Significance
The directed evolution of proteins in vitro has generated highly functional proteins and has contributed to elucidating the sequence–function relationship of proteins. However, the available methods consider globular proteins, not membrane proteins, despite the biological and pharmaceutical importance of the latter. We report the development of a method called liposome display, which enables the in vitro evolution of membrane proteins. We applied the method to evolve the pore-forming activity of α-hemolysin from Staphylococcus aureus and obtained a mutant with 30-fold higher activity than the WT. Given its high degree of controllability, liposome display allows for the rapid and efficient evolution of a wide range of membrane proteins, thereby improving the field of membrane protein engineering.
Keywords: directed evolution, PURE system, FACS, Giant unilamellar vesicles, in vitro synthetic biology
Abstract
In vitro methods have enabled the rapid and efficient evolution of proteins and successful generation of novel and highly functional proteins. However, the available methods consider only globular proteins (e.g., antibodies, enzymes) and not membrane proteins despite the biological and pharmaceutical importance of the latter. In this study, we report the development of a method called liposome display that can evolve the properties of membrane proteins entirely in vitro. This method, which involves in vitro protein synthesis inside liposomes, which are cell-sized phospholipid vesicles, was applied to the pore-forming activity of α-hemolysin, a membrane protein derived from Staphylococcus aureus. The obtained α-hemolysin mutant possessed only two point mutations but exhibited a 30-fold increase in its pore-forming activity compared with the WT. Given the ability to synthesize various membrane proteins and modify protein synthesis and functional screening conditions, this method will allow for the rapid and efficient evolution of a wide range of membrane proteins.
An in vitro translation (IVT) system produces proteins without using living cells (1, 2). Protein synthesis with IVT is initiated by adding a DNA or RNA template to the reaction mixture and is completed after a few hours of incubation. Based on these characteristics, IVT has been used for various applications, including in vitro protein evolution (3, 4). In vitro evolution is the iteration of mutant library generation and selection of the fittest specimens, which leads to the creation of proteins with the desired properties. Compared with living cell-based methods, IVT-based methods allow for more rapid and efficient protein evolution. Such methods allow for the generation of a large gene library with a diversity of up to 1013 (5). In most cases, a single round of selection can be achieved within 1 d (3, 4). In addition, a wide range of proteins can be targeted because protein synthesis with IVT is disconnected from cell growth. However, all the proteins evolved to date using in vitro evolutionary methods have been globular proteins (e.g., antibodies, enzymes), and there have been no previous reports of the in vitro evolution of membrane proteins.
Membrane proteins account for 20–25% of all ORFs in the genome, and more than 50% of current pharmaceutical targets are membrane proteins (6, 7). Therefore, membrane proteins are a high-priority research topic. Directed evolution that uses living cells has been applied to membrane proteins to investigate the properties of these molecules. For example, multidrug transporter was evolved to pump a novel pharmaceutical drug out of the cell (8), G protein-coupled receptor was evolved to be synthesized in larger amounts in Escherichia coli cells without losing its original function (9), and caveolin was evolved for soluble expression (10). These results identified the mutations required to change the properties of membrane proteins, and consequently revealed the sequence–function relationships of the target membrane proteins (11). However, these examples share a common limitation: The target membrane protein should not affect and/or inhibit the growth of the host cell. In addition, in many cases, the function of the target membrane protein (e.g., its transporter activity) and the growth of the cell often have to be coupled (8, 12, 13). The development of a method for in vitro evolution of membrane proteins will circumvent these restrictions, and thus enable more rapid and efficient evolution of a greater variety of membrane proteins.
In this paper, we report the development of a method called liposome display, which is used to evolve the properties of membrane proteins entirely in vitro. This method is then applied to evolve the pore-forming activity of α-hemolysin (AH) from Staphylococcus aureus, which is known to be expressed as a water-soluble monomer (14). The expressed monomer binds to the membrane surface; assembles into a heptamer; and finally generates a pore 2.6 nm in diameter, allowing for the penetration of molecules smaller than 3 kDa (14). With liposome display, the DNA-encoding membrane protein is encapsulated at the single-molecule level together with the protein synthesis using recombinant elements (PURE) system (15), a reconstituted IVT system, inside giant unilamellar vesicles (GUVs) (16). The membrane protein synthesized from the DNA is then displayed on the surface of the GUVs, thereby establishing the genotype–phenotype linkage that is required for all directed evolution methods. Starting from a random mutagenized DNA library of AH, we obtained a mutant with 30-fold higher pore-forming activity compared with the WT after 20 rounds of selection using liposome display.
Results
Schematic of Liposome Display.
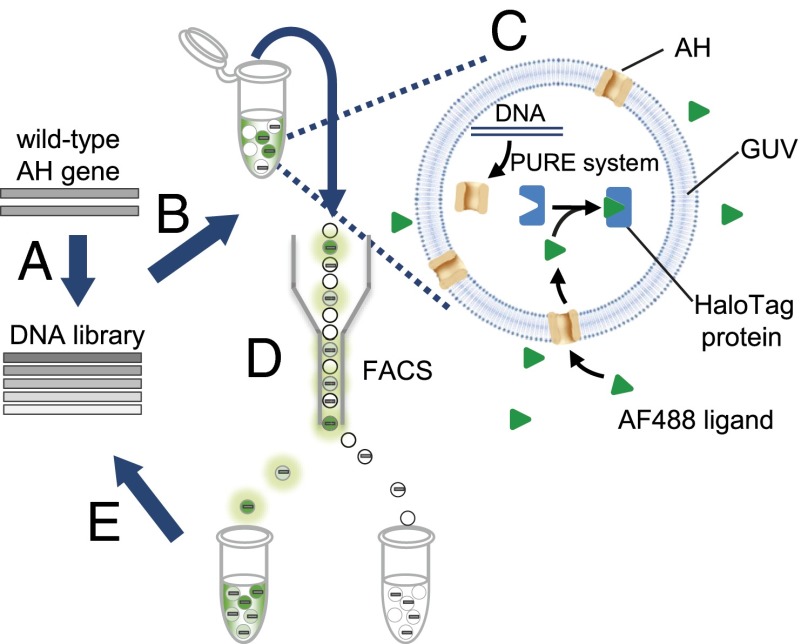
Directed evolution of AH was performed using liposome display, as shown schematically in Fig. 1. A randomly mutagenized gene library of WT AH was prepared (Fig. 1A). The gene was encapsulated into GUVs at the single-molecule level (i.e., one DNA molecule per GUV), together with the reconstituted IVT system (the PURE system) and a HaloTag protein (17) (Fig. 1B). We used GUVs with a lipid composition of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC)/cholesterol at a 1:1 ratio (Fig. S1A). The single-gene encapsulation enabled genotype–phenotype linkage, a property required for directed evolution. The HaloTag protein is a modified haloalkane dehalogenase designed to bind covalently to synthetic ligands (HaloTag ligands) (17), and it was used to quantify the activity of the synthesized AH. After synthesizing the AH, one of the HaloTag ligands, the Alexa Fluor 488-labeled (AF488) ligand (Promega), was added to the exterior of the GUVs (Fig. 1C). Depending on the pore-forming activity of the AH, influx of the AF488 ligand into the GUVs occurred. The pore-forming activity is a function of multiple parameters, including expression level, protein stability, membrane-binding affinity, and heptamer assembly. The penetrating AF488 ligands were trapped inside the vesicles by the HaloTag protein, resulting in enrichment of the fluorescent signal. By collecting GUVs with strong fluorescent signals using FACS, we obtained the AH genes with higher activities (Fig. 1D). The recovered genes were amplified and brought to the next round of selection (Fig. 1E).
Fig. 1.
Schematic of the in vitro evolution of AH using liposome display. (A) Gene encoding WT AH was subjected to mutagenesis to generate a randomly mutagenized gene library. (B) Gene was encapsulated in GUVs at the single-molecule level (i.e., one DNA molecule per GUV), together with the IVT system and HaloTag protein. (C) After GUV preparation, AH was synthesized and the fluorescence ligand (AF488) was added to the exterior of the GUVs. The ligand was cell-impermeable due to its negative charge and was expected to penetrate the membrane only through the AH pores. (D) GUVs with high-fluorescence signals were collected via FACS. (E) Finally, the recovered gene was amplified and included in the next round of selection.
To demonstrate that the strategy presented the above functions properly, the following section describes three experimental observations: (i) the pore-forming activity of the AH can be visualized using the HaloTag system, (ii) the function of AH synthesized from a single copy of DNA in the GUV is detected by FACS, and (iii) an active gene can be enriched from a mixture of active and inactive genes.
Synthesis and Evaluation of AH in GUVs.
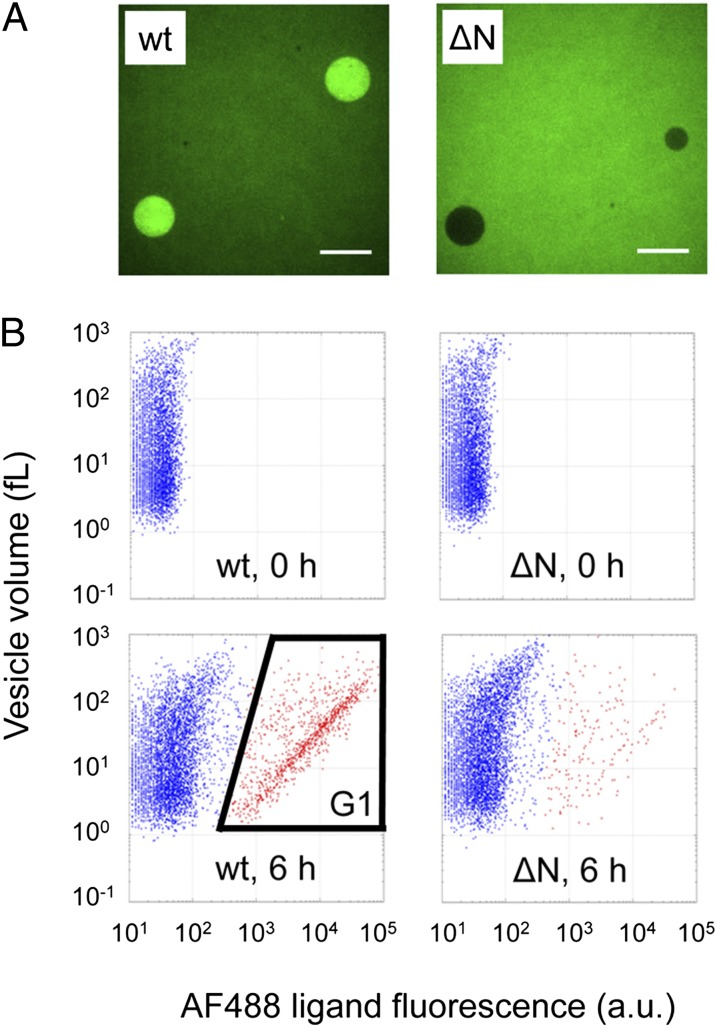
We first investigated whether the synthesized AH can function inside the GUVs. We synthesized AH protein from 5 nM DNA of WT AH or the ΔN mutant [an inactive mutant of AH prepared by deleting 66 residues from the N terminus (18)] inside the GUVs, added a sufficient amount of AF488 ligand to the exterior, and observed the resulting GUVs by fluorescence microscopy (Fig. 2A). We observed clear enrichment of the fluorescent signal; this level was close to that obtained when HaloTag protein was saturated with AF488 ligand with WT AH, but no such enrichment was observed with the ΔN mutant. These results indicated that the synthesized AH was functional, as shown in the schematic diagram in Fig. 1.
Fig. 2.
Detection of AH activity inside the GUVs. (A) Fluorescence images of the WT (wt) or ΔN mutant (ΔN) gene-encapsulated GUVs. The DNA concentration used for protein synthesis was 5 nM. The concentration of AF488 ligand was 200 nM. (Scale bar = 10 μm.) (B) 2D FACS data of WT or ΔN mutant gene-encapsulated GUVs; 5 pM DNA and 200 nM AF488 ligand were used. The vertical axis shows the aqueous volume of each GUV, and the horizontal axis shows the fluorescence intensity of the HaloTag A488 ligand. GUVs containing WT DNA appeared in the G1 region. GUVs in G1 are plotted in red. The aqueous volume of each vesicle was estimated by encapsulating 1 μM TA647, as described in Materials and Methods. The a.u. represents the raw value of FITC-A obtained by FACS.
Second, we investigated whether the function of the AH synthesized from a single DNA copy in the GUV was detectable by FACS. We encapsulated the DNA encoding the WT or ΔN mutant, added the AF488 ligand to the exterior of the GUVs, and observed the resulting GUVs by FACS. A population with increased fluorescence was only observed in WT AH (Fig. 2B). Because the GUV volume was between 1 and 100 fL (Fig. 2B, vertical axis) and based on the DNA concentration used (5 pM = 0.003 copies per femtoliter), we expected 0.003–0.3 copies per GUV, on average. These numbers suggested that most of the GUVs would have no DNA and that those with DNA would have only a single copy. In fact, the 2D plot (Fig. 2B) displays two distinct populations, with low (outside gate G1) and high (inside gate G1) AF488 fluorescent signals, which we assumed did not and did carry DNA, respectively. To confirm this assumption, we investigated the GUV volume dependency of the GUV fraction inside G1 (Fig. S1B) and found that the DNA encapsulation can be described as a Poisson process (Fig. S1C). Using the Poisson distribution (Eq. S1 in the legend for Fig. S1C), the average number of DNA molecules inside the GUV in G1 was estimated to be very close to unity (Table S1). Therefore, the AH synthesized from a single DNA copy in the GUV can be detected by FACS. With the fluorescent vesicles in Fig. 2B, a linear correlation between AF488 fluorescence and the vesicle volume was observed. This correlation can be explained with the HaloTag protein, whose number in each vesicle is proportional to the volume, being saturated with AF488 ligand.
Finally, we performed a model enrichment experiment to confirm the validity of the schematic presented in Fig. 1. The DNAs encoding WT and ΔN mutant were mixed at a ratio of 1:9, and this gene mixture was subjected to the procedure depicted in Fig. 1. GUVs exhibiting high AF488 fluorescence intensity but with a different volume were sorted by FACS, and the DNA recovered was purified and subjected to a quantitative PCR assay to estimate the enrichment factor (Fig. S2A). After a single round of the liposome display experiment, the ratios of WT to ΔN mutant genes had increased in all sorted vesicle volumes (Fig. S2B). The enrichment factor was higher with smaller GUVs, as expected (19, 20). This trend occurs because larger GUVs tend to encapsulate multiple genes, thus resulting in a reduction of the enrichment factor. These results indicated that the selection system functioned as expected.
The GUV that we used showed a widely varying volume (1–100 fL; Fig. 2B). The difference in the vesicle volume may affect the intravesicular protein synthesis, for example, by creating a difference in the ratio of the surface area to the volume. This was not the case, because our previous report indicated that GFP synthesis using 1 nM DNA inside the GUV proceeded similarly regardless of the vesicle volume (21). Furthermore, the volume distribution of the vesicle did not change significantly during the protein synthesis (5-h incubation at 37 °C) (Fig. S1D), indicating that GUVs did not rupture during the experiments. These properties allowed us to perform the above proof-of-principle experiments of the liposome display method.
Directed Evolution of AH.
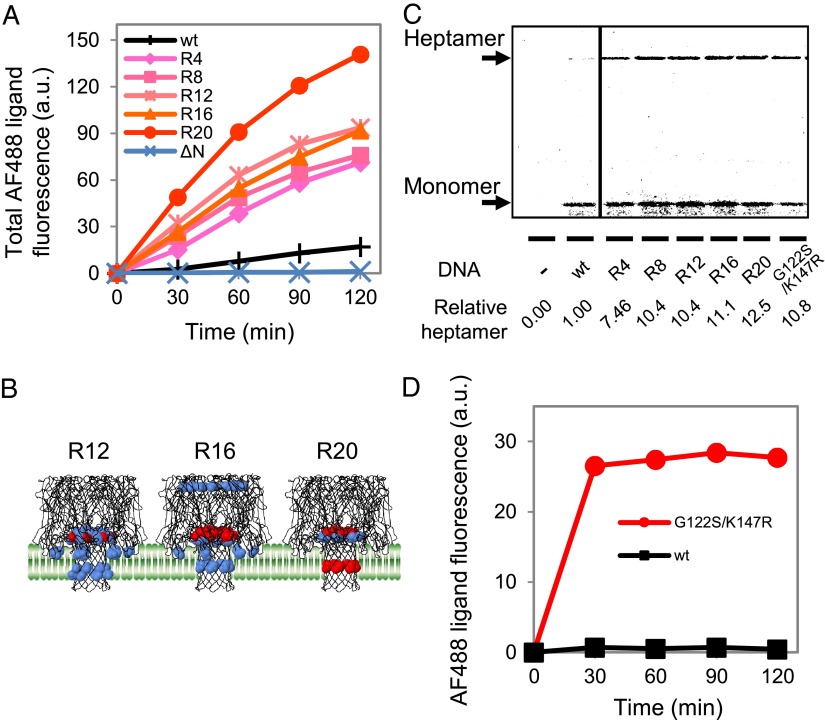
Next, we attempted to evolve the pore-forming activity of WT AH. Starting from a randomly mutagenized DNA library, we repeated the liposome display process 20 times. The DNA concentration used was fixed at 5 pM for every round. For each round, FACS was used to sort ∼10,000 GUVs representing the 1% with the highest AF488 ligand fluorescence among those with a size between 6 and 30 fL, and the obtained gene pool was brought to the next round of screening. The sorting was performed at the time point at which the HaloTag proteins were far from being saturated with the AF488 ligand. The GUV size used for sorting was chosen, despite the lower enrichment factor compared with smaller GUVs (Fig. S2), because larger numbers of GUVs were present. Consequently, the rate of AF488 ligand incorporation of the gene pool increased over the rounds, and the rate was increased by 11.8-fold over that with the WT after the 20th iteration [round 20 (R20)] (Fig. 3A and Fig. S3A).
Fig. 3.
Properties of the AH mutants obtained after multiple rounds of liposome display. (A) Time course of AF488 ligand accumulation for the various AH gene pools. R4 represents the gene pool obtained after R4 of screening (similarly for R8 and others). The vertical axis is the sum of the AF488 ligand fluorescence intensity of GUVs measured by FACS (Fig. S3A). The values at time 0 were set to zero. (B) Locations of the fixed mutations among the selected clones mapped on the 3D structure of the WT AH. A model membrane is also depicted. The mutations found in more than 10% and 25% of the clones are shown in blue and red, respectively. (C) Binding of the AH variants to the GUVs. The AH variants were synthesized on the exterior of the GUVs with the PURE system supplemented with [35S]methionine and 15 pM DNA. After removing the unbound proteins, the GUV fraction was subjected to SDS/PAGE without boiling, allowing for the quantitative analysis of both heptamers and monomers (18, 22, 23). (D) Pore-forming activities of the WT AH and G122S/K147R mutant with purified proteins. Purified AH (150 nM) was added to the HaloTag protein-encapsulated GUVs, and the AF488 ligand was added to the exterior of the GUVs. The measurement and analysis were performed as described for A.
We then determined the clone sequence from R12, R16, and R20 and identified the fixed mutations (Fig. S3B). We defined the fixed mutations as those observed in more than 10% of the sequenced clones, and we mapped these on the 3D structure of the WT AH (Fig. 3B). The fixed mutations were enriched in the stem domain and at the interface between the phospholipid membranes. We also observed an increase in the occupancy of fixed mutations over the rounds. The fixed mutation with the highest occupancy at R12 showed a value of 36%, which increased to 50% at R16 and to 76% at R20 (Fig. S3B). These results indicated that enrichment of the mutations occurred during the rounds, suggesting that the directed evolution experiments worked as desired.
We then analyzed the activities of the individual clones obtained after R20. For this purpose, 21 clones were subjected to an AH activity assay. We found that most of the clones exhibited 3- to 11-fold higher activity than the WT (Fig. S4A). From these results, we concluded that liposome display generated highly functional AH mutants with fixed mutations in the stem domain.
Characterization of AH Mutants.
Next, we investigated the cause of the high activity observed in the mutants. The increased rates of AF488 ligand accumulation by the mutants indicated that more pores (heptameric AH) were present on the GUVs of the mutants compared with those of the WT. This outcome may have been due to an increase in protein expression and/or an improved ability to form heptameric pores on the phospholipid membranes. To test the first assumption, the gene pools obtained after R4, R8, R12, R16, and R20 were synthesized using the PURE system containing [35S]methionine, and the quantity of synthesized protein was estimated from the band intensity of the corresponding band obtained from the autoradiography of the SDS polyacrylamide gel. We found no detectable differences between the samples tested (Fig. S4B). To test the second assumption, various AHs were synthesized using the PURE system containing [35S]methionine outside the GUVs. After synthesis, GUVs were washed to remove any unbound proteins and the GUV suspension was subjected to SDS/PAGE (Fig. 3C). The samples were not boiled before SDS/PAGE; thus, the amounts of both heptamer and monomer were analyzed (18, 22, 23). Although the monomer quantity differed little among the samples, the heptamer quantity increased during the rounds and had increased to 12-fold over that of the WT by R20. This result corresponded well with the 11.8-fold difference in activity of AF488 ligand influx (Fig. 3A and Fig. S3A). These results indicated that the as-obtained mutants did not exhibit an altered expression level but did exhibit an increased ability to form heptameric pores on the GUV membrane.
To investigate the properties of the identified mutants further, we constructed a mutant that harbored G122S/K147R mutations, two of the three fixed mutations shown in Fig. 3B, at R20 (red mutation). The expression level of the G122S/K147R mutant was not markedly different from that of the WT (Fig. S4B). In contrast, the G122S/K147R mutant showed a 10.8-fold increase in heptamer formation compared with the WT (Fig. 3C). As with the gene pools, this mutant exhibited improved activity by means of an increased ability to form heptameric pores on the GUV membrane.
We further confirmed the improved pore-forming activity of the mutant using purified proteins. The G122S/K147R mutant and WT AH were overexpressed and purified from E. coli, and their pore-forming activities were compared. The purified protein was added to the exterior of GUVs carrying the HaloTag protein, and the rate of AF488 ligand accumulation was measured by FACS. We found that the rate was 30-fold higher with the mutant (Fig. 3D). The improved properties of the G122S/K147R mutant were confirmed by analysis of the purified protein.
Discussion
We developed a method to evolve membrane proteins entirely in vitro; this method, called liposome display, was applied to obtain AH mutants with high pore-forming activity. The IVT system has been shown to be applicable for synthesizing membrane proteins (24) but has not been adapted for the directed evolution of membrane proteins.
From the liposome display rounds, we obtained a highly active AH mutant with two mutations, G122S/K147R, in the stem domain. Here, we discuss the possible roles of these mutations. Because K147 is located within the pore and the side chain of the Lys residue faces the pore (Fig. S3C), we first postulated that the mutant may exhibit increased influx of AF488 ligand but not other substances. We tested the efflux of carboxyfluorescein (Fig. S5A) and the influx of fluorescently labeled trinucleotides (Fig. S5 B and C) and found that the G122S/K147R mutant also penetrated substrates other than AF488 ligand more than the WT, suggesting that the improved activity of the mutant was not specific to the AF488 ligand.
The increase in the amount of heptamer on the membrane of the G122S/K147R mutant relative to the WT (Fig. 3C) could be due to the improved affinity of the monomeric AH to the membrane and/or heptameric pore formation on the membrane. We postulated that the increased ability of the mutant was due to the latter reason, because previous reports suggested the involvement of the rim domain in the initial lipid recognition (14, 25), whereas the two mutations were located in the stem domain. In addition, there was an increase in the amount of membrane-bound heptamer but not in that of monomer. Therefore, although we could not measure the on-membrane heptamerization directly, the two mutations, G122S/K147R, are likely to have improved this step, thereby increasing the amount of heptamer bound to the membrane (Fig. 3C). However, determination of the detailed mechanism will require further studies, including determination of the 3D structures of the mutants.
Liposome display can engineer a greater variety of membrane proteins compared with living cell-based methods due to the following features:
i) The membrane proteins that affect cell growth can also be engineered because the protein is synthesized in vitro. AH is a toxin, and engineering this protein in a high-throughput manner using living cells remains difficult.
ii) Conditions appropriate for the target membrane protein can be adopted because the lipid composition of the GUVs is adjustable. We have shown previously that GUVs with different lipid compositions can be used without affecting intravesicular protein synthesis (21), indicating that a wide range of membranes may be used for liposome display.
iii) Conditions appropriate for the target membrane protein can be adopted because the intravesicular solution can be adjusted as needed. For example, a reporter system, the HaloTag system in our case, can be encapsulated inside the GUVs to detect the activity of the target membrane protein, thereby directly detecting the influx or efflux of particular substances. We can also add other systems, such as membrane translocase (26), when needed.
iv) Because the PURE system is composed only of the components involved in protein synthesis, no membrane proteins other than the target protein are present. Therefore, selection and screening steps can be performed under well-controlled conditions.
v) In addition to the aforementioned features, liposome display shares the properties possessed by all in vitro evolutionary methods: rapid and efficient directed evolution. Liposome display can accommodate a library with a diversity of 107 (Materials and Methods), and a single round of selection can be performed in 1 d.
Liposome display has features different from other in vitro evolutionary methods. In vitro display technologies, including mRNA display (27, 28), ribosome display (29), and CIS display (30), aim to screen or select proteins based on the ligand-binding property of the displayed protein. However, one of the major functions of membrane proteins is the transport of substances across the membrane, which cannot be screened with the aforementioned techniques. In addition, membrane proteins often require oligomerization for their function (31). The aforementioned techniques cannot be used for oligomerizing proteins because monomers with different sequences may assemble and disrupt the proper genotype–phenotype linkage. Liposome display enables the evolution of both monomeric and oligomeric membrane proteins based on the transporter activity of the displayed protein. Liposome display should also be applicable for the evolution based on the ligand-binding properties of the displayed protein by using FACS or standard panning procedures.
We demonstrated that liposome display is applicable for the in vitro evolution of AH. AH is expressed in the soluble fraction and assembles on the membrane to form a pore. These properties differ from those of typical membrane proteins. Therefore, whether our method is applicable to display other membrane proteins remains unclear. Thus, we chose seven membrane proteins from E. coli, which has between 3 and 10 transmembrane domains, and investigated whether these bacterial membrane proteins could be displayed on the GUV surface (Fig. S6). First, the seven genes were fused to a myc-tag sequence at both the N and C termini. Then, each membrane protein was synthesized inside the GUV composed of POPC, stained with fluorescence-labeled anti–myc-tag antibody, and subjected to FACS analysis (Fig. S6A). Five of the seven genes showed a detectable signal (Fig. S6B), indicating the feasibility of displaying membrane proteins other than AH on the surface of the vesicle.
The IVT system has been shown to be applicable for synthesizing membrane proteins (32). By adding phospholipids in the form of a unilamellar vesicle whose size is typically 30–200 nm to the IVT, a number of membrane proteins have been shown to be integrated into the membrane from the outside of the vesicle (24). Living cells use translocases to integrate membrane proteins into the lipid bilayer (26, 33). For the IVT synthesis of several membrane proteins, the efficiency of membrane insertion has been reported to increase significantly with the translocases (34, 35). However, many of the membrane proteins have been reported to be integrated into the membrane without them, although the yield may not be as good as with the translocases. Membrane protein synthesis outside of the small vesicles (<1 μm) cannot be used for the liposome display method because the small vesicles are too small to be detected by FACS and proper genotype–phenotype linkage cannot be established. Nevertheless, the previous reports suggest the potential for successful membrane protein integration using membrane protein synthesis inside the GUV. The previous reports, together with the data shown in Fig. S6, strongly suggest the feasibility of expanding our liposome display method to other membrane proteins.
In this study, we developed an entirely in vitro liposome display method to evolve membrane proteins and we applied this method to obtain AH mutants with high pore-forming activity. The nanopore structure of AH has found recent use in emerging DNA sequencing techniques (36, 37) and as a molecule transporter (38–40). Liposome display may be used to produce AH mutants suitable for such applications. Given its high degree of controllability, ability to modify protein synthesis, and functional screening conditions, liposome display allows for the rapid and efficient evolution of a wide range of membrane proteins, including transporters and signaling proteins, thereby improving the field of membrane protein engineering.
Materials and Methods
Details of the plasmid construction and preparation of protein are provided in SI Materials and Methods.
IVT System.
The IVT system used in this study involved a reconstituted IVT system (the PURE system) prepared in the laboratory (41). The composition of the components is shown in Table S2. The template DNA used for the IVT was prepared by PCR using pIVEX2.3d-AH (42) as a template and P1 and P2 as primers. The primer sequences are shown in Table S3. The PCR assay was performed using KOD FX Neo DNA polymerase (Toyobo) according to the manufacturer’s instructions unless otherwise noted.
Preparation of GUVs.
GUVs containing the PURE system were prepared using the water-in-oil (w/o) emulsion/transfer method (16, 43), as described in our previous report (21). Briefly, 30 μL of the PURE system supplemented with the template DNA, 200 mM sucrose, 0.8 U/μL RNase inhibitor (RNasIn Plus; Promega), 1 μM transferrin Alexa Fluor 647 conjugate (TA647; Life Technologies), and 3 μM HaloTag protein (SI Materials and Methods) was added to 300 μL of liquid paraffin (Wako Pure Chemical Industries) containing 3 mg of POPC (Avanti Polar Lipids) and 3 mg of cholesterol (Nacalai Tesque). TA647 was used to estimate the aqueous volume of each vesicle by FACS (see below). The mixtures were vortexed for 30 s to form w/o emulsions, which were then equilibrated on ice for 10 min. An aliquot of 250 μL of this solution was gently placed on top of 150 μL of the outer solution (see below for composition) and centrifuged at 9,000 × g for 30 min at 4 °C. The pelleted GUVs were collected through an opening at the bottom of the tube. The GUVs were pelleted via centrifugation at 6,000 × g for 5 min at 4 °C, and the supernatant was replaced with the new outer solution. Protein synthesis inside the GUVs was conducted at 37 °C for 12 h. The outer solution contained the low-molecular weight components of the PURE system [0.3 mM of each amino acid, 3.75 mM ATP, 2.5 mM GTP, 1.25 mM CTP and UTP, 1.5 mM spermidine, 25 mM creatine phosphate, 1.5 mM DTT, 0.01 μg/μL 10-formyl-5,6,7,8-tetrahydrofolic acid, 280 mM potassium glutamate, 24.5 mM Mg[OAc]2, and 100 mM Hepes (pH 7.6)] supplemented with 200 mM glucose. After synthesis, the GUV suspension was diluted fivefold in dilution buffer [50 mM Hepes⋅KOH (pH 7.6), 280 mM potassium glutamate, 24.5 mM Mg[OAc]2, 1.5 mM DTT, and 200 mM glucose]. Following preparation, the AF488 ligand (Promega) was added for ligand influx analysis.
FACS Analysis.
The fluorescent signals from the AF488 ligand and TA647 were measured by FACS (FACSAria2; BD Biosciences). The nozzle size used was 70 μm; the flow rate was set to ∼3,000 events per second; and fluorescent detection voltages were set to 25, 400, 550, and 600 for forward scatter (FSC)-A, side scatter (SSC)-A, FITC-A, and allophycocyanin (APC)-A, respectively. The AF488 ligand was excited with a 488-nm semiconductor laser, and emission was detected through a 530 ± 15-nm bandpass filter. TA647 was excited with a HeNe laser (633 nm), and emission was detected through a 660 ± 10-nm bandpass filter. The total fluorescence intensity of the 50,000 GUVs was measured and subjected to analysis. The threshold of the detection was set to 200 for both FSC-A and SSC-A. We included 1 μM TA647 in the GUV. By measuring the TA647 fluorescence intensities of each vesicle by FACS, we estimated the number of TA647 molecules in each vesicle, which could be converted to the vesicle volume by knowing the concentration of TA647 (1 μM). The conversion was performed using the equation V = 0.0038 FI647, where V (femtoliters) is the volume of the GUVs and FI647 is the TA647 fluorescence intensity. Analysis and sorting were conducted with vesicles that satisfied log(FSC-A) > 1.5 × log(SSC-A) − 1, which is the population defined as the GUVs in our previous reports (16, 21).
Procedure of Liposome Display Rounds.
To construct a randomly mutagenized gene library, mutations were introduced into the WT AH via error-prone PCR using Mutazyme II DNA polymerase according to the manufacturer’s instructions (GeneMorph II random mutagenesis kit; Agilent Technologies). A 100-ng sample of template DNA (pIVEX2.3d-AH) and 0.5 μM P1 and P2 primers were used for PCR and amplified for 25 cycles. Under these conditions, zero to four mutations were expected per gene according to the instruction manual. Mutations were introduced only in the ORF region of the gene. The amplified gene library (5 pM) was used for protein synthesis inside the GUVs. After in vitro protein synthesis, 2 nM AF488 ligand was added to the GUVs at 37 °C and incubated for 1 h. The labeling reaction was terminated by adding 500 nM nonfluorescent HaloTag biotin ligand (Promega). Because this ligand is cell-permeable, all unlabeled HaloTag proteins were labeled with the biotin ligand and no longer reacted with the AF488 ligand. Then, FACS was used to sort the ∼10,000 GUVs that had the 1% highest fluorescence of AF488 ligand among those with a size of 6–30 fL. The DNA was purified from the sorted GUVs using a MinElute PCR Purification kit (Qiagen) and amplified by PCR using the primers P1 and P2 for inclusion in the next round of screening. Gel purification of the DNA was used to eliminate any PCR side products. Although we did not perform a mutagenic PCR assay between each round, mutations could be introduced during the PCR amplification steps. The P1 and P2 primer-amplified PCR fragments were cloned into pET-gusA (44) using an In-Fusion HD kit (TAKARA BIO) for DNA sequencing when necessary. The concentration of T7 RNA polymerase in the PURE system was reduced from 100 nM to 50 nM after the 16th cycle to increase the selection pressure.
Our gene-screening system was able to screen a genetic diversity of ∼107 in 1 d (20). Although FACS can analyze and sort more than 109 vesicles per day, each GUV should contain only a single copy of the gene. To achieve this goal, the DNA concentration should be lowered such that only 1 in 100 compartments actually contains DNA, limiting the manageable level of diversity to 107.
Quantification of Membrane-Bound AH.
GUVs were prepared as described previously above without template DNA, pelleted by centrifugation at 6,000 × g for 5 min at 4 °C, and suspended with the PURE system, including 15 pM various AH DNA and [35S]methionine (NEG009T; PerkinElmer). After incubation at 37 °C for 4 h, GUVs were washed three times by centrifugation at 6,000 × g for 5 min at 25 °C and the supernatant was replaced with dilution buffer. The washed GUVs were applied to SDS/PAGE without boiling, and the as-obtained gel was analyzed using a Typhoon FLA7000 laser scanner (GE Healthcare).
Supplementary Material
Acknowledgments
We thank Ms. Hitomi Komai, Tomomi Sakamoto, and Ryoko Otsuki for their technical assistance. We also thank Yoshikazu Tanaka (Hokkaido University) for discussions regarding the AH structure and Vincent Noireaux (University of Minnesota) for his kind gift of the pIVEX2.3d-AH plasmid. This research was supported in part by the “Global Centers of Excellence Program” of the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314585110/-/DCSupplemental.
References
- 1.Endo Y, Sawasaki T. Cell-free expression systems for eukaryotic protein production. Curr Opin Biotechnol. 2006;17(4):373–380. doi: 10.1016/j.copbio.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 2.Carlson ED, Gan R, Hodgman CE, Jewett MC. Cell-free protein synthesis: Applications come of age. Biotechnol Adv. 2012;30(5):1185–1194. doi: 10.1016/j.biotechadv.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsuura T, Yomo T. In vitro evolution of proteins. J Biosci Bioeng. 2006;101(6):449–456. doi: 10.1263/jbb.101.449. [DOI] [PubMed] [Google Scholar]
- 4.Amstutz P, Forrer P, Zahnd C, Plückthun A. In vitro display technologies: Novel developments and applications. Curr Opin Biotechnol. 2001;12(4):400–405. doi: 10.1016/s0958-1669(00)00234-2. [DOI] [PubMed] [Google Scholar]
- 5.Keefe AD, Szostak JW. Functional proteins from a random-sequence library. Nature. 2001;410(6829):715–718. doi: 10.1038/35070613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yildirim MA, Goh KI, Cusick ME, Barabási AL, Vidal M. Drug-target network. Nat Biotechnol. 2007;25(10):1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 7.Stevens TJ, Arkin IT. Do more complex organisms have a greater proportion of membrane proteins in their genomes? Proteins. 2000;39(4):417–420. doi: 10.1002/(sici)1097-0134(20000601)39:4<417::aid-prot140>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 8.Brill S, Falk OS, Schuldiner S. Transforming a drug/H+ antiporter into a polyamine importer by a single mutation. Proc Natl Acad Sci USA. 2012;109(42):16894–16899. doi: 10.1073/pnas.1211831109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarkar CA, et al. Directed evolution of a G protein-coupled receptor for expression, stability, and binding selectivity. Proc Natl Acad Sci USA. 2008;105(39):14808–14813. doi: 10.1073/pnas.0803103105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hajduczki A, Majumdar S, Fricke M, Brown IA, Weiss GA. Solubilization of a membrane protein by combinatorial supercharging. ACS Chem Biol. 2011;6(4):301–307. doi: 10.1021/cb1001729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlinkmann KM, et al. Critical features for biosynthesis, stability, and functionality of a G protein-coupled receptor uncovered by all-versus-all mutations. Proc Natl Acad Sci USA. 2012;109(25):9810–9815. doi: 10.1073/pnas.1202107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bokma E, Koronakis E, Lobedanz S, Hughes C, Koronakis V. Directed evolution of a bacterial efflux pump: Adaptation of the E. coli TolC exit duct to the Pseudomonas MexAB translocase. FEBS Lett. 2006;580(22):5339–5343. doi: 10.1016/j.febslet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Young EM, Comer AD, Huang H, Alper HS. A molecular transporter engineering approach to improving xylose catabolism in Saccharomyces cerevisiae. Metab Eng. 2012;14(4):401–411. doi: 10.1016/j.ymben.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 14.Song L, et al. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science. 1996;274(5294):1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 15.Shimizu Y, et al. Cell-free translation reconstituted with purified components. Nat Biotechnol. 2001;19(8):751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 16.Nishimura K, et al. Population analysis of structural properties of giant liposomes by flow cytometry. Langmuir. 2009;25(18):10439–10443. doi: 10.1021/la902237y. [DOI] [PubMed] [Google Scholar]
- 17.Los GV, et al. HaloTag: A novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol. 2008;3(6):373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 18.Walker B, Krishnasastry M, Zorn L, Bayley H. Assembly of the oligomeric membrane pore formed by Staphylococcal alpha-hemolysin examined by truncation mutagenesis. J Biol Chem. 1992;267(30):21782–21786. [PubMed] [Google Scholar]
- 19.Sunami T, et al. Femtoliter compartment in liposomes for in vitro selection of proteins. Anal Biochem. 2006;357(1):128–136. doi: 10.1016/j.ab.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 20.Nishikawa T, Sunami T, Matsuura T, Ichihashi N, Yomo T. Construction of a gene screening system using giant unilamellar liposomes and a fluorescence-activated cell sorter. Anal Chem. 2012;84(11):5017–5024. doi: 10.1021/ac300678w. [DOI] [PubMed] [Google Scholar]
- 21.Nishimura K, et al. Cell-free protein synthesis inside giant unilamellar vesicles analyzed by flow cytometry. Langmuir. 2012;28(22):8426–8432. doi: 10.1021/la3001703. [DOI] [PubMed] [Google Scholar]
- 22.Valeva A, et al. Evidence that clustered phosphocholine head groups serve as sites for binding and assembly of an oligomeric protein pore. J Biol Chem. 2006;281(36):26014–26021. doi: 10.1074/jbc.M601960200. [DOI] [PubMed] [Google Scholar]
- 23.Palmer M, Weller U, Messner M, Bhakdi S. Altered pore-forming properties of proteolytically nicked staphylococcal alpha-toxin. J Biol Chem. 1993;268(16):11963–11967. [PubMed] [Google Scholar]
- 24.Katzen F, Peterson TC, Kudlicki W. Membrane protein expression: No cells required. Trends Biotechnol. 2009;27(8):455–460. doi: 10.1016/j.tibtech.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 25.Kawate T, Gouaux E. Arresting and releasing Staphylococcal α-hemolysin at intermediate stages of pore formation by engineered disulfide bonds. Protein Sci. 2003;12(5):997–1006. doi: 10.1110/ps.0231203 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papanikou E, Karamanou S, Economou A. Bacterial protein secretion through the translocase nanomachine. Nat Rev Microbiol. 2007;5(11):839–851. doi: 10.1038/nrmicro1771. [DOI] [PubMed] [Google Scholar]
- 27.Nemoto N, Miyamoto-Sato E, Husimi Y, Yanagawa H. In vitro virus: Bonding of mRNA bearing puromycin at the 3′-terminal end to the C-terminal end of its encoded protein on the ribosome in vitro. FEBS Lett. 1997;414(2):405–408. doi: 10.1016/s0014-5793(97)01026-0. [DOI] [PubMed] [Google Scholar]
- 28.Roberts RW, Szostak JW. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc Natl Acad Sci USA. 1997;94(23):12297–12302. doi: 10.1073/pnas.94.23.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanes J, Plückthun A. In vitro selection and evolution of functional proteins by using ribosome display. Proc Natl Acad Sci USA. 1997;94(10):4937–4942. doi: 10.1073/pnas.94.10.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Odegrip R, et al. CIS display: In vitro selection of peptides from libraries of protein-DNA complexes. Proc Natl Acad Sci USA. 2004;101(9):2806–2810. doi: 10.1073/pnas.0400219101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodsell DS, Olson AJ. Structural symmetry and protein function. Annu Rev Biophys Biomol Struct. 2000;29:105–153. doi: 10.1146/annurev.biophys.29.1.105. [DOI] [PubMed] [Google Scholar]
- 32.Schwarz D, Dötsch V, Bernhard F. Production of membrane proteins using cell-free expression systems. Proteomics. 2008;8(19):3933–3946. doi: 10.1002/pmic.200800171. [DOI] [PubMed] [Google Scholar]
- 33.Xie K, Dalbey RE. Inserting proteins into the bacterial cytoplasmic membrane using the Sec and YidC translocases. Nat Rev Microbiol. 2008;6(3):234–244. doi: 10.1038/nrmicro3595. [DOI] [PubMed] [Google Scholar]
- 34.Kuruma Y, Nishiyama K, Shimizu Y, Müller M, Ueda T. Development of a minimal cell-free translation system for the synthesis of presecretory and integral membrane proteins. Biotechnol Prog. 2005;21(4):1243–1251. doi: 10.1021/bp049553u. [DOI] [PubMed] [Google Scholar]
- 35.Wuu JJ, Swartz JR. High yield cell-free production of integral membrane proteins without refolding or detergents. Biochim Biophys Acta. 2008;1778(5):1237–1250. doi: 10.1016/j.bbamem.2008.01.023. [DOI] [PubMed] [Google Scholar]
- 36.Cherf GM, et al. Automated forward and reverse ratcheting of DNA in a nanopore at 5-Å precision. Nat Biotechnol. 2012;30(4):344–348. doi: 10.1038/nbt.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci USA. 1996;93(24):13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Movileanu L, Cheley S, Bayley H. Partitioning of individual flexible polymers into a nanoscopic protein pore. Biophys J. 2003;85(2):897–910. doi: 10.1016/S0006-3495(03)74529-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Rapid nanopore discrimination between single polynucleotide molecules. Proc Natl Acad Sci USA. 2000;97(3):1079–1084. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Microsecond time-scale discrimination among polycytidylic acid, polyadenylic acid, and polyuridylic acid as homopolymers or as segments within single RNA molecules. Biophys J. 1999;77(6):3227–3233. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsuura T, Kazuta Y, Aita T, Adachi J, Yomo T. Quantifying epistatic interactions among the components constituting the protein translation system. Mol Syst Biol. 2009;5:297. doi: 10.1038/msb.2009.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noireaux V, Libchaber A. A vesicle bioreactor as a step toward an artificial cell assembly. Proc Natl Acad Sci USA. 2004;101(51):17669–17674. doi: 10.1073/pnas.0408236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pautot S, Frisken BJ, Weitz DA. Production of unilamellar vesicles Using an inverted emulsion. Langmuir. 2003;19(7):2870–2879. [Google Scholar]
- 44.Matsuura T, Hosoda K, Ichihashi N, Kazuta Y, Yomo T. Kinetic analysis of β-galactosidase and β-glucuronidase tetramerization coupled with protein translation. J Biol Chem. 2011;286(25):22028–22034. doi: 10.1074/jbc.M111.240168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.