Significance
Natural killer (NK) cell function is critically regulated by inhibitory receptors for MHC class I. This work shows that peptides derived from both host and viruses can engage CD94 in the absence of a signalling partner and augment inhibition of NK cells expressing NK cell receptor gene 2A. This result establishes CD94 as a receptor that binds HLA-E in a peptide-dependent fashion. It also demonstrates that NK cells expressing inhibitory receptors from the killer cell immunoglobulin-like receptor or C-type lectin-like receptor families respond with different stoichiometries to changes in the levels of cell-surface MHC class I. Thus these two receptor families may have distinct but complementary functions in recognizing changes in MHC class I.
Keywords: innate immunity, MHC-I peptides
Abstract
Peptide selectivity is a feature of inhibitory receptors for MHC class I expressed by natural killer (NK) cells. CD94–NKG2A operates in tandem with the polymorphic killer cell Ig-like receptors (KIR) and Ly49 systems to inhibit NK cells. However, the benefits of having two distinct inhibitory receptor–ligand systems are not clear. We show that noninhibitory peptides presented by HLA-E can augment the inhibition of NKG2A+ NK cells mediated by MHC class I signal peptides through the engagement of CD94 without a signaling partner. Thus, CD94 is a peptide-selective NK cell receptor, and NK cells can be regulated by nonsignaling interactions. We also show that KIR+ and NKG2A+ NK cells respond with differing stoichiometries to MHC class I down-regulation. MHC-I–bound peptide functions as a molecular rheostat controlling NK cell function. Selected peptides which in isolation do not inhibit NK cells can have different effects on KIR and NKG2A receptors. Thus, these two inhibitory systems may complement each other by having distinct responses to bound peptide and surface levels of MHC class I.
Natural killer (NK) cells play an important role in the immune response to viral infections and cancer. Their responses are determined by signals integrated from activating and inhibitory receptor–ligand interactions (1). In many situations inhibitory signals dominate activating signals. Therefore, releasing NK cells from inhibition is an important mechanism of enhancing their response to target cells. Inhibitory interactions are mediated by receptors for self-MHC class I. Most species have at least two discrete gene families of inhibitory receptors for MHC class I: the CD94–NKG2A C-type lectin-like receptor system and either the related Ly49 family of receptors or the unrelated killer cell Ig-like receptors (KIR) (2). The KIR family is important in humans and other primates, having undergone extensive diversification under positive selection. In contrast, the CD94–NKG2A system has remained relatively well conserved across the species with orthologous genes in primates and a closely related functional homolog in rodents (3, 4). Consistent with the coevolution of these families and their MHC class I ligands, KIR bind polymorphic MHC class I, HLA-A, -B, and -C molecules, whereas CD94–NKG2A binds the conserved oligomorphic HLA-E molecule or the rodent homolog Qa-1 (5, 6).
Both receptor families are important in the immune response to viral infections. KIR are genetic determinants in the outcome of both HIV and hepatitis C virus (HCV) infection (7–10). Expression of CD94–NKG2A is up-regulated on NK cells in HIV and HCV infection and in the latter has been associated with a poor response to treatment (11, 12). Furthermore NKG2A+ NK cell clones lyse vaccinia-infected targets (13), and CD94 is important in clearing mouse pox infection (14). Both KIR and CD94–NKG2A respond to MHC class I down-regulation. One hypothesis is that the KIR have evolved to recognize MHC class I-specific down-regulation (15). However, because the majority of MHC class I leader peptides bind HLA-E and are inhibitory for NKG2A, the CD94–NKG2A system also is able to recognize down-regulation of most MHC class I alleles. It has been shown that KIR+ NK cells can be modulated by changes in the peptide bound by MHC class I, which confers additional functionality on the KIR system (16–18). In particular peptide antagonism is a potent mechanism for activating KIR+ NK cells (19, 20). The CD94–NKG2A receptor also is peptide selective, with receptor binding being particularly influenced by residues 5, 6, and 8 of the peptide bound by HLA-E (21–23). These residues interact primarily with the nonsignaling CD94 moiety, which occupies the majority of the HLA-E–binding interface. CD94–NKG2A seems to be a target for viral escape, with peptides derived from CMV, HCV, HIV, and EBV binding HLA-E and subsequently inhibiting NK cells (24–27). Viral peptides that inhibit at KIR also are identifiable (28), but their relevance likely is limited to the subset of individuals who have the relevant peptide-binding MHC class I allele. Understanding differences in how the KIR and NKG2 systems respond to peptide may be important for interpreting their roles in the immune response to viral infections and tumors. Therefore we explored how HLA-E–bound peptide can influence NK cell reactivity.
Results
HCV Core35–44 Stabilizes HLA-E but Requires HLA Class I Leader Peptides to Inhibit NKG2A+ NK Cells.
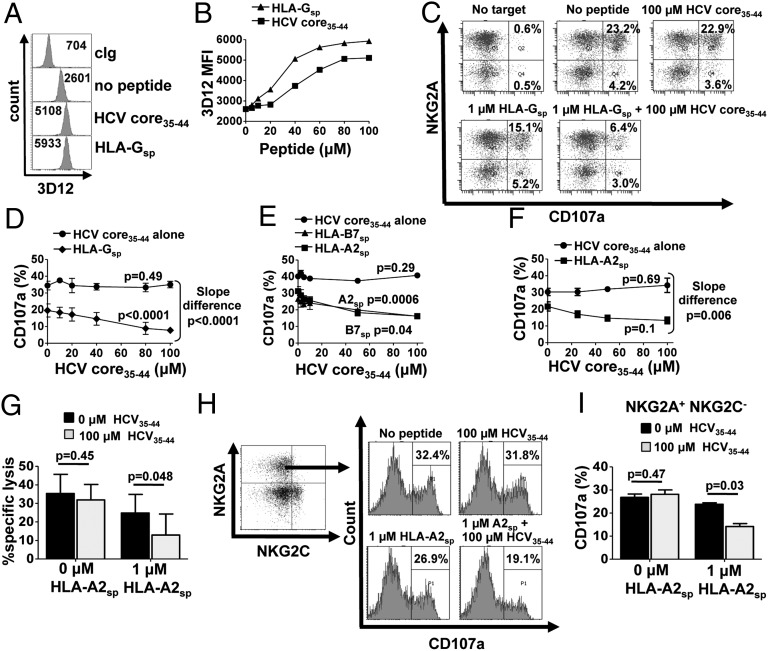
HCV core35–44 (YLLPRRGPRL) is a known HLA-A2 CTL epitope that also up-regulates HLA-E expression (27). Therefore we investigated whether HCV core35–44 could inhibit NK cells via NKG2A. To avoid the confounding effects of endogenous peptides, we used the transporters associated with antigen processing (TAP)-deficient cell line 721.174 (.174), which constitutively expresses HLA-E. The .174 cells were incubated overnight with 0–100 μM of HCV core35–44 or with the HLA-E–binding HLA-G signal peptide (HLA-Gsp,VMAPRTLFL) and were stained for cell-surface HLA-E expression. In the absence of exogenous peptide, HLA-E expression was detectable on the cell surface. Both HLA-Gsp and HCV core35–44 enhanced cell-surface HLA-E, with expression saturating at ∼100 µM (Fig. 1 A and B). To determine the inhibitory effect of HLA-E stabilization, degranulation assays were performed using IL-15–stimulated NK cells and gating on the NKG2A+ subset (Fig. 1C). Just 1 µM of HLA-Gsp was sufficient to inhibit about 35% of the .174 reactive NKG2A+ NK cells. However, no inhibition was noted with HCV core35–44 peptide, even at saturating levels of peptide. In vivo HLA-E presents a mixture of peptides derived from the leader sequences of HLA class I molecules, and during infection viral peptides also may be presented (26). To rationalize our findings with previous work, we performed further degranulation assays using combinations of HCV core35–44 and MHC class I leader peptides (27). In the presence of HCV core35–44, inhibition of killing mediated by 1 µM HLA-Gsp was enhanced twofold (Fig. 1 C and D). Titrating HCV core35–44 with 1 µM HLA-Gsp demonstrated that this synergistic inhibitory effect was dose dependent (P < 0.0001; ANOVA) (Fig. 1D). Furthermore there was a significant difference in the slope of the HCV core35–44 peptide titration in the presence or absence of HLA-Gsp (P < 0.0001).
Fig. 1.
HCV core35–44 inhibits NKG2A+ NK cells only in the presence of an MHC class I leader peptide. (A and B) Stabilization of HLA-E on .174 cells, either in the absence of peptide or loaded with 0–100 µM HCV core35–44 or HLA-Gsp, as determined by flow cytometry. The mean fluorescence intensity (MFI) of 3D12 staining is shown in the histograms (A) and is plotted against peptide concentration (B). (C) CD107a assay of NKG2A+ NK cells in response to 0–100 µM HCVcore35–44 or HLA-Gsp alone or in combination. Dot plots are gated on CD3− CD56+ CD158b− lymphocytes. (D) CD3− CD56+ NKG2A+ CD158b− NK cell degranulation in response to increasing concentrations of HCV core35–44 in the presence or absence of 1 µM HLA-Gsp. Data shown are the mean ± SEM of three independent experiments. (E and F) CD3− CD56+ NGK2A+ CD158b− NK cell degranulation to .174 cells loaded with HCV core35–44 in the presence or absence of HLA-A2sp or HLA-B7sp for a healthy donor (E) and three HCV donors (F). (G) Cytotoxicity assay of sorted NKG2A+ NK cells using .174 targets incubated with the indicated peptides at an E:T ratio of 10:1. Data shown are the mean and SEM of three independent experiments from three different donors. (H and I) CD107a assay gating on NKG2A+ NKG2C− NK cells stimulated with .174 cells loaded with the indicated peptides. One representative experiment is shown in H, and the mean + SEM from three independent experiments is shown in I. Slope difference P values in D, E, and F were calculated using linear regression comparing the difference the between the two lines. All other P values were calculated using one-way ANOVA.
Because HLA-G is expressed primarily in the fetal trophoblast, we repeated experiments with the HCV core35–44 peptide mix using leader peptides derived from the more widely expressed classical HLA class I molecules HLA-A2 (HLA-A2sp, VMAPRTLVL) and HLA-B7 (HLA-B7sp, VMAPRTVLL). As was observed with HLA-Gsp, the presence of HCV core35–44 peptide significantly enhanced NKG2A-mediated inhibition of NK cells by HLA-A2sp (P = 0.0006) or HLA-B7sp (P = 0.04) (Fig. 1E). The synergy of HCV core35–44 with HLA-A2sp was confirmed in peripheral blood mononuclear cells (PBMCs) from HCV-infected individuals (Fig. 1F) and from three additional healthy donors (Fig. S1) and in cytotoxicity assays using sorted NKG2A+ NK cells (Fig. 1G and Fig. S2).
CD94–NKG2C is an activating NK cell receptor that is triggered by HLA-E, although it binds with a much lower affinity than CD94–NKG2A (29). Synergistic inhibition of NK cells was observed on NKG2A+NKG2C− NK cells (Fig. 1 H and I). Therefore, NKG2C is not required for peptide synergism. In addition, as is consistent with the lower affinity of NKG2C for HLA-E, we observed no activation of NKG2C+ NK cells at these low levels of cognate MHC class I leader peptide in the presence or absence of HCV core35–44. Nor did NKG2C have any significant effect on the synergy of inhibition of NKG2A+ NK cells (Fig. S3).
Synergistic Inhibition at CD94–NKG2A Occurs with a Broad Range of Unrelated Peptides.
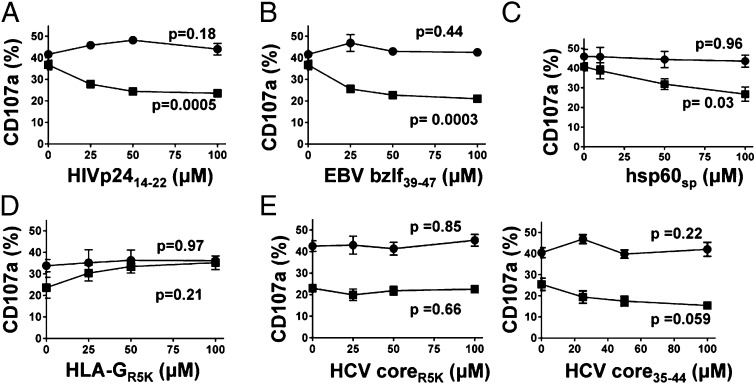
Peptides derived from a variety of viruses, including EBV, HIV, and CMV, are known to bind to HLA-E (24–26). The HIV p24 peptide residues 14–22 (HIVp2414–22,AISPRTLNA) was identified by a motif-based approach, and in chronic HIV infection the up-regulation of HLA-E on CD4+ T cells results in increased inhibition of NKG2A+ NK cells (25, 30). The peptide SQAPLPCVL from EBV BZLF-1 protein residues 39–47 (EBVbzlf39–47) also has been shown to bind to HLA-E (24). Although both these viral peptides stabilized cell-surface HLA-E in the .174 cell line (Fig. S4), in isolation they did not inhibit NKG2A+ NK cells (Fig. 2 A and B). However, in the presence of 1 µM HLA-A2sp, these peptides had a synergistic inhibitory effect in a dose-dependent manner. Similarly, hsp60sp, which also is known to bind to HLA-E but does not inhibit at CD94–NKG2A, had a synergistic effect on the inhibition mediated by HLA-A2sp (Fig. 2C). Thus, synergy at CD94–NKG2A can be induced by both host and viral peptides and is a general mechanism governing NK- cell reactivity.
Fig. 2.
Synergy at NKG2A occurs with a broad range of peptides. Degranulation of CD3− CD56+ NKG2A+ CD158b− NK cells in response to increasing concentrations of the following peptides in the presence (■) or absence (●) of 1 µM HLA-A2sp: (A) HIV p2414–22; (B) EBV BZLF39–47; (C) hsp60sp; (D) the HLA-E–binding mutant HLA-GR5K; and (E) the HCV coreR5K mutant (Left) and the control HCVcore35–44 for this experiment (Right). Data are shown as mean ± SEM of three independent experiments. All P values were calculated using one-way ANOVA.
Previous studies have shown that that replacing the P5 arginine residue of HLA-Gsp with lysine reduces the affinity of the peptide/HLA-E complex for CD94–NKG2A by more than 10-fold (23). The P5 residue of wild-type peptide is in direct contact with CD94 and NKG2A (22). We synthesized the HLA-Gsp peptide with this lysine substitution, VMAPKTLFL (HLA-GR5K). HLA-GR5K stabilized HLA-E to a similar extent as the wild-type peptide but did not induce inhibition of NKG2A+ NK cells (Fig. 2D and Fig. S4). However, the addition of HLA-GR5K to HLA-A2sp did not result in synergy; instead, increasing concentrations of HLA-GR5K relieved HLA-A2sp–mediated NK cell inhibition. Furthermore making an equivalent substitution in the HCV core35–44 peptide (HCV coreR5K) failed to augment inhibition in the presence of HLA-A2sp (Fig. 2E), in contrast to the wild-type HCV core35–44. Thus, synergy at HLA-E is peptide specific and requires an interaction with either CD94 or NKG2A, an interaction which may be abrogated by P5 lysine substitution.
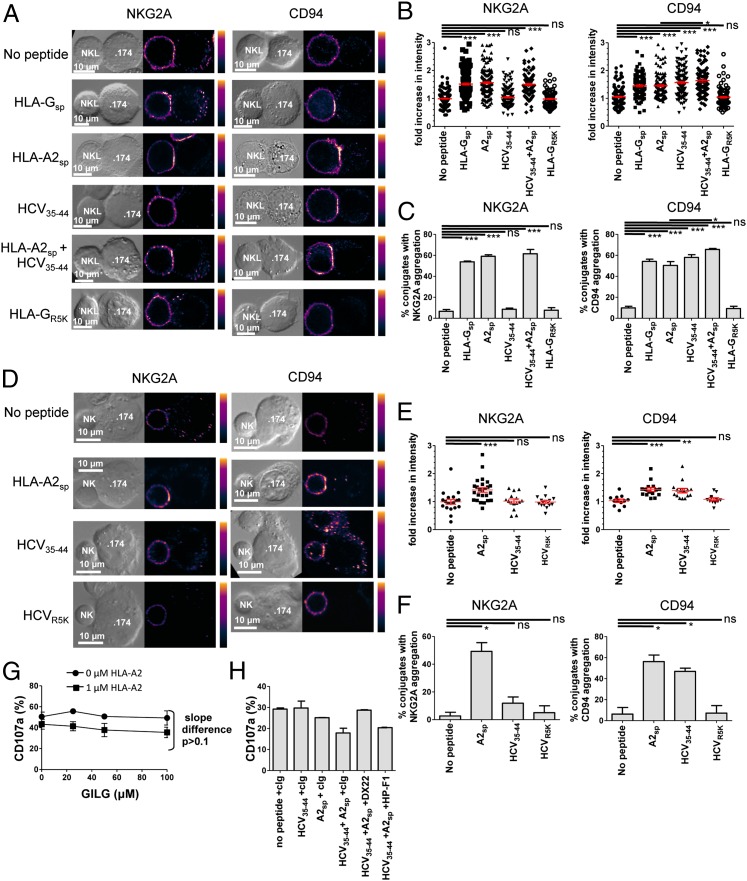
CD94 Clustering at the Immune Synapse Is Peptide Specific and Occurs in the Absence of a Signaling Partner.
The signaling component of the CD94–NKG2A heterodimer is NKG2A, which possesses two immunoreceptor tyrosine-based inhibitory motifs (ITIMs) within its cytoplasmic tail, whereas CD94 contains no signaling motifs (31, 32). We used confocal microscopy to examine the effect of peptides presented by HLA-E on the clustering of CD94 and NKG2A at the immunological synapse formed between effector and target cells. NKL cells were coincubated with .174 cells that had been loaded with peptide (HLA-Gsp, HLA-A2sp, HCV core35–44, HLA-A2sp+HCV core35–44, or HLA-GR5K). The target and effector-cell complexes were stained for CD94 or NKG2A. The MHC class I inhibitory peptides HLA-Gsp and HLA-A2sp induced aggregation of both CD94 and NKG2A (Fig. 3 A–C). However, HCV core35–44 induced clustering of CD94 (P < 0.0001; one-way ANOVA) but not of NKG2A at the immune synapse (P > 0.05). The combination of HCV core35–44 and HLA-A2sp resulted in greater CD94 aggregation (both fold-increase intensity and as percentage of total aggregates, P < 0.05) compared with HLA-A2sp alone, implying recruitment of additional CD94-associated complexes. In contrast, HLA-GR5K did not induce aggregation of either CD94 or NKG2A. Confocal microscopy also was performed using primary NK cells (Fig. 3 D–F). Again, HCV core35–44 promoted clustering of CD94 but not of NKG2A, in contrast to HCV coreR5K, which did not induce aggregation of either subunit. Conversely HLA-A2sp induced clustering of both CD94 and NKG2A. These data therefore demonstrate that, like NKL cells, primary NK cells can aggregate CD94 without clustering NKG2A. Thus, a synergistic peptide induces aggregation of the nonsignaling molecule CD94 in the absence of the signaling moiety NKG2A, indicative of binding to CD94 homodimers. Consistent with this model, we observed no synergy between HLA-A2sp and GILGFVFTL which stabilizes HLA-A2 molecules but not HLA-E and therefore does not interact with CD94 (Fig. 3G and Fig. S4). Furthermore in blocking experiments, preincubation of PBMCs with anti-CD94 completely abrogated the synergistic inhibitory effect of HCV core35–44 in the presence of 1 µM HLA-A2sp, but no effect was observed with the LILRB1-specific antibody HP-F1 (Fig. 3H).
Fig. 3.
HCV core35–44 induces aggregation of CD94 but not NKG2A at the interface between NK and target cells. (A) Comparison of NKG2A (Left) and CD94 (Right) clustering at the interface between NKL effector and .174 target cells, loaded with the indicated peptides. (B) Fold increase of fluorescence intensity at the interface between NKL and .174 target cells compared with a noncontact area of the NKL plasma membrane. (C) The percentage of conjugates with aggregation at the interface between NK cells and target cells. For B and C 100–150 conjugates were analyzed for each condition. (D–F) Clustering of NKG2A (Left) and CD94 (Right) between primary NK cells and .174 target cells as in A–C. E shows the fold increase in intensity, and F shows the percentage of conjugates with aggregation at the interface between the NK cells and .174 target cells. For each condition 20–30 conjugates were analyzed. (G) Degranulation of CD3− CD56+ NKG2A+ CD158b− NK cells in response to increasing concentrations of the HLA-A2–binding peptide GILGFVFTL (GILG) in the presence or absence of 1 µM HLA-A2sp. (H) Degranulation assay of CD3− CD56+ CD158b− NK cells incubated with .174 cells in the presence or absence of the indicated peptides and antibodies to CD94 (DX22), LILRB-1 (HP-F1), or an isotype control antibody (cIg). Data are shown as mean ± SEM of three independent experiments. Statistical analyses were performed using ANOVA with Dunnett’s post test to compare individual pairs of conditions. *P < 0.05; **P < 0.001; ***P < 0.001; ns, nonsignificant.
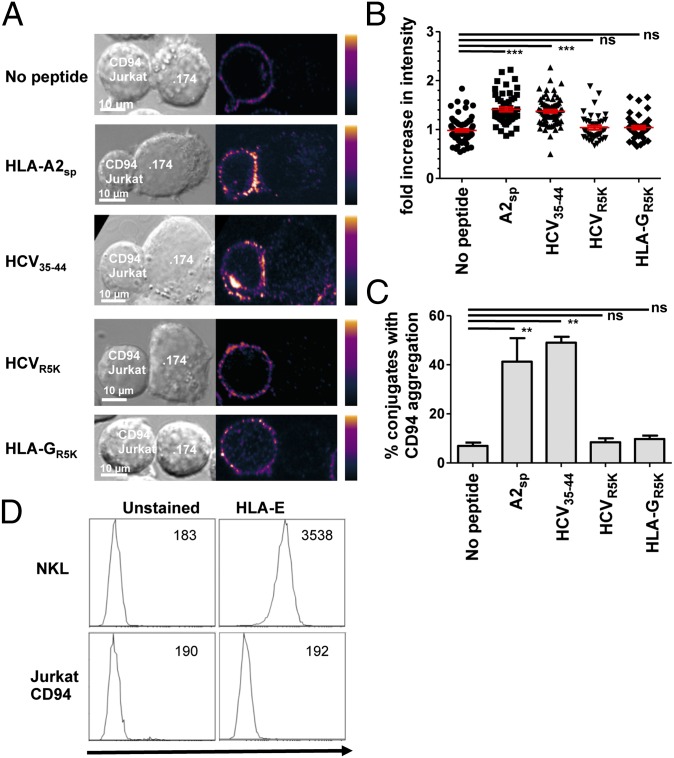
To confirm that HLA-E loaded with HCV core35–44 could engage cell-surface CD94 in the absence of NKG2A, we tested clustering of CD94 on Jurkat-CD94 transfectants. These express high levels of CD94 as a homodimer on the cell surface (33). HCV core35–44 and HLA-A2sp induced clustering of CD94 on the cell surface, whereas HLA-GR5K and HCV coreR5K did not (Fig. 4 A–C). Furthermore pentamers of HLA-E and the signal peptide VMAPRTLIL, which binds to CD94–NKG2A, do not bind to CD94 on Jurkat, indicating that the avidity of the HLA-E+HLA-A2sp for CD94 alone is lower than for the CD94–NKG2A heterodimer (Fig. 4D). Thus, the nature of the bound peptide determines whether the CD94–NKG2A heterodimer or CD94 in the absence of NKG2A is preferentially engaged.
Fig. 4.
CD94 is a peptide-specific receptor for HLA-E. (A) Clustering of CD94 on the surface of Jurkat–CD94 transfectants in the presence of the indicated peptides. (B and C) Fold increase in intensity (B) and percentage of aggregates with conjugates from the Jurkat–CD94 clustering experiments (C). Overall, 40–60 conjugates were analyzed for each condition. Data are shown as mean + SEM of three independent experiments. (D) Pentamer binding of HLA-E and HLA leader peptide to NKL but not to Jurkat–CD94 transfectants. The mean fluorescence intensity of each histogram is indicated. Statistical analyses were performed using ANOVA with Dunnett’s post test to compare individual pairs of conditions. **P < 0.01; ***P < 0.001; ns, nonsignificant.
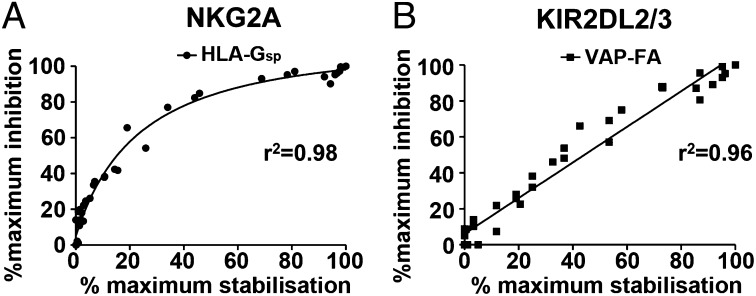
Our previous data showed peptide antagonism for KIR2DL2/3+ NK cells, in contrast to our observation of peptide synergy for NKG2A NK cells (19). These data imply that KIR and NKG2A may have different roles in recognizing changes in MHC class I. We therefore hypothesized that, in addition to responding differently to changes in MHC-bound peptide, KIR+ and NKG2A+ NK cells also may respond differently to MHC class I down-regulation. To investigate this premise, we studied two potent inhibitory peptides; the HLA-G leader peptide for NKG2-mediated inhibition and the peptide VAPWNSFAL, which had the highest affinity for KIR2DL2 and KIR2DL3 in a peptide screen (19, 21). Degranulation assays using peptide titrations showed that inhibition of NK cells by MHC class I exhibited saturation kinetics for NKG2A+ NK cells (r2 = 0.98) but was linear for KIR+ NK cells (r2 = 0.96) (Fig. 5). Thus, more NKG2A+ NK cells respond to changes in MHC class I at low HLA-E levels than at high levels. However, the level of MHC does not appear to influence the fraction of KIR+ NK cells that respond to a given change in HLA-C expression. Thus, NKG2A+ NK cells are tuned to respond to changes in MHC class I at low levels of MHC expression, but KIR+ NK cells are not.
Fig. 5.
NKG2A+ but not KIR+ NK cells are tuned to low cell-surface levels of MHC class I. .174 cells were incubated with increasing concentrations of HLA-Gsp or VAPWNSFAL (VAP-FA) before the stabilization of HLA-E (A) or HLA-Cw*0102 (B) was measured. They were then used as targets in degranulation assays for (A) NKG2A+ CD158b− (NKG2A+) or (B) NKG2A− CD158b+ (KIR2DL2/3+) NK cells. The levels of cell-surface MHC class as determined by flow cytometry and the fraction of degranulating NK cells were then correlated. The data for MHC class I stabilization are normalized to “no peptide” (0% stabilization) and “100 μM peptide” (100% stabilization) concentrations. For degranulation, data are normalized to CD107a expression for .174 cells incubated in the absence of peptide (0% inhibition) or the presence of 100 μM peptide (100% inhibition).
Discussion
Our data demonstrate that CD94 in the absence of NKG2A has a distinct specificity for HLA-E–peptide complexes. The combination of peptides that bind the CD94–NKG2A heterodimer with those binding only CD94 results in a synergistic inhibition of NKG2A+ NK cells. The CD94-binding synergistic peptides can be either host- and virus-derived and hence represent a diverse potential source. These peptides do not comply to a specific canonical motif but, consistent with the HLA-E peptide-binding groove, contain mainly hydrophobic residues. Peptide elution studies have shown that when TAP function is impaired the repertoire of peptides presented by HLA-E can be expanded substantially to present peptides other than leader sequences (34). Thus it may be difficult to predict the range of peptides that can act as CD94 binders.
In our model system we have shown that peptides that stabilize HLA-C, but do not themselves inhibit NK cells through KIR, can antagonize rather than synergize the inhibition of NK cells (19, 20). Thus, we have demonstrated differences in the ways in which the KIR and the NKG2A systems respond to changes in the peptide content of MHC class I; these observations could be generalizable to other KIR– or CD94–NKG2A HLA–peptide interactions. Changes in the peptide repertoire of HLA-E can favor the inhibition of NKG2A+ NK cells, but peptide repertoire changes at HLA-C favor the activation of KIR+ NK cells. This difference is related to the ability of CD94 to bind to HLA-E–peptide complexes in the absence of the inhibitory signaling moiety NKG2A. Viruses therefore could exploit the tolerance of the HLA-E–CD94–NKG2A system by generating HLA-E–binding peptides to enhance the inhibition of NKG2A+ NK cells. The evolution of additional receptor families, either KIR or Ly49, provides the host with a second inhibitory receptor family for MHC class I for which peptide changes are more likely to lead to NK cell activation. It has been proposed that the major function of KIR is to recognize the down-regulation of specific MHC class I allotypes. However, most HLA class I allotypes express leader peptides cognate for CD94–NKG2A. Therefore we propose that, given the sensitivity of CD94–NKG2A+ NK cells to changes in MHC class I at low levels of expression, this function also can be subserved by CD94–NKG2A. Conversely KIR+ NK cells can respond efficiently to changes in peptide repertoire and thus provide the host with selective advantage over the NKG2A system (19).
The observed synergistic effect of HLA-E–binding peptides involves engagement of CD94 in the absence of NKG2A, most likely related to CD94 homodimers. HLA-E binding to CD94–NKG2A is dominated by the CD94 moiety, and expression of CD94 on the Jurkat transfectants is predominantly in the homodimeric form (22, 32). X-ray crystallography suggests that noncovalently linked CD94 homodimers can form spontaneously (35). Furthermore CD94 homodimers have been demonstrated in transfection studies, and CD94 is also expressed on the cell surface in the absence of signaling NKG2 partners (32, 36). Thus, there is potential for CD94 to interact with HLA-E in the absence of a signaling partner, and the relatively low sequence identity between CD94 and NKG2A (∼30%) may lead to an alteration in peptide specificity. However, some peptide specificity is conserved, because the HLA-GR5K and the mutated HCV coreR5K peptides did not induce CD94 aggregation.
Inhibitory signaling by CD94–NKG2A is complex at both structural and signaling levels (37). Thus, the inhibitory synapse may consist of several different receptor–ligand interactions including ICAM–LFA1 and 2B4–CD48 in addition to those of KIR or CD94–NKG2A with MHC class I (38, 39). Kirwan and Burshtyn (40) previously have shown that KIR2DL1 with mutated ITIMs can cooperate with LILRB1 to facilitate inhibitory signaling, indicating that a nonsignaling receptor can augment inhibitory signaling. Thus, inhibitory signaling may be modulated by noninhibitory receptor–ligand interactions in addition to an HLA-E–peptide–dependent effect. The mechanism for synergy is not clear, but one potential model could be based on receptor oligomerization. In this model, for signaling to occur, inhibitory NK cell receptors may need to be clustered at a critical density. At low concentrations of MHC-class I signal peptides there may be insufficient HLA-E expression to achieve adequate CD94–NKG2A clustering at the immune synapse, leading to a relatively weak inhibitory signal. Synergistic peptides induce up-regulation of HLA-E and may permit recruitment and clustering of CD94 homodimers cognate for the peptide–HLA-E complex but not CD94–NKG2A heterodimers. If these CD94 molecules bind noncovalently to either an adjacent CD94 or NKG2A molecule or a second HLA-E molecule, then they may form higher-order macromolecular structures capable of signaling. The formation of macromolecular aggregates of inhibitory receptors would be similar to the model for KIR–KIR aggregation proposed on the basis of the KIR2DL2–HLA-Cw*0304 cocrystal structure (41). Overall, however, our data imply that that interactions between HLA-E–peptide and CD94 may enhance the interaction between NK cells and target cells to augment inhibitory signaling at CD94–NKG2A and that the peptide specificity of this interaction differs from that of HLA-E with the CD94–NKG2A heterodimer.
The stoichiometry of inhibition of NKG2A relative to MHC class I expression implies that NKG2A+ NK cells are modulated more easily at low levels of HLA-E expression. Thus, small changes in HLA-E expression result in a greater change in the fraction of responding NK cells when cell-surface levels of HLA-E are low than when they are high. This response is different from the inhibition by KIR, for which the stoichiometry is linear across the tested range and which in our experiments reflects endogenous MHC levels. We thus propose that NK cells expressing the NKG2A receptor respond efficiently to changes in cell-surface MHC class I expression but not to bound peptide, and those expressing KIR respond more efficiently to changes in peptide repertoire (19). Most species tested to date have two distinct inhibitory receptor systems for MHC class I, one of which is CD94–NKG2A. In primates there has been an expansion of the KIR locus, which is under strong positive selection. Although we have studied selected peptides, the differences between these two receptor–ligand systems in response to changes in MHC class I level or bound peptide suggest complementary functions in terms of MHC class I recognition that may provide a rationale for their maintenance in extant species.
Methods
Cell Lines and Culture.
721.174 cells were cultured in R10 medium (RPMI 1640 supplemented with 1% penicillin streptomycin and glutamine) (Invitrogen) and 10% (vol/vol) FCS (Lonza). NKL cell lines were cultured in R10 medium supplemented with 100 IU/mL recombinant human IL-2 (National Cancer Institute Biometric Research Branch; Fisher BioServices, Bishop’s Stortford, United Kingdom). Jurkat cells transfected with CD94 were obtained from Miguel López-Botet (University Pompeu Fabra, Barcelona) and cultured in R10 medium. PBMCs were isolated from whole blood using Ficoll-Hypaque (Amersham Biosciences) density centrifugation and were frozen until use. Purified untouched NK cells were isolated from fresh PBMCs using a negative selection kit (Dynabeads; Invitrogen).
Antibodies and Staining Reagents.
The following monoclonal antibodies were used for flow cytometry: phycoerythrin (PE)-conjugated anti-human HLA-E (3D12; eBioscience), anti-human HLA-Cw*0102 (VP6G3), FITC-conjugated goat anti-human IgM antibody (AbD Serotec), FITC-conjugated anti–HLA-A2 (BB7.2; Abcam), anti-CD3 eFluor 450 (OKT3; eBioscience), anti–CD56 Cy7-PE (HCD56; Biolegend), anti–CD159a-PE (Z199; Beckman Coulter), anti–CD158b-FITC (CH-L; BD Pharmingen), Alexa Fluor 647-conjugated antibody to human CD107a (H4A3; eBioscience), and anti–NKG2C-PerCP (134591; R&D Systems). For blocking experiments and microscopy, we used anti-human CD94 (DX22; eBioscience), anti-human CD85j (HP-F1; eBioscience), anti-CD159a (Z199; Beckman Coulter), and Alexa Fluor-488 goat anti-mouse IgG (Invitrogen). The HLA-E–VMAPRTLIL pentamers were obtained from ProImmune.
MHC Class I Stabilization Assays.
Peptides were purchased from Peptide Protein Research Ltd, with identity confirmed by MS (>95% purity). They were dissolved in DMSO and then were added at a final concentration of 0–100 μM. .174 cells were incubated overnight at 26 °C in R10 medium alone or in R10 medium containing 0–100 μM of the specified peptide. The cells then were stained for HLA-E (3D12) or HLA-A2 (BB7.2) and were maintained at 4 °C in the dark for 30 min before washing with FACS wash (PBS with 0.002% NaN3 and 1% BSA). For HLA-Cw*0102 detection, HLA-Cw*0102 (VP6G3) was used at room temperature for 30 min, followed by washing. Then an FITC-conjugated goat anti-human IgM antibody was added for 30 min before further washes. All samples were fixed in 1% paraformaldehyde (PFA) and were analyzed on a BD FACS Canto II analyzer (Becton Dickinson).
NK Cell Assays.
PBMCs were incubated in R10 medium supplemented with 1 ng/mL recombinant human IL-15 (rhIL-15; R&D Systems) overnight at 37 °C. .174 cells (4 × 104) were incubated in R10 medium alone or in R10 plus peptide at 26 °C overnight. .174 cells then were washed and were resuspended with the PBMCs at an effector-to-target (E:T) ratio of 5:1 in fresh R10 medium with or without peptide and anti-CD107a PE-conjugated or control PE-conjugated IgG1. Cells were incubated at 26 °C for 4 h; 7 μg/mL Golgi-Stop (BD Biosciences) was added after 1 h. Cells were stained, washed, and fixed in 1% PFA before analysis on a FACS Canto II analyzer (Becton Dickinson). For the blocking experiments, the above protocol was followed except that PBMCs were incubated with 25 μg/mL DX22 (eBioscience), HP-F1 (eBioscience), or IgG1 control antibody (Invitrogen) for 30 min at 4 °C before the addition of .174 cells.
Cytotoxicity assays were performed on NKG2A+ NK cells sorted by flow cytometry and then stimulated for 48 h with 100 IU/mL IL-2 and 1 ng/mL rhIL-15. Cells then were incubated with peptide-loaded .174 targets at 26 °C for 4 h at an E:T ratio of 10:1, stained with Live/Dead Fixable Aqua Dead Cell Stain (Invitrogen), and fixed in 1% PFA. Then the cells were analyzed by flow cytometry.
Microscopy.
.174 cells (0.5 × 106) were incubated overnight at 26 °C alone or with 100 μM peptide and then were coincubated with NKL cells, primary NK cells, or CD94–Jurkat transfectants at an E:T ratio of 2:1 at 37 °C for 10 min. Cells were fixed in prewarmed 2% (wt/vol) PFA for 30 min at 37 °C and then were permeabilized with 0.01% Triton X-100 for 1 min. Cells then were stained with CD94 (DX22) or NKG2A (Z199), washed, stained with Alexa Fluor-488 goat anti-mouse IgG (Invitrogen) at 4 °C in the dark, followed by a further wash. Cells were imaged using a Leica SP2 upright microscope (Leica Microsystems) with transmission images and FITC emission collected in separate channels. Data were processed using Leica imaging software (Leica Microsystems) and ImageJ (National Institutes of Health). The increase in fluorescence intensity at the immune synapse was calculated as a ratio of the average fluorescence intensity along the effector–.174 interface compared with the average fluorescence intensity along a noncontact area of the effector cell plasma membrane, with both values corrected for background fluorescence as measured within a cellular region of the image. The percentage of conjugates with >1.4-fold increased fluorescence at the synapse was calculated also.
Statistical Analysis.
All statistical analyses were performed using GraphPad Prism, version 5 (GraphPad Software).
Supplementary Material
Acknowledgments
We thank Miguel López-Botet for the Jurkat-CD94 cells. This work was funded by Clinical Training Fellowships (to K.S.C. and K.M.J.), a Senior Clinical Fellowship (to S.I.K.) and Project Grant 093465/Z/10/Z (to M.A.P.), all from The Wellcome Trust.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304366110/-/DCSupplemental.
References
- 1.Lanier LL. Up on the tightrope: Natural killer cell activation and inhibition. Nat Immunol. 2008;9(5):495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parham P, et al. Primate-specific regulation of natural killer cells. J Med Primatol. 2010;39(4):194–212. doi: 10.1111/j.1600-0684.2010.00432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shum BP, et al. Conservation and variation in human and common chimpanzee CD94 and NKG2 genes. J Immunol. 2002;168(1):240–252. doi: 10.4049/jimmunol.168.1.240. [DOI] [PubMed] [Google Scholar]
- 4.Guethlein LA, Flodin LR, Adams EJ, Parham P. NK cell receptors of the orangutan (Pongo pygmaeus): A pivotal species for tracking the coevolution of killer cell Ig-like receptors with MHC-C. J Immunol. 2002;169(1):220–229. doi: 10.4049/jimmunol.169.1.220. [DOI] [PubMed] [Google Scholar]
- 5.Vance RE, Kraft JR, Altman JD, Jensen PE, Raulet DH. Mouse CD94/NKG2A is a natural killer cell receptor for the nonclassical major histocompatibility complex (MHC) class I molecule Qa-1(b) J Exp Med. 1998;188(10):1841–1848. doi: 10.1084/jem.188.10.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vance RE, Raulet DH. Toward a quantitative analysis of the repertoire of class I MHC-specific inhibitory receptors on natural killer cells. Curr Top Microbiol Immunol. 1998;230:135–160. doi: 10.1007/978-3-642-46859-9_10. [DOI] [PubMed] [Google Scholar]
- 7.Martin MP, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31(4):429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 8.Alter G, et al. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med. 2007;204(12):3027–3036. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khakoo SI, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305(5685):872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 10.Romero V, et al. Interaction of NK inhibitory receptor genes with HLA-C and MHC class II alleles in Hepatitis C virus infection outcome. Mol Immunol. 2008;45(9):2429–2436. doi: 10.1016/j.molimm.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golden-Mason L, et al. Virahep-C Study Group Natural killer inhibitory receptor expression associated with treatment failure and interleukin-28B genotype in patients with chronic hepatitis C. Hepatology. 2011;54(5):1559–1569. doi: 10.1002/hep.24556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang R, et al. Increased NKG2A found in cytotoxic natural killer subset in HIV-1 patients with advanced clinical status. AIDS. 2007;21(Suppl 8):S9–S17. doi: 10.1097/01.aids.0000304691.32014.19. [DOI] [PubMed] [Google Scholar]
- 13.Brooks CR, Elliott T, Parham P, Khakoo SI. The inhibitory receptor NKG2A determines lysis of vaccinia virus-infected autologous targets by NK cells. J Immunol. 2006;176(2):1141–1147. doi: 10.4049/jimmunol.176.2.1141. [DOI] [PubMed] [Google Scholar]
- 14.Fang M, et al. CD94 is essential for NK cell-mediated resistance to a lethal viral disease. Immunity. 2011;34(4):579–589. doi: 10.1016/j.immuni.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen GB, et al. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity. 1999;10(6):661–671. doi: 10.1016/s1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- 16.Malnati MS, et al. Peptide specificity in the recognition of MHC class I by natural killer cell clones. Science. 1995;267(5200):1016–1018. doi: 10.1126/science.7863326. [DOI] [PubMed] [Google Scholar]
- 17.Rajagopalan S, Long EO. The direct binding of a p58 killer cell inhibitory receptor to human histocompatibility leukocyte antigen (HLA)-Cw4 exhibits peptide selectivity. J Exp Med. 1997;185(8):1523–1528. doi: 10.1084/jem.185.8.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajagopalan S, Long EO. Antagonizing inhibition gets NK cells going. Proc Natl Acad Sci USA. 2010;107(23):10333–10334. doi: 10.1073/pnas.1005636107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fadda L, et al. Peptide antagonism as a mechanism for NK cell activation. Proc Natl Acad Sci USA. 2010;107(22):10160–10165. doi: 10.1073/pnas.0913745107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borhis G, et al. A peptide antagonist disrupts NK cell inhibitory synapse formation. J Immunol. 2013;190(6):2924–2930. doi: 10.4049/jimmunol.1201032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Llano M, et al. HLA-E-bound peptides influence recognition by inhibitory and triggering CD94/NKG2 receptors: Preferential response to an HLA-G-derived nonamer. Eur J Immunol. 1998;28(9):2854–2863. doi: 10.1002/(SICI)1521-4141(199809)28:09<2854::AID-IMMU2854>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 22.Petrie EJ, et al. CD94-NKG2A recognition of human leukocyte antigen (HLA)-E bound to an HLA class I leader sequence. J Exp Med. 2008;205(3):725–735. doi: 10.1084/jem.20072525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaiser BK, Pizarro JC, Kerns J, Strong RK. Structural basis for NKG2A/CD94 recognition of HLA-E. Proc Natl Acad Sci USA. 2008;105(18):6696–6701. doi: 10.1073/pnas.0802736105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ulbrecht M, Modrow S, Srivastava R, Peterson PA, Weiss EH. Interaction of HLA-E with peptides and the peptide transporter in vitro: Implications for its function in antigen presentation. J Immunol. 1998;160(9):4375–4385. [PubMed] [Google Scholar]
- 25.Nattermann J, et al. HIV-1 infection leads to increased HLA-E expression resulting in impaired function of natural killer cells. Antivir Ther. 2005;10(1):95–107. doi: 10.1177/135965350501000107. [DOI] [PubMed] [Google Scholar]
- 26.Tomasec P, et al. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science. 2000;287(5455):1031. doi: 10.1126/science.287.5455.1031. [DOI] [PubMed] [Google Scholar]
- 27.Nattermann J, et al. The HLA-A2 restricted T cell epitope HCV core 35-44 stabilizes HLA-E expression and inhibits cytolysis mediated by natural killer cells. Am J Pathol. 2005;166(2):443–453. doi: 10.1016/S0002-9440(10)62267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alter G, et al. HIV-1 adaptation to NK-cell-mediated immune pressure. Nature. 2011;476(7358):96–100. doi: 10.1038/nature10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valés-Gómez M, Reyburn HT, Erskine RA, López-Botet M, Strominger JL. Kinetics and peptide dependency of the binding of the inhibitory NK receptor CD94/NKG2-A and the activating receptor CD94/NKG2-C to HLA-E. EMBO J. 1999;18(15):4250–4260. doi: 10.1093/emboj/18.15.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martini F, Agrati C, D’Offizi G, Poccia F. HLA-E up-regulation induced by HIV infection may directly contribute to CD94-mediated impairment of NK cells. Int J Immunopathol Pharmacol. 2005;18(2):269–276. doi: 10.1177/039463200501800209. [DOI] [PubMed] [Google Scholar]
- 31.Lazetic S, Chang C, Houchins JP, Lanier LL, Phillips JH. Human natural killer cell receptors involved in MHC class I recognition are disulfide-linked heterodimers of CD94 and NKG2 subunits. J Immunol. 1996;157(11):4741–4745. [PubMed] [Google Scholar]
- 32.Carretero M, et al. The CD94 and NKG2-A C-type lectins covalently assemble to form a natural killer cell inhibitory receptor for HLA class I molecules. Eur J Immunol. 1997;27(2):563–567. doi: 10.1002/eji.1830270230. [DOI] [PubMed] [Google Scholar]
- 33.Pérez-Villar JJ, et al. Biochemical and serologic evidence for the existence of functionally distinct forms of the CD94 NK cell receptor. J Immunol. 1996;157(12):5367–5374. [PubMed] [Google Scholar]
- 34.Lampen MH, et al. Alternative peptide repertoire of HLA-E reveals a binding motif that is strikingly similar to HLA-A2. Mol Immunol. 2013;53(1-2):126–131. doi: 10.1016/j.molimm.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 35.Boyington JC, et al. Structure of CD94 reveals a novel C-type lectin fold: Implications for the NK cell-associated CD94/NKG2 receptors. Immunity. 1999;10(1):75–82. doi: 10.1016/s1074-7613(00)80008-4. [DOI] [PubMed] [Google Scholar]
- 36.Orr MT, et al. Development and function of CD94-deficient natural killer cells. PLoS ONE. 2010;5(12):e15184. doi: 10.1371/journal.pone.0015184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Long EO, Kim HS, Liu D, Peterson ME, Rajagopalan S. Controlling natural killer cell responses: Integration of signals for activation and inhibition. Annu Rev Immunol. 2013;31:227–258. doi: 10.1146/annurev-immunol-020711-075005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis DM, et al. The human natural killer cell immune synapse. Proc Natl Acad Sci USA. 1999;96(26):15062–15067. doi: 10.1073/pnas.96.26.15062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schleinitz N, March ME, Long EO. Recruitment of activation receptors at inhibitory NK cell immune synapses. PLoS ONE. 2008;3(9):e3278. doi: 10.1371/journal.pone.0003278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kirwan SE, Burshtyn DN. Killer cell Ig-like receptor-dependent signaling by Ig-like transcript 2 (ILT2/CD85j/LILRB1/LIR-1) J Immunol. 2005;175(8):5006–5015. doi: 10.4049/jimmunol.175.8.5006. [DOI] [PubMed] [Google Scholar]
- 41.Boyington JC, Motyka SA, Schuck P, Brooks AG, Sun PD. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature. 2000;405(6786):537–543. doi: 10.1038/35014520. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.