Abstract
Aberrant EGFR signaling strongly promotes glioma malignancy and treatment resistance. The most prevalent mutation, ΔEGFR/EGFRvIII, is an in-frame deletion of the extracellular domain, which occurs in more than 25% of glioblastomas and enhances growth and survival of tumor cells. Paradoxically, the signaling of the potent oncogene ΔEGFR is of low intensity, raising the question of whether it exhibits preferential signaling to key downstream targets. We have observed levels of phosphorylation of STAT5 at position Y699 in cells expressing ΔEGFR that are similar or higher than in cells that overexpress EGFR and are acutely stimulated with EGF, prompting us to investigate the role of STAT5 activation in glioblastoma. Here, we show that in human glioblastoma samples, pSTAT5 levels correlated positively with EGFR expression and were associated with reduced survival. Interestingly, the activation of STAT5b downstream of ΔEGFR was dependent on SFKs, while the signal from acutely EGF-stimulated EGFR to STAT5b involved other kinases. Phosphorylated STAT5b and ΔEGFR associated in the nucleus, bound DNA and were found on promoters known to be regulated by STAT5 including that of the Aurora A gene. ΔEGFR cooperated with STAT5b to regulate the Bcl-XL promoter and knockdown of STAT5b suppressed anchorage independent growth, reduced the levels of Bcl-XL and sensitized glioblastoma cells to cisplatin. Together these results delineate a novel association of nuclear ΔEGFR with STAT5b, which promotes oncogenesis and treatment resistance in glioblastoma by direct regulation of anti-apoptotic gene, Bcl-XL.
Keywords: nuclear EGFRvIII/ΔEGFR, STAT5b, Bcl-XL, glioma transcriptional regulation
Dysregulated signaling by receptor tyrosine kinases (RTK) is a major contributor to cancer, including gliomas, and is an important target for therapy. Gene amplification and consequent high-level expression of the epidermal growth factor receptor (EGFR) is frequently observed in glioma and is often accompanied by DNA rearrangement.1 The most common of the rearrangements leads to the deletion of exons 2–7 in the EGFR mRNA, causing an in-frame deletion of 801 bp in the extracellular domain2,3 and results in a protein, deleted-(2-7) EGFR (also known as EGFRvIII, EGFR* and ΔEGFR). ΔEGFR overexpression together with EGFR amplification has been shown to be a strong indicator of poor survival.4 Recent research has shown an unfavorable prognostic relationship between EGFR amplification and overall survival in patients with GBM.5 More recently, analyses of over 600 patients identified ΔEGFR as one marker of a group of patients with poorer outcomes.6 This association with poorer outcomes in patients is reflected in ΔEGFR’s ability to confer enhanced tumorigenicity on glioma cells in vivo by reducing apoptosis and increasing proliferation, as well as reducing sensitivity to several chemotherapeutics.7,8
Several characteristics associated with GBM malignancies including invasion, treatment resistance and immunosuppression have recently been associated with signaling pathways converging on a small number of transcription factors including the STAT (signal transducer and activator of transcription) family. The role of STAT3 in glioma has been well characterized by several laboratories. We recently identified STAT5b as another important member of this family in glioblastoma, particularly in ΔEGFR expressing cells.9 STAT5 is a latent cytoplasmic protein, which comprises two highly homologous isoforms, STAT5a and STAT5b, encoded by separate genes10 and constitutive activation of STAT5 has been reported to be associated with malignant transformation of hematological malignancies, breast cancer and prostate cancer. 11–13 STAT5a and STAT5b proteins exert not only overlapping but also distinct functions that can be attributed to cell-specific differences in mRNA levels,14 slightly different DNA binding specificities,15 altered half life of pYSTAT5 isoforms, nucleo-cytoplasmic shuttling16,17 or differential activation by serine phosphorylation.18 Furthermore, mammary-directed expression of only wild-type STAT5a resulted in mammary tumors,19 and both isoforms have differential activities in association with the ERα or ERβ isoforms.15 In contrast, other tumors such as HCC or glioblastoma rely on STAT5b activation for tumor aggressiveness and increased cell invasion.20,21 These studies indicate that downstream factors controlled by distinct STAT5 isoforms may also modulate organ specific oncogenic functions. However, the distinct roles of STAT5a and STAT5b in human GBM have not been elucidated.
Here, we present data related to the function of STAT5 activation in glioblastoma. Analysis of human tumors showed STAT5b as the predominant isoform in glioblastoma and demonstrated a positive correlation between EGFR expression levels and STAT5b phosphorylation at Y699 in glioblastoma. The presence of pY699-STAT5b was associated with poor outcome in this disease. STAT5b interacted with ΔEGFR in the nucleus of glioma cells, and this complex associated with promoter sequences and regulated gene expression. Importantly, we found that STAT5b is a key regulator of Bcl-XL, long recognized as a major effector of ΔEGFR’s ability to confer resistance to DNA-damaging chemotherapeutic agents.22 Accordingly, knockdown of STAT5b suppressed transformation by ΔEGFR and sensitized glioblastoma cells to cisplatin-induced apoptotic death.
Material and Methods
Cell culture, retrovirus infection, and transfection
The human glioblastoma cell lines, LNZ308 and LN428, were cultured as described earlier.9 STAT5a/b double knockout mouse embryo fibroblasts (MEFs) were purchased from St. Jude’s Children’s Research Hospital and grown in DMEM medium as described earlier. Cell lines were fingerprinted for identity using a PCR-based analysis (GenomeLab Human STR Primer set from Beckman Coulter) (Supporting Information Table 7).
Co-immunoprecipitation and immunoblotting assays
All primary antibodies used in this study were from Cell Signaling Technologies, except anti-pEGFR pY845, which was from Upstate Biotechnology, and anti-STAT5, which was from Santa Cruz Biotechnology, Inc. Secondary antibody used was goat-anti rabbit from Pierce (Thermo Scientific).
For the co-immunoprecipitation assay, total lysates or nuclear extracts were incubated with anti-EGFR antibodies We used a mix of two EGFR antibodies, 1 µg of anti-EGFR (AB-1, mouse mAb528 from Calbiochem) and 2 µg of anti-EGFR (LA22, Upstate Biotechnology). The immunoprecipitation complexes were resolved by NuPage gels and subjected to immunoblotting analysis.
Plasmid construction and mutagenesis
To clone ΔEGFR into pENTR/TEV/D-TOPO vector (Invitrogen), we PCR-amplified the gene from pcDNA 3.1/ΔEGFR using primers EGFR-F (5′-CACCATGCGACCCTCCGGGACGGCC-3′) & EGFR-R (5′-TGCTCCAATAAATTCACTGCT-3′). The gene fragment was then TOPO-cloned into pENTR/TEV/D-TOPO vector and sequence verified. Mutagenesis of the NLS (378RRR380 to 378AAA380) was performed using QuikChange mutagenesis kit (Stratagene) using primers ΔEGFR-NLS1-F (5′-CGGCCTCTTCATGGCAGCGGCCCACATCGTTC-3′) and ΔEGFR-NLS1-R (5′-GAACGATGTGGGCCGCTGCCATGAAGAGGCCG- 3′) as per manufacturer’s instructions. Both ΔEGFR and ΔEGFR-NLS mutant were subcloned into pcDNA-DEST47 via Gateway cloning technology (Invitrogen) to generate C-terminal GFP-tagged protein.
Chromatin immunoprecipitation (ChIP)
ChIP in U87 and mouse astroctyes were performed as published before.23,24 For the sequential ChIP assay, the DNA–protein complex was eluted from the protein A Sepharose beads by incubating with elution buffer containing 10 mM DTT in 37°C for 30 min twice. After 10-fold dilution with dilution buffer, the eluted mixture was used to perform the second immunoprecipitation assay. For the ChIP real-time PCR, the resulting DNA from the ChIP assay was used for PCR using SYBR Green Supermix, and each reaction was performed in duplicate, and the experiment was done in triplicate. Specific sequences of the human Aurora A promoter in the immunoprecipitates were detected by qPCR with forward (5′-CTGTTGCTTCACCGATAAATGGC-3′) and reverse (5′-CTCTAGCTAGAAAGCCGATTGGC-3′) primers. Sequences of the human Bcl-XL promoter in the immunoprecipitates were detected with forward (5′-CACTGGTGCTTTCGATTTGA-3′) and reverse (5′-GGGAGAGAAAGAGCTTCAGGA-3′) primers.
Immunohistochemistry
Immunohistochemistry for phospho-STAT5 was performed using methods similar to those previously described.25 A block with representative tumor tissue was chosen for study, and 5-µm sections were placed on positively charged slides. Multiple serial sections from each case were stored for later staining. An antibody from Cell Signaling against pSTAT5 (Y699) was used at a concentration of 1:50. While some cytoplasmic staining was noted, staining was predominately nuclear and scoring was based on nuclear expression. Scoring was performed using a semi-quantitative system and divided into scores of 0 (no staining), 1 (light-moderate staining) or 2 (strong staining). Staining for EGFR was performed as previously described.26
Immunofluorescence
The cellular localization of GFP, GFP-tagged ΔEGFR and GFP-tagged ΔEGFR-NLS1 as well as phospho-STAT5 was determined using confocal immunofluorescence. Cells grown on coverslips were fixed in phosphate-buffered 4% paraformaldehyde and incubated with anti-phospho-STAT5 antibody (Cell Signaling) followed by Alexa-546-labeled goat anti-rabbit antibody (red color, Molecular Probes). Nucleus was localized using TOPRO3 (blue color, Molecular Probes). Images were captured using the Olympus LSM 510 microscope and a 60× objective. For calculating the percentage of cells with increased pSTAT5 signal, at least 100 transfected cells were inspected manually for increased signal. To calculate the signal intensity, pSTAT5 signal was quantified in transfected and non-transfected cells using Image J software and the generated signal intensities were computed.
Promoter assay
Cells were transfected with wild type or mutated Aurora-A promoter (−968 + 124) luciferase construct (pGL2-Aurora-A promoter) by LipofectAMINE 2000 (Invitrogen) as described earlier.27 For investigating the transcriptional activity of STAT5b/mutants and EGFR/ΔEGFR, we used the STAT5a/b MEFs and the Sp2.1 containing reporter plasmid that is specific for Stat5b. Cells were transfected with either the His-STAT5b/mutants and/or the EGFR expression vector-pcDNA3-EGFR/ΔEGFR along with the Renilla luciferase in 6-well plates and subjected to luciferase assay as described earlier. The same method was followed with respect to the Bcl-XL promoter assay.
DNA constructs and siRNA
pMX-puro-STAT5a1*6 and pMX-puro-STAT5b1*6 were a gift from Dr. Kitamura T (University of Tokyo, Japan). Lentiviruses expressing shRNA-GFP and -STAT5b were obtained from the RNAi Consortium shRNA Library of the Broad Institute (Boston, MA). STAT1 and STAT3 siRNAs were purchased from Santa Cruz Biotechonolgy (Santa Cruz, CA).
Electrophoretic mobility shift assay (EMSA)
Nuclear extracts were prepared and EMSAs were performed on 4% native polyacrylamide gels. Complementary duplex β-casein probes (5′-AGATTTCTAGGAATTCAATTC-3′) and Bcl-XL probes (5′-TTCCGAGGAAGGCATTTCGGAGAAGACGGGG-3′) were synthesized and labeled with 5 µCi of [γ-32P]ATP (PerkinElmer Life Sciences) using T4 polynucleotide kinase (Promega). For supershift experiments, extracts were preincubated with Stat5b polyclonal antibody (Milliopore, Billerica, MA).
Results
STAT5b is the predominant STAT5 isoform in GBM
A tyrosine-directed, mass spectrometry based screen identified phosphorylation of STAT5 at Y694/9 as being elevated in LN428 and LNZ308 cells expressing ΔEGFR over the wild-type receptor and acutely stimulated with EGF.9 In cultured U87 cells, expression of ΔEGFR at high levels also resulted in an increase in STAT5 phosphorylation at this residue (Supporting Information Fig. 1). Phosphorylation at Y694/9 is an obligatory and dominant activation step and is required for formation of the STAT5 dimer,28 suggesting that ΔEGFR may functionally activate this transcription factor. We first determined which STAT5 isoform was present in human GBM, since previous studies have not distinguished the key functional role of the STAT5 isoforms though there are reports implicating varied expression patterns of STAT5a and STAT5b in cancer.12,29 We analyzed a collection of GBM tumor samples and cell lines by western blot analysis and found STAT5b to be the predominant STAT5 isoform expressed in eight clinical samples (Fig. 1a), as well as in several cell lines (Fig. 1b). These observations are in line with an earlier report that showed STAT5b to be the most predominant isoform in glioblastoma cells.21 Expression of ΔEGFR led to a statistically significant increase in STAT5 phosphorylation in the cell lines used in the initial phosphoproteomic analysis, LNZ308 and LN428, and a trend was observed in U87 and U251 cells.9 Hence we decided to use the LNZ308, LN428 and U87 cells for further studies throughout this paper.
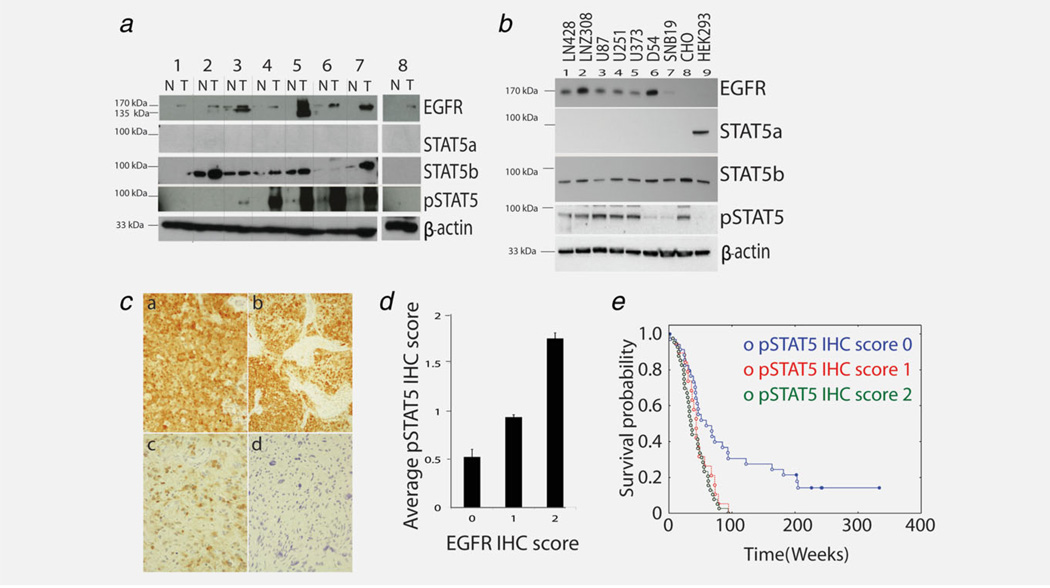
Figure 1.
STAT5b is the predominant STAT5 molecule in gliomas and its phosphorylation is associated with elevated EGFR and poor outcome (a) Western blot analysis of STAT5a, STAT5b, pSTAT5 and EGFR expression in GBM patients. N and T denote normal and tumor tissue, respectively. (b) Western blot analysis of STAT5a, STAT5b, pSTAT5 and EGFR in glioma cell lines. (c) Representative photo-micrographs (×400 magnification) of staining for pSTAT5 in glioblastoma, with examples showing (a, b) strong staining in tumor cells; (c) moderate staining; and (d) no staining. (d) Correlation of pSTAT5 staining with EGFR expression, the spearman rank order correlation is 0.62 with a p value < 0.001. (e) Kaplan Meier curve comparing outcomes of patients (total n = 97) with glioblastoma whose tumors showed no staining (upper curve) compared with moderate or strong staining (lower curves) (p value = 0.018).
Phosphorylation of STAT5b is associated with elevated EGFR and poor outcome
To examine whether activation of STAT5b was related to either EGFR status or outcomes in human tumors, we performed immunohistochemistry on a set of glioblastoma samples. Staining was predominately nuclear, but similar to previous reports12 some cytoplasmic staining was also seen (Fig. 1c). We found a significant correlation (p value <0.001) between pSTAT5 and EGFR expression (Fig. 1d) which is consistent with Fig. 1a showing that pSTAT5 is positively correlated with EGFR expression in five of eight GBMs. Furthermore, semi-quantitative scoring showed that patients whose tumors had detectable pSTAT5 (moderate or strong staining, scores = 1 or 2) had a worse clinical outcome (p value = 0.018, log rank test) when compared with patients whose tumors had no detectable pSTAT5 (no staining, score = 0) (Fig. 1e). It has been previously reported that STAT5b expression is markedly increased in glioblastoma compared with normal cortex and diffuse astrocytoma.21 These data suggest that increased EGFR signaling is associated with STAT5b activation and that this activation is in turn prognostic of a worse clinical outcome in glioblastoma.
Nuclear ΔEGFR associates directly with STAT5
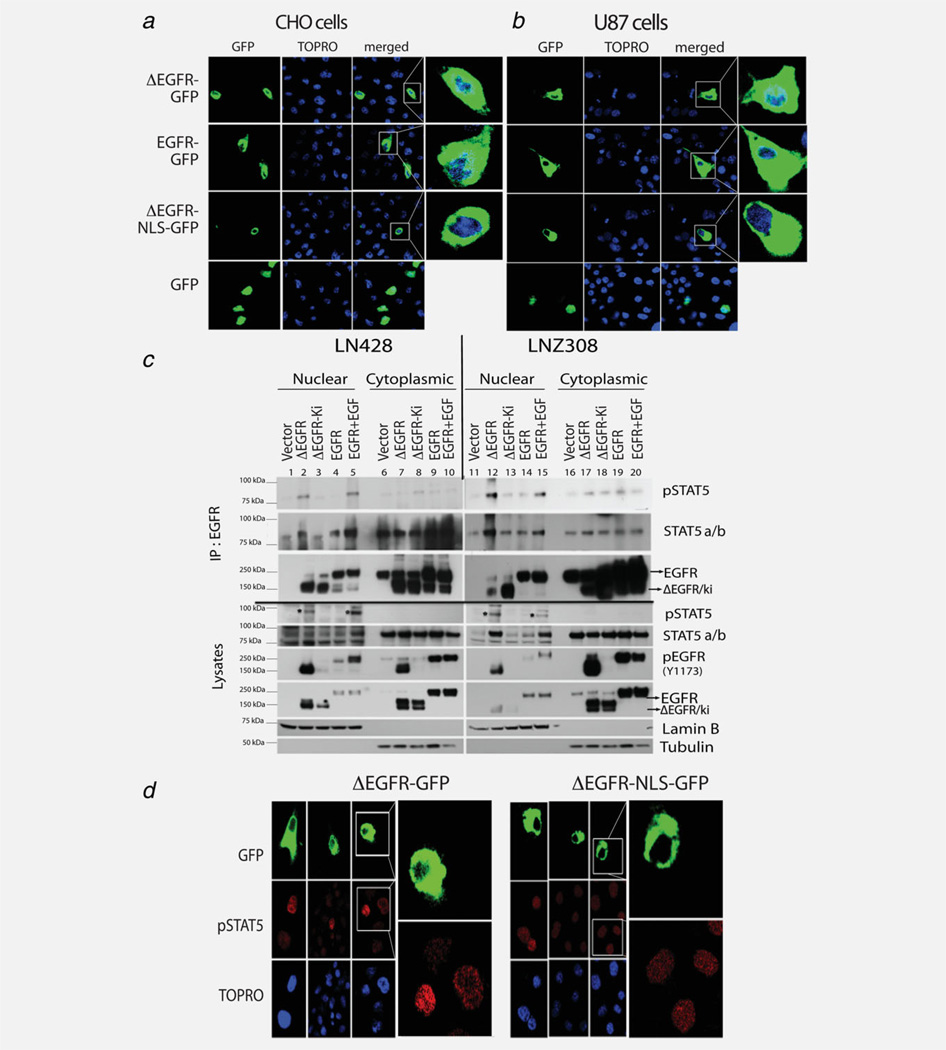
The classical model of STAT activation proposes that the phosphorylation by RTK occurs near the plasma membrane and results in an active, receptor-independent STAT dimer that translocates to the nucleus to execute its function.28 In view of the growing evidence that activated EGFR is capable of translocating to the nucleus30 and some indications that this may also be true for ΔEGFR,31,24 we investigated whether ΔEGFR is found in the nucleus of our cells, which could then prompt us to examine whether ΔEGFR and pSTAT5b interacted there. As glioma cells express endogenous EGFR, confocal analysis to explore this required the use of a tagged ΔEGFR and EGFR. GFP-fusion constructs of EGFR and ΔEGFR showed nuclear localization in both CHO cells and U87 glioma cells (Figs. 2a and 2b). Significantly, a ΔEGFR-GFP fusion construct in which the nuclear localization signal32 had been mutated (378RRR380 to 378AAA380), named ΔEGFR-NLS-GFP, failed to show any nuclear translocation (Figs. 2a and 2b), implying that nuclear localization is a process governed by molecular recognition. Fractionation of lysates from glioma cell lines stably transfected with ΔEGFR, ΔEGFR-ki (a kinase inactive mutant of ΔEGFR) or EGFR revealed that all forms of the EGFR could be identified in the nucleus (Fig. 2c, lysates). Interestingly, only in the presence of ΔEGFR or with the stimulation of EGFR-overexpressing cells with EGF, measurable elevation of nuclear pSTAT5 was observed. These data suggested that nuclear translocation of EGFR and ΔEGFR is a regulated process in glioma cells and raised the possibility of interaction between the receptor and STAT5a/b in the nucleus. To investigate this directly, we immunoprecipitated the EGFR from the nuclear and cytoplasmic extracts shown in Figure 2c (lysates), revealing an association between pSTAT5 and EGFR in all venues (Fig. 2c, IP). Though both LN428 and LNZ308 glioma cell lines have a basal level of endogenous EGFR, overexpression of ΔEGFR and EGFR led to an increase in the level of pSTAT5 recovered in nuclear extract immunoprecipitates (Fig. 2c, IP). Therefore, active EGFR associates with phosphorylated STAT5 in the nucleus of glioma cells. To examine whether the nucleus could be a site at which EGFR induces STAT5 phosphorylation, we compared the impact on pSTAT5 staining of ΔEGFR-GFP with that of ΔEGFR-NLS-GFP, which has an inactive nuclear localization signal (Fig. 2d). When ΔEGFR-GFP was transiently transfected into U87 cells, a small proportion of the ΔEGFR was found in the nucleus, and in these green cells there was a visible increase in the pSTAT5 signal, when compared wit nearby cells that were not transfected (Fig. 2d, left). In contrast, when ΔEGFR-NLS-GFP was transfected, no increase was detected in pSTAT5 signal in the green cells (Fig. 2d, right). These results were corroborated with quantification of the number of cells demonstrating increased pSTAT5 signal in the ΔEGFR-GFP overexpressing cells over their non-overexpressing neighbor (Supporting Information Fig. 2a) as well as the intensity of pSTAT5 signaling between the ΔEGFR and ΔEGFR-NLS cells (Supporting Information Fig. 2b). These findings suggest that an intact NLS and access to the nucleus is required for ΔEGFR to mediate an increase in STAT5 phosphorylation. Interestingly, studies from our laboratory show that reducing ΔEGFR-transport to the nucleus attenuates its oncogenic signal, while promoting nuclear localization enhances this, including in 3D colony formation and intracranial tumor growth assays (unpublished data). Together, these data raise the possibility that nuclear signaling by ΔEGFR is a key element in its oncogenic signal and, thus, provides new opportunities for therapeutic intervention.
Figure 2.
ΔEGFR is a nuclear protein and associates directly with STAT5. (a, b) Confocal microscope images of cells transiently overexpressing GFP-tagged EGFR, ΔEGFR or ΔEGFR-NLS as indicated in CHO (a) and U87 (b) cells showed that EGFR-GFP and ΔEGFR-GFP translocated to the nucleus, while ΔEGFR-NLS-GFP did not. (c) Cytoplasmic and nuclear fractions of lysates from two glioma cell lines LNZ308, LN428 were analyzed by western blot as indicated. Expression of ΔEGFR and stimulation of EGFR expressing cells with EGF (EGFR + EGF) was associated with more pEGFR and more pSTAT5 (Y699) in the nucleus (indicated by asterisk in the WB). Immunoprecipitates of EGFR from the lysates recovered STAT5 and pSTAT5, with the highest levels of pSTAT5 being found in complexes with ΔEGFR and EGFR+EGF. (d) Confocal analysis of U87 cells transiently transfected with ΔEGFR-GFP or ΔEGFR-NLS-GFP and stained for pSTAT5 (Y699).
ΔEGFR signals mainly through SFKs to phosphorylate STAT5
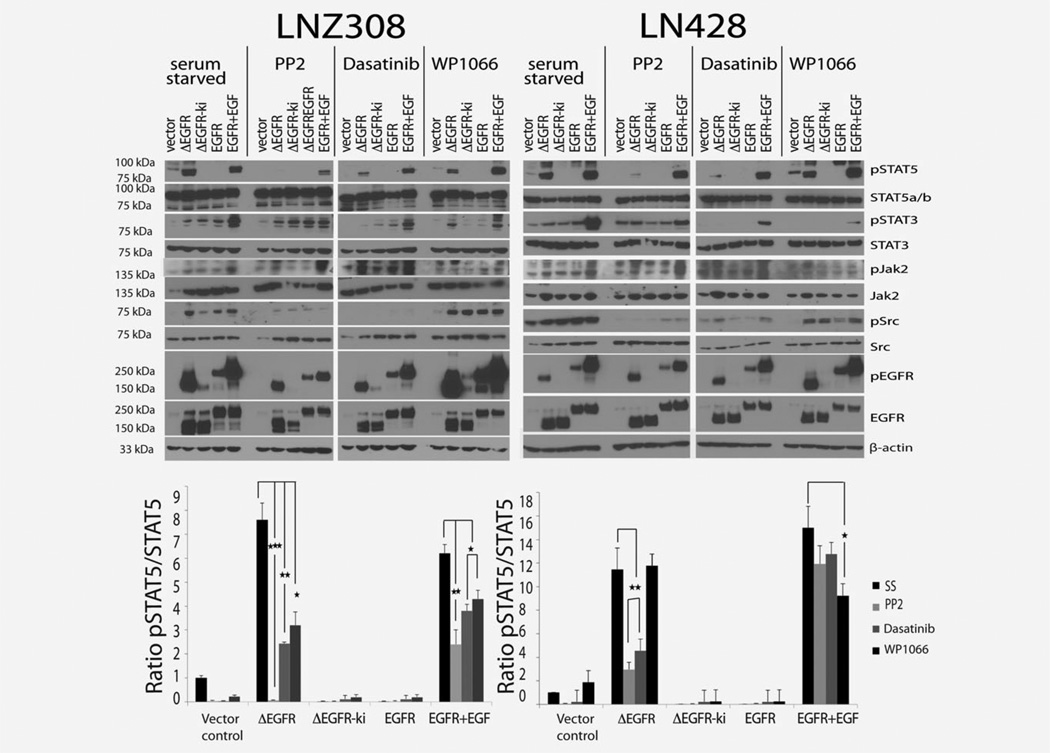
Although the association of ΔEGFR or EGFR and STAT5 would allow for a direct phosphorylation of STAT5 by the RTK, it does not preclude the involvement of other kinases. Phosphorylation of the tyrosine residue Y699 in STAT5b appears to be achieved by growth factor receptors as well as JAK and Src kinases, depending on the cell type and the nature of the ligand/receptor interactions.28,33 The primary candidates for transmission of the signal from EGFR to STAT5 are Src and Jak2,28,34 and so we tested its dependence on these two kinases using inhibitors. LNZ308 and LN428 glioma cells were serum starved and treated with the Src inhibitor PP2, the BCR/ABL and Src inhibitor Dasatinib or the Jak2 inhibitor WP1066. There are numerous reports corroborating the use of PP2 as an effective Src inhibitor and of Dasatinib as a potent orally-active inhibitor of SFKs (Src Family Kinases) and a less potent inhibitor of other tyrosine kinases, including PDGFR and BCR-ABL.35,36 WP1066 is a novel analogue of the Jak2 inhibitor AG490,37 which inhibited phosphorylation of Jak2 and also blocked further downstream signaling of Jak2, including STAT3. Assessment of pSTAT5 by western blot revealed a significant reduction in the ΔEGFR-stimulated pSTAT5 levels in cells exposed to PP2 or Dasatinib, and to a lesser extent with WP1066, in both cell lines (Fig. 3), implicating a predominantly Src-mediated ΔEGFR phosphorylation of pSTAT5. Interestingly, EGFR+EGF stimulated pSTAT5 was not suppressed as efficiently by PP2 or Dasatinib, and also to a lesser extent with WP1066, suggesting that other kinases maybe involved in this signaling. In parallel, phosphorylation of STAT3 was significantly suppressed in the presence of Dasatinib and WP1066 in both ΔEGFR and EGF-stimulated EGFR cells (as revealed by quantification, Supporting Information Fig. 3). Src family kinases (SFKs) namely, Fyn and Src, have been shown to be effectors of oncogenic EGFR signaling, enhancing invasion and tumor cell survival in vivo, while gene silencing of Fyn and Src limited EGFR and ΔEGFR dependent tumor cell motility in GBM.38 As expected, genetic silencing of Fyn and Src by shRNA dramatically reduced pSTAT5 levels in ΔEGFR cells (Supporting Information Fig. 4a), while Jak2 shRNA effectively reduced pSTAT5 levels in the EGFR cells (Supporting Information Fig. 4b), corroborating our hypothesis of preferential reliance on SFKs for ΔEGFR signaling to STAT5 in glioblastoma cells.
Figure 3.
ΔEGFR signals primarily through Src and not Jak2 to pSTAT5. Glioma cell lines LNZ308 and LN428 expressing ΔEGFR, ΔEGFR-ki or EGFR were cultured in SS (serum free medium (for 24 hrs) and treated with Src inhibitor PP2 (10 µM/ml for 24 hrs), BCR/ABL and Src inhibitor Dasatinib (1 µM/ml for 4 hrs) or the Jak2 inhibitor WP1066 (10 µM/ml for 3 hrs). Dose choice of inhibitors was determined from previously published reports. Phosphorylation of STAT5 and STAT3 as well as the drug targets Src and Jak2 were monitored by western blot. Each experiment was done in triplicates and the densitometric quantification of the western blots represented as pSTAT5 signal relative to the total STAT5 signal, shown as mean and SEM (*p < 0.05; **p < 0.01; ***p < 0.001; t-test) are given below the western blots of the respective cell lines.
ΔEGFR induces pSTAT5 binding with DNA and regulates Aurora A
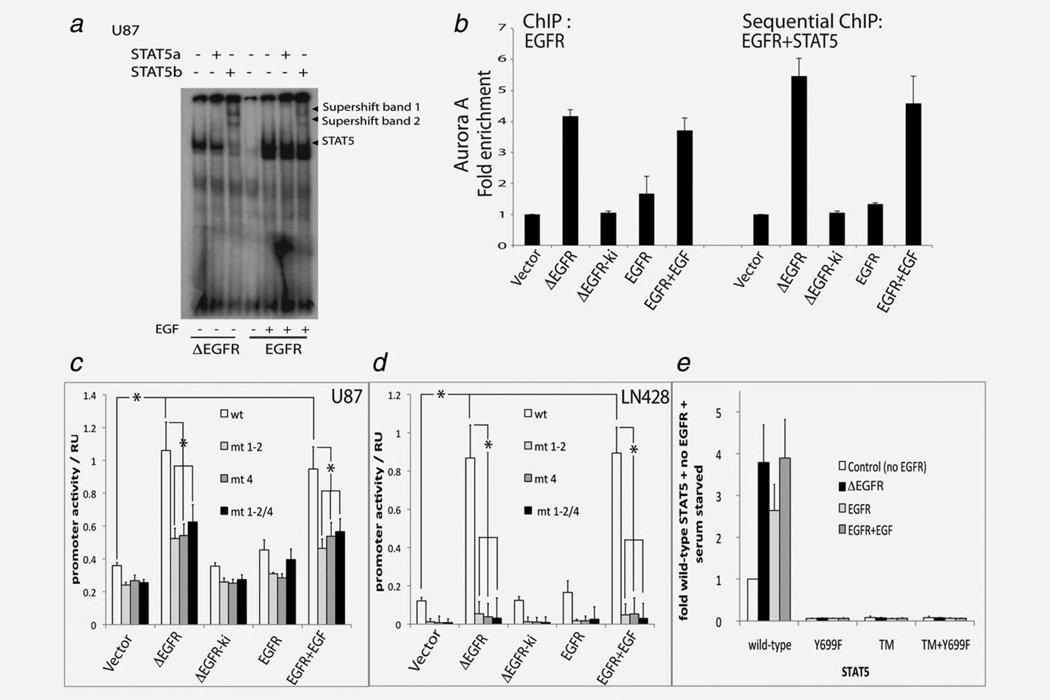
Next, we measured DNA binding of STAT5 complexes in ΔEGFR-expressing cells by EMSA (Fig. 4a) to determine whether ΔEGFR could mediate functional activation of STAT5 in association with phosphorylation. U87 cells used for the EMSA show an increased pSTAT5 expression only in the ΔEGFR/EGFR+EGF overexpressing cells (Supporting Information Fig. 5). STAT-DNA complexes were detected with a β-casein probe that binds to STAT5b in U87 cells, and more importantly ΔEGFR and EGFR activity induced STAT5b:STAT5b homodimer formation as demonstrated by an anti-STAT5 supershifted band (Fig. 4a). The association of ΔEGFR and STAT5 in the nucleus raised the possibility that this complex could be associated with DNA and specifically with promoters regulated by STAT5. We first analyzed the Aurora A promoter, as its regulation by STAT5 and EGFR is characterized.27 We performed ChIP on cross-linked extracts of U87 cells and detected recruitment of STAT5a/b on Aurora A promoter in cells overexpressing ΔEGFR or EGFR acutely stimulated with EGF, thus confirming that activation of STAT5a/b by EGFR drove its association with this promoter (Fig. 4b). To confirm that these two proteins co-associated on the Aurora A promoter, we performed sequential ChIP, by first recovering all EGFR/ΔEGFR containing complexes, and then recovering the STAT5a/b containing complexes from this mixture. Results from this experiment provided further evidence that EGFR or ΔEGFR were found on the Aurora A promoter together with STAT5a/b (Fig. 4b).
Figure 4.
ΔEGFR and EGF-stimulated EGFR induce pSTAT5 binding to DNA and the regulation of the Aurora A promoter (a) EMSA analysis of STAT5a and STAT5b binding activity in U87-ΔEGFR and U87-EGFR cells using a β-casein probe (b) ChIP-qPCR assays were performed on cultured cells using antibodies against STAT5 and EGFR, and the recovery of the Aurora A promoter was monitored. Sequential ChIP assays were done using the anti-EGFR antibody followed by the anti-STAT5a/b antibody (c, d). Transcriptional activity of the Aurora A promoter in U87 and LN428 glioma cells measured using a dual promoter assay system is shown as relative units (RU). Glioma cells stably expressing vector, ΔEGFR, ΔEGFR-ki, EGFR or EGFR and stimulated with EGF were transiently transfected with reporter plasmids containing either the Aurora A promoter sequence mediating STAT5 responsiveness or ATRs mutants of this sequence which no longer respond to STAT5 (mt 1-2, mt 4, and the combination mt1-2/4). Promoter activity was measured by a dual assay system. Data are form three independent triplicate experiments and are shown as mean ± SEM (*p < 0.05; t-test between data series as indicated). (e) Transcriptional activity of the STAT5 specific promoter, sp2.1 in STAT5a/STAT5b null MEFs is shown relative to the signal obtained in cells expressing wild-type STAT5 but no EGFR and serum starved (=1). Empty vector control, ΔEGFR or EGFR were transfected with wt-STAT5b (wild-type), Y699F STAT5b (Y699F), Y725F/740F/743F STAT5b triple mutant (TM) or Y699F/Y725F/740F/743F STAT5b combination mutant (TM+Y699F). Data are from three individual triplicate experiments.
As association with the promoter is not direct evidence of activation, we next performed reporter assays using the Aurora A promoter in glioma cells (Figs. 4c and 4d). In addition to the wild-type Aurora A promoter, we tested several mutant forms in which the STAT5 binding sites had been inactivated. In these experiments, the wild-type Aurora A promoter responded positively to the co-transfection of ΔEGFR or the stimulation of EGFR-transfected cells with EGF, but not under conditions of mutant promoters where STAT5 is not able to bind (Figs. 4c and 4d). Therefore, regulation of the Aurora A promoter by ΔEGFR is dependent on the presence of functional STAT5-binding sites.
To examine the ability of ΔEGFR to act directly via STAT5, we next combined various EGFR states with wild-type and mutant STAT5 proteins in STAT5a/b null MEFs and measured the activity of a STAT5 reporter plasmid. As stated earlier, the Y699 residue of STAT5b, which we identified in our phosphoproteomics screen, is tyrosine phosphorylated in response to cytokine treatment and is a key regulator of dimerization, translocation, DNA binding and transcriptional activation.14 Three additional EGF-induced tyrosine phosphorylation sites, Y725, Y740 and Y743, have also been identified using metabolic labeling studies in breast cancer cells.39 To test whether ΔEGFR regulates STAT5-responsive promoters in a activity dependent manner, we transfected wild-type STAT5b as well as the Y699F mutant and a triple mutant (Y725F/Y740F/Y743F abbreviated as TM in Fig. 4e) alone and together into MEF cells to test their activity on a STAT5b-specific reporter, Sp2.1. Transfection of STAT5b alone resulted in a measurable baseline of reporter activity, and this level was used to normalize the experiments (Fig. 4e). Cotransfection of EGFR resulted in an increase of reporter activity, with ΔEGFR and with EGF-stimulated EGFR resulting in higher levels than EGFR under serum starved condition. In contrast, the Y699F and the other STAT5b mutants showed no transcriptional activity even when EGFR or ΔEGFR were co-transfected, suggesting that these sites are important for EGFR and ΔEGFR to activate STAT5b in these assays. In agreement with the observation that ΔEGFR positively regulates the Aurora A promoter, we saw upregulation of Aurora A protein in the U87 cell model where ΔEGFR is expressed at medium, high or super-high levels (Supporting Information Fig. 1).
ΔEGFR activates Bcl-XL expression mediated by STAT5b activity
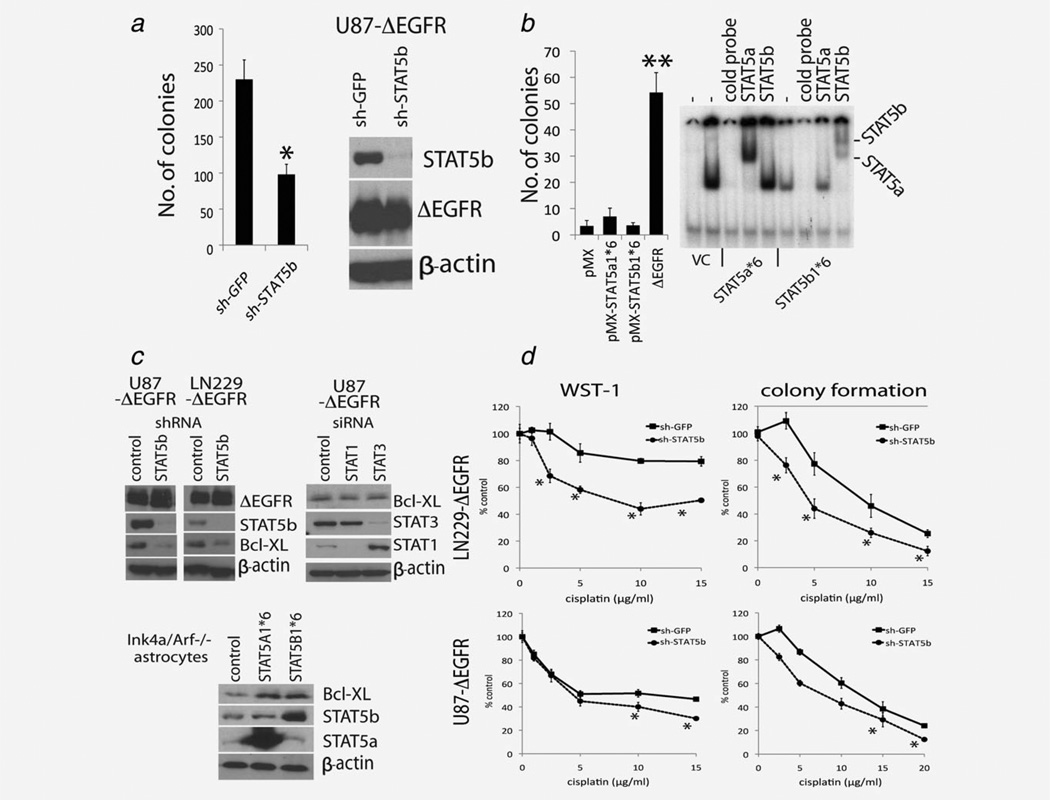
Next, we characterized the potential biological function of STAT5b in ΔEGFR-expressing glioblastoma cells. To accomplish this, loss-of-function studies were performed with glioma cells in which STAT5b expression was ablated by shRNA-mediated silencing. Elimination of STAT5b significantly inhibited ΔEGFR-mediated colony formation in U87 cells in soft agar (Fig. 5a), implying that STAT5b plays a necessary role in ΔEGFR-mediated cell transformation. However, overexpression of constitutively active STAT5 mutants (STAT5a1*6 or STAT5b1*6) was not able to substitute for ΔEGFR in transforming mouse astrocytes (Fig. 5b), suggesting that activation of STAT5 alone is not sufficient to rescue transforming capabilities of ΔEGFR. We previously reported that a central component of ΔEGFR’s oncogenic signal is mediated by the upregulation of Bcl-XL.22 Further, based on the fact that Bcl-XL can be regulated by STAT5,40 we hypothesized that maybe ΔEGFR modulates BcL-XL expression via STAT5b in glioma cells. Knockdown of STAT5b but not STAT1 or STAT3 dramatically decreased Bcl-XL protein expression in ΔEGFR expressing U87 and LN229 cells (Fig. 5c). Conversely, overexpression of STAT5a1*6 or STAT5b1*6 led to an upregulation of Bcl-XL protein expression in mouse astrocytes (Fig. 5c). Again, the U87 cell model where ΔEGFR is expressed at medium, high and super-high levels also showed a positive correlation between expression of the mutated receptor and Bcl-XL (Supporting Information Fig. 1). One measurable consequence of ΔEGFR expression that is mediated by positive regulation of Bcl-XL levels is increased resistance to cisplatin.22 Importantly, knockdown of STAT5b significantly sensitized LN229-ΔEGFR and U87-ΔEGFR cells to cisplatin treatment as demonstrated by WST-1 and colony formation assays (Fig. 5d). Consistent with the in vitro data, in vivo experiments also showed sensitization of U87-ΔEGFR-shSTAT5b cells to cisplatin treatment (Supporting Information Fig. 6). Together these data suggests that ΔEGFR and STAT5b concomitantly confers resistance to the chemotherapeutic drug cisplatin by increased expression of the anti-apoptotic gene, Bcl-XL.
Figure 5.
ΔEGFR activates Bcl-XL expression mediated by STAT5 activity in glioma cells. (a) Left, colony formation analysis in soft agar of U87-ΔEGFR cells expressing lentivirus shRNA-STAT5b or shRNA-GFP (n = 4, *p < 0.05; t-test). Right, Western blot analysis of EGFR, STAT5b expression in the lentivirus infected U87--ΔEGFR cells. (B) Left, colony formation analysis in soft agar of mouse astrocytes (Ink4a/Arf-/-) expressing constitutive mutants of STAT5A1*6 or STAT5B1*6 (n = 4, **p < 0.01; t-test). Right, shown are the EMSA analysis of mouse astrocytes (Ink4a/Arf-/-) expressing constitutive mutants of STAT5A1*6 or STAT5B1*6. (c) Western blot analysis of Bcl-XL expression in ΔEGFR-expressing glioma cells (U87-ΔEGFR and LN229-ΔEGFR) transduced with lentiviral shRNA-STAT5b or control shRNA-GFP (Upper left), in U87-ΔEGFR cells transduced with control siRNA, siRNA-STAT1 or -STAT3 (Upper right), and in mouse astrocytes (Ink4a/Arf-/-) expressing constitutive mutants of vector control, STAT5A1*6 or STAT5B1*6 (Bottom). (d) WST-1 and colony formation assays were performed with U87-ΔEGFR and LN229-ΔEGFR cells transduced with lentiviral- STAT5b shRNA and treated with cisplatin for indicated concentration either for 2 days (WST) or for 14 days (colony formation) (*p < 0.05; t-test).
ΔEGFR regulates the Bcl-XL promoter via STAT5b
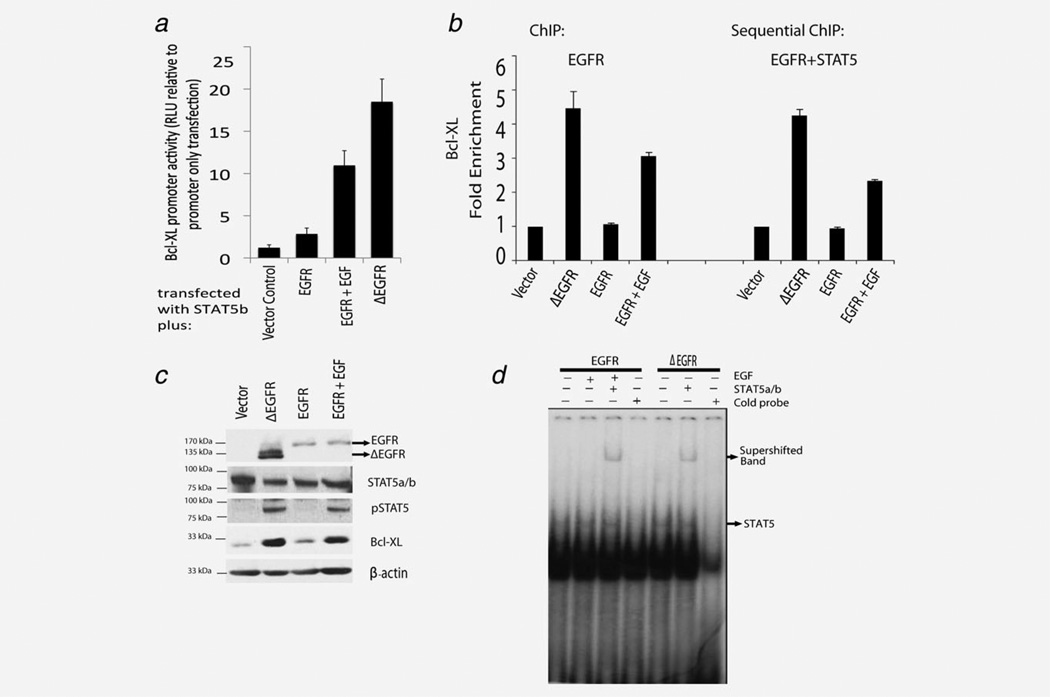
Finally, we examined the regulation of the Bcl-XL expression and promoter activity in STAT5a/STAT5b null MEFs (Fig. 6a). Transfection of STAT5b alone caused a modest increase in promoter activity. Co-transfection of STAT5b and EGFR resulted in a measurable baseline of the Bcl-XL reporter activity when no growth factors were added. Stimulation of EGFR-transfected cells with EGF caused a marked increase in promoter activity, but transfection of ΔEGFR in the absence of EGF stimulation caused a nearly 20-fold increase in Bcl- XL promoter activity (Fig. 6a). Consistent with the reporter assay, ChIP showed that nuclear ΔEGFR and nuclear STAT5 associate with the Bcl-XL gene promoter (Fig. 6b). Sequential ChIP analysis revealed that ΔEGFR/EGFR and STAT5 are co-recruited to the Bcl-XL promoter (Fig. 6b) suggesting their existence in the same complex. Western blot analysis of the astrocytes used in the ChIP assay also showed an increase in Bcl-XL protein expression only in the ΔEGFR or EGFR+EGF cells (Fig. 6c). Furthermore, EMSA using the Bcl-XL probe also showed that ΔEGFR and EGFR+EGF mediated direct binding to STAT5 (Fig. 6d). These results together support the conclusion that drug resistance long associated with the ΔEGFR/Bcl-XL signaling axis in glioblastoma cells is regulated via STAT5b.
Figure 6.
ΔEGFR mediated by STAT5 regulates the Bcl-XL promoter. (a) Transcriptional activity of the Bcl-XL promoter was measured in STAT5a/STAT5b knockdown MEFs. Empty vector control, ΔEGFR or EGFR were transfected with STAT5b, the Bcl-XL promoter and with the control Renilla luciferase as detailed in Methods. Data are mean and SEM from three, triplicate experiments. (b) ChIP assays for Bcl-XL were performed on mouse astrocytes (Ink4a/Arf-/-) using EGFR and EGFR+STAT5a/b antibodies and analyzed by qPCR. (c) Western blot analysis of mouse astrocytes expressing ΔEGFR or EGFR stimulated with or without EGF stimulation (100 ng/ml, 30 min). (d) EMSA analysis of Bcl-XL binding activity in mouse astrocytes (Ink4a/arf-/-) expressing ΔEGFR and EGFR with or without EGF stimulation (100 ng/ml, 30 min).
Discussion
ΔEGFR is a potent oncogene in glioblastoma exhibiting ligand-independent low-intensity signaling, which does not involve prevalent or prolonged receptor dimerization. We recently performed a tyrosine-directed search for signaling events downstream of ΔEGFR and identified STAT5 phosphorylation at Y699 as a key event,9 prompting us to investigate its significance, the pathway that transmits the signal and the events that STAT5 mediates downstream of ΔEGFR.
Although constitutive activation of STAT5 at residue Y694 has been implicated in prostate and breast cancer, it was only recently reported that STAT5 regulates glioma cell invasion and specifically that inhibiting STAT5b suppresses tumor cell invasion in human GBM.21,41 Furthermore, immmunohistochemical staining showed that STAT5b expression is markedly increased in GBM when compared with normal cortex and diffuse astrocytoma implicating its significant role in astrocytoma progression.21 Our analysis of human samples showed a correlation between EGFR protein expression and pSTAT5 levels, and more importantly a correlation between pSTAT5 and poorer survival, suggesting that this pathway is clinically significant in glioblastoma. At the molecular level, we saw that STAT5b is phosphorylated at Y699 in ΔEGFR-expressing cells, which given the relatively weaker activity of ΔEGFR when compared with overexpressed, acutely activated EGFR, implies a strong coupling between ΔEGFR and STAT5b. Interestingly, we observed direct interaction of STAT5a/b with EGFR and ΔEGFR, including in the nucleus and on DNA. The association of ΔEGFR with STAT5a/b in the nucleus is consistent with previous reports showing that activated EGFR can be found in transcription factor complexes.27 Furthermore, when we expressed a mutant of ΔEGFR that was incapable of entering the nucleus, we did not detect an increase in pY699 signal on STAT5, suggesting that the activation event actually occurs in the nucleus. Accumulating reports reveal a new mode of EGFR signaling involving direct shuttling of activated EGFR into the nucleus. However, the nuclear functions of EGFR are still being defined at the molecular level. Within the nucleus, EGFR serves as a transcriptional co-activator for a series of tumor-promoting genes such as cyclin D1, iNOS, B-myb, Aurora A and Cox-2.30 Consistent with this observation, our data showing that ΔEGFR interacts with STAT5b and regulates the transcription of Aurora A and Bcl-XL promoters is the first demonstration of a role for this pathway in glioma. Two recent studies have shown that ΔEGFR also interacts with STAT3 in the nucleus of gliomas24,31 and that this is important for the ΔEGFR oncogenic signal, suggesting that multiple STAT family members may be subject to a similar mechanism of activation by ΔEGFR.
It is well established that SFKs are necessary for full activation of EGFR. It has been reported in breast cancer cells that Src kinase modulates STAT5 activation in at least two ways: (i) by direct phosphorylation of STAT5b (at Tyr699) and (ii) by phosphorylating EGFR at Tyr845. Although overexpression of Src alone does not lead to the activation of STAT5, it has been shown that the kinase activity of Src is required for maximal tyrosine phosphorylation of STAT5b in response to EGF.22 It has been demonstrated that ER, c-Src, and EGFR impinge on the STAT5b-signaling pathway in breast cancer cells.42 Furthermore, mutant EGFRs were constitutively associated with Src and STAT5 while EGFR bound to JAK2 in the absence of added ligand in NSCLC.43 Indeed, we found that the phosphorylation of STAT5b at Y699 by ΔEGFR is largely dependent on SFKs, namely, Src and Fyn, and much less so on Jak2 signaling. Interestingly, Iida et al.44 recently implicated the SFKs Yes and Lyn in the nuclear translocation of EGFR, in agreement with our finding that they influence the nuclear localization of the ΔEGFR-STAT5b complex. This suggests that investigating the translational impact of suppressing SFK activity to sensitize glioma cells to EGFR-targeted therapies is worthwhile.
ΔEGFR is associated with radioresistance and chemoresistance.45,46 An important mechanism by which ΔEGFR confers this resistance is the upregulation of anti-apoptotic proteins, with the clearest connection being to Bcl-XL.7 More recently it was shown that STAT5 also regulated the response to the chemotherapy agent, camptothecin (CAM), which induced apoptosis and did so by regulating Bcl-XL expression in rat C6-glioma cells.47 We found that the Bcl-XL promoter is regulated by ΔEGFR in a STAT5b-dependent manner, and that knockdown of STAT5b reduces levels of Bcl-XL and sensitizes glioblastoma cells to cisplatin both in vitro and in vivo. Significantly we also observed a strong correlation between the detection of pSTAT5 in glioblastoma patient samples and poor survival, suggesting that treatment resistance promoted by STAT5 activity plays an important role in clinical outcomes. These findings are consistent with two recent studies that showed that nuclear EGFR contributes to resistance to cisplatin48 and Cetuximab.49 These results may have important clinical implications for the use of cisplatin in the treatment of those malignant gliomas expressing ΔEGFR.
In summary, our results demonstrate for the first time that ΔEGFR activates STAT5b, forms complexes with activated pSTAT5b on the promoters of genes including Bcl-XL, and together directly increases Bcl-XL gene expression to promote tumor growth and treatment resistance in glioblastoma. Ongoing studies are directed at these genes to gain further insights into their role in the malignant biology of tumors with active ΔEGFR nuclear pathways.
Supplementary Material
What’s new?
EGFRvIII (ΔEGFR), characterized by deletion of exons 2-7 in the epidermal growth factor receptor transcript, occurs in more than 25 percent of glioblastomas and is associated with enhanced tumor growth and survival. In this study, EGFRvIII was found to interact with the transcription factor STAT5b, forming a nuclear complex that promoted oncogenesis and treatment resistance in glioblastoma through direct regulation of the anti-apoptotic gene Bcl-XL. These findings may have important implications for the treatment of glioblastoma.
Acknowledgements
This work was supported by the National Cancer Institute of the National Institutes of Health: RO1CA108500 (O.B.), SPORE P50CA127001 (O.B.), PO1 CAO95616 (W.K.C., F.B.F.); the University of Texas M.D. Anderson’s Cancer Centre Support Grant CA016672; The Goldhirsh Foundation (F.F.). M.L. was supported in part by an American Brain Tumor Association Fellowship. W.K.C. is a Fellow of the National Foundation for Cancer Research. We thank the Anthony Bullock III Foundation for their generous support of the proteomics capabilities in the Brain Tumor Center at The University of Texas MD Anderson Cancer Center. We would like to thank our colleagues at MD Anderson: Dr. Mien-Chie Hung for helpful discussions, Dr. John de Groot for Dasatinib, Dr. Waldemar Priebe for WP1066, Dr. Zhen Fan for Jak2 shRNA constructs and Mrs. Laura Gibson for technical assistance. We also thank Dr. Liang-Yi Hung, National Cheng Kung University, Taiwan, for pGL2-Aurora-A promoter constructs, Dr. Corinne M. Silva, University of Virginia, for mutant STAT5 constructs, and Dr. Sylvia L. Asa, University of Toronto, for Bcl-XL promoter constructs.
References
- 1.Wong AJ, Bigner SH, Bigner DD, et al. Increased expression of the epidermal growth factor receptor gene in malignant gliomas is invariably associated with gene amplification. Proc Natl Acad Sci USA. 1987;84:6899–6903. doi: 10.1073/pnas.84.19.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frederick L, Eley G, Wang XY, et al. Analysis of genomic rearrangements associated with EGRFvIII expression suggests involvement of Alu repeat elements. Neuro Oncol. 2000;2:159–163. doi: 10.1093/neuonc/2.3.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong AJ, Ruppert JM, Bigner SH, et al. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc Natl Acad Sci USA. 1992;89:2965–2969. doi: 10.1073/pnas.89.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shinojima N, Tada K, Shiraishi S, et al. Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res. 2003;63:6962–6970. [PubMed] [Google Scholar]
- 5.Heimberger AB, Hlatky R, Suki D, et al. Prognostic effect of epidermal growth factor receptor and EGFRvIII in glioblastoma multiforme patients. Clin Cancer Res. 2005;11:1462–1466. doi: 10.1158/1078-0432.CCR-04-1737. [DOI] [PubMed] [Google Scholar]
- 6.Pelloski CE, Ballman KV, Furth AF, et al. Epidermal growth factor receptor variant III status defines clinically distinct subtypes of glioblastoma. J Clin Oncol. 2007;25:2288–2294. doi: 10.1200/JCO.2006.08.0705. [DOI] [PubMed] [Google Scholar]
- 7.Nagane M, Coufal F, Lin H, et al. A common mutant epidermal growth factor receptor confers enhanced tumorigenicity on human glioblastoma cells by increasing proliferation and reducing apoptosis. Cancer Res. 1996;56:5079–5086. [PubMed] [Google Scholar]
- 8.Nishikawa R, Ji XD, Harmon RC, et al. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci USA. 1994;91:7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chumbalkar V, Latha K, Hwang Y, et al. Analysis of phosphotyrosine signaling in glioblastoma identifies STAT5 as a novel downstream target of DeltaEGFR. J Proteome Res. 2011;10:1343–1352. doi: 10.1021/pr101075e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X, Robinson GW, Gouilleux F, et al. Cloning and expression of Stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue. Proc Natl Acad Sci USA. 1995;92:8831–8835. doi: 10.1073/pnas.92.19.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coffer PJ, Koenderman L, de Groot RP. The role of STATs in myeloid differentiation and leukemia. Oncogene. 2000;19:2511–2522. doi: 10.1038/sj.onc.1203479. [DOI] [PubMed] [Google Scholar]
- 12.Nevalainen MT, Xie J, Torhorst J, et al. Signal transducer and activator of transcription-5 activation and breast cancer prognosis. J Clin Oncol. 2004;22:2053–2060. doi: 10.1200/JCO.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 13.Li H, Ahonen TJ, Alanen K, et al. Activation of signal transducer and activator of transcription 5 in human prostate cancer is associated with high histological grade. Cancer Res. 2004;64:4774–4782. doi: 10.1158/0008-5472.CAN-03-3499. [DOI] [PubMed] [Google Scholar]
- 14.Grimley PM, Dong F, Rui H. Stat5a and Stat5b: fraternal twins of signal transduction and transcriptional activation. Cytokine Growth Factor Rev. 1999;10:131–157. doi: 10.1016/s1359-6101(99)00011-8. [DOI] [PubMed] [Google Scholar]
- 15.Frasor J, Park K, Byers M, et al. Differential roles for signal transducers and activators of transcription 5a and 5b in PRL stimulation of ERalpha and ERbeta transcription. Mol Endocrinol. 2001;15:2172–2181. doi: 10.1210/mend.15.12.0745. [DOI] [PubMed] [Google Scholar]
- 16.Zeng R, Aoki Y, Yoshida M, et al. Stat5B shuttles between cytoplasm and nucleus in a cytokine-dependent and -independent manner. J Immunol. 2002;168:4567–4575. doi: 10.4049/jimmunol.168.9.4567. [DOI] [PubMed] [Google Scholar]
- 17.Iyer J, Reich NC. Constitutive nuclear import of latent and activated STAT5a by its coiled coil domain. FASEB J. 2008;22:391–400. doi: 10.1096/fj.07-8965com. [DOI] [PubMed] [Google Scholar]
- 18.Yamashita H, Xu J, Erwin RA, et al. Differential control of the phosphorylation state of proline-juxtaposed serine residues Ser725 of Stat5a and Ser730 of Stat5b in prolactin-sensitive cells. J Biol Chem. 1998;273:30218–30224. doi: 10.1074/jbc.273.46.30218. [DOI] [PubMed] [Google Scholar]
- 19.Iavnilovitch E, Cardiff RD, Groner B, et al. Deregulation of Stat5 expression and activation causes mammary tumors in transgenic mice. Int J Cancer. 2004;112:607–619. doi: 10.1002/ijc.20484. [DOI] [PubMed] [Google Scholar]
- 20.Lee TK, Man K, Poon RT, et al. Signal transducers and activators of transcription 5b activation enhances hepatocellular carcinoma aggressiveness through induction of epithelial-mesenchymal transition. Cancer Res. 2006;66:9948–9956. doi: 10.1158/0008-5472.CAN-06-1092. [DOI] [PubMed] [Google Scholar]
- 21.Liang QC, Xiong H, Zhao ZW, et al. Inhibition of transcription factor STAT5b suppresses proliferation, induces G1 cell cycle arrest and reduces tumor cell invasion in human glioblastoma multiforme cells. Cancer Lett. 2009;273:164–171. doi: 10.1016/j.canlet.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 22.Nagane M, Levitzki A, Gazit A, et al. Drug resistance of human glioblastoma cells conferred by a tumor-specific mutant epidermal growth factor receptor through modulation of Bcl-XL and caspase-3-like proteases. Proc Natl Acad Sci USA. 1998;95:5724–5729. doi: 10.1073/pnas.95.10.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lo HW, Ali-Seyed M, Wu Y, et al. Nuclear-cytoplasmic transport of EGFR involves receptor endocytosis, importin beta1 and CRM1. J Cell Biochem. 2006;98:1570–1583. doi: 10.1002/jcb.20876. [DOI] [PubMed] [Google Scholar]
- 24.Lo HW, Cao X, Zhu H, et al. Cyclooxygenase-2 is a novel transcriptional target of the nuclear EGFR-STAT3 and EGFRvIII-STAT3 signaling axes. Mol Cancer Res. 2010;8:232–245. doi: 10.1158/1541-7786.MCR-09-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 26.Burton EC, Lamborn KR, Forsyth P, et al. Aberrant p53, mdm2, and proliferation differ in glioblastomas from long-term compared with typical survivors. Clin Cancer Res. 2002;8:180–187. [PubMed] [Google Scholar]
- 27.Hung LY, Tseng JT, Lee YC, et al. Nuclear epidermal growth factor receptor (EGFR) interacts with signal transducer and activator of transcription 5 (STAT5) in activating Aurora-A gene expression. Nucleic Acids Res. 2008;36:4337–4351. doi: 10.1093/nar/gkn417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quesnelle KM, Boehm AL, Grandis JR. STAT-mediated EGFR signaling in cancer. J Cell Biochem. 2007;102:311–319. doi: 10.1002/jcb.21475. [DOI] [PubMed] [Google Scholar]
- 29.Teglund S, McKay C, Schuetz E, et al. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93:841–850. doi: 10.1016/s0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- 30.Wang YN, Yamaguchi H, Hsu JM, et al. Nuclear trafficking of the epidermal growth factor receptor family membrane proteins. Oncogene. 2010;29:3997–4006. doi: 10.1038/onc.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de la Iglesia N, Konopka G, Puram SV, et al. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 2008;22:449–462. doi: 10.1101/gad.1606508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu SC, Hung MC. Characterization of a novel tripartite nuclear localization sequence in the EGFR family. J Biol Chem. 2007;282:10432–10440. doi: 10.1074/jbc.M610014200. [DOI] [PubMed] [Google Scholar]
- 33.Rane SG, Reddy EP. Janus kinases: components of multiple signaling pathways. Oncogene. 2000;19:5662–5679. doi: 10.1038/sj.onc.1203925. [DOI] [PubMed] [Google Scholar]
- 34.Silva CM. Role of STATs as downstream signal transducers in Src family kinase-mediated tumorigenesis. Oncogene. 2004;23:8017–8023. doi: 10.1038/sj.onc.1208159. [DOI] [PubMed] [Google Scholar]
- 35.Ahluwalia MS, de Groot J, Liu WM, et al. Targeting SRC in glioblastoma tumors and brain metastases: rationale and preclinical studies. Cancer Lett. 2010;298:139–149. doi: 10.1016/j.canlet.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Araujo J, Logothetis C. Dasatinib: a potent SRC inhibitor in clinical development for the treatment of solid tumors. Cancer Treat Rev. 2010;36:492–500. doi: 10.1016/j.ctrv.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verstovsek S, Manshouri T, Quintas-Cardama A, et al. WP1066, a novel JAK2 inhibitor, suppresses proliferation and induces apoptosis in erythroid human cells carrying the JAK2 V617F mutation. Clin Cancer Res. 2008;14:788–796. doi: 10.1158/1078-0432.CCR-07-0524. [DOI] [PubMed] [Google Scholar]
- 38.Lu KV, Zhu S, Cvrljevic A, et al. Fyn and SRC are effectors of oncogenic epidermal growth factor receptor signaling in glioblastoma patients. Cancer Res. 2009;69:6889–6898. doi: 10.1158/0008-5472.CAN-09-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kloth MT, Catling AD, Silva CM. Novel activation of STAT5b in response to epidermal growth factor. J Biol Chem. 2002;277:8693–8701. doi: 10.1074/jbc.M111884200. [DOI] [PubMed] [Google Scholar]
- 40.Turkson J. STAT proteins as novel targets for cancer drug discovery. Expert Opin Ther Targets. 2004;8:409–422. doi: 10.1517/14728222.8.5.409. [DOI] [PubMed] [Google Scholar]
- 41.Cao S, Wang C, Zheng Q, et al. STAT5 regulates glioma cell invasion by pathways dependent and independent of STAT5 DNA binding. Neurosci Lett. 2011;487:228–233. doi: 10.1016/j.neulet.2010.10.028. [DOI] [PubMed] [Google Scholar]
- 42.Fox EM, Bernaciak TM, Wen J, et al. Signal transducer and activator of transcription 5b, c-Src, and epidermal growth factor receptor signaling play integral roles in estrogen-stimulated proliferation of estrogen receptor-positive breast cancer cells. Mol Endocrinol. 2008;22:1781–1796. doi: 10.1210/me.2007-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang J, Stark GR. Roles of unphosphorylated STATs in signaling. Cell Res. 2008;18:443–451. doi: 10.1038/cr.2008.41. [DOI] [PubMed] [Google Scholar]
- 44.Iida M, Brand TM, Campbell DA, et al. Yes and Lyn play a role in nuclear translocation of the epidermal growth factor receptor. Oncogene. 2012 doi: 10.1038/onc.2012.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chakravarti A, Dicker A, Mehta M. The contribution of epidermal growth factor receptor (EGFR) signaling pathway to radioresistance in human gliomas: a review of preclinical and correlative clinical data. Int J Radiat Oncol Biol Phys. 2004;58:927–931. doi: 10.1016/j.ijrobp.2003.09.092. [DOI] [PubMed] [Google Scholar]
- 46.Huang PH, Cavenee WK, Furnari FB, et al. Uncovering therapeutic targets for glioblastoma: a systems biology approach. Cell Cycle. 2007;6:2750–2754. doi: 10.4161/cc.6.22.4922. [DOI] [PubMed] [Google Scholar]
- 47.Qian YH, Xiao Q, Chen H, et al. Dexamethasone inhibits camptothecin-induced apoptosis in C6-glioma via activation of Stat5/Bcl-xL pathway. Biochim Biophys Acta. 2009;1793:764–771. doi: 10.1016/j.bbamcr.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hsu SC, Miller SA, Wang Y, et al. Nuclear EGFR is required for cisplatin resistance and DNA repair. Am J Transl Res. 2009;1:249–258. [PMC free article] [PubMed] [Google Scholar]
- 49.Li C, Iida M, Dunn EF, et al. Nuclear EGFR contributes to acquired resistance to cetuximab. Oncogene. 2009;28:3801–3813. doi: 10.1038/onc.2009.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.