Abstract
APE1/Ref-1 protects cells from oxidative stress by acting as a central enzyme in base excision repair pathways of DNA lesions and through its independent activity as a redox transcriptional co-activator. Dysregulation of this protein has been associated to cancer development. At present, contrasting data have been published regarding the biological relevance of the two functions as well as the molecular mechanisms involved. Here, we combined both mRNA expression profiling and proteomic analysis to determine the molecular changes associated with APE1 loss-of-expression induced by siRNA technology. This approach identified a role of APE1 in cell growth, apoptosis, intracellular redox state, mitochondrial function and cytoskeletal structure. Thus, overall, our data show that APE1 acts as a hub in coordinating different and vital functions in mammalian cells, highlighting the molecular determinants of the multifunctional nature of APE1 protein.
Keywords: Cell signaling, Difference analysis, Transcription regulation, Two-dimensional gel electrophoresis, Microarrays, APE1, Ref-1, proteomics, siRNA, oxidative stress, apoptosis, mitochondria
1. Introduction
APE1/Ref-1 (henceforth referred to as APE1) is a master regulator of cellular response to oxidative stress and plays a central role in the maintenance of the genome stability. The vital effects of APE1 appear to be attributable to its role in the base excision repair (BER) pathways of DNA lesions [1]. However, the biological relevance of APE1 in eukaryotic transcriptional regulation of gene expression as a redox co-activator of different transcription factors, such as Egr-1, NF-κB, p53, AP-1 and others [2], has yet to be elucidated. APE1 biological activities are exerted by two functionally distinct domains [2–4]. The N-terminus, containing the nuclear localization signal sequence, is principally devoted to the redox transcriptional co-activation activity [5], while the C-terminus exerts the enzymatic activity on the abasic sites of DNA [6].
Several reports indicated that dysregulation of APE1 expression is associated to different tumorigenic processes [2]. In particular, a higher intracellular expression and a robust cytoplasmic localization of APE1 have been described in lung, ovarian, thyroid and breast cancers [2], where they correlate with a higher aggressiveness of the tumors. The possible causal role played by this particular distribution in tumor progression is currently unknown. However, APE1 ability to activate transcription factors, such as p53 and Egr-1 [2, 7–9], which are mainly involved in controlling cell-cycle-arrest and apoptotic programs, leaves the debate open about the mechanisms responsible for controlling its different functions in several contexts.
APE1 is essential for cell viability. Genetic knockout of APE1 in mice causes post-implantation embryonic lethality [10, 11] and any attempt to isolate stable APE1-knockout cell lines has been so far unsuccessful. For these reasons, achieving a comprehensive understanding of the molecular targets mediating APE1 functions has been very difficult. In the last two years, however, conditional knockout and knockdown strategies [1, 12] confirmed the essentiality of this protein for cell life and allowed establishment of cell models to inspect and characterize the major functions of APE1. Nonetheless, complete knowledge of molecular effectors regulated by APE1 in determining its biological roles is still scanty. The essentiality of APE1 for mammalian cells seems to be mainly due to its DNA-repair activity in BER pathway. However, attempts to restore this activity in cells not expressing APE1 by using the yeast homologous Apn1 [1], which lacks the redox-transcriptional activation domain, or an APE1 mutant lacking the acetylation sites but not the DNA-repair activity [12], were unsuccessful. By specifically blocking the APE1 redox activity with the drug E3330, but not its DNA repair activity, very recently it has been shown that cytokine-mediated hemangioblast development in vitro was significantly impaired [13]. These observations led us to hypothesize that essentiality of APE1 may be due to its pleiotropic biological effects rather than merely to its DNA-repair activity.
In this work, we have identified changes at the proteome and transcriptome level associated with the loss of APE1 expression in HeLa cells by using conditional RNAi technology and an unbiased comparative approach of proteomic and microarray analysis. Our data shed light on some of the molecular aspects mediating the biological effects of the multifunctional APE1. This is the first large-scale attempt to dissect the roles of APE1 both at gene expression and protein level and provide the basis for future studies on the comprehension of the role played by this protein in regulating different biological mechanisms through the control of signal transduction pathways and gene expression.
2. Materials and methods
2.1. Inducible siRNA of APE1 in HeLa cell line
The oligonucleotides used for siRNA of APE1 were as follows: sense, 5’-GATCCCCCCTGCCACACTCAAGATCTGCTTCAAGAGAGCAGATCTTGAGTGTGGCAGGTTTTTGGAAA-3’; antisense, 5’-AGCTTTTCCAAAAACCTGCCACACTCAAGATCTGCTCTCTTGAAGCAGATCTTGAGTGTGGCAGGGGG-3’. These sequences were drawn following the empirical rules of Mittal [14] and were designed to recognize and bind to a 21 base sequence (underlined) placed 175 nucleotides after the AUG initiation codon of APE1 gene. As a control, we used the scrambled oligonucleotide sequences: sense, 5’-GATCCCCAGTCTAACTCGCCACCCCGTATTCAAGAGATACGGGGTGGCGAGTTAGA CTTTTTTGGAAA-3’; antisense, 5’-AGCTTTTCCAAAAAAGTCTAACTCGCCACCCCGTATCTCTTGAATACGGGGTGGCG AGTTAGACTGGG-3’. These sequences were checked with BLAST (http://www.ncbi.nlm.nih.gov/blast/) for their inability to pair any human cDNA sequence. The sequences were cloned into BglII and HindIII restriction sites of the pTer vector [15], which presents a tetracycline- (doxycycline-) responsive promoter to form the so called pTer/APE1 vector and pTer/Scr, respectively. In a first step, HeLa cells were transfected with pcDNA6/TR to generate stable Tet-repressor expressing cell clones that were selected for the acquired resistance by incubation with blasticidin (5 µg/ml) (Invitrogen, Milan, Italy) for 14 days. Individual clones were isolated by using cell cloning cylinders (Sigma, Milan, Italy) transferred and grown stepwise into 24-well, 12-well, 6-well plates for expansion to 107 cells. Clone TR5 was shown to express the Tet-repressor at higher levels and was therefore selected for transfection with pTer/APE1 vector previously linearized with Bst1107I (Fermentas, St.Leon Rot, UK) and subjected to selection with zeocine (200 µg/ml) (Invitrogen) for 14–21 days. Thirty single clones were isolated by using cell cloning cylinders transferred and grown stepwise for expansion to 107 cells. As a control, we used cell clones transfected with the empty pTer vector or with pTer/Scr vector. For inducible siRNA experiments, doxycycline (Sigma) was added to the cell culture medium at the final concentration of 1 µg/ml and cells were grown for the indicated times. Total cellular extracts were analyzed for APE1 expression by immunoblotting.
All biological data were reproduced in at least two different cell clones for each model.
2.2 Cell growth assays, cell cycle and apoptosis studies
For proliferation assays, cells were harvested at indicated times, stained with Trypan blue (Sigma), plated in triplicate and counted. Clonogeneic assay was performed according to Plumb [16]. Briefly, an equal number (500) of control and siRNA cells were plated in petri dishes and grown with medium containing or not doxycycline (1 µg/ml). On day 10, the medium was removed and colonies were stained for 2 min with 2 ml of cristal violet solution (10% w/v in 70% aqueous ethanol). The dye was then poured off. Then, plates were rinsed with tap water and allowed to dry. Colonies were counted by using ImageQuant TL software (GE Healthcare, Milan, Italy). For each experimental point, the mean, SD and statistical significance were calculated performing three independent experiments of cell colony count.
Cell cycle studies were performed by flow cytometry by using a FACScan (Becton Dickinson, Franklin Lakes, NJ, USA). The number of apoptotic cells in control and APE1 siRNA cells after 0 and 10 days of treatment with 1 µg/ml of doxycycline was determined using the method of Nicoletti et al. [17] by evaluating the number of cells with subdiploid DNA content through flow cytometric determination. Briefly, 2 × 106 cells were harvested and washed once with cold PBS/0.1% w/v sodium azide solution, resuspended in 1 ml of low-salt stain solution [4 mM sodium citrate, 3% v/v polyethylene glycol 8000, 1 mg/ml propidium iodide solution (Invitrogen, Carlsbad, CA, USA), 180 U/ml RNAse A (Sigma) and 1% w/v Triton X-100 in PBS/sodium azide solution] and then incubated, in the dark, at 37 °C, for 20 min, with gentle mixing every 5 min. Then, 1 ml of high-salt stain solution (0.4 M sodium chloride, 3% v/v polyethylene glycol 8000, 1 mg/ml propidium iodide solution and 1% w/v Triton X-100 in PBS/sodium azide solution) was added by gentle pipetting and samples were stored at 4 °C overnight. Cells were centrifuged at 5000 × g for 5 min, at 4 °C, the supernatant was removed, pellet resuspended in 500 µl of low-salt solution and then cells were analyzed on a Becton-Dickinson Canto using an Ar laser (excitation 488 nm). For each sample, 25,000 single events were detected and data analysis was performed using both WinMDI 2.8 (written by Joseph Trotter, Scripps Research Institute, La Jolla, CA) and ModFitLT V3.0 software program.
2.5 Preparation of cell extracts and Western blot analysis
To obtain total protein extracts, 107 HeLa cells were directly lysed into lysis buffer containing 7.0 M urea, 2.0 M thiourea, 2% w/v 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid, 10 mM DTT, 1% v/v carrier ampholytes pH 3–10 (GE Healthcare), 1% (v/v) β-mercaptoethanol and 40 mM Tris-HCl, by using a potter. Samples were lysed in a total volume of 500 µl. Homogenates were then centrifuged at 10,000 × g for 30 min, at 4 °C, and supernatants collected for the following analysis or stored at −80 °C. Protein quantification was performed by the Bradford colorimetric method [18] and SDS-PAGE analysis.
Cell nuclear and cytoplasmic extracts were prepared as previously described [19]. Briefly, 107 cells were washed once with PBS and resuspended in 100 µl of hypotonic lysis buffer A (10 mM Hepes, 10 mM KCl, 0.1 mM MgCl2, 0.1 mM EDTA, 2 µg/ml leupeptin, 2 µg/ml pepstatin, 0.5 mM PMSF, pH 7.9). After 10 min, cells were homogenized by ten strokes with a loose-fitting Dounce homogenizer. Nuclei were collected by centrifugation at 500 × g, at 4 °C, for 5 min, in a microcentrifuge. The supernatant was considered as the cytoplasmic fraction. Nuclei were then washed three times with the same volumes of buffer A to minimize cytoplasmic contamination. Nuclear proteins were extracted with 100 µl of buffer B (10 mM Hepes, 400 mM NaCl, 1.5 mM MgCl2, 0.1 mM EDTA, 2 µg/ml leupeptin, 2 µg/ml pepstatin, 0.5 mM PMSF, pH 7.9). After incubating for 30 min at 4 °C, samples were centrifuged at 12,000 × g for 20 min, at 4 °C. Nuclear extracts were then analyzed for protein content [18] and stored at −80 °C in aliquots.
Western blot analysis were performed on the indicated amounts of cell extracts as described previously [19]. Normalization was performed with the polyclonal anti-actin or anti-β-tubulin antibodies (Sigma). Blots were quantified by using a Gel Doc 2000 videodensitometer (Bio-Rad, Hercules, CA, USA).
2.6 Two-dimensional polyacrylamide gel electrophoresis
Thirty µg of total cell extracts were loaded onto 13 cm, pH 3–10 L IPG strips (GE Healthcare). IEF was conducted using an IPGPhor II system (GE Healthcare) according to the manufacturer’s instructions. Focused strips were equilibrated with 6 M urea, 26 mM DTT, 4% w/v SDS, 30% v/v glycerol in 0.1 M Tris-HCl (pH 6.8) for 15 min, followed by 6 M urea, 0.38 M iodoacetamide, 4% w/v SDS, 30% v/v glycerol, and a dash of bromophenol blue in 0.1 M Tris-HCl, pH 6.8, for 10 min. The equilibrated strips were applied directly to 10% w/v SDS-polyacrylamide gels and separated at 130 V. Gels were fixed and stained by ammoniacal silver [20]. Gels were scanned with an Image Master 2-D apparatus and analyzed by the Melanie 5 software (GE Healthcare), which allowed estimation of relative differences in spot intensities for each represented protein.
2.7 Evaluation of differentially-represented spots and mass spectrometry analysis
Each sample was analyzed in triplicate; then, a comparative analysis was carried out as previously reported [21] (see Supplementary Material for details). Differential spots from 2-DE were excised from the gel, triturated and washed with water. Proteins were in-gel reduced, S-alkylated and digested with trypsin as previously reported [21]. Digested aliquots were removed and subjected to a desalting/concentration step on µZipTipC18 (Millipore Corp., Bedford, MA, USA) using acetonitrile as eluent before MALDI-TOF-MS analysis. Peptide mixtures were loaded on the MALDI target, using the dried droplet technique and α-cyano-4-hydroxycinnamic acid as matrix, and analyzed by using Voyager-DE PRO mass spectrometer (Applied Biosystems, Framingham, MA, USA). Spectra were elaborated using the DataExplorer 5.1 software (Applied Biosystems) and manually inspected to get the peak lists. Internal mass calibration was performed with peptides deriving from enzyme autoproteolysis. PROWL software package was used to identify spots unambiguously from updated all taxa non-redundant sequence database (NCBI nr 2008/05/03) by using a mass tolerance value of 40–60 ppm, trypsin as enzyme specificity, a missed cleavages maximum value of 2 and cysteine carbamidomethyl and methionine oxidation as fixed and variable modification, respectively. Candidates with ProFound’s Est’d Z scores > 2.0 were further evaluated by the comparison with their calculated mass and pI using the experimental values obtained from 2-DE. Detailed PMF analysis is reported in the supplemental data.
2.8 Microarray analysis
Total RNA was extracted in triplicate by using TRIzol reagent kit (Invitrogen), and quantified. The total RNA quality was determined using the Agilent Bioanalyser (Agilent Technologies, Stockport, Cheshire, UK). Human genome HG U133 PLUS2 microarrays were run according to manufacturer’s instructions (Affymetrix Inc., Santa Clara, CA, USA) and submitted in MIAME (Minimum Information About a Microarray Experiment)-compliant format to the ArrayExpress database (www.mged.org/Workgroups/MIAME/miame.html, accession number E-MEXP-1315). Array data were normalized and summarized using RMAExpress [22]. Differential expression in response to siRNA treatment was calculated using Cyber-T [23]. A false-discovery correction was applied to these p-values to produce a q-value [24].
2.9 Reconstruction of the APE1 response network
Network construction was performed with MetaCore software (Genego Inc, St. Joseph, MI, USA). This software builds networks based on human annotated relationships between proteins based on peer reviewed publications.
2.10 Statistical analysis
Statistical analysis was performed using the Microsoft Excel data analysis program for Student’s t-test analysis. P<0.05 was considered as statistically significant.
3. Results
3.1 Biological effects of endogenous APE1 downregulation by conditional siRNA
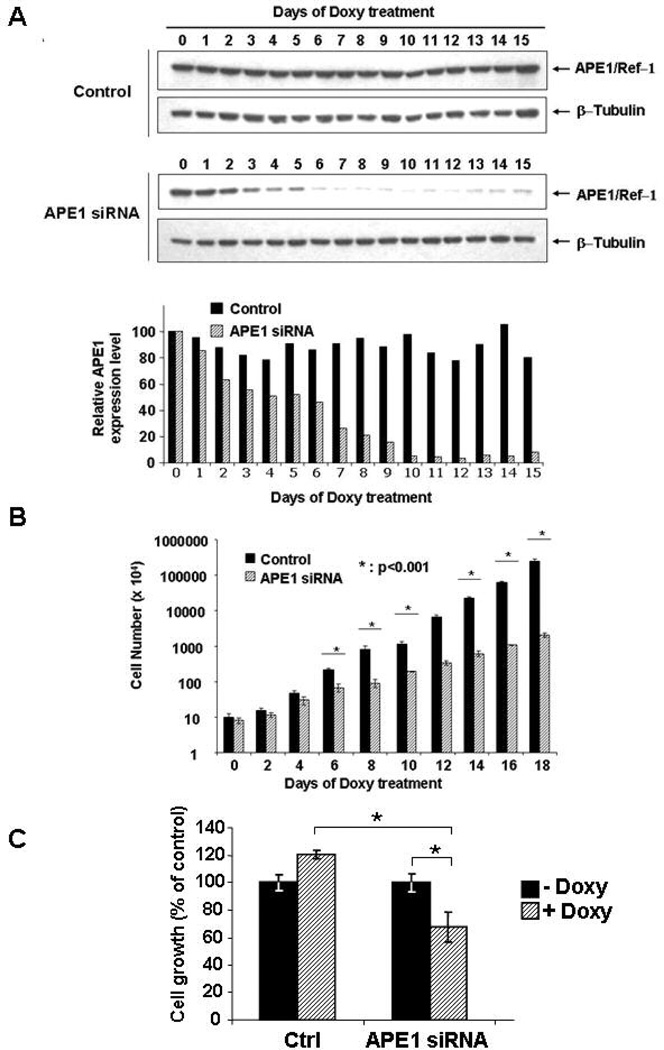
Conditional knock-down by RNAi technology was chosen to study molecular changes, at the protein and gene expression levels, associated to loss of APE1 functions. We used HeLa cells as a general cellular model for this study. Endogenous expression of APE1 was knocked-down by siRNA technology in a conditional manner through a doxycycline-responsive promoter [15]. Control HeLa cells, obtained after stable transfection with the control siRNA empty vector (pTer) or with a scrambled sequence (pTer/Scr), or silenced HeLa cells bearing the APE1 specific siRNA plasmid (pTer/APE1), were treated with doxycycline for different times up to fifteen days and the expression levels of APE1 were evaluated each day (Figure 1A). Neither stable transfection with the empty vector nor with the scrambled sequence affected the endogenous expression level of APE1 protein. On the contrary, inducible expression of APE1 siRNA sequences efficiently promoted APE1 down-regulation in a time-dependent manner. At ten days of treatment, endogenous APE1 expression was almost undetectable as compared to both controls. This time point was therefore chosen for further gene expression and proteomics analysis.
Figure 1. Effects of APE1 downregulation on cell proliferation.
Panel A. HeLa cells were stably transfected with the inducible siRNA vectors, as described in the methodological section. Suppression of endogenous APE1 expression was assayed after different days of treatment with doxycycline (Doxy), by Western blot analysis on total cellular extracts. Fifteen µg of total protein extracts were separated by 10% SDS-PAGE, and then transferred onto a nitrocellulose membrane. The membrane was immunoblotted with anti-APE1 or anti-β-tubulin antibodies (upper panel). The lower panel shows normalized expression levels of APE1 protein obtained after densitometric analysis of the bands. Values were reported as histograms of the ratio between APE1 and β-tubulin bands intensities. Black boxes represent APE1 normalized expression in the control cell clone (control) and dashed boxes represent APE1 expression in the silenced clone (APE1 siRNA).
Panel B. Effect of APE1 silencing on cell proliferation. HeLa cells were seeded in 60 mm petri dishes. Growth was followed by measuring cell number at various times after doxycycline treatment. Data, expressed as the percent change with respect to control cultures, are the mean ± SD of three independent experiments. See Materials and Methods section for details.
Panel C. Effect of APE1 silencing on cell growth by clonogeneic assay. Five-hundred cells of control and siRNA clones were seeded in petri dishes and then treated or not, as indicated, with 1 µg/ml of doxycycline for 10 days. Data, expressed as percentage of survival with respect to control cultures, are the mean ± SD of three independent experiments and show the inhibition of colony formation due to APE1 loss of expression in siRNA clone. (*, p < 0.05)
Progressive reduction of endogenous APE1 levels was paralleled by: i) a concomitant loss of the AP-endonuclease activity of nuclear extracts in silenced cells and ii) a parallel increase in AP-sites accumulation on genomic DNA was observed (Supplementary Figs. 1A and B). These data were confirmatory of the role of APE1 as the major AP-endonuclease enzyme in eukaryotic cells.
Next, we evaluated the effects of loss of APE1 expression on cell proliferation, cell cycle and apoptosis. While direct cell counting and clonogeneic proliferation assays clearly showed a strong inhibition of cell growth in the silenced clone that paralleled the reduction in the endogenous APE1 levels, control clones were not affected (Fig. 1A, 1B and 1C). These results suggest that, similarly to previously reported evidence using different cell lines [1], APE1 levels affect cell division. In order to test whether the observed reduction of cell proliferation following APE1 down-regulation was the result of induction of apoptosis or of inhibition of the cell cycle, apoptosis and cell cycle analyses were performed.
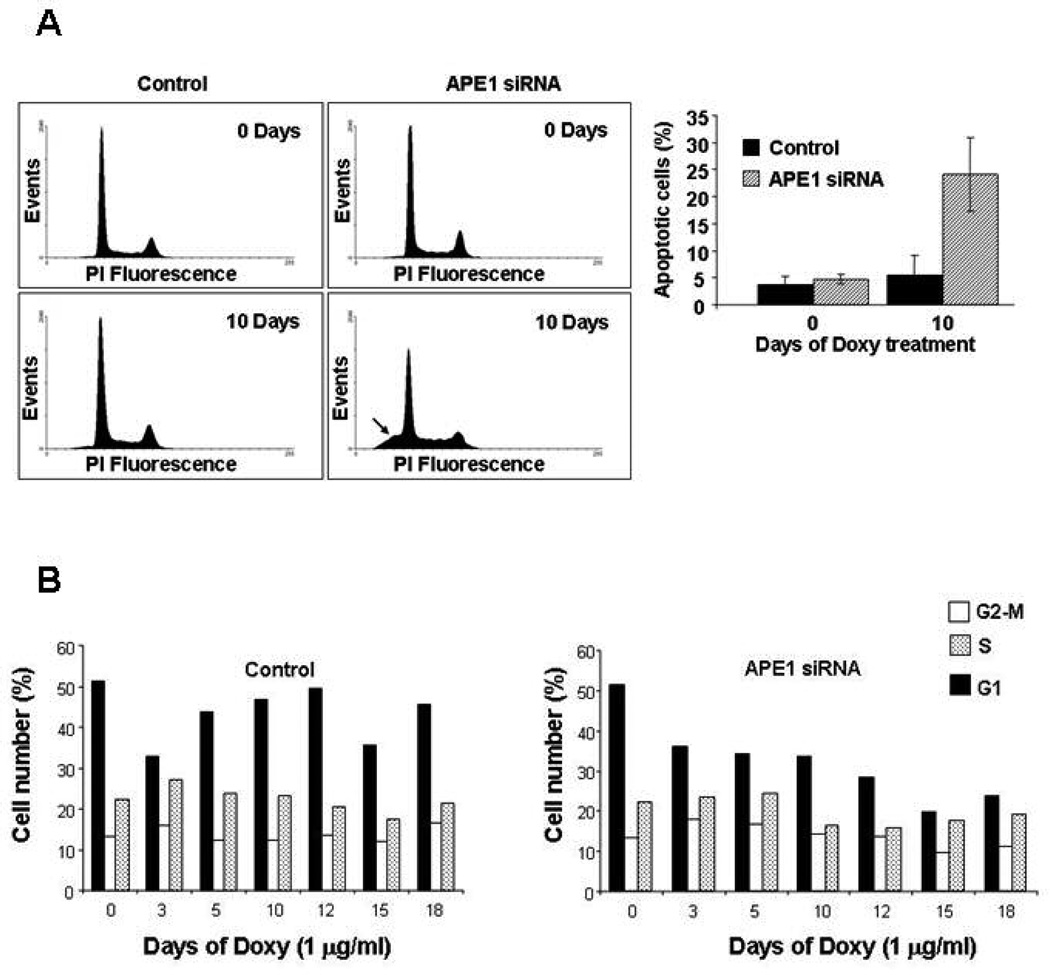
3.2 APE1 knocked-down cells undergo apoptosis and bear impaired cell cycle
Silencing of APE1 caused an increase in cell death by apoptosis. Flow cytometry analysis performed on control and silenced clones showed a strong correlation between down-regulation of APE1 expression and increased apoptosis (Fig. 2A). In particular, at 10 days of treatment with doxycycline (when APE1 expression level in the silenced clone is minimal), 25% of the cell population appeared in the flow histogram as sub-G1 peak corresponding to apoptotic cells possessing a sub-diploid DNA content (Fig. 2A, right). Apoptosis analysis performed by Annexin-V staining confirmed these results (data not shown).
Figure 2. Impaired proliferation in APE1 silenced cells is due to altered cell cycling and induced apoptosis.
Panel A. Left, evaluation of apoptosis, on isolated nuclei, on control and on APE1 siRNA cells at 10 days of treatment with doxycycline by flow cytometry. Apoptosis is expressed as the percentage of sub-G1 DNA containing cells (indicated by arrow), as detected by propidium iodide (PI) DNA staining. Right, each bar of the histogram represents the mean of three separate experiments ± SD.
Panel B. Analysis of cell cycle distribution. Control and APE1 siRNA HeLa cells were grown in the presence of doxycycline for the indicated times. Cell cycle distribution was evaluated at the indicated time points by FACS analysis, as described in Materials and Methods. APE1 siRNA cells show a time-of-induction dependent decrease in the cycling cells population, and a parallel increase in the S-G2/M versus G1 ratio.
Given these findings, we concentrated on the relationship between cell-death and cell cycle progression following APE1 silencing. Increased cell-death in APE1-deficient cells was associated to an increase in S-G2/M versus G1 population ratio (Fig. 2B), as measured by FACS-analysis. In fact, in the APE1-silenced clone, the percentage of cycling cells decreases according with the observed increase in apoptosis upon induction with doxycycline (time 0 of induction: 87.05% cycling cells; 10 days and 12 days: 64.62% and 57.99% of cycling cells, respectively). This phenomenon did not occur for the control clone (time 0: 85.75% of induction; time 10 days: 81.85%, time 12 days 83.11%). Among the cycling population, we observed a decrease in the ratio between G1 and S+G2-M phases upon APE1 silencing. In the APE1-silenced clone, at time 0 of induction with doxycycline, the G1/S+G2-M ratio was 1.43, whereas at time 10 days this ratio was 0.96 and at time 12 days it was 0.73. For the control clone, the G1/S+G2-M ratios were almost constant, being 1.46 at time 0 of induction and 1.33 and 1.46 at time 10 and 12 days, respectively. This evidence led us to hypothesize that APE1 silencing impairs the passage from S-G2/M phases to the following G1 phase. This hypothesis was verified by synchronization experiments (Supplementary Fig. 2). These data were suggestive for a pivotal role of APE1 in controlling cell cycle progression and the importance of its functions in cell surviving, opening also different scenarios in which APE1 plays a role not only in DNA repair but also in the redox transcriptional co-activation of genes involved in cell-cycle progression and in the redox-mediated cell signalling, functions still largely unexplored.
3.3 APE1 loss-of-expression impairs cell growth, signal transduction, cytoskeletal structure, oxidative stress and DNA-repair-related genes
In order to identify the molecular changes associated to the loss of APE1 expression, we adopted an unbiased differential proteomic approach, based on 2-DE gel quantitative analysis coupled to MALDI-TOF-MS identification of the up/down-regulated proteins, and a differential gene expression approach based on microarray analysis.
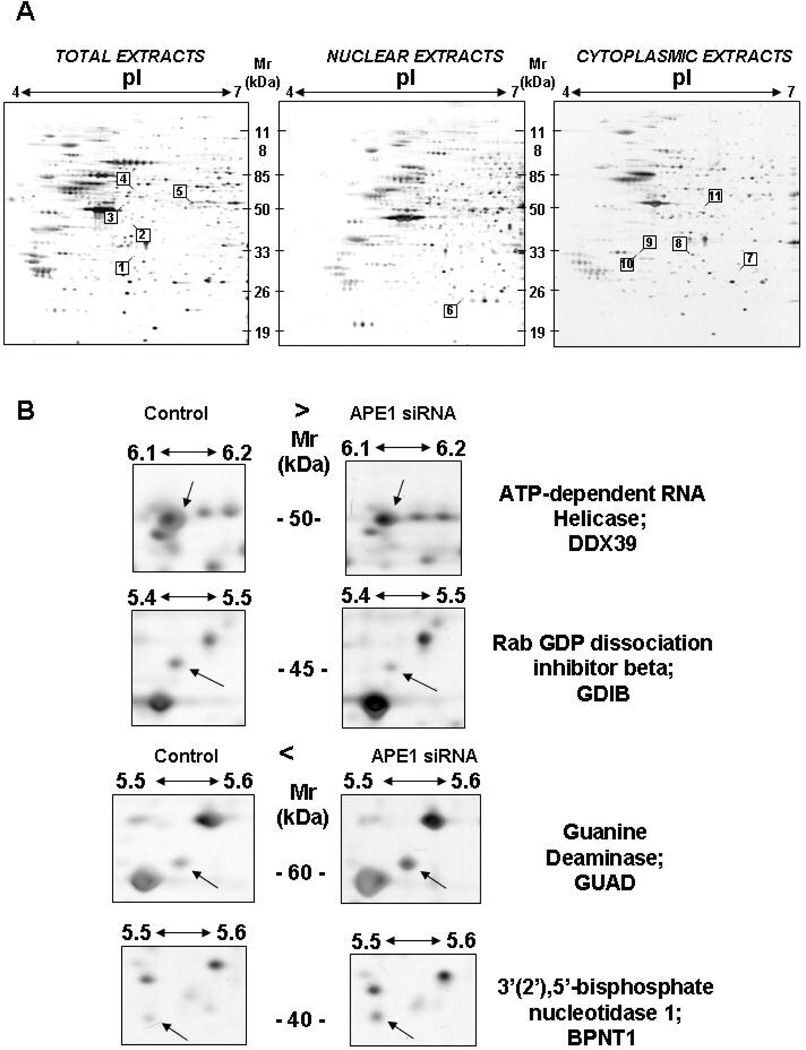
To obtain more information during the proteomic investigation, the analysis was performed on total, nuclear and cytoplasmic extracts, respectively. Silver staining of 2-DE allowed for the visualization and simultaneous quantitative evaluation of about 600–700 protein spots in each gel (Fig. 3). A total of 10 different proteins displayed a statistically significant change in knocked-down cells as compared to the control cells (Table 1 and Supplementary Material Fig. 3). Peptide mass fingerprint analysis and non-redundant sequence database searching allowed the non-ambiguous identification of all the analyzed species. Table 1 reports the nature of each identified spot, the measured 2-DE coordinates and relative sequence coverage, together with their relative abundance. Six of them were up-regulated (ANXA3, BPNT1, DDX39, GRB2, ACTB, CLIC3) and four (GDIB, GUAD, COPE, MARE1) were down-regulated in the APE1 knocked-down cell clone. It is noteworthy that all the proteins belong to functional categories involved in signal transduction (ANXA3, GRB2, GDIB, CLIC3), cytoskeletal structure and intracellular trafficking (DDX39, COPE, MARE1, ACTB) and metabolism (BPNT1 and GUAD).
Figure 3. Reference 2-DE maps and differential proteomic analysis.
Both control and APE1 siRNA HeLa cells were stimulated with doxycycline for 10 days and then collected for proteomic analysis of total, nuclear and cytoplasmic extracts. All spots showing significant changes between the two cell lines are highlighted. The identification numbers of the spots correspond to numbering in Table 1. These spots were identified by MALDI-TOF-MS. Protein spots numbered 1 and 8 correspond to the same protein species.
Panel B. Representative gel regions comprising some of the statistically significant changes in proteome repertoire were cropped
Table 1. List of up-/down-regulated proteins upon APE1 silencing, as detected by 2-DE and identified by peptide mass fingerprint analysis.
The spot number, protein description, accession number (SwissProt entry), experimental and theoretical (in parenthesis) molecular mass and pI values, sequence coverage, fold increase with respect to control, and the known function and subcellular localization are listed.
| Protein No. |
Protein identity | Swiss Prot entry |
MW, kDa |
pI | Num. Peptides Matched/Searched |
Sequence coverage |
Est’d Z Profoud Score |
APE1 siRNA/Control Fold ratio |
Function | |
|---|---|---|---|---|---|---|---|---|---|---|
| Total Extracts | 1 | Annexin A3 ANXA3 | P12429 | 33 (36) | 5.5 (5.6) | 10/11 | 35% | 2.39 | 2.94 | Inhibitor of phospholipase A2 |
| 2 | 3’(2’),5’-bisphosphate nucleotidase 1 BPNT1 | O95861 | 40 (33) | 5.5 (5.5) | 10/13 | 38% | 2.30 | 2.17 | Adenosine metabolism | |
| 3 | Rab GDP dissociation inhibitor beta GDIB | P50395 | 45 (51) | 5.4 (6.1) | 13/14 | 35% | 2.31 | −1.94 | Regulates the GDP/GTP exchange reaction of most Rab proteins by inhibiting the dissociation of GDP from them, and the subsequent binding of GTP to them | |
| 4 | Guanine deaminase GUAD | Q9Y2T3 | 60 (51) | 5.5 (5.4) | 9/14 | 30% | 2.30 | −1.67 | Catalyzes the hydrolytic deamination of guanine, producing xanthine and ammonia | |
| 5 | ATP-dependent RNA helicase DDX39 | O00148 | 50 (49) | 6.1 (5.5) | 9/11 | 24% | 1.98 | 1.66 | Involved in pre-mRNA splicing; required for the export of mRNA out of the nucleus | |
| Nuclear Extracts | 6 | Growth factor receptor-bound protein 2 GRB2 | P62993 | 26 (25) | 5.9 (5.9) | 8/10 | 47% | 2.32 | 1.58 | Adapter protein involved in Ras signaling pathway |
| Cytoplasmic Extracts | 7 | Chloride intracellular channel protein 3 CLIC3 | O95833 | 27 (27) | 6.3 (6.0) | 9/13 | 56% | 2.41 | Q (new appearance) | Mediates chloride ion transport. It may participate in cellular growth control |
| 8 | Annexin A3 ANXA3 | P12429 | 33 (36) | 5.5 (5.6) | 10/13 | 36% | 2.34 | 1.78 | Inhibitor of phospholipase A2 | |
| 9 | Coatomer subunit epsilon COPE | O14579 | 33 (34) | 5.2 (5.0) | 6/12 | 18% | 2.33 | −1.47 | Is required for budding from Golgi membranes, and is essential for the retrograde Golgi-to-ER transport of dilysine-tagged proteins | |
| 10 | Microtubule associated protein RP/EB family member 1 MARE1 | Q15691 | 33 (30) | 5.2 (5.0) | 9/15 | 41% | 2.34 | −1.47 | Involved in microtubule polymerization and in cell migration | |
| 11 | Actin, cytoplasmic 1 ACTB | P60709 | 40 (41) | 5.9 (5.3) | 7/10 | 28% | 1.67 | 1.53 | Cytoskeleton |
To identify genes whose expression changed as a consequence of APE1 silencing, we compared the gene expression profiles of WT and APE1 knocked-down cell clones by using the human Affimetrix GeneChip (HG-U133 PLUS2), which comprises over 20,000 genes. Three replicated RNA preparations from independent individual cultures of WT and APE1 silenced HeLa cells were hybridized to six arrays. In general, a gene was considered to be differentially expressed in APE1-deficient cells when the q-value was less than 0.05, the fold change was greater than or less than 1.5 and the mean expression level intensity was greater than 100 in either sample. By using these criteria, 1126 genes were identified as differentially expressed. Of these, 550 were identified as up-regulated and 576 down-regulated in APE1 deficient HeLa cells. Some of the differentially expressed genes corresponded to eight of the differentially expressed proteins already found by proteomic analysis (i.e. ANXA3, BPNT1, GDIB, GUAD, DDX39, GRB2, CLIC3, ACTB), suggesting that changes found at the protein level were due to transcriptional regulation. Interestingly, an already hypothesized target gene, i.e. that of the transcription factor Egr-1 [9], was identified by our analysis. Other notable genes are listed in Supplementary Table 1 (see the Supplementary Table 2 for the complete list of differentially expressed genes).
To confirm the microarray gene expression data, the expression level of 10 differentially expressed genes was also determined by Q-PCR as well as by Western blotting analysis (Supplementary Fig. 3). The fold induction/repression calculated by Q-PCR and Western blot analysis was in remarkable agreement with the corresponding ratio determined in the Affimetrix GeneChip analysis. Overall, a concordant behaviour between gene expression and proteomic data was proved for 20 different genes (see Supplementary Table 3 and Table 4).
Functional annotation of the microarray data was performed with DAVID [25]. Genes up-regulated by APE1 silencing showed a strong over-representation of cytoskeleton and microtubule components. There was also over-representation of extracellular matrix components, lipid metabolism, positive regulation of I-kappaB kinase/NF-kappaB cascade, and cell cycle arrest genes. Genes down-regulated by APE1 silencing included genes involved in cell growth, protein biosynthesis, DNA repair, cell cycle, mitochondria location, and oxidoreductase activity.
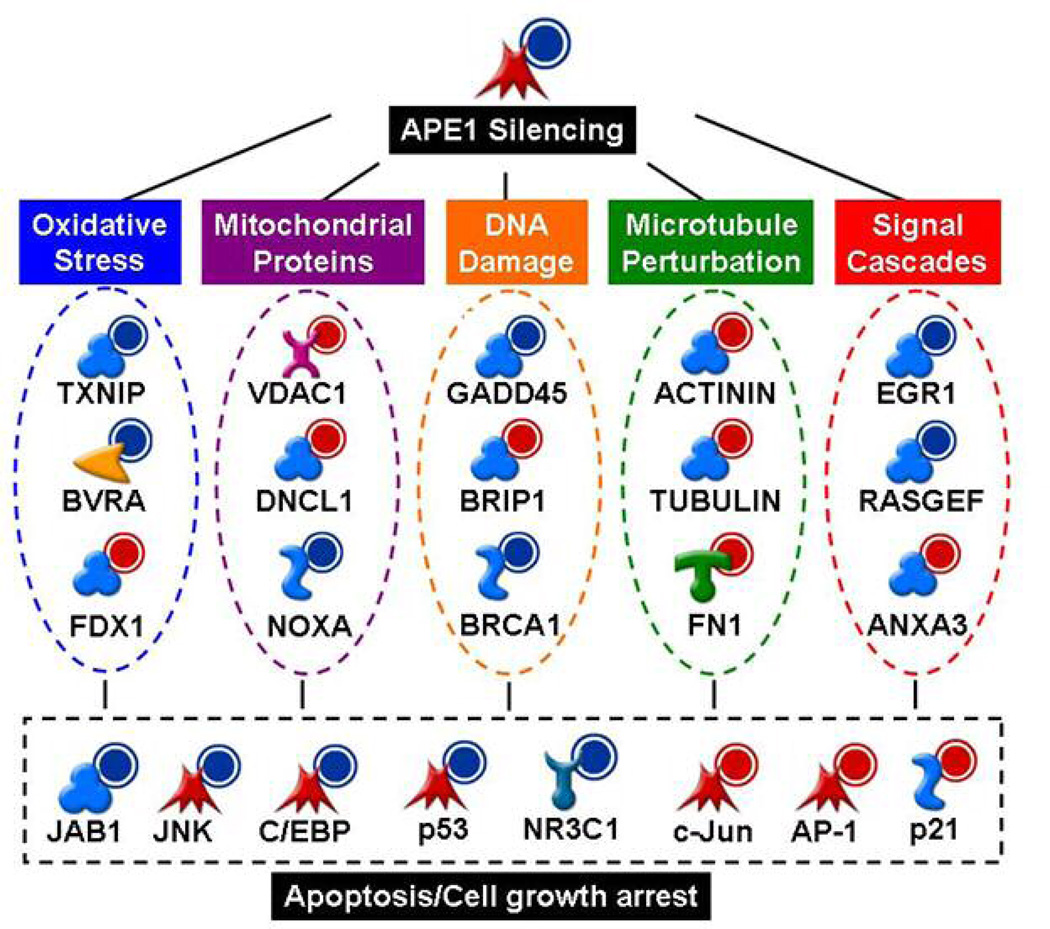
We sought to integrate the disparate consequences of APE1 silencing into a series of relationships. Networks were constructed based on published relationships between proteins using MetaCore software. A summary of these relationships is shown in Fig. 4. APE1 functionally interacts with molecular components of the cell responsible for dealing with oxidative stress, mitochondria, DNA damage, microtubules and signal cascades. APE1 silencing could lead to apoptosis and/or cell growth arrest through any one or several of these cellular perturbations.
Figure 4. Inference of APE1 molecular network.
Summarized networks built with MetaCore software (Genego Inc). Based on microarray data, genes which were significantly up-regulated or down-regulated upon APE1 silencing are shown with a red or a blue halo, respectively.
We performed a series of experiments to evaluate the validity of these molecular findings at the biological level (Fig. 5 and Supplementary Material Figs. 5 and 6). To define the role of mitochondria in cell death due to APE1-loss-of-expression, the mitochondrial membrane potential (Δψm), and the release of mitochondrial apoptosis effector Cytochrome-c (Cyt-c) and Apoptosis Inducing Factor (AIF) were studied. We clearly observed that APE1 silencing caused impairment of mitochondrial function, as measured by Δψm membrane potential depolarization, which was associated with Cyt-c and AIF release. These results may account for apoptotic triggering through the intrinsic pathway (Fig. 5A and Supplementary Material Fig.5).
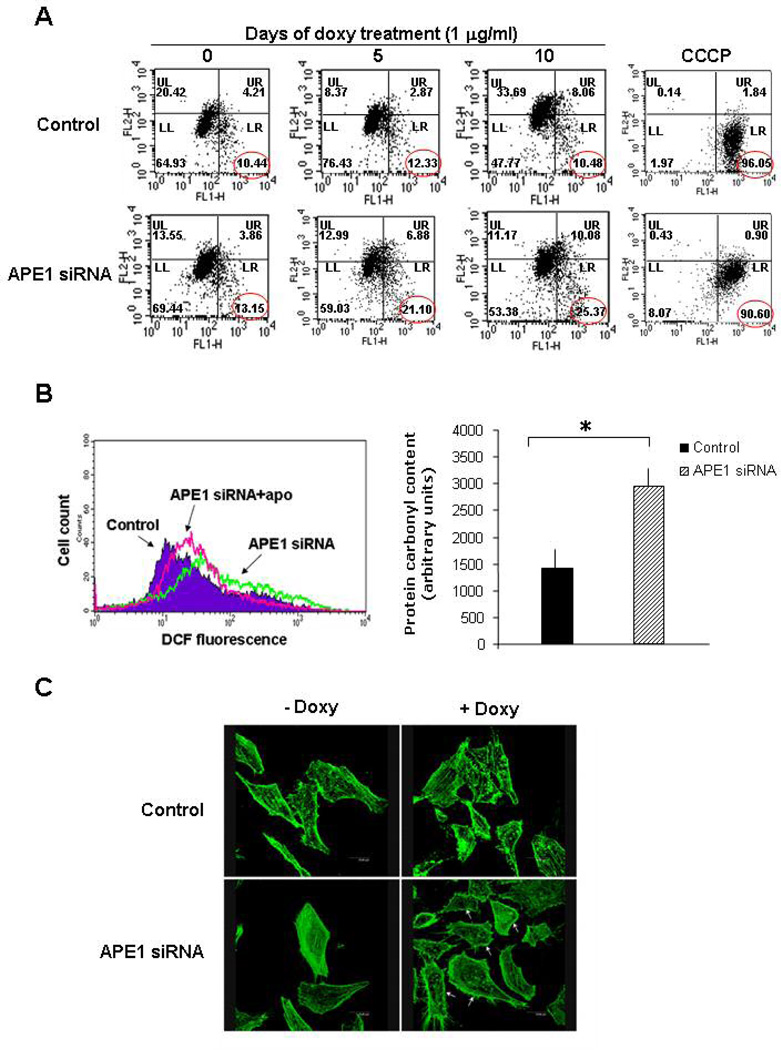
Figure 5. APE1 silenced cells show mitochondrial depolarization, increased oxidative stress and disturbation of cytoskeleton organization.
Panel A. Control and siRNA cells (after 10 days of doxycycline treatement) were treated with the mitochondrion specific probe JC-1. Mitochondrial depolarization was monitored by FACS and it was measured as a red to green shift in the fluorescent emission of the dye. FACS plots of control and siRNA cells at several times of silencing induction with doxycycline. To define the depolarization region of the plot, cells treated with carbonyl cyanide 3-chlorophenyl hydrazone (CCCP) were used as depolarization control. The percentage of cell population falling in this region is marked.
Panel B. To monitor endogenous ROS concentration, APE1 silenced-cells and control cells were treated with the profluorophore 2’,7’-dichlorodihydrofluorescein diacetate (H2DCFDA), which is converted in the fluorescent molecule 2’,7’-dichlorodihydrofluorescein (DCF) upon oxidation by intracellular ROS. ROS dependent staining was evaluated by FACS analysis. Control and APE1 silenced cells were treated for 1 h with 1 mM apocynin (apo), a specific blocker of NAD(P)H oxidase. The treatment reduced the endogenous ROS concentration evaluated as DCF fluorescence only in APE1 siRNA clone. Left: FACS profiles of control cells (Control), siRNA cells (APE1siRNA) and apocynin treated siRNA cells (APE1siRNA + apo) upon silencing induction. Right. To monitor oxidative damage, protein carbonylation was quantified in APE1-deficient and control cell extracts by derivatization with dinitrophenilhydrazine (DNPH), as described in Supplementary Material and Methods. The derivatizated samples were subjected to dot blot followed by recognition with anti-dinitrophenilhydrazone antibody. Histogram reporting the densitometric values of dinitrophenilhydrazone specific signals, normalized for β-tubulin content, in three independent experiments is shown on the right. The intensity of dinitrophenilhydrazone staining was significantly higher in APE1 silenced than in control cell line (*, p<0.05).
Panel C. Reorganization of F-actin in Hela cells. The effects on actin stress fibers organization were observed by confocal microscopy after phalloidin staining. F-actin was stained with Alexa 488-conjugated phalloidin and visualized through a Leica confocal microscope. The amount of stress fibers in silenced clone treated with doxycycline was clearly diminished compared with the control clones. The actin network reorganization was associated with membrane ruffling (white arrow). Original magnification, 1000×.
Based on the alteration in redox signalling, as derived from genome-wide and proteomics data, we measured both the intracellular ROS and the protein carbonyl content upon APE1 loss of expression. We found that APE1 silencing is associated to a pro-oxidant cellular status, as measured by increased ROS production, as a consequence of NADPH oxidase hyperactivation and increase in the protein carbonyl content (Fig. 5B and Supplementary Material Fig.6).
Finally, we asked whether cytoskeletal organization was disturbed in APE1 silenced cells. Confocal microscopic observations revealed a substantial disorganization of the actin stress fibers associated with membrane ruffling in APE1 knocked-down cells. In all the clones, with the exception of APE1 siRNA clone treated with doxycycline, the cells appeared flattened and well spread, with actin arranged in parallel arrays of long and almost continuous stress fibers running through the cytosol (Fig. 5C). Notably, upon APE1 silencing, a drastic reorganization of the F-actin network was observed in a way that the cytosolic stress fibers almost completely disappeared and actin aggregates were accumulated in a cortical mantle.
4. Discussion
Despite the central role played by APE1 in mammalian cell viability, comprehension of the molecular signalling networks underlying the multifaceted action of APE1 is yet elusive. Achieving such an understanding may be of great relevance in explaining why attempts to revert the apoptosis due to APE1 suppression via sole targeting of the DNA-repair enzymatic activity of APE1 in knocked-down or knocked-out mammalian cells were unsuccessful. With this goal in mind, the aim of this study was to use an unbiased strategy, by combining both mRNA expression profiling and proteomic analysis, to investigate the molecular changes associated to APE1 loss-of-expression by siRNA technology. A series of molecular evidences came out from this study, some of them providing a more solid base to already published data, which led us to build up the molecular networks reported in Fig. 6. However, the main message of this work is that APE1 represents a hub in coordinating different functions in mammalian cells. Thus, particular care must be taken in the future studies to understand the fine-tuning mechanisms responsible for its functional regulation.
Figure 6. Different model networks for APE1-mediated signalling as derived by gene expression and proteomic data.
Direct interaction network for genes dysregulated upon APE1 knock-down, involved in redox control, transcription regulators and chaperones (Panel A, https://portal.genego.com/pub/network/n-839812191.html) or in Egr-1-signaling (Panel B, https://portal.genego.com/pub/network/n-860173808.html). Colours of the symbols indicate inhibition (blue) and activation (red). Here, the name APEX is used to refer to APE1.
APE1 is strongly associated with the Thioredoxin system and with the COP9 signalosome complex
One interesting aspect of the network derived from this work is the Thioredoxin (TRX) system. APE1 directly binds to TRX in the nucleus [26]. The TRX/APE1 complex then regulates the activity of DNA-binding proteins by redox modification(s) of specific cysteine residue(s); these include Jun/Fos (AP-1), nuclear factor-kappaB (NF-κB) and p53 [27–30]. We realized that several of these transcription factors display altered expression (and possible impaired functional activity) in our APE1 silenced cells. This finding perfectly matches with the alteration of expression of a number of their target genes (Fig. 6A). After abolishment of APE1 expression by siRNA, a significant perturbation of the redox cellular environment occurs, consistent with our findings of increased cellular oxidative stress condition. Expression of TRX, however, did not change significantly. Nonetheless, we found a relevant down-regulation of TXNIP (VDUP1), which directly binds to reduced TRX, forming a stable disulfide-linked complex [31]. The involvement of APE1 in the TRX system can explain, at least in part, the extensive biological consequences on cell growth that we observed after APE1 down-regulation and may also explain APE1 implication in different pathogenetic events (see below) such as cancer [32].
Our gene expression data showed a significant down-regulation of the Jun activation domain-binding protein 1 (JAB1), another protein known to directly bind to TRX and TXNIP [33]. JAB1 binds the nuclear progesterone receptor (PR) and binds to and thus coactivates transcription activation factors AP-1 and Src-1 [34]. Interestingly, both AP-1 and Src-1 have been already suggested as APE1-regulated proteins [6, 35]. Nuclear receptors, their coactivators and coregulators are present in large multimeric complexes [36], whereas JAB1 is localized in the COP9 signalosome complex. COPS5, one of the 8 proteins in the COP9 signalosome complex, also showed a significant down-regulation in our experiments after silencing of APE1 expression (Fig. 6A). Therefore, we can speculate that APE1 takes part to important general processes of intracellular signalling transduction pathways.
The induction of stress proteins gene expression, such as Hsp70 (Fig. 6A), may be ascribable to adaptive cellular response mechanisms to the stress condition due to APE1 loss-of-expression. Similarly, up-regulation of NF-κB signalling cascade could be interpreted as a pro-survival program, triggered to escape apoptosis due to APE1 loss-of-expression.
APE1 silencing leads to mitochondrial-mediated apoptosis
APE1 silencing induces apoptosis. Our data provide no definitive evidence in favor of a single model of induction of apoptosis (Fig. 4), even though a major involvement of mitochondrial-mediated apoptosis is plausible. Mitochondrial localization of APE1 may be associated to a potential role in DNA repair of oxidized bases in mitochondrial genome [2, 37, 38]. Accumulation of mtDNA oxidative damage may negatively impact the expression of proteins involved in respiratory chain activity, which leads to impairment in oxidative phosphorylation, and in mitochondrial malfunctioning, causing, in turn, enhanced ROS production. These events would perpetuate the damage and constitute a ‘vicious cycle’ exacerbating the original oxidative offence. In accordance with these hypotheses, we were able to demonstrate a significant impairment in mitochondrial functionality upon APE1 silencing, as determined by mitochondrial membrane potential depolarization assays. At present, our data are suggestive of a major involvement of mitochondrial pathways in the apoptotic process associated to APE1 loss-of-expression, as demonstrated by release of Cyt-c and AIF perinuclear accumulation in APE1 silenced cells. Further experiments are needed to address this issue. Of interest, is the observation that mitochondrial antioxidant proteins (i.e. PRDX3, NOXA) were also found down-regulated upon APE1 gene silencing, suggesting that the role of APE1 within mitochondria could be also indirect.
APE1 controls the intracellular redox state
Mitochondria are the major source of ROS within eukaryotic cells. However, other multienzymatic protein complexes may affect intracellular ROS production. This is the case of the non-phagocytic NADPH oxidase system, which is an enzyme composed of multiple membrane-associated (Mox and p22phox) and cytosolic components (p67 phox, p47phox, Rac1), which catalyzes the transformation of the molecular oxygen to the superoxide anion by transferring an electron from the substrates NADH or NADPH [39]. It has been recently suggested that APE1 may inhibit Rac1 activity in vivo, thus possibly controlling intracellular ROS production [40, 41]. However, those findings were obtained by over-expressing ectopic APE1 in cell models, and therefore did not definitely demonstrate a causal role for APE1 in the process due to the presence of non-physiological amount of APE1. Here, by using the specific NADPH oxidase inhibitor apocynin, we ultimately showed that indeed APE1 plays an inhibitory role on ROS production by the membrane NADPH system. Since we recently demonstrated that NADPH-mediated ROS production induced by P2Y triggering was able to promote APE1 functional activation [19], we propose the existence of an autoregulatory loop between these two systems. This observation will be of therapeutical relevance for endothelial, fibroblastic and smooth muscle cells in vascular system pathologies, where an over-activation of the NADPH oxidase system is involved [42], as well as in the angiogenesis process [43], where a further hypothesis of autoregulatory loop between APE1 and VEGF may be inferred [44]. This observation could be therapeutically relevant also for treatment of tumor progression and cancer metastasis.
APE1 controls the cytoskeletal structure
During mitosis, complete disassembly and reassembly of all the cytoskeletal components is required. The pathways that link cell activation to cytoskeletal changes are the key to the transmission of signals for the pleiotropic effects necessary for appropriate cellular response. A number of genes associated with cytoskeletal structures were identified through our microarrays analysis. This is suggestive of a major role of cytoskeleton downstream of APE1. Actin, an element of the microfilaments, exerts important functions in cell motility, mitosis and intracellular trafficking and changes in the dynamics/distribution of this cytoskeletal protein are observed during apoptosis. Though the involvement of cytoskeletal actin in apoptosis is still poorly understood, its causal role seems to be clear [45]. In this study, a major disorganization of actin stress fibers associated with membrane ruffling paralleled the loss of APE1 expression. Thus, it seems likely that many of the morphologic manifestations of apoptosis, as well as the general effects on signal transduction pathways, are linked to the disassembly of the rigid cytoskeleton. A further perspective of this observation would be a possible role of APE1 in controlling cell migration via cytoskeleton.
Egr-l is a direct functional target of APE1
The unbiased approach we used in identifying new genes directly or indirectly regulated by APE1 was a useful tool to confirm the multifaceted nature of APE1. Based on gene expression data, we found that the expression levels of Egr-1 strictly depend on the presence of APE1. The fact that APE1 is also able to regulate Egr-1 expression levels, in addition to act as a redox coactivator over Egr-1 transcriptional activity [8, 9], reinforces our previous hypothesis of a functional regulatory loop between APE1 and Egr-1 [9]. This finding also underlines the biological complexity typical of this multi-functional protein and could constitute a means through which APE1 may regulate the expression of DNA repair genes such as GADD45 [46]. Egr-1 activates the transcription of cell-growth associated genes in response to a variety of mitogenic and non-mitogenic stimuli, including growth factors and hypoxia [47, 48] and is able to form redox-modulated transcriptional complexes with the above mentioned transcription factors (i.e. AP-1), as well as with APE1 and TRX [49]. Thus, in light of the new evidence presented by our results, the role of Egr-1 seems pivotal. The central effect of APE1 on Egr-1 biological functions is also reinforced by the concomitant down-regulation of additional Egr-1 target genes, such as C/EBPβ, VEGF and c-Myc (see Fig. 5B) [50].
Future perspectives for understanding APE1 roles in human pathologies
Overall, our data convincingly show a leading role for APE1 in controlling the cellular redox state both through the intracellular ROS production level in the cytoplasm and through the nuclear redox state by means of the TRX system, resulting in the fine tuning of specific gene expression. Modulation of the cellular redox state may affect different biological processes involved in cell growth, inhibition of apoptosis and modulation of inflammation. Therefore, the role of APE1 in a wide range of human diseases and conditions, including cancer, ischemia-reperfusion injury, cardiac conditions, neurodegenerative diseases, aging [2] is not surprising. However, whether APE1 contributes to or prevents the pathological progression of a particular condition is not always clear. For example, in several cancers, the expression levels of APE1 are increased and the protein is often found in the cytoplasmic compartment [2]. However, the reasons for this change in expression patterns are still poorly understood. It can be hypothesized that under these conditions, APE1 mainly exerts antiapoptotic and growth-promoting effects, thus playing a leading role in cancer progression. In ischemia-reperfusion injury and in neurodegenerative disorders, in which apoptosis contributes to the pathology, APE1 might protect against injurious insults by promoting cell-survival programs [41, 51–53]. This dichotomous role of APE1 needs to be explained at the molecular level by investigating the role of post-translational modifications in controlling the different functions of the protein as well as the different interactomes in which APE1 is enrolled. The cellular model that we characterized in this work, will be helpful in elucidating these lines.
In summary, our data represent the first attempt to gain a global view of the molecular role of APE1 in the cell. Combining large-scale studies with classical biochemical approaches, we clearly showed that the wild-type APE1 protein plays a central role in different biological processes and provides a molecular basis for explaining its multifunctional biological activity. The dataset coming from this work also offers several scientific leads for future studies aiming to shed light on the subtle mechanisms of this control.
Supplementary Material
Acknowledgements
This work was supported by grants from MIUR (FIRB #RBRN07BMCT) and Telethon (Grant #GGP05062 and #GGP06268) to G.T. and from CNR (AG.P04.015, MERIT and RSTL 862) to A.S.. DD was supported by a NERC Advanced Fellowship.
List of abbreviations
- APE1/Ref-1
Apurinic apyrimidinic Endonuclease/Redox effector factor 1
- BER
base excision repair
- RNAi
RNA interference
- ROS
reactive oxygen species
Footnotes
Gene expression data have been loaded into ArrayExpress database and are accessible for review purposes by using the following link: www.ebi.ac.uk/aerep/login; Username: Reviewer_E-MEXP-1315; Password: 1194526322909.
References
- 01.Fung H, Demple B. A vital role for APE1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol. Cell. 2005;17:463–470. doi: 10.1016/j.molcel.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 02.Tell G, Damante G, Caldwell D, Kelley MR. The intracellular localization of APE1/Ref-1: more than a passive phenomenon? Antioxid. Redox Signal. 2005;7:367–384. doi: 10.1089/ars.2005.7.367. [DOI] [PubMed] [Google Scholar]
- 03.Luo M, Delaplane S, Jiang A, Reed A, et al. Role of the multifunctional DNA repair and redox signaling protein Ape1/Ref-1 in cancer and endothelial cells: Small molecule inhibition of Ape1’s redox function. Antioxid. Redox Signal. 2008 doi: 10.1089/ars.2008.2120. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 04.McNeill DR, Wilson DM., 3rd. A dominant-negative form of the major human abasic endonuclease enhances cellular sensitivity to laboratory and clinical DNA-damaging agents. Mol Cancer Res. 2007;5:61–70. doi: 10.1158/1541-7786.MCR-06-0329. [DOI] [PubMed] [Google Scholar]
- 05.Xanthoudakis S, Miao GG, Curran T. The redox and DNA-repair activities of Ref-1 are encoded by nonoverlapping domains. Proc Natl Acad Sci USA. 1994;91:23–27. doi: 10.1073/pnas.91.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 06.Xanthoudakis S, Curran T. Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. EMBO J. 1992;11:653–665. doi: 10.1002/j.1460-2075.1992.tb05097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 07.Gaiddon C, Moorthy NC, Prives C. Ref-1 regulates the transactivation and pro-apoptotic functions of p53 in vivo. EMBO J. 1999;18:5609–5621. doi: 10.1093/emboj/18.20.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 08.Huang RP, Adamson ED. Characterization of the DNA-binding properties of the early growth response-1(Egr-1) transcription factor: evidence for modulation by a redox mechanism. DNA Cell Biol. 1993;12:265–273. doi: 10.1089/dna.1993.12.265. [DOI] [PubMed] [Google Scholar]
- 09.Pines A, Bivi N, Romanello M, Damante G, et al. Cross-regulation between Egr-1 and APE/Ref-1 during early response to oxidative stress in the human osteoblastic HOBIT cell line: evidence for an autoregulatory loop. Free Radic. Res. 2005;39:269–281. doi: 10.1080/10715760400028423. [DOI] [PubMed] [Google Scholar]
- 10.Ludwig DL, MacInnes MA, Takiguchi Y, Purtymun PE, et al. A murine AP-endonuclease gene-targeted deficiency with post-implantation embryonic progression and ionizing radiation sensitivity. Mutat. Res. 1998;409:17–29. doi: 10.1016/s0921-8777(98)00039-1. [DOI] [PubMed] [Google Scholar]
- 11.Xanthoudakis S, Smeyne RJ, Wallace JD, Curran T. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc Natl Acad Sci USA. 1996;93:8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Izumi T, Brown DB, Naidu CV, Bhakat KK, et al. Two essential but distinct functions of the mammalian abasic endonuclease. Proc Natl Acad Sci USA. 2005;102:5739–5743. doi: 10.1073/pnas.0500986102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou GM, Luo MH, Reed A, Kelley MR, Yoder MC. APE1 regulates hematopoietic differentiation of embryonic stem cells through its redox functional domain. Blood. 2007;109:1917–22. doi: 10.1182/blood-2006-08-044172. [DOI] [PubMed] [Google Scholar]
- 14.Mittal V. Improving the efficiency of RNA interference in mammals. Nat. Rev. Genet. 2004;5:355–365. doi: 10.1038/nrg1323. [DOI] [PubMed] [Google Scholar]
- 15.van de Wetering M, Oving I, Muncan V, Pon Fong MT, et al. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 2003;4:609–615. doi: 10.1038/sj.embor.embor865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plumb JA. In: Cytotoxic drug resistance mechanisms. Boger-Brown, editor. Totowa: Humana Press, NJ; 1999. pp. 17–23. [Google Scholar]
- 17.Nicoletti I, Migliorati G, Pagliacci MC, Grignani F, Riccardi C. A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J. Immunol. Methods. 1991;139:271–279. doi: 10.1016/0022-1759(91)90198-o. [DOI] [PubMed] [Google Scholar]
- 18.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 19.Pines A, Perrone L, Bivi N, Romanello M, et al. Activation of APE1/Ref-1 is dependent on reactive oxygen species generated after purinergic receptor stimulation by ATP. Nucleic Acids Res. 2005;33:4379–4394. doi: 10.1093/nar/gki751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 21.Vascotto C, Cesaratto L, D’Ambrosio C, Scaloni A, et al. Proteomic analysis of liver tissues subjected to early ischemia/reperfusion injury during human orthotopic liver transplantation. Proteomics. 2006;6:3455–3465. doi: 10.1002/pmic.200500770. [DOI] [PubMed] [Google Scholar]
- 22.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A Comparison of Normalization Methods for High Density Oligonucleotide Array Data Based on Bias and Variance. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 23.Baldi P, Long AD. A Bayesian Framework for the Analysis of Microarray Expression Data: Regularized t-Test and Statistical Inferences of Gene Changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 24.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dennis G, Jr., Sherman BT, Hosack DA, Yang J, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 26.Hirota K, Matsui M, Iwata S, Nishiyama A, et al. AP-1 transcriptional activity is regulated by a direct association between thioredoxin and Ref-1. Proc Natl Acad Sci USA. 1997;94:3633–3638. doi: 10.1073/pnas.94.8.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karimpour S, Lou J, Lin LL, Rene LM, et al. Thioredoxin reductase regulates AP-1 activity as well as thioredoxin nuclear localization via active cysteines in response to ionizing radiation. Oncogene. 2002;21:6317–6327. doi: 10.1038/sj.onc.1205749. [DOI] [PubMed] [Google Scholar]
- 28.Seemann S, Hainaut P. Roles of thioredoxin reductase 1 and APE/Ref-1 in the control of basal p53 stability and activity. Oncogene. 2005;24:3853–3863. doi: 10.1038/sj.onc.1208549. [DOI] [PubMed] [Google Scholar]
- 29.Ueno M, Masutani H, Arai RJ, Yamauchi A, et al. Thioredoxin-dependent redox regulation of p53-mediated p21 activation. J. Biol. Chem. 1999;274:35809–35815. doi: 10.1074/jbc.274.50.35809. [DOI] [PubMed] [Google Scholar]
- 30.Wei SJ, Botero A, Hirota K, Bradbury CM, et al. Thioredoxin nuclear translocation and interaction with redox factor-1 activates the activator protein-1 transcription factor in response to ionizing radiation. Cancer Res. 2000;60:6688–6695. [PubMed] [Google Scholar]
- 31.Patwari P, Higgins LJ, Chutkow WA, Yoshioka J, Lee RT. The interaction of thioredoxin with Txnip. Evidence for formation of a mixed disulfide by disulfide exchange. J. Biol. Chem. 2006;281:21884–21891. doi: 10.1074/jbc.M600427200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burke-Gaffney A, Callister ME, Nakamura H. Thioredoxin: friend or foe in human disease? Trends Pharmacol. Sci. 2005;26:398–404. doi: 10.1016/j.tips.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 33.Hwang CY, Ryu YS, Kim KD, Park SS, et al. Thioredoxin modulates activator protein 1 (AP-1) activity and p27Kip1 degradation through direct interaction with Jab1. Oncogene. 2004;23:8868–8875. doi: 10.1038/sj.onc.1208116. [DOI] [PubMed] [Google Scholar]
- 34.Chauchereau A, Georgiakaki M, Perrin-Wolff M, Milgrom E, Loosfelt H. JAB1 interacts with both the progesterone receptor and SRC-1. J. Biol. Chem. 2000;275:8540–8548. doi: 10.1074/jbc.275.12.8540. [DOI] [PubMed] [Google Scholar]
- 35.Gray MJ, Zhang J, Ellis LM, Semenza GL, et al. HIF-1alpha, STAT3, CBP/p300 and Ref-1/APE are components of a transcriptional complex that regulates Src-dependent hypoxia-induced expression of VEGF in pancreatic and prostate carcinomas. Oncogene. 2005;24:3110–3120. doi: 10.1038/sj.onc.1208513. [DOI] [PubMed] [Google Scholar]
- 36.McKenna NJ, Nawaz Z, Tsai SY, Tsai MJ, O’Malley BW. Distinct steady-state nuclear receptor coregulator complexes exist in vivo. Proc Natl Acad Sci USA. 1998;95:11697–11702. doi: 10.1073/pnas.95.20.11697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chattopadhyay R, Wiederhold L, Szczesny B, et al. Identification and characterization of mitochondrial abasic (AP)-endonuclease in mammalian cells. Nucleic Acids Res. 2006;34:2067–2076. doi: 10.1093/nar/gkl177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitra S, Izumi T, Boldogh I, Bhakat KK, et al. Intracellular trafficking and regulation of mammalian AP-endonuclease 1 (APE1), an essential DNA repair protein. DNA Repair. 2007;6:461–469. doi: 10.1016/j.dnarep.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 39.Babior BM. NADPH oxidase: an update. Blood. 1999;93:1464–1476. [PubMed] [Google Scholar]
- 40.Angkeow P, Deshpande SS, Qi B, Liu YX, et al. Redox factor-1: an extra-nuclear role in the regulation of endothelial oxidative stress and apoptosis. Cell Death Differ. 2002;9:717–725. doi: 10.1038/sj.cdd.4401025. [DOI] [PubMed] [Google Scholar]
- 41.Ozaki M, Suzuki S, Irani K. Redox factor-1/APE suppresses oxidative stress by inhibiting the rac1 GTPase. FASEB J. 2002;16:889–890. doi: 10.1096/fj.01-0664fje. [DOI] [PubMed] [Google Scholar]
- 42.De Keulenaer GW, Chappell DC, Ishizaka N, Nerem RM, et al. Oscillatory and steady laminar shear stress differentially affect human endothelial redox state: role of a superoxide-producing NADPH oxidase. Circ. Res. 1998;82:1094–1101. doi: 10.1161/01.res.82.10.1094. [DOI] [PubMed] [Google Scholar]
- 43.Ushio-Fukai M. Redox signaling in angiogenesis: role of NADPH oxidase. Cardiovasc. Res. 2006;71:226–235. doi: 10.1016/j.cardiores.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 44.Ziel KA, Campbell CC, Wilson GL, Gillespie MN. Ref-1/Ape is critical for formation of the hypoxia-inducible transcriptional complex on the hypoxic response element of the rat pulmonary artery endothelial cell VEGF gene. FASEB J. 2004;18:986–988. doi: 10.1096/fj.03-1160fje. [DOI] [PubMed] [Google Scholar]
- 45.Gourlay CW, Ayscough KR. The actin cytoskeleton: a key regulator of apoptosis and ageing? Nat. Rev. Mol. Cell. Biol. 2005;6:583–589. doi: 10.1038/nrm1682. [DOI] [PubMed] [Google Scholar]
- 46.Thyss R, Virolle V, Imbert V, Peyron JF, et al. NF-kappaB/Egr-1/Gadd45 are sequentially activated upon UVB irradiation to mediate epidermal cell death. EMBO J. 2005;24:128–137. doi: 10.1038/sj.emboj.7600501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khachigian LM, Lindner V, Williams AJ, Collins T. Egr-1-induced endothelial gene expression: a common theme in vascularinjury. Science. 1996;271:1427–1431. doi: 10.1126/science.271.5254.1427. [DOI] [PubMed] [Google Scholar]
- 48.Adamson E, de Belle I, Mittal S, Wang Y, et al. Egr1 signaling in prostate cancer. Cancer Biol Ther. 2003;2:617–622. [PubMed] [Google Scholar]
- 49.Khomenko T, Deng X, Jadus MR, Szabo S. Effect of cysteamine on redox-sensitive thiol-containing proteins in the duodenal mucosa. Biochem. Biophys. Res. Commun. 2003;309:910–916. doi: 10.1016/j.bbrc.2003.08.092. [DOI] [PubMed] [Google Scholar]
- 50.Vidal F, Aragones J, Arantzazu A, de Landazuri MO. Upregulation of vascular endothelial growth factor receptor Flt-1 after endothelial denudation: role of transcription factor Egr-1. Blood. 2000;95:3387–3394. [PubMed] [Google Scholar]
- 51.Gurusamy N, Malik G, Gorbunov NV, Das DK. Redox activation of Ref-1 potentiates cell survival following myocardial ischemia reperfusion injury. Free Radic. Biol. Med. 2007;43:397–407. doi: 10.1016/j.freeradbiomed.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 52.Malik G, Gorbounov N, Das S, Gurusamy N, et al. Ischemic preconditioning triggers nuclear translocation of thioredoxin and its interaction with Ref-1 potentiating a survival signal through the PI-3-kinase-Akt pathway. Antioxid. Redox Signal. 2006;8:2101–2109. doi: 10.1089/ars.2006.8.2101. [DOI] [PubMed] [Google Scholar]
- 53.Tan Z, Sun N, Schreiber SS. Immunohistochemical localization of redox factor-1 (Ref-1) in Alzheimer's ippocampus. Neuroreport. 1998;9:2749–2752. doi: 10.1097/00001756-199808240-00012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.