reiner SL. Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science. 2008;321:408–411. doi: 10.1126/science.1159806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 9.Moazed D. Mechanisms for the inheritance of chromatin states. Cell. 2011;146:510–518. doi: 10.1016/j.cell.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 11.De Carvalho DD, You JS, Jones PA. DNA methylation and cellular reprogramming. Trends Cell Biol. 2010;20:609–617. doi: 10.1016/j.tcb.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 13.Wade PA. Methyl CpG-binding proteins and transcriptional repression. Bioessays. 2001;23:1131–1137. doi: 10.1002/bies.10008. [DOI] [PubMed] [Google Scholar]
- 14.Bird AP, Wolffe AP. Methylation-induced repression--belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 15.Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, Cherry SR, Tsai JH, Tucker SM, Weaver WM, Kelso A, Jaenisch R, Wilson CB. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 16.Thomas RM, Gamper CJ, Ladle BH, Powell JD, Wells AD. De novo DNA methylation is required to restrict T helper lineage plasticity. The Journal of biological chemistry. 2012;287:22900–22909. doi: 10.1074/jbc.M111.312785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chappell C, Beard C, Altman J, Jaenisch R, Jacob J. DNA methylation by DNA methyltransferase 1 is critical for effector CD8 T cell expansion. Journal of immunology. 2006;176:4562–4572. doi: 10.4049/jimmunol.176.8.4562. [DOI] [PubMed] [Google Scholar]
- 18.Kersh EN. Impaired memory CD8 T cell development in the absence of methyl-CpG-binding domain protein 2. Journal of immunology. 2006;177:3821–3826. doi: 10.4049/jimmunol.177.6.3821. [DOI] [PubMed] [Google Scholar]
- 19.Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, Lee H, Aryee MJ, Irizarry RA, Kim K, Rossi DJ, Inlay MA, Serwold T, Karsunky H, Ho L, Daley GQ, Weissman IL, Feinberg AP. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shearstone JR, Pop R, Bock C, Boyle P, Meissner A, Socolovsky M. Global DNA demethylation during mouse erythropoiesis in vivo. Science. 2011;334:799–802. doi: 10.1126/science.1207306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidl C, Klug M, Boeld TJ, Andreesen R, Hoffmann P, Edinger M, Rehli M. Lineage-specific DNA methylation in T cells correlates with histone methylation and enhancer activity. Genome research. 2009;19:1165–1174. doi: 10.1101/gr.091470.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohkura N, Hamaguchi M, Morikawa H, Sugimura K, Tanaka A, Ito Y, Osaki M, Tanaka Y, Yamashita R, Nakano N, Huehn J, Fehling HJ, Sparwasser T, Nakai K, Sakaguchi S. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity. 2012;37:785–799. doi: 10.1016/j.immuni.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 23.Youngblood B, Oestreich KJ, Ha SJ, Duraiswamy J, Akondy RS, West EE, Wei Z, Lu P, Austin JW, Riley JL, Boss JM, Ahmed R. Chronic virus infection enforces demethylation of the locus that encodes PD-1 in antigen-specific CD8(+) T cells. Immunity. 2011;35:400–412. doi: 10.1016/j.immuni.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kersh EN, Fitzpatrick DR, Murali-Krishna K, Shires J, Speck SH, Boss JM, Ahmed R. Rapid demethylation of the IFN-gamma gene occurs in memory but not naive CD8 T cells. Journal of immunology. 2006;176:4083–4093. doi: 10.4049/jimmunol.176.7.4083. [DOI] [PubMed] [Google Scholar]
- 25.Oestreich KJ, Yoon H, Ahmed R, Boss JM. NFATc1 regulates PD-1 expression upon T cell activation. Journal of immunology. 2008;181:4832–4839. doi: 10.4049/jimmunol.181.7.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schoenborn JR, Dorschner MO, Sekimata M, Santer DM, Shnyreva M, Fitzpatrick DR, Stamatoyannopoulos JA, Wilson CB. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nature immunology. 2007;8:732–742. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, Tiwari VK, Schubeler D. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 28.Pircher H, Burki K, Lang R, Hengartner H, Zinkernagel RM. Tolerance induction in double specific T-cell receptor transgenic mice varies with antigen. Nature. 1989;342:559–561. doi: 10.1038/342559a0. [DOI] [PubMed] [Google Scholar]
- 29.Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y, Olshen A, Ballinger T, Zhou X, Forsberg KJ, Gu J, Echipare L, O’Geen H, Lister R, Pelizzola M, Xi Y, Epstein CB, Bernstein BE, Hawkins RD, Ren B, Chung WY, Gu H, Bock C, Gnirke A, Zhang MQ, Haussler D, Ecker JR, Li W, Farnham PJ, Waterland RA, Meissner A, Marra MA, Hirst M, Milosavljevic A, Costello JF. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nature biotechnology. 2010;28:1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nature genetics. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 31.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chavez L, Jozefczuk J, Grimm C, Dietrich J, Timmermann B, Lehrach H, Herwig R, Adjaye J. Computational analysis of genome-wide DNA methylation during the differentiation of human embryonic stem cells along the endodermal lineage. Genome research. 2010;20:1441–1450. doi: 10.1101/gr.110114.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Consortium EP. A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS biology. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quinlan AR, I, Hall M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, Ahmed R. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol. 2003;77:4911–4927. doi: 10.1128/JVI.77.8.4911-4927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Down TA, V, Rakyan K, Turner DJ, Flicek P, Li H, Kulesha E, Graf S, Johnson N, Herrero J, Tomazou EM, Thorne NP, Backdahl L, Herberth M, Howe KL, Jackson DK, Miretti MM, Marioni JC, Birney E, Hubbard TJ, Durbin R, Tavare S, Beck S. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nature biotechnology. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nature genetics. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 40.Hughes T, Webb R, Fei Y, Wren JD, Sawalha AH. DNA methylome in human CD4+ T cells identifies transcriptionally repressive and non-repressive methylation peaks. Genes and immunity. 2010;11:554–560. doi: 10.1038/gene.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Illingworth RS, Gruenewald-Schneider U, Webb S, Kerr AR, James KD, Turner DJ, Smith C, Harrison DJ, Andrews R, Bird AP. Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS genetics. 2010:6. doi: 10.1371/journal.pgen.1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hatcher VB, Oberman MS, Lazarus GS, Grayzel AI. A cytotoxic proteinase isolated from human lymphocytes. Journal of immunology. 1978;120:665–670. [PubMed] [Google Scholar]
- 43.Miaw SC, Choi A, Yu E, Kishikawa H, Ho IC. ROG, repressor of GATA, regulates the expression of cytokine genes. Immunity. 2000;12:323–333. doi: 10.1016/s1074-7613(00)80185-5. [DOI] [PubMed] [Google Scholar]
- 44.Yoon HS, Scharer CD, Majumder P, Davis CW, Butler R, Zinzow-Kramer W, Skountzou I, Koutsonanos DG, Ahmed R, Boss JM. ZBTB32 is an early repressor of the CIITA and MHC class II gene expression during B cell differentiation to plasma cells. Journal of immunology. 2012;189:2393–2403. doi: 10.4049/jimmunol.1103371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Acosta JC, O’Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, Fumagalli M, Da Costa M, Brown C, Popov N, Takatsu Y, Melamed J, d’Adda di Fagagna F, Bernard D, Hernando E, Gil J. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–1018. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 46.Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, van de Wetering M, Oosterwegel M, Wilson A, MacDonald HR, Clevers H. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature. 1995;374:70–74. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]
- 47.Willinger T, Freeman T, Herbert M, Hasegawa H, McMichael AJ, Callan MF. Human naive CD8 T cells down-regulate expression of the WNT pathway transcription factors lymphoid enhancer binding factor 1 and transcription factor 7 (T cell factor-1) following antigen encounter in vitro and in vivo. Journal of immunology. 2006;176:1439–1446. doi: 10.4049/jimmunol.176.3.1439. [DOI] [PubMed] [Google Scholar]
- 48.Yu S, Zhou X, Steinke FC, Liu C, Chen SC, Zagorodna O, Jing X, Yokota Y, Meyerholz DK, Mullighan CG, Knudson CM, Zhao DM, Xue HH. The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity. 2012;37:813–826. doi: 10.1016/j.immuni.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quigley M, Pereyra F, Nilsson B, Porichis F, Fonseca C, Eichbaum Q, Julg B, Jesneck JL, Brosnahan K, Imam S, Russell K, Toth I, Piechocka-Trocha A, Dolfi D, Angelosanto J, Crawford A, Shin H, Kwon DS, Zupkosky J, Francisco L, Freeman GJ, Wherry EJ, Kaufmann DE, Walker BD, Ebert B, Haining WN. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nature medicine. 2010;16:1147–1151. doi: 10.1038/nm.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuroda S, Yamazaki M, Abe M, Sakimura K, Takayanagi H, Iwai Y. Basic leucine zipper transcription factor, ATF-like (BATF) regulates epigenetically and energetically effector CD8 T-cell differentiation via Sirt1 expression. Proc Natl Acad Sci U S A. 2011;108:14885–14889. doi: 10.1073/pnas.1105133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lecomte V, Meugnier E, Euthine V, Durand C, Freyssenet D, Nemoz G, Rome S, Vidal H, Lefai E. A new role for sterol regulatory element binding protein 1 transcription factors in the regulation of muscle mass and muscle cell differentiation. Molecular and cellular biology. 2010;30:1182–1198. doi: 10.1128/MCB.00690-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, Noshiro M, Kato Y, Honma K. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- 53.Shen M, Yoshida E, Yan W, Kawamoto T, Suardita K, Koyano Y, Fujimoto K, Noshiro M, Kato Y. Basic helix-loop-helix protein DEC1 promotes chondrocyte differentiation at the early and terminal stages. The Journal of biological chemistry. 2002;277:50112–50120. doi: 10.1074/jbc.M206771200. [DOI] [PubMed] [Google Scholar]
- 54.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, Khatun J, Lajoie BR, Landt SG, Lee BK, Pauli F, Rosenbloom KR, Sabo P, Safi A, Sanyal A, Shoresh N, Simon JM, Song L, Trinklein ND, Altshuler RC, Birney E, Brown JB, Cheng C, Djebali S, Dong X, Dunham I, Ernst J, Furey TS, Gerstein M, Giardine B, Greven M, Hardison RC, Harris RS, Herrero J, Hoffman MM, Iyer S, Kelllis M, Khatun J, Kheradpour P, Kundaje A, Lassman T, Li Q, Lin X, Marinov GK, Merkel A, Mortazavi A, Parker SC, Reddy TE, Rozowsky J, Schlesinger F, Thurman RE, Wang J, Ward LD, Whitfield TW, Wilder SP, Wu W, Xi HS, Yip KY, Zhuang J, Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M, Pazin MJ, Lowdon RF, Dillon LA, Adams LB, Kelly CJ, Zhang J, Wexler JR, Green ED, Good PJ, Feingold EA, Bernstein BE, Birney E, Crawford GE, Dekker J, Elinitski L, Farnham PJ, Gerstein M, Giddings MC, Gingeras TR, Green ED, Guigo R, Hardison RC, Hubbard TJ, Kellis M, Kent WJ, Lieb JD, Margulies EH, Myers RM, Snyder M, Starnatoyannopoulos JA, Tennebaum SA, Weng Z, White KP, Wold B, Khatun J, Yu Y, Wrobel J, Risk BA, Gunawardena HP, Kuiper HC, Maier CW, Xie L, Chen X, Giddings MC, Bernstein BE, Epstein CB, Shoresh N, Ernst J, Kheradpour P, Mikkelsen TS, Gillespie S, Goren A, Ram O, Zhang X, Wang L, Issner R, Coyne MJ, Durham T, Ku M, Truong T, Ward LD, Altshuler RC, Eaton ML, Kellis M, Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Batut P, Bell I, Bell K, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena HP, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Li G, Luo OJ, Park E, Preall JB, Presaud K, Ribeca P, Risk BA, Robyr D, Ruan X, Sammeth M, Sandu KS, Schaeffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Hayashizaki Y, Harrow J, Gerstein M, Hubbard TJ, Reymond A, Antonarakis SE, Hannon GJ, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR, Rosenbloom KR, Sloan CA, Learned K, Malladi VS, Wong MC, Barber GP, Cline MS, Dreszer TR, Heitner SG, Karolchik D, Kent WJ, Kirkup VM, Meyer LR, Long JC, Maddren M, Raney BJ, Furey TS, Song L, Grasfeder LL, Giresi PG, Lee BK, Battenhouse A, Sheffield NC, Simon JM, Showers KA, Safi A, London D, Bhinge AA, Shestak C, Schaner MR, Kim SK, Zhang ZZ, Mieczkowski PA, Mieczkowska JO, Liu Z, McDaniell RM, Ni Y, Rashid NU, Kim MJ, Adar S, Zhang Z, Wang T, Winter D, Keefe D, Birney E, Iyer VR, Lieb JD, Crawford GE, Li G, Sandhu KS, Zheng M, Wang P, Luo OJ, Shahab A, Fullwood MJ, Ruan X, Ruan Y, Myers RM, Pauli F, Williams BA, Gertz J, Marinov GK, Reddy TE, Vielmetter J, Partridge EC, Trout D, Varley KE, Gasper C, Bansal A, Pepke S, Jain P, Amrhein H, Bowling KM, Anaya M, Cross MK, King B, Muratet MA, Antoshechkin I, Newberry KM, McCue K, Nesmith AS, Fisher-Aylor KI, Pusey B, DeSalvo G, Parker SL, Balasubramanian S, Davis NS, Meadows SK, Eggleston T, Gunter C, Newberry JS, Levy SE, Absher DM, Mortazavi A, Wong WH, Wold B, Blow MJ, Visel A, Pennachio LA, Elnitski L, Margulies EH, Parker SC, Petrykowska HM, Abyzov A, Aken B, Barrell D, Barson G, Berry A, Bignell A, Boychenko V, Bussotti G, Chrast J, Davidson C, Derrien T, Despacio-Reyes G, Diekhans M, Ezkurdia I, Frankish A, Gilbert J, Gonzalez JM, Griffiths E, Harte R, Hendrix DA, Howald C, Hunt T, Jungreis I, Kay M, Khurana E, Kokocinski F, Leng J, Lin MF, Loveland J, Lu Z, Manthravadi D, Mariotti M, Mudge J, Mukherjee G, Notredame C, Pei B, Rodriguez JM, Saunders G, Sboner A, Searle S, Sisu C, Snow C, Steward C, Tanzer A, Tapanan E, Tress ML, van Baren MJ, Walters N, Washieti S, Wilming L, Zadissa A, Zhengdong Z, Brent M, Haussler D, Kellis M, Valencia A, Gerstein M, Raymond A, Guigo R, Harrow J, Hubbard TJ, Landt SG, Frietze S, Abyzov A, Addleman N, Alexander RP, Auerbach RK, Balasubramanian S, Bettinger K, Bhardwaj N, Boyle AP, Cao AR, Cayting P, Charos A, Cheng Y, Cheng C, Eastman C, Euskirchen G, Fleming JD, Grubert F, Habegger L, Hariharan M, Harmanci A, Iyenger S, Jin VX, Karczewski KJ, Kasowski M, Lacroute P, Lam H, Larnarre-Vincent N, Leng J, Lian J, Lindahl-Allen M, Min R, Miotto B, Monahan H, Moqtaderi Z, Mu XJ, O’Geen H, Ouyang Z, Patacsil D, Pei B, Raha D, Ramirez L, Reed B, Rozowsky J, Sboner A, Shi M, Sisu C, Slifer T, Witt H, Wu L, Xu X, Yan KK, Yang X, Yip KY, Zhang Z, Struhl K, Weissman SM, Gerstein M, Farnham PJ, Snyder M, Tenebaum SA, Penalva LO, Doyle F, Karmakar S, Landt SG, Bhanvadia RR, Choudhury A, Domanus M, Ma L, Moran J, Patacsil D, Slifer T, Victorsen A, Yang X, Snyder M, White KP, Auer T, Centarin L, Eichenlaub M, Gruhl F, Heerman S, Hoeckendorf B, Inoue D, Kellner T, Kirchmaier S, Mueller C, Reinhardt R, Schertel L, Schneider S, Sinn R, Wittbrodt B, Wittbrodt J, Weng Z, Whitfield TW, Wang J, Collins PJ, Aldred SF, Trinklein ND, Partridge EC, Myers RM, Dekker J, Jain G, Lajoie BR, Sanyal A, Balasundaram G, Bates DL, Byron R, Canfield TK, Diegel MJ, Dunn D, Ebersol AK, Ebersol AK, Frum T, Garg K, Gist E, Hansen RS, Boatman L, Haugen E, Humbert R, Jain G, Johnson AK, Johnson EM, Kutyavin TM, Lajoie BR, Lee K, Lotakis D, Maurano MT, Neph SJ, Neri FV, Nguyen ED, Qu H, Reynolds AP, Roach V, Rynes E, Sabo P, Sanchez ME, Sandstrom RS, Sanyal A, Shafer AO, Stergachis AB, Thomas S, Thurman RE, Vernot B, Vierstra J, Vong S, Wang H, Weaver MA, Yan Y, Zhang M, Akey JA, Bender M, Dorschner MO, Groudine M, MacCoss MJ, Navas P, Stamatoyannopoulos G, Kaul R, Dekker J, Stamatoyannopoulos JA, Dunham I, Beal K, Brazma A, Flicek P, Herrero J, Johnson N, Keefe D, Lukk M, Luscombe NM, Sobral D, Vaquerizas JM, Wilder SP, Batzoglou S, Sidow A, Hussami N, Kyriazopoulou-Panagiotopoulou S, Libbrecht MW, Schaub MA, Kundaje A, Hardison RC, Miller W, Giardine B, Harris RS, Wu W, Bickel PJ, Banfai B, Boley NP, Brown JB, Huang H, Li Q, Li JJ, Noble WS, Bilmes JA, Buske OJ, Hoffman MM, Sahu AO, Kharchenko PV, Park PJ, Baker D, Taylor J, Weng Z, Iyer S, Dong X, Greven M, Lin X, Wang J, Xi HS, Zhuang J, Gerstein M, Alexander RP, Balasubramanian S, Cheng C, Harmanci A, Lochovsky L, Min R, Mu XJ, Rozowsky J, Yan KK, Yip KY, Birney E EN CODEProject Consortium . An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 56.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, Cheng X, Bestor TH. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, Bollen M, Esteller M, Di Croce L, de Launoit Y, Fuks F. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 59.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F, Lee K, Canfield T, Weaver M, Sandstrom R, Thurman RE, Kaul R, Myers RM, Stamatoyannopoulos JA. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome research. 2012;22:1680–1688. doi: 10.1101/gr.136101.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim HP, Leonard WJ. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. The Journal of experimental medicine. 2007;204:1543–1551. doi: 10.1084/jem.20070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Polansky JK, Schreiber L, Thelemann C, Ludwig L, Kruger M, Baumgrass R, Cording S, Floess S, Hamann A, Huehn J. Methylation matters: binding of Ets-1 to the demethylated Foxp3 gene contributes to the stabilization of Foxp3 expression in regulatory T cells. J Mol Med (Berl) 2010;88:1029–1040. doi: 10.1007/s00109-010-0642-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prendergast GC, Ziff EB. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991;251:186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- 65.Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, Munzel M, Wagner M, Muller M, Khan F, Eberl HC, Mensinga A, Brinkman AB, Lephikov K, Muller U, Walter J, Boelens R, van Ingen H, Leonhardt H, Carell T, Vermeulen M. Dynamic Readers for 5-(Hydroxy)Methylcytosine and Its Oxidized Derivatives. Cell. 2013 doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed]
- 66.Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 67.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK, Clarke ND, Wei CL, Ng HH. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 68.Calnan BJ, Szychowski S, Chan FK, Cado D, Winoto A. A role for the orphan steroid receptor Nur77 in apoptosis accompanying antigen-induced negative selection. Immunity. 1995;3:273–282. doi: 10.1016/1074-7613(95)90113-2. [DOI] [PubMed] [Google Scholar]
- 69.Corre S, Galibert MD. Upstream stimulating factors: highly versatile stress-responsive transcription factors. Pigment Cell Res. 2005;18:337–348. doi: 10.1111/j.1600-0749.2005.00262.x. [DOI] [PubMed] [Google Scholar]
- 70.Thompson MR, Xu D, Williams BR. ATF3 transcription factor and its emerging roles in immunity and cancer. J Mol Med (Berl) 2009;87:1053–1060. doi: 10.1007/s00109-009-0520-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Douglas NC, Jacobs H, Bothwell AL, Hayday AC. Defining the specific physiological requirements for c-Myc in T cell development. Nature immunology. 2001;2:307–315. doi: 10.1038/86308. [DOI] [PubMed] [Google Scholar]
- 73.Jain J, McCaffrey PG, Valge-Archer VE, Rao A. Nuclear factor of activated T cells contains Fos and Jun. Nature. 1992;356:801–804. doi: 10.1038/356801a0. [DOI] [PubMed] [Google Scholar]
- 74.Epsztejn-Litman S, Feldman N, Abu-Remaileh M, Shufaro Y, Gerson A, Ueda J, Deplus R, Fuks F, Shinkai Y, Cedar H, Bergman Y. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nature structural & molecular biology. 2008;15:1176–1183. doi: 10.1038/nsmb.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Velasco G, Hube F, Rollin J, Neuillet D, Philippe C, Bouzinba-Segard H, Galvani A, Viegas-Pequignot E, Francastel C. Dnmt3b recruitment through E2F6 transcriptional repressor mediates germ-line gene silencing in murine somatic tissues. Proc Natl Acad Sci U S A. 2010;107:9281–9286. doi: 10.1073/pnas.1000473107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Makar KW, Perez-Melgosa M, Shnyreva M, Weaver WM, Fitzpatrick DR, Wilson CB. Active recruitment of DNA methyltransferases regulates interleukin 4 in thymocytes and T cells. Nature immunology. 2003;4:1183–1190. doi: 10.1038/ni1004. [DOI] [PubMed] [Google Scholar]
- 78.Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nature immunology. 2003;4:235–240. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- 79.Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, Abramowitz LK, Bartolomei MS, Rambow F, Bassi MR, Bruno T, Fanciulli M, Renner C, Klein-Szanto AJ, Matsumoto Y, Kobi D, Davidson I, Alberti C, Larue L, Bellacosa A. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Costa Y, Ding J, Theunissen TW, Faiola F, Hore TA, Shliaha PV, Fidalgo M, Saunders A, Lawrence M, Dietmann S, Das S, Levasseur DN, Li Z, Xu M, Reik W, Silva JC, Wang J. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature. 2013;495:370–374. doi: 10.1038/nature11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.