Summary
Telomerase is a ribonucleoprotein reverse transcriptase that replicates the ends of chromosomes thus maintaining genome stability. Telomerase ribonucleoprotein assembly is primarily mediated by the RNA binding domain (TRBD) of the enzyme. Here we present the high-resolution structure of the vertebrate, Takifugu rubripes TRBD (trTRBD). The structure shows that with the exception of the N-terminal linker, the trTRBD is conserved with the Tribolium castaneum and Tetrahymena thermophila TRBDs suggesting evolutionary conservation across species. The structure provides a view of the structural organization of the vertebrate-specific VSR motif that binds the activation domain (CR4/5) of the RNA component of telomerase. It also reveals a motif (TFLY) that forms part of the T-CP pocket implicated in template boundary element (TBE) binding. Mutant proteins of conserved residues that consist part of the T and TFLY motifs disrupt trTRBD-TBE binding and telomerase activity and processivity, supporting an essential role of these motifs in telomerase RNP assembly and function.
Keywords: Telomerase, RNA Binding, Aging, Cancer
Introduction
Telomeres are nucleoprotein complexes that serve to protect and maintain the integrity of the ends of chromosomes (Baumann et al., 2002; Blackburn and Gall, 1978; Grandin et al., 2001; Miyake et al., 2009; Song et al., 2008). Telomerase, the enzyme that replicates telomeres is a stable ribonucleoprotein (RNP) reverse transcriptase consisting of a protein (TERT) that comprises the catalytic subunit of the enzyme and a large RNA component (TER), which contains the template TERT uses to replicate telomeres (Blackburn, 2000; Lamond, 1989; Miller and Collins, 2002; Shippen-Lentz and Blackburn, 1990). TERT is evolutionarily conserved across species with high degree of conservation within a family of organisms. Differences across species primarily localize to the N-terminal portion of the protein. The most notable difference is the absence of the TEN domain from insects and worms and the diversity of the linker connecting it to the TRBD (Malik et al., 2000; Osanai et al., 2006). The core TERT protein consists of four distinct domains, including the RNA binding domain (TRBD), which is involved in telomerase RNP assembly, template boundary definition and repeat addition processivity (Cunningham and Collins, 2005; Lai et al., 2002; Miller et al., 2000). Structures of the Tetrahymena thermophila and Tribolium castaneum TRBD (ttTRBD and tcTRBD respectively) revealed an almost all helical nucleic acid binding fold containing several conserved motifs implicated in TER binding (Bley et al., 2011; Bryan et al., 2000; Lai et al., 2001; Moriarty et al., 2002; O’Connor et al., 2005). These include the T and CP motifs, which localize at the interface of the TRBD and fingers domains of TERT. The two motifs are located adjacent to each other and form an extended pocket (T-CP pocket) on the surface of the protein implicated in TBE binding (Gillis et al., 2008; Rouda and Skordalakes, 2007). In vertebrates, additional contacts between TERT and TER are mediated by the VSR motif, a conserved element of the vertebrate N-terminal linker of TERT (Moriarty et al., 2002), which binds the activation domain of TER (Bley et al., 2011).
Here we present the structure of the vertebrate trTRBD, which has high sequence identity to the human enzyme. The structure provides evidence of structural conservation of the core TRBD across species. It also reveals the structural organization of the conserved portion of the N-terminal linker of vertebrate TRBDs, which contains motifs implicated in TER binding. These include the VSR motif, which binds the activation domain of TER (CR4/5) and a motif (TFLY), which contributes to the T-CP pocket formation and TBE binding. We also show that mutations of conserved residues that form part of the T-TFLY pocket disrupt TBE binding and reduce telomerase processivity.
Results
Structure of the trTRBD domain
We engineered a telomerase, trTRBD construct consisting of residues 294–545, initially based on sequence alignment and structure prediction software the boundaries of which were subsequently confirmed by limited proteolysis of the TEN-TRBD N-terminal portion of trTERT (Kelley and Sternberg, 2009; Larkin et al., 2007). The construct contains the core TRBD domain of telomerase and the conserved region of the vertebrate N-terminal linker that connects it to the TEN domain (Figure 1A). We subsequently solved the structure of trTRBD to 2.37 Å resolution using a single mercury derivative and the method of single isomorphous replacement with anomalous signal (SIRAS) (Table 1). The structure, which revealed density for the entire construct, is an elongated helical bundle (Figures 1B and C) with clear structural homology to the Tribolium castaneum and Tetrahymena thermophila TRBDs (Figures 2A–F). Earlier structural and biochemical studies of this portion of TERT identified a number of RNA binding elements, which include the universally CP, T and vertebrate VSR conserved motifs (Bley et al., 2011; Moriarty et al., 2002; Rouda and Skordalakes, 2007) (Figures 1B and C). Like in Tribolium castaneum and Tetrahymena thermophila TRBDs, the trTRBD CP and T motifs localize at the interface of the TRBD and fingers domains, forming an extended pocket on the surface of the protein implicated in TBE binding (Gillis et al., 2008; Rouda and Skordalakes, 2007) (Figures 1B and C).
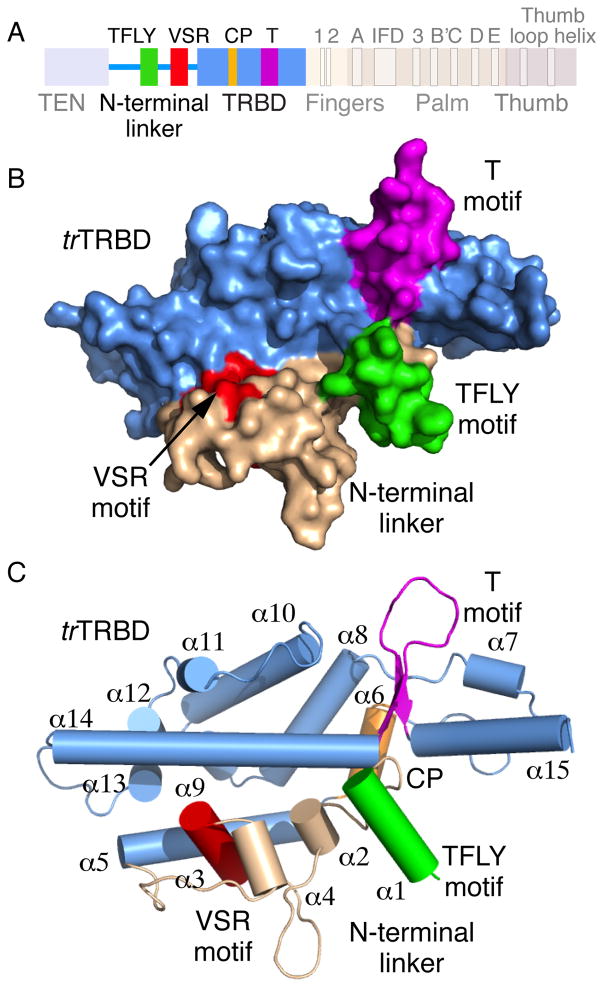
Figure 1. Primary and tertiary structure of vertebrate TRBD and N-terminal linker.
A) Primary structure of vertebrate TERT highlighting the TRBD and N-terminal linker motifs. B) Surface representation of the 3D structure of the Takifugu rubripes TRBD (trTRBD) and conserved portion of the N-terminal linker. The N-terminal linker (wheat) the TFLY (green), T (magenta) and VSR (red) motifs are shown. C) Cartoon representation of the trTRBD shown the N-terminal linker and TFLY, T and VSR motifs in the same orientation and color as in B.
Table 1.
Data collection, phasing and refinement statistics
| Native | Phasing
|
|||
|---|---|---|---|---|
| Hg Derivative 1 | Hg Derivative 2 | Native | ||
| Data collection | ||||
|
| ||||
| Wavelength (Å) | 1.1 | 1.008 | 1.008 | 1.1 |
| Space group | P21 | P212121 | P212121 | P212121 |
| Cell dimensions | ||||
| a, b, c (Å) | 48.6 84.5 137.2 | 48.8 84.6 | 48.6 84.7 | 48.7 84.7 |
| α, β, γ (°) | 94.3 | 139.2 | 138.8 | 139.4 |
| Resolution (Å) | 40-2.37 (2.51-2.37)* | 40-3.4 (3.58-3.4) | 40-3.3 (3.48-3.30) | 40-3.0 (3.16-3.0) |
| Rsym or Rmerge | 7.0 (32.0) | 13.2 (38.9) | 18.9 (41.5) | 8.4 (39.9) |
| I/σI | 9.5 (2.3) | 9.7 (2.7) | 9.7 (2.1) | 13 (2.5) |
| Completeness (%) | 98.6 (92.3) | 99.7 (99.6) | 97.3 (91.1) | 99.4 (99.2) |
| Redundancy | 3.0 (2.4) | 3.1 (3.2) | 3.3 (3.0) | 3.0 (3.0) |
| Phasing Analysis | ||||
| Resolution (Å) | 40-3.5 | |||
| Number of sites | 4 | |||
| Z score | 20 | |||
| Mean figure of merit (FOM) | 0.50 | |||
| Refinement | ||||
| Resolution (Å) | 40-2.37 | |||
| No. reflections | 42409 | |||
| Rwork/Rfree | 21.9/25.8 | |||
| No. atoms | ||||
| Protein | 8446 | |||
| Water | 148 | |||
| B-factors | ||||
| Protein | 35.2 | |||
| Water | 37.6 | |||
| R.m.s deviations | ||||
| Bond lengths (Å) | 0.005 | |||
| Bond angles (°) | 0.943 | |||
| Ramachandran plot (%) | ||||
| Most favored | 91.5 | |||
| Allowed | 8.5 | |||
Parentheses represent highest resolution shell
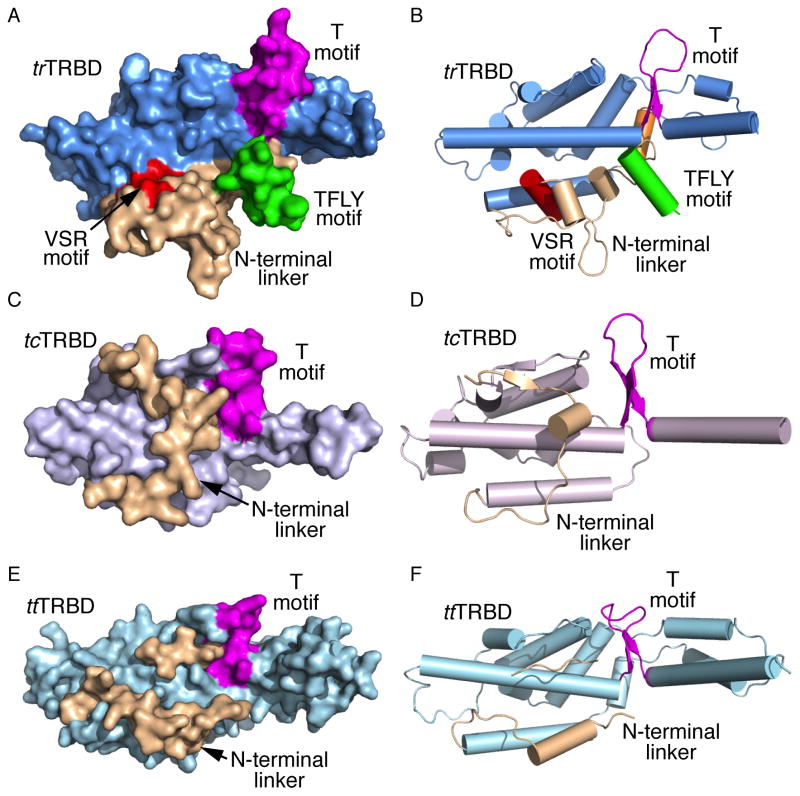
Figure 2. Structural comparison of the Takifugu rubripes, Tribolium castaneum and Tetrahymena thermophila TRBD and portions of their N-terminal linkers.
Surface and cartoon representation of A, B) trTRBD, C, D) tcTRBD and E, F) ttTRBD respectively showing the N-terminal linker (wheat) and the T (magenta) and TFLY (green) motifs.
The vertebrate conserved portion of the N-terminal linker of trTRBD is comprised of a complex network of loops and four short α-helices (Figures 1B and C). This portion of TERT is stably anchored on the surface of TRBD via extensive interactions with helices α5 and α14 that form part of the core trTRBD (Figure 1C). In turn, the N-terminal linker creates an extended surface area that contains several conserved and solvent accessible residues that may be important for TER binding. For example, the VSR motif is a short helix (α4) that sits perpendicular to helices α5 and α14, which run parallel to each other and span the length of the core of this domain. Interestingly, part of helix α4 is buried under the coils and helices flanking the VSR motif with conserved residues shown to bind CR4/5 (Bley et al., 2011) partially solvent exposed for RNA binding.
Another interesting feature of the trTRBD N-terminal linker is a short α-helix (α1) located upstream of the VSR motif (Figures 1B and C). Interestingly, α1 is located in close proximity of the CP and T motifs thus contributing to the formation of the previously identified T-CP pocket (Figures 1B and C) and proposed to bind the TBE (Rouda and Skordalakes, 2007). We will refer to it as the TFLY motif after the conserved residues (TxxFLY) that comprise this structural element.
trTRBD and CR4/5 binding
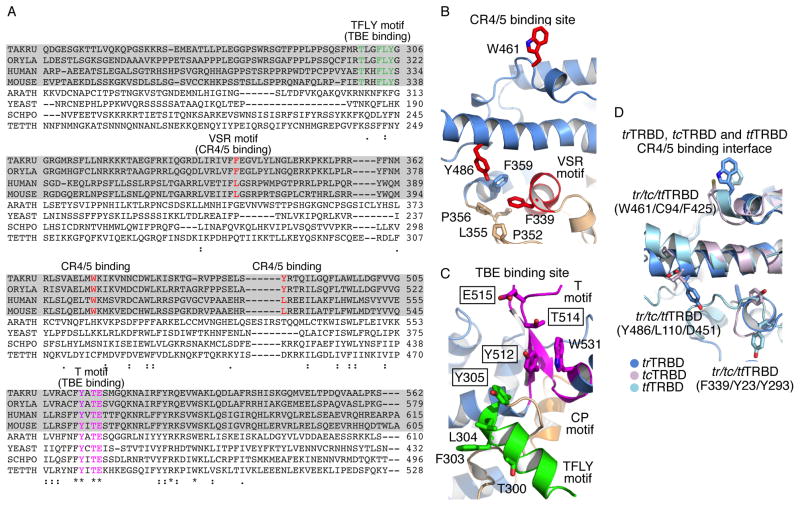
The Chen lab recently identified three vertebrate conserved residues (F339, W461 and Y486 in trTRBD – Figures 3A and B) on the surface of Oryzias latipies TRBD (olTRBD), a close structural homolog of trTRBD, involved in CR4/5 binding (Figure S1B) (Bley et al., 2011). F339 (F355 in olTERT), which forms part of the VSR motif, is partially buried forming extensive interactions with P352, L355 and P356 of the coil that connects α4 with α5 as well as F359 of helix α5 (Figure 3B). However, part of F339 is solvent accessible for contacts with the incoming nucleotide of CR4/5. W461 (W477 in olTERT) forms part of the linker that connects helices α11 and α12 (Figure 3B) and is located in proximity to the interface of the TRBD-thumb domain of TERT (Gillis et al., 2008; Mitchell et al., 2010). The third residue, Y486 (Y503 in olTERT), is located toward the N-terminal portion of helix α14 and is centered between F339 (16.8 Å) and W461 (16.5 Å) (Figure 3B). Together the three residues form an extensive protein-CR4/5, binding surface that spans the entire width of TRBD.
Figure 3. TRBD and N-terminal linker sequence and structural conservation.
A) Sequence alignment of the N-terminal linker and TRBD with the conserved TFLY (green), VSR (red) and T (magenta) motifs shown in color. B and C) Portions of the trTRBD structure highlighting B) the VSR (red) and adjacent residues involved in CR4/5 binding and C) the T (magenta) and TFLY (green) motifs involved in TBE binding. Conserved residues mutated in this study are shown in boxes. D) Structural overlay of trTRBD (blue), ttTRBD (cyan) and tcTRBD (violet) CR4/5 binding surface showing structural and amino acid conservation.
trTRBD and TBE binding
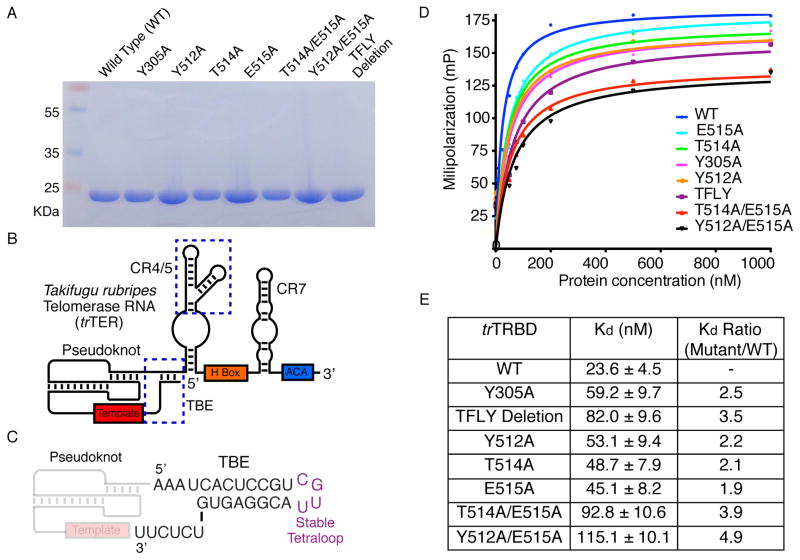
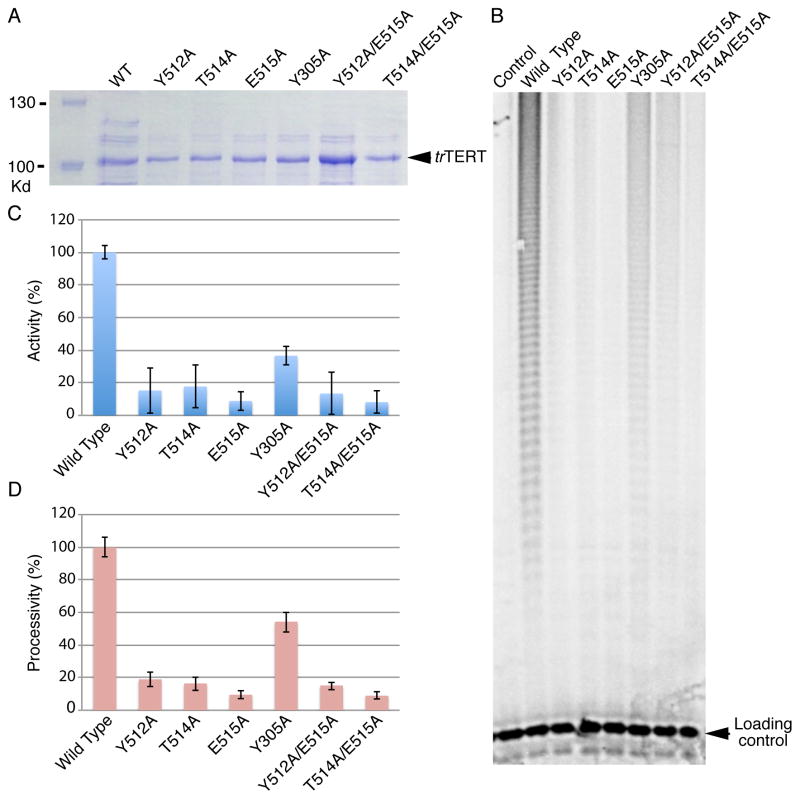
We had previously proposed that the T and CP motifs of ttTRBD are involved in TBE binding (Rouda and Skordalakes, 2007). The structure presented here shows that the TFLY motif comprises part of the T-CP pocket and contains several conserved and solvent accessible hydrophobic residues for TBE binding (Figure 3C). To test this hypothesis, we engineered a host of single and double alanine mutants of conserved and solvent exposed residues of the T (Y512A, E514A, E515A, Y512A/T514A and T514A/E5151A) and TFLY (Y305A) motifs (Figures 3A and C). We expressed and purified the proteins to homogeneity (Figure 4A) and tested their ability to bind the TBE (Figures 4B, C and Table S1) using fluorescence polarization (FP) (Figure 4D and E) and electrophoretic mobility shift (EMSA) (Figure S1C–K) assays in the presence of 30-fold excess of cold tRNA competitor. We also tested the recombinant trTRBD for CR4/5 (Table S1) binding as a control (Figure S1B). The wild type trTRBD has a Kd of ~24 nM for the TBE, while the mutant proteins displayed an overall 1.9 – 4.9 fold reduction in TBE binding (Figure 4E). The Y305A mutant and deletion of the TFLY motif showed a 2.5 and 3.5 fold reduction in TBE binding respectively suggesting that additional residues of this motif, other than Y305, are involved in TERT-TER association. For example, the vertebrate conserved T300, F303 and L304 residues (Figure 3A) of the TFLY motif face away from the T-CP-TFLY pocket, forming a solvent exposed hydrophobic patch on the surface of the protein, which may be involved in protein or nucleic acid binding. Mutations of residues that comprise the T motif (Y512A, T514A and E515A) also showed approximately 2 fold reduction in affinity for TBE. The least effect was observed for the E515A mutant (1.9 fold), which forms part of the T motif and is most proximal to the interior cavity of the TERT ring and where the active site of the enzyme is located (Gillis et al., 2008; Mitchell et al., 2010).
Figure 4. trTER primary structure and trTRBD-TBE binding data.
A) SDS PAGE gel of the wild type (WT) and mutant trTRBD proteins used in this study. B) Schematic of the trTER RNA showing conserved motifs in color. The CR4/5 and TBE motifs are highlighted with blue dashed line C) Nucleotide sequence of trTBE used for the FP and EMSAs in this study; the template and pseudoknot regions are also shown faded. D) Fluorescence polarization (FP) data of the TBE with WT, Y305A, Y512A, T514A, E515A, T514A/E515A, Y512A/E515A and TFLY deletion trTRBD proteins. E) Table of WT and mutant trTRBD-TBE binding constants calculated using Prism5 (GraphPad Software).
Disruption of trTRBD - TBE binding results in loss of telomerase processivity
To test the effects of trTRBD-TBE binding in telomerase function we assembled the minimal telomerase holoenzyme in vitro using E. coli overexpressed wild type and mutant (Y305A, Y512A, T514A, E515A, Y512A/E515A and T514A/E515A) trTERT proteins (Figure 5A) and in vitro transcribed trTER. The proteins were isolated by affinity chromatography and the RNP complex assembled by mixing trTERT with trTER at 1:1 ratio in a buffer containing 25mM Hepes pH 7.5, 5% glycerol, 100 mM KCl, and 1 mM TCEP. We then tested the wild type and T-TFLY mutant proteins for telomerase activity and processivity using direct assays as described in the methods section of this manuscript. The wild type in vitro assembled Takifugu rubripes telomerase showed robust telomerase activity (Figures 5B and C), while the single and double alanine T-TFLY mutant proteins that disrupt TBE binding (Figures 3C, 4D and E) showed significant loss of telomerase activity and processivity (Figures 5B–D) consistent with the important role of these motifs in telomerase function.
Figure 5. Direct assays of the wild type (WT) and mutant trTRBD proteins that disrupt trTRBD-TBE binding.
A) SDS PAGE gel of WT and mutant trTERT proteins expressed in E. coli and used for direct assays with in vitro transcribed RNA. B) Direct telomerase assays of the WT and mutant trTERTs used in this study. C) Percent (%) telomerase activity or total product intensity of telomeric DNA produced by the WT and mutant trTERT telomerases; The percent (%) telomerase activity of each telomerase RNP (WT and mutant trTERT) were measured as the total intensity of all telomeric bands (Using ImageJ), normalized for loading with the intensity of the loading control and divided by the activity of the WT trTERT telomerase. D) Number of total repeats produced by the WT and mutant trTERT telomerases. The percent (%) telomerase processivity was calculated by measuring the approximate total number of repeats of the WT and mutant telomerase proteins and divided by the total number of WT telomerase repeats. Each experiment was carried out in triplicate, and the error bar represents the standard deviation of the average difference between the three measurements.
Discussion
Structural conservation of TRBD
Striking differences in the primary structure of the telomerase protein and RNA subunits have raised significant questions regarding structural and functional conservation of the enzyme across species. For instance, insect and worm TERTs lack the TEN domain while TERTs of evolutionarily distant species have sequence identity below 20%. Despite these differences, the structure presented here demonstrates that the core of telomerase TRBD is evolutionarily conserved from ciliates to vertebrates with an average RMSD of 3.0 Å. The inflated RMSD observed among the trTRBD, ttTRBD and tcTRBD is primarily due to the flexibility of helix (α15) that connects the TRBD to the fingers domain of TERT (Figure 1C). Additional structural differences in the fold of TRBD are observed in the N-terminal region of the protein (Figures 2A–F), which is the least conserved portion of TERT. For example, insects and worms contain only a portion of this linker, while it is widely variable both in length and sequence even within a family of organisms (Malik et al., 2000). Despite the diversity of the N-terminal linker of telomerase this structural element is far from merely a passive component of the enzyme. In vertebrates, the TEN-TRBD linker contains motifs specific to higher eukaryotes, such as the VSR and TFLY, which are involved in RNA binding. It is therefore likely that the N-terminal region of TERT has co-evolved with the RNA component of telomerase to maintain high-affinity binding and stable RNP assembly. The structural and biochemical data presented here provides novel insights into the role of the vertebrate TERT N-terminal linker motifs VSR and TFLY in TERT - TER binding and telomerase function.
trTRBD CR4/5 binding and implications for telomerase RNP assembly
The Chen lab showed recently that the vertebrate TRBDs use the VSR motif (F339) and proximal surface exposed residues (W461 and Y486) to bind the activation domain of TER (CR4/5) (Bley et al., 2011; Moriarty et al., 2002) (Figure 3B). While W461 and F339 are located on opposite ends of the surface of TRBD, Y486 is located at the center of the protein and is equidistant from these two residues. Together Y486 with W461 and F339 form an extensive and conserved area on the surface of TRBD involved in stable interactions with the activation domain of TER thus facilitating TERT-TER assembly. Surprisingly, the VSR motif is mostly buried under a network of coils and helices, consisting of several conserved residues among vertebrate telomerases with the F339 partially solvent exposed for CR4/5 binding. The coils and helices of this portion of the N-terminal linker form a structural protrusion on the side of TRBD, located opposite to the TRBD-thumb interface (Figure 6A and B). Examination of the surface charge, residue conservation and the proximity of this portion of the N-terminal linker to the CR4/5 binding site suggests that additional components of this structural element may be involved in RNA or protein (TEN domain) binding. For example, F339 is located near a cluster of conserved and solvent exposed basic residues, such as R349, K350, K353, and K354, which may be involved in interactions with the backbone of CR4/5. Interestingly, W461 is located adjacent to the TRBD - thumb domain interface of TERT (Figure 6A and B) (Gillis et al., 2008; Mitchell et al., 2010). The proximity of W461 to the thumb domain suggests that the telomerase RNA and in particular the CR4/5 domain, may also be making contacts with the C-terminal domain of the enzyme. Contacts between TER and the thumb domain may further stabilize the closed TERT ring configuration, a process required for full telomerase activity and processivity.
Figure 6. Model of tcTERT and trTRBD showing the structural organization of the N-terminal linker and TFLY motifs in the context of the TERT ring.
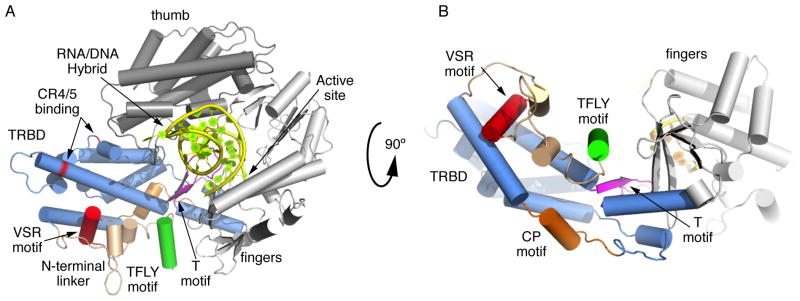
A) Structural overlay of the trTRBD with the Tribolium castaneum TERT structures (cartoon). The model shows the structural organization of the CR4/5 (VSR, W461 and Y368 - red) and TBE (T and TFLY motifs – magenta and green color respectively) binding interfaces. It shows the RNA/DNA hybrid (yellow) docked in the interior cavity of the TERT ring. It also shows the position of the T motif with respect to the active site of the enzyme. B) Same as (A) rotated 90° along the X axis.
Despite the low sequence identity of TERT and TER genes and the different structural composition of their N-terminal linker, the CR4/5 binding interface of TRBD appears to be in part conserved across species. For example, structural comparison of trTRBD, ttTRBD and tcTRBD shows that the VSR motif, a short helix in vertebrates (Figures 1C and 2B), is a loop in tcTRBD (Figures 2C and D) and ttTRBD (Figures 2E and F). However, the proximal spatial position of F339, involved in CR4/5 binding, is occupied in all three structures with Y23 in tcTRBD (Figure 3D) and Y293 in ttTRBD (Figure 3D). It is therefore possible that Y23 and Y293 of tcTRBD and ttTRBD respectively, like F339 of trTRBD are involved in stacking interactions with an incoming nucleotide of the CR4/5 motif. In contrast, the tertiary structural organization of W461 and Y368 positions is highly conserved (Figure 3D), while the amino acid composition varies considerably. The W461 of trTRBD is a C94 in tcTRBD and a F425 in ttTRBD, while Y368 is a L110 in tcTRBD and a D451 in ttTRBD (Figure 3D). The sequence diversity observed in these two positions could be due to significant differences of the TER nucleotide sequence across species.
The TFLY motif of vertebrate TERTs contributes to T-CP pocket formation and TBE binding
The data presented here provides direct evidence of telomerase TRBD-TBE binding a process mediated by the universally and vertebrate conserved CP-T and TFLY motifs respectively. A host of additional structural and biochemical data supports T-CP-TFLY/TBE binding, an interaction important for telomerase assembly, template boundary definition and repeat addition processivity. For example, the structure of the tcTERT-RNA/DNA complex showed that the 5′ end of the RNA template is located at the entry of the T-CP pocket, which positions the TBE within coordinating distance of the T and CP motifs (Figures 6A and B) (Mitchell et al., 2010). Mutations of conserved residues in the T and CP motifs resulted in reduced full length TER binding and telomerase activity and processivity (Bryan et al., 2000; Friedman and Cech, 1999; Lai et al., 2002; Moriarty et al., 2002). Moreover, the recent EM structure of Tetrahymena thermophila telomerase supports T-CP/TBE binding (Jiang et al., 2013). The newly identified TFLY motif is a short helix positioned above the CP pocket and forms an extension of the T motif. The organization and residue composition of the TFLY motif creates an extended channel on the surface of TRBD (Figures 1B, C and 6A, B). The width and chemical composition of this channel suggest that this portion of TRBD is most likely involved in binding the single stranded RNA portion of TBE (Figure 4C). Single and double alanine mutants of conserved residues that form the T and TFLY motifs partially disrupt TBE binding (Figure 4D, E and S1C–K), which suggests that the T, CP and TFLY motifs act in concert for proper TBE binding. Interestingly, the CP2 motif of ciliate telomerases, also present within the N-terminal linker of the protein, was recently shown to localize in proximity to the T-CP pocket and being involved in TBE binding (Akiyama et al., 2013). As we proposed previously the double stranded stem loop of the TBE would fit into the electropositive groove formed by the CP motif (Rouda and Skordalakes, 2007). The interaction of the CP motif with the stem loop would position the single stranded portion of the TBE into the pocket formed by the TFLY and T motifs. Within this pocket there is a series of several conserved aromatic residues, such as the F303, Y305 of the TFLY and the Y512 and W531 of the T motifs (Figure 3C), which likely form base stacking interactions with the TBE. Interestingly, the T and TFLY motif mutants show modest disruption of TBE binding suggesting that these residues may have more of a regulatory role in telomerase function. This is consistent with our direct assays, which show that single and double alanine mutants of these residues undergo significant loss of telomerase activity and processivity (Figures 5B and C). A striking example is the single alanine mutant E515A. While this mutant showed almost wild type TBE binding (1.9 fold – Figures 4D, E and S1D, H), it resulted in almost complete loss of telomerase activity and processivity (Figure 5C and D). E515A is an invariant residue, located near the tip of the T-motif, which extends toward the interior cavity of the TERT ring and is in proximity to the active site of the enzyme (Figure 6A and B). It is therefore possible that this residue and its interaction with the single stranded portion of the TBE guide the RNA template toward the active site of the enzyme (Gillis et al., 2008; Mitchell et al., 2010). Elimination of the E515-TBE interaction would misplace the RNA template thus affecting telomerase activity and processivity. We therefore propose that the T and TFLY motifs are important effectors of telomerase activity and processivity by guiding the RNA template in the interior cavity of the TERT ring and were the active site of the enzyme is located for nucleotide binding and selectivity.
Experimental Procedures
Construct design and protein expression and purification
We identified a stable construct of T. rubripes TRBD via limited proteolysis of the full length N-terminal portion of trTERT. We subsequently cloned the trTRBD construct consisting of residues 294–545 in a modified version of the pET28b vector expressing a hexahistidine tag followed by a TEV cleavable small ubiquitin-like modifier (SUMO) fusion protein at its N-terminus. We overexpressed the protein in BL21-CodonPlus(DE3)-RIPL cells (Stratagene) for 5 hours at 20° C. We lysed the cells via sonication and purified the protein using Ni-NTA resin (Qiagen), followed by TEV cleavage of the His-SUMO tag. We further purified the protein to homogeneity using Poros HS (PerSpective Biosystems) and a Superdex S200 size exclusion chromatography (GE Healthcare).
Protein Crystallization and Data Collection
For crystallization studies, we concentrated the purified trTRBD to ~15 mg/ml and dialyzed it in a buffer containing 5 mM Tris-HCl, 200 mM KCl, and 1 mM TCEP (pH 7.5) prior to crystallization trials. Sparse matrix crystal screening using the Hampton Research crystal screens produced two crystal forms (Natrix condition G1) that belong to the P21 and P212121 space groups and diffracted to 2.37 Å and 3.0 Å resolution respectively. Data were collected at the National Synchrotron Light Source (NSLS), beam line X25 and processed with MOSFLM as implemented in Elves. Phases were calculated using the method of single isomorphous replacement with anomalous signal (SIRAS) and a mercury derivative (MeHgCl2). We located the heavy atom sites, calculated phases and built a partial model in SOLVE (Brunger et al., 1998) using four fold NCS. The protein model was subsequently completed manually in COOT (Emsley and Cowtan, 2004) and refined in REFMAC5 (Murshudov et al., 2011). Structural overlays were performed in COOT and figures were made in pymol.
Electrophoretic Mobility Shift Assays
The TBE and CR4/5 RNA substrates (Table S1) were purchased from Dharmacon and were 32P labeled using 10 μCi/μl of γ-ATP and T4 polynucleotide kinase (New England Biolabs). The wild type and mutant trTRBD proteins were incubated with 30 nM cold tRNA (Life technologies - Ambion yeast tRNA) competitor and 5 μg BSA for 30 minutes prior to addition of 1 nM of the 32P labeled TBE to increasing concentrations of protein on ice and in a buffer containing 20 mM Tris.HCl pH 8.0, 100 mM KCl, 2 mM DTT, 1 mM MgCl2, 5% glycerol, 1 mM EDTA and 0.01% Triton-X 100 for 30 minutes. trTRBD concentrations were calculated using a standard Bradford assay as follows: 50 μL of samples were added to 1.5 mL of Bradford dye and absorbance was measured at A600. Results of the Bradford assay were corroborated with UV spec at A280 absorbance. The reactions were then loaded onto 6% DNA retardation PAGE gels (Invitrogen) and run at 100V for one hour. The gels were vacuum dried and exposed using a storage phosphor screen (GE Healthcare).
Fluorescence Polarization (FP)
We performed FP, trTRBD-TBE binding reactions in 15 μl samples using a Wallac EnVision Plate reader (PerkinElmer). The binding reactions were carried out in a buffer containing 20 mM Tris.HCl pH 8.0, 100 mM KCl, 2 mM MgCl2, 1 mM EDTA, 2 mM DTT, 5% v/v glycerol and 30 nM cold tRNA competitor (Life technologies - Ambion yeast tRNA). The TBE probe was purchased with a 5′ labeled Cy5™ tag from IDT. The final probe concentration used was 1 nM, while the trTRBD protein concentration ranged from 0 to 1 μM. The reactions were incubated at room temperature for five minutes and pipetted in triplicate into a black 384 well ProxiPlate (PerkinElmer). The reactions were excited with 650 nm light and the emissions were measured at 690 nm. The milipolarization values were calculated by the Envision operating software (PerkinElmer). The data was fit and the binding constants were determined with a one site binding linear regression model using PRISM 5.0 (GraphPad Software, San Diego California USA, www.graphpad.com).
trTER, in vitro transcription and purification
The full-length trTER gene was cloned into the pBSK vector (Addgene) using the KpnI and SacI restriction enzyme sites. The pBSK vector carrying the trTER gene was linearized using SacI-HF (NEB). We then extracted the linearized plasmid from a 1% agarose gel and further purified it using the QIAquick Gel Extraction kit (Qiagen). The trTER was in vitro transcribed using a reaction mix containing 1x transcription buffer (Promega) containing 10 μM DTT, 0.01% Triton X-100, 2.2 U inorganic pyrophosphatase, 0.5 mM rNTPs, 1 μg of linearized trTER-pBSK, and 80 U of T7 RNA polymerase (Promega). The transcription reaction mix was incubated at 37° C for 1.5 hours, and then 2.2 U of RNase-free DNase (Promega) was added to the mix and incubated for another hour at 37° C to digest the linear trTER-pBSK plasmid. The transcribed product was purified using Trizol extraction (Invitrogen), and a sample was run on a 1% agarose gel to check for RNA quality.
trTERT expression and purification
The trTERT wild type and mutant genes were cloned into pET28b vector containing a N-terminal hexahistidine tag followed by a TEV cleavable small ubiquitin-like modifier (SUMO) fusion protein. The trTERT proteins were over-expressed in the Scarab E. coli strain (Scarab Genomics) overnight at 14° C. The cells were harvested by centrifugation and lysed by sonication in buffer containing 25 mM Tris-HCl (pH 7.5), 5% glycerol, 0.5 M KCl, 0.1 mM benzamidine, 15 mM imidazole and 0.1 mM phenylmethyl sulfonyl flouride (PMSF). The proteins were purified using Ni-NTA resin (Qiagen) and samples were run on a 12% SDS-PAGE gel to test expression levels and purity. We then determined the concentration of the trTERT proteins by accurately calculating the intensity of the TERT band of each sample using ImageJ.
Direct telomerase activity assay
We tested the activity and processivity of the in vitro reconstituted wild type and mutant telomerase proteins using direct telomerase activity assays. The concentration of the Ni-NTA purified trTERT proteins used in each assay was normalized by first loading equal volumes of the Ni-NTA eluted trTERT samples on a SDS-PAGE gel (Figure 5A), and comparing the protein band intensities using ImageJ. Approximately 2 μl or 0.1 μg of the Ni.NTA purified trTERT proteins (volume was adjusted so that identical concentrations of trTERT proteins were used in all reactions) were incubated for 1.5 hours at 30° C in 20 μL of direct assay buffer consisting of 0.1 μg trTER, 50 mM Tris-HCl pH 8.0, 50 mM KCl, 1 mM MgCl2, 5 mM beta-mercaptoethanol, 1 mM spermidine, 1 μM TS primer (5′-AATCCGTCGAGCAGAGTT-3′), 0.5 mM dGTP, 0.5 mM dTTP, 2 μM dATP, and 1.25 μM [α-32P] dATP. Reactions were stopped by adding 100 μL of 5 M NH4OAc, 20 μg of glycogen, 450 μL of 2-propanol, and 0.5 μL of a 200 nM 32P-labeled loading control (budding yeast telomeric 11-mer substrate). Samples were precipitated for 1.5 hours at room temperature then were pelleted by centrifugation, washed with 75% 2-propanol and then dried. Pelleted DNA products were resuspended in 25 μL gel loading buffer (10 mM Tris-HCl pH 8.0, 10 mM EDTA, 40% formamide, and 0.05% xylene cyanol) and heat denatured at 95° C for 10 minutes. Samples were run on a 12% polyacrylamide (19:1), 7 M Urea gel and exposed overnight to a phosphor screen for imaging. The percent (%) telomerase activity of each protein (WT and Mutant trTERT) were measured as the total intensity of all telomeric bands, normalized for loading with the intensity of the loading control and divided by the activity of the WT trTERT. The percent (%) processivity was calculated by measuring the approximate total number of repeats of the WT and mutant telomerase proteins and divided by the total number of WT telomerase repeats.
Supplementary Material
Highlights.
Structure of a vertebrate telomerase TRBD
Identification of a conserved motif (TFLY) in vertebrate telomerase N-terminal linker
The TFLY motif contributes to the T-CP pocket formation and TBE binding
T and TFLY motif TBE binding required for telomerase activity and processivity
Acknowledgments
This project was funded by the Pennsylvania Department of Health and the NIGMS (5 R01 GM088332-03).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akiyama BM, Gomez A, Stone MD. A conserved motif in Tetrahymena thermophila telomerase reverse transcriptase is proximal to the RNA template and is essential for boundary definition. J Biol Chem. 2013 doi: 10.1074/jbc.M113.452425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, Podell E, Cech TR. Human Pot1 (protection of telomeres) protein: cytolocalization, gene structure, and alternative splicing. Mol Cell Biol. 2002;22:8079–8087. doi: 10.1128/MCB.22.22.8079-8087.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH. The end of the (DNA) line. Nat Struct Biol. 2000;7:847–850. doi: 10.1038/79594. [DOI] [PubMed] [Google Scholar]
- Blackburn EH, Gall JG. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J Mol Biol. 1978;120:33–53. doi: 10.1016/0022-2836(78)90294-2. [DOI] [PubMed] [Google Scholar]
- Bley CJ, Qi X, Rand DP, Borges CR, Nelson RW, Chen JJ. RNA-protein binding interface in the telomerase ribonucleoprotein. Proc Natl Acad Sci U S A. 2011;108:20333–20338. doi: 10.1073/pnas.1100270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Bryan TM, Goodrich KJ, Cech TR. Telomerase RNA bound by protein motifs specific to telomerase reverse transcriptase. Mol Cell. 2000;6:493–499. doi: 10.1016/s1097-2765(00)00048-4. [DOI] [PubMed] [Google Scholar]
- Cunningham DD, Collins K. Biological and biochemical functions of RNA in the tetrahymena telomerase holoenzyme. Mol Cell Biol. 2005;25:4442–4454. doi: 10.1128/MCB.25.11.4442-4454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Friedman KL, Cech TR. Essential functions of amino-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes Dev. 1999;13:2863–2874. doi: 10.1101/gad.13.21.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–637. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- Grandin N, Damon C, Charbonneau M. Cdc13 prevents telomere uncapping and Rad50-dependent homologous recombination. Embo J. 2001;20:6127–6139. doi: 10.1093/emboj/20.21.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Miracco EJ, Hong K, Eckert B, Chan H, Cash DD, Min B, Zhou ZH, Collins K, Feigon J. The architecture of Tetrahymena telomerase holoenzyme. Nature. 2013 doi: 10.1038/nature12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Lai CK, Miller MC, Collins K. Template boundary definition in Tetrahymena telomerase. Genes Dev. 2002;16:415–420. doi: 10.1101/gad.962602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CK, Mitchell JR, Collins K. RNA binding domain of telomerase reverse transcriptase. Mol Cell Biol. 2001;21:990–1000. doi: 10.1128/MCB.21.4.990-1000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI. Tetrahymena telomerase contains an internal RNA template. Trends Biochem Sci. 1989;14:202–204. doi: 10.1016/0968-0004(89)90022-4. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Malik HS, Burke WD, Eickbush TH. Putative telomerase catalytic subunits from Giardia lamblia and Caenorhabditis elegans. Gene. 2000;251:101–108. doi: 10.1016/s0378-1119(00)00207-9. [DOI] [PubMed] [Google Scholar]
- Miller MC, Collins K. Telomerase recognizes its template by using an adjacent RNA motif. Proc Natl Acad Sci U S A. 2002;99:6585–6590. doi: 10.1073/pnas.102024699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MC, Liu JK, Collins K. Template definition by Tetrahymena telomerase reverse transcriptase. Embo J. 2000;19:4412–4422. doi: 10.1093/emboj/19.16.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol. 2010;17:513–518. doi: 10.1038/nsmb.1777. [DOI] [PubMed] [Google Scholar]
- Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, Saito M, Ishikawa F. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell. 2009;36:193–206. doi: 10.1016/j.molcel.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Moriarty TJ, Huard S, Dupuis S, Autexier C. Functional multimerization of human telomerase requires an RNA interaction domain in the N terminus of the catalytic subunit. Mol Cell Biol. 2002;22:1253–1265. doi: 10.1128/MCB.22.4.1253-1265.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor CM, Lai CK, Collins K. Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. J Biol Chem. 2005;280:17533–17539. doi: 10.1074/jbc.M501211200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osanai M, Kojima KK, Futahashi R, Yaguchi S, Fujiwara H. Identification and characterization of the telomerase reverse transcriptase of Bombyx mori (silkworm) and Tribolium castaneum (flour beetle) Gene. 2006;376:281–289. doi: 10.1016/j.gene.2006.04.022. [DOI] [PubMed] [Google Scholar]
- Rouda S, Skordalakes E. Structure of the RNA-binding domain of telomerase: implications for RNA recognition and binding. Structure. 2007;15:1403–1412. doi: 10.1016/j.str.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Shippen-Lentz D, Blackburn EH. Functional evidence for an RNA template in telomerase. Science. 1990;247:546–552. doi: 10.1126/science.1689074. [DOI] [PubMed] [Google Scholar]
- Song X, Leehy K, Warrington RT, Lamb JC, Surovtseva YV, Shippen DE. STN1 protects chromosome ends in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2008;105:19815–19820. doi: 10.1073/pnas.0807867105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.