Abstract
Aims/hypothesis
Hyperglycaemia disproportionately affects African-Americans (AfAs). We tested the transferability of 18 single-nucleotide polymorphisms (SNPs) associated with glycaemic traits identified in European ancestry (EuA) populations in 5,984 non-diabetic AfAs.
Methods
We meta-analysed SNP associations with fasting glucose (FG) or insulin (FI) in AfAs from five cohorts in the Candidate Gene Association Resource. We: (1) calculated allele frequency differences, variations in linkage disequilibrium (LD), fixation indices (Fsts) and integrated haplotype scores (iHSs); (2) tested EuA SNPs in AfAs; and (3) interrogated within ±250 kb around each EuA SNP in AfAs.
Results
Allele frequency differences ranged from 0.6% to 54%. Fst exceeded 0.15 at 6/16 loci, indicating modest population differentiation. All iHSs were <2, suggesting no recent positive selection. For 18 SNPs, all directions of effect were the same and 95% CIs of association overlapped when comparing EuA with AfA. For 17 of 18 loci, at least one SNP was nominally associated with FG in AfAs. Four loci were significantly associated with FG (GCK, p=5.8 × 10-8; MTNR1B, p=8.5 × 10-9; and FADS1, p=2.2 × 10-4) or FI (GCKR, p=5.9 × 10-4). At GCK and MTNR1B the EuA and AfA SNPs represented the same signal, while at FADS1, and GCKR, the EuA and best AfA SNPs were weakly correlated (r2<0.2), suggesting allelic heterogeneity for association with FG at these loci.
Conclusions/interpretation
Few glycaemic SNPs showed strict evidence of transferability from EuA to AfAs. Four loci were significantly associated in both AfAs and those with EuA after accounting for varying LD across ancestral groups, with new signals emerging to aid fine-mapping.
Keywords: African ancestry, Genetics, Genome-wide association, LD mapping, Minorities, Type 2 diabetes
In the USA, over 23 million people have type 2 diabetes, but minority groups such as African-Americans (AfAs) are disproportionately affected. In 2005, the prevalence of type 2 diabetes was about 28% higher in AfAs than in individuals of European ancestry (EuA), and risk for diabetes (defined by either elevated fasting glucose or HbA1c) was about twice as high. By 2050, this racial disparity is projected to increase even further [1]. Racial/ethnic variation in health-related behaviours and obesity accounts for a lot of this disparity [2, 3], but genetic factors could also contribute [4-6].
Until recently, the majority of genetic studies of type 2 diabetes and related quantitative traits (e.g. levels of fasting glucose [FG] and fasting insulin [FI]) have been conducted in EuA populations [7, 8]. There are strong scientific reasons to expand diabetes-related genetic analyses to AfA, quite apart from the disparity caused by their exclusion from this important research frontier. No single ancestral population is sufficient to fully uncover variants contributing to disease in humans, and of all continental ancestral groups, those of African ancestry contain the richest range of genetic variation. Trans-ethnic genetic analyses can elucidate ancestral differences in common risk-allele frequencies, linkage disequilibrium (LD) patterns and variant effect size differences, as well as guide locus fine-mapping to uncover functional variants identified in EuA genetic studies [4-6, 9, 10].
Study of AfA populations is likely to be especially important to identify diabetes-related genetic variants [4, 11]. Although one recent study examined whether type 2 diabetes associations found in EuA were also associated in about 2,500 AfA individuals [12], analyses of transferability of type-2-diabetes-associated variants from EuA to AfA populations have not so far been especially instructive, probably because of limited sample sizes and testing of tagging variants rather than functional variants in the setting of reduced LD [13-16]. On the other hand, analyses of continuously distributed quantitative traits such as FG or FI, elevations of which herald type 2 diabetes [17], offer the advantage of increased power and reduced misclassification by disease status [18]. One recent study of 927 unrelated AfAs confirmed the association of six EuA-identified variants with FG [19]. Another study of 3024 adults reported that allele frequencies of 16 novel FG-associated single-nucleotide polymorphisms (SNPs) varied across non-Hispanic whites, non-Hispanic blacks and Mexican Americans [9]. In the present study, we investigated whether testing associations of EuA-identified variants in the large Candidate Gene Association Resource (CARe) study would confirm wide risk-allele frequency variation and LD differences in AfA vs EuA and show limited EuA-associated SNP associations in AfA, but when accounting for LD, would show that most FG- or FI-associated loci are associated with diabetes-related quantitative traits in AfAs at different, generally independent, variants.
Methods
Candidate Gene Association Resource study samples
The Candidate Gene Association Resource (CARe) is a National Heart, Lung, and Blood Institute (NHLBI) shared resource for genotype association analyses comprising five parent studies with AfA participants and traits of interest. All individuals provided written informed consent for DNA studies as part of their parent study. CARe study samples are described in greater detail in Lettre et al [15]. Briefly, the studies were as follows.
Atherosclerosis Risk in Communities
The Atherosclerosis Risk in Communities (ARIC) study is an ongoing prospective cohort study originally designed to investigate risk factors of subclinical and clinical atherosclerosis. It includes measurement of cardiovascular and diabetes risk factors. ARIC enrolled 15,792 participants, aged 45-64 years, from four field centres, and has been described in detail elsewhere [20]. This study includes only self-reported AfA participants, and all participants provide written informed consent.
Coronary Artery Risk Development in young Adults
The Coronary Artery Risk Development in young Adults (CARDIA) study is a prospective multicentre investigation of the development and determinants of clinical and subclinical cardiovascular disease (CVD) and its risk factors. The participants were recruited from four sites and appropriate informed consent was provided [21].
Cleveland Family Study
The Cleveland Family Study (CFS) is a family-based longitudinal study originally designed to investigate genetic and non-genetic risk factors for sleep apnoea [22]. For this analysis, only data from AfAs participating in the most recent examination that included a morning fasting blood draw were analysed. All participants provided written informed consent.
Jackson Heart Study
The Jackson Heart Study (JHS) is a prospective population-based study of CVD among non-institutionalised AfA adults aged 21–95 years and residing in the Jackson, MS metropolitan statistical area (MSA) [23]. The final JHS cohort included 5,301 participants, equivalent to 7% of all AfAs aged 21–95 residing in the Jackson MSA17. Details of the study design and recruitment protocol have been described elsewhere [23-26]. Appropriate informed consent was provided.
Multi-Ethnic Study of Atherosclerosis
The Multi-Ethnic Study of Atherosclerosis (MESA) is a community-based cohort study designed to investigate characteristics and risk factors for subclinical CVD. Participants were recruited from six centres and appropriate informed consent was provided. From 2000 to 2002, 6,814 participants (38% non-Hispanic white, 28% black, 12% Chinese, 22% Hispanic; aged 45–84 years) without existing clinical CVD were enrolled [27]. Only data from AfA individuals were included in this analysis.
In all studies, FG was measured using hexokinase (in mmol/l) and FI was measured using RIA or ELISA methods in serum or plasma (in μU/ml, converted into pmol/l using a conversion factor of 6).
Genotyping, imputation and principal components analysis
All samples were genotyped at the Broad Institute using the Affymetrix Genome-Wide Human SNP Array 6.0 (Affy6.0, Santa Clara, CA, USA). Quality-control steps were performed centrally by the CARe analytical group at the Broad Institute [15]. Imputation was conducted centrally at the Broad Institute with MACH 1.0.16 using the HapMap 2 reference panel of Utah residents of northern and western European ancestry (CEU) and the Yoruba in Ibadan, Nigeria (YRI). The allelic concordance rate, defined as 1–1/2*|imputed_dosage – genotyped_dosage| based on the CEU+YRI panel, was 95.6%, comparable with rates for individuals of African descent imputed with HapMap 2 YRI individuals [15]. Filters applied to the imputation data were based on the imputation quality measure RSQ_HAT with a threshold of >0.3 and a minor allele frequency threshold of >0.01. Ancestry principal components (PCs) were generated using EIGENSTRAT (http://genepath.med.harvard.edu/∼reich/EIGENSTRAT.htm).
Allele frequency differences and natural selection
We used several approaches to evaluate population differentiation and natural selection at index SNPs. First, we assessed index SNP allele-frequency differences in CARe comparing individuals of EuA vs AfA by calculating the absolute value of (index-risk-allele frequency in EuA – index-risk-allele frequency in AfA), using the trait-raising allele in EuA as the risk allele. Second, we calculated Wright's fixation index (Fst) to assess the degree of population differentiation as reflected by divergent allele frequencies. We calculated Fst using CARe risk-allele frequencies for AfAs, and the Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) [8] risk-allele frequencies for those of EuA, but because the EuA sample size (46,186) was about seven times the AfA sample size, potentially obscuring population differentiation at any locus, we also calculated the Fst using CEU and YRI HapMap 2 data for which the sample sizes are similar. Third, we calculated the integrated haplotype score (iHS) in HapMap 2 data to assess evidence for recent positive selection at the index SNP. Fst and iHS were calculated using Haplotter [28]. We used varLD (A program for quantifying variation in linkage disequilibrium patterns between populations) to compare EuA vs AfA regional LD patterns. The approach quantifies genomic regions carrying dissimilar patterns of LD, regions at which different haplotypes, structural variants and signs of positive selection may be particularly common [29].
Statistical analysis to generate SNP-trait-association results
We tested SNP associations with quantitative traits in each cohort according to a uniform protocol. The primary study traits were FG and FI; in secondary analyses we also tested associations with HOMA-insulin resistance (HOMA-IR) and HOMA-beta cell function (HOMA-B) [30]. FI and HOMA were analysed on the natural log scale. Ancestry PCs were created in each study; as just the first two PCs were associated with FG (p<0.005) these were used as covariates in each study to control population stratification. Genomic control correction based on median χ2 value was calculated across all loci in the genome-wide association study (GWAS) data within each study to adjust for inflation of test statistics. Traits were analysed using linear regression under an additive genetic model, adjusted for age, age2, sex, study site (if applicable) and the first two PCs; relatedness was accounted for when necessary using linear mixed effect (LME) models. PLINK (http://pngu.mgh.harvard.edu/∼purcell/PLINK/) and R (v2.9.0) were used for data management and statistical analyses. We performed fixed-effect meta-analyses on study-specific genome-wide association results to generate a combined association effect estimate for each SNP using the inverse-variance weighted approach in METAL (a tool for meta-analysis genome-wide association scans) [31]. Nominal significance was defined as a SNP association p value < 0.05. We estimated the power of the sample to detect additive SNP-FG (in mmol/l) associations, using the detectable effect size as the 25th, 50th and 75th percentiles of the distribution of β coefficients seen in Dupuis et al [8]. At n∼6,000 and α= 4.05 × 10-5, we had low power to detect all but the largest effect sizes (electronic supplementary material [ESM] Fig. 1).
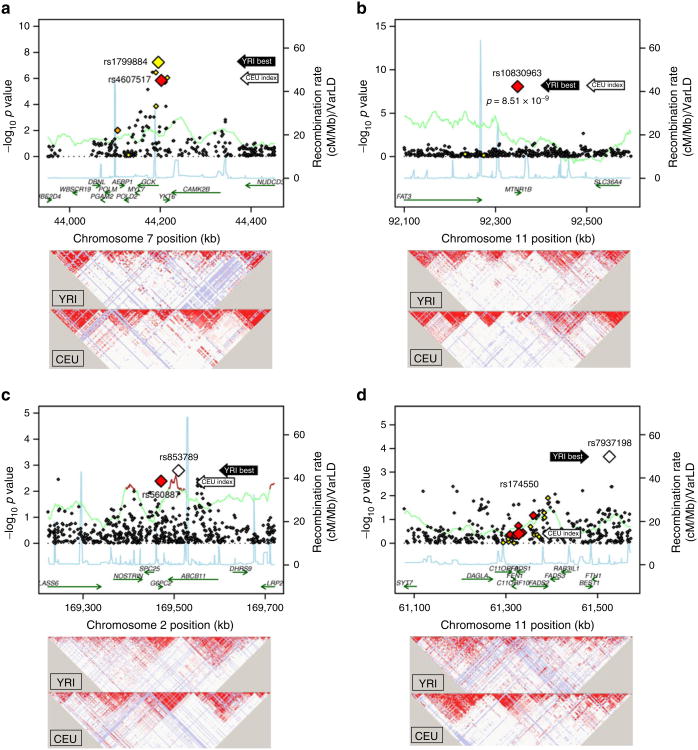
Fig. 1.
500 kb regional association plots centred at the index SNPs identified from EuA samples at GCK, MTNR1B, FADS1 and G6PC2. The x-axis denotes genomic position and the y-axis denotes the –log(p value) for each SNP. The two larger-sized data points represent the best SNPs within the region in the AfA CARe sample (black arrows) and the index SNPs (white arrows, locating the centre of the region) previously reported from EuA population samples. The colour of each data point indicates its LD value (r2) with the index SNP based on HapMap 2 YRI: white, r2=0.0–0.2; yellow, r2=0.2–0.5; orange, r2=0.5–0.8; red, r2=0.8–1.0. The blue line represents the recombination rate. The green line shows the varLD score at each SNP and is highlighted with dark brown if the varLD score is ≥95th percentile of the genome-wide varLD score, comparing LD information between YRI and CEU HapMap 2 samples [29]. (a)GCK region: the index SNP rs4607517 has a p value of 1.42 × 10-6 in AfA CARe individuals, while the best SNP rs1799884 has a p value of 5.79 × 10-8. Their r2 values are 0.469 in HapMap 2 YRI and 1.0 in HapMap 2 CEU. (b) MTNR1B region: rs10830963 is the index SNP and the best SNP within the region in AfA CARe individuals, with a p value of 8.51 × 10-9. (c) G6PC2 region: the index SNP rs560887 has a p value of 4.08 × 10-3 in AfA CARe individuals and the best SNP rs853789 has a p value of 1.62 × 10-3. The r2 value is not known in HapMap 2 YRI and is 0.692 in HapMap 2 CEU. A known functional SNP in the region, rs13431652 [38], has r2=0.923 with rs560887 and r2=0.595 with rs853789 in HapMap 2 CEU, but in HapMap 2 YRI the r2 of rs13431652 with rs853789 is 0.004. (d) FADS1 region: the index SNP rs174550 has a p value of 0.399 in AfA CARe individuals and the best SNP rs7937198 has a p value of 2.22 × 10-4. Their r2 values are 0.00 in HapMap 2 YRI and 0.24 in HapMap 2 CEU
Interrogation of transferability
We defined SNP transferability as an EuA index SNP having the same direction of association with the quantitative trait and a p value <0.05 in AfA individuals, and locus transferability as a locus containing any SNPs significantly associated with the quantitative trait in AfAs, with significance defined as any SNP with a p value <0.05 after accounting for the effective number of tests at the locus. We took two approaches to investigate transferability of EuA quantitative trait-associated SNPs and loci in AfA. First, we tested index SNPs reported to be associated with FG or FI at genome-wide significance (5 × 10-8) in EuA individuals in the MAGIC study [8]. We also tested replicated significant type 2 diabetes SNPs [32-36] that had consistent nominal associations with FG or FI in the Diabetes Genetics Replication and Meta-analysis Consortium (DIAGRAM) or MAGIC studies [7, 8]. Second, we searched for the best SNPs in AfAs accounting for differences in regional LD structure in AfAs vs those with EuA by interrogating the flanking 250 kb regions on each side of the index SNP, with the best SNP defined as the SNP with the smallest FG or FI association p value in the region in AfA individuals. Note that we treated each region independently in this follow-up study of regions that had a higher prior probability of association with the trait of interest than the genomic average, based on previously reported associations in Europeans. As the results for any one region had no inferential bearing on the results for any other region, we made region-wide (not genome-wide) corrections to adjust for multiple comparisons. For a locus, the significance of the identified best SNPs was evaluated using a Bonferroni correction adjusting for the effective number of tests within that locus, estimated using the Li and Ji algorithm [37].
Results
We analysed data from 5,984 (FG) and 5,969 (FI) non-diabetic AfA individuals from five CARe participating cohorts (Table 1). About 40% within each cohort were men and cohorts had a similar mean FG levels (range from 4.5 to 5.4 mmol/l). Mean age ranged more widely (from 24.4 to 61.7 years) across cohorts, as did mean FI (from 40.6 to 97.8 pmol/l).
Table 1. Characteristics of five CARe cohorts.
| Cohort | ARIC (n=1734) | CARDIA (n=930) | CFS (n=272) | JHS (n=1707) | MESA (n=1341) |
|---|---|---|---|---|---|
| Men, n (%) | 649 (37.4) | 362 (38.9) | 104 (38.2) | 682 (40.0) | 596 (44.4) |
| Age (years) | 53±5.8 | 24±3.9 | 36±17.7 | 49±12.1 | 62±10.3 |
| FG (mmol/l) | 5.4±0.5 | 4.5±0.48 | 5.1±0.54 | 5±0.5 | 5±0.6 |
| FI (pmol/l)a | 89.1±65.7 | 75.1±52.7 | 84.2±69.5 | 97.8±55.6 | 40.6±27.8 |
| HOMA-Bb | 160.7±110.1 | 290.6±248.3 | 187.4±193.1 | 229.9±129.7 | 95.8±65.6 |
| HOMA-IRb | 3.6±2.9 | 2.6±1.9 | 3.3±2.8 | 3.7±2.4 | 1.6±1.2 |
Data are mean±SD unless otherwise specified
1 pmol/l=6 μU/ml for insulin
Numbers of missing individuals for HOMA-B and HOMA-IR: ARIC, 9; CARDIA, 17; CFS, 1; JHS, 0; MESA, 4
Allele frequency differences and natural selection
As observed for quantitative glycaemic traits, index SNP allele-frequency differences between those of EuA and AfAs had a wide range (Table 2). Fst exceeded 0.15 at 1/18 loci in CARe but at 6/18 loci in HapMap 2, indicating modest population differentiation based on allele frequency differences at ADRA2A, CRY2, PROX1, FADS1, IGF1 and C2CD4B. The absolute values of all iHSs were <2, suggesting no recent strong positive selection at any locus. For the FADS1 and G6PC2 loci at which the index and best SNPs were weakly correlated, the varLD scores were elevated in AfAs, suggesting allelic heterogeneity of tag SNPs for causal variants or other forms of variation, such as copy number variations (Fig. 1).
Table 2. Allele frequency differences, Fsts and iHSs in AfA and EuA populations.
| Nearest gene(s) | SNP | Allelesa | EUA EAFb | AfA EAFb | CARe (EAF EuA– AfA) | p value for EAF differencec | Fst CARe | EAF HapMap 2 CEU | EAF HapMap 2 YRI | Fst HapMap 2 CEU vs YRI | his HapMap 2 CEU | his HapMap 2 YRI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FG-associated loci | ||||||||||||
| PROX1 | rs340874 | C/T | 0.520 | 0.169 | 0.351 | < 1 × 10-384 | 0.051 | 0.508 | 0.092 | 0.207 | −0.06 | −0.09 |
| G6PC2 | rs560887 | C/T | 0.700 | 0.943 | 0.243 | < 1 × 10-384 | 0.031 | 0.700 | 1.000 | − | −0.26 | − |
| GCKR | rs780094 | C/T | 0.620 | 0.817 | 0.197 | 8.78 × 10-196 | 0.017 | 0.617 | 0.883 | 0.095 | −0.39 | 0.14 |
| ADCY5 | rs11708067 | A/G | 0.780 | 0.843 | 0.063 | 3.68 × 10-29 | 0.002 | 0.783 | 0.917 | 0.035 | −0.36 | −0.79 |
| SLC2A2 | rs11920090 | T/A | 0.870 | 0.645 | 0.225 | < 1 × 10-384 | 0.040 | 0.853 | 0.667 | 0.051 | 1.17 | 0.60 |
| GCK | rs4607517 | A/G | 0.160 | 0.101 | 0.059 | 6.49 × 10-33 | 0.003 | 0.200 | 0.059 | 0.045 | −0.26 | −0.20 |
| DGKB-TMEM195 | rs2191349 | T/G | 0.520 | 0.593 | 0.073 | 1.60 × 10-26 | 0.002 | 0.467 | 0.593 | 0.016 | −0.13 | −0.07 |
| SLC30A8 | rs13266634 | C/T | 0.680 | 0.908 | 0.228 | 2.96 × 10-290 | 0.026 | 0.750 | 0.942 | 0.070 | −1.87 | −0.55 |
| GLIS3 | rs7034200 | A/C | 0.490 | 0.630 | 0.140 | 3.79 × 10-92 | 0.008 | 0.525 | 0.585 | 0.003 | 0.47 | −0.24 |
| ADRA2A | rs10885122 | G/T | 0.870 | 0.334 | 0.537 | < 1 × 10-384 | 0.191 | 0.900 | 0.217 | 0.484 | −0.23 | −0.25 |
| TCF7L2 | rs7903146 | T/C | 0.290 | 0.284 | 0.006 | 3.20 × 10-1 | 0.000 | 0.250 | 0.292 | 0.002 | 0.09 | −0.27 |
| MTNR1B | rs10830963 | G/C | 0.300 | 0.073 | 0.227 | < 1 × 10-384 | 0.027 | 0.300 | 0.050 | 0.108 | −0.05 | −0.55 |
| MADD | rs7944584 | A/T | 0.750 | 0.950 | 0.200 | 2.27 × 10 -254 | 0.024 | 0.692 | 1.000 | − | −0.31 | − |
| FADS1 | rs174550 | T/C | 0.640 | 0.916 | 0.276 | < 1 × 10-384 | 0.036 | 0.633 | 0.983 | 0.198 | −0.62 | − |
| CRY2 | rs11605924 | A/C | 0.490 | 0.867 | 0.377 | < 1 × 10-384 | 0.059 | 0.542 | 0.941 | 0.209 | −1.50 | −1.02 |
| C2CD4B | rs11071657 | A/G | 0.630 | 0.868 | 0.238 | 4.61 × 10-292 | 0.026 | 0.592 | 0.942 | 0.171 | 1.07 | −1.65 |
| FI-associated loci | ||||||||||||
| GCKR | rs780094 | C/T | 0.620 | 0.817 | 0.197 | 2.70 × 10-192 | 0.020 | 0.617 | 0.883 | 0.095 | −0.39 | 0.14 |
| IGF1 | rs35767 | G/A | 0.850 | 0.570 | 0.280 | < 1 × 10-384 | 0.061 | 0.900 | 0.525 | 0.172 | −1.290 | 0.374 |
The loci in this table were obtained from Dupuis et al (2010) [8]
Effect (trait-raising)/other allele
EAF, effect allele frequency
P-value for testing whether the effect allele frequency EAF in AfAs is the same as the effect allele frequency in EuA populations
Interrogation of EuA FG- and FI-associated loci in AfAs
Trait-raising allele frequencies of EuA index SNPs in AfAs varied widely, from a minimum of a 0.6% difference (TCF7L2) up to a 54% difference (ADRA2) between ancestry groups (Table 3). Of EuA index SNPs tested directly in AfAs, all 18 were directionally consistent; three of the 18 were nominally (p<0.05) associated with FG and one with FI. In addition, all of the published 95% CIs around point estimates for association of index SNPs in those of EuA overlapped with those tested here in AfAs. We also performed formal heterogeneity tests comparing effects in EuAs with effects in AfAs. Heterogeneity tests yielded only one nominally significant result for rs10830963 (p value = 4.26 × 10-2 at MTNR1B), indicating that the effect sizes in those of EuA vs AfAs for at least 17 index SNPs were not statistically different.
Table 3. Transferability to AfAs of FG- and FI-related loci previously reported in EuA populations.
| Nearest gene(s) | Index SNP | Chr. | Position | Allelesb | EuA association | AfA association for index SNP | Heterogeneitya | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||
| EAF | Effect ([mmol/l] per effect allele) | 95% CI | EAF | Effect ([mmol/l] per effect allele) | 95% CI | p value | p value | |||||
| FG-associated loci | ||||||||||||
| PROX1 | rs340874 | 1 | 212225879 | C/T | 0.520 | 0.013 | 0.007, 0.019 | 0.169 | 0.006 | −0.020, 0.031 | 6.63 × 10-1 | 6.00 × 10 1 |
| G6PC2 | rs560887 | 2 | 169471394 | C/T | 0.700 | 0.075 | 0.069, 0.081 | 0.943 | 0.060 | 0.019, 0.1000 | 4.08 × 10-3 | 4.80 × 10-1 |
| GCKR | rs780094 | 2 | 27594741 | C/T | 0.620 | 0.029 | 0.023, 0.035 | 0.817 | 0.022 | −0.002, 0.046 | 7.05 × 10-2 | 5.72 × 10-1 |
| ADCY5 | rs11708067 | 3 | 124548468 | A/G | 0.780 | 0.027 | 0.021, 0.033 | 0.843 | 0.015 | −0.010, 0.041 | 2.31 × 10-1 | 3.68 × 10-1 |
| SLC2A2 | rs11920090 | 3 | 172200215 | T/A | 0.870 | 0.020 | 0.012, 0.028 | 0.645 | 0.005 | −0.014, 0.025 | 5.81 × 10-1 | 1.64 × 10-1 |
| GCK | rs4607517 | 7 | 44202193 | A/G | 0.160 | 0.062 | 0.054, 0.070 | 0.101 | 0.084 | 0.050, 0.118 | 1.42 × 10-6 | 2.088 × 10-1 |
| DGKB-TMEM195 | rs2191349 | 7 | 15030834 | T/G | 0.520 | 0.030 | 0.024, 0.036 | 0.593 | 0.016 | −0.003, 0.036 | 1.02 × 10-1 | 1.80 × 10-1 |
| SLC30A8 | rs13266634 | 8 | 118253964 | C/T | 0.680 | 0.027 | 0.019, 0.035 | 0.908 | 0.004 | −0.030, 0.038 | 8.16 × 10-1 | 1.88 × 10-1 |
| GLIS3 | rs7034200 | 9 | 4279050 | A/C | 0.490 | 0.018 | 0.012, 0.024 | 0.630 | 0.018 | −0.001, 0.037 | 6.15 × 10-2 | 1.00 × 10-0 |
| ADRA2A | rs10885122 | 10 | 113032083 | G/T | 0.870 | 0.022 | 0.014, 0.030 | 0.334 | 0.006 | −0.014, 0.026 | 5.68 × 10-1 | 1.37 × 10-1 |
| TCF7L2 | rs7903146 | 10 | 114748339 | T/C | 0.290 | 0.023 | 0.015, 0.031 | 0.284 | 0.013 | −0.010, 0.035 | 2.63 × 10-1 | 3.93 × 10-1 |
| MTNR1B | rs10830963 | 11 | 92348358 | G/C | 0.300 | 0.067 | 0.061, 0.073 | 0.073 | 0.104 | 0.069, 0.139 | 8.51 × 10-9 | 4.26 × 10-2 |
| MADD | rs7944584 | 11 | 47292896 | A/T | 0.750 | 0.021 | 0.015, 0.027 | 0.950 | 0.017 | −0.024, 0.059 | 4.18 × 10-1 | 8.50 × 10-1 |
| FADS1 | rs174550 | 11 | 61328054 | T/C | 0.640 | 0.017 | 0.011, 0.023 | 0.916 | 0.014 | −0.019, 0.048 | 3.99×10-1 | 8.62×10-1 |
| CRY2 | rs11605924 | 11 | 45829667 | A/C | 0.490 | 0.015 | 0.009, 0.021 | 0.867 | 0.016 | −0.012, 0.043 | 2.71×10 1 | 9.44×10 1 |
| C2CD4B | rs11071657 | 15 | 60221254 | A/G | 0.630 | 0.008 | 0.002, 0.014 | 0.868 | 0.006 | −0.027, 0.038 | 7.37×10-1 | 9.08×10-1 |
| FI-associated loci | ||||||||||||
| GCKR | rs780094 | 2 | 27594741 | C/T | 0.620 | 0.032 | 0.024, 0.040 | 0.817 | 0.025 | −0.00, 0.053 | 7.67 ×10-2 | 6.31×10-1 |
| IGF1 | rs35767 | 12 | 101399699 | G/A | 0.850 | 0.010 | −0.002, 0.022 | 0.570 | 0.001 | −0.022, 0.023 | 9.49×10-1 | 5.02×10-1 |
The loci in this table were all obtained from Dupuis et al (2010) [8]
Heterogeneity test of the β coefficients for EuA populations and AfAs
Effect (trait-raising)/other allele
Chr chromosome; EAF effect allele frequency
Further, in the ±250 kb flanking regions around index SNPs (Table 4), nearly all (17 of 18) had a best SNP nominally significant in AfAs, and four (at GCK, MTNR1B and FADS1 for FG and GCKR for FI) had SNPs that remained significantly associated after adjusting for the effective number of SNPs tested within the regions. At 15 of 18 loci in AfAs, the index SNP and the best SNP were not in substantial LD (all r2< 0.2 in AfAs). At GCK, MTNR1B and CRY2, the index and best SNPs were in strong LD or were the same SNP. Of the best AfA SNPs, five of 18 (at G6PC2, GCKR, GCK, MTNR1B and CRY2) were also at least nominally associated (p<0.05) with FG or FI in those of EuA in the MAGIC data (ESM Table 1).
Table 4. Regional interrogation in AfAs for FG- and FI-related loci previously reported in EuA populations.
| Nearest gene(s) FG-associated loci | Index SNP in EuA | Interrogation in AfA within 500 kb region of index SNP | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||
| Best SNP in AfA | Position | LDa in YRI r2 (D′) | LDb in CEU r2 (D′) | Allelesc | EAF | Effect ([mmol/l] per effect allele) | SE | 95% CI | p value | Eff. Nd | Adjusted p value | ||
| FG-associated loci | |||||||||||||
| PROX1 | rs340874 | rs7519597 | 212261507 | 0.004 (1.000) | N/A | T/C | 0.038 | −0.0879 | 0.0276 | −0.142, −0.034 | 1.42×10-3 | 90 | 1.28×10-1 |
| G6PC2 | rs560887 | rs853789 | 169509734 | N/A | 0.692 (0.916) | A/G | 0.105 | −0.0494 | 0.0157 | −0.080, −0.019 | 1.62×10-3 | 105 | 1.70×10-1 |
| GCKR | rs780094 | rs7586601 | 27438170 | 0.169 (0.491) | 0.549 (0.956) | A/G | 0.239 | −0.0303 | 0.0111 | −0.052, −0.009 | 6.64×10-3 | 22 | 1.46×10-1 |
| ADCY5 | rs11708067 | rs10934645 | 124552510 | 0.009 (1.000) | 0.022 (1.000) | A/G | 0.091 | 0.0573 | 0.0184 | 0.021, 0.093 | 1.86×10-3 | 66 | 1.23 ×10-1 |
| SLC2A2 | rs11920090 | rs9827202 | 172296993 | 0.005 (0.105) | 0.000 (0.118) | T/C | 0.443 | 0.0274 | 0.0100 | 0.008, 0.047 | 5.86×10-3 | 65 | 3.81×10-1 |
| GCK | rs4607517 | rs1799884 | 44195593 | 0.469 (1.000) | 0.003 (0.196) | T/C | 0.177 | 0.0659 | 0.0121 | 0.042, 0.090 | 5.79×10-8 | 34 | 1.97×10-6 |
| DGKB-TMEM195 | rs2191349 | rs17327498 | 15111235 | 0.001 (0.184) | 0.003 (0.196) | A/G | 0.972 | 0.1132 | 0.0343 | 0.046, 0.180 | 9.66×10-4 | 90 | 8.69×10-2 |
| SLC30A8 | rs13266634 | rs2649098 | 118304453 | 0.008 (1.000) | 0.004 (0.088) | T/C | 0.106 | 0.0477 | 0.0153 | 0.018, 0.078 | 1.83×10-3 | 82 | 1.50×10-1 |
| GLIS3 | rs7034200 | rs3895473 | 4319209 | 0.003 (0.098) | 0.006 (0.158) | A/G | 0.800 | −0.0417 | 0.0117 | −0.065, −0.019 | 3.70×10-4 | 169 | 6.25 ×10-2 |
| ADRA2A | rs10885122 | rs7908674 | 112913361 | 0.070 (0.426) | 0.028 (0.500) | A/G | 0.159 | 0.0345 | 0.0141 | 0.007, 0.062 | 1.42×10-2 | 83 | 1.00×10-0 |
| TCF7L2 | rs7903146 | rs7909517 | 114852926 | 0.006 (0.255) | 0.003 (1.000) | C/G | 0.103 | 0.0606 | 0.0182 | 0.025, 0.096 | 8.42×10-4 | 72 | 6.06 ×10-2 |
| MTNR1B | rs10830963 | rs10830963 | 92348358 | Same SNP | Same SNP | C/G | 0.927 | −0.1038 | 0.018 | −0.139, −0.069 | 8.51×10-9 | 86 | 7.32×10-7 |
| MADD | rs7944584 | rs1685404 | 47200241 | N/A | 0.258 (1.000) | C/G | 0.241 | −0.032 | 0.0109 | −0.053, −0.011 | 3.35×10-3 | 27 | 9.03 ×10-2 |
| FADS1 | rs174550 | rs7937198 | 61525549 | 0.000 (0.016) | 0.003 (0.055) | A/G | 0.491 | −0.0341 | 0.0092 | −0.052, −0.016 | 2.22×10-4 | 65 | 1.44×10-2 |
| CRY2 | rs11605924 | rs11605924 | 45829667 | Same SNP | Same SNP | A/C | 0.867 | 0.0155 | 0.0141 | −0.012, 0.043 | 2.71×10-1 | 58 | 1.00×10-0 |
| C2CD4B | rs11071657 | rs893157 | 60165933 | 0.019 (0.488) | 0.174 (0.527) | T/C | 0.584 | −0.0264 | 0.0097 | −0.045, −0.007 | 6.30×10-3 | 60 | 3.78×10-1 |
| FI-associated loci | |||||||||||||
| GCKR | rs780094 | rs7586601 | 27438170 | 0.169 (0.491) | 0.549 (0.956) | A/G | 0.239 | −0.0452 | 0.0131 | −0.071, −0.020 | 5.87×10 4 | 22 | 1.29×10 2 |
| IGF1 | rs35767 | rs7970320 | 101508605 | 0.011 (0.126) | 0.001 (0.067) | T/C | 0.557 | −0.03 | 0.011 | −0.052, −0.008 | 6.60×10-3 | 59 | 3.89×10-1 |
The loci in this table were all obtained from Dupuis et al (2010) [8]
LD in YRI between the best SNP in AfAs and top SNP in EuA populations
LD in CEU between the best SNP in AfAs and top SNP in EuA populations
Effect (trait-raising)/other allele
Effective number of independent (typed) SNPs interrogated in CARe samples of AfAs
N/A, not available
Associations with HOMA-B (for FG loci) and with HOMA-IR (for FI loci) for both the index SNPs and the best FG or FI SNPs are shown in ESM Tables 2 and 3. Seven SNPs out of 16 EuA FG index SNPs showed nominally significant association with HOMA-B in AfAs, while three best AfA SNPs reached nominal significance. Neither of two EuA FI index SNPs were nominally significant, while the best AfA FI SNPs at both loci were nominally significant (at GCKR and IGF1).
Interrogation of FG or FI-associated type 2 diabetes loci in AfAs
Fourteen of the ∼40 known type 2 diabetes loci have also been nominally associated with FG or FI in individuals of EuA [7, 8]. Of these 14, only TP53INP1 was directionally consistent and nominally associated with FG in AfAs (Table 5). Overall, 8/14 index EuA SNPs had directionally consistent FG associations in AfAs, ten out of 13 best AfA SNPs (one SNP was monomorphic in AfA) showed 95% CIs that overlapped with EuA index SNPs in AfA and eight out of 13 best AfA SNPs did not reach nominal significance level in formal heterogeneity tests comparing those of EuA with AfAs. Note that the loci MTNR1B (p value = 1.38 × 10-6 for index SNP rs1381753) is again among the loci (THADA, ADAMTS9, HHEX and KCNQ1) with heterogeneous effect (Table 5) and its allelic effect of 0.060 is remarkably larger in the sample of EuA compared with 0.008 in the sample of AfAs. After accounting for ancestral LD by querying SNPs flanking the index SNP, all 14 best SNPs around the 14 index SNPs showed nominally significant associations in AfAs, and four of these best SNPs (at KCNQ1, MTNR1B, ADAMTS9 and CDC123/CAMK1D) remained significantly associated with FG after adjusting for multiple comparisons (Table 6 and ESM Fig. 2). At all 14 loci the best SNP was in low LD with the index SNP (all r2 <0.2). The best SNPs at ZBED3, TP53INP1, KCNQ1 and MTNR1B in AfAs were nominally associated with FG in EuA individuals in the MAGIC data (ESM Table 1).
Table 5. Association with FG in AfAs of FG-associated type 2 diabetes risk SNPs previously reported in EuA populations.
| Nearest gene(s) | Index SNPd | Chr. | Position | Allelesb | EuA FG result for index SNP | AfA FG result for index SNP | Heterogeneitya | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||||
| EAF | Effect ([mmol/l] per effect allele) | 95% CI | p value | EAF | Effect ([mmol/l] per effect allele) | 95% CI | p value | p value | |||||
| THADA | rs7578597 | 2 | 43586327 | C/T | 0.9 | 0.026 | 0.014, 0.037 | 2.40×10-5 | 0.72 | −0.003 | −0.023, 0.017 | 7.81×10-1 | 1.29×10-2 |
| ADAMTS9 | rs4607103 | 3 | 64686944 | T/C | 0.76 | −0.010 | −0.018, −0.001 | 2.64×10-2 | 0.71 | 0.028 | 0.008, 0.048 | 6.85×10-3 | 4.18×10-4 |
| IGF2BP2 | rs4402960 | 3 | 18699438 1 | T/G | 0.29 | 0.011 | 0.003, 0.018 | 8.34×10-3 | 0.51 | −0.003 | −0.021, 0.015 | 7.63×10-1 | 1.55×10-1 |
| ZBED3 | rs4457053 | 5 | 76460705 | G/A | 0.26 | 0.018 | 0.009, 0.026 | 9.15×10-5 | N/A | N/A | N/A | N/A | N/A |
| CDKAL1 | rs7754840 | 6 | 20769229 | C/G | 0.31 | 0.010 | 0.002, 0.017 | 1.41×10-2 | 0.57 | 0.008 | −0.010, 0.027 | 3.74×10-1 | 8.39×10-1 |
| TP53INP1 | rs896854 | 8 | 96029687 | T/C | 0.48 | 0.012 | 0.005, 0.019 | 8.79×10-4 | 0.69 | 0.022 | 0.002, 0.042 | 3.40×10-2 | 3.53×10-1 |
| SLC30A8 | rs1326663 4 | 8 | 11825396 4 | C/T | 0.75 | 0.027 | 0.018, 0.036 | 5.46×10-10 | 0.91 | 0.004 | −0.030, 0.038 | 8.16×10-1 | 1.88×10-1 |
| CDKN2A/B | rs1081166 1 | 9 | 22124094 | T/C | 0.79 | 0.019 | 0.009, 0.028 | 9.99×10-5 | 0.93 | 0.026 | −0.011, 0.063 | 1.68×10-1 | 7.22×10-1 |
| CDC123/CAMK1 D | rs1277979 0 | 10 | 12368016 | A/G | 0.18 | −0.016 | −0.025, −0.006 | 1.20×10-3 | 0.87 | −0.014 | −0.044, 0.015 | 3.40×10-1 | 8.99×10-1 |
| HHEX | rs1111875 | 10 | 94452862 | C/T | 0.56 | 0.009 | 0.002, 0.016 | 1.41×10-2 | 0.77 | −0.017 | −0.039, 0.004 | 1.16×10-1 | 2.63×10-2 |
| KCNQ1 | rs231362 | 11 | 2648047 | G/A | 0.52 | 0.019 | 0.011, 0.027 | 5.29×10-6 | 0.78 | −0.019 | −0.044, 0.006 | 1.46×10-1 | 5.21×10-3 |
| CENTD2c | rs1552224 | 11 | 72110746 | A/C | 0.88 | 0.019 | 0.010, 0.029 | 1.20×10-4 | 0.97 | −0.005 | −0.065, 0.055 | 8.73×10-1 | 4.45×10-1 |
| MTNR1B | rs1387153 | 11 | 92313476 | T/C | 0.28 | 0.060 | 0.052, 0.069 | 6.59×10-45 | 0.38 | 0.008 | −0.012, 0.028 | 4.38×10-1 | 1.38×10-6 |
| PRC1 | rs8042680 | 15 | 89322341 | A/C | 0.22 | 0.010 | 0.002, 0.017 | 1.30×10-2 | 0.84 | 0.019 | −0.007, 0.045 | 1.50×10-1 | 5.08×10-1 |
Heterogeneity test of the β coefficients for EuA populations and AfAs
Effect/other allele
Also known as ARAP1
Voight et al (2010) [7]; Zeggini et al (2008) [32]; Rung et al (2009) [53]; Altshuler (2000) [54]; Zeggini et al (2007) [33]; Scott et al (2007) [34], Diabetes Genetics Initiative of Broad Institute et al (2007) [55]; Sladek (2007) [36]; Gloyn (2003) [56]; Winckler et al (2007) [37]; Gudmundsson (2007) [58]
Chr., chromosome; N/A, not available
Table 6. Regional interrogation in AfAs for FG-associated type 2 diabetes loci previously reported in EuA populations.
| Nearest gene(s) | Index SNP | Interrogation for FG in AfA within 500 kb region of type 2 diabetes index SNP | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Best SNP | Position | LDa in YRI r2 (D′) | LDb in CEU r2 (D′) | Allelesc | EAF | Effect ([mmol/l] per effect allele) | 95% CI | p value | Eff. Nd | Adjusted p value | ||
| THADA | rs7578 597 | rs6724 325 | 43529 402 | 0.066 (0.322) | N/A | T/C | 0.1 89 | 0.036 | 0.012, 0.059 | 2.80×103 | 62 | 1.74×10-1 |
| ADAMTS9 | rs4607 103 | rs4688 504 | 64659 981 | 0.020 (0.394) | 0.003 (0.080) | T/C | 0.319 | 0.040 | 0.018, 0.062 | 3.82×10-4 | 125 | 4.78×10-2 |
| IGF2BP2 | rs4402960 | rs7627308 | 186919528 | 0.031 (1.000) | N/A | A/G | 0.013 | 0.166 | 0.052, 0.279 | 4.19×1-3 | 55 | 2.30×10-1 |
| ZBED3 | rs4457053 | rs12517269 | 76454133 | 0.001 (0.029) | 0.003 (0.080) | A/G | 0.883 | −0.049 | −0.077, −0.021 | 6.06×10-4 | 85 | 5.15×10-2 |
| CDKAL1 | rs7754840 | rs9368255 | 21008207 | 0.002 (0.250) | 0.019 (1.000) | T/C | 0.949 | 0.057 | 0.015, 0.099 | 7.88×10-3 | 72 | 5.67×10-1 |
| TP53INP1 | rs896854 | rs509594 | 96036332 | 0.069 (1.000) | 0.737 (1.000) | C/G | 0.795 | −0.035 | −0.058, −0.012 | 2.54×10-3 | 61 | 1.55×10-1 |
| SLC30A8 | rs13266634 | rs2649098 | 118304453 | 0.008 (1.000) | 0.004 (0.088) | T/C | 0.1 06 | 0.048 | 0.018, 0.078 | 1.83×10-3 | 82 | 1.50×10-1 |
| CDKN2A/B | rs10811661 | rs10965319 | 22293465 | 0.092 (1.000) | 0.008 (0.216) | A/T | 0.8 40 | −0.041 | −0.068, −0.014 | 2.86×10-3 | 72 | 2.06×10-1 |
| CDC123/CA MK1D | rs12779790 | rs7074893 | 12446905 | 0.000 (1.000) | 0.001 (0.068) | T/G | 0.9 77 | −0.209 | −0.312, −0.105 | 8.29×10-5 | 89 | 7.38×10-3 |
| HHEX | rs1111875 | rs11187186 | 94576008 | 0.000 (0.017) | 0.000 (0.017) | T/C | 0.457 | −0.029 | −0.048, −0.010 | 2.24×10-3 | 42 | 9.41×10-2 |
| KCNQ1 | rs231362 | rs2011766 | 2490838 | 0.000 (0.033) | 0.001 (0.085) | T/C | 0.3 32 | −0.044 | −0.068, −0.021 | 2.02×10-4 | 91 | 1.84×10-2 |
| CENTD2e | rs1552224 | rs6592481 | 72067275 | N/A | 0.001 (0.273) | T/C | 0.220 | 0.038 | 0.012, 0.064 | 4.79×10-3 | 51 | 2.44×10-1 |
| MTNR1B | rs1387153 | rs10830963 | 92348358 | 0.036 (0.710) | 0.705 (0.874) | C/G | 0.927 | −0.104 | −0.139, −0.069 | 8.51×10-9 | 87 | 7.40×10-7 |
| PRC1 | rs8042680 | rs10083635 | 89195519 | 0.006 (1.000) | N/A | A/G | 0.145 | 0.060 | 0.025, 0.094 | 6.53×10-4 | 98 | 6.40×10-2 |
Index SNP data are from Voight et al (2010) [7]; Zeggini et al (2008) [32]; Rung et al (2009) [53]; Altshuler (2000) [54]; Zeggini et al (2007) [33]; Scott et al (2007) [34]; Diabetes Genetics Initiative of Broad Institute et al (2007) [55]; Sladek (2007) [36]; Gloyn (2003) [56]; Winckler et al (2007) [57]; Gudmundsson (2007) [58]
LD in YRI between the best SNP in AfAs and top SNP in EuA populations
LD in CEU between the best SNP in AfAs and top SNP in EuA populations
Effect/other allele
Effective number of independent (typed) SNPs interrogated in CARe samples of AfA
Also known as ARAP1
N/A, not available
Discussion
We found that among 18 index SNPs robustly associated with glycaemic quantitative traits in the MAGIC EuA populations, just three SNPs (at GCK, MTNR1B and G6PC2 for FG) were directly transferrable to the ∼6,000 AfA individuals from five separate cohorts in CARe, but all 18 published 95% CIs around association-point estimates in MAGIC overlapped with the CIs based on associations in AfAs. However, after accounting for ancestral differences in LD by searching more broadly in regions flanking the EuA index SNP, 17 of 18 loci were nominally associated with FG or FI in AfAs, and SNPs at four of these loci (GCK, MTNR1B and FADS1 for FG and GCKR for FI) remained significantly associated with FG or FI in AfAs after accounting for multiple comparisons. For FG-associated type 2 diabetes SNPs discovered in EuA populations, only one (TP53INP1) was directly transferable to AfAs, but ten of 13 published 95% CIs overlapped with the CIs based on associations in AfA. These overlapping intervals indicate that the trait-raising allelic effects for glycaemic loci are similar in individuals of EuA and AfAs. Four FG-associated type 2 diabetes loci (ADAMTS9, CDC123/CAMK1D, KCNQ1 and MTNR1B) had a best AfA SNP significantly associated with FG in AfAs after adjusting for multiple comparisons. As expected, allele frequencies and regional LD varied widely: there was evidence of modest population differentiation for six index SNPs (including FADS1), but none showed evidence of recent positive selection. These results provide evidence that some genetic loci associated with glycaemic regulation are reproducibly shared across human populations, with a suggestion of potential transferability for most loci, at least for individuals of European or African ancestry. The data also provide evidence for allelic heterogeneity at many loci and evidence for shared haplotypes with proximity of an index SNP with a functional variant at several loci.
These results illustrate the value of examining AfA cohorts for fine-mapping. Where there is strong LD between the index SNP and the best SNP, association with FG in an ancestral group with reduced LD increases confidence that the index signal is on the same haplotype or is otherwise close to a functional mutation (GCK, MTNR1B and CRY2). For instance, at MTNR1B, the FG–type 2 diabetes signal is the same for both those of EuA and AfAs (rs10830963). Where there is weak LD between the index SNP and the best SNP (all other loci in AfAs), there are two alternative explanations: [1] these SNPs represent independent signals where the index and best SNPs were significant in only one population (population-specific allelic heterogeneity); or [2] they share LD with a tagged untyped functional SNP in the region where the differential LD pattern between those of EuA and AfAs can focus fine-mapping of the association signal. For instance, at G6PC2 in EuA the FG index SNP (rs560887) and best SNP (rs853789) are in strong LD with a known functional SNP (rs13431652; r2 with rs560887 in CEU = 0.923, r2 with rs853789 in CEU = 0.595) [38], but AfA rs853789 is not correlated with rs13431652 (r2 in YRI = 0.004). KCNQ1 also demonstrates multiple independent signals identified in different ancestry groups [7, 39, 40]. Variation in LD structure across these ancestral groups has focused regions of interest and identified new signals that may aid future fine-mapping to identify functional variants involved in FG and FI regulation.
Just one paper has investigated the transferability of EuA diabetes-related quantitative trait loci in AfAs. Ramos et al examined the index and correlated SNPs (r2 ≥ 0.3 in CEU) for 16 fasting glucose loci reported by MAGIC in 927 AfAs [19], demonstrating associations of index and/or correlated SNPs at TMEM195 (also known as AGMO), SLC30A8, TCF7L2, G6PC2, GCKR and MTNR1B. This is consistent with our index SNP association results at G6PC2 and MTNR1B and regional association at GCKR. In addition, the direction and magnitude of effect of the T allele in rs2191349 at TMEM195 were comparable (β = 0.014 [Ramos] vs 0.016 [this study]), despite not reaching significance in this study. Taken together with the significant association of AfA best SNPs at GCKR, GCK and MTNR1B with FG or FI in EuA populations in MAGIC, the loci GCKR, GCK and MTNR1B appear likely to share the same causal variant(s) in populations with EuA and AfAs. Differences in LD such as those found at GCK and GCKR between those with EuA and AfAs also facilitate the fine-mapping of causal variants in these loci. The associations of independent SNPs at G6PC2 and FADS1 in EuA populations and AfAs probably point to distinct signals requiring further study.
Our data add to the list of glycaemic loci confirmed and informatively fine-mapped by cross-ancestry analyses [41-43]. For instance, at KCNQ1, studies in South and East Asian populations identified the type-2-diabetes-associated SNPs rs2237892, rs2237895 and rs2237897 [39, 40], weakly correlated with the signal seen in EuA populations (rs231362) [7]. Here, we add an FG-associated signal in AfAs (rs2011766), bringing to five the total number of apparently independent signals at KCNQ1 associated with glycaemia in humans. SNP rs2011766 is intronic and is in very weak LD with the other SNPs in the 1000 Genomes Pilot 1 reference panel: rs231362 vs rs2011766, r2 = 0.06; rs2011766 vs rs2237895 r2 = 0.03; and rs2011766 vs rs2237892 r2 = 0.02. However, rs2011766 does not appear to be a type-2-diabetes-risk locus, with an odds ratio for type 2 diabetes in the DIAGRAM discovery meta-analysis of 1.04 (p=0.19) [7]. Multi-ethnic allelic heterogeneity in association with FG or type 2 diabetes has also been reported for the C2CD4A/B locus [44]. Other loci at which cross-ancestry fine-mapping has confirmed the index SNP to be the same in EuA populations and AfAs are TCF7L2 [10, 13, 45, 46], PPARG, IGF2BP2, JAZF1 [12] and FTO [47].
Type 2 diabetes disproportionately affects AfAs. This differential risk may be due partly to genetic factors [5]. Most AfAs have differential mixed ancestry from Western Africa and Europe [48]. AfA genetic architectures are substantially different and generally have lower LD compared with other US populations [5], which is theoretically able to assist fine-mapping of causal variants. Our observation of large differences in risk-allele frequency and high Fst between AfAs and EuA populations at some SNPs is consistent with studies of other complex-disease-associated SNPs [4, 49]. Differences in risk-allele frequencies or in ability to increase FG (ancestral vs derived allele) did not seem to have strong impact on the association as supported by many of the loci showing transferability with similar effect sizes in AfAs [16]; of 18 glycaemic loci, only SNPs at FADS1 had evidence of population differentiation and significant association with FG at weakly correlated EuA and AfA signals. A limitation of our study is that lower risk-allele frequencies at some loci may be due to insufficient study power. In addition, lack of transferability may arise from the confounding effects of social and environmental risk exposures that vary across populations and that have not been considered here [50-52].
Conclusions
Cross-ancestry studies can show new population-specific risk variants and confirm shared risk variants. Allele frequencies and LD variation influence power and the ability to resolve the signal to a smaller region containing possible functional variants. In the AfA CARe cohort of ∼6,000 person, there was substantial allele frequency variation but modest EuA index SNP transferability. However, most glycaemia-associated loci showed at least nominal evidence of similar association in AfA and EuA groups after accounting for LD differences. Four FG loci (G6PC2, GCK, MTNR1B and FADS1) and three type 2 diabetes-FG loci (KCNQ1, ADAMTS9 and CDC123/CAMK1D) were convincingly transferable from EuA populations to AfAs. Cross-ancestry analyses that account for and leverage varying LD across ancestral groups can identify signals that may aid fine-mapping. In the near future one can expect that new or confirmatory variants associated with type 2 diabetes and diabetes-related quantitative traits are likely to be revealed by more extensive sequencing and genome-wide analysis ongoing in other ancestral population groups, further illuminating the genetic architecture of type 2 diabetes risk and glycaemic regulation in all humans.
Supplementary Material
Acknowledgments
The authors wish to acknowledge the support of the NHLBI and the contributions of the research institutions, study investigators and field staff involved, and the study participants of ARIC, CARDIA, CFS, JHS and MESA in creating the CARe for biomedical research (CARe; http://public.nhlbi.nih.gov/GeneticsGenomics/home/care.aspx). The following five parent studies have contributed parent study data, ancillary study data and DNA samples through the Massachusetts Institute of Technology - Broad Institute (N01-HC-65226) to create this genotype/phenotype database for wide dissemination to the biomedical research community: ARIC; CFS; JHS; CARDIA; and MESA.
Funding: This work was supported by NIDDK R01 2 DK078616 and NIDDK K24 DK080140 to J. B. Meigs, a Doris Duke Charitable Foundation Clinical Scientist Development Award to J. C. Florez, RO1 DK066358 to D. W. Bowden, 5RC1HL099911-02 5UC2HL103010-02 and the NHLBI MESA contract N01HC95165-21-0-1 to S. S. Rich.
ARIC is carried out as a collaborative study supported by NHBLI contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, and N01-HC-55022. CFS: Case Western Reserve University (NIH HL 46380, M01RR00080).
JHS is carried out as a collaborative study supported by Jackson State University (N01-HC-95170), University of Mississippi (N01-HC-95171), Tougaloo College (N01-HC-95172);
MESA is conducted and supported by the NHLBI in collaboration with MESA investigators. Support is provided by grants and contracts N01-HD-95159 through N01-HC-95169, RR-024156, and R01-HL-071205.
CARDIA is carried out as a collaborative study supported by University of Alabama at Birmingham (N01-HC-48047), University of Minnesota (N01-HC-48048), Northwestern University (N01-HC-48049), Kaiser Foundation Research Institute (N01-HC-48050), University of Alabama at Birmingham (N01-HC-95095), Tufts-New England Medical Center (N01-HC-45204), Wake Forest University (N01-HC-45205), Harbor-UCLA Research and Education Institute (N01-HC-05187), University of California Irvine (N01-HC-45134, N01-HC-95100).
Abbreviations
- AfA
African-American
- ARIC
Atherosclerosis Risk in Communities
- CARDIA
Coronary Artery Risk Development in young Adults
- CARe
Candidate Gene Association Resource
- CEU
HapMap 2 reference panel of Utah residents of northern and western European ancestry
- CFS
Cleveland Family Study
- CVD
Cardiovascular disease
- DIAGRAM
The Diabetes Genetics Replication And Meta-analysis Consortium
- EuA
European ancestry
- FG
Fasting glucose
- FI
Fasting insulin
- Fst
Fixation index
- HOMA-B
HOMA-beta cell function
- HOMA-IR
HOMA- insulin resistance
- iHS
Integrated haplotype score
- JHS
Jackson Heart Study
- LD
Linkage disequilibrium
- MAGIC
The Meta-Analyses of Glucose and Insulin-related traits Consortium
- MESA
Multi-Ethnic Study of Atherosclerosis
- MSA
Metropolitan statistical area
- NHLBI
National Heart, Lung, and Blood Institute
- SNP
Single-nucleotide polymorphism
- varLD
A program for quantifying variation in linkage disequilibrium patterns between populations
- YRI
HapMap 2 reference panel of residents of Yoruba in Ibadan, Nigeria
Footnotes
Electronic supplementary material: The online version of this article (doi:10.1007/s00125-012-2656-4) contains peer-reviewed but unedited supplementary material, which is available to authorised users.
Contribution statement: The experiments were conceived and designed by SR, WHK, DS, JCF, JIR, JGW, DWB and JBM. C-TL, DR, JD, MCYN, SRP, WHK and KLL analysed the data. The paper was written by: C-TL, MCYN, JCF, JD, and JBM. SR, WHK, DS, JCF, JIR, JGW, DWB and JBM recruited individuals. Analysis and interpretation of data and critical review of manuscript were performed by C-TL, MCYN, DR, AA, SJB, EB, IB, BC, YDIC, LD, MF, MOG, SFAG, XG, TH, EK, JRK, YL, KLL, KM, JAN, JSP, SRP, ER, LR-T, SSR, CNR, DS, DS, MS, JMZ, SR, WHK, DS, JCF, JIR, JD, JGW, DWB and JBM.
JBM and C-TL are the guarantors of this work, had full access to all the data and take full responsibility for the integrity of data and the accuracy of data analysis. All authors have approved the final version of manuscript.
Duality of interest: The authors declare that there is no duality of interest associated with this manuscript.
Contributor Information
C.-T. Liu, Department of Biostatistics, Boston University School of Public Health, Boston, MA, USA
M. C. Y. Ng, Center for Genomics and Personalized Medicine Research, Center for Diabetes Research, Wake Forest University School of Medicine, Winston-Salem, NC, USA
D. Rybin, Boston University Data Coordinating Center, Boston, MA, USA
A. Adeyemo, National Human Genome Research Institute, Bethesda, MD, USA
S. J. Bielinski, Mayo Clinic, Rochester, MN, USA
E. Boerwinkle, University of Texas Health Science Center at Houston, Houston, TX, USA
I. Borecki, Washington University, St Louis, MO, USA
B. Cade, Brigham and Women's Hospital, Boston, MA, USA
Y. D. I. Chen, Cedars Sinai Medical Center, Los Angeles, CA, USA
L. Djousse, Email: jmeigs@partners.org, Brigham and Women's Hospital, Boston, MA, USA; Department of Medicine, Harvard Medical School, Boston, MA, USA; Boston VA Healthcare System, Boston, MA, USA;.
M. Fornage, University of Texas Health Science Center at Houston, Houston, TX, USA
M. O. Goodarzi, Cedars Sinai Medical Center, Los Angeles, CA, USA
S. F. A. Grant, Children's Hospital of Philadelphia, Philadelphia, PA, USA
X. Guo, Cedars Sinai Medical Center, Los Angeles, CA, USA
T. Harris, National Institute on Aging, Bethesda, MD, USA
E. Kabagambe, University of Alabama, Birmingham, AL, USA
J. R. Kizer, Cornell University, New York, NY, USA
Y. Liu, Center for Genomics and Personalized Medicine Research, Center for Diabetes Research, Wake Forest University School of Medicine, Winston-Salem, NC, USA; Department of Epidemiology and Prevention, Wake Forest University, Winston-Salem, North Carolina, USA
K. L. Lunetta, Department of Biostatistics, Boston University School of Public Health, Boston, MA, USA; National Heart, Lung, and Blood Institute' Framingham Heart Study, Framingham, MA, USA
K. Mukamal, Department of Medicine, Harvard Medical School, Boston, MA, USA
J. A. Nettleton, University of Texas Health Science Center at Houston, Houston, TX, USA
J. S. Pankow, University of Minnesota, Minneapolis, MN, USA
S. R. Patel, Brigham and Women's Hospital, Boston, MA, USA
E. Ramos, National Human Genome Research Institute, Bethesda, MD, USA
L. Rasmussen-Torvik, Northwestern University, Chicago, IL, USA
S. S. Rich, University of Virginia, Charlottesville, VA, USA
C. N. Rotimi, National Human Genome Research Institute, Bethesda, MD, USA
D. Sarpong, Jackson State University, Jackson, MS, USA
D. Shriner, National Human Genome Research Institute, Bethesda, MD, USA
M. Sims, University of Mississippi Medical Center, Jackson, MS, USA
J. M. Zmuda, University of Pittsburgh, Graduate School of Public Health, Pittsburgh, PA, USA
S. Redline, Brigham and Women's Hospital, Boston, MA, USA
W. H. Kao, Johns Hopkins University, Baltimore, MD, USA
D. Siscovick, University of Washington, Seattle, WA, USA
J. C. Florez, Department of Medicine, Harvard Medical School, Boston, MA, USA; Diabetes Unit and Center for Human Genetic Research, Massachusetts General Hospital, Boston, MA, USA; Program in Medical and Population Genetics, Broad Institute, Cambridge, MA, USA
J. I. Rotter, Cedars Sinai Medical Center, Los Angeles, CA, USA
J. Dupuis, Department of Biostatistics, Boston University School of Public Health, Boston, MA, USA; National Heart, Lung, and Blood Institute's Framingham Heart Study, Framingham, MA, USA
J. G. Wilson, University of Mississippi Medical Center, Jackson, MS, USA
D. W. Bowden, Center for Genomics and Personalized Medicine Research, Center for Diabetes Research, Wake Forest University School of Medicine, Winston-Salem, NC, USA; Departments of Biochemistry and Internal Medicine, Wake Forest University School of Medicine, Winston-Salem, NC, USA
J. B. Meigs, Department of Medicine, Harvard Medical School, Boston, MA, USA; General Medicine Division, Massachusetts General Hospital, 50 Staniford Street, 9th Flr, Boston, MA, USA
References
- 1.Cowie CC, Rust KF, Ford ES, et al. Full accounting of diabetes and pre-diabetes in the U.S. population in 1988-1994 and 2005-2006. Diabetes Care. 2009;32:287–394. doi: 10.2337/dc08-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mokdad AH, Bowman BA, Ford ES, et al. The continuing epidemics of obesity and diabetes in the United States. JAMA. 2001;286:1195–1200. doi: 10.1001/jama.286.10.1195. [DOI] [PubMed] [Google Scholar]
- 3.Mokdad AH, Ford ES, Bowman BA, et al. Prevalence of obesity, diabetes, and obesity-related health risk factors, 2001. JAMA. 2003;289:76–79. doi: 10.1001/jama.289.1.76. [DOI] [PubMed] [Google Scholar]
- 4.Myles S, Davison D, Barrett J, et al. Worldwide population differentiation at disease-associated SNPs. BMC Med Genome. 2008;1:22. doi: 10.1186/1755-8794-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guthery SL, Salisbury BA, Pungliya MS, et al. The structure of common genetic variation in United States populations. Am J Hum Genet. 2007;81:1221–1231. doi: 10.1086/522239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adeyemo A, Rotimi C. Genetic variants associated with complex human diseases show wide variation across multiple populations. Pub Health Genome. 2010;13:72–79. doi: 10.1159/000218711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voight BF, Scott LJ, Steinthorsdottir V, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Q, Liu T, Shrader P, Yesupriya A, et al. Racial/ethnic differences in association of fasting glucose-associated genomic loci with fasting glucose, HOMA-B, and impaired fasting glucose in the U.S. adult population. Diabetes Care. 2010;33:2370–2377. doi: 10.2337/dc10-0898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmer ND, Hester JM, An SS, et al. Resequencing and analysis of variation in the TCF7L2 gene in African Americans suggests that SNP rs7903146 is the causal diabetes susceptibility variant. Diabetes. 2011;60:662–668. doi: 10.2337/db10-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casto AM, Feldman MW, Casto AM, et al. Does selection affect unlinked SNPs with shared trait associations? PLoS Genet. 2011;7:e1001266. doi: 10.1371/journal.pgen.1001266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waters KM, Stram DO, Hassanein MT, et al. Consistent association of type 2 diabetes risk variants found in Europeans in diverse racial and ethnic groups. PLoS Genet. 2010;6:e1001078. doi: 10.1371/journal.pgen.1001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis JP, Palmer ND, Hicks PJ, et al. Association analysis in African Americans of European-derived type 2 diabetes single nucleotide polymorphisms from whole-genome association studies. Diabetes. 2008;57:2220–2225. doi: 10.2337/db07-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sale MM, Smith SG, Mychaleckyj JC, et al. Variants of the transcription factor 7-like 2 (TCF7L2) gene are associated with type 2 diabetes in an African-American population enriched for nephropathy. Diabetes. 2007;56:2638–2642. doi: 10.2337/db07-0012. [DOI] [PubMed] [Google Scholar]
- 15.Lettre G, Palmer CD, Young T, et al. Genome-wide association study of coronary heart disease and its risk factors in 8,090 African Americans: the NHLBI CARe Project. PLoS Genet. 2011;7:e1001300. doi: 10.1371/journal.pgen.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ioannidis JP, Ntzani EE, Trikalinos TA, et al. ‘Racial’ differences in genetic effects for complex diseases. Nat Genet. 2004;36:1312–1318. doi: 10.1038/ng1474. [DOI] [PubMed] [Google Scholar]
- 17.Wilson PW, Meigs JB, Sullivan L, et al. Prediction of incident diabetes mellitus in middle-aged adults: the Framingham Offspring Study. Arch Int Med. 2007;167:1068–1074. doi: 10.1001/archinte.167.10.1068. [DOI] [PubMed] [Google Scholar]
- 18.Rasmussen-Torvik LJ, Alonso A, Li M, et al. Impact of repeated measures and sample selection on genome-wide association studies of fasting glucose. Genet Epidemiol. 2010;34:665–673. doi: 10.1002/gepi.20525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramos E, Chen G, Shriner D, et al. Replication of genome-wide association studies (GWAS) loci for fasting plasma glucose in African-Americans. Diabetologia. 2011;54:783–788. doi: 10.1007/s00125-010-2002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The ARIC Investigators. The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. Am J Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 21.Friedman GD, Cutter GR, Donahue RP, et al. CARDIA: study design, recruitment, and some characteristics of the examined subjects. J Clin Epidemiol. 1988;41:1105–1116. doi: 10.1016/0895-4356(88)90080-7. [DOI] [PubMed] [Google Scholar]
- 22.Buxbaum SG, Elston RC, Tishler PV, et al. Genetics of the apnea hypopnea index in Caucasians and African Americans: I. Segregation analysis. Genet Epidemiol. 2002;22:243–253. doi: 10.1002/gepi.0170. [DOI] [PubMed] [Google Scholar]
- 23.Taylor HA, Jr, Wilson JG, Jones DW, et al. Toward resolution of cardiovascular health disparities in African Americans: design and methods of the Jackson Heart Study. Ethn Dis. 2005;15(S6):4–17. [PubMed] [Google Scholar]
- 24.Fuqua SR, Wyatt SB, Andrew ME, et al. Recruiting African-American research participation in the Jackson Heart Study: methods, response rates, and sample description. Ethn Dis. 2005;15(S6):18–29. [PubMed] [Google Scholar]
- 25.Wilson JG, Rotimi CN, Ekunwe L, et al. Study design for genetic analysis in the Jackson Heart Study. Ethn Dis. 2005;15(S6):30–37. [PubMed] [Google Scholar]
- 26.Taylor HA., Jr The Jackson Heart Study: an overview. Ethn Dis. 2002;15(S6):1–3. [PubMed] [Google Scholar]
- 27.Bild DE, Bluemke DA, Burke GL, et al. Multi-ethnic study of atherosclerosis: objectives and design. Am J Epidemiol. 2002;156:871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 28.Voight BF, Kudaravalli S, Wen X, et al. A map of recent positive selection in the human genome. Plos Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ong RT, Teo YY, Ong RTH, et al. varLD: a program for quantifying variation in linkage disequilibrium patterns between populations. Bioinformatics. 2010;26:1269–1270. doi: 10.1093/bioinformatics/btq125. [DOI] [PubMed] [Google Scholar]
- 30.Matthews DR, Hosker JP, Rudenski AS, et al. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 31.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeggini E, Scott LJ, Saxena R, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 36.Sladek R, Rocheleau G, Rung J, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 37.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 38.Bouatia-Naji N, Bonnefond A, Baerenwald DA, et al. Genetic and functional assessment of the role of the rs13431652-A and rs573225-A alleles in the G6PC2 promoter that are strongly associated with elevated fasting glucose levels. Diabetes. 2010;59:2662–2671. doi: 10.2337/db10-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yasuda K, Miyake K, Horikawa Y, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet. 2008;40:1092–1097. doi: 10.1038/ng.207. [DOI] [PubMed] [Google Scholar]
- 40.Unoki H, Takahashi A, Kawaguchi T, et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet. 2008;40:1098–1102. doi: 10.1038/ng.208. [DOI] [PubMed] [Google Scholar]
- 41.Sim X, Ong RT, Suo C, et al. Transferability of type 2 diabetes implicated loci in multi-ethnic cohorts from Southeast Asia. PLoS Genet. 2011;7:e1001363. doi: 10.1371/journal.pgen.1001363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu C, Zhang R, Wang C, et al. Variants from GIPR, TCF7L2, DGKB, MADD, CRY2, GLIS3, PROX1, SLC30A8 and IGF1 are associated with glucose metabolism in the Chinese. PLoS One. 2010;5:e15542. doi: 10.1371/journal.pone.0015542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ng MC, Tam CH, So WY, et al. Implication of genetic variants near NEGR1, SEC16B, TMEM18, ETV5/DGKG, GNPDA2, LIN7C/BDNF, MTCH2, BCDIN3D/FAIM2, SH2B1, FTO, MC4R, and KCTD15 with obesity and type 2 diabetes in 7705 Chinese. J Clin Endocrinol Metab. 2010;95:2418–2425. doi: 10.1210/jc.2009-2077. [DOI] [PubMed] [Google Scholar]
- 44.Yamauchi T, Hara K, Maeda S, et al. A genome-wide association study in the Japanese population identifies susceptibility loci for type 2 diabetes at UBE2E2 and C2CD4A-C2CD4B. Nat Genet. 2010;42:864–868. doi: 10.1038/ng.660. [DOI] [PubMed] [Google Scholar]
- 45.Yan Y, North KE, Ballantyne CM, et al. Transcription factor 7-like 2 (TCF7L2) polymorphism and context-specific risk of type 2 diabetes in African American and Caucasian adults: the Atherosclerosis Risk in Communities study. Diabetes. 2009;58:285–289. doi: 10.2337/db08-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dabelea D, Dolan LM, D'Agostino R, Jr, et al. Association testing of TCF7L2 polymorphisms with type 2 diabetes in multi-ethnic youth. Diabetologia. 2011;54:535–539. doi: 10.1007/s00125-010-1982-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grant SF, Li M, Bradfield JP, et al. Association analysis of the FTO gene with obesity in children of Caucasian and African ancestry reveals a common tagging SNP. PLoS One. 2008;3:e1746. doi: 10.1371/journal.pone.0001746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tishkoff SA, Reed FA, Friedlaender FR, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Myles S, Tang K, Somel M, et al. Identification and analysis of genomic regions with large between-population differentiation in humans. Ann Hum Genet. 2008;72:99–110. doi: 10.1111/j.1469-1809.2007.00390.x. [DOI] [PubMed] [Google Scholar]
- 50.Martinez-Marignac VL, Valladares A, Cameron E, et al. Admixture in Mexico City: implications for admixture mapping of type 2 diabetes genetic risk factors. Hum Genet. 2007;120:807–819. doi: 10.1007/s00439-006-0273-3. [DOI] [PubMed] [Google Scholar]
- 51.Florez JC, Price AL, Campbell D, et al. Strong association of socioeconomic status with genetic ancestry in Latinos: implications for admixture studies of type 2 diabetes. Diabetologia. 2009;52:1528–1536. doi: 10.1007/s00125-009-1412-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCarthy MI. Casting a wider net for diabetes susceptibility genes. Nat Genet. 2008;40:1039–1040. doi: 10.1038/ng0908-1039. [DOI] [PubMed] [Google Scholar]
- 53.Rung J, Cauchi S, Albrechtsen A, et al. Genetic variant near IRS1 is associated with type 2 diabetes, insulin resistance and hyperinsulinemia. Nat Genet. 2009;41:1110–1115. doi: 10.1038/ng.443. [DOI] [PubMed] [Google Scholar]
- 54.Altshuler D, Hirschhorn JN, Klannemark M, et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 55.Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 56.Gloyn AL, Weedon MN, Owen KR, et al. Large-scale association studies of variants in genes encoding the pancreatic beta-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) confirm that the KCNJ11 E23K variant is associated with type 2 diabetes. Diabetes. 2003;52:568–572. doi: 10.2337/diabetes.52.2.568. [DOI] [PubMed] [Google Scholar]
- 57.Winckler W, Weedon MN, Graham RR, et al. Evaluation of common variants in the six known maturity-onset diabetes of the young (MODY) genes for association with type 2 diabetes. Diabetes. 2007;56:685–693. doi: 10.2337/db06-0202. [DOI] [PubMed] [Google Scholar]
- 58.Gudmundsson J, Sulem P, Steinthorsdottir V, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.