“Their fruit will serve for food and their leaves for healing.” (Ezekiel 47:12)
“What the eyes perceive in herbs or stones or trees is not yet a remedy; the eyes see only the dross.” (Paracelsus)
1.0 Introduction and background
1.1 Natural product-derived drugs
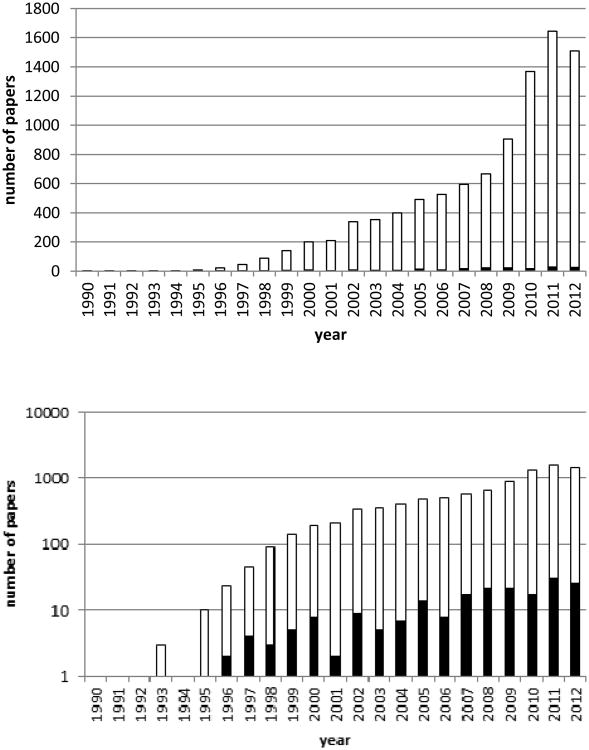
The search for natural product modulators of disease states goes back millennia; drugs have been purified from natural sources for many decades. Even today, natural products or their derivatives continue to comprise a large proportion of drugs including, for example, the majority of anti-cancer drugs.1 Since the 1990s, development of high-throughput methods for evaluation of potentially bioactive samples has accelerated in parallel with high throughput chemistry methods which have supplied large libraries of synthetic compounds. The history, state of the art, and effectiveness of these methods, particularly high-throughput screening (HTS), has been the subject of a recent review.2 However, application of HTS for identification of biologically active natural products remains a relatively uncommon activity (Figure 1). In contrast to screening libraries of synthetic compounds, high-throughput assessment of natural products is fraught with a variety of obstacles. This review addresses the challenges of applying HTS methodology to identification of biologically active crude natural product mixtures (extracts from organisms) and characterization of active components from those mixtures.
Figure 1. High throughput screening publications indexed in MedLine.
The number of publications combining “high throughput screening” and “natural products” (black bars) is a small fraction of the total references indexed under “high throughput screening” (open bars).
1.2 Traditional natural product drug discovery
The vast chemical diversity available in nature3 along with the proven record of natural products as sources for drugs4 has resulted in natural product research continuing to be an important contributor to drug discovery as well as bioprobe development.5,6 The starting materials for natural product drug discovery are crude solvent extracts from source organisms.7 An outstanding example of the richness of the resources available is the Natural Products Repository of the National Cancer Institute, comprising >180,000 extracts from >50,000 organisms. Other organizations maintain similar, often focused libraries of natural product extracts, including the Spanish Fundación Medina, with a large collection of microbial samples8 the Korean Research Institute of Bioscience and Biotechnology, with a Korean plant extract bank (http://www.kribb.re.kr/eng/sub02/sub02_07_02.jsp), and the Eskitis Institute in Australia, with Australian plants and marine invertebrates.9 Identification of active extracts may occur by a variety of methods. For example, organisms used in folk medicine to treat particular disease phenomena have been useful sources of biologically active compounds.10 Similarly, mining of existing data sets such as the anti-cancer cell growth inhibitory/cytotoxic activities of compounds against the NCI60 panel of cell lines11,12 has led to identification of natural product extracts likely to contain compounds active against specific cancer targets which has proven to be a fruitful approach resulting in identification and characterization of a number of potentially valuable anti-cancer compounds.13,14,15 Typically, once bioactive extracts from source organisms are identified, active components are isolated and their structures characterized by a variety of standard approaches, nicely summarized in a recent review.16
1.3 High throughput screening (HTS)
The development and effectiveness of high-throughput screening programs have been extensively reviewed in recent years.17,18,19,20 Tens of thousands to millions of samples are tested in HTS campaigns for their ability to modulate biochemical targets in cell-free assays and/or phenotypic or targeted cell-based assays.21,22 Assays are typically performed at a single concentration followed by additional testing to confirm active “hits” and to estimate potency and target or phenotype specificity. HTS thus requires extremely reproducible, relatively inexpensive assays in small volumes (most commonly ≤50 μl in 384-well plates) and the use of reliable and accurate high-throughput liquid handling systems. Each of these requirements brings its own set of challenges, including maintenance of sample integrity (sample precipitation, degradation in assay buffers, etc.), potential for interference of samples with assay reagents or readouts, and non-specific/off-target effects. Most of these potential problems can be addressed by careful design and management of all aspects of assays, liquid handling, and process flow.
The targets and cellular phenomena amenable to HTS include nuclear receptors,23 GPCRs,24 ion channels,25 protein kinases,26 proteases,27 signaling pathways,28 cell death mechanisms,29 etc. These have been applied to a wide range of disease states such as cancer,30,31 Fabry disease,32 atherosclerosis,33 parasitic 34 and autoimmune diseases,35 among others. Increasingly, HTS technologies have begun to be applied to traditionally “non-druggable” targets (i.e., generally thought to be inaccessible to modulation by small molecules) such as protein-protein interactions.36,37
The effectiveness of HTS campaigns is only as good as the quality of the libraries screened and most high throughput screening programs have focused on synthetic compounds. As a result, HTS development has occurred in parallel with high-throughput chemistry methods for synthesis of large libraries of compounds. Application of these techniques led to large libraries of synthetic compounds designed to mimic the chemical characteristics of existing drugs.38,39,40 However, there has been increasing recognition that even the largest, most carefully designed synthetic compound libraries access only a tiny fraction of possible chemical diversity.41,42 A number of approaches have been taken to increase the effectiveness of compound libraries, including development of fragment-based libraries,43,44 and increased inclusion of pure natural products and natural product-like compounds in screening libraries.45,46 The use of natural product extracts in HTS has been particularly relevant to ongoing efforts to effectively address “non-druggable” targets.47 Once obtained, pure natural products (isolated or synthesized) and synthetic natural product-like compounds are assessed in the same manner as are any other sets of defined compounds.
1.4 Application of HTS to natural product extracts and bioassay-guided purification
For each of the categories of targets commonly subjected to HTS, natural product modulators have been identified.4 This fact suggests that application of HTS technologies to natural products will continue to be fruitful. Although pure natural products have increasingly been used in HTS, this still represents only a very small sampling of the natural product chemical diversity available. It has been estimated that only about 250,000 natural products have been isolated and characterized from nature and many of these have been obtained in insufficient quantity to allow for commercial distribution and widespread assessment of possible biological activities. In order to access the wealth of chemical diversity available, a different approach must be taken, namely application of HTS to large numbers of natural product extracts followed by characterization of their active components. Although this approach has been successfully applied, it remains significantly challenging. Addressing these challenges is the subject of the remainder of this review, which primarily covers reports published from our laboratory and others over the past decade or so.
2.0 Challenges of natural product samples for HTS
2.1 Common “problem” compounds and phenomena
2.1.1 Fluorescent molecules and quenchers
The use of fluorescent endpoints in HTS is widespread, either through small fluorescent tags such as fluorescein or chimeric fluorescent proteins (e.g., green fluorescent protein). Thus, the interference of sample fluorophores with the fluorescent endpoint is a common problem.48 While following any primary screen with an orthogonal counter screen (i.e., a non-fluorescent endpoint) is good screening practice regardless of endpoint, a high hit rate in the primary screen may be an inefficient use of resources. There are many ways to limit the effect of fluorescent test samples. The simplest is to avoid use of UV-range tags such as 4-methyl umbelliferone or Alexafluor 350 and green tags such as fluorescein, and give preference to red-shifted tags such as rhodamine or Texas Red.49 Endpoints other than direct fluorescence can be useful, with time-resolved fluorescence (TRF) the most popular. The majority of test samples exhibit very short fluorescence lifetimes, while TRF tags such as lanthanide chelates or cryptands have much longer lifetimes, permitting simple gated observation protocols.50
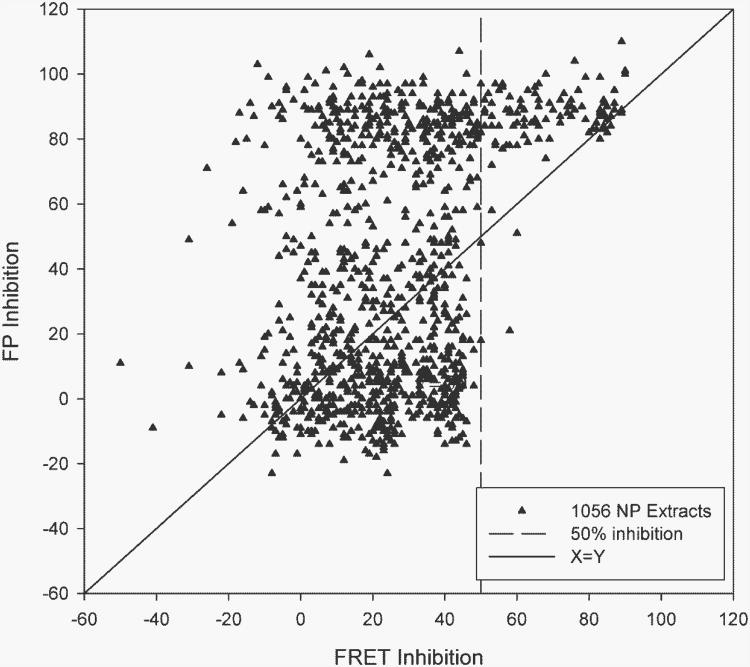
While fluorescence anisotropy endpoints are useful for screening pure compounds if suitable controls are implemented, screening of complex natural product matrices is fraught with difficulty. A natural product extract is likely to contain a variety of fluorescent or fluorescence quenching materials of unknown molecular weight, which may lead to very high apparent hit rates and poor confirmation in counter screens. In our laboratory's experience, this was highlighted by development of several cell-free assay formats for identification of inhibitors of HIV-1 RNase H activity. We chose to use a FRET assay as the primary screen,51 and also demonstrated that capillary electrophoresis with fluorescence detection was useful as a secondary assay, since the fluorescent substrate could be separated from sample fluorophores.52 However, a fluorescence anisotropy assay which we validated for pure compounds proved unsuitable for natural product samples, since the hit rate was ∼40% for plant extracts (Figure 2).
Figure 2.
Comparison of RNase H extract screening of extracts using fluorescence polarization and FRET assay designs.
2.1.2 Nuisance compounds
In a screening program, any class of compounds which occur frequently enough to consume significant resources for dereplication or secondary screening beyond their value to the program can be considered a nuisance. Well known examples of common compounds that we have found problematic include phorbol esters in cellular assays,53 acidic polysaccharides in antiviral assays,54 and polyphenolics in cell free assays.55 Much effort has been expended to remove nuisance compounds from screening samples,54,56 or to modify the HTS assay to avoid detecting them.57 Several specific examples are discussed below.
One recent development is the finding that metal impurities in synthetic samples (e.g., residues from catalysts) can cause false positives in screening assays.58 Given that plants and fungi are capable of concentrating various metals from their environment,59 and that contamination with metals has been problematic with commercial herbal samples,60 it is possible that erratic bioassay results from extract testing might be due to varied metal content.
2.1.3 Toxicity
In cellular assays in which the endpoint does not involve cell death, the presence of toxic constituents in an extract can mask the presence of other compounds with the desired activity. Thus, detergent compounds such as saponins can rapidly lyse assay cells before the target of interest can be interrogated.61 Other classes of cytotoxins may require a longer time frame to exert their effects. While many current cell based assays are conducted on a relatively short time scale (e.g., 6-8 hrs for reporter gene assays) compared to typical cell viability assays (48-72 hrs), thereby reducing the effects of cytotoxicity, the separation of cytotoxic materials from other constituents by extract prefractionation is nonetheless advantageous.
2.2 Difficulty of liquid handling with extracts in HTS mode
Among the primary problems inherent in the use of natural product extracts in HTS are issues related to sample handling, e.g., sample reconstitution, dilutions, and liquid transfer steps. These include a number of characteristics inherent in highly complex mixtures such as sample variability, viscosity, and precipitation.62,63 One of the most effective approaches increasingly taken to address natural product-specific liquid handling issues is partial purification (or “prefractionation”) of crude extracts for removal and/or sequestration of problem constituents as well as increasing consistency of samples.64,63,65
Of course, many of the issues that affect HTS of natural products are inherent in any high throughput sample handling process and the first step in their resolution is to apply standard approaches. For example, variations in solvents for sample solubilization,66 dilution schemes,66,67 order of reagent addition,68 and methods for delivery of samples to assay plates69 can often resolve sample-related issues, independent of the source of the samples. Variations in sample preparation procedures, including alternative solvents and use of carrier macromolecules for stabilization have been successfully applied to problems arising in the preparation of natural product extracts for HTS.62 Application of standard sample management techniques70 as well as proper use, maintenance, and calibration of liquid handling equipment67 can often forestall sample handling problems. Sample storage and management are further discussed below as focused on natural product extracts.
2.3 Library development: How should we select samples and process them?
2.3.1 Selection
One goal of a screening library is to provide a high level of structural diversity in order to give the screen its best chance of identifying lead structures. In terms of natural products, this can be accomplished by sampling a wide variety of taxonomy (i.e., higher plants, marine invertebrates, bacteria and fungi), or by collecting in unexploited ecological niches, or by selecting samples with known ecological or traditional medicinal information. In addition, the method of extraction and level of purification of the samples can be an important factor in the effectiveness of screening.
2.3.2 Biodiversity
Chemical diversity of natural products can reflect the biodiversity of their source organisms. One approach is to utilize genetic and chemical methods to dereplicate microbial cultures prior to screening. Since soil bacteria have been the subject of screening for decades, it is important, even in novel habitats, to maximize the isolation of novel strains while minimizing duplication of strains already available for study in other libraries. Thus, Goodfellow and Fiedler described a combination of selective isolation with 16S rRNA sequencing to genus, followed by taxon-specific probes, to dereplicate marine actinomycetes.71 Liu et al. constructed a marine microbe library with low redundancy using a similar dereplication system.72,73 ITS sequencing was used by the Strobel group to identify unique cultures of endophytic fungi prior to screening.74 Third, LC-MS chemical profiling of microbial cultures has been used to identify common metabolites to avoid chemical redundancy.75 A tantalizing combination of the last two methods would incorporate detection of biosynthetic clusters by sequencing, and prediction of the potentially produced natural product structure, as envisioned by Gerwick and Moore, though technology is not yet sufficient to perform this in high throughput.76 Many screening programs seek chemical diversity by exploiting novel ecological niches, for example, microbes found at deep-sea hydrothermal vents,77 fungal endophytes74,78 or plants from poorly sampled areas such as central Asia.79
2.3.3 Traditional Chinese Medicine (TCM) and other ethnomedical information
The Eisenberg group at Harvard and their Chinese collaborators established a prototype library of >200 authenticated TCM plant and fungal samples represented in the PRC Pharmacopeia.60 The origin of all samples was rigorously documented, taxonomy was verified, and pharmacopeial standard testing was performed. Tannins were removed from the bulk extract, followed by preparative HPLC, which generated ∼45 fractions per extract.60 A group at Zhejiang University developed a different strategy for production of fractionated libraries from Chinese herbal formulas, in which decoction of the polyherbal mixtures was followed by polystyrene resin extraction, medium pressure liquid chromatography over silica gel, then semipreparative C-18 HPLC.80 A group at the Shanghai Institute of Materia Medica has reviewed other approaches suitable for screening TCM materials.81
2.3.4 Organism identification and documentation
Accurate identification of the biological source of an active sample is obviously required in order to be able to recollect enough biomass to support further development. In the early days of microbial drug discovery, a culture was rarely identified until a hit was confirmed, but this is no longer the case. To avoid examination of duplicate microbial cultures, screening groups use genetic profiling,74 chemical profiling,75 and classical taxonomic characters.71 For plants, dried herbarium vouchers still constitute the gold standard for documentation, however, ribosomal DNA ITS region sequences may be of use when fine taxonomic distinctions are required at the species level.82 Properly prepared voucher specimens are also critical for marine invertebrate specimen identification and documentation, due to the relatively poor state of knowledge of marine invertebrate taxonomy. For example, a recent review of sponge diversity noted that there are many new species already collected but awaiting formal description, though genetic methods and systematic databases may address this problem.83 DNA bar coding is also under study for identification of samples.84 With all types of sources, it is also important to record location of collection (GPS), elevation above sea level/depth, and date, for future reference.
2.3.5 Extraction solvents and polarity
Many approaches have been used to prepare extracts of natural materials for screening. Actinomycte broths are most often filtered and the marc extracted separately, though whole broth extracts with EtOAc or MTBE are also used. Plant extracts have traditionally been made using ethanol, while the diversity of marine invertebrates made it difficult to settle on a single extraction method. The NCI extraction protocol, which addresses all three of these sources, was thoroughly documented in a recent publication by McCloud.85
Highly polar and highly lipophilic compounds may be problematic drug candidates, and it may be desirable to exclude them from screening samples. Lipids can be removed from dry material using hexane extraction, while other polar materials such as tannins and polysaccharides can be removed from extracts using polyamide cartridges86 and ethanol precipitation,56 respectively. The Eskitis Institute group in Australia has developed a protocol based on elution of the extract from Oasis HLB solid phase cartridges with 70% aqueous methanol, the goal of which is to provide screening samples with intermediate polarity.87
2.3.6 Prefractionation
A trend in sample preparation that is gaining acceptance is to fractionate the extract prior to screening. This has several advantages, including removal or separation of highly polar and nonpolar materials, concentration of minor metabolites, and separation of interfering compounds (e.g., cytotoxins, fluorophores) from other compounds. There is a spectrum of prefractionation methods, ranging from near-complete purification to rough polarity separation into a small number of fractions. Wagenaar performed a cost-benefit analysis for microbial sample prefractionation, finding that 10 samples per extract provided advantages while limiting costs.88 Methanol extracts of microbial cultures were separated into only four fractions by HPLC by Butler's group,89 with demonstrated improvement in hit rates and separation of interfering compounds. Marine invertebrate extracts were efficiently fractionated using HP20SS resin, then characterized by mass spectroscopy.90,91 Predictably, the cost for more complete purification is high, limiting the number of primary extracts processed. Automated chromatography systems are commercially available for this process. An automated high throughput system to generate prefractionated libraries from plant materials has been reported.65,92 Even with simple fractionation schemes, automated liquid handling and weighing capabilities are essential if large numbers of samples are to be processed. An interesting variant extraction method was reported by Yuliana, et al., in which ground plant material was continuously extracted with solvents of increasing polarity to generate 20 fractions, which were then tested for adenosine A1 receptor binding activity and profiled by 1H-NMR. Multivariate analysis allowed identification of two flavonoids as the active compounds responsible for the receptor binding activity.93 The method is thus a combination of extraction, prefractionation, and metabolomics.
2.4 How should samples be stored?
Besides structural diversity, another important consideration for library quality is assessment of sample integrity as the compound enters the library, when it is selected as a screening hit, and last, when re-supplied from its original source. Maintaining sample integrity involves utilization of storage and retrieval conditions which avoid compound precipitation from DMSO solution due to adsorption of atmospheric moisture, or degradation.
2.4.1 2D bar coded tubes
As the number of samples in a screening library increases, it becomes desirable to employ automation to store and retrieve samples. Many commercial automated freezer storage systems are available which can handle a wide variety of sample containers including plates, vials, and polypropylene tubes. The use of bar code labeling for these containers is critical throughout the screening process.
2.4.2 Cold! Dry!
A major concern in sample storage is to preserve the integrity of the sample, whether a pure compound, an extract, or a partially pure fraction. It is most convenient to store samples in DMSO solution, however, the hygroscopic nature of DMSO can lead to precipitation of the sample as water is absorbed from the air.94 Samples stored at −20°C must be warmed before they are unsealed at ambient temperature and humidity, and tubes or plates should not be left open to the air more than necessary. To achieve these ends, our laboratory stores samples at −20°C in multiple formats. Bulk samples are stored dry in vials, while a modest aliquot of each sample (0.5-2.0 mg) is stored in DMSO in a 2D bar coded tube. From those tubes, source plates (384 well, 50 μL per well) are prepared for direct transfer to screening assays. When hits are obtained from a screen, confirmation samples are then obtained from the stored tubes. If the hit is confirmed, dry stores of the bulk samples are accessed for further biological and chemical testing. Checks of chemical integrity of purified compounds are made by LC-MS at this point. To assist in resolubilization of samples, small magnetic disks are routinely added to sample tubes so that they can be stirred in the tube using a 96-position magnetic stirrer.
If screening samples are to be stored dry, efficient resolubilization is particularly important. To that end, Waybright et al.95 explored alternative strategies for pure compounds. They found that addition of glycerol as nonvolatile cosolvent facilitated resolubilization of samples. In addition, they proposed that a solvent combination of DMSO/glycerol/water (45:45:10, v/v) would avoid changes in water content in samples, though it did not allow high stock concentrations.95
Others have proposed storage of samples under dry inert gas, or at −80°C. However, these conditions have not been widely adopted due to the high cost and requirements for specialized equipment.
3.0: Adaptation of HTS Assays for Natural Products
An important general principle illustrated by many of the examples cited in the following section is the need to understand the effect(s) of samples on assay systems. This is important for any HTS application, but critical for screening of natural product extracts. The availability and application of parallel, non-specific or off-target, assays can quickly identify spurious apparent activities that actually affect detection enzymes or reagents, cell survival, or other critical assay parameters.
3.1: Biochemical assays
Constituents of natural product extracts can cause significant problems that manifest themselves in increased false positive or false negative rates in biochemical assays. For example, polyphenols such as tannins and flavonoids can affect assays by quenching fluorescence,96 by non-specifically binding and/or reacting with proteins in assay mixtures,97,98 or by generation of spurious results due to aggregation.99 Similarly, highly fluorescent or colored compounds common to extracts may interfere with colorometric or fluorescent endpoints in enzymatic or protein binding assays.96 Fatty acid constituents of extracts can also interfere with biochemical assays.100
The most obvious approach to alleviating the problems associated with polyphenols is to remove them from the library samples, typically as part of a prefractionation process (discussed in section 2.3.6 above). However, some phenolic compounds may be relevant modulators of enzymatic activities. Thus, removing them prior to screening may result in loss of potentially important activities. As a result, multiple approaches have been taken to reduce (or at least identify) non-specific effects while maintaining the ability to detect specific responses, even in the presence of potentially interfering compounds. Relatively simple, logical, straightforward modification of assay protocols can often significantly ameliorate nonspecific assay interference by non-selective compounds in extracts. Among the strategies successfully applied to biochemical screening assays are: addition of or increased rigor of wash steps for capture assays, testing at multiple concentrations, choice of detection reagents based on fluorescence, luminescence, or colorimetric characteristics, and addition of disaggregating agents or additives to reduce non-specific binding of components. These approaches are illustrated by a series of examples from the literature and detailed in the following paragraphs.
Cataloging effects of samples in “mock” assays by measuring intrinsic fluorescence of samples in the excitation and emission ranges required for detection of fluorescent probes, or quenching of probe fluorescence by samples can identify problem substances. Zou, et al.96 used this approach to identify interfering compounds in cytochrome P450 assays using fluorogenic substrates. They observed a high proportion of problematic plant-derived samples (4 of 25 tested) and concluded that the assay was unsuitable for use with natural products. This example used pure natural products and was not an HTS assay, but it illustrates the point. Application of this approach can conceivably be used to eliminate categories of compounds or extracts and/or to identify particularly problematic detection systems. In general, ensuring that the detection system chosen is robust with reference to the range of samples to be screened is a requirement for successful HTS, particularly when applied to natural products. Testing samples at multiple concentrations can provide useful dose response data for identification of true active samples as well as for prioritization of active extracts for follow-up studies. Cruz, et al. applied this philosophy to identify luciferase inhibitors in a natural product extract library.101 This proof of principal study suggested that assaying at multiple concentrations can increase the probability of identifying active samples as well as providing baseline data for the effects of a library of natural product extracts on commonly used bioluminescence enzymes such as luciferase.
Interference with assays dependent on fluorescence detection can often be reduced or eliminated by changing to a colorimetric or luminescence detection system. Fluorescence-based assays of proteolytic enzymes often depend on detection of peptide substrates conjugated to coumarin derivatives. Grant, et al.102 noted that use of alternative, red-shifted fluorescent tags such as rhodamine can often reduce interference by natural products. However, they additionally reported that using a mixture of substrate peptides labeled with either rhodamine or a coumarin derivative with non-overlapping excitation and emission wavelengths allowed for distinguishing true actives from interfering samples. They recognized that some level of interference remained possible and recommended that samples be tested against each fluorophore in order to identify potential problematic compounds and extracts. Similarly, use of red-shifted dyes (Cy3B and Cy5) in fluorescence polarization-based kinase assays significantly reduced assay interference as compared to use of a fluorescein tag in an AKT kinase HTS assay.103 A variety of compounds, including many polyphenols, interfere with a commonly used assay for detection of amyloid fibrils, based on thioflavin T fluorescence which increases upon binding.104 An alternative, colorimetric dye binding assay using Congo red was suggested by the authors of the cited study. Although some polyphenols still had some effect on the results, they could be directly corrected by measuring absorbance in the presence and absence of the dye. As applied to natural product extracts colorimetric detection might also be problematic when screening extracts containing highly colored compounds. This can limit the high test concentration that is feasible without leading to high background absorbance. A fairly simple approach to dealing with fluorescence interference is illustrated by fluorescent assays for inhibitors of influenza neuraminidase which are also affected by polyphenols.105,106 In an analysis of the quenching effects of flavonoids, the authors concluded that assessing samples for quenching effects and subtracting these effects from the assay signal might be a fruitful approach.105 Alternatively, parallel application of a neuraminidase assay using a chemiluminescence detection system might be of value in identifying false positives and false negatives due to interference.107
Alterations in assay configurations are often required for successful HTS with natural product extracts (as compared to HTS of pure compound libraries), whether they are prefractionated or crude extracts. An enzymatic assay for the E3 ubiquitin ligase, MDM2, was developed based on antibody binding to immobilized polyubiquitinated MDM257 and illustrates two common approaches to dealing with extract-related issues. First, to avoid problems related to fluorescent or fluorescence quenching components in extracts, an electrochemiluminescence detection system was employed. Second, assay re-optimization was required to reduce non-specific interference by extracts as compared to pure compounds. In this case, relatively straightforward modifications were sufficient. Additional BSA in the reaction mixture, increased wash steps, and alterations in incubation times and order of addition of components reduced the apparent hit rate in a pilot experiment from ∼40% to < 2%. The mechanism(s) of non-specific interference was not further investigated. In addition to non-specific binding to proteins, aggregation of compounds leading to non-specific inhibition is a common phenomenon.99,108 Disaggregation can be accomplished by addition of detergents or inert proteins or by simply increasing the concentration of the target enzyme in an enzymatic assay.99,108 Reassessing apparent hits at multiple enzyme concentrations (incubation times adjusted to remain in the linear range for the assay), can identify potential non-stoichiometric inhibitors, typically due to aggregation.108 In an enzymatic assay of the proprotein convertase furin, based on a fluorogenic peptide substrate, polyphenols were found to appear as false positives, i.e. apparent inhibitors without effects on enzymatic activity in follow-up assays.98 Interference was not due to aggregation, but a result of autoxidation of compounds and reaction of the resulting product with protein components of the assay mixture. Although this study, like many of those focused on aggregation, employed a model system with pure compounds, the authors' conclusions and suggestions to include disaggregating agents along with the reducing agent glutathione to reduce background due to compound reactivity, are appropriate to HTS applications with polyphenol-containing natural product extracts.
Application of orthogonal assays to active samples, particularly using cell-based assays for compounds found to be active in biochemical screens, can often reduce false positives. It has been shown that naturally occurring fatty acids can provide false positive results in cell-free aromatase assays and that these can be identified and eliminated from further consideration by reassaying hits in a cell-based assay utilizing a cell line overexpressing aromatase.100 Other alternative detection systems such as surface plasmon resonance (SPR) have been used, for example, to distinguish specific from nonspecific binding of polyphenols to topoisomerase.97 This technique is not high throughput and requires pure components, including putative ligands, but can be applied to assessment of binding of compounds to protein targets.
In order to understand mechanisms of interference by classes of problematic natural products, many of the experiments designed to assess and control assay interference use purified compounds rather than extracts. However, the principles derived from these studies and from more empirical assessment of extracts have been applied via careful reagent, detection, configuration, and process development to successful HTS assays employing natural product extracts. As a result, in the last 10-12 years, HTS of natural product extracts has identified modulators of a number of enzymatic targets including: E3 ubiquitin ligase MDM2,57 luciferase,101 proteases,102 AKT kinase,103 asparaginyl-tRNA synthetase,109 PPARgamma,110 aromatase,100 influenza neuraminidases,111,112,113 tyrosinase,114 acetylcholinesterase,115 phosphodiesterase,116,117 HDAC and SIRT enzymes,118 MRSA pyruvate kinase119 and lipases.116 Protein-protein interaction screens have included HIV gp41 binding120 and Hes1 dimerization.121
3.2 Cell-based assays
One of the most significant confounding factors in cell-based HTS is the need to identify and eliminate non-specifically cytotoxic compounds. For targeted screens (e.g. reporter gene assays, cellular enzymatic activities) or phenotypic assays (e.g. growth inhibition or differentiation), the presence of cytotoxic compounds in a sample may contribute to false positive or false negative rates. Cell-based assays generally fall into one of two general categories, either targeted (e.g. reporter gene assays, cellular enzymatic activities) or phenotypic (e.g. growth inhibition or differentiation). In the case of targeted assays, the HTS readout typically reflects an enzymatic activity such as luciferase for reporter gene assays or enzymatic conversion, accumulation, or depletion of target-specific substrates. Examples of reporter gene assays reflecting modulation of signaling pathways and applied to natural product screening include AP1,122 HIF1,123 HIF2α,124 interferon regulatory factor 1,125 PPARγ,126 and heat shock response.127 Reporter constructs have also been used to monitor stability of target proteins. An example of this approach applied to screening of natural product extracts is a luciferase-Pdcd4 fusion protein that retained the regulatory regions of the Pdcd4 molecule, a tumor suppressor protein.128 In this assay, the goal was to identify stabilizers of Pdcd4 by measuring luciferase activity as a surrogate for cellular Pdcd4 protein levels. A luciferase reporter was also developed for identification of modulators of adaptive and apoptotic unfolded protein responses (UPR). In this case, activation of the pathways leads to accumulation of the luciferase reporters.129 Another luciferase reporter was constructed to assess effects of compounds and natural product extracts on the ubiquitin-proteasome pathway using a ubiquitin-luciferase construct.130 Other targeted screens used to identify active natural products include receptor-dependent calcium influx using a fluorescent Ca2+ sensitive dye,131 multidrug resistance transporter ABCG2132 (inhibition of transporter in over expressing cells leads to accumulation of a fluorescent substrate) and modulation of processing of amyloid precursor protein (APP) in cultured cells by ELISA-based detection of amyloid-beta peptides,133 and estrogen receptor (ER) by fluorescence detection of nuclear localization of an ER-GFP fusion protein.134
Phenotypic screens applied to natural products include primarily growth inhibition/cytotoxicity assays. An outstanding example as applied to natural products is the aforementioned NCI60 cell line panel which has been used to screen approximately 50,000 natural product extracts.135 In this case, patterns of growth inhibition and/or cytotoxicity against specific tumor-derived cell lines can guide discovery of active agents as well as identification of possible new targets.136 Although most commonly applied in cancer research, there are examples of the use of growth inhibition/cytotoxicity/cell death assays in a variety of other fields as well,29 including to identify antiparasitic137,138 and antibacterial139,140 activities of natural product extracts. Clearly, generally cytotoxic compounds in samples can be problematic. In the latter cases, cytotoxicity against mammalian cell lines is relevant in that it may affect the assay itself (if host cells are present in the assay), and/or as a potential complicating factor for further development (i.e. searching for agents that will not affect host cells).
In addition to these two main categories, a “hybrid” approach has been applied in which the HTS readout is phenotypic (often cytotoxicity or growth inhibition), but where the phenotype is dependent on expression of a specific target or targets. In this situation, the target is often expressed in one cell line and not in another (ideally similar cell background). Alternatively, the target may be activated by a specific known modulator and the effects of samples on cells in the presence or absence of the activating agent are assessed. HTS of natural product extracts using this approach has not been extremely common, but several examples appear in the literature. Targeting pathways is illustrated by a screen developed for identification of substances able to synergize with the death ligand TRAIL in killing of TRAIL-resistant cells. In this assay, effects of samples on cell growth and viability were assessed in the presence or absence of TRAIL.141 Other efforts to find natural products affecting TRAIL signaling have been reviewed by Ishibashi.142 Effects of natural product extracts on ligand-independent growth of cells expressing constitutively active c-KIT was assessed as compared to cells expressing wild-type c-KIT by a differential cytotoxicity assay.143 An assay to identify glycolysis inhibitors employed a differential growth inhibition/cytotoxicity assay in the presence or absence of rotenone which results in exclusive dependence of the cells on glycolysis to survive.144
In each of these types of assays, reduction in a readout can occur due to targeted effects (e.g. inhibition of a signaling cascade or differential cytotoxicity against multiple cell lines) or non-specifically due to reduction in cell numbers as they begin to die. This becomes particularly problematic in working with complex mixtures of multiple compounds such as natural product extracts in which the activity of a constituent may be masked by a non-specific effect. For the examples cited above, the potential problems are obvious. For targeted assays, cell death will lead to a decreased signal for the readout. For example, cytotoxicity will lead to reduction in reporter gene activity, not because the signaling pathway is affected, but because the cells are dying. An apparently active extract must then be further assessed to determine if the effect is due to targeted activity or non-specific cell death. Similarly, for differential toxicity assays, generally toxic compounds in an extract may mask the presence of specifically active compounds by overshadowing the differential activity.
Although dead cells tell no tales, several approaches have been successfully used to identify and/or to mitigate the effects of non-specific toxicity. Some of the commonly found generally cytotoxic compounds in natural product extracts are known and can be removed or sequestered by prefractionation. For example, saponins have long been recognized as potentially interfering agents due to toxicity, but as is the case with polyphenols in biochemical assays, removal of saponins from test samples may result in the loss of interesting and important activities.145,146 Other approaches to identification of generally cytotoxic samples are similar to those employed for biochemical assays. For reporter assays, cytotoxicity assays are often applied in order to identify samples that are apparently active as a result of signal reduction due to cell killing. This can be assessed by running a cytotoxicity assay in parallel with the reporter assay.122,124 Alternatively, cell toxicity in reporter assays can be assessed by inclusion of a second reporter as exemplified by a dual luciferase assay to simultaneously assess cytostatic and cytotoxic effects of samples (subsequently applied to natural product extracts) on NF1-null astrocytoma cells.147 Cataloging libraries for cytotoxic activity has also been applied to identify generally toxic extracts or fractions.148 Similarly, retaining data for multiple cell lines as they are amassed in a laboratory provides a baseline of toxicity against which new toxicity data can be compared. These approaches allow identification of potentially problematic extracts (or other samples) which can then be excluded from analysis or further fractionated for subsequent screening. Understanding mechanisms of cell death via high content imaging has also been successfully applied to understand and identify cytotoxic natural product extract samples in screening assays.134,148 Assessment of samples at multiple concentrations allowed for identification of several active extracts in a c-KIT screen.143 The apparent differential effects of some extracts were diminished or disappeared when tested over a broader concentration range. Given that potentially specific target modulators and non-specific cytotoxic compounds are both present at unknown concentrations, secondary assays at multiple concentrations are always desirable where possible.
Other nonspecific effects of natural product extracts, unrelated to cytotoxicity are often similar to those discussed for biochemical assays. Most prominent among these are intrinsic fluorescence or fluorescence quenching, nonspecific effects on reporter enzymes, and effects on general cellular machinery (e.g. non-specific transcription or translation inhibitors in the case of reporter assays). These can be addressed by prescreening for interference as discussed for luciferase.101 Careful choice of detection systems can also alleviate non-specific effects. For example, the AP1 assay referenced above122 employed a β lactamase reporter rather than the usual luciferase. The choice of a fluorescent ABCG2 substrate, pheophorbide a, with a wide Stokes shift reduced the probability of interference, but since pheophorbide a is a plant-derived chlorophyll derivative, there was significant interference from similar compounds in plant extracts. In this case, entire classes of plant extracts could not be accurately assessed, thus reducing the pool of extracts that could be screened.132 As with cytotoxicity, the proliferation of reporter assays allows for the development of databases identifying non-specific effects on transcription or translation (i.e. modulatory toward multiple gene expression systems). This approach was applied on a small scale in that the assays for inhibitors of the transcription factors AP1 and HIF2α were performed in our laboratory at approximately the same time.122,124 As a result, one of the criteria for hit identification with one transcription factor was lack of inhibition in the other reporter assay. For reporter assays, “off target” false positive effects can also be assessed by using unregulated and/or constitutively active reporters. Examples include constructs driven by the CMV promoter to help identify general transcription/translation/cell health effects and, in the assay for stabilizers of Pdcd4,128 inclusion of a control construct without the Pdcd4 regulatory regions as a parallel assay to confirm specificity.
Finally, orthogonal assays focused on activities related to the putative target(s) can help identify true active samples. Often this is not possible until pure compounds are obtained, but ideally it should be applied during or immediately after screening. Examples from the above referenced assays include biochemical assays for kinases involved in c-KIT signaling143 and cell death-specific assays in the case of the assay for TRAIL synergizers (in this example, caspase assays and assessment of mitochondrial potential in treated cells).141
An important general requirement illustrated by many of the examples above is the need to understand the effect(s) of samples on assay systems. This is important for any HTS application, but critical for screening of natural product extracts. The availability and application of parallel, non-specific assays can quickly identify spurious apparent activities that actually affect detection enzymes or reagents, cell survival, or other critical assay parameters.
3.3 Novel potential high throughput screening modes
The following sections summarize work that may have the potential for becoming a high throughput method with further development. In most cases, proof of the assay principle has been obtained for using natural product samples.
3.3.1 Capillary electrophoresis(CE)
Capillary electrophoretic endpoints offer the benefit of separating enzymatic substrates from products, as well as potentially avoiding sample interference. We demonstrated the utility of CE with crude extracts in an assay for HIV-1 RNase H inhibition.52 As noted above, this was used as a secondary assay due to limitations of speed using a single CE column. Current efforts using this format in our laboratory for other enzymatic assays utilize “lab-on-a-chip” multiplexed CE equipment capable of increased throughput. Electrophoretic endpoints have also been explored for HTS of tyrosine kinases and other enzymes, though in those cases, natural product samples were not tested.149,150
3.3.2 Mass spectral HTS endpoints
A useful counterpoint to the conventional use of fluorescent tags as HTS assay endpoints is to utilize the power and sensitivity of mass spectrometry. This has been done in a variety of different modes, including direct measurement of bound complexes, indirect measurement of separated bound ligand (e.g. quinone reductase-2 inhibitors by ultrafiltration LC-MS151), or by detection of enzymatic substrates, as outlined in a recent review.152
3.3.3 Immobilized receptors
Protein receptors which are naturally embedded in lipid bilayer membranes have historically proven difficult to utilize in cell-free assays. However, new approaches involving immobilization of intact proteins on fused silica columns have been applied to acetylcholinesterase153 and both cannabinoid receptors.154 These methods were applied in proof of principle to natural product extracts, with the former method using capillary electrophoretic separation of substrate and product, and the latter using displacement of a tritium labeled ligand. Neither method has been perfected for high throughput analysis at this time, but there is clear potential to do so. Another group used vascular smooth muscle cell membranes to affinity purify active molecules from TCM extracts that were then characterized in a second LC-MS step.155 The Arai and Ishibashi groups developed an affinity method where the vitamin D receptor (VDR) was covalently attached to magnetic beads. Immobilization on beads permitted convenient washing to remove unbound extract components, followed by elution of bound ligands with ethanol, and LC-MS analysis.156 Screening of several plant extracts with this method yielded a pair of novel labdane glycosides which were characterized as weak antagonists of the VDR. Bead based methods were also used by the Arai group to discover hedgehog inhibitors.157 Similarly, an assay for detecting triplex DNA binding compounds from plant extracts used agarose bead-immobilized DNA to capture compounds from the extract, followed by HPLC-ESMS.158 These have not yet been developed into high-throughput methods, but with advances in miniaturization and sample processing, there is potential for higher throughput.
3.3.4 Novel antibacterial HTS screens
Classical antibiotic discovery relied on simple whole cell assays of bacterial growth. With the genomic revolution, it appeared that sequencing bacterial strains would provide abundant drug targets for HTS. While many antibacterial targets were identified, cell free assay screening was notably unsuccessful in finding drug candidates.159 The Merck group developed a novel strategy for antibacterial discovery based on antisense methods. This was applied to the discovery of fatty acid synthesis inhibitors. Xylose-inducible antisense constructs were designed to suppress fatty acid synthesis enzymes; inhibition of essential enzymes made the organism hypersensitive to growth inhibition by compounds which target that enzyme. Implemented in Staphylococcus aureus, this fitness test approach led to the discovery of FabH/FabF inhibitors from microbial broth samples.160 A separate screen for bacterial topoisomerase inhibitors using the same approach led to the kibdelomycins, a potent and novel antibacterial class.161 Another novel approach to antibacterial screens was developed by the Clardy and Watnick groups, who carried out a whole cell screen of fungal endophyte extracts against Vibrio cholera, in which sugar metabolism was used to report on bacterial growth. A novel compound, mirandamycin, was identified with broad spectrum activity inclusive of MRSA and Mycobacterium tuberculosis.162
3.3.5 Yeast methods
Yeast assays are an attractive screening platform due to the relative ease of genetic manipulation, however, early efforts were limited by poor compound penetration into yeast cells, and by active drug export systems integral to the yeast.163 Strains in which exporters are deleted have been developed to address active efflux, and addition of low concentrations of detergents has also been found to improve results.164 Using these methods, Fernández-Acero et al. engineered a human phosphatidylinositol 3-kinase (PI3K) system in yeast and used it to screen a microbial natural product extract library. The assay relied on the toxicity of PI3K to yeast, with rescue of growth the desired endpoint for PI3K inhibition.164 A phenotypic screening approach with budding yeast classified cell cycle effects by observing changes in cell morphology caused by different natural product extracts.165
3.3.6 High content methods
Many HTS groups have wished to increase the information content of screening assays by using high throughput microscopy, also known as high content screening (HCS). While widely applied as a secondary technique, our group has utilized HCS in a primary screen for inhibitors of nuclear receptor translocation from the cytoplasm to the nucleus, with a chimeric glucocorticoid-estrogen receptor construct driven by the ER ligand binding domain. The assay was validated for both pure compounds and natural product extracts.134 The Linington group has reported the development of an image-based screening protocol using nuclear (histone H3) and cytoskeletal protein (tubulin, actin) histochemistry to distinguish compound and extract mechanisms of action.148 At this point it is not clear that high content methods will be sufficiently rapid for primary screening, however major improvements in speed and convenience have already been achieved in a few years.
3.3.7 Zebrafish
Using whole vertebrate animals for screening purposes has largely been abandoned due to the high cost of maintaining and breeding animals, as well as for ethical concerns. The zebrafish Danio rerio, however, is a model organism for which the complete genome has been sequenced. In addition, the small size of the larvae is compatible with 384 well plate assays, and drug metabolism is relatively similar to that of humans.166 Diverse patterns of locomotor behavior can be monitored to identify neuroactive compounds, while vascular development can be captured by high content microscopy. A Belgian group screened East African medicinal plant extracts for angiogenesis inhibitors using zebrafish, and found several lead compounds.167 A review of zebrafish screening of natural products has recently appeared.166
4.0 Trends and prospects
4.1 Rapid dereplication
As the number of isolated natural product structures passes 250,000 (cf. CRC Press Dictionary of Natural Products), it becomes ever more important to efficiently identify known compounds active in HTS screening of extracts. This demands a tight integration of internal chemistry and biology information with external databases and effective use of the available information. To build a user friendly interface and curate the huge amount of data is a daunting task, but one which is well worth the effort. It is also a challenge to directly link internal data systems with external systems such as SciFinder and the CRC Dictionary of Natural Products, though we have found the public PubChem system to be amenable to linkage. The combination of informatics with microgram scale separation, bioassay, and dereplication is changing the landscape of compound identification.168 New mass spectral databases and techniques promise to provide substantial leverage to rapidly recognize analogues of known structural types.169 A recent review by Potterat and Hamburger has summarized techniques for rapid deconvolution of active samples using hyphenated methods.170
4.2 Increased regulation by source countries and limited access
Effective application of HTS to natural products and the development of hits from screening campaigns into leads for drug development clearly requires continued access to source organisms. One of the drawbacks cited by pharma for natural products discovery has been the necessity for coping with source country regulations and permits, particularly for plant specimens. It should be noted that these regulations are in response to widespread exploitation of source country biological resources dating back to the colonial period. The trend of increased regulation continues, however, as countries establish defined protocols for access to biodiversity, it may be hoped that some of the previous uncertainty can be mitigated.
4.2.1 NCI letter of collection
Many years ago, the U.S. National Cancer Institute implemented a “letter of collection” (LOC) with several natural product source countries, notably Gabon, Ghana, Madagascar, Tanzania, New Zealand and Malaysia.171,172 The LOC states NCI's willingness to collaborate with local scientists and/or authorities in the discovery and development of novel drugs from organisms collected in their countries and/or territorial waters. If requested, the NCI will enter into formal agreements based on the LOC with relevant source country government agencies or organizations. The intent is to share benefits of discovery work with the source country at all stages of the process initially by supporting training and local development, and later by sharing licensing royalties. For countries in which this agreement was not signed, the NCI nonetheless observed and continues to observe the same principles.
4.2.2 Nagoya protocols
The 1992 Convention on Biodiversity (CBD) is a treaty intended to promote conservation of biological diversity, sustainable use of its components, and fair and equitable sharing of benefits arising from genetic resources. Its original provisions are known as the Rio protocol. More recently, the Nagoya Protocol, adopted in 2010, was drafted to create greater legal certainty and transparency for providers and users of genetic resources by designating points of access to genetic resources in each country, and by assuring benefit sharing when genetic resources leave the country which provided the genetic resources. By helping to ensure benefit sharing, the Nagoya Protocol creates incentives for conservation and sustainable use of genetic resources, and therefore enhances the contribution of biodiversity to development and human needs. The full impact of the Nagoya Protocol has yet to be seen. More details on the CBD are contained in a recent review.172
4.3 Which groups are currently doing HTS of Natural Products?
With the de-emphasis of active natural product discovery programs in many pharmaceutical companies,5 academic, government and biotech groups have become more prominent. In the following section, we mention a few of them.
Our own group, the Molecular Targets Laboratory, part of the Center for Cancer Research of the U.S. National Cancer Institute, has been conducting HTS of natural products for more than 15 years.173 We have relied on the many intramural labs of the NCI Center for Cancer Research for validated cancer and HIV screening targets,120,132,124,143,122,57,141,128,134 and have benefitted by the proximity of the Natural Products Repository, which holds one of the largest plant and marine invertebrate extract collections in the world,1 and the Natural Products Support Group, which prepares the extracts.174
The Eskitis Institute of Griffith University conducts drug discovery research, including HTS of natural products derived primarily from Australian plants and marine invertebrates.87,9 Natural products with a wide range of biological targets, including antiparasitic175 and hormonal176,177 activity have been reported. A focus on producing extract fractions with drug like log P characteristics has been implemented by the group.87
The Ishibashi group at Chiba University, Japan has developed novel techniques for natural product isolation in the context of HTS.156 Their work has focused on plant and myxomycete products, and has included a variety of assays for apoptotic targets.142,178,179,180
Many large pharmaceutical companies have made legacy natural product sample collections available. Among those taking advantage of this opportunity are a number of start-up biotechnology companies. One such is Warp Drive Bio, a venture backed by Sanofi and two venture capital partners.181,182 Its business plan is based on fully sequencing >100,000 microbial strains, primarily from Sanofi's microbial collection, to identify biosynthetic gene clusters, a strategy reminiscent of that used by Ecopia Biosciences,183 or Kosan Biosciences,184 but massively scaled up. Once clusters are identified, means must be found to express them and their products productively in either the natural source organism (if known) or in a heterologous host, and only then can screening commence.
A second example is the Fundación Medina, which has possession of the vast Merck microbial culture collection. Based in Granada, Spain, the group has strain development, screening, and natural product isolation capabilities.8,181 A second group at the Natural Products Discovery Institute in Doylestown, PA also has Merck and Schering-Plough microbial and plant collections and screening capabilities (http://www.npdi-us.org/).
5.0 Opportunity
Natural products continue to have great potential in screening for new drug leads. New approaches to screening libraries, greater facility in screen design, and more rapid prosecution of hit samples to pure compounds all will increase success in modern drug discovery from natural products. The future of natural products discovery is unlikely to look very much like the past, and as long as natural product scientists continue to creatively adapt new technology to their needs, that future will be bright.
Acknowledgments
We thank all former and current members of our laboratory for their contributions, and James McMahon, Barry O'Keefe and Kirk Gustafson for their thoughtful comments on the manuscript. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E, and also was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Contributor Information
Curtis J. Henrich, Email: henrichcj@mail.nih.gov.
John A. Beutler, Email: beutlerj@mail.nih.gov.
References
- 1.Cragg GM, Grothaus PG, Newman DJ. chem Rev. 2009;109:3012. doi: 10.1021/cr900019j. [DOI] [PubMed] [Google Scholar]
- 2.Mayr LM, Fuerst P. J Biomol Screen. 2008;13:443. doi: 10.1177/1087057108319644. [DOI] [PubMed] [Google Scholar]
- 3.Bade R, Chan HF, Reynisson J. Eur J Med Chem. 2010;45:5646. doi: 10.1016/j.ejmech.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 4.Cragg GM, Newman DJ. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbagen.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter GT. Nat Prod Rep. 2011;28:1783. doi: 10.1039/c1np00033k. [DOI] [PubMed] [Google Scholar]
- 6.Li JW, Vederas JC. Science. 2009;325:161. doi: 10.1126/science.1168243. [DOI] [PubMed] [Google Scholar]
- 7.Beutler JA. Curr Protoc Pharmacol. 2009;46:9. doi: 10.1002/0471141755.ph0911s46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Genilloud O, Gonzalez I, Salazar O, Martin J, Tormo JR, Vicente F. J Ind Microbiol Biotechnol. 2011;38:375. doi: 10.1007/s10295-010-0882-7. [DOI] [PubMed] [Google Scholar]
- 9.Camp D, Davis RA, Evans-Illidge EA, Quinn RJ. Future Med Chem. 2012;4:1067. doi: 10.4155/fmc.12.55. [DOI] [PubMed] [Google Scholar]
- 10.Harris ES, Erickson SD, Tolopko AN, Cao S, Craycroft JA, Scholten R, Fu Y, Wang W, Liu Y, Zhao Z, Clardy J, Shamu CE, Eisenberg DM. J Ethnopharmacol. 2011;135:590. doi: 10.1016/j.jep.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Covell DG, Huang R, Wallqvist A. Mol Cancer Ther. 2007;6:2261. doi: 10.1158/1535-7163.MCT-06-0787. [DOI] [PubMed] [Google Scholar]
- 12.Shoemaker RH. Nat Rev Cancer. 2006;6:813. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 13.Beutler JA, Cardellina JH, II, Lin CM, Hamel E, Cragg GM, Boyd MR. Bioorg Med Chem Lett. 1993;3:581. [Google Scholar]
- 14.Bowman EJ, Gustafson KR, Bowman BJ, Boyd MR. J Biol Chem. 2003;278:44147. doi: 10.1074/jbc.M306595200. [DOI] [PubMed] [Google Scholar]
- 15.Chan J, Khan SN, Harvey I, Merrick W, Pelletier J. RNA. 2004;10:528. doi: 10.1261/rna.5200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarker SD, Nahar L. Methods Mol Biol. 2012;846:1. doi: 10.1007/978-1-61779-624-1_1. [DOI] [PubMed] [Google Scholar]
- 17.Carnero A. Clin Transl Oncol. 2006;8:482. doi: 10.1007/s12094-006-0048-2. [DOI] [PubMed] [Google Scholar]
- 18.Guido RV, Oliva G, Andricopulo AD. Comb Chem High Throughput Screen. 2011;14:830. doi: 10.2174/138620711797537067. [DOI] [PubMed] [Google Scholar]
- 19.Mayr LM, Bojanic D. Curr Opin Pharmacol. 2009;9:580. doi: 10.1016/j.coph.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 20.Pereira DA, Williams JA. Br J Pharmacol. 2007;152:53. doi: 10.1038/sj.bjp.0707373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.An WF, Tolliday N. Mol Biotechnol. 2010;45:180. doi: 10.1007/s12033-010-9251-z. [DOI] [PubMed] [Google Scholar]
- 22.Inglese J, Johnson RL, Simeonov A, Xia M, Zheng W, Austin CP, Auld DS. Nat Chem Biol. 2007;3:466. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- 23.Raucy JL, Lasker JM. Curr Drug Metab. 2010;11:806. doi: 10.2174/138920010794328896. [DOI] [PubMed] [Google Scholar]
- 24.Frimurer TM, Hogberg T. Curr Top Med Chem. 2011;11:1882. doi: 10.2174/156802611796391258. [DOI] [PubMed] [Google Scholar]
- 25.Trivedi S, Liu J, Liu R, Bostwick R. Expert Opin Drug Discov. 2010;5:995. doi: 10.1517/17460441.2010.513377. [DOI] [PubMed] [Google Scholar]
- 26.Jia Y, Quinn CM, Kwak S, Talanian RV. Curr Drug Discov Technol. 2008;5:59. doi: 10.2174/157016308783769414. [DOI] [PubMed] [Google Scholar]
- 27.Deu E, Verdoes M, Bogyo M. Nat Struct Mol Biol. 2012;19:9. doi: 10.1038/nsmb.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan F, Wood KV. Assay Drug Dev Technol. 2007;5:127. doi: 10.1089/adt.2006.053. [DOI] [PubMed] [Google Scholar]
- 29.Kepp O, Galluzzi L, Lipinski M, Yuan J, Kroemer G. Nat Rev Drug Discov. 2011;10:221. doi: 10.1038/nrd3373. [DOI] [PubMed] [Google Scholar]
- 30.Canaani D. Br J Cancer. 2009;100:1213. doi: 10.1038/sj.bjc.6605000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia X, Wong ST. Stem Cells. 2012;30:1800. doi: 10.1002/stem.1168. [DOI] [PubMed] [Google Scholar]
- 32.Motabar O, Sidransky E, Goldin E, Zheng W. Curr Chem Genomics. 2010;4:50. doi: 10.2174/1875397301004010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y, Wang L, Si S, Hong B. Expert Opin Drug Discov. 2010;5:1175. doi: 10.1517/17460441.2010.529896. [DOI] [PubMed] [Google Scholar]
- 34.Muskavitch MA, Barteneva N, Gubbels MJ. Comb Chem High Throughput Screen. 2008;11:624. doi: 10.2174/138620708785739989. [DOI] [PubMed] [Google Scholar]
- 35.Brooks WH. Clin Rev Allergy Immunol. 2012;42:58. doi: 10.1007/s12016-011-8290-y. [DOI] [PubMed] [Google Scholar]
- 36.Makley LN, Gestwicki JE. Chem Biol Drug Des. 2013;81:22. doi: 10.1111/cbdd.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wells JA, McClendon CL. Nature. 2007;450:1001. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 38.Gillet VJ. Curr Opin Chem Biol. 2008;12:372. doi: 10.1016/j.cbpa.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 39.Muegge I. Med Res Rev. 2003;23:302. doi: 10.1002/med.10041. [DOI] [PubMed] [Google Scholar]
- 40.Schuffenhauer A, Popov M, Schopfer U, Acklin P, Stanek J, Jacoby E. Comb Chem High Throughput Screen. 2004;7:771. doi: 10.2174/1386207043328238. [DOI] [PubMed] [Google Scholar]
- 41.Campbell JB. IDrugs. 2010;13:874. [PubMed] [Google Scholar]
- 42.Drewry DH, Macarron R. Curr Opin Chem Biol. 2010;14:289. doi: 10.1016/j.cbpa.2010.03.024. [DOI] [PubMed] [Google Scholar]
- 43.Gozalbes R, Carbajo RJ, Pineda-Lucena A. Curr Med Chem. 2010;17:1769. doi: 10.2174/092986710791111224. [DOI] [PubMed] [Google Scholar]
- 44.Schulz MN, Hubbard RE. Curr Opin Pharmacol. 2009;9:615. doi: 10.1016/j.coph.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 45.Lachance H, Wetzel S, Kumar K, Waldmann H. J Med Chem. 2012;55:5989. doi: 10.1021/jm300288g. [DOI] [PubMed] [Google Scholar]
- 46.Lopez-Vallejo F, Giulianotti MA, Houghten RA, Medina-Franco JL. Drug Discov Today. 2012;17:718. doi: 10.1016/j.drudis.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 47.Bauer RA, Wurst JM, Tan DS. Curr Opin Chem Biol. 2010;14:308. doi: 10.1016/j.cbpa.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gul S, Gribbon P. Exp Opin Drug Discov. 2010;5:681. doi: 10.1517/17460441.2010.495748. [DOI] [PubMed] [Google Scholar]
- 49.Simeonov A, Jadhav A, Thomas CJ, Wang Y, Huang R, Southall NT, Shinn P, Smith J, Austin CP, Auld DS, Inglese J. J Med Chem. 2008;51:2363. doi: 10.1021/jm701301m. [DOI] [PubMed] [Google Scholar]
- 50.Dufau I, Lazzari A, Samson A, Pouny I, Ausseil F. Assay Drug Dev Technol. 2008;6:673. doi: 10.1089/adt.2008.143. [DOI] [PubMed] [Google Scholar]
- 51.Parniak MA, Min KL, Budihas SR, Le Grice SFJ, Beutler JA. Anal Biochem. 2003;322:33. doi: 10.1016/j.ab.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 52.Chan KC, Budihas SR, Le Grice SFJ, Parniak MA, Crouch RJ, Gaidamakov SA, Isaaq HJ, Wamiru A, McMahon JB, Beutler JA. Anal Biochem. 2004;331:296. doi: 10.1016/j.ab.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 53.Beutler JA, Alvarado-Lindner AB, McCloud TG. Ann Missouri Bot Garden. 1996;83:530. [Google Scholar]
- 54.Beutler JA, McKee TC, Fuller RW, Tischler M, Cardellina JH, II, Snader KM, McCloud TG, Boyd MR. Antiviral Chem Chemother. 1993;4:167. [Google Scholar]
- 55.Zhu M, Phillipson JD, Greengrass PM, Bowery NG, Cai Y. Phytochemistry. 1997;44:441. doi: 10.1016/s0031-9422(96)00598-5. [DOI] [PubMed] [Google Scholar]
- 56.Cardellina JH, II, Munro MHG, Fuller RW, Manfredi KP, McKee TC, Tischler M, Bokesch HR, Gustafson KR, Beutler JA, Boyd MR. J Nat Prod. 1993;56:1123. doi: 10.1021/np50097a016. [DOI] [PubMed] [Google Scholar]
- 57.Sasiela CA, Stewart DH, Kitagaki J, Safiran Y, Yang Y, Weissman AM, Oberoi P, Davydov I, Goncharova E, Beutler JA, McMahon JB, O'Keefe BR. J Biomol Screen. 2008;13:229. doi: 10.1177/1087057108315038. [DOI] [PubMed] [Google Scholar]
- 58.Hermann JC, Chen Y, Wartchow C, Menke J, Gao L, leason GSK, Haynes NE, Scott N, Petersen A, Gabriel S, Vu B, George KM, Narayanan A, Li SH, Qian H, Beatini N, Niu L, Gan QF. ACS Med Chem Lett. 2013;4:197. doi: 10.1021/ml3003296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fernando DR, Marshall AT, Forster PI, Hoebee SE, Siegele R. Am J Bot. 2013 doi: 10.3732/ajb.1200545. [DOI] [PubMed] [Google Scholar]
- 60.Eisenberg DM, Harris ES, Littlefield BA, Cao S, Craycroft JA, Scholten R, Bayliss P, Fu Y, Wang W, Qiao Y, Zhao Z, Chen H, Liu Y, Kaptchuk T, Hahn WC, Wang X, Roberts T, Shamu CE, Clardy J. Fitoterapia. 2011;82:17. doi: 10.1016/j.fitote.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hostettmann K, Marston A. Saponins. Cambridge University Press; Cambridge, UK: 1995. pp. 232–237. [Google Scholar]
- 62.Maes J, Verlooy L, Buenafe OE, de Witte PA, Esguerra CV, Crawford AD. PLoS One. 2012;7:e43850. doi: 10.1371/journal.pone.0043850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmid I, Sattler I, Grabley S, Thiericke R. J Biomol Screen. 1999;4:15. doi: 10.1177/108705719900400104. [DOI] [PubMed] [Google Scholar]
- 64.Johnson TA, Sohn J, Inman WD, Estee SA, Loveridge ST, Vervoort HC, Tenney K, Liu J, Ang KK, Ratnam J, Bray WM, Gassner NC, Shen YY, Lokey RS, McKerrow JH, Boundy-Mills K, Nukanto A, Kanti A, Julistiono H, Kardono LB, Bjeldanes LF, Crews P. J Nat Prod. 2011;74:2545. doi: 10.1021/np200673b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tu Y, Jeffries C, Ruan H, Nelson C, Smithson D, Shelat AA, Brown KM, Li XC, Hester JP, Smillie T, Khan IA, Walker L, Guy K, Yan B. J Nat Prod. 2010;73:751. doi: 10.1021/np9007359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Di L, Kerns EH. Drug Discov Today. 2006;11:446. doi: 10.1016/j.drudis.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 67.Popa-Burke I, Lupotsky B, Boyer J, Gannon W, Hughes R, Kadwill P, Lyerly D, Nichols J, Nixon E, Rimmer D, Saiz-Nicolas I, Sanfiz-Pinto B, Holland S. J Biomol Screen. 2009;14:1017. doi: 10.1177/1087057109339938. [DOI] [PubMed] [Google Scholar]
- 68.Liu Y, Beresini MH, Johnson A, Mintzer R, Shah K, Clark K, Schmidt S, Lewis C, Liimatta M, Elliott LO, Gustafson A, Heise CE. J Biomol Screen. 2012;17:225. doi: 10.1177/1087057111421525. [DOI] [PubMed] [Google Scholar]
- 69.Smith T, Ho PI, Yue K, Itkin Z, MacDougall D, Paolucci M, Hill A, Auld DS. J Biomol Screen. 2013;18:14. doi: 10.1177/1087057112455434. [DOI] [PubMed] [Google Scholar]
- 70.Janzen WP, Popa-Burke IG. J Biomol Screen. 2009;14:444. doi: 10.1177/1087057109335262. [DOI] [PubMed] [Google Scholar]
- 71.Goodfellow M, Fiedler HP. Antonie Van Leeuwenhoek. 2010;98:119. doi: 10.1007/s10482-010-9460-2. [DOI] [PubMed] [Google Scholar]
- 72.Liu X, Ashforth E, Ren B, Song F, Dai H, Liu M, Wang J, Xie Q, Zhang L. J Antibiot. 2010;63:415. doi: 10.1038/ja.2010.56. [DOI] [PubMed] [Google Scholar]
- 73.Liu X, Bolla K, Ashforth EJ, Zhuo Y, Gao H, Huang P, Stanley SA, Hung DT, Zhang L. Antonie Van Leeuwenhoek. 2012;101:55. doi: 10.1007/s10482-011-9671-1. [DOI] [PubMed] [Google Scholar]
- 74.Bascom-Slack CA, Ma C, Moore E, Babbs B, Fenn K, Greene JS, Hann BD, Keehner J, Kelley-Swift EG, Kembaiyan V, Lee SJ, Li P, Light DY, Lin EH, Schorn MA, Vekhter D, Boulanger LA, Hess WM, Vargas PN, Strobel GA, Strobel SA. Microb Ecol. 2009;58:374. doi: 10.1007/s00248-009-9494-z. [DOI] [PubMed] [Google Scholar]
- 75.Ito T, Odake T, Katoh H, Yamaguchi Y, Aoki M. J Nat Prod. 2011;74:983. doi: 10.1021/np100859a. [DOI] [PubMed] [Google Scholar]
- 76.Gerwick WH, Moore BS. Chem Biol. 2012;19:85. doi: 10.1016/j.chembiol.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thornburg CC, Zabriskie TM, McPhail KL. J Nat Prod. 2010;73:489. doi: 10.1021/np900662k. [DOI] [PubMed] [Google Scholar]
- 78.Yu H, Zhang L, Li L, Zheng C, Guo L, Li W, Sun P, Qin L. Microbiol Res. 2010;165:437. doi: 10.1016/j.micres.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 79.Dey M, Ripoll C, Pouleva R, Dorn R, Aranovich I, Zaurov D, Kurmukov A, Eliseyeva M, Belolipov I, Akimaliev A, Sodombekov I, Akimaliev D, Lila MA, Raskin I. Phytother Res. 2008;22:929. doi: 10.1002/ptr.2427. [DOI] [PubMed] [Google Scholar]
- 80.Liu L, Li YF, Cheng YY. J Chromatogr B. 2008;862:196. doi: 10.1016/j.jchromb.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 81.Zhu Y, Zhang Z, Zhang M, Mais DE, Wang MW. Comb Chem High Throughput Screen. 2010;13:837. doi: 10.2174/138620710793360257. [DOI] [PubMed] [Google Scholar]
- 82.Manissorn J, Sukrong S, Ruangrungsi N, Mizukami H. Biol Pharm Bull. 2010;33:1723. doi: 10.1248/bpb.33.1723. [DOI] [PubMed] [Google Scholar]
- 83.Van Soest RW, Boury-Esnault N, Vacelet J, Dohrmann M, Erpenbeck D, de Voogd NJ, Santodomingo N, Vanhoorne B, Kelly M, Hooper JN. PLoS One. 2012;7:e35105. doi: 10.1371/journal.pone.0035105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Puillandre N, Bouchet P, Boisselier-Dubayle MC, Brisset J, Buge B, Castelin M, Chagnoux S, Christophe T, Corbari L, Lambourdiere J, Lozouet P, Marani G, Rivasseau A, Silva N, Terryn Y, Tillier S, Utge J, Samadi S. Mol Ecol Resour. 2012;12:396. doi: 10.1111/j.1755-0998.2011.03105.x. [DOI] [PubMed] [Google Scholar]
- 85.McCloud TG. Molecules. 2010;15:4526. doi: 10.3390/molecules15074526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Collins RA, Ng TB, Fong WP, Wan CC, Yeung HW. Biochem Mol Biol Int. 1998;45:791. doi: 10.1080/15216549800203212. [DOI] [PubMed] [Google Scholar]
- 87.Camp D, Campitelli M, Carroll AR, Davis RA, Quinn RJ. Chem Biodivers. 2013;10:524. doi: 10.1002/cbdv.201200302. [DOI] [PubMed] [Google Scholar]
- 88.Wagenaar MM. Molecules. 2008;13:1406. doi: 10.3390/molecules13061406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Appleton DR, Buss AD, Butler MS. Chimia. 2007;61:327. [Google Scholar]
- 90.Bugni TS, Richards B, Bhoite L, Cimbora D, Harper MK, Ireland CM. J Nat Prod. 2008;71:1095. doi: 10.1021/np800184g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bugni TS, Harper MK, McCulloch MW, Reppart J, Ireland CM. Molecules. 2008;13:1372. doi: 10.3390/molecules13061372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tu Y, Yan B. Methods Mol Biol. 2012;918:117. doi: 10.1007/978-1-61779-995-2_9. [DOI] [PubMed] [Google Scholar]
- 93.Yuliana ND, Khatib A, Verpoorte R, Choi YH. Anal Chem. 2011;83:6902. doi: 10.1021/ac201458n. [DOI] [PubMed] [Google Scholar]
- 94.Ellson R, Stearns R, Mutz M, Brown C, Browning B, Harris D, Qureshi S, Shieh J, Wold D. Comb Chem High Throughput Screen. 2005;8:489. doi: 10.2174/1386207054867382. [DOI] [PubMed] [Google Scholar]
- 95.Waybright TJ, Britt JR, McCloud TG. J Biomol Screen. 2009;14:708. doi: 10.1177/1087057109335670. [DOI] [PubMed] [Google Scholar]
- 96.Zou L, Harkey MR, Henderson GL. Phytomedicine. 2002;9:263. doi: 10.1078/0944-7113-00121. [DOI] [PubMed] [Google Scholar]
- 97.Douat-Casassus C, Chassaing S, Di PC, Quideau S. Chembiochem. 2009;10:2321. doi: 10.1002/cbic.200900287. [DOI] [PubMed] [Google Scholar]
- 98.Zhu J, Van de Ven WJ, Verbiest T, Koeckelberghs G, Chen C, Cui Y, Vermorken AJ. Curr Med Chem. 2013;20:840. [PubMed] [Google Scholar]
- 99.Coan KE, Ottl J, Klumpp M. Expert Opin Drug Discov. 2011;6:405. doi: 10.1517/17460441.2011.561309. [DOI] [PubMed] [Google Scholar]
- 100.Balunas MJ, Su B, Landini S, Brueggemeier RW, Kinghorn AD. J Nat Prod. 2006;69:700. doi: 10.1021/np050513p. [DOI] [PubMed] [Google Scholar]
- 101.Cruz PG, Auld DS, Schultz PJ, Lovell S, Battaile KP, MacArthur R, Shen M, Tamayo-Castillo G, Inglese J, Sherman DH. Chem Biol. 2011;18:1442. doi: 10.1016/j.chembiol.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Grant SK, Sklar JG, Cummings RT. J Biomol Screen. 2002;7:531. doi: 10.1177/1087057102238627. [DOI] [PubMed] [Google Scholar]
- 103.Turek-Etienne TC, Lei M, Terracciano JS, Langsdorf EF, Bryant RW, Hart RF, Horan AC. J Biomol Screen. 2004;9:52. doi: 10.1177/1087057103259346. [DOI] [PubMed] [Google Scholar]
- 104.Hudson SA, Ecroyd H, Kee TW, Carver JA. FEBS J. 2009;276:5960. doi: 10.1111/j.1742-4658.2009.07307.x. [DOI] [PubMed] [Google Scholar]
- 105.Chamni S, De-Eknamkul W. Expert Opin Ther Pat. 2013;23:409. doi: 10.1517/13543776.2013.765861. [DOI] [PubMed] [Google Scholar]
- 106.Kongkamnerd J, Milani A, Cattoli G, Terregino C, Capua I, Beneduce L, Gallotta A, Pengo P, Fassina G, Monthakantirat O, Umehara K, De-Eknamkul W, Miertus S. J Biomol Screen. 2011;16:755. doi: 10.1177/1087057111409221. [DOI] [PubMed] [Google Scholar]
- 107.Okomo-Adhiambo M, Hurt AC, Gubareva LV. Methods Mol Biol. 2012;865:95. doi: 10.1007/978-1-61779-621-0_6. [DOI] [PubMed] [Google Scholar]
- 108.Habig M, Blechschmidt A, Dressler S, Hess B, Patel V, Billich A, Ostermeier C, Beer D, Klumpp M. J Biomol Screen. 2009;14:679. doi: 10.1177/1087057109336586. [DOI] [PubMed] [Google Scholar]
- 109.Danel F, Caspers P, Nuoffer C, Hartlein M, Kron MA, Page MG. Curr Drug Discov Technol. 2011;8:66. doi: 10.2174/157016311794519947. [DOI] [PubMed] [Google Scholar]
- 110.Wu B, Gao J, Wang MW. Acta Pharmacol Sin. 2005;26:339. doi: 10.1111/j.1745-7254.2005.00040.x. [DOI] [PubMed] [Google Scholar]
- 111.Dao TT, Dang TT, Nguyen PH, Kim E, Thuong PT, Oh WK. Bioorg Med Chem Lett. 2012;22:3688. doi: 10.1016/j.bmcl.2012.04.028. [DOI] [PubMed] [Google Scholar]
- 112.Grienke U, Schmidtke M, von GS, Kirchmair J, Liedl KR, Rollinger JM. Nat Prod Rep. 2012;29:11. doi: 10.1039/c1np00053e. [DOI] [PubMed] [Google Scholar]
- 113.Liu AL, Liu B, Qin HL, Lee SM, Wang YT, Du GH. Planta Med. 2008;74:847. doi: 10.1055/s-2008-1074558. [DOI] [PubMed] [Google Scholar]
- 114.Azizuddin, Khan AM, Choudhary MI. Nat Prod Res. 2011;25:750. doi: 10.1080/14786419.2010.513684. [DOI] [PubMed] [Google Scholar]
- 115.Carpinella MC, Andrione DG, Ruiz G, Palacios SM. Phytother Res. 2010;24:259. doi: 10.1002/ptr.2923. [DOI] [PubMed] [Google Scholar]
- 116.Lee YM, Kim YS, Lee Y, Kim J, Sun H, Kim JH, Kim JS. Phytother Res. 2012;26:778. doi: 10.1002/ptr.3644. [DOI] [PubMed] [Google Scholar]
- 117.Schenk T, Breel GJ, Koevoets P, van den Berg S, Hogenboom AC, Irth H, Tjaden UR, van der Greef J. J Biomol Screen. 2003;8:421. doi: 10.1177/1087057103255973. [DOI] [PubMed] [Google Scholar]
- 118.Halley F, Reinshagen J, Ellinger B, Wolf M, Niles AL, Evans NJ, Kirkland TA, Wagner JM, Jung M, Gribbon P, Gul S. J Biomol Screen. 2011;16:1227. doi: 10.1177/1087057111416004. [DOI] [PubMed] [Google Scholar]
- 119.Zoraghi R, Worrall L, See RH, Strangman W, Popplewell WL, Gong H, Samaai T, Swayze RD, Kaur S, Vuckovic M, Finlay BB, Brunham RC, McMaster WR, Davies-Coleman MT, Strynadka NC, Andersen RJ, Reiner NE. J Biol Chem. 2011;286:44716. doi: 10.1074/jbc.M111.289033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Beutler JA, McMahon JB, Johnson TR, O'Keefe BR, Buzzell RA, Robbins D, Gardella R, Wilson J, Boyd MR. J Biomolecular Screening. 2002;7:105. doi: 10.1177/108705710200700202. [DOI] [PubMed] [Google Scholar]
- 121.Arai MA, Masada A, Ohtsuka T, Kageyama R, Ishibashi M. Bioorg Med Chem Lett. 2009;19:5778. doi: 10.1016/j.bmcl.2009.07.146. [DOI] [PubMed] [Google Scholar]
- 122.Ruocco KM, Goncharova EI, Young MR, Colburn NH, McMahon JB, Henrich CJ. J Biomol Screen. 2007;12:133. doi: 10.1177/1087057106296686. [DOI] [PubMed] [Google Scholar]
- 123.Hattori H, Okuda K, Murase T, Shigetsura Y, Narise K, Semenza GL, Nagasawa H. Bioorg Med Chem. 2011;19:5392. doi: 10.1016/j.bmc.2011.07.060. [DOI] [PubMed] [Google Scholar]
- 124.Woldemichael GM, Vasselli JR, Gardella RS, McKee TC, Linehan WM, McMahon JB. J Biomol Screen. 2006;11:678. doi: 10.1177/1087057106289234. [DOI] [PubMed] [Google Scholar]
- 125.Gao J, Wang Y, Xing Q, Yan J, Senthil M, Akmal Y, Kowolik CM, Kang J, Lu DM, Zhao M, Lin Z, Cheng CH, Yip ML, Yim JH. Mol Cancer Ther. 2011;10:1774. doi: 10.1158/1535-7163.MCT-11-0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mora FD, Jones DK, Desai PV, Patny A, Avery MA, Feller DR, Smillie T, Zhou YD, Nagle DG. J Nat Prod. 2006;69:547. doi: 10.1021/np050397q. [DOI] [PubMed] [Google Scholar]
- 127.Santagata S, Xu YM, Wijeratne EM, Kontnik R, Rooney C, Perley CC, Kwon H, Clardy J, Kesari S, Whitesell L, Lindquist S, Gunatilaka AA. ACS Chem Biol. 2012;7:340. doi: 10.1021/cb200353m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Blees JS, Schmid T, Thomas CL, Baker AR, Benson L, Evans JR, Goncharova EI, Colburn NH, McMahon JB, Henrich CJ. J Biomol Screen. 2010;15:21. doi: 10.1177/1087057109351028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fribley AM, Cruz PG, Miller JR, Callaghan MU, Cai P, Narula N, Neubig RR, Showalter HD, Larsen SD, Kirchhoff PD, Larsen MJ, Burr DA, Schultz PJ, Jacobs RR, Tamayo-Castillo G, Ron D, Sherman DH, Kaufman RJ. J Biomol Screen. 2011;16:825. doi: 10.1177/1087057111414893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ausseil F, Samson A, Aussagues Y, Vandenberghe I, Creancier L, Pouny I, Kruczynski A, Massiot G, Bailly C. J Biomol Screen. 2007;12:106. doi: 10.1177/1087057106296494. [DOI] [PubMed] [Google Scholar]
- 131.Zhang R, Yan PK, Zhou CH, Liao JY, Wang MW. Acta Pharmacol Sin. 2007;28:125. doi: 10.1111/j.1745-7254.2007.00451.x. [DOI] [PubMed] [Google Scholar]
- 132.Henrich CJ, Bokesch HR, Dean M, Bates SE, Robey RW, Goncharova EI, Wilson JA, McMahon JB. J Biomol Screen. 2006;11:176. doi: 10.1177/1087057105284576. [DOI] [PubMed] [Google Scholar]
- 133.Findeis MA, Schroeder F, McKee TD, Yager D, Fraering PC, Creaser SP, Austin WF, Clardy J, Wang R, Selkoe D, Eckman CB. ACS Chem Neurosci. 2012;3:941. doi: 10.1021/cn3000857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dull A, Goncharova E, Hager G, McMahon JB. J Steroid Biochem Mol Biol. 2010;122:341. doi: 10.1016/j.jsbmb.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Holbeck SL, Collins JM, Doroshow JH. Mol Cancer Ther. 2010;9:1451. doi: 10.1158/1535-7163.MCT-10-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Souza AC, de FA, da Silveira RB, Justo GZ. Curr Drug Targets. 2012;13:1072. doi: 10.2174/138945012802008991. [DOI] [PubMed] [Google Scholar]
- 137.Fernandez LS, Jobling MF, Andrews KT, Avery VM. Phytother Res. 2008;22:1409. doi: 10.1002/ptr.2510. [DOI] [PubMed] [Google Scholar]
- 138.Gonzalez-Coloma A, Reina M, Saenz C, Lacret R, Ruiz-Mesia L, Aran VJ, Sanz J, Martinez-Diaz RA. Parasitol Res. 2012;110:1381. doi: 10.1007/s00436-011-2638-3. [DOI] [PubMed] [Google Scholar]
- 139.Guzman JD, Gupta A, Evangelopoulos D, Basavannacharya C, Pabon LC, Plazas EA, Munoz DR, Delgado WA, Cuca LE, Ribon W, Gibbons S, Bhakta S. J Antimicrob Chemother. 2010;65:2101. doi: 10.1093/jac/dkq313. [DOI] [PubMed] [Google Scholar]
- 140.Zhou YM, Shao L, Li JA, Han LZ, Cai WJ, Zhu CB, Chen DJ. Biosci Biotechnol Biochem. 2011;75:1746. doi: 10.1271/bbb.110290. [DOI] [PubMed] [Google Scholar]
- 141.Booth NL, Sayers TJ, Brooks AD, Thomas CF, Jacobsen K, Goncharova EI, McMahon JB, Henrich CJ, Henrich CJ. Cancer Immunol Immunother. 2009;58:1229. doi: 10.1007/s00262-008-0637-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ishibashi M, Ohtsuki T. Med Res Rev. 2008;28:688. doi: 10.1002/med.20123. [DOI] [PubMed] [Google Scholar]
- 143.Henrich CJ, Goncharova EI, Wilson JA, Gardella RS, Johnson TR, McMahon JB, Takada K, Bokesch HR, Gustafson KR. Chemical Biology & Drug Design. 2007;69:321. doi: 10.1111/j.1747-0285.2007.00508.x. [DOI] [PubMed] [Google Scholar]
- 144.Datta S, Li J, Mahdi F, Jekabsons MB, Nagle DG, Zhou YD. J Nat Prod. 2012;75:2216. doi: 10.1021/np300711e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ali R, Mirza Z, Ashraf GM, Kamal MA, Ansari SA, Damanhouri GA, Abuzenadah AM, Chaudhary AG, Sheikh IA. Anticancer Res. 2012;32:2999. [PubMed] [Google Scholar]
- 146.Coleman JJ, Okoli I, Tegos GP, Holson EB, Wagner FF, Hamblin MR, Mylonakis E. ACS Chem Biol. 2010;5:321. doi: 10.1021/cb900243b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hawes JJ, Nerva JD, Reilly KM. J Biomol Screen. 2008;13:795. doi: 10.1177/1087057108321085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Schulze CJ, Bray WM, Woerhmann MH, Stuart J, Lokey RS, Linington RG. Chem Biol. 2013;20:285. doi: 10.1016/j.chembiol.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Li Y, Liu D, Bao JJ. J Chromatogr B Analyt Technol Biomed Life Sci. 2011;879:107. doi: 10.1016/j.jchromb.2010.10.034. [DOI] [PubMed] [Google Scholar]
- 150.Liu DN, Li L, Lu WP, Liu YQ, Wehmeyer KR, Bao JJ. Anal Sci. 2008;24:333. doi: 10.2116/analsci.24.333. [DOI] [PubMed] [Google Scholar]
- 151.Choi vY, Jermihov K, Nam SJ, Sturdy M, Maloney K, Qiu X, Chadwick LR, Main M, Chen SN, Mesecar AD, Farnsworth NR, Pauli GF, Fenical W, Pezzuto JM, van Breemen RB. Anal Chem. 2011;83:1048. doi: 10.1021/ac1028424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Xin GZ, Zhou JL, Qi LW, Li P. Comb Chem High Throughput Screen. 2011;14:93. doi: 10.2174/138620711794474060. [DOI] [PubMed] [Google Scholar]
- 153.Min W, Wang W, Chen J, Wang A, Hu Z. Anal Bioanal Chem. 2012;404:2397. doi: 10.1007/s00216-012-6333-8. [DOI] [PubMed] [Google Scholar]
- 154.Moaddel R, Rosenberg A, Spelman K, Frazier J, Frazier C, Nocerino S, Brizzi A, Mugnaini C, Wainer IW. Anal Biochem. 2011;412:85. doi: 10.1016/j.ab.2010.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yang X, Wang Y, Zhang X, Chang R, Li X. J Sep Sci. 2011;34:2586. doi: 10.1002/jssc.201100366. [DOI] [PubMed] [Google Scholar]
- 156.Arai MA, Kobatake E, Koyano T, Kowithayakorn T, Kato S, Ishibashi M. Chem Asian J. 2009;4:1802. doi: 10.1002/asia.200900357. [DOI] [PubMed] [Google Scholar]
- 157.Arai MA. Chem Pharm Bull. 2011;59:417. doi: 10.1248/cpb.59.417. [DOI] [PubMed] [Google Scholar]
- 158.Xu N, Yang H, Cui M, Wan C, Liu S. Anal Chem. 2012;84:2562. doi: 10.1021/ac202796v. [DOI] [PubMed] [Google Scholar]
- 159.Singh SB, Young K, Miesel L. Expert Rev Anti Infect Ther. 2011;9:589. doi: 10.1586/eri.11.81. [DOI] [PubMed] [Google Scholar]
- 160.Young K, Jayasuriya H, Ondeyka JG, Herath K, Zhang C, Kodali S, Galgoci A, Painter R, Brown-Driver V, Yamamoto R, Silver LL, Zheng Y, Ventura JI, Sigmund J, Ha S, Basilio A, Vicente F, Tormo JR, Pelaez F, Youngman P, Cully D, Barrett JF, Schmatz D, Singh SB, Wang J. Antimicrob Agents Chemother. 2006;50:519. doi: 10.1128/AAC.50.2.519-526.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Phillips JW, Goetz MA, Smith SK, Zink DL, Polishook J, Onishi R, Salowe S, Wiltsie J, Allocco J, Sigmund J, Dorso K, Lee S, Skwish S, de la Cruz M, Martin J, Vicente F, Genilloud O, Lu J, Painter RE, Young K, Overbye K, Donald RG, Singh SB. Chem Biol. 2011;18:955. doi: 10.1016/j.chembiol.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 162.Ymele-Leki P, Cao S, Sharp J, Lambert KG, McAdam AJ, Husson RN, Tamayo G, Clardy J, Watnick PI. PLoS One. 2012;7:e31307. doi: 10.1371/journal.pone.0031307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Norcliffe JL, Alvarez-Ruiz E, Martin-Plaza JJ, Steel PG, Denny PW. Parasitology. 2013:1. doi: 10.1017/S0031182013000425. [DOI] [PubMed] [Google Scholar]
- 164.Fernandez-Acero T, Rodriguez-Escudero I, Vicente F, Monteiro MC, Tormo JR, Cantizani J, Molina M, Cid VJ. J Biomol Screen. 2012;17:1018. doi: 10.1177/1087057112450051. [DOI] [PubMed] [Google Scholar]
- 165.Qaddouri B, Guaadaoui A, Bellirou A, Hamal A, Melhaoui A, Brown GW, Bellaoui M. Evid Based Complement Alternat Med. 2011;2011:954140. doi: 10.1093/ecam/nep069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hung MW, Zhang ZJ, Li S, Lei B, Yuan S, Cui GZ, Man HP, Chan K, Lee SM. Evid Based Complement Alternat Med. 2012;2012:605303. doi: 10.1155/2012/605303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Crawford AD, Liekens S, Kamuhabwa AR, Maes J, Munck S, Busson R, Rozenski J, Esguerra CV, de Witte PA. PLoS One. 2011;6:e14694. doi: 10.1371/journal.pone.0014694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lang G, Mayhudin NA, Mitova MI, Sun L, van der SS, Blunt JW, Cole AL, Ellis G, Laatsch H, Munro MH. J Nat Prod. 2008;71:1595. doi: 10.1021/np8002222. [DOI] [PubMed] [Google Scholar]
- 169.Kersten RD, Yang YL, Xu Y, Cimermancic P, Nam SJ, Fenical W, Fischbach MA, Moore BS, Dorrestein PC. Nat Chem Biol. 2011;7:794. doi: 10.1038/nchembio.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Potterat O, Hamburger M. Nat Prod Rep. 2013;30:546. doi: 10.1039/c3np20094a. [DOI] [PubMed] [Google Scholar]
- 171.Cragg GM, Boyd MR, Grever MJ, Schepartz SA. 1994:83. [Google Scholar]
- 172.Cragg GM, Katz F, Newman DJ, Rosenthal J. Nat Prod Rep. 2012;29:1407. doi: 10.1039/c2np20091k. [DOI] [PubMed] [Google Scholar]
- 173.McMahon JB, Beutler JA, O'Keefe BR, Goodrum CBB, Myers MA, Boyd MR, Biomolecular Screening J. 2000;5:169. doi: 10.1177/108705710000500309. [DOI] [PubMed] [Google Scholar]
- 174.McCloud TG. Molecules. 2010;15:4526. doi: 10.3390/molecules15074526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Carroll AR, Fechner GA, Smith J, Guymer GP, Quinn RJ. J Nat Prod. 2008;71:1479. doi: 10.1021/np8002707. [DOI] [PubMed] [Google Scholar]
- 176.Carroll AR, Lamb J, Moni R, Guymer GP, Forster PI, Quinn RJ. J Nat Prod. 2008;71:1564. doi: 10.1021/np800247u. [DOI] [PubMed] [Google Scholar]
- 177.Carroll AR, Lamb J, Moni R, Hooper JN, Quinn RJ. J Nat Prod. 2008;71:884. doi: 10.1021/np070658r. [DOI] [PubMed] [Google Scholar]
- 178.Kikuchi H, Ohtsuki T, Koyano T, Kowithayakorn T, Sakai T, Ishibashi M. J Nat Prod. 2010;73:452. doi: 10.1021/np900404e. [DOI] [PubMed] [Google Scholar]
- 179.Kikuchi H, Ohtsuki T, Koyano T, Kowithayakorn T, Sakai T, Ishibashi M. J Nat Prod. 2007;70:1910. doi: 10.1021/np0703904. [DOI] [PubMed] [Google Scholar]
- 180.Toume K, Nakazawa T, Hoque T, Ohtsuki T, Arai MA, Koyano T, Kowithayakorn T, Ishibashi M. Planta Med. 2012;78:1370. doi: 10.1055/s-0032-1314975. [DOI] [PubMed] [Google Scholar]
- 181.Monteiro MC, de la Cruz M, Cantizani J, Moreno C, Tormo JR, Mellado E, De Lucas JR, Asensio F, Valiante V, Brakhage AA, Latge JP, Genilloud O, Vicente F. J Biomol Screen. 2012;17:542. doi: 10.1177/1087057111433459. [DOI] [PubMed] [Google Scholar]
- 182.Sheridan C. Nature Biotechnology. 2012;30:385. doi: 10.1038/nbt.2208. [DOI] [PubMed] [Google Scholar]
- 183.Banskota AH, McAlpine JB, Sorensen D, Ibrahim A, Aouidate M, Piraee M, Alarco AM, Farnet CM, Zazopoulos E. J Antibiotics. 2006;59:533. doi: 10.1038/ja.2006.74. [DOI] [PubMed] [Google Scholar]
- 184.Menzella HG, Reeves CD. Curr Opin Microbiol. 2007;10:238. doi: 10.1016/j.mib.2007.05.005. [DOI] [PubMed] [Google Scholar]