Summary
Botulinum neurotoxins (BoNTs) can cause paralysis at exceptionally low concentrations and include seven serotypes (BoNT/A-G). The chimeric BoNT/DC toxin has a receptor binding domain similar to the same region in BoNT/C. However, BoNT/DC does not share protein receptor with BoNT/C. Instead, it shares synaptotagmin (Syt) I and II as receptors with BoNT/B, despite their low sequence similarity. Here we present the crystal structures of the binding domain of BoNT/DC in complex with the recognition domains of its protein receptors, Syt-I and Syt-II. The structures reveal that BoNT/DC possesses a novel Syt binding site, distinct from the established Syt-II binding site in BoNT/B. Structure-based mutagenesis further show that hydrophobic interactions play a key role in Syt binding. The structures suggest that the BoNT/DC ganglioside binding sites are independent of the protein receptor binding site. Our results reveal the remarkable versatility in the receptor recognition of the BoNTs.
Introduction
Botulinum neurotoxins (BoNTs) are a family of bacterial toxins. They target neurons and block synaptic vesicle exocytosis at nerve terminals, thereby causing paralysis and death in humans and animals (Montal, 2010; Schiavo et al., 2000; Swaminathan, 2011). These toxins are the most potent toxins known and are classified as one of the six most dangerous potential bioterrorism agents (Arnon et al., 2001; Gill, 1982). Due to their ability to block overactive neurons and relax muscles, BoNTs are also widely utilized to treat many human diseases, as well as for cosmetic purposes (Dolly et al., 2009; Johnson, 1999).
Seven distinct BoNT serotypes exist (denominated as BoNT/A-G (Montal, 2010; Schiavo et al., 2000; Swaminathan, 2011)). These toxins share the same overall structure and mode of actions, but differ significantly on both protein sequences and receptors/substrates. BoNTs are composed of a proteolytically active light chain (~ 50 kDa) and a heavy chain (~ 100 kDa), connected via a disulfide bond. The heavy chain contains two functional domains, a membrane translocation domain at the N-terminal half (HN, ~ 50 kDa) and a receptor binding domain at the C-terminal half (HC, ~ 50 kDa). The HC is composed of two sub-domains, the N-terminal HCN domain, and the C-terminal HCC domain. HCN is fairly conserved across BoNTs, and may facilitate the attachment of toxins to plasma membranes by binding to phosphatidylinositol phosphates (Muraro et al., 2009; Zhang and Varnum, 2012). HCC is one of the most diverse parts among BoNTs and contains the receptor binding sites. BoNTs target and enter neurons via receptor-mediated endocytosis. Once inside neurons, the HN domain facilitates the translocation and release of the toxin light chain into the cytosol, where they cleave neuronal proteins required for synaptic vesicle exocytosis.
The guiding model for BoNT receptor recognition has been the “double-receptor” model proposed by Montecucco in 1986, which states that BoNTs recognize neurons by binding to both gangliosides as well as specific protein receptors (Montecucco, 1986). Gangliosides are complex glycolipids that have been demonstrated to be an essential co-receptor for all seven BoNTs. A conserved ganglioside binding site (GBS) with a SXWY motif has been identified in BoNT/A, B, E, F, G, as well as the related tetanus neurotoxin (Fotinou et al., 2001; Fu et al., 2009; Rummel et al., 2009; 2004b; Schmitt et al., 2010; Stenmark et al., 2010; 2008; Swaminathan and Eswaramoorthy, 2000). Crystal structures of BoNT/A and BoNT/F with bound complex gangliosides (GT1b and GD1a, respectively), as well as of BoNT/B bound to sialyllactose, have provided further structural evidence to establish this conserved GBS at the C-terminus of HCC domain (Benson et al., 2011; Stenmark et al., 2008; Swaminathan and Eswaramoorthy, 2000).
Ganglioside binding in BoNT/C and D is more complicated. They lack the conserved GBS, but analogous sites at the same area of the proteins have been shown to bind gangliosides (Karalewitz et al., 2012; Strotmeier et al., 2011; 2010; Zhang et al., 2010), suggesting a distinct binding model. Furthermore, both BoNT/C and D contain a solvent exposed hydrophobic loop, in contrast to other serotypes. Mutations in this loop clearly disrupt binding of gangliosides, suggesting that it is involved in binding gangliosides (Karalewitz et al., 2010; Kroken et al., 2011b; Strotmeier et al., 2010; 2011). Therefore, this loop has been termed as the ganglioside-binding loop (GBL, also known as WY-loop), although how the GBL mediates ganglioside binding still remains unknown. In addition, a recent structure of BoNT/C has also revealed a binding site for sialic acid, termed as Sia-1 site, which is located at a different region from GBS in HCC (Strotmeier et al., 2011). Mutations within the Sia-1 site reduce ganglioside binding, demonstrating that it is a potential ganglioside binding site. It is possible that the sialic acid binding pocket and the GBL are different parts of the same ganglioside interaction area.
Recent studies have also identified that BoNT/B and G utilize synaptic vesicle protein synaptotagmin (Syt, including two isoforms Syt-I and Syt-II) as their protein receptors (Chai et al., 2006; Dong et al., 2003; Jin et al., 2006; Nishiki et al., 1994; 1993; 1996; Rummel et al., 2004a), while BoNT/A, D and E use another vesicle protein, SV2, as receptor (Dong et al., 2008; 2006; Mahrhold et al., 2006; Peng et al., 2011). In addition, BoNT/F may also use SV2 as its receptor (Fu et al., 2009; Rummel et al., 2009), although it has not been confirmed at the functional level (Peng et al., 2011; Yeh et al., 2010). Among these toxins, the crystal structure of BoNT/B bound to its receptor Syt-II has been solved (Chai et al., 2006; Jin et al., 2006). BoNT/G shares the highest sequence identity within the HCC domain with BoNT/B among the seven BoNTs, and this Syt-II binding site is also conserved in BoNT/G (Schmitt et al., 2010; Stenmark et al., 2010). How BoNT/A, D, E, and F bind SV2 still remains to be determined at the structural level. It has also been suggested that BoNT/C may not need a protein receptor (Tsukamoto et al., 2005). Instead, it may bind neurons via multiple gangliosides (Karalewitz et al., 2010; 2012; Strotmeier et al., 2011).
In addition to the seven major serotypes, recent studies have revealed a growing number of subtypes and mosaic toxins with significant sequence differences from their parental serotypes (Hill et al., 2007; Jacobson et al., 2008; Moriishi et al., 1996; Smith et al., 2005; Zhang et al., 2011). In particular, we focused on a mosaic toxin known as BoNT/DC (also known as BoNT/D South Africa) (Moriishi et al., 1996). Its light chain and HN shares high sequence identity with BoNT/D, but its HC is similar to BoNT/C-HC. However, we previously reported that BoNT/DC requires Syt-I/II as its protein receptor and identified the approximate receptor interaction area (Peng et al., 2012). This is the first case of a mosaic toxin that uses a different receptor than its parental toxin. Furthermore, the established Syt-II binding site found in BoNT/B and G is not conserved in the apo structure of BoNT/DC-HC, raising the question how BoNT/DC diverges from BoNT/C and achieves the recognition of Syt-II. To understand the molecular mechanism underlying these divergence/convergence changes on receptor recognition in the BoNT family, we solved the crystal structure of BoNT/DC-HC in complex with the Syt-I and Syt-II recognition domains to 2.65 Å and 2.6 Å, respectively. The structures reveal that BoNT/DC possesses a novel protein receptor binding site, distinct from the established Syt-II binding site in BoNT/B. Mutational analysis verifies and highlights the importance of hydrophobic interactions in the receptor binding. The structures suggest that the BoNT/DC ganglioside binding sites are independent of the protein receptor binding site and that BoNT/DC could bind three receptors on the neuronal membrane.
Results
Structure determination of BoNT/DC-HC in complex with synaptotagmin
Co-crystallization studies of BoNT/DC with Syt-I and Syt-II were initiated using purified recombinant His6-tagged BoNT/DC-HC and a synthesized peptide bearing the toxin binding site of human Syt-I and rat Syt-II (residues 33–53 and 40–60, respectively). This part of the receptor has been shown to disordered in solution and to contain the toxin binding region (Chai et al., 2006; Jin et al., 2006; Peng et al., 2012). The crystals grew in space group P21 and diffracted to 2.65 Å and 2.6 Å resolution, respectively (Table 1). The structures of the complexes were determined by molecular replacement using the previously determined structure of apo-BoNT/DC-HC (PDB code: 3N7L) as a search model (Karalewitz et al., 2010). The asymmetric unit contains, in both cases, three molecules that were all similar (rmsd of ca 0.5 Å2 in between the molecules). Chain b displayed limited differences in the distal part of the protein, with differences in the loops 923–930 and 1030–1037, likely induced by the crystal packing. However, this face of the protein does not interact with any ligand, and the differences can thus be neglected. The electron density accounted for most of the BoNT/DC-HC sequence, with all residues between 863 – 1263 visible, with the exception of a flexible loop between residues 1052 – 1059 (Fig. 1a). For both structures well-defined electron density, not part of BoNT/DC-HC protein, appeared directly after molecular replacement, and became even clearer after refinement. This density was unambiguously assigned to the Syt-I and Syt-II recognition domains, respectively (Fig. 1b–c). With the exception of 3 residues on both ends for Syt-I, and 2 and 3 on the N- and C-terminal ends for Syt-II, the entire peptide was visible in the electron density. The bound regions correspond to residues 36–50 in Syt-I and 42–57 in Syt-II.
Table 1.
Data collection and refinement statistics
| Data collection | BoNT/DC•Syt-I | BoNT/DC•Syt-II |
|---|---|---|
| Space group | P21 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 164.2, 57.6, 165.3 | 165.2, 57.8, 169.5 |
| α, β, γ (°) | 90, 118.1, 90 | 90.0, 118.5, 90.0 |
| Resolution (Å) | 48.5 – 2.65 (2.79-2.65) | 48.3 – 2.6 (2.69-2.6) |
| Rsym (%) | 12.5 (65.8) | 11.3 (63.1) |
| I/σ (I) | 9.3 (2.3) | 12.2 (2.3) |
| Completeness (%) | 100 (100) | 99.5 (98.5) |
| Redundancy | 3.6 (3.6) | 3.9 (3.9) |
| Refinement | ||
| Resolution | 48.6 – 2.65 | 47.7 – 2.6 |
| No. unique reflections | 76148 (12157) | 88524 (14062) |
| Rwork/Rfree | 20.5/22.2 | 22.0/23.5 |
| No. atoms | ||
| Protein | 10175 | 10146 |
| Ligand | 384 | 414 |
| Water | 260 | 430 |
| B-factors | ||
| Protein | 46.4 | 40.9 |
| Ligand | 59.8 | 49.2 |
| Water | 32 | 31.8 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.004 | 0.005 |
| Bond angles (°) | 0.79 | 0.89 |
| Ramachandran plot, residues in (%) | ||
| most favorable region | 95.7 | 94.8 |
| additional allowed region | 4.2 | 5.2 |
| outliers | 0.1 | 0 |
Values in parenthesis are for the highest-resolution shell.
Figure 1. Structure of the BoNT/DC-HC•Syt-II complex.
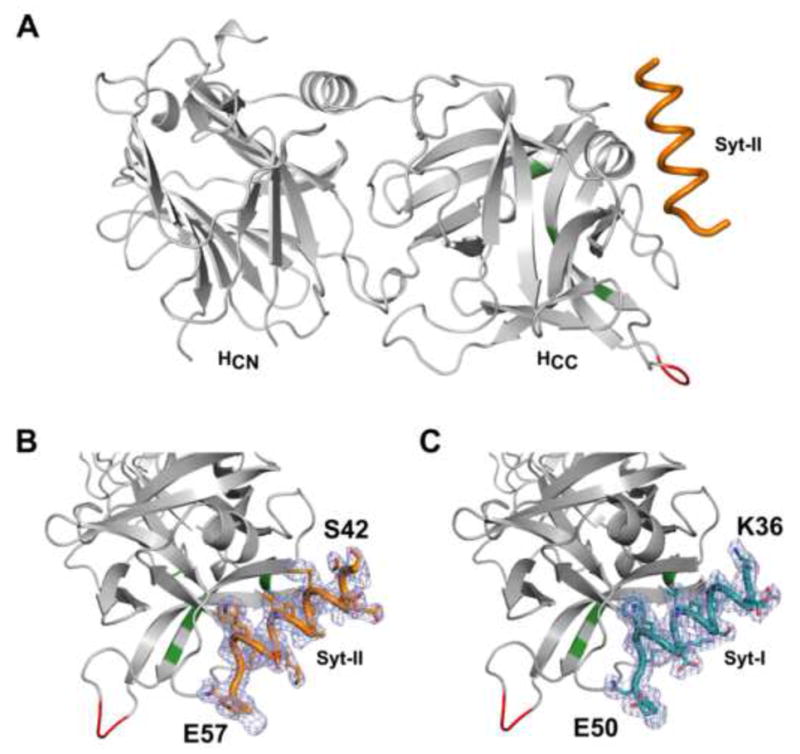
(a) Cartoon representation of the overall structure of BoNT/DC-HC bound with the Syt-II peptide containing the toxin binding site. The bound Syt-II peptide (orange) forms an α-helix and docks to the C-terminal region of BoNT/DC-HCC. The residues involved in the GBS and GBL are shown in green and red, respectively. (b–c) Enlargement of the Syt-II (b) and of the Syt-I (c) binding sites. Electron density for Syt is shown in light blue mesh (SigmaA-weighted 2FO – FC map, contoured at 1 σ), with the modeled Syt-II and Syt-I (orange and teal, respectively). The start and end of each peptide is marked in the figure. The remaining residues of Syt-II and Syt-I are disordered, and are not visible in the structure.
The overall structure of BoNT/DC-HC bound to Syt-I, or Syt-II, is similar to the previously reported apo-structure of BoNT/DC-HC (Karalewitz et al., 2010), with an rmsd of 0.8 Å. Binding of Syt does not induce any significant structural changes in BoNT/DC, instead the binding pocket is preset for binding of both Syt-I and Syt-II. The luminal domain of Syt-II, which contains the toxin binding site, has previously been shown to be disordered in solution (Jin et al., 2006). Upon binding to BoNT/DC, residues 37–48 in Syt-I, and 43–54 in Syt-II, form an amphipathic helix, with the rest of the ordered residues in an extended conformation (Fig. 1b–c). The Syt-I and Syt-II molecules are positioned virtually identical within the binding site (Fig 2a–b). Syt-I and Syt-II are both bound in a defined register, verifying that they have one defined binding mode. The BoNT/DC•Syt-I complex reported here represents the first toxin•Syt-I structure, confirming at the structural level that Syt-I and -II are recognized by the same receptor binding pockets in a highly similar way. This is very likely the case also for Syt-I and Syt-II binding to BoNT/B and BoNT/G.
Figure 2. Interactions between BoNT/DC and Syt-II.
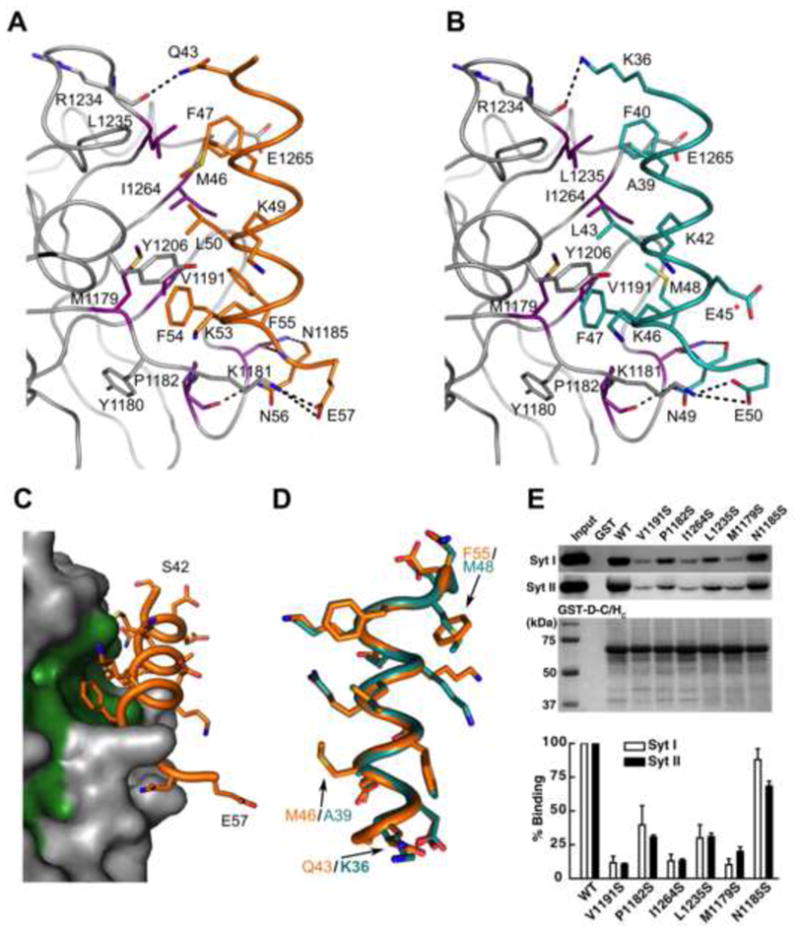
(a) A close-up view of the binding interface between BoNT/DC (gray) and Syt-II (orange). All directly interacting residues are shown as sticks. Violet residues at the Syt binding site were selected for mutagenesis studies as described in the following panels. (b) Close-up view of the binding interface between BoNT/DC and Syt-I (teal), with the same coloring as in a. E45, marked with a red star, is the only residue that is different between human and rat Syt-I. E45 is, as seen here, not involved in the binding to the toxin. (c) Surface representation of the Syt-II binding site of BoNT/DC (gray), with bound Syt II (orange) and the hydrophobic residues of BoNT/DC that form the Syt-II binding site shown in green. (d) Comparison of Syt-I and Syt-II bound to BoNT/DC, as seen when superimposing only the BoNT/DC protein chains upon each other. The hydrophobic face of the amphipathic Syt helices, as seen from the BoNT/DC binding site, with the three differences in the binding region between Syt-I and Syt-II highlighted. The other differences between the two isoforms are located in the hydrophilic face of the helix that facing away from the binding site, and are not involved in the binding to BoNT/DC (e) Upper panel: Mutated BoNT/DC-HC were used to pull down native Syt-I/II from rat brain detergent (Triton X-100) extracts. Wild type (WT) BoNT/DC-HC and GST protein were assayed in parallel as controls. Bound Syt-I/II were detected via immunoblot using their specific antibodies. GST-tagged BoNT/DC-HCs used in pull-down assays were visualized by Coomassie blue staining to indicate equal loading. Lower panel: binding of Syt-I/II to BoNT/DC-HC mutants was quantified and normalized to the amount of binding to WT BoNT/DC-HC. See also Figure S1 for a sequence alignment of Syt isoforms. See also Figure S1 for a) CD spectra illustrating that the mutations in BoNT/DC-HC does not induce any major structural changes and b) for a schematic overview of the two binding sites.
BoNT/DC interactions with Syt
The hydrophobic side of the Syt amphipathic helix docks into a hydrophobic patch at the C-terminal region of BoNT/DC-HCC (Fig. 2a–c). The interface between BoNT/DC and Syt buries a solvent-accessible surface area of 652 Å2 and 668 Å2 for Syt-I and Syt-II, respectively, slightly larger than the 608 Å2 that gets buried on BoNT/B upon Syt-II binding (Chai et al., 2006). The contacts are mainly constituted by hydrophobic and van der Waals interactions, with residues M1179, V1191, L1196, L1226, L1235 and I1264 forming the hydrophobic patch where Syt binds (Fig. 2a–c). Y1180 and K1181, despite being overall hydrophilic, also provide hydrophobic contacts, both to F54 on Syt-II (F47 in Syt-I). A total of four hydrogen bonds are formed to Syt, with three hydrogen bonds formed to N56 on Syt-II (N49 in Syt-I), two coming from N1185 and one from the backbone of P1182. The last hydrogen bond is formed between the backbone of R1234 and Q43 on Syt-II (K36 in Syt-I). Finally one salt bridge is also formed between E57 of Syt-II (E50 in Syt-I) and K1181 of BoNT/DC. There are only three differences between human Syt-I and rat Syt-II that are involved in the toxin binding, namely Q43, M46 and F55 in Syt-II being exchanged to lysine, alanine and methionine in Syt-I, respectively (Fig. 2d).
To further characterize the binding, we mutated six residues located along the binding interface at the surface of BoNT/DC, (M1179, P1182, N1185, V1191, L1235, I1264) (Fig. 2a–b), replacing them with serine to abolish the hydrophobic interactions that cover the entire binding site (Fig. 2c). No major structural changes were induced by the mutations, as verified by circular dichroism (CD) experiments (Suppl. Fig. S1a). These mutants were purified as glutathione-S-transferase (GST) tagged recombinant proteins, immobilized on beads, and were used to pull down native Syt-I and -II from rat brain detergent extracts. As shown in Fig. 2e, mutating residues in the center of the binding interface, M1179, V1191, and I1264, abolished binding of both Syt-I and –II. Mutating a residue at the periphery of the binding interface (L1235) decreased the binding significantly (~3-fold). A similar decrease is seen in the P1182S mutant, which does not remove the H-bond. In this case it is likely due to changes in the loop region, also containing N1185, which causes the decrease in binding. Mutating N1185 to a serine, thereby removing two hydrogen bonds to Syt, hardly disturbed the binding, as it retained 88% and 68% of the binding to Syt-I and Syt-II, respectively. The results for the site-directed mutagenesis are similar between the binding of both Syt-I and Syt-II for all mutants, consistent with the high similarity between the Syt-I and Syt-II complexes reported here. These results are consistent with our structural data and further show that the hydrophobic interactions are an important force in Syt binding to BoNT/DC. Human and rat Syt-I differs only in one residue (E45Q) that is facing away from the toxin, and can thus be considered virtually identical with respect to toxin binding (Fig. 2b).
We previously demonstrated that human Syt-II did not have detectable binding to BoNT/DC in pull-down assays (Peng et al., 2012). This is due to a single residue change at the position F54 of Syt-II (rat sequence number), the F residue is highly conserved across a majority of vertebrates, but changes to L in humans (Suppl Fig. S2). The crystal structure of the BoNT/DC•Syt-II complexes now provided a structural explanation: F54 is located at the core of the binding interface and forms multiple hydrophobic interactions with residues in BoNT/DC (Fig. 2a, Suppl Fig. S1b). The F54L mutation leads to the loss of a π-stacking interaction with Y1206, as well as a (minor) hydrophobic mismatch when the smaller leucine residue cannot cover the entire hydrophobic patch previously covered by F54. Therefore, the high affinity receptor for BoNT/DC in humans is restricted to Syt-I. This makes the binding domain of BoNT/DC a potential tool to specifically target human neurons expressing Syt-I. Furthermore, the protein receptor and ganglioside co-receptor binding sites could be altered to modify the affinity and specificity of the BoNT/DC binding domain.
BoNT/DC has a novel Syt binding site in comparison to BoNT/B
We next compared the structure of BoNT/DC•Syt-II versus BoNT/B•Syt-II (Chai et al., 2006; Jin et al., 2006). As shown in Fig. 3a, the overall structure of BoNT/B-HC and BoNT/DC-HC are fairly similar, with an r.m.s.d. of 2.3 Å. However, there are large differences in surface regions of the proteins. Syt-II does not bind at the same site as in BoNT/B, but instead binds in a novel binding pocket in BoNT/DC located on another face of the HCC domain. Furthermore the binding is shifted ~90° to the Syt-II binding site in BoNT/B (Fig. 3a). Furthermore, the region of Syt-II bound to each toxin is shifted. BoNT/DC-binding involves residues 43–57, while BoNT/B-binding involves residues 44–60 of Syt-II (Fig. 3a, enlarged panel). It is interesting to note that the C-terminal end of Syt-II (E57 of Syt-II in BoNT/DC and K60 of Syt-II in BoNT/B) ends up at a similar position in both toxins, despite the perpendicular orientation of the two Syt-II binding sites (Fig. 3a, right panel). This would put both toxins at similar position to the membrane, as the C-terminal of Syt-II recognition sequence directly precedes its transmembrane domain. In addition, BoNT/DC might have slightly higher flexibility than BoNT/B, as there are three extra residues between the bound region of Syt-II and its anchoring transmembrane helix in BoNT/DC.
Figure 3. BoNT/DC utilizes a novel Syt-II binding site in comparison to BoNT/B.
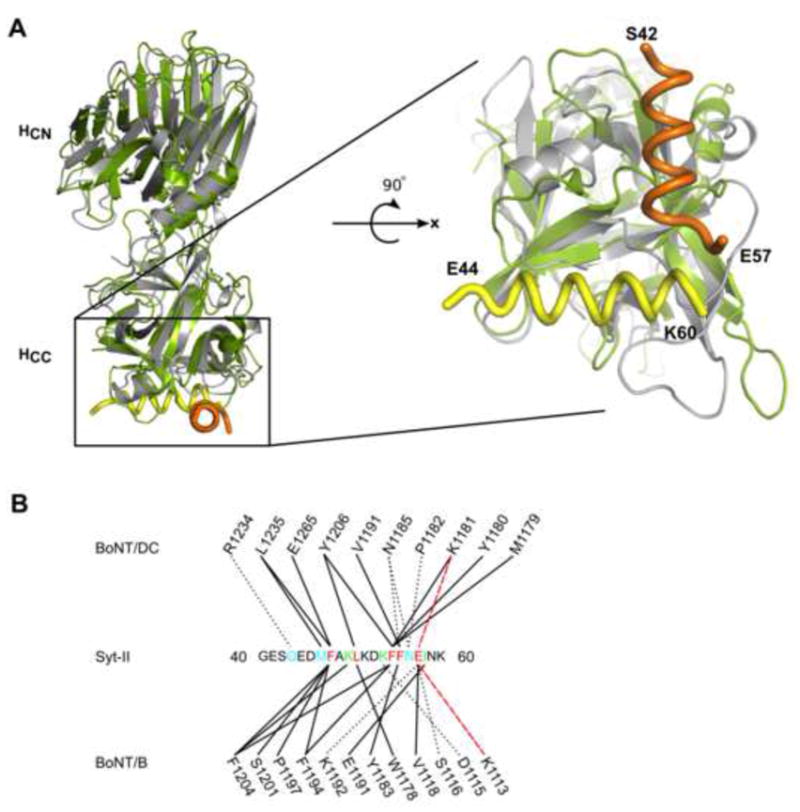
(a) The structure comparison between BoNT/DC(gray)•Syt-II (orange) complex and BoNT/B (green)•Syt-II (yellow). The right panel is an enlarged view of the Syt-II binding sites in BoNT/DC (gray) and BoNT/B (green), with Syt-II peptide bound to BoNT/DC labeled in orange and Syt-II peptide bound to BoNT/B labeled in yellow. The N- and C-terminal residues of the toxin binding site in Syt-II are marked. The Syt-II binding sites in BoNT/DC and BoNT/B are rotated approximately 90° from each other. The C-terminal ends of Syt-II (E57 in BoNT/DC•Syt-II and K60 in BoNT/B•Syt-II) are in close vicinity of each other. (b) Schematic overview of the interactions between BoNT/DC and BoNT/B to Syt-II. Solid lines represent hydrophobic contacts, dotted black lines represent hydrogen bonds and red dashed lines represent salt bridges. Syt-II residues are color coded: cyan indicates binding to BoNT/DC only, green indicates binding to BoNT/B only, and red indicates residues contribute to binding of both BoNT/DC and B. See also Figure S2 for a sequence comparison between Syt isoforms from different species.
The structural comparison also revealed that both BoNT/DC and BoNT/B recognize Syt-II in a similar manner (Fig. 3b, Suppl. Fig. S1b,c). The hydrophobic interactions play the key role, with the core residues F47, L50, F54, and F55 in Syt-II forming the hydrophobic contacts to both toxins (Fig. 3b, black line). In addition, the residue E57 also forms salt-bridge with different lysine residues in both toxins (Fig. 3b, red dashed line). The interactions to Syt-II are further strengthened by similar numbers of hydrogen bonds, four in BoNT/DC and three in BoNT/B (Fig. 3b, black dashed line). On the other hand, the toxin residues forming the binding site are not conserved, and located at a different area of the protein (Fig. 3). For instance, F47 from Syt-II stacks with Y1181 and F1194 in BoNT/B, whereas the same residue does not have any π interactions in BoNT/DC, only hydrophobic interactions with L1235 and E1265.
Comparison of BoNT/DC to BoNT/C
Among the seven BoNTs, the receptor binding domain of BoNT/DC has the highest sequence identity with BoNT/C, 92% in the HCN and 61% in the HCC domains (Peng et al., 2012). However, BoNT/C does not use Syt-I/II as receptors. To understand the molecular basis for this divergence, we compared the structure of BoNT/DC Syt-II with BoNT/C-HC in complex with sialic acids (PDB code: 3R4S). Overall the structures of the two toxins are highly similar, with an r.m.s. deviation of mere 0.9 Å, but we found that BoNT/C lacks a hydrophobic patch in the BoNT/DC Syt binding region, leading to an incompatible Syt binding site (Fig. 4a, Suppl Fig. S3).
Figure 4. BoNT/DC comparison with BoNT/C.
(a) Surface representation of BoNT/DC (left panel) and BoNT/C (right panel), with hydrophilic surface in blue and hydrophobic in green. The Syt-II recognition sequence bound to BoNT/DC is shown in both figures. It is clear that Syt-II cannot bind to BoNT/C, due to a lack of the important hydrophobic patch found in BoNT/DC, as well as steric clashes. (b) Superimposing BoNT/C-HC (cyan)•Sialic acid complex with BoNT/DC-HC(gray)•Syt-II(orange) complex. The ganglioside GT1b (green) was also modeled onto established ganglioside binding site (GBS) based on the previously determined BoNT/A-GT1b structure (PDB code: 2VU9). The biologically relevant sialic acid moiety, co-crystallized together with BoNT/C, is shown in blue. The ganglioside binding loop (GBL) is shown in red. (c) Close up of the Sia-1 sites in BoNT/C (cyan) and BoNT/DC (gray). Conserved residues in BoNT/DC are marked in the figure, and possible hydrogen bonds between the sialic acid and BoNT/DC are shown as dashed lines. The Sia-1 site is highly conserved between BoNT/C and BoNT/DC. See also Figure S3 for a structure based sequence alignment of BoNT/DC-HC and BoNT/C-HC.
As mentioned earlier, no protein receptor has been identified for BoNT/C, rather it has been proposed that BoNT/C has at least two different ganglioside binding sites (Karalewitz et al., 2012; Strotmeier et al., 2011), involving three regions: (1) the GBS found in other BoNTs, (2) the GBL, and (3) a novel sialic acid binding pocket (Sia-1, Fig. 4b). The GBL and the sialic acid binding pocket could be part of one ganglioside interaction area, as complex gangliosides can reach both simultaneously (Fig. 5). The ability to bind multiple gangliosides may allow BoNT/C to bind neurons without protein receptors. As BoNT/DC shares high sequence identity to BoNT/C in their receptor binding domain, are all these ganglioside binding sites conserved in BoNT/DC?
Figure 5. A model of BoNT/DC binding to its neuronal receptors.
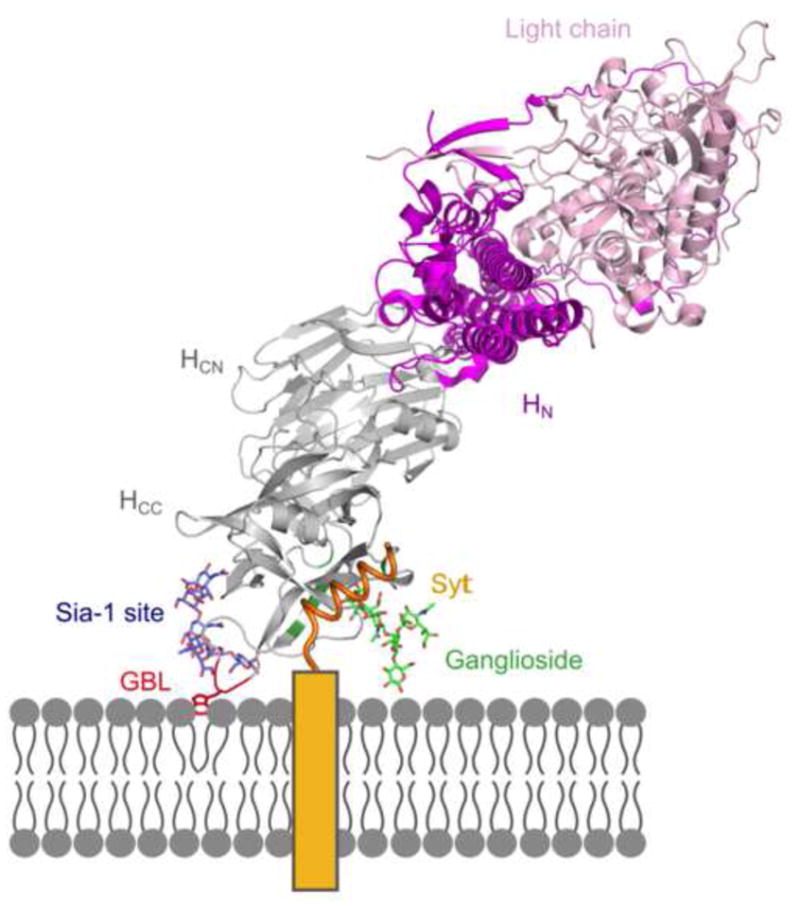
BoNT/DC-HC (gray) and the Syt-II peptide (orange) come from data presented here. The ganglioside binding loop (GBL) is shown in red. The rest of the BoNT structure, HN (purple) and Light chain (pink), is modeled onto the DC structure using the previously determined BoNT/B structure (PDB code: 2NP0). The ganglioside (green) is modeled based on the BoNT/A – GT1b structure (PDB code: 2VU9), and the potential second ganglioside (blue) is superimposed to the sialic acid moiety in the Sia-1 site of BoNT/C (PDB code: 3R4S). The transmembrane section of Syt-II is shown as a yellow box.
Previous biochemical characterization and crystal structure of BoNT/DC have demonstrated that the GBS is conserved, and located in a similar position as in BoNT/A (Nuemket et al., 2011). The GBL is also conserved between BoNT/C and DC (Karalewitz et al., 2010; Nuemket et al., 2011). Mutations in both the GBS and GBL of BoNT/DC abolished its binding to purified gangliosides and to cell surfaces (Nuemket et al., 2011), demonstrating their role for binding gangliosides.
We compared the Sia-1 site of BoNT/DC with BoNT/C (Strotmeier et al., 2011), this shows that the Sia-1 site from BoNT/C is largely intact in BoNT/DC, with most of the important residues and contacts conserved (Fig. 4c, Suppl Fig. S3). Most hydrogen bonds are conserved in this region, with residues Y1142, I1170, D1171 and Y1175 (numbering from BoNT/DC) forming the conserved core (Fig. 4c). Furthermore, the backbone of K1123 and G1124 are in a similar position as their corresponding residues in BoNT/C, and can form hydrogen bonds to the sialic acid. D1171 is conserved in BoNT/DC, and although it cannot form a hydrogen bond directly to the Sia-1 molecule, a bridging water molecule could form the bridge, similar to what has been observed in the BoNT/C•Sialic acid complex structure (Strotmeier et al., 2011). For BoNT/DC to form the Sia-1 site, the side-chain of K1122 would have to change its conformation. That, together with a small change in the backbone of Gly1124 and Asn1125, situated in a flexible loop, would form a virtually identical sialic acid binding site as in BoNT/C.
Discussion
It is puzzling how BoNT/DC, which has a rather low sequence identity to BoNT/B and does not have the well-established Syt binding site found in BoNT/B, could recognize the same toxin binding site on Syt-II as BoNT/B. Here we solved the co-crystal structures of BoNT/DC-HC bound to either the Syt-I or Syt-II peptide containing the toxin binding domain. The structures revealed that BoNT/DC possesses a novel Syt binding site located adjacent, but rotated approximately 90°, to the Syt-II binding site in BoNT/B. Structure based mutagenesis studies further confirmed the location of this new Syt binding site. Although the Syt binding sites in BoNT/DC and BoNT/B are not conserved and located at different positions, they both have a large hydrophobic patch that allow binding of the hydrophobic side of the Syt amphipathic helix. The interactions to Syt-II, in both proteins, are mainly mediated by hydrophobic forces, with additional 3–4 hydrogen bonds and one salt bridge. The key residues of Syt-II involved in the binding, such as F47, F54, and F55, are utilized by both BoNT/DC and BoNT/B. This provides a structural explanation for the previous finding that mutations at these residues in Syt-II reduce binding of both BoNT/DC and BoNT/B to Syt-II (Peng et al., 2012).
The fact that the BoNT toxin family have developed two non-conserved binding domains to recognize the same stretch of Syt suggests an intriguing possibility of convergent evolution, where BoNT/DC and BoNT/B have taken two different routes to end up binding very similar regions of Syt. The short stretch of the toxin binding site (< 20 residues) in both Syt-I and Syt-II certainly possesses attractive properties for mediating protein-protein interactions. This region is unstructured in solution, but can form an amphipathic helix and bind to hydrophobic binding sites of toxins (Chai et al., 2006; Jin et al., 2006; Peng et al., 2012). In both isoforms, it is located adjacent to the transmembrane domain of Syt, therefore providing a way to anchor toxins within the proximity of the membrane. In fact, the C-terminus of the toxin binding site in Syt-II, which precedes the transmembrane domain, is at approximately the same position in both the BoNT/DC and BoNT/B complexes. This is very likely the case also for Syt-I, however, no structure is available of BoNT/B bound to Syt-I. Both BoNT/DC and BoNT/B are anchored at similar distance to membranes. This convergence may reflect a spatial constraint in order to maintain simultaneous ganglioside binding and/or to facilitate the later translocation step.
The toxin binding site in Syt-II is also highly conserved across vertebrates (Craxton, 2010), although whether it has any physiological function remains to be determined. Interestingly, seemingly minor residue changes within this region in different species have significant impact on binding of BoNTs. For instance, a single residue change of F54 (in rat/mouse sequence) to a leucine (as in human Syt-II), dramatically reduces binding of BoNT/B, G, and DC (Peng et al., 2012; Strotmeier et al., 2012).
The receptor binding domain of BoNT/DC has the highest sequence identity to BoNT/C among the seven BoNTs, yet the Syt binding site in BoNT/DC is not conserved in BoNT/C. Indeed, it has been proposed that BoNT/C does not use a protein receptor, but utilizes at least two gangliosides, with the GBS, GBL as well as a Sia-1 site involved in ganglioside binding (Karalewitz et al., 2012; Strotmeier et al., 2011). Structural comparison between the BoNT/DC•Syt complexes and the BoNT/C•Sialic acid complex revealed that the Sia-1 site is mostly conserved in BoNT/DC (Strotmeier et al., 2011). Because both the GBS and the GBL are also conserved in BoNT/DC and their involvement for ganglioside binding has been further confirmed via mutagenesis studies (Nuemket et al., 2011), BoNT/DC could have two ganglioside binding sites, like BoNT/C (Karalewitz et al., 2012; Strotmeier et al., 2011). Because the newly identified Syt binding site is independent of both the GBS and the GBL/Sia-1 site, BoNT/DC has the potential to bind at three anchoring points on the neuronal membrane. (Fig. 4b, Fig. 5). These three sites give us sterical restrictions, allowing us to model the entire complex on the membrane via a triple binding site model (Fig. 5). It is currently not clear whether the GBL binds gangliosides independently, or whether it acts in a dependent manner with the Sia-1 site or GBS (Nuemket et al., 2011; Strotmeier et al., 2011). The GBL is however close in space to where a potential complex ganglioside would be if bound to the Sia-1 site (Fig. 5). With the Sia-1 binding the sialic acid moiety of a ganglioside, as previously suggested for BoNT/C (Karalewitz et al., 2012; Strotmeier et al., 2011), a second GBS could then be formed consisting of the Sia-1 site and the GBL together, a GBL/Sia-1 site. The tryptophan of the GBL would be in a suitable location to interact and stack with a sugar moiety further down an oligosaccharide bound at the Sia-1 site (Fig. 5). Furthermore, the exposed hydrophobic residues of the GBL are well positioned for a direct interaction with the membrane, as seen in Fig. 5. The exposed nature of the GBL is interesting also from a pharmacological perspective. As previously noted, it could be a potential site for targeting serotype specific antibodies (Kroken et al., 2011a). One biological reason for two ganglioside binding sites could be the possibility to compensate for the lower affinity BoNT/DC has towards Syt-I/II compared to BoNT/B (Peng et al., 2012). By having three independent binding sites BoNT/DC increases its binding affinity to the neuronal membrane. The receptor binding sites are well aligned for simultaneous interaction with the membrane and could be suitable targets for anti-toxins, perhaps utilizing the possibility to block several sites simultaneously. Although the mechanism of ganglioside binding of BoNT/DC requires further experimental validation, the structure presented here suggests that BoNT/DC binds to three sites on the membrane, an extension of the original “double-receptor” model (Montecucco, 1986).
In conclusion
The structures presented here reveals that BoNT/DC recognizes Syt-I and Syt-II via a novel Syt binding region. The Syt binding sites in BoNT/B and BoNT/DC are formed by distinct residues. The non-conserved nature of these two Syt binding sites and their distinct locations suggest that they likely evolved independently to recognize the same recognition domain of Syt. The binding of Syt-I and Syt-II to BoNT/DC is highly similar and does not induce any structural changes in the toxin. Mutational studies highlight the key hydrophobic interactions in Syt binding. The structures reveal that the ganglioside binding sites in BoNT/DC are independent of the Syt binding site. Therefore, the structure reported here suggests that BoNT/DC may bind both a protein receptor and two separate gangliosides.
Experimental Procedures
Constructs, peptides, and antibodies
The cDNAs encoding BoNT/DC-HC (residues 859 – 1285, GenBank: AB461915.1) was synthesized by GenScript Inc. (New Brunswick, NJ) with codon optimized for E.coli expression, and was sub-cloned into pET28a vector using Nhe I/Xho I sites for expression as recombinant proteins with a His6 tag fused to the N-terminus. BoNT/DC-HC was also subcloned into pGEX-4T vector for expression as GST-tagged recombinant proteins, which was used for pull-down assays. Mutagenesis of BoNT/DC-HC was carried out using Quickchange site-directed mutagenesis kit following manufacturer’s instructions (Agilent Tech, Santa Clara, CA). Peptides corresponding to the toxin binding site in rat Syt-II (residue 40–60) and human Syt-I (residue 33–53) were synthesized (>95% purity) by JPT Peptide Technologies Gmgh (Germany). Mouse monoclonal antibodies for Syt-I (Cl41.1) was generously provided by E. Chapman (Madison, WI). Mouse monoclonal anti-Syt II was purchased from BD transduction (San Jose, CA).
Protein expression and purification
His6 tagged BoNT/DC-HC was expressed in E. coli (BL21 strain). The cells were grown in EPCM1 medium containing 50 μg/mL kanamycin, and grown in a 7L fermentor (Applikon Biotechnology, Foster City CA) at 37°C, pH 7.0 with mixing and addition of air and oxygen to maintain an excess of oxygen. The temperature was lowered to 20°C at an OD600 of 5–7. Upon reaching the set temperature the expression was induced upon the addition of IPTG with a final concentration of 0.8 mM. GST-tagged BoNT/DC-HC was also expressed in E. coli (BL21), and grown in LB medium containing 50 μg/mL carbenicillin. The cells were grown at 37 °C until an OD600 of ca 0.5, whereupon the temperature was lowered to 18°C and expression induced with 0.25 mM IPTG. After over-night induction the cells were harvested, and the pelleted cells frozen in −80 °C. For His6 tagged BoNT/DC-HC purification, pellets were thawed, and resuspended in 20 mM phosphate buffer, pH 7.4, 500 mM NaCl and 20 mM imidazole (lysis buffer), and sonicated at 95% amplitude on ice for 3 rounds of 3×10 s with 5 s rest between each 10 s sonication. Cell debris was removed by centrifugation at 23000g for 20 min at 4°C. The soluble fraction was loaded to a HisTrap HP Ni column (GE Healthcare). The column was subsequently washed for 5 column volumes (CV) with lysis buffer. Contaminating proteins were washed away with 5 CV of lysis buffer containing 90 mM imidazole. Elution was performed with lysis buffer containing 250 mM imidazole. The purified protein was subsequently dialyzed against 20 mM Hepes, pH 7.5, 150 mM NaCl, 10% (v/v) glycerol and 1 mM DTT, and subsequently flash frozen in liquid nitrogen and stored at −80 °C. For binding and circular dichroism analysis, WT and mutant BoNT/DC-HC were purified as GST-tagged recombinant proteins. The samples were analyzed by SDS-PAGE.
Brain detergent extracts, GST pull down assay, and immunoblot analysis
Rat brain detergent extracts were prepared as previously described (Peng et al., 2011). 10 μg of GST-BoNT/DC-HC or GST were immobilized on glutathione-Sepharose beads (GE bioscience, Piscataway, NJ) and were subsequently incubated with 0.5 ml rat brain detergent extracts for 1 hr at 4 °C. Beads were washed three times using Tris buffer (20 mM Tris, 150 mM NaCl, pH 7.4) plus 0.5% Triton X-100. Ten percent of bound materials were subjected to SDS-PAGE, followed by either immunoblot analysis using the enhanced chemiluminescence (ECL) method (Pierce, Rockford, IL) to detect Syt-I/II, or Coomassie blue staining to visualize GST-BoNT/DC-HCs.
Circular dichroism experiments
CD measurements were done on a Chirascan circular dichroism spectrometer (Applied Photophysics). 400 μL of protein sample at a protein concentration of 0.04 mg/mL in 50 mM KPi, pH 7.5, was used in cuvette with 2 mm path length. The protein concentration was based on the calculated extinction coefficient for the GST-tagged BoNT/DC-HC (127340 M−1cm−1), on samples that were concentrated to ca 1 mg/mL and subsequently diluted to 0.04 mg/mL for the CD measurement. Samples were measured between 194 and 260 nm at 20 °C. Measurements were done in quadruplets and subsequently averaged.
Crystallization and structure determination
BoNT/DC-HC was thawed, human Syt-I or rat Syt-II peptide was added to a final concentration of 0.8 mM, and was crystallized using the vapor diffusion technique. The crystals were grown from a solution containing 1.8–2.2 M NaSO4 and 0.1 M Na-cacodylate pH 7.0, at 18 °C. Crystals appeared within 5 days and grew to full size after 2–4 weeks. They were cryoprotected with mother liquor supplemented with 18% glycerol.
Diffraction data of the BoNT/DC•Syt-II complex were collected at 0.95 Å at beamline 14.1, BESSY, Berlin, Germany, and for the BoNT/DC•Syt-I complex on the PX1 beamline at the Swiss Light Source, Paul Scherrer Institute, Villigen, Switzerland. Data processing and reduction were carried out using XDS (Kabsch, 2010) and programs from the CCP4 suite (Collaborative Computational Project 4, 1994). Relevant statistics for data processing and refinement are shown in Table 1. The phase problem was solved using molecular replacement, using an apo structure of BoNT/DC-HC (PDB code: 3N7L). A few cycles of refinement in Refmac5 (Murshudov et al., 1997) using TLS refinement (Winn et al., 2001), interspersed with manual model building using Coot (Emsley and Cowtan, 2004), were necessary to complete the full model. The final model contains residues 863–1284, with the exception of residues 1052–1059 that resides in a loop region and are presumed to be disordered. With the exception of 2–3 residues on both termini, the entire peptide was visible in the electron density, corresponding to residues 36–50 in human Syt-I, and to 42–57 in rat Syt-II. The coordinates and structure factors of the BoNT/DC complexes with human Syt-I and rat Syt-II were deposited in the PDB with the accession codes 4ISQ and 4ISR respectively. PDB validation reports can be found in supplementary materials.
Supplementary Material
Highlights.
A novel Syt binding site within the BoNT family is established in BoNT/DC.
The first crystal structure of a BoNT in complex with Syt-I.
BoNT/DC has a distinctly different binding site for Syt than BoNT/B.
The structures suggest three potential anchoring points to the neuronal membrane.
Acknowledgments
This work was supported by grants from the Swedish Research Council (2010-5200), the Wenner-Gren foundations, the Magnus Bergvalls foundation and the Swedish Foundation for Strategic research to P.S, by an EMBO Long Term Fellowship and Marie Curie Actions (EMBOCOFUND2010, GA-2010-267146) to R.P-A.B., and by the NIH Grant 8P51OD011103-51 (to the New England Primate Research Center), 1R56AI097834-01, and 1R01NS080833-01 to M.D. We thank the Biomolecule Production Core at the NERCE (New England Regional Center for Excellence, NIAID U54 AI057159) for their assistance on protein purification. We acknowledge Biostruct-X for support. We also thank the beamline scientists at BESSY, Berlin, Max-Lab, Lund and SLS, Villigen for their support in data collection.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnon SS, Schechter R, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Hauer J, Layton M, et al. Botulinum toxin as a biological weapon: medical and public health management. 2001. pp. 1059–1070. [DOI] [PubMed] [Google Scholar]
- Benson MA, Fu Z, Kim JJP, Baldwin MR. Unique ganglioside recognition strategies for clostridial neurotoxins. J Biol Chem. 2011;286:34014–34022. doi: 10.1074/jbc.M111.272054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Q, Arndt JW, Dong M, Tepp WH, Johnson EA, Chapman ER, Stevens RC. Structural basis of cell surface receptor recognition by botulinum neurotoxin B. Nature. 2006;444:1096–1100. doi: 10.1038/nature05411. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Craxton M. A manual collection of Syt, Esyt, Rph3a, Rph3al, Doc2, and Dblc2 genes from 46 metazoan genomes - an open access resource for neuroscience and evolutionary biology. BMC Genomics. 2010;11:37. doi: 10.1186/1471-2164-11-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolly JO, Lawrence GW, Meng J, Wang J, Ovsepian SV. Neuro-exocytosis: botulinum toxins as inhibitory probes and versatile therapeutics. Curr Opin Pharmacol. 2009;9:326–335. doi: 10.1016/j.coph.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Dong M, Liu H, Tepp WH, Johnson EA, Janz R, Chapman ER. Glycosylated SV2A and SV2B mediate the entry of botulinum neurotoxin E into neurons. Mol Biol Cell. 2008;19:5226–5237. doi: 10.1091/mbc.E08-07-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M, Richards DA, Goodnough MC, Tepp WH, Johnson EA, Chapman ER. Synaptotagmins I and II mediate entry of botulinum neurotoxin B into cells. J Cell Biol. 2003;162:1293–1303. doi: 10.1083/jcb.200305098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M, Yeh F, Tepp WH, Dean C, Johnson EA, Janz R, Chapman ER. SV2 is the protein receptor for botulinum neurotoxin A. Science. 2006;312:592–596. doi: 10.1126/science.1123654. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fotinou C, Emsley P, Black I, Ando H, Ishida H, Kiso M, Sinha K, Fairweather N, Isaacs N. The crystal structure of tetanus toxin Hc fragment complexed with a synthetic GT1b analogue suggests cross-linking between ganglioside receptors and the toxin. J Biol Chem. 2001;276:32274–32281. doi: 10.1074/jbc.M103285200. [DOI] [PubMed] [Google Scholar]
- Fu Z, Chen C, Barbieri JT, Kim JJP, Baldwin MR. Glycosylated SV2 and gangliosides as dual receptors for botulinum neurotoxin serotype F. Biochemistry. 2009;48:5631–5641. doi: 10.1021/bi9002138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill DM. Bacterial toxins: a table of lethal amounts. Microbiol Rev. 1982;46:86–94. doi: 10.1128/mr.46.1.86-94.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill KK, Smith TJ, Helma CH, Ticknor LO, Foley BT, Svensson RT, Brown JL, Johnson EA, Smith LA, Okinaka RT, et al. Genetic diversity among botulinum neurotoxin-producing clostridial strains. J Bacteriol. 2007;189:818–832. doi: 10.1128/JB.01180-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson MJ, Lin G, Raphael B, Andreadis J, Johnson EA. Analysis of neurotoxin cluster genes in Clostridium botulinum strains producing botulinum neurotoxin serotype A subtypes. Appl Environ Microbiol. 2008;74:2778–2786. doi: 10.1128/AEM.02828-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R, Rummel A, Binz T, Brunger AT. Botulinum neurotoxin B recognizes its protein receptor with high affinity and specificity. Nature. 2006;444:1092–1095. doi: 10.1038/nature05387. [DOI] [PubMed] [Google Scholar]
- Johnson EA. Clostridial toxins as therapeutic agents: benefits of nature’s most toxic proteins. Annu Rev Microbiol. 1999;53:551–575. doi: 10.1146/annurev.micro.53.1.551. [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karalewitz APA, Fu Z, Baldwin MR, Kim JJP, Barbieri JT. Botulinum neurotoxin serotype C associates with dual ganglioside receptors to facilitate cell entry. J Biol Chem. 2012;287:40806–40816. doi: 10.1074/jbc.M112.404244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karalewitz APA, Kroken AR, Fu Z, Baldwin MR, Kim JJP, Barbieri JT. Identification of a unique ganglioside binding loop within botulinum neurotoxins C and D-SA. Biochemistry. 2010;49:8117–8126. doi: 10.1021/bi100865f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroken AR, Karalewitz APA, Fu Z, Baldwin MR, Kim JJP, Barbieri JT. Unique ganglioside binding by botulinum neurotoxins C and D-SA. FEBS J. 2011a;278:4486–4496. doi: 10.1111/j.1742-4658.2011.08166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroken AR, Karalewitz APA, Fu Z, Kim JJP, Barbieri JT. Novel Ganglioside-mediated Entry of Botulinum Neurotoxin Serotype D into Neurons. J Biol Chem. 2011b;286:26828–26837. doi: 10.1074/jbc.M111.254086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahrhold S, Rummel A, Bigalke H, Davletov B, Binz T. The synaptic vesicle protein 2C mediates the uptake of botulinum neurotoxin A into phrenic nerves. FEBS Lett. 2006;580:2011–2014. doi: 10.1016/j.febslet.2006.02.074. [DOI] [PubMed] [Google Scholar]
- Montal M. Botulinum Neurotoxin: A Marvel of Protein Design. Annu Rev Biochem. 2010;79:591–617. doi: 10.1146/annurev.biochem.051908.125345. [DOI] [PubMed] [Google Scholar]
- Montecucco C. How do tetanus and botulinum toxins bind to neuronal membranes? Trends Biochem Sci. 1986;11:314–317. [Google Scholar]
- Moriishi K, Koura M, Abe N, Fujii N, Fujinaga Y, Inoue K, Ogumad K. Mosaic structures of neurotoxins produced from Clostridium botulinum types C and D organisms. Biochim Biophys Acta. 1996;1307:123–126. doi: 10.1016/0167-4781(96)00006-1. [DOI] [PubMed] [Google Scholar]
- Muraro L, Tosatto S, Motterlini L, Rossetto O, Montecucco C. The N-terminal half of the receptor domain of botulinum neurotoxin A binds to microdomains of the plasma membrane. Biochem Biophys Res Commun. 2009;380:76–80. doi: 10.1016/j.bbrc.2009.01.037. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson E. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nishiki T, Kamata Y, Nemoto Y, Omori A, Ito T, Takahashi M, Kozaki S. Identification of protein receptor for Clostridium botulinum type B neurotoxin in rat brain synaptosomes. J Biol Chem. 1994;269:10498–10503. [PubMed] [Google Scholar]
- Nishiki T, Ogasawara J, Kamata Y, Kozaki S. Solubilization and Characterization of the Acceptor for Clostridium botulinum Type B Neurotoxin From Rat Brain Synaptic Membranes. Biochim Biophys Acta. 1993;1158:333–338. doi: 10.1016/0304-4165(93)90032-4. [DOI] [PubMed] [Google Scholar]
- Nishiki T, Tokuyama Y, Kamata Y, Nemoto Y, Yoshida A, Sekiguchi M, Takahashi M, Kozaki S. Binding of botulinum type B neurotoxin to Chinese hamster ovary cells transfected with rat synaptotagmin II cDNA. Neurosci Lett. 1996;208:105–108. doi: 10.1016/0304-3940(96)12557-x. [DOI] [PubMed] [Google Scholar]
- Nuemket N, Tanaka Y, Tsukamoto K, Tsuji T, Nakamura K, Kozaki S, Yao M, Tanaka I. Structural and mutational analyses of the receptor binding domain of botulinum D/C mosaic neurotoxin: Insight into the ganglioside binding mechanism. Biochem Biophys Res Commun. 2011;411:433–439. doi: 10.1016/j.bbrc.2011.06.173. [DOI] [PubMed] [Google Scholar]
- Peng L, Berntsson RP-A, Tepp WH, Pitkin RM, Johnson EA, Stenmark P, Dong M. Botulinum neurotoxin D-C uses synaptotagmin I/II as receptors and human synaptotagmin II is not an effective receptor for type B, D-C, and G toxins. J Cell Sci. 2012;125:3233–3242. doi: 10.1242/jcs.103564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Tepp WH, Johnson EA, Dong M. Botulinum Neurotoxin D Uses Synaptic Vesicle Protein SV2 and Gangliosides as Receptors. PLoS Pathog. 2011;7:e1002008. doi: 10.1371/journal.ppat.1002008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rummel A, Häfner K, Mahrhold S, Darashchonak N, Holt M, Jahn R, Beermann S, Karnath T, Bigalke H, Binz T. Botulinum neurotoxins C, E and F bind gangliosides via a conserved binding site prior to stimulation-dependent uptake with botulinum neurotoxin F utilising the three isoforms of SV2 as second receptor. J Neurochem. 2009;110:1942–1954. doi: 10.1111/j.1471-4159.2009.06298.x. [DOI] [PubMed] [Google Scholar]
- Rummel A, Karnath T, Henke T, Bigalke H, Binz T. Synaptotagmins I and II act as nerve cell receptors for botulinum neurotoxin G. J Biol Chem. 2004a;279:30865–30870. doi: 10.1074/jbc.M403945200. [DOI] [PubMed] [Google Scholar]
- Rummel A, Mahrhold S, Bigalke H, Binz T. The HCC-domain of botulinum neurotoxins A and B exhibits a singular ganglioside binding site displaying serotype specific carbohydrate interaction. Mol Microbiol. 2004b;51:631–643. doi: 10.1046/j.1365-2958.2003.03872.x. [DOI] [PubMed] [Google Scholar]
- Schiavo G, Matteoli M, Montecucco C. Neurotoxins affecting neuroexocytosis. Physiol Rev. 2000;80:717–766. doi: 10.1152/physrev.2000.80.2.717. [DOI] [PubMed] [Google Scholar]
- Schmitt J, Karalewitz A, Benefield DA, Mushrush DJ, Pruitt RN, Spiller BW, Barbieri JT, Lacy DB. Structural analysis of botulinum neurotoxin type G receptor binding. Biochemistry. 2010;49:5200–5205. doi: 10.1021/bi100412v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TJ, Lou J, Geren IN, Forsyth CM, Tsai R, Laporte SL, Tepp WH, Bradshaw M, Johnson EA, Smith LA, et al. Sequence variation within botulinum neurotoxin serotypes impacts antibody binding and neutralization. Infect Immun. 2005;73:5450–5457. doi: 10.1128/IAI.73.9.5450-5457.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark P, Dong M, Dupuy J, Chapman ER, Stevens RC. Crystal Structure of the Botulinum Neurotoxin Type G Binding Domain: Insight into Cell Surface Binding. J Mol Biol. 2010;397:1287–1297. doi: 10.1016/j.jmb.2010.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark P, Dupuy J, Imamura A, Kiso M, Stevens RC. Crystal structure of botulinum neurotoxin type A in complex with the cell surface co-receptor GT1b-insight into the toxin-neuron interaction. PLoS Pathog. 2008;4:e1000129. doi: 10.1371/journal.ppat.1000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strotmeier J, Gu S, Jutzi S, Mahrhold S, Zhou J, Pich A, Eichner T, Bigalke H, Rummel A, Jin R, et al. The biological activity of botulinum neurotoxin type C is dependent upon novel types of ganglioside binding sites. Mol Microbiol. 2011;81:143–156. doi: 10.1111/j.1365-2958.2011.07682.x. [DOI] [PubMed] [Google Scholar]
- Strotmeier J, Lee K, Völker AK, Mahrhold S, Zong Y, Zeiser J, Zhou J, Pich A, Bigalke H, Binz T, et al. Botulinum neurotoxin serotype D attacks neurons via two carbohydrate-binding sites in a ganglioside-dependent manner. Biochem J. 2010;431:207–216. doi: 10.1042/BJ20101042. [DOI] [PubMed] [Google Scholar]
- Strotmeier J, Willjes G, Binz T, Rummel A. Human synaptotagmin-II is not a high affinity receptor for botulinum neurotoxin B and G: increased therapeutic dosage and immunogenicity. FEBS Lett. 2012;586:310–313. doi: 10.1016/j.febslet.2011.12.037. [DOI] [PubMed] [Google Scholar]
- Swaminathan S, Eswaramoorthy S. Structural analysis of the catalytic and binding sites of Clostridium botulinum neurotoxin B. Nat Struct Biol. 2000;7:693–699. doi: 10.1038/78005. [DOI] [PubMed] [Google Scholar]
- Swaminathan S. Molecular structures and functional relationships in clostridial neurotoxins. FEBS J. 2011;278:4467–4485. doi: 10.1111/j.1742-4658.2011.08183.x. [DOI] [PubMed] [Google Scholar]
- Tsukamoto K, Kohda T, Mukamoto M, Takeuchi K, Ihara H, Saito M, Kozaki S. Binding of Clostridium botulinum type C and D neurotoxins to ganglioside and phospholipid - Novel insights into the receptor for clostridial neurotoxins. J Biol Chem. 2005;280:35164–35171. doi: 10.1074/jbc.M507596200. [DOI] [PubMed] [Google Scholar]
- Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- Yeh FL, Dong M, Yao J, Tepp WH, Lin G, Johnson EA, Chapman ER. SV2 Mediates Entry of Tetanus Neurotoxin into Central Neurons. PLoS Pathog. 2010;6:e1001207. doi: 10.1371/journal.ppat.1001207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Varnum SM. The receptor binding domain of botulinum neurotoxin serotype C binds phosphoinositides. Biochimie. 2012;94:920–923. doi: 10.1016/j.biochi.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Buchko GW, Qin L, Robinson H, Varnum SM. Structural analysis of the receptor binding domain of botulinum neurotoxin serotype D. Biochem Biophys Res Commun. 2010;401:498–503. doi: 10.1016/j.bbrc.2010.09.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Buchko GW, Qin L, Robinson H, Varnum SM. Crystal structure of the receptor binding domain of the botulinum C-D mosaic neurotoxin reveals potential roles of lysines 1118 and 1136 in membrane interactions. Biochem Biophys Res Commun. 2011;404:407–412. doi: 10.1016/j.bbrc.2010.11.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


