Significance
Maize was domesticated from teosinte in Southern Mexico roughly 9,000 years ago. Maize originally was sensitive to photoperiod and required short-day conditions to flower. Thus, the reduced sensitivity to photoperiod is prerequisite for maize spread to long-day temperate regions. A gene encoding a CCT domain-containing protein, ZmCCT, was found by many researchers to modulate photoperiod sensitivity. The current study shows that insertion of a CACTA-like transposon into the ZmCCT promoter can suppress the ZmCCT expression remarkably and thus attenuates maize sensitivity under long-day conditions. The transposable element (TE) insertion event occurred in a tropical maize plant and has been selected for and accumulated as maize adapted to vast long-day environments. This selection leaves behind a TE-related linkage disequilibrium block with the very-low-nucleotide variations.
Abstract
The postdomestication adaptation of maize to longer days required reduced photoperiod sensitivity to optimize flowering time. We performed a genome-wide association study and confirmed that ZmCCT, encoding a CCT domain-containing protein, is associated with the photoperiod response. In early-flowering maize we detected a CACTA-like transposable element (TE) within the ZmCCT promoter that dramatically reduced flowering time. TE insertion likely occurred after domestication and was selected as maize adapted to temperate zones. This process resulted in a strong selective sweep within the TE-related block of linkage disequilibrium. Functional validations indicated that the TE represses ZmCCT expression to reduce photoperiod sensitivity, thus accelerating maize spread to long-day environments.
Maize (Zea mays L.) was domesticated in Southern Mexico roughly 9,000 y ago from Balsas teosinte (Zea mays ssp. parviglumis) (1), which requires short-day conditions to flower (2). Therefore the spread of maize from tropical to temperate regions required the postdomestication adaptation of maize to longer days (1, 3, 4). As such, temperate maize is largely day-length insensitive, whereas tropical maize lines are generally sensitive to longer day lengths.
To modulate the timing of flowering, plants integrate signals from the environment and from endogenous regulatory pathways (5). Most genes known to regulate maize flowering (6–12) are part of the autonomous pathway, such as id1 (6, 7), ZCN8 (8), dlf1 (9), zfl1 (10), conz1 (11), and Vgt1 (12). Flowering time in maize is extremely variable (ranging from 35–120 d) (13) and is controlled primarily by a large number of quantitative trait loci (QTLs), each with a small effect (14). Relatively few of these flowering-time QTLs affect the photoperiod response, although ZmCCT, encoding a CCT domain-containing protein, appears to be the most important locus in these contexts (15–18). As such, molecular details concerning the photoperiodic control of maize flowering remain unclear.
Transposable elements (TEs) played a key role in adaptive plant evolution and phenotypic variation by altering gene expression and function (19–23). In fact, TEs often served as targets of selection during evolution (24). Insertion of the Rider retrotransposon into the tomato genome increased expression of the gene SUN, which led to an elongated fruit shape (25). Similarly, insertion of a miniature inverted-repeat TE (MITE) into Vgt1, which is a cis-regulatory element located ∼70 kb upstream of the flowering-time repressor ZmRap2.7, is tightly associated with flowering-time variation in maize (12). Finally, insertion of a Hopscotch retrotransposon upstream of the maize-domestication gene tb1 increased apical dominance in maize (26, 27).
Here we performed a genome-wide association study (GWAS) using a diverse panel of maize lines (28, 29) to identify genetic variants near ZmCCT that associate with flowering time. Using an overlapping PCR approach, we detected a CACTA-like TE within the ZmCCT regulatory region. Genetic effects of this TE on flowering time were investigated by ZmCCT-based association mapping and biparental linkage analysis. The CACTA-like TE appeared to be a causative factor in reducing photoperiod sensitivity under long-day conditions and was the target of a strong selective sweep during the postdomestication spread of maize. Functional validations demonstrate that ZmCCT is involved in the photoperiod response and that the CACTA-like TE within ZmCCT represses gene expression, rendering maize insensitive to long days.
Results
The ZmCCT Locus Is Associated with Photoperiod Sensitivity.
We grew the CAM508 panel of maize lines (Table S1) in eight environments at seven different latitudes (SI Appendix, Table S2). Substantial variation in flowering time was observed among these different lines (SI Appendix, Table S3). Days to anthesis (DTA) or silking (DTS) differed by >20 d (180 growing degree days; GDD) under both long- and short-day environments, whereas anthesis photoperiod response (APR) or silking photoperiod response (SPR) differed by >265 GDD. Repeatability was estimated to be 88.9–95.4% for DTA, 86.5–93.2% for DTS, 75.1% for APR, and 72.8% for SPR. Pairwise correlation coefficients of these four flowering-time traits ranged from 0.50 to 0.94 (P < 0.0001) (SI Appendix, Table S4).
We performed a GWAS using 368 inbred lines of maize (a subset of the CAM508 panel) to identify loci associated with flowering time. For each line, 557,955 polymorphic sites with a minor allele frequency (MAF) ≥0.05 and a missing rate <25% were used for the GWAS. This analysis identified 48 polymorphic sites, 41 SNPs, and seven insertions or deletions (InDels) that were significantly associated with flowering time (P ≤ 1.8 × 10−6) and an additional 20 SNPs and one InDel with marginal significance (1.8 × 10−6 < P ≤ 1.8 × 10−5) (SI Appendix, Table S5 and Figs. S1 and S2). Of the 48 highly significant sites, 42 were located within the ZmCCT promoter region. This finding is consistent with previous reports that ZmCCT is a major determinant of maize photoperiod sensitivity (15, 17, 18). The 21 less significant sites were scattered across 16 loci besides the ZmCCT locus. Eleven of these loci were significantly associated with photoperiod response traits, APR/SPR, and seemed to influence maize flowering time through photoperiod pathways, whereas the remaining five were significantly associated only with flowering time, DTA/DTS, and seemed to function through other endogenous pathways or environmental signals.
Discovery of a CACTA-like TE in the ZmCCT Promoter.
Our GWAS, together with previous findings (17, 18), indicated that sequences within the ZmCCT regulatory region modulate photoperiod sensitivity in maize. Because of dramatic sequence divergence, however, the longest promoter region that can be obtained consistently for GWAS contains only ∼1.8 kb of sequence upstream of ZmCCT. Therefore it is critical to characterize the genomic architecture of the entire ZmCCT regulatory region. We isolated genomic DNA from both early- and late-flowering inbred lines of maize and subjected them to PCR amplification using overlapping primer pairs (SI Appendix, Table S6 and Fig. S3A). Two PCR products, 5UCCT2 and 5UCCT3, were obtained from most early-flowering lines, such as B73 and Mo17, but not from late-flowering lines, such as 1145 and Qi319. In contrast, 5UCCT1 and 5UCCT4 were amplified from all inbred lines. This result suggests that a presence/absence variant (PAV) may reside within the ZmCCT regulatory region. We next used the primer pair TED, which covers the entire 5UCCT2 and 5UCCT3 region, to amplify the putative PAV, resulting in a 372-bp fragment from 1145 and Qi319, but no product from B73 and Mo17. We assumed that sequences associated with B73 and Mo17 were too long for PCR amplification. Overlapping PCR fragments 5UCCT1, 5UCCT2, 5UCCT3, and 5UCCT4 were assembled to construct a 9,491-bp segment from B73. Similarly, overlapping fragments 5UCCT1, TED, and 5UCCT4 were assembled to construct a 4,487-bp segment from 1145. Alignment of these two sequences revealed a 5,122-bp CACTA-like TE in B73 (SI Appendix, Fig. S3 B and C). This element was 2,543 bp upstream of the ORF (SI Appendix, Fig. S4) and had a 3-bp (5′-GCT-3′) target-site duplication and 13-bp (5′-CACTACAGGAAAA-3′) terminal inverted repeats SI Appendix, Fig. S3 B and C). Based on the TE and flanking sequences, we designed two additional TE-related markers, TELB and TERB (SI Appendix, Table S6). These markers, when coupled with TED, detected the presence/absence of this CACTA-like TE, which we designated the “TE-related PAV.”
TE-Related PAV Is Associated with Photoperiod Sensitivity.
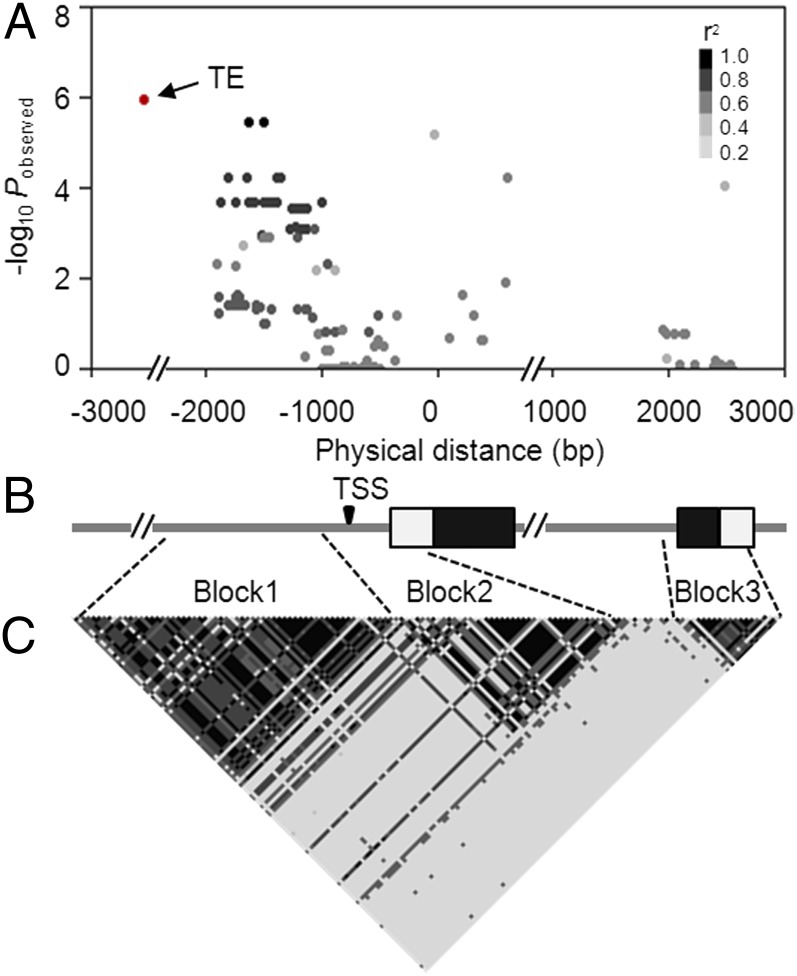
After identifying the TE-related PAV, we resequenced the ZmCCT genomic locus within a diverse group of temperate (107) and tropical (73) lines of maize (Table S1). Sequences from the TE-related PAV to the 3′UTR of ZmCCT were analyzed. We discovered 136 variants with MAF ≥0.05 across the ZmCCT locus, with only eight in the coding region. Of these 136 variants, 29 were significantly associated with photoperiod sensitivity (P ≤ 2.8 × 10−4, n = 180) (Fig. 1A and SI Appendix, Table S7). The TE-related PAV showed the strongest association with flowering-time traits (P = 1.1 × 10−6 for APR, P = 7.5 × 10−6 for SPR, P = 4.1 × 10−6 for DTA, and P = 1.8 × 10−5 for DTS, n = 180). In addition, two SNPs at −1,636 bp and −1,498 bp were in almost complete linkage disequilibrium (LD) with the TE-related PAV (r2 = 0.97). These SNPs were identified previously to associate with photoperiod sensitivity (18). LD analysis revealed three clear blocks of LD within the resequenced fragments of ZmCCT (Fig. 1C). The TE-related PAV was in strong LD (r2 > 0.6) with all significant polymorphic sites in the first block of LD (bp −1,904 to −1,003).
Fig. 1.
ZmCCT-based association mapping and LD analysis of 180 diverse maize lines. (A) Associations between polymorphic sites within the ZmCCT locus (MAF ≥0.05) and APR. Each dot represents a polymorphic site, and the color reflects the level of LD (r2) with the TE (except for the TE, which is red). (B) Structure of the ZmCCT locus. Black rectangles represent exons, and white rectangles represent UTRs. The transcription start site (TSS) is indicated. (C) The pattern of LD for all polymorphic sites within the ZmCCT locus. All polymorphic sites (MAF ≥0.05) excluding the TE were used.
We also analyzed temperate and tropical germplasms within the CAM508 panel to detect genetic effects of the TE-related PAV on flowering time in multiple environments (SI Appendix, Figs. S5 and S6). For each flowering-time trait, the difference between maize lines with and without the TE insertion was larger in tropical germplasms than in temperate germplasms. Moreover, this difference increased gradually as maize was grown at higher latitudes. When plants were grown under long-day conditions in Beijing, for example, differences between maize lines with and without the TE were 87.5 GDD for APR, 111.2 GDD for SPR, 5.3 d (88.3 GDD) for DTA, and 6.3 d (104.6 GDD) for DTS for temperate germplasms. These same differences were 182.5 GDD, 209.3 GDD, 12.1 d (192.7 GDD), and 13.9 d (218.8 GDD) for tropical germplasms.
TE-Related Haplotypes Displayed Variable Photoperiod Sensitivities.
In addition to single-variant investigation, we used the CAM508 panel to test associations between TE-related haplotypes and flowering time. Among 62 variants (MAF ≥0.05) within the ZmCCT regulatory region, we identified 13 variants that captured all 15 haplotypes (SI Appendix, Table S8). Seven of these haplotypes had a sample size >10 lines (frequencies >0.01) (Fig. 2 and SI Appendix, Tables S8 and S9), of which only two (Hap1 and Hap2) had the TE insertion. These seven haplotypes had heterogeneous effects on photoperiod responses in long-day environments. Hap2 showed the weakest photoperiod response and had the earliest flowering time, followed by Hap1 (n = 300), which included the majority of tested lines. For haplotypes that lacked the TE insertion, Hap4 exhibited the weakest photoperiod responses, which were similar to those of Hap1, whereas Hap3, Hap5, Hap6, and Hap7 showed strong photoperiod responses. When TE-positive haplotypes (Hap1 and Hap2) were compared with TE-negative haplotypes (Hap3–7), a highly significant difference in APR was observed (P = 8.3 × 10−15, n = 439) (Fig. 2A). Similar results were obtained for SPR, DTA, and DTS (SI Appendix, Table S9). Analysis of the TE-related PAV and other variants suggested relatedness between the seven haplotypes (Fig. 2B). Hap1 evolved from Hap3 by a TE insertion at 2,543 bp upstream of ZmCCT. This single TE insertion reduced APR by 35.7 GDD, SPR by 41.2 GDD, DTA by 2.8 d (42.9 GDD), and DTS by 2.7 d (43.2 GDD). Hap1 turned into Hap2 by an A-to-T transversion at −1,722 bp, which resulted in mild reductions in APR (14.3 GDD), SPR (16.9 GDD), DTA (0.2 d, 4.3 GDD), and DTS (0.7 d, 12.1 GDD) (SI Appendix, Table S9). Hap3 developed from Hap4, and the Hap4 progenitor may have been Hap7, based on the eight low-frequency (< 0.01) haplotypes (SI Appendix, Table S8). Hap5, Hap6, and Hap7 are closely related with similar genetic distances between them.
Fig. 2.
Genetic effects and relatedness of ZmCCT-promoter haplotypes. (A) Estimated effects of haplotypes with MAF ≥0.01 on APR in the CAM508 panel after correcting for population structure and kinship. When a string of polymorphic sites are in complete LD, only one is shown. Insensitive alleles are in bold text. #The number of lines for each haplotype (N) and the P values are indicated. S1–S7 correspond to sites at −1,983, −1,884, −1,875, −1,722, −1,518, −1,341, and −1,206 bp, respectively. (B) Proposed relatedness of the seven haplotypes. Each circle represents a haplotype, and the size of the circle is proportional to the number of lines within the haplotype: smallest circles <20, small circles 20–30, large circles 30–300, and largest circles ≥300 lines. Green and yellow represent temperate and tropical germplasms, respectively.
To determine the effect of different haplotypes on photoperiod sensitivity, we developed four F2 populations from crosses between sensitive (Hap6, Qi319, and Tian77) and insensitive (Hap1, Mo17, and Zheng58) lines. In these F2 populations, photoperiod sensitivity tended to be higher for Hap6 individuals when grown at higher latitudes (SI Appendix, Figs. S7 and S8). Compared with homozygous Hap1, Hap6 delayed DTA by 2–6 d and DTS by 1–7 d at high latitudes (Jilin and Beijing). This effect was greatly reduced at low latitudes (Hubei and Hainan). Furthermore, Hap1 and Hap6 also show significant phenotypic variation in other morphological and yield-related traits, including ear height and node number, in Jilin and Beijing (SI Appendix, Fig. S8).
Evolution of the ZmCCT Locus in Response to Photoperiod Changes.
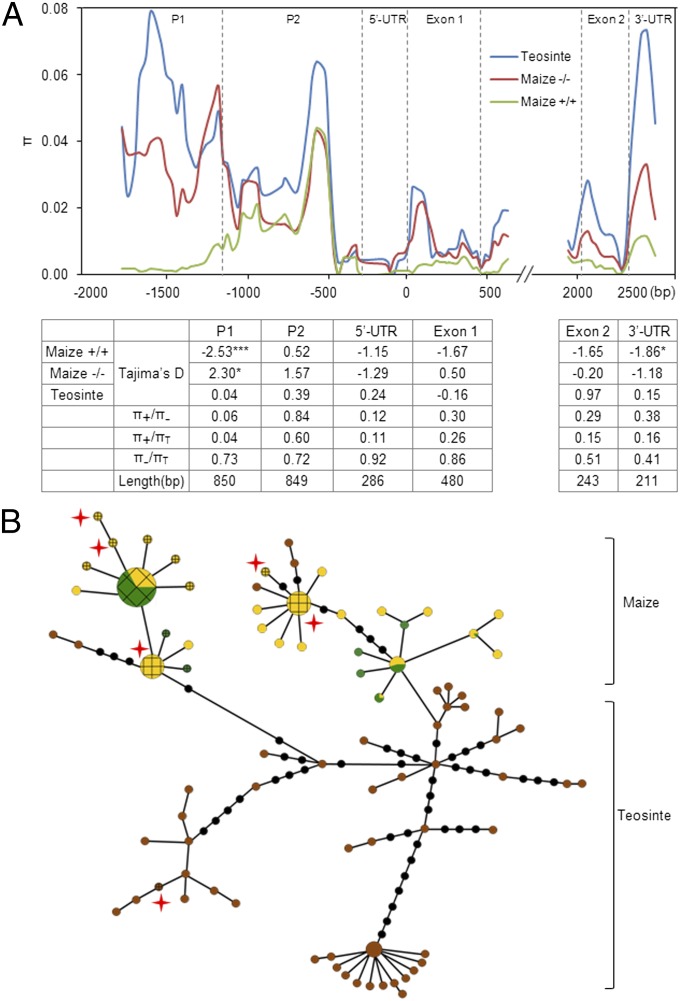
We next sequenced six regions of the ZmCCT gene from 32 teosinte entries. Together with sequences from 143 lines of maize, we analyzed evolutionary changes at the ZmCCT locus (Fig. 3A). We found no evidence of a selective sweep across ZmCCT in maize lines that lacked the TE insertion, because the ratio of diversity in maize lacking the TE insertion (π−, n = 37) to teosinte (πT, n = 32) ranged from 0.41 to 0.92 within the six regions that were sequenced. Maize lines with the TE insertion (π+, n = 106), however, exhibited dramatically reduced diversity in the P1 region (π+/πT = 0.04, π+/π− = 0.06). P1 extended from −1.98 kb to −1.14 kb and corresponded to the LD block 1. Significant reductions in diversity were not detected in the other five regions. It seems that the TE insertion is at the root of this selective sweep, which extended only to a region ∼1.14 kb upstream of the ZmCCT ORF. The distal border of this sweep (i.e., upstream of the TE insertion) remains unclear. Low levels of diversity associated with both maize and teosinte coding regions suggest that the ZmCCT region is evolving under functional constraint.
Fig. 3.
Sequence diversity of the ZmCCT locus between maize and teosinte. (A) Nucleotide diversity revealed by comparisons between 143 maize lines and 32 teosinte entries across the ZmCCT locus. Nucleotide diversity (π) for teosinte (blue), TE-positive maize (green), and TE-negative maize (red) was calculated using a 100-bp sliding window with a 25-bp step. Results from the Tajima’s D test and three π ratios, TE-positive maize (π+) to TE-negative maize (π−), π+ to teosinte (πT), and π− to πT, are shown. *P < 0.05; ***P < 0.0001. (B) A minimum-spanning tree for the ZmCCT promoter region including 481 diverse maize sequences and 93 diverse teosinte sequences. Each haplotype group is represented by a circle, and circle sizes represent the number of lines within the haplotype, as in Fig. 2. Green, yellow, and brown represent temperate, tropical, and teosinte germplasms, respectively. The circles with grids indicate haplotypes that have TE insertions; the red stars near the haplotypes show that their TE genotypes are heterozygous.
To gain deeper insights into the evolution of the CACTA-like TE, we genotyped the ZmCCT promoter region (including the TE-related PAV) for 61 additional teosinte entries and 338 maize lines (Table S1). We identified 30 haplotypes for 481 diverse maize lines and 53 haplotypes for 93 teosinte entries. Similar to the findings described above (Fig. 3A), the nucleotide diversity was much higher in teosinte than in maize that contained the TE insertion, supporting a history of positive selection in the TE-related PAV region. Furthermore, a minimum-spanning tree of these haplotypes (Fig. 3B) revealed two distinct clusters—a maize haplotype cluster and a teosinte haplotype cluster—in the TE-related PAV region. The maize haplotype cluster contained all the maize lines and a few teosinte entries and could be divided further into two discrete subgroup, whereas teosinte haplotype cluster was composed entirely of teosinte entries. Within the maize haplotype cluster, temperate lines had more TE insertions than tropical lines, whereas only one teosinte entry (Z. mays ssp. mexicana) contained a TE insertion (Table S1). These results suggest that the TE insertion occurred after domestication and accumulated during the adaptation of maize to temperate regions. The teosinte TE insertion may have resulted from postdomestication gene flow between tropical maize and Z. mays ssp. mexicana. Although such gene flow is rare (30), it has been described before (1, 4, 31–33).
Transformation-Mediated Validation of ZmCCT.
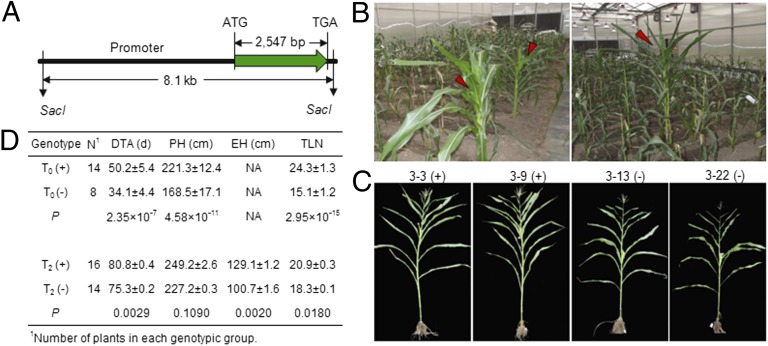
To assess ZmCCT function, we isolated this locus from the late-flowering line, 1145 (Hap6). This fragment, which included a 5.4-kb promoter, 2,547 bp of coding sequence, and a 500-bp 3′ UTR, was transferred into the maize receptor, HiII (Hap1) (Fig. 4A). Fourteen T0 transgenic plants were derived from four independent transgenic events. These plants exhibited delayed anthesis, greater plant height, and elevated total leaf number under long-day conditions (16 h light/8 h dark) compared with nontransgenic plants (Fig. 4 B and D). T2 families of transgenic plants also exhibited delayed anthesis (by 5.5 d), increased plant and ear height (by 22.0 cm and 28.4 cm, respectively), and a higher total leaf number (by 2.6 leaves) relative to sibling controls (Fig. 4 C and D). This experiment clearly demonstrated that ZmCCT affects photoperiod sensitivity, as does its rice homolog, Ghd7 (34).
Fig. 4.
Functional validation of ZmCCT via transformation. (A) The DNA fragment used for the ZmCCT complementation test is shown. (B) The greenhouse performance of T0 transgenic plants. Red arrows indicate transgenic plants. (C) The greenhouse performance of T2 transgenic plants. Transgenic plants and sibling controls are indicated by (+) and (−), respectively. (D) Comparison between transgenic lines and controls in T0 and T2 generations under long-day greenhouse conditions. DTA, days to anthesis (for T0 plants, from the date of transplantation to anthesis); EH, ear height; PH, plant height; TLN, total leaf number. Data are shown as mean ± SE.
Genomic Architecture, Subcellular Localization, and Phylogenetic Analysis of ZmCCT.
Both 5′ and 3′ RACE were used to isolate ZmCCT cDNA from the late-flowering inbred line, 1145. A full-length ZmCCT cDNA was constructed by joining these two RT-PCR products together. The full-length cDNA consisted of a 274-bp 5′ UTR, a 717-bp ORF (two exons), and a 239-bp 3′ UTR (SI Appendix, Fig. S9). ZmCCT encodes a 238-aa protein with a C-terminal CCT domain (amino acids 194–236), similar to its rice homolog, Ghd7 (34).
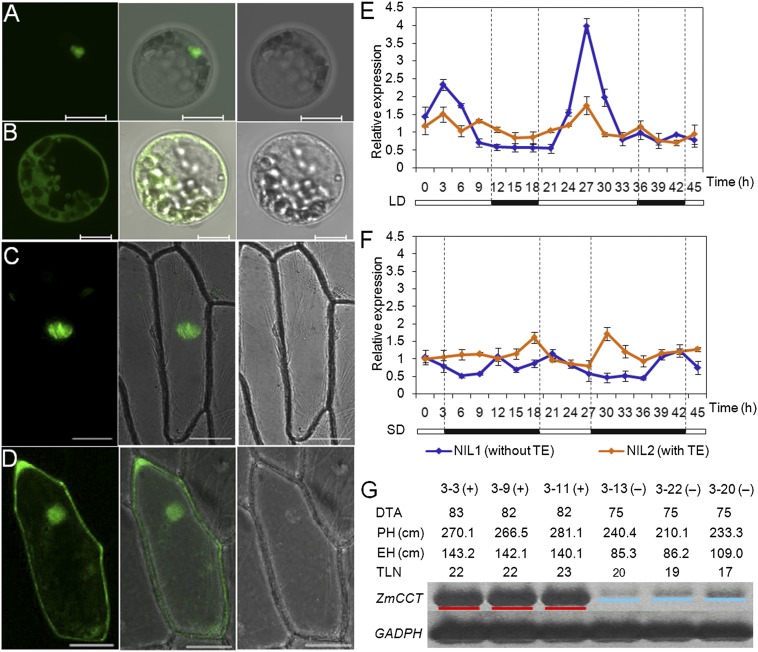
CCT domains are sufficient for the nuclear localization of proteins (35). Therefore we created a construct that used the CaMV35S promoter to drive expression of ZmCCT-GFP (CaMV35S: ZmCCT-GFP). This construct then was introduced into onion epidermal cells and maize protoplasts. In both cases a GFP signal was detected in the nucleus (Fig. 5 A–D).
Fig. 5.
Expression and subcellular localization of ZmCCT and its encoded protein. (A–D) A CaMV35S:ZmCCT-GFP construct was used to assess protein localization. CaMV35S:GFP was used as a control. (A) Nuclear localization of ZmCCT-GFP in maize protoplast. (Scale bar, 10 μm.) (B) GFP localization in maize protoplast. (Scale bar, 10 μm.) (C) Nuclear localization of ZmCCT-GFP in onion epidermal cells. (Scale bar, 20 μm.) (D) GFP localization in onion epidermal cells. (Scale bar, 20 μm.) (E and F) Diurnal rhythms of expression for ZmCCT in NIL1 (TE-negative, blue) and NIL2 (TE-positive, orange) in long-day (E) and short-day (F) environments. Expression was normalized to GADPH. Black bars represent dark periods, and white bars represent light periods. Data are shown as mean ± SE. (G) Allele-specific ZmCCT expression in transgenic and nontransgenic plants (T2). RT-PCR analysis of transgenic-plant samples yielded a 235-bp band (underlined in red), whereas nontransgenic siblings yielded a 241-bp band (underlined in blue). Phenotypic data associated with each plant are indicated. Transgenic plants and sibling controls are indicated by (+) and (−), respectively. GADPH was used as the control.
The maize genome encodes 35 proteins with a CCT domain, but phylogenetic analysis indicated that ZmCCT has low homology with most of these proteins. Intriguingly, ZmCCT is most similar to Sb06g000570 in sorghum and Ghd7 in rice (34); this similarity may explain why ZmCCT and Ghd7 are functionally related (SI Appendix, Fig. S10).
Transcription and Methylation of ZmCCT.
In the photoperiod-insensitive line, B73, ZmCCT is expressed at very low levels in all organs and at all developmental stages (36). Therefore we used RT-PCR to measure ZmCCT expression levels in leaf tissue isolated from two near-isogenic lines (NILs), Hap1 and Hap6. We analyzed different time points under both long- and short-day conditions. Expression of the photoperiod-sensitive ZmCCT allele (Hap6) exhibited a circadian rhythm under long-day conditions, with higher levels of transcription in the light (Fig. 5E). This circadian rhythm was not apparent under short-day conditions (Fig. 5F). The insensitive ZmCCT allele (Hap1) maintained a constant level of expression regardless of the light/dark cycle (Fig. 5E). Furthermore, ZmCCT expression levels were extremely high in transgenic plants as compared with sibling controls because of the expression from the exogenous ZmCCT gene (Hap6) (Fig. 5G). This allele-specific expression indicates that the CACTA-like transposon acts in cis to suppress ZmCCT transcription under long-day conditions, resulting in photoperiod insensitivity.
We further characterized the role of the CACTA-like TE in gene expression through promoter analysis (SI Appendix, Fig. S11C) using the promoter from the Huangzaosi (HZS, Hap1) line. Two overlapping HZS promoter fragments (bp −641 to −1 and −2,304 to −1) were capable of driving GUS expression in tobacco leaves under long-day but not under short-day conditions. In contrast, a 4,204-bp HZS promoter fragment (bp −4,204 to −1), which included some of the TE, did not drive GUS expression in tobacco leaves, even under long-day conditions.
We next sought to determine whether the CACTA-like TE participated in the epigenetic regulation of gene expression by affecting DNA-methylation levels (37, 38). Levels of methylation within an ∼2.5-kb segment upstream of the ZmCCT ORF were measured for 1145 (TE-negative, Hap6) and HZS (TE-positive, Hap1). For HZS, high levels of cytosine methylation were detected between −700 and −2,100 bp. This same region was less methylated in 1145 samples (SI Appendix, Fig. S11 A and B). Interestingly, methylated cytosines were not detected within the ∼700 bp upstream of the start codon in either line, consistent with the observation that the 641-bp HZS promoter drove high levels of GUS expression (SI Appendix, Fig. S11C). Several light-response motifs were identified in the ZmCCT promoter in both 1145 and HZS (SI Appendix, Table S10). One motif, INRNTPSADB, exhibited cytosine methylation in HZS (SI Appendix, Fig. S11B). These studies suggest that elevated levels of cytosine methylation within the ZmCCT promoter, particularly methylation of the INRNTPSADB motif, may contribute to low levels of ZmCCT transcription in HZS.
A Conceptual Model of the Maize Photoperiod Pathway Under Long-Day Conditions.
Components of the circadian clock and their functions are likely conserved in plants (39), and a comprehensive photoperiod pathway that involves ZmCCT was proposed recently in maize (40). To demonstrate further the function of ZmCCT in the maize photoperiod pathway, RNA-seq data generated from HZS (TE-positive, Hap1) and its NIL (TE-negative, Hap6) allowed comprehensive screening of genes involved in the ZmCCT regulatory network. Under long-day conditions the highest levels of ZmCCT expression were detected in leaf tissue at the three-leaf stage. It is clear that ZmCCT regulates many genes, because 1,117 genes were differentially expressed (≥1.67-fold) between HZS and its NIL. When these data are combined with those of previous studies (41, 42), a conceptual model of the photoperiod pathway under long-day conditions can be proposed in maize (SI Appendix, Fig. S12). Some critical steps in this pathway were validated using RT-PCR (SI Appendix, Fig. S13). We identified a gene that encodes a CCT domain-containing protein, GRMZM2G367834 (GO:0005515, IPR010402), and has the same annotation and expression pattern as PRR37 and -73, TOC1, and ZmCCT, which may represent a node in the loop. Four maize genes (18), ZmSRR1 (GRMZM2G178795), ZmFKF1 (GRMZM2G106363), ZmELF4 (GRMZM2G025646), and ZmELF9 (GRMZM2G171660), which are homologous to the respective Arabidopsis genes SRR1, FKF1, ELF4, and ELF9, also may participate in this pathway. In the next step, detailed study on these genes may help explain the photoperiod pathway in plants, thus enhancing crop breeding.
Discussion
Recent studies concerning the genetics of flowering time in maize have determined that the chromosomal region bin 10.04, which contains ZmCCT, is a hot spot for QTLs associated with the photoperiod response (14–18, 43, 44). However, these studies failed to identify the causative variant within the ZmCCT locus. Here we identified a CACTA-like TE inserted upstream of the ZmCCT ORF in early-flowering maize. The TE-related PAV was strongly associated with flowering time (Figs. 1 and 2 and SI Appendix, Table S7) and caused large variations in flowering time (SI Appendix, Figs. S6–S8). We also checked the 26 diverse nested association mapping founders and found TE insertions in 22 of these founders but no TE insertion in CML228, CML277, Ki11, or Ky21 (SI Appendix, Table S11). CML228, CML277, and Ki11 display the strongest photoperiod response, and Ky21 exhibits an increased DTA (18). TE insertions within a conserved noncoding sequence can cause dramatic functional variations. These types of insertions include a Hopscotch element ∼60 kb upstream of tb1 (26, 27) and a MITE associated with Vgt1 (12). TEs themselves often are capable of cis-regulation, influencing nearby functional genes (23). In addition, TEs can affect DNA methylation and thereby repress transcription from nearby genes (SI Appendix, Fig S11 A and B).
ZmCCT regulates maize flowering time under long-day conditions (17, 18). For sensitive (TE-negative) ZmCCT alleles, we observed a distinct diurnal pattern of expression, which is the same pattern observed with rice Ghd7 (34). We provide indisputable biological evidence that ZmCCT is involved in the maize response to long days through transformation-mediated functional validation. We also show that TE represses gene transcription (Fig. 5 and SI Appendix, Fig. S11). Our findings provide important details concerning the molecular basis of photoperiod sensitivity. Under long-day conditions the CACTA-like TE in ZmCCT acts in cis to suppress ZmCCT expression. This action in turn up-regulates the expression of the floral activator ZCN8, causing maize to flower early.
In the ZmCCT-based association mapping a single TE-related LD block contains most of the highly significant association variants, including two SNPs that were identified previously (18). Within the TE-related LD block, maize lines lacking the TE captured 73% of the nucleotide diversity of teosinte, which is higher than the 57% genomic average in maize (45). In contrast, maize lines containing the TE insertion had much lower nucleotide diversity (π+/πT = 0.04, π+/π− = 0.06). Thus, we conducted separate association analyses for maize lines with (n = 328) and without (n = 133) the TE insertion. To our surprise, no significant variant (P < 0.05) with MAF >0.05 was identified in either association study despite the many sequence variants across ZmCCT. Consistent with these findings, teosinte alleles of ZmCCT consistently express at high levels under long-day conditions despite extremely high nucleotide diversity within their regulatory regions (18). Therefore a single SNP (or small InDel) in ZmCCT regulatory sequences is not sufficient to affect the photoperiod response. Only when genetic variants are in strong LD with the TE are they associated with the photoperiod response, because the TE-related PAV is such a causative factor. The allele substitution at ZmCCT via marker-assisted selection enables early flowering for tropical maize in temperate regions. This rapid adaptation could accelerate exploitation of tropical maize to breed diverse, high-yielding maize varieties for sustainable maize production.
The TE was selected during the postdomestication spread of maize, and this strong selective sweep resulted in the TE-related LD block, which was characterized with very low nucleotide diversity (Fig. 3A). At the same time, numerous recombination events took place downstream of the TE-related LD block, resulting in numerous ZmCCT alleles. This inference may explain why the selective sweep was restricted to the TE-related block of LD and why the five other sequenced regions exhibited a neutral model.
We now can draw a vivid picture of the evolutionary history of ZmCCT. Before the TE insertion, the germplasm of tropical maize carried numerous ZmCCT alleles, which were characterized by high levels of nucleotide diversity within their regulatory regions. At some point in time, a CACTA-like transposon was inserted 2,543 bp upstream of ZmCCT in a tropical plant. This TE repressed ZmCCT expression and attenuated photoperiod sensitivity under long-day conditions. As a result, maize lines that contained the TE were selected for and accumulated as maize adapted to a diverse array of long-day environments. This selective sweep finally created a TE-related LD block in maize.
Materials and Methods
A collection of 508 maize inbred lines was investigated for their flowering-related traits in eight environments. All 508 lines were genotyped using the MaizeSNP50 BeadChip, and a subset of 368 lines was further genotyped by RNA-seq for GWAS. Details of all materials and methods used are listed in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Jinsheng Lai and Weibin Song for maize transformation, Dr. Edward S. Buckler and two anonymous reviewers for helpful comments on the manuscript. This research was supported by Grants 2009CB118402 and 2011CB111509 from the National Basic Research ‘973’ Program of China and by Grants 2012AA10A306, 2012AA10A307, and 2012AA101104 from the National High-Tech and Development Program of China.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KF575627–KF576205).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310949110/-/DCSupplemental.
References
- 1.Matsuoka Y, et al. A single domestication for maize shown by multilocus microsatellite genotyping. Proc Natl Acad Sci USA. 2002;99(9):6080–6084. doi: 10.1073/pnas.052125199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Emerson RA. A genetic view of sex expression in the flowering plants. Science. 1924;59(1521):176–182. doi: 10.1126/science.59.1521.176. [DOI] [PubMed] [Google Scholar]
- 3.Piperno DR, Ranere AJ, Holst I, Iriarte J, Dickau R. Starch grain and phytolith evidence for early ninth millennium B.P. maize from the Central Balsas River Valley, Mexico. Proc Natl Acad Sci USA. 2009;106(13):5019–5024. doi: 10.1073/pnas.0812525106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Heerwaarden J, et al. Genetic signals of origin, spread, and introgression in a large sample of maize landraces. Proc Natl Acad Sci USA. 2011;108(3):1088–1092. doi: 10.1073/pnas.1013011108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colasanti J, Coneva V. Mechanisms of floral induction in grasses: Something borrowed, something new. Plant Physiol. 2009;149(1):56–62. doi: 10.1104/pp.108.130500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colasanti J, Yuan Z, Sundaresan V. The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell. 1998;93(4):593–603. doi: 10.1016/s0092-8674(00)81188-5. [DOI] [PubMed] [Google Scholar]
- 7.Colasanti J, et al. The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants. BMC Genomics. 2006;7:158. doi: 10.1186/1471-2164-7-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meng X, Muszynski MG, Danilevskaya ON. The FT-like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. Plant Cell. 2011;23(3):942–960. doi: 10.1105/tpc.110.081406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muszynski MG, et al. delayed flowering1 Encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiol. 2006;142(4):1523–1536. doi: 10.1104/pp.106.088815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bomblies K, Doebley JF. Pleiotropic effects of the duplicate maize FLORICAULA/LEAFY genes zfl1 and zfl2 on traits under selection during maize domestication. Genetics. 2006;172(1):519–531. doi: 10.1534/genetics.105.048595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller TA, Muslin EH, Dorweiler JE. A maize CONSTANS-like gene, conz1, exhibits distinct diurnal expression patterns in varied photoperiods. Planta. 2008;227(6):1377–1388. doi: 10.1007/s00425-008-0709-1. [DOI] [PubMed] [Google Scholar]
- 12.Salvi S, et al. Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc Natl Acad Sci USA. 2007;104(27):11376–11381. doi: 10.1073/pnas.0704145104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colasanti J, Muszynski M. In: Handbook of Maize: Its Biology. Bennetzen J, Hake S, editors. New York: Springer; 2009. pp. 41–55. [Google Scholar]
- 14.Buckler ES, et al. The genetic architecture of maize flowering time. Science. 2009;325(5941):714–718. doi: 10.1126/science.1174276. [DOI] [PubMed] [Google Scholar]
- 15.Coles ND, McMullen MD, Balint-Kurti PJ, Pratt RC, Holland JB. Genetic control of photoperiod sensitivity in maize revealed by joint multiple population analysis. Genetics. 2010;184(3):799–812. doi: 10.1534/genetics.109.110304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coles ND, Zila CT, Holland JB. Allelic effect variation at key photoperiod response quantitative trait loci in maize. Crop Sci. 2011;51(3):1036–1049. [Google Scholar]
- 17.Ducrocq S, et al. Fine mapping and haplotype structure analysis of a major flowering time quantitative trait locus on maize chromosome 10. Genetics. 2009;183(4):1555–1563. doi: 10.1534/genetics.109.106922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hung HY, et al. ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc Natl Acad Sci USA. 2012;109(28):E1913–E1921. doi: 10.1073/pnas.1203189109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biémont C, Vieira C. Genetics: Junk DNA as an evolutionary force. Nature. 2006;443(7111):521–524. doi: 10.1038/443521a. [DOI] [PubMed] [Google Scholar]
- 20.Bourque G. Transposable elements in gene regulation and in the evolution of vertebrate genomes. Curr Opin Genet Dev. 2009;19(6):607–612. doi: 10.1016/j.gde.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 21.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10(10):691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lisch D. Epigenetic regulation of transposable elements in plants. Annu Rev Plant Biol. 2009;60:43–66. doi: 10.1146/annurev.arplant.59.032607.092744. [DOI] [PubMed] [Google Scholar]
- 23.Lisch D. How important are transposons for plant evolution? Nat Rev Genet. 2013;14(1):49–61. doi: 10.1038/nrg3374. [DOI] [PubMed] [Google Scholar]
- 24.Britten RJ. Transposable element insertions have strongly affected human evolution. Proc Natl Acad Sci USA. 2010;107(46):19945–19948. doi: 10.1073/pnas.1014330107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao H, Jiang N, Schaffner E, Stockinger EJ, van der Knaap E. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science. 2008;319(5869):1527–1530. doi: 10.1126/science.1153040. [DOI] [PubMed] [Google Scholar]
- 26.Studer A, Zhao Q, Ross-Ibarra J, Doebley J. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat Genet. 2011;43(11):1160–1163. doi: 10.1038/ng.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou L, Zhang J, Yan J, Song R. Two transposable element insertions are causative mutations for the major domestication gene teosinte branched 1 in modern maize. Cell Res. 2011;21(8):1267–1270. doi: 10.1038/cr.2011.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, et al. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat Genet. 2013;45(1):43–50. doi: 10.1038/ng.2484. [DOI] [PubMed] [Google Scholar]
- 29.Yang X, et al. Characterization of a global germplasm collection and its potential utilization for analysis of complex quantitative traits in maize. Mol Breed. 2011;28(4):511–526. [Google Scholar]
- 30.Hufford MB, et al. The genomic signature of crop-wild introgression in maize. PLoS Genet. 2013;9(5):e1003477. doi: 10.1371/journal.pgen.1003477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fukunaga K, et al. Genetic diversity and population structure of teosinte. Genetics. 2005;169(4):2241–2254. doi: 10.1534/genetics.104.031393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gallavotti A, et al. The role of barren stalk1 in the architecture of maize. Nature. 2004;432(7017):630–635. doi: 10.1038/nature03148. [DOI] [PubMed] [Google Scholar]
- 33.Ross-Ibarra J, Tenaillon M, Gaut BS. Historical divergence and gene flow in the genus Zea. Genetics. 2009;181(4):1399–1413. doi: 10.1534/genetics.108.097238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xue W, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40(6):761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- 35.Wenkel S, et al. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell. 2006;18(11):2971–2984. doi: 10.1105/tpc.106.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sekhon RS, et al. Genome-wide atlas of transcription during maize development. Plant J. 2011;66(4):553–563. doi: 10.1111/j.1365-313X.2011.04527.x. [DOI] [PubMed] [Google Scholar]
- 37.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8(4):272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 38.Parisod C, et al. Rapid structural and epigenetic reorganization near transposable elements in hybrid and allopolyploid genomes in Spartina. New Phytol. 2009;184(4):1003–1015. doi: 10.1111/j.1469-8137.2009.03029.x. [DOI] [PubMed] [Google Scholar]
- 39.Song YH, Ito S, Imaizumi T. Similarities in the circadian clock and photoperiodism in plants. Curr Opin Plant Biol. 2010;13(5):594–603. doi: 10.1016/j.pbi.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong Z, et al. A gene regulatory network model for floral transition of the shoot apex in maize and its dynamic modeling. PLoS ONE. 2012;7(8):e43450. doi: 10.1371/journal.pone.0043450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jarillo J, et al. Review. Photoperiodic control of flowering time. Span J Agric Res. 2008;6:221–244. [Google Scholar]
- 42.Sawa M, Nusinow DA, Kay SA, Imaizumi T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science. 2007;318(5848):261–265. doi: 10.1126/science.1146994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang CL, et al. Genetic analysis of photoperiod sensitivity in a tropical by temperate maize recombinant inbred population using molecular markers. Theor Appl Genet. 2008;117(7):1129–1139. doi: 10.1007/s00122-008-0851-y. [DOI] [PubMed] [Google Scholar]
- 44.Wang C, et al. Mapping QTL associated with photoperiod sensitivity and assessing the importance of QTL×environment interaction for flowering time in maize. PLoS ONE. 2010;5(11):e14068. doi: 10.1371/journal.pone.0014068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wright SI, et al. The effects of artificial selection on the maize genome. Science. 2005;308(5726):1310–1314. doi: 10.1126/science.1107891. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.