Abstract
The adaptive immune system (AIS) in mammals, which is centred on lymphocytes bearing antigen receptors that are generated by somatic recombination, arose approximately 500 million years ago in jawed fish. This intricate defence system consists of many molecules, mechanisms and tissues that are not present in jawless vertebrates. Two macroevolutionary events are believed to have contributed to the genesis of the AIS: the emergence of the recombination-activating gene (RAG) transposon, and two rounds of whole-genome duplication. It has recently been discovered that a non-RAG-based AIS with similarities to the jawed vertebrate AIS — including two lymphoid cell lineages — arose in jawless fish by convergent evolution. We offer insights into the latest advances in this field and speculate on the selective pressures that led to the emergence and maintenance of the AIS.
The adaptive immune system (AIS) is fascinating to both scientists and laymen: we have a specific yet incredibly diverse system that can fight myriad pathogens and has a ‘memory’ — the basis of vaccination — that enables a rapid response to previously encountered pathogens. The complexity of immune response regulation rivals that of the nervous system in terms of the connections forged and suppressed between cells, but immune cells must also traverse the body through blood, lymph and tissue until they encounter invading organisms. How did such a system arise, and can studies of non-mammalian vertebrates help us to understand the immunity gestalt?
Antibodies were discovered over 100 years ago, and major questions relating to the generation of diversity were solved in the 1970s with the detection of somatic hypermutation1 and variable–diversity–joining rearrangement (VDJ rearrangement)2 of antibody (or immunoglobulin (Ig) or B cell receptor (BCR)) genes. In the 1980s, T cell receptors (TCRs) were discovered and there was universal agreement that they shared a common ancestor with BCR genes, based on their similar domain organization and reliance on the same rearrangement mechanism to generate diversity3. After the discovery of enzymes that are involved in the rearrangement of BCR and TCR genes4 and of hypermutation of BCR genes5, attention shifted to asking how a system that is capable of generating such diversity evolved. Jawed fish were found to have almost all of these genes and mechanisms, but jawless fish (Agnathans) apparently had none. This mystifying finding led to the ‘big bang’ theory of AIS emergence6, which is one of the main topics of this Review.
The discovery in jawless fish of a lymphoid cell-based system of adaptive immunity that is strikingly similar to the system in jawed fish was a total surprise7. The system in jawless fish is similar to VDJ rearrangement but has rearranging receptors that are encoded by another gene family — the variable lymphocyte receptor (VLR) family. We discuss the compelling possibility that the VLR rearrangement process is under the control of the same family of enzymes that are responsible for somatic hypermutation of BCR genes8, which pushes the origins of acquired immunity back to the ancestor of all vertebrates. However, despite this new finding of adaptive immunity in jawless fish, the ‘big bang’ theory concerning the origin and rapid development of the highly complex BCR- and TCR-based AIS remains intact.
In this Review, we describe the basic features of BCRs, TCRs and the major histocompatibility complex (MHC) and stress their preservation and modification over evolutionary time. We then discuss the two catastrophic, innovative events behind the ‘big bang’ origins of the BCR–TCR–MHC-based AIS — the recombinationactivating gene (RAG) transposon invasion and genomewide duplications early in vertebrate history — and we discuss the new type of AIS that has been discovered in jawless fish. Finally, we speculate on the origins of adaptive immunity in all vertebrates, emphasizing the lessons we have learned from extant immune systems.
The AIS of jawed vertebrates
The most evolutionarily ancient extant organisms in which the AIS, as defined in humans, is found are the cartilaginous fish. It is believed to have arisen in the first jawed vertebrates (gnathostomes) — the placoderms (FIG. 1). Components of the innate immune system, for example, pattern-recognition receptors (PRRs) — including the Toll-like receptors (TLRs), nod-like receptors (nLRs) and scavenger receptors (SRs) — are found throughout the animal kingdom (FIG. 1). In some invertebrates, such as the sea urchin9, and in many plants10 there has been a great expansion of PRR families, which suggests that in the absence of an AIS, complex innate mechanisms might be required for defence.
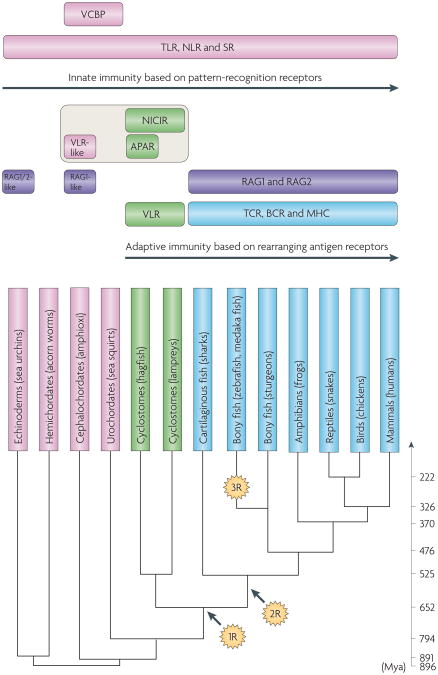
Figure 1. Overview of the evolution of the immune system in deuterostomes.
The stages in phylogeny at which the immune molecules referred to in this Review emerged. Molecules restricted to jawed and jawless vertebrates are indicated in blue and green, respectively. Molecules that emerged at the stage of invertebrates are in pink. Recombination-activating gene (RAG)-like genes (indicated in purple) are of viral or bacterial origin (from the transib transposon family) and are also present in the genomes of sea urchins and amphioxi. Agnathan paired receptors resembling antigen receptors (APAR) and novel immunoreceptor tyrosine-based activation motif-containing immunoglobulin superfamily receptor (NICIR, also known as T cell receptor (TCR)-like) are agnathan immunoglobulin superfamily (IgSF) molecules that are thought to be related to the precursors of TCRs and B cell receptors (BCRs). 1R and 2R indicate the two rounds of whole-genome duplication (WGD). Whether the 2R, the second round of WGD, occurred before or after the divergence of jawed and jawless vertebrates is controversial; the figure places it after the divergence according to the commonly held view (BOX 2). An ancestor of the majority of ray-finned fish is thought to have experienced an additional, lineage-specific WGD (designated as 3R) ∼320 million years ago134,135. The divergence time of animals (shown in Mya (million years ago)) is based on Blair and Hedges136. MHC, major histocompatibility complex; NLR, Nod-like receptor; SR, scavenger receptor; TLR, Toll-like receptor; VCBP, V-region containing chitin-binding protein; VLR, variable lymphocyte receptor.
Lymphoid cells — the core of the AIS — are first found in pre-vertebrate deuterostomes (that is, these are the most evolutionarily ancient living phylogenetic group in which lymphoid cells can be found). The AIS requires a large population of cells in each individual to permit clonal selection of cells with receptors specific for a particular pathogen. Large-bodied vertebrates were the first organisms to fulfil this requirement (FIG. 1). Here, we discuss important features of the AIS in jawed vertebrates and consider their conservation or plasticity over evolutionary time. The concept of plasticity was first proposed by Klein and colleagues in the context of MHC molecules11, but can be extended to all features of the AIS.
B cell receptors
Immunoglobulin M
Like most molecules of the AIS as defined in humans, among living animals Igs are first found in cartilaginous fish (sharks, skates, rays and chimaeras or ratfish) and are generated by a somatic recombination mechanism. Their evolutionary features are summarized in FIG. 2a. IgM is the most ancient antibody class and has the same function in all gnathostomes12.The transmembrane form of IgM defines the B cell lineage. After B cell stimulation — by an antigen and cognate interactions from T cells — this isotype is secreted into the plasma as a multimeric protein. In bony fish, IgM is secreted as a tetramer in different reduction or oxidation states that seem to modify the binding strength of the antibodies13. In cartilaginous fish, IgM is an abundant secreted monomer; this secretion correlates with a switch to a higher affinity response14. It is not known whether a cell programmed to produce multimeric IgM switches to produce the monomeric form or whether sharks have lineages of B cells that are specialized to make either the multimeric or monomeric Ig. Despite these modifications in fish, IgM is generally evolutionarily conserved.
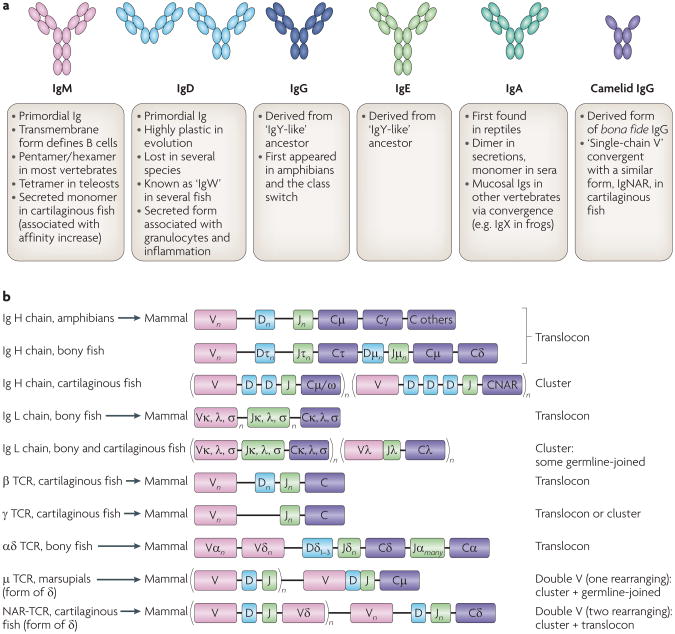
Figure 2. Antigen receptor proteins and genes in jawed vertebrates.
a | The different B cell receptor (BCR) isotypes in mammals and their features in other vertebrates. Each oval represents an immunoglobulin superfamily (IgSF) domain consisting of 90-100 amino acids. Immunoglobulin D (IgD) is represented in two forms, mouse (left) and human (right); note that even in mammals the IgD structure is highly plastic. Major phylogenetic features are described in the text. b | BCR genes (which are defined by their heavy (H) chain) and T cell receptor (TCR) genes throughout phylogeny Recombination signal sequences are located at the 3′ end of all variable (V) loci, and the 5′ end of joining (J) seqments and on both sides of diversity (D) segments. Note that in both the bony fish Ig H chain and vertebrate αδ TCRs, entire antigen receptor families are deleted upon rearrangement of the V segment to the downstream D and J segments (τ genes are deleted in bony fish and δ is deleted at the TCR locus). Note also that, despite its name, μ TCR in marsupials is related to δ TCR and not the IgM constant region. L chain, light chain; NAR, new antigen receptor.
Immunoglobulin D
The function of IgD remains poorly understood. It had generally been thought to be an evolutionary newcomer, probably arising in mammals (FIG. 2a), but the detection in bony fish15 and frogs16,17 of an Ig isotype that resembles IgD showed that this is not the case (FIG. 2b). Furthermore, phylogenetic analyses suggest that another isotype in cartilaginous fish and some bony fish, IgW18–20, is related to IgD. Therefore, an IgD-like isotype was present in the ancestor of all gnathostomes. In frogs and mammals, IgD/IgW occurs predominantly in the transmembrane form, but in bony fish there are high levels of secreted IgD, which is mostly bound to the surface of granulocytes. This work in fish prompted a recent re-examination of the function of IgD in humans, and it was found that the secreted form of IgD is bound to the surface of basophils and has strong proinflammatory properties21. Therefore, although this new study does not address the function of cell-surface IgD, the work in fish heralded an entirely new understanding of secretory IgD function in humans. Like IgM, IgD is an ancient antibody class, but unlike IgM, it has been extraordinarily malleable in structure and (presumably) in function over evolutionary time.
Other immunoglobulins
Besides IgM and IgD, mammals have three Ig isotypes: IgG, which is involved in high-affinity memory responses; Ige, which functions in inflammatory (and allergic) responses at epithelial surfaces; and the mucosal antibody IgA (FIG. 2a). IgG and Ige originate from an isotype known as IgY, which is first found in amphibians and has the same function as IgG (at least in frogs and birds17,22). Shorter forms of IgY, which are formed either by alternative splicing or from separate genes, are present in multiple taxa; these short forms are believed to neutralize pathogens without activation of inflammatory responses. IgA is found first in reptiles and seems to serve the same functions in all animals. Interestingly, in lower taxa, other isotypes — for example, IgX in frogs23 — provide mucosal immunity. Finally, in camelids, there is a modified form of IgG that does not associate with light (L) chains, and its V regions are free to interact with antigens; a similar form called immunoglobulin new antigen receptor (IgnAR) is found in cartilaginous fish; the IgnAR V region is also conserved in some TCRs24,25 (see below). In summary, in evolutionary terms all vertebrates have multiple isotypes that provide distinct effector functions. Amphibians are the most primitive class, with a classical memory response that is based on isotype switching to IgG and IgY after antigen stimulation.
B cell receptor light chains
It was previously believed that the two isotypes of the BCR L chains in humans —λ and κ — arose late in evolution, probably in the mammalian lineage (FIGS 2,3a). However, genome and eST projects and studies of model vertebrate species have shown that these two isotypes emerged in the common ancestor of all living vertebrates26–29. Furthermore, another L chain — identified in frogs and named σ30 — also arose early in evolution and has been retained in all cold-blooded gnathostomes. The reason(s) for the emergence and maintenance of multiple L chains is (are) not known, but it might be related to the formation of specific binding sites by particular combinations of L chains and heavy (H) chains.
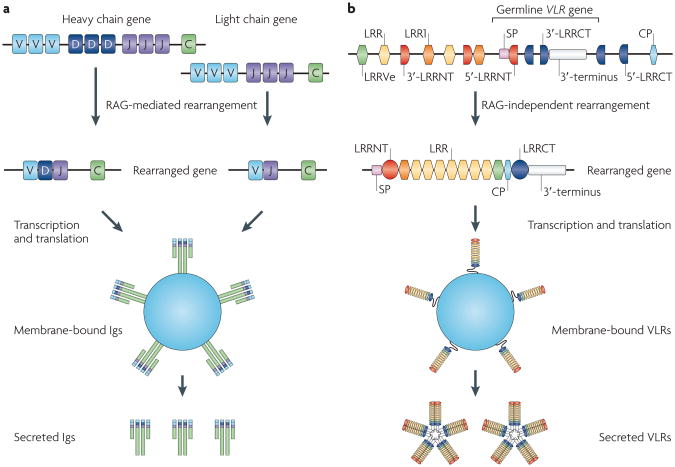
Figure 3. Two distinct forms of adaptive immunity in vertebrates. a.
a | Immunoglobulin (Ig) genes in jawed vertebrates. Ig genes generate repertoire diversity by recombining variable (V), diversity (D) and joining (J) gene segments with the help of the recombination-activating gene (RAG) recombinase. D gene segments occur only in the heavy chain genes. T cell receptors generate diversity in a similar manner (not shown). They occur only in a membrane-bound form. b | Variable lymphocyte receptors (VLRs) in jawless vertebrates. Multiple amino-terminal leucine-rich repeat (LRRNT)-, leucine-rich repeat (LRR)-, connecting peptide (CP)- and carboxy-terminal LRR (LRRCT)-encoding modules are located adjacent to the germline VLR gene. During the development of lymphoid cells, these modules are incorporated into the VLR gene. The rearranged VLR gene encodes a membrane-bound protein. VLRB has a secreted form and functions as an antibody, whereas VLRA apparently occurs only in a membrane-bound form. Whether membrane-bound VLRs occur as monomers or multimers is not known. LRR1 and LRRVe denote LRR modules located at the N- and C-termini, respectively. The organization of the VLR locus shows minor variations depending on loci and species. The figure is intended to emphasize the essential features of VLR assembly and does not accurately reproduce the organization of a specific VLR locus. SP, signal peptide.
B cell receptor heavy chains
From bony fish to mammals, BCR H chain genes are arranged in a so-called translocon organization (FIGS 2b,3a): multiple V genes occur upstream of multiple D and J segments, followed by single genes that encode the different constant (C) regions. The primary H chain repertoire is generated by rearrangement in the primary lymphoid tissues (for example, bone marrow), which involves a single V gene (out of tens to hundreds of V genes) being rearranged to the downstream D and J segments31. In some species, such as chickens and rabbits, after the rearrangement of a single V gene, gene conversion events involving nearby non-functional V genes generate diversity in the primary repertoire32,33. In cartilaginous fish, the Ig genes are in a cluster configuration with a single V, D, J and C gene in each cluster34. RAG-mediated rearrangement occurs within a cluster, but diversity is generated by junctional diversity at the V–D, D–D and D–J rearrangement joints and by somatic hypermutation after antigenic stimulation of B cells35–37. The situation is similar for the L chain genes, except they lack D segments (FIG. 2b). Also, in bony fish the L chain genes can occur in translocon organization or cluster organization; this suggests that extensive receptor editing — that is, re-rearrangement of BCR genes — can occur for L chain genes in all fish38–41 (FIG. 2b).
The H chain class switch first evolved in amphibians, which have a mammalian-like Ig H chain locus. In bony fish, the Ig H chain isotypes IgM and IgD are generated by alternative splicing. The recently discovered IgT isotype is generated by alternative rearrangement: the IgT D, J and C gene segments are located between the V genes and the IgM and IgD D, J and C segments42,43 (FIG. 2b), and rearrangement that joins V genes to the IgM D segments deletes the IgT genes, a situation similar to that at the αδ TCR locus. There is no switching apparent between clusters in cartilaginous fish, neither within the IgM class nor between the IgM and cartilaginous fish-specific IgnAR/IgW classes.
Amphibians, which have class switching and IgY, mark a major transition from the AIS in fish. A single B cell has the capacity to modify the effector function of its secreted molecule and to provide a memory response. In sharks, a ‘switch’ at the biochemical level from multimeric to monomeric IgM provides at least some of the functions of an IgG — for example, the capacity to enter extravascular sites — and an increase in affinity, but the memory response is clearly inferior to that of the higher vertebrates.
T cell receptors
αβ T cell receptors
αβ TCRs recognize both the antigen (in the form of a peptide) and the MHC class I or II molecule to which it is bound. This is known as major histocompatibility complex restriction (MHC restriction)44. T cells develop in the thymus, and the basic evolution of the thymus and T cell development is conserved for all gnathostomes, except in some species in which the system seems to have degenerated (FIG. 1 and see below). A translocon organization of both the α and β TCR genes is found in all animals (FIG. 2b). The presence of a D segment in the β TCR locus and the absence of D segments and the presence of a large number of J segments in the α TCR locus are also evolutionarily conserved, at least from mammals to bony fish45 (FIG. 2b). The many J segments in the α locus allow extensive receptor editing in developing T cells, resulting in greater opportunities for modification of the antigen receptor, followed by potential selection of self-MHC-restricted TCRs in the thymus46.
γδ T cell receptors
γδ TCRs are quite different from αβ TCRs. The basic gene organization is conserved in gnathostomes, with only J segments in the γ gene, usually two D segments in the δ gene, and a close linkage of the δ and α TCR genes. However, the receptor can be adapted in several ways. It was shown in mice that γδ T cells that arose early in ontogeny bear invariant receptors and home to sites, such as the skin, where they form a first line of defence and self-renew for the life of the organism47. Similarly, in humans, T cells with canonical γδ receptors make up a large percentage of lymphocytes in the blood and become highly expanded upon infection. These cells respond to the metabolite isoprenylphosphate, which is produced by all cells but is produced in a more immunogenic form by unicellular pathogens48. The ‘innate-like’ γδ T cells can make up the majority of T cells in some species, but in others γδ T cells do not bear canonical receptors, are a large proportion of the T cells in the body, and seem to be more adaptive in nature49. The binding sites of these receptors (even of some of the innate receptors) are more like BCR binding sites: they have one long and one short complementarity-determining region 3 (CDR3), whereas αβ TCRs have similarly sized CDR3s for both chains50.
Recently, new types of δ TCR genes have been discovered in different vertebrate taxa. In about a quarter of their δ TCR genes, sharks have a three-domain receptor chain with two amino-terminal V domains and a membrane-proximal C domain called nAR-TCR51 (FIG. 2b). Both V domains are generated by VDJ rearrangement: the N-terminal domain is encoded by an IgnAR-like cluster, and the other is generated by rearrangement of V to canonical D and J elements. It is likely that this TCR chain recognizes antigens in a similar way to the bona fide IgnAR, which is presumed to have a single-domain binding site. In marsupials, the δ TCR locus has been duplicated. An apparently ancient trans rearrangement of IgV regions to this locus in germ cells52 has resulted in one set of V, D and J segments that rearrange in a cluster that is upstream of a pre-rearranged (that is, germline-joined) VDJ gene. This is predicted to result in a three-domain TCR chain that has two V domains and one C domain and is similar to the TCR chain in sharks (FIG. 2b). Although the function of these receptors is not known, it is proposed that — unlike the MHC-restricted αβ TCRs — they recognize soluble antigens, which would give them a similar function to Igs53. Consistent with this idea, shark γ TCR genes have recently been shown to hypermutate like BCR genes but unlike αβ TCR genes54. Therefore, a new paradigm emerging from the comparative studies is that γδ TCRs probably have the capacity to interact with free antigens in a similar way to Igs. Additionally, when comparing αβ TCRs and γδ TCRs, the former clearly are the more evolutionarily conserved, probably because they are constrained by MHC restriction. By contrast, γδ TCRs are more evolutionarily labile and have adaptive immune and innate immune functions that differ even between vertebrates in the same class.
Major histocompatibility complex
The MHC is defined by: MHC class I and class II genes; genes that encode transporter associated with antigen processing (TAP) and tapasin, which are involved in processing antigens for presentation by MHC class I molecules; and genes that are involved in general immunity (see below). The MHC, like TCRs and BCRs, is first found in cartilaginous fish55. Genes involved in class I processing and presentation are closely linked in almost all vertebrates (except in placental mammals, in which there is close linkage of the class I processing genes and class II genes) and are believed to have co-evolved55,56. As described below, genes that encode natural killer (nK) cell receptors are associated with the MHC of various living vertebrates, and these serve as connections to what seems to have been a primordial gene complex composed of receptors and ligands55,57,58. Therefore, it seems likely that nK receptors (and perhaps also the forerunners of antigen receptors) were physically linked to the primordial MHC in a similar way to the links between receptors and ligands in the myriad histocompatibility systems of modern-day lower deuterostomes and protostomes59; these links were subsequently fragmented in gnathostomes.
How did the jawed vertebrate AIS arise?
As discussed above, the BCR–TCR–MHC-based AIS is present in cartilaginous fish but absent in lower chordates. Two macroevolutionary events — the invasion of the RAG transposon and two whole-genome duplications (WGDs) — are believed to have provided the innovation and raw material, respectively, for the relatively rapid emergence of the AIS in jawed vertebrates. In this section we discuss the potential contribution of these two events.
The recombination-activating gene transposon
In the late 1970s, Sakano and Tonegawa discovered recombination signal sequences (RSSs) flanking V, D and J rearranging segments60, and they noticed that the inverted repeats within the RSSs were reminiscent of a transposon. This suggested that a gene encoding an immunoglobulin superfamily (IgSF) exon was invaded by a transposon. Therefore, the gene could not be expressed unless it was stitched back together through the action of a recombinase. There are many IgSF genes in lower chordates that are related to BCR and TCR V regions (the so-called ‘VJ’ type of IgSF gene; FIG. 1), so there are many candidate genes that could have been invaded by the RAG transposon in a vertebrate ancestor61. For example, the gene family encoding the variable region containing chitin-binding proteins (VCBPs) in amphioxus is presumed to be involved in defence and could be related to the non-rearranging ancestor of the BCR and TCR genes62. Other genes that might be related to the ancestral VJ gene (for example, ReFS 63–65) are described below.
The RAG1 and RAG2 genes have no homologous regions and no introns but are closely linked to each other, which suggests that they could have been acquired by vertebrates as part of the original transposon4,66. Complete RAG genes have been isolated from all gnathostomes studied, but not from jawless fish. This is consistent with the idea that acquisition of the RAG transposon helped to trigger BCR- and TCR-based immunity. Surprisingly, linked RAG1 and RAG2 genes were found in the genome of the sea urchin (an echinoderm)67 (FIG. 1), which implies either that the RAG transposon invaded the genome 100 million years before the emergence of adaptive immunity and remained dormant, or that it had invaded at multiple times. The authors suggested that a RAG1-like gene invaded the genome and inserted near a RAG2-like gene that was subsequently included in the original transposon. The RAG1-like gene could have originated from transib — a transposon that encodes the RAG1 core and that has been found in many organisms68 — or, as Dreyfus has recently suggested69, from a viral genetic element. RAG2 then lost its original function and was co-opted to the VDJ rearrangement process. This proposition is consistent with recent work showing that RAG2 does not contribute to rearrangement per se, but instead interacts with modified histones at sites of active transcription70. We propose a working model in which RAG2 binds to active regions of transcription and RAG1 binds to the exposed RSS. RAG1 interacts with, and is dependent on, RAG2 to initiate VDJ rearrangement, but RAG1 performs all of the major functions associated with recombination.
Regardless of the route of its emergence in chordates, the RAG transposon invasion must have been crucial for adaptive immunity, as all of the BCR and TCR genes use the same mechanism of VDJ rearrangement. Furthermore, despite arguments to the contrary71, RAG activity was likely to have been an innovative event for adaptive immunity, as jawed vertebrates have many novel mechanisms and molecules that are involved in immunity in general, and adaptive immunity in particular, compared with jawless fish (discussed further below)72.
Whole-genome duplication
Nearly 40 years ago, Susumu Ohno proposed that the vertebrate genome underwent one or two rounds of WGD at the stage of fish or amphibians through a tetraploidization process73. Although this proposal was a matter of hot debate until recently (BOX 1), it is now widely accepted that the vertebrate genome experienced two rounds of WGD after the emergence of urochordates and before the radiation of jawed vertebrates74–77 (FIGS 1,4). WGDs seem to have had a crucial role in the emergence of jawed vertebrate-type adaptive immunity78,79. This is indicated by the fact that many ohnologues, which arose as a result of WGDs, are essential components of the jawed vertebrate-type AIS80,81.
Box 1. Controversies surrounding whole-genome duplication.
The idea that the vertebrate genome underwent two rounds of whole-genome duplication (WGD) at a time close to the origin of vertebrates — known as the two-round (2R) hypothesis — caused hot debate for more than a decade. The major evidence in favour of this hypothesis was that paralogues that emerged close to the origin of vertebrates are not distributed at random in the human genome, but have a strong tendency to occur in clusters, known as paralogons, on multiple — typically four — chromosomes. The major arguments against this hypothesis were twofold127,128. First, ohnologues (for example, A, B, C and D) generated by two rounds of WGD should display the tree topology (that is, A and B together and C and D together), which is not always the case127. Second, paralogons occasionally contain paralogues that duplicated much earlier than the origin of vertebrates. We now know that incongruent tree topologies can occur if two waves of genome doubling occurred in close succession129, if paralogues have evolved at different evolutionary rates130 or if the resolving power of phylogenetic trees is not sufficient129. Also, the divergence time of paralogues can be much older than the age of WGD if tandem duplication preceded WGD and the duplicated genes were lost differentially after WGD. Therefore, neither of these two counterarguments contradicts the 2R hypothesis.
Recently, comparison of the human and amphioxus (Branchiostoma floridae) genomes revealed widespread quadruple conserved synteny, in which four sets of human paralogons corresponded to one set of linked genes in B. floridae131. This observation provided definitive evidence for the 2R hypothesis and settled the long-standing debate. The exact timing of WGD relative to the emergence of jawless vertebrates (FIG. 1) is still controversial. A commonly held view, which we also favour, is that the first round of WGD occurred in a common ancestor of jawed and jawless vertebrates, and the second round in a common ancestor of jawed vertebrates76. However, recent analysis of 55 gene families in jawless vertebrates suggested that both rounds of WGD might have taken place in a common ancestor of all vertebrates, although this analysis could not discount the idea that the second round of WGD occurred in a common ancestor of jawed vertebrates132, as depicted in FIG. 1.
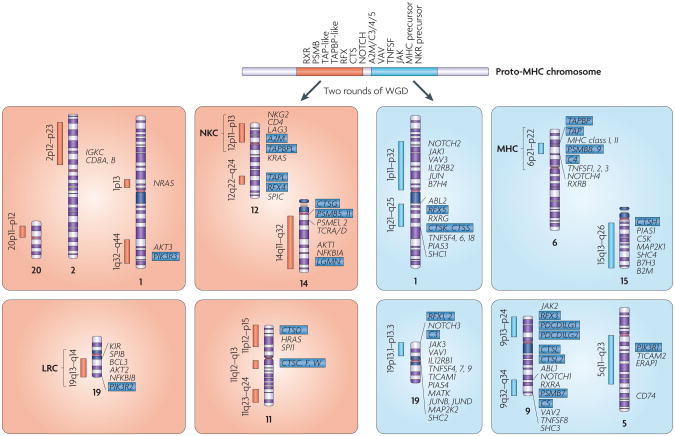
Figure 4. The major histocompatibility complex paralogy group and the neurotrophin paralogy group in the human genome.
Four sets of major histocompatibility complex (MHC) paralogons are located on chromoso mes 1, 6, 9 and 19 (ReFS 55,137,138). A number of smaller-sized MHC paralogons, which presumably originated from fragmentation and subsequent translocation of the major paralogons, have been identified55. Among them, those located on 15q13-q26 and 5q11–q23 seem to have broken off from the paralogons on chromosomes 6 and 9, respectively130. Four sets of major neurotrophin paralogons are located on chromosomes 1, 11, 12 and 19 (REF. 89).The MHC paralogy group (right) and neurotrophin paralogy group (left) are thought to have descended from neighbouring regions on a single ancestral chromosome. Paralogues that are distributed across the two paralogy groups are indicated by blue highlighted text. In addition to genes that share paralogues among the relevant paralogons, immunologically important genes, such as those encoding MHC class I, MHC class II, T cell receptors (TCRs), and immunoglobulins (Igs) are shown. A2M, α2-macroglobulin; B2M, β2-microglobulin; BCL, B cell leukaemia/lymphoma; CSK, cytoplasmic tyrosine kinase; CTS, cathepsin; ERAP, endoplasmic reticulum aminopeptidase; IL12RB, interleukin 12 receptor, β-subunit; JAK, Janus kinase; KIR, killer Ig-like receptor; LAG3, lymphocyte activation gene 3; LGMN, legumain; LRC, leukocyte receptor complex; MAP2K, mitogen-activated protein kinase kinase; MATK, megakaryocyte-associated tyrosine kinase; NFKBIA, nuclear factor-κB inhibitor; NKC, natural killer complex; NKR, NK receptor; NRAS, neuroblastoma RAS; PD, programmed cell death 1 ligand; PIAS, protein inhibitors of activated STAT; PIK3R, phosphatidylinositol 3-kinase, regulatory subunit; PSMB, proteasome subunits, β-type; PSME, proteasome activator subunits; RFX, regulatory factor X; RXR, retinoid X receptor; SPI1, spleen focus forming virus proviral integration oncogene; TAP, transporter associated with antigen processing; TAPBP, TAP-binding protein (also known as tapasin); TAPBPL, TAPBP-like; TAPL, TAP-like (also known as ABCB9); TICAM, TIR domain-containing adaptor molecule; TNFSF, tumour necrosis factor ligand superfamily; WGD, whole-genome duplication.
The MHC paralogy group (FIG. 4a) and the HOX paralogy group (Supplementary information S1 (figure)) are illustrative examples. In the case of the MHC paralogy group82–84, paralogues of more than 100 gene families are distributed in four sets of paralogons (for a list of gene families, see REF. 85). As would be expected from Ohno's theory of WGD, the genomes of invertebrates, such as amphioxi84,86 and Ciona intestinalis87, do not have multiple sets of MHC paralogons and contain only one MHC-like region, which is devoid of class I and II genes (a proto‑MHC).
Recently, Olinski et al.85 suggested that the neurotrophin paralogy group (FIG. 4, left) and the MHC paralogy group (FIG. 4, right) were derived from neighbouring regions on the same ancestral chromosome. This suggestion is based on the observation that several genes of the MHC- and neurotrophin-paralogy groups are linked in the genomes of C. intestinalis85 and amphioxi88. Interestingly, chromosomal regions 12p13.2–p12.3 and 19q13.4, which encode the human nK complex (nKC) and leukocyte receptor complex (LRC), respectively (BOX 2), are embedded in the neurotrophin paralogons89. Therefore, Olinski et al.'s suggestion provides a logical basis for the earlier proposals that the ancestors of nK receptor genes encoded in the nKC and LRC were located in the proto-MHC55,57,90,91.
Box 2. Natural killer cell receptors.
Natural killer (NK) cells are part of the innate immune system. They can produce cytokines and kill virally infected cells without a clonal proliferation phase. NK cells share many characteristics with T cells, including most cell-surface markers, elaboration of the same effector functions and origins from a similar haematopoietic precursor111. However, the NK cell receptor (NKR) recognition strategy is different from that of T cells (although the signalling cascades are homologous). When cells become virally infected, major histocompatibility complex (MHC) class I molecules are often downregulated, making the cells ‘invisible’ to cytotoxic T cells. Inhibitory NKRs on NK cells recognize MHC class I molecules; when the inhibitory NKRs are not engaged, the NK cells can be stimulated by activating receptors that often recognize stress-induced molecules that are upregulated on viral infection. This is known as the ‘missing self hypothesis133.
Unlike T cell receptors (TCRs), NKRs are not generated by recombination-activating gene (RAG)-mediated recombination, but rather through stochastic, clonal expression of a few receptors from a cluster of genes encoding activating and inhibitory NKRs. These receptors are encoded by the NK complex (NKC) and leukocyte receptor complex (LRC), found on different chromosomes in all mammals examined. The NKRs that recognize classical (polymorphic) class I molecules are encoded by genes in different gene families in various species, for example the C-type lectin Ly49 genes found in the NKC in mice and the killer immunoglobulin superfamily (IgSF) receptor (KIR) genes found in the LRC of humans. This is the only known case in the immune system in which receptors from different gene families perform precisely the same functions in closely related species, and it is a testament to the rapid evolution of the NK system of defence. Evidence from comparative studies of the MHC, LRC and NKC strongly suggest that the three complexes were originally linked and that their polymorphic receptors and ligands have co-evolved, as has been found in many histocompatibility systems in plants and invertebrates59.
Strikingly, the integrated MHC/neurotrophin paralogy group encodes many ohnologues that have important functions in the AIS, particularly in antigen presentation by MHC class I and II molecules (FIG. 4; Supplementary information S2 (table)). In addition to TAP and tapasin, three proteasome subunits that are specialized for antigen presentation are encoded in this paralogy group, together with some of their paralogues. Similarly, the integrated paralogy group encodes 10 of the 11 cathepsin ohnologues, including cathepsins S and L, which have crucial roles in antigen presentation by class II molecules and in thymic selection. Other examples include: retinoid X receptor-β (RXRB) and regulatory factor X5 (RFX5), which encode proteins that regulate the expression of class I and II molecules, respectively; NOTCH1, which is involved in lymphocyte differentiation; and VAV1, which is required for T and B cell development and activation (Supplementary information S2 (table)).
The convergent AIS of jawless vertebrates
With some exceptions, and despite the involvement of the well-conserved and rapidly evolving molecular features described above, the AIS of jawed vertebrates has been remarkably uniform over 500 million years. Functional assays of jawless vertebrates (lampreys and hagfish) have shown signs of adaptive immunity, but neither adaptive immune molecular characteristics nor tissues, such as a thymus or spleen, were detected in these animals. For example, jawless vertebrates produce specific agglutinins when immunized with particulate and soluble antigens92–95, but extensive transcriptome analysis of jawless fish leukocytes provided no evidence for the presence of MHC, BCR or TCR molecules96–98. Their absence has been recently confirmed by the analysis of the sea lamprey (Petromyzon marinus) draft genome sequence. How can they mount adaptive immune responses in the absence of BCR, TCR and MHC molecules? The discovery of a VLR gene, which generates diversity by somatically rearranging leucinerich repeat (LRR) modules7, provided an answer99. The germline VLR gene lacks gene segments coding for LRR modules and is not capable of encoding functional proteins (FIG. 3b). During the development of lymphoid cells, LRR modules that flank the gene are sequentially incorporated into VLR by a process called ‘copy choice’100. Incorporation is presumably mediated by cytidine deaminases of the AID-APOBeC family8. The mature VLR gene encodes a glycosyl-phosphatidylinositol-anchored polypeptide. The sequence of individual LRR modules assembled into the VLR gene is highly diverse, and the number of LRR modules present in a rearranged gene varies from one to nine. Therefore a single VLR gene can have combinatorial diversity comparable to that of Igs (∼1014 combinations)101. The crystal structures of hagfish VLR monomers indicate that they adopt a horseshoe-like solenoid structure that is characteristic of LRR family proteins. The majority of variable residues in the VLR are located on the concave surface, which suggests that this surface is involved in antigen binding102. This was recently confirmed by in vitro mutagenesis experiments103 and the crystal structure analysis of VLR in complex with the human blood group H-trisaccharide104 or with hen egg lysozyme105.
Lampreys have two VLR genes: VLRA and VLRB8. Both seem to be expressed exclusively in lymphoid cells, and they usually exhibit allelic exclusion. Stimulation with antigens causes lymphoid cells expressing specific VLRB molecules to undergo clonal expansion and to secrete VLRs in a manner analogous to the secretion of Igs by B cells7,101. Secreted VLRB molecules occur as pentamers or tetramers of dimers and have eight to ten antigenbinding sites103. Therefore, the subunit organization of secreted VLRB molecules resembles that of IgM antibodies. The multivalent structure of VLRB enables it to bind antigens carrying repetitive epitopes with high avidity and specificity, and it displays strong agglutinating activities106. It has been suggested that, like subclasses of IgM, VLRB might preferentially or exclusively recognize thymus-independent antigens106. By contrast, VLRA seems to occur only in a cell-bound form and is expressed on lymphoid cells that functionally resemble T cells. Therefore lampreys have two populations of lymphoid cells that seem to be functionally similar to the T and B cells of jawed vertebrates107.
Hagfish also have two VLR genes108, VLRA and VLRB, which are presumed to be orthologous to lamprey VLRA and VLRB, based on sequence comparison. They map to the same chromosome but are distant from each other, which indicates that they function as separate units109. Consistent with this, the rearranged VLRA and VLRB genes do not share an LRR module with an identical nucleotide sequence. It remains to be determined whether hagfish VLRA and VLRB are functionally similar to the corresponding lamprey genes.
Origin of the two distinct forms of AIS
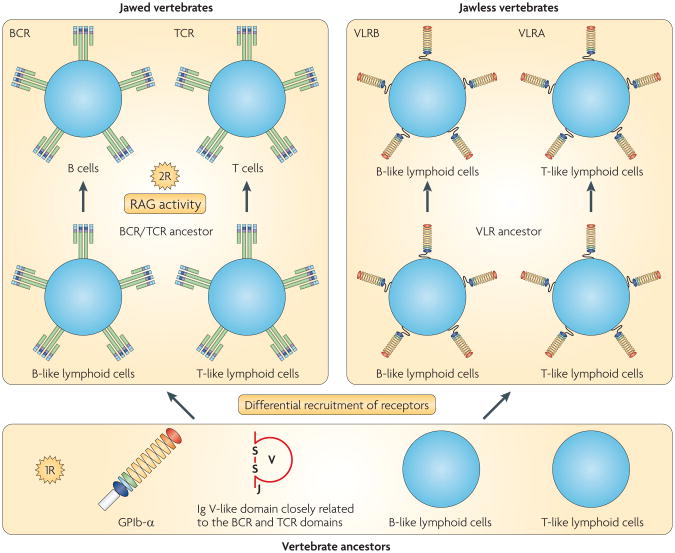
Both VLRs and BCRs/TCRs rely on combinatorial diver-sity to generate a vast repertoire of binding specificities; however, the genes of VLRs show no structural similarity to those of BCRs or TCRs (FIG. 3). Their recombination machineries must differ because RAG1 and RAG2 are required only for BCR/TCRs. These observations suggest that the ancestors of jawed and jawless vertebrates developed their antigen receptors independently.
Although VLRs probably exist only in lampreys and hagfish, the origin of LRR modules is ancient; LRR module repeats are found in many animal and plant proteins, in which they seem to mediate protein interactions or microbial recognition. It has been proposed, on the basis of the similarity of gene structures and a characteristic insertion in the LRR carboxy-terminal (LRRCT) domain, that an ancestor of VLR might have emerged from a glycoprotein Ib-α (GPIb-α)-like protein (GPIb-α is a component of the platelet glycoprotein receptor complex that is conserved in all vertebrates)8. Furthermore, searches for IgSF proteins in jawless vertebrates have identified molecules that are thought to be related to the potential evolutionary precursors of BCRs and TCRs63–65. These are: the agnathan paired receptors resembling antigen receptors (APAR) family of innate immune receptors, which encode a group of membrane proteins carrying a single extracellular V-type Ig-like domain with a canonical J segment64; and an immunoreceptor tyrosine-based inhibition motif-bearing membrane protein, named ‘TCR-like’65, which has V-C2-type domain organization and seems to be closely related to the novel immunoreceptor tyrosine-based activation motif-containing IgSF receptor (nICIR) family of activating innate immune receptors110. The occurrence of these receptors in jawless vertebrates suggests that a common ancestor of all vertebrates had V-type Ig-like domains that evolved into the Ig-like domains of BCRs and TCRs. Hence, as mentioned above, it seems that a vertebrate ancestor had both BCR/TCR and VLR precursors (FIG. 5). In a jawed vertebrate lineage in which RAG insertion occurred, an IgSF protein was ‘chosen’ as an antigen receptor, which led to the BCRs and TCRs. In a jawless vertebrate lineage with no appropriate RAG insertion, an LRR protein family that could mediate microbial recognition was ‘chosen’ as an antigen receptor, which led to the VLRs.
Figure 5. A hypothetical model for the origin of the two major forms of adaptive immune system.
The occurrence of B- and T-like lymphoid cells in jawless vertebrates indicates that they were likely to be present in a common ancestor of all vertebrates107. Similarly, the occurrence of a V-type immunoglobulin (Ig)-like domain with a canonical joining (J) segment in jawless vertebrates indicates that a common ancestor of all vertebrates had V-type Ig-like domains that could be converted to B cell receptor (BCR) or T cell receptor (TCR) Ig-like domains. Amphioxus, a basal chordate, has a glycoprotein Ib-α (GPIb-α)-like protein, which is the most likely precursor to the variable lymphocyte receptor (VLR)8. Therefore, a common ancestor of all vertebrates presumably had all of the key structural elements of the two radically different antigen receptors. Acquisition of recombination-activating gene (RAG) recombinase activities and the second round of whole-genome duplication (WGD; indicated as 2R) are thought to have had a crucial role in the emergence of the BCR–TCR–MHC-based adaptive immune system (AIS). Little is known about the genetic events that contributed to the emergence of the VLR-based AIS in jawless vertebrates. It is possible that the first round of WGD (indicated as 1R) contributed to the emergence of the two lineages of lymphoid cells; however, we cannot rule out the possibility that their origin predates the emergence of vertebrates.
The occurrence of structurally unrelated receptors that are used for the same or similar purposes is not unprecedented (BOX 2). For example, in primates the nK receptors that interact with classical MHC class I molecules are members of the killer cell Ig-like receptor (KIR) family, but in rodents the corresponding receptors are C-type lectin-like molecules known as Ly49 receptors111. A common ancestor of primates and rodents is thought to have had precursor genes for both types of nK receptors; the KIR gene family expanded in the primate lineage and the Ly49 family expanded in the rodent lineage112,113. We suggest that the ancestors of jawed and jawless vertebrates adopted distinct molecules as their antigen receptors in an analogous manner.
The observation that jawless vertebrates have B- and T-like lymphoid cells107 indicates that the two lineages of lymphoid cells emerged before the divergence of jawed and jawless vertebrates (FIG. 5). Therefore, the origin of B- and T-like lymphoid cells is likely to be more ancient than previously thought. This is compatible with recent observations that B and T cells do not share an immediate common progenitor, but differentiate from myeloid B progenitors and myeloid T progenitors, respectively88,113–115. This suggests that B- and T-like lymphoid cells diverged before the divergence of myeloid and lymphoid cells. Although we must be cautious when using cell lineage information as a proxy for the order of evolutionary emergence, it seems likely that the building blocks for both types of antigen receptors and also the two lineages of lymphoid cells were present in a common ancestor of all vertebrates (FIG. 5).
Pressures for adaptive immunity
Why did the AIS arise? As noted above, invertebrates and plants manage with innate mechanisms of defence (however, it should be mentioned that primitive adaptive systems have been described in several invertebrate taxa116 and that an RnAi-like mechanism provides a potent adaptive response in many organisms117). Indeed, it has been argued that an anticipatory system of defence is more trouble than it is worth, as it necessitates the generation of tolerance to self during the ontogeny of lymphocytes, and failure to regulate these defence mechanisms can result in autoimmunity118. Furthermore, the adaptive system might only be maintained when under strong selective pressure: organisms such as cod119, seahorses120 and axolotl121, which might not be under strong pressure from pathogens, seem to have lost the capacity to mount effective immune responses, at least as far as they can be measured experimentally121.
However, we think that at least the BCR–TCR–MHC system arose as a potent system of defence. It is believed that this system emerged in animals with jaws, which are likely to have been predatory. Large predators generally do not have many offspring, thereby maintaining a ratio of predator-to-prey that perpetuates the status quo122. Sharks, the oldest living vertebrates with this adaptive system, have many new mechanisms of reproduction to protect young offspring — such as impenetrable eggcases and modes of viviparity that have converged on the mechanisms in mammals. Therefore it can be proposed that a potent system of defence arose in conjunction with the need to protect offspring. In jawless fish, the AID-APOBEC family members might have enabled the diversification of a preimmune repertoire of IgSF genes, as seems to be the case for the VLR system. The addition of the RAG transposon to this adaptive system would have provided a great advantage, causing a break during rearrangement that would have created the potential for large variations in the size of one of the CDR loops that interact with antigens37. This advantage, along with the raw material of a large number of newly duplicated genes that could be co-opted for immune functions, probably resulted in the highly complex AIS seen today in gnathostomes.
Conclusions
There are many characteristics of the AIS that can generally be agreed upon: jawless and jawed fish both have an AIS based on different gene families, but the BCR–TCR– MHC-based system is more complex; the RAG transposon and WGDs had major roles in the emergence of the jawed vertebrate AIS, and the major features of the system arose early in a ‘big bang’ and have been modified only slightly over evolutionary time; and the jawed vertebrate AIS displays both conserved and plastic features, the former presumably to allow for a general functioning of the system and the latter to permit rapid changes over evolutionary time to respond to pathogens11.
Many questions still need to be addressed, most of which will require incremental progress. For example, what is the nature of the RAG transposon? What are the functions of antigen receptors in different organisms, especially the γδ TCR? And how is the VLR system regulated? However, there are two glaring questions: did T-and B-like lymphoid cells really emerge in the common ancestor of jawless and jawed vertebrates, and what were the pressures that selected for the emergence of the AIS? Regarding the first of these questions, besides what we have discussed above concerning the antigen receptors, it is essential to determine whether, like gnathostome αβ T cells, the ‘T-like lymphoid cells’ of jawless vertebrates recognize processed antigens, perhaps through presentation on a convergent type of ‘MHC’ molecule. Instead, they might recognize native antigens in a cellbound form, perhaps in a way similar to the recognition that occurs for some subsets of gnathostome γδ T cells. Because the lamprey ‘T cell’ antigen receptor VLRA has been shown to bind soluble antigens and because its gene apparently mutates after antigenic stimulation, our prediction is that the γδ TCR-like mechanism will prove to be correct123. Regarding the second question, we will never be able to resolve this problem experimentally. new ideas concerning selection pressures suggest that the capacity of the immune system to distinguish between commensal and pathogenic microorganisms resulted in the great complexity of the gnathostome AIS124,125. The understanding of the immune system's interactions with commensal organisms is in its infancy, and perhaps we can develop defined hypotheses as this field develops. One hypothesis, which emphasizes the importance of the gut, suggests that with the advent of jaws, digested material could injure the intestine and therefore result in massive infections. The seahorse, which has seemingly lost features of adaptive immunity, is a filter-feeding animal, so it is speculated to have reverted to a pre-adaptive state120. Our view is that the AIS arose as a system of defence and became more crucial and more sophisticated with the emergence of the large predatory jawed vertebrates that bore few offspring. We think that our position has been strengthened by recent evidence that has shown that the jawed vertebrates in which it is believed that the AIS first emerged — the placoderms — had internal fertilization126. We hope that this Review will spark further interest in this area, especially among non-immunologists who might see the problem from a fresh perspective.
Supplementary Material
Acknowledgments
M.F.F. has been funded by the US National Institutes of Health grants AI027877 and RR006603. M.K. has been funded by a KAKENHI grant for the priority area ‘Comparative Genomics’ from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Glossary
- Somatic hypermutation
Mutation of the variable gene after mature B cells are stimulated. It results in affinity maturation of the antibody response. Like the class switch, it requires activation-induced cytidine deaminase
- Variable–diversity–joining rearrangement
(VDJ rearrangement.) The recombination-activating gene (RAG)-mediated ligation of T cell receptor or B cell receptor variable (V), diversity (D) and joining (J) gene segments during lymphocyte ontogeny, which generates the antigen receptor repertoire.
- Immunoglobulins
Also known as antibodies or B cell receptors, they are composed of two identical heavy (H) and light (L) chains that are covalently linked by disulphide bonds. Monomeric immunoglobulins are bivalent, and the binding site is made up of the amino-terminal variable domains of one H and one L chain. The class or isotype of an immunoglobulin is defined by its H chain.
- Jawless fish
Primordial vertebrates without jaws, of which the only two extant forms are lampreys and hagfish.
- Major histocompatibility complex
A large complex of tightly linked genes, many of which are involved in immunity. It encodes the polymorphic class I and class II molecules, which present antigens in the form of peptides to cytotoxic and helper T cells, respectively.
- Jawed vertebrates
Vertebrates from cartilaginous fish to mammals. The first class of vertebrates with jaws, the placoderms, all became extinct.
- Mucosal immunity
Immune responses made across epithelial surfaces, such as the gut and lung.
- Immunoglobulin new antigen receptor
(IgNAR.) A specialized antibody in sharks that is composed of disulphide-linked heavy chains and no associated light chains.
- Translocon organization
An organization of immunoglobulin genes in which there are multiple variable (V), diversity (D) and joining (J) segments upstream of a single constant gene.
- Cluster organization
An organization of immunoglobulin genes in which there are single variable (V), diversity (D), joining (J) and constant gene segments, although sometimes two or three diversity segments occur together. Also known as minilocus organization.
- Receptor editing
The re-rearrangement of antigen receptor genes to avoid self reactivity in the case of B cells or to express new receptors that can be positively selected in the case of T cells.
- Class switch
The rearrangement of an existing variable–diversity– joining (VDJ) exon from the 5′ end of the immunoglobulin M gene to downstream constant (C) genes. Like somatic hypermutation, it requires activation-induced cytidine deaminase.
- Major histocompatibility complex restriction
T cell receptors recognize antigens in the form of small peptides ranging from 9–22 amino acids that are bound to major histocompatibility complex (MHC) class I or class II molecules. The T cell receptor recognizes both the MHC protein and the peptide antigen — this is known as ‘MHC restriction.’.
- Complementarity-determining region 3
A loop in the variable (V) region of the B cell receptor and T cell receptor chains that is encoded by the variable– diversity–joining (VDJ) intersection that is generated by recombination-activating gene (RAG)-mediated somatic rearrangement. It is the part of the antigen-binding site that is most diverse in amino acid sequence and length.
- Transporter associated with antigen processing
(TAP). A transporter of the ATP-binding cassette superfamily that is involved in the transport of peptides from the cytosol into the lumen of the endoplasmic reticulum. It is a heterodimer composed of TAP1 and TAP2 subunits and has a crucial role in the transport of major histocompatibility complex class I-binding peptides.
- Tapasin
A protein that facilitates the binding of peptides to major histocompatibility complex (MHC) class I molecules by forming a bridge between MHC class I molecules and transporter associated with antigen processing (TAP).
- Recombination signal sequences
Conserved nucleotide sequences flanking variable (V), diversity (D) and joining (J) segments that are recognized by the recombination-activating gene (RAG) proteins to induce rearrangement.
- Immunoglobulin superfamily
Domains of 90–100 amino acids composed of 7–9 β strands forming two sheets, generally stabilized by a disulphide bond. Found in B cell receptor, T cell receptor and major histocompatibility complex molecules.
- Ohnologues
Paralogues, named after Susumu Ohno, that are thought to have emerged close to the origin of vertebrates by whole-genome duplication.
- Paralogy group
A set of paralogons that are derived from a single ancestral region.
- Paralogues
(Also known as paralogous genes.) Genes within a single species that belong to the same gene family. In contrast to ‘paralogues’, ‘orthologues’ refers to genes that diverged by speciation events.
- Paralogons
Chromosomal segments that contain closely linked sets of paralogues. Also known as paralogous regions.
- Agglutinin
Any substance that can clump particles together.
- AID-APOBEC
A family of cytidine deaminases that are involved in the hypermutation of variable–diversity–joining (VDJ) segments, in the immunoglobulin class switch and in defence against viruses. Two members of the family are implicated in generating diversity in variable lymphocyte receptors.
- Allelic exclusion
The expression of a single receptor in cells with the potential to express more than two receptors.
Footnotes
Competing interests statement: The authors declare no competing financial interests.
Databases: Entrez Gene: http://www.ncbi.nlm.nih.gov/gene RAG1 | RAG2
Further Information: Masanori Kasahara's homepage: http://www.med.hokudai.ac.jp/en/dept/outline/path/index.html
Pre-Ensembl genome database: http://pre.ensembl.org Lamprey (Petromyzon marinus) draft genome sequence
Supplementary Information: See online article: S1 (figure) | S2 (table)
Contributor Information
Martin F. Flajnik, Email: mflajnik@som. umaryland.edu.
Masanori Kasahara, Email: mkasaha@med.hokudai.ac.jp.
References
- 1.Weigert MG, Cesari IM, Yonkovich SJ, Cohn M. Variability in the λ light chain sequences of mouse antibody. Nature. 1970;228:1045–1047. doi: 10.1038/2281045a0. [DOI] [PubMed] [Google Scholar]
- 2.Tonegawa S. Reiteration frequency of immunoglobulin light chain genes: further evidence for somatic ge neration of antibody diversity. Proc Natl Acad Sci USA. 1976;73:203–207. doi: 10.1073/pnas.73.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis MM, Chien YH, Gascoigne NR, Hedrick SM. A murine T cell receptor gene complex: isolation, structure and rearrangement. Immunol Rev. 1984;81:235–258. doi: 10.1111/j.1600-065x.1984.tb01113.x. [DOI] [PubMed] [Google Scholar]
- 4.Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 5.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 6.Schluter SF, Bernstein RM, Bernstein H, Marchalonis JJ. ‘Big Bang’ emergence of the combinatorial immune system. Dev Comp Immunol. 1999;23:107–111. doi: 10.1016/s0145-305x(99)00002-6. [DOI] [PubMed] [Google Scholar]
- 7.Pancer Z, et al. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature. 2004;430:174–180. doi: 10.1038/nature02740. This is a seminal paper that described a novel rearranging gene in the lamprey. This work suggested strongly that jawless vertebrates have an alternative form of adaptive immunity that is not dependent on BCRs, TCRs or the MHC. [DOI] [PubMed] [Google Scholar]
- 8.Rogozin IB, Iyer LM, et al. Evolution and diversification of lamprey antigen receptors: evidence for involvement of an AID-APOBEC family cytosine deaminase. Nature Immunol. 2007;8:647–656. doi: 10.1038/ni1463. This paper suggests that the diversity of the VLR gene is generated by a gene conversion-like process that is presumably mediated by cytidine deaminases of the AID-APOBEC family. It also describes the identification of the lamprey VLRA gene. [DOI] [PubMed] [Google Scholar]
- 9.Hibino T, Loza-Coll M, et al. The immune gene repertoire encoded in the purple sea urchin genome. Dev Biol. 2006;300:349–365. doi: 10.1016/j.ydbio.2006.08.065. [DOI] [PubMed] [Google Scholar]
- 10.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 11.Klein J, Satta Y, O’hUigin C, Takahata N. The molecular descent of the major histocompatibility complex. Annu Rev Immunol. 1993;11:269–295. doi: 10.1146/annurev.iy.11.040193.001413. [DOI] [PubMed] [Google Scholar]
- 12.Flajnik MF. Comparative analyses of immunoglobulin genes: surprises and portents. Nature Rev Immunol. 2002;2:688–698. doi: 10.1038/nri889. [DOI] [PubMed] [Google Scholar]
- 13.Kaattari S, Evans D, Klemer J. Varied redox forms of teleost IgM: an alternative to isotypic diversity? Immunol R ev. 1998;166:133–142. doi: 10.1111/j.1600-065x.1998.tb01258.x. [DOI] [PubMed] [Google Scholar]
- 14.Dooley H, Flajnik MF. Shark immunity bites back: affinity maturation and memory response in the nurse shark, Ginglymostoma cirratum. Eur J Immunol. 2005;35:936–945. doi: 10.1002/eji.200425760. [DOI] [PubMed] [Google Scholar]
- 15.Wilson M, et al. A novel chimeric Ig heavy chain from a teleost fish shares similarities to IgD. Proc Natl Acad Sci USA. 1997;94:4593–4597. doi: 10.1073/pnas.94.9.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohta Y, Flajnik M. IgD, like IgM, is a primordial immunoglobulin class perpetuated in most jawed vertebrates. Proc Natl Acad Sci USA. 2006;103:10723–10728. doi: 10.1073/pnas.0601407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Y, et al. Identification of IgF, a hinge-region-containing Ig class, and IgD in Xenopus tropicalis. Proc Natl Acad Sci USA. 2006:103, 12087–12092. doi: 10.1073/pnas.0600291103. References 16 and 17 confirm that IgD is much older than previously realized and that it has evolved rapidly over evolutionary time. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenberg AS, et al. A novel ‘chimeric’ antibody class in cartilaginous fish: IgM may not be the primordial immunoglobulin. Eur J Immunol. 1996;26:1123–1129. doi: 10.1002/eji.1830260525. [DOI] [PubMed] [Google Scholar]
- 19.Berstein RM, Schluter SF, Shen S, Marchalonis JJ. A new high molecular weight immunoglobulin class from the carcharhine shark: implications for the properties of the primordial immunoglobulin. Proc Natl Acad Sci USA. 1996;93:3289–3293. doi: 10.1073/pnas.93.8.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ota T, Rast JP, Litman GW, Amemiya CT. Lineage-restricted retention of a primitive immunoglobulin heavy chain isotype within the Dipnoi reveals an evolutionary paradox. Proc Natl Acad Sci USA. 2003;100:2501–2506. doi: 10.1073/pnas.0538029100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen K, et al. Immunoglobulin D enhances immune surveillance by activating antimicrobial, proinflammatory and B cell-stimulating programs in basophils. Nature Immunol. 2009;10:889–898. doi: 10.1038/ni.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warr GW, Magor KE, Higgins DA. IgY: clues to the origins of modern antibodies. Immunol Today. 1995;16:392–398. doi: 10.1016/0167-5699(95)80008-5. [DOI] [PubMed] [Google Scholar]
- 23.Mussmann R, Du Pasquier L, Hsu E. Is Xenopus IgX an analog of IgA? Eur J Immunol. 1996;26:2823–2830. doi: 10.1002/eji.1830261205. [DOI] [PubMed] [Google Scholar]
- 24.Desmyter A, et al. Crystal structure of a camel single-domain VH antibody fragment in complex with lysozyme. Nature Struct Biol. 1996;3:803–811. doi: 10.1038/nsb0996-803. [DOI] [PubMed] [Google Scholar]
- 25.Greenberg AS, et al. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature. 1995;374:168–173. doi: 10.1038/374168a0. [DOI] [PubMed] [Google Scholar]
- 26.Greenberg AS, Steiner L, Kasahara M, Flajnik MF. Isolation of a shark immunoglobulin light chain cDNA clone encoding a protein resembling mammalian κ light chains: implications for the evolution of light chains. Proc Natl Acad Sci USA. 1993;90:10603–10607. doi: 10.1073/pnas.90.22.10603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Criscitiello MF, Flajnik MF. Four primordial immunoglobulin light chain isotypes, including λ and κ, identified in the most primitive living jawed vertebrates. Eur J Immunol. 2007;37:2683–2694. doi: 10.1002/eji.200737263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hohman VS, Schuchman DB, Schluter SF, Marchalonis JJ. Genomic clone for sandbar shark λ light chain: generation of diversity in the absence of gene rearrangement. Proc Natl Acad Sci USA. 1993;90:9882–9886. doi: 10.1073/pnas.90.21.9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rast JP, et al. Immunoglobulin light chain class multiplicity and alternative organizational forms in early vertebrate phylogeny. Immunogenetics. 1994;40:83–99. doi: 10.1007/BF00188170. [DOI] [PubMed] [Google Scholar]
- 30.Schwager J, Burckert N, Schwager M, Wilson M. Evolution of immunoglobulin light chain genes: analysis of Xenopus IgL isotypes and their contribution to antibody diversity. EMBO J. 1991;10:505–511. doi: 10.1002/j.1460-2075.1991.tb07976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 32.Reynaud CA, Anquez V, Grimal H, Weill JC. A hyperconversion mechanism generates the chicken light chain preimmune repertoire. Cell. 1987;48:379–388. doi: 10.1016/0092-8674(87)90189-9. [DOI] [PubMed] [Google Scholar]
- 33.Knight KL, Becker RS. Molecular basis of the allelic inheritance of rabbit immunoglobulin VH allotypes: implications for the generation of antibody diversity. Cell. 1990;60:963–970. doi: 10.1016/0092-8674(90)90344-e. [DOI] [PubMed] [Google Scholar]
- 34.Hinds KR, Litman GW. Major reorganization of immunoglobulin VH segmental elements during vertebrate evolution. Nature. 1986;320:546–549. doi: 10.1038/320546a0. [DOI] [PubMed] [Google Scholar]
- 35.Hinds-Frey KR, Nishikata H, Litman RT, Litman GW. Somatic variation precedes extensive diversification of germline sequences and combinatorial joining in the evolution of immunoglobulin heavy chain diversity. J Exp Med. 1993;178:815–824. doi: 10.1084/jem.178.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diaz M, Greenberg AS, Flajnik MF. Somatic hypermutation of the new antigen receptor gene (NAR) in the nurse shark does not generate the repertoire: possible role in antigen-driven reactions in the absence of germinal centers. Proc Natl Acad Sci USA. 1998;95:14343–14348. doi: 10.1073/pnas.95.24.14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee SS, Tranchina D, Ohta Y, Flajnik MF, Hsu E. Hypermutation in shark immunoglobulin light chain genes results in contiguous substitutions. Immunity. 2002;16:571–582. doi: 10.1016/s1074-7613(02)00300-x. [DOI] [PubMed] [Google Scholar]
- 38.Daggfeldt A, Bengten E, Pilstrom L. A cluster type organization of the loci of the immunoglobulin light chain in Atlantic cod (Gadus morhua L.) and rainbow trout (Oncorhynchus mykiss Walbaum) indicated by nucleotide sequences of cDNAs and hybridization analysis. Immunogenetics. 1993;38:199–209. doi: 10.1007/BF00211520. [DOI] [PubMed] [Google Scholar]
- 39.Hsu E, Criscitiello MF. Diverse immunoglobulin light chain organizations in fish retain potential to revise B cell receptor specificities. J Immunol. 2006;177:2452–2462. doi: 10.4049/jimmunol.177.4.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lange MD, Waldbieser GC, Lobb CJ. Patterns of receptor revision in the immunoglobulin heavy chains of a teleost fish. J Immunol. 2009;182:5605–5622. doi: 10.4049/jimmunol.0801013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zimmerman AM, Yeo G, Howe K, Maddox BJ, Steiner LA. Immunoglobulin light chain (IgL) genes in zebrafish: genomic configurations and inversional rearrangements between (VL-JL-CL) gene clusters. Dev Comp Immunol. 2008;32:421–434. doi: 10.1016/j.dci.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Danilova N, Bussmann J, Jekosch K, Steiner LA. The immunoglobulin heavy-chain locus in zebrafish: identification and expression of a previously unknown isotype, immunoglobulin Z. Nature Immunol. 2005;6:295–302. doi: 10.1038/ni1166. [DOI] [PubMed] [Google Scholar]
- 43.Hansen JD, Landis ED, Phillips RB. Discovery of a unique Ig heavy-chain isotype (IgT) in rainbow trout: implications for a distinctive B cell developmental pathway in teleost fish. Proc Natl Acad Sci USA. 2005;102:6919–6924. doi: 10.1073/pnas.0500027102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zinkernagel RM, Doherty PC. Immunological surveillance against altered self components by sensitised T lymphocytes in lymphocytic choriomeningitis. Nature. 1974;251:547–548. doi: 10.1038/251547a0. [DOI] [PubMed] [Google Scholar]
- 45.Charlemagne J, Fellah JS, De Guerra A, Kerfourn F, Partula S. T-cell receptors in ectothermic vertebrates. Immunol Rev. 1998;166:87–102. doi: 10.1111/j.1600-065x.1998.tb01255.x. [DOI] [PubMed] [Google Scholar]
- 46.Guo J, et al. Regulation of the TCRα repertoire by the survival window of CD4 + CD8+ thymocytes. Nature Immunol. 2002;3:469–476. doi: 10.1038/ni791. [DOI] [PubMed] [Google Scholar]
- 47.Havran WL, et al. Limited diversity of T-cell receptor y-chain expression of murine Thy-1+ dendritic epidermal cells revealed by V γ 3-specific monoclonal antibody. Proc Natl Acad Sci USA. 1989;86:4185–4189. doi: 10.1073/pnas.86.11.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morita CT, et al. Direct presentation of nonpeptide prenyl pyrophosphate antigens to human γδ T cells. Immunity. 1995;3:495–507. doi: 10.1016/1074-7613(95)90178-7. [DOI] [PubMed] [Google Scholar]
- 49.Thedrez A, et al. Self/non-self discrimination by human γδ T cells: simple solutions for a complex issue? Immunol Rev. 2007;215:123–135. doi: 10.1111/j.1600-065X.2006.00468.x. [DOI] [PubMed] [Google Scholar]
- 50.Rock EP, Sibbald PR, Davis MM, Chien YH. CDR3 length in antigen-specific immune receptors. J Exp Med. 1994;179:323–328. doi: 10.1084/jem.179.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Criscitiello MF, Saltis M, Flajnik MF. An evolutionary mobile antigen receptor variable region gene: doubly rearranging NAR-TcR genes in sharks. Proc Natl Acad Sci USA. 2006;103:5036–5041. doi: 10.1073/pnas.0507074103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parra ZE, et al. A unique T cell receptor discovered in marsupials. Proc Natl Acad Sci USA. 2007;104:9776–9781. doi: 10.1073/pnas.0609106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sciammas R, Bluestone JA. TCRγδ cells and viruses. Microbes Infect. 1999;1:203–212. doi: 10.1016/s1286-4579(99)80035-5. [DOI] [PubMed] [Google Scholar]
- 54.Chen H, et al. Characterization of arrangement and expression of the T cell receptor y locus in the sandbar shark. Proc Natl Acad Sci USA. 2009;106:8591–8596. doi: 10.1073/pnas.0811283106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flajnik MF, Kasahara M. Comparative genomics of the MHC: glimpses into the evolution of the adaptive immune system. Immunity. 2001;15:351–362. doi: 10.1016/s1074-7613(01)00198-4. [DOI] [PubMed] [Google Scholar]
- 56.Kaufman J. Co-evolving genes in MHC haplotypes: the ‘rule’ for nonmammalian vertebrates? Immunogenetics. 1999;50:228–236. doi: 10.1007/s002510050597. [DOI] [PubMed] [Google Scholar]
- 57.Trowsdale J. Genetic and functional relationships between MHC and NK receptor genes. Immunity. 2001;15:363–374. doi: 10.1016/s1074-7613(01)00197-2. [DOI] [PubMed] [Google Scholar]
- 58.Rogers SL, Viertlboeck BC, Gobel TW, Kaufman J. Avian NK activities, cells and receptors. Semin Immunol. 2008;20:353–360. doi: 10.1016/j.smim.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 59.Nyholm SV, et al. fester, a candidate allorecognition receptor from a primitive chordate. Immunity. 2006;25:163–173. doi: 10.1016/j.immuni.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 60.Sakano H, Huppi K, Heinrich G, Tonegawa S. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature. 1979;280:288–294. doi: 10.1038/280288a0. [DOI] [PubMed] [Google Scholar]
- 61.Du Pasquier L, Zucchetti I, De Santis R. Immunoglobulin superfamily receptors in protochordates: before RAG time. Immunol Rev. 2004;198:233–248. doi: 10.1111/j.0105-2896.2004.00122.x. [DOI] [PubMed] [Google Scholar]
- 62.Hernández Prada JA, et al. Ancient evolutionary origin of diversified variable regions demonstrated by crystal structures of an immune-type receptor in amphioxus. Nature Immunol. 2006;7:875–882. doi: 10.1038/ni1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu C, et al. Genes ‘waiting’ for recruitment by the adaptive immune system: the insights from amphioxus. J Immunol. 2005;174:3493–3500. doi: 10.4049/jimmunol.174.6.3493. [DOI] [PubMed] [Google Scholar]
- 64.Suzuki T, Shin I, Fujiyama A, Kohara Y, Kasahara M. Hagfish leukocytes express a paired receptor family with a variable domain resembling those of antigen receptors. J Immunol. 2005;174:2885–2891. doi: 10.4049/jimmunol.174.5.2885. [DOI] [PubMed] [Google Scholar]
- 65.Pancer Z, Mayer WE, Klein J, Cooper MD. Prototypic T cell receptor and CD4-like coreceptor are expressed by lymphocytes in the agnathan sea lamprey. Proc Natl Acad Sci USA. 2004;101:13273–13278. doi: 10.1073/pnas.0405529101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thompson CB. New insights into V(D)J recombination and its role in the evolution of the immune system. Immunity. 1995;3:531–539. doi: 10.1016/1074-7613(95)90124-8. [DOI] [PubMed] [Google Scholar]
- 67.Fugmann SD, Messier C, Novack LA, Cameron RA, Rast JP. An ancient evolutionary origin of the Rag1/2 gene locus. Proc Natl Acad Sci USA. 2006;103:3728–3733. doi: 10.1073/pnas.0509720103. This paper demonstrates that RAG1 and RAG2 were present in echinoderms (sea urchins) approximately 100 million years before the emergence of the AIS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kapitonov VV, Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. This work shows that a transposable element, transib, contains the RAG1 core element and is found in many animal species, including invertebrates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dreyfus DH. Paleo-immunology: evidence consistent with insertion of a primordial herpes virus-like element in the origins of acquired immunity. PLoS ONE. 2009;4:e5778. doi: 10.1371/journal.pone.0005778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds hypermethylated lysine 4 of histone H3 is necessary for efficient antigen–receptor–gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Klein J, Nikolaidis N. The descent of the antibody-based immune system by gradual evolution. Proc Natl Acad Sci USA. 2005;102:169–174. doi: 10.1073/pnas.0408480102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Flajnik MF, Du Pasquier L. Evolution of innate and adaptive immunity: can we draw a line? Trends Immunol. 2004;25:640–644. doi: 10.1016/j.it.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 73.Ohno S. Evolution by Gene Duplication. Springer; 1970. [Google Scholar]
- 74.Furlong RF, Holland PW. Were vertebrates octoploid? Phil Trans R Soc Lond B. 2002;357:531–544. doi: 10.1098/rstb.2001.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Panopoulou G, Poustka AJ. Timing and mechanism of ancient vertebrate genome duplications — the adventure of a hypothesis. Trends Genet. 2005;21:559–567. doi: 10.1016/j.tig.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 76.Kasahara M. The 2R hypothesis: an update. Curr Opin Immunol. 2007;19:547–552. doi: 10.1016/j.coi.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 77.Van de Peer Y, Maere S, Meyer A. The evolutionary significance of ancient genome duplications. Nature R ev Genet. 2009;10:725–732. doi: 10.1038/nrg2600. [DOI] [PubMed] [Google Scholar]
- 78.Kasahara M, Nakaya J, Satta Y, Takahata N. Chromosomal duplication and the emergence of the adaptive immune system. Trends Genet. 1997;13:90–92. doi: 10.1016/s0168-9525(97)01065-2. [DOI] [PubMed] [Google Scholar]
- 79.Kasahara M, Suzuki T, Du Pasquier L. On the origins of the adaptive immune system: novel insights from invertebrates and cold-blooded vertebrates. Trends Immunol. 2004;25:105–111. doi: 10.1016/j.it.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 80.Okada K, Asai K. Expansion of signaling genes for adaptive immune system evolution in early vertebrates. BMC Genomics. 2008;9:218. doi: 10.1186/1471-2164-9-218. This paper provides a comprehensive analysis of ohnologues that are involved in the AIS of jawed vertebrates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kasahara M. What do the paralogous regions in the genome tell us about the origin of the adaptive immune system? Immunol Rev. 1998;166:159–175. doi: 10.1111/j.1600-065x.1998.tb01261.x. [DOI] [PubMed] [Google Scholar]
- 82.Kasahara M, et al. Chromosomal localization of the proteasome Z subunit gene reveals an ancient chromosomal duplication involving the major histocompatibility complex. Proc Natl Acad Sci USA. 1996;93:9096–9101. doi: 10.1073/pnas.93.17.9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Katsanis N, Fitzgibbon J, Fisher EM. Paralogy mapping: identification of a region in the human MHC triplicated onto human chromosomes 1 and 9 allows the prediction and isolation of novel PBX and NOTCH loci. Genomics. 1996;35:101–108. doi: 10.1006/geno.1996.0328. [DOI] [PubMed] [Google Scholar]
- 84.Vienne A, et al. Evolution of the proto-MHC ancestral region: more evidence for the plesiomorphic organisation of human chromosome 9q34 region. Immunogenetics. 2003;55:429–436. doi: 10.1007/s00251-003-0601-x. [DOI] [PubMed] [Google Scholar]
- 85.Olinski RP, Lundin LG, Hallbook F. Conserved synteny between the Ciona genome and human paralogons identifies large duplication events in the molecular evolution of the insulin-relaxin gene family. Mol Biol Evol. 2006;23:10–22. doi: 10.1093/molbev/msj002. Based on the comparative analysis of the insulin-relaxin family in humans and C. intestinalis, this paper proposes that the MHC paralogy group and the neurotrophin paralogy group were derived from a single contiguous region on an invertebrate proto-chromosome. [DOI] [PubMed] [Google Scholar]
- 86.Abi-Rached L, Gilles A, Shiina T, Pontarotti P, Inoko H. Evidence of en bloc duplication in vertebrate genomes. Nature Genet. 2002;31:100–105. doi: 10.1038/ng855. [DOI] [PubMed] [Google Scholar]
- 87.Azumi K, et al. Genomic analysis of immunity in a Urochordate and the emergence of the vertebrate immune system: ‘waiting for Godot’. Immunogenetics. 2003;55:570–581. doi: 10.1007/s00251-003-0606-5. [DOI] [PubMed] [Google Scholar]
- 88.Holland LZ, et al. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 2008;18:1100–1111. doi: 10.1101/gr.073676.107. This paper provides a comprehensive analysis of the genes that encode the major biological systems in amphioxi. Among the genes analysed are those involved in host defence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hallbook F, Wilson K, Thorndyke M, Olinski RP. Formation and evolution of the chordate neurotrophin and Trk receptor genes. Brain Behav Evol. 2006;68:133–144. doi: 10.1159/000094083. [DOI] [PubMed] [Google Scholar]
- 90.Belov K, et al. Reconstructing an ancestral mammalian immune supercomplex from a marsupial major histocompatibility complex. PLoS Biol. 2006;4:e46. doi: 10.1371/journal.pbio.0040046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kasahara M. Genome dynamics of the major histocompatibility complex: insights from genome paralogy. Immunogenetics. 1999;50:134–145. doi: 10.1007/s002510050589. [DOI] [PubMed] [Google Scholar]
- 92.Marchalonis JJ, Edelman GM. Phylogenetic origins of antibody structure. III Antibodies in the primary immune response of the sea lamprey, Petromyzon marinus. J Exp Med. 1968;127:891–914. doi: 10.1084/jem.127.5.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Finstad J, Good RA. The evolution of the immune response. III Immunologic responses in the lamprey. J Exp Med. 1964;120:1151–1168. doi: 10.1084/jem.120.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Litman GW, Finstad FJ, Howell J, Pollara BW, Good RA. The evolution of the immune response. III Structural studies of the lamprey immunoglobulin. J Immunol. 1970;105:1278–1285. [PubMed] [Google Scholar]
- 95.Linthicum DS, Hildemann WH. Immunologic responses of Pacific hagfish. III Serum antibodies to cellular antigens. J Immunol. 1970;105:912–918. [PubMed] [Google Scholar]
- 96.Mayer WE, et al. Isolation and characterization of lymphocyte-like cells from a lamprey. Proc Natl Acad Sci USA. 2002;99:14350–14355. doi: 10.1073/pnas.212527499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Uinuk-Ool T, et al. Lamprey lymphocyte-like cells express homologs of genes involved in immunologically relevant activities of mammalian lymphocytes. Proc Natl Acad Sci USA. 2002;99:14356–14361. doi: 10.1073/pnas.212527699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Suzuki T, Shin I, Kohara Y, Kasahara M. Transcriptome analysis of hagfish leukocytes: a framework for understanding the immune system of jawless fishes. Dev Comp Immunol. 2004;28:993–1003. doi: 10.1016/j.dci.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 99.Pancer Z, Cooper MD. The evolution of adaptive immunity. Annu Rev Immunol. 2006;24:497–518. doi: 10.1146/annurev.immunol.24.021605.090542. [DOI] [PubMed] [Google Scholar]
- 100.Nagawa F, et al. Antigen-receptor genes of the agnathan lamprey are assembled by a process involving copy choice. Nature Immunol. 2007;8:206–213. doi: 10.1038/ni1419. This paper is the first to analyse the genetic mechanisms involved in VLR gene assembly It proposes that LRR modules are tethered by a copy-choice mechanism. [DOI] [PubMed] [Google Scholar]
- 101.Alder MN, et al. Diversity and function of adaptive immune receptors in a jawless vertebrate. Science. 2005;310:1970–1973. doi: 10.1126/science.1119420. [DOI] [PubMed] [Google Scholar]
- 102.Kim HM, et al. Structural diversity of the hagfish variable lymphocyte receptors. J Biol Chem. 2007;282:6726–6732. doi: 10.1074/jbc.M608471200. [DOI] [PubMed] [Google Scholar]
- 103.Herrin BR, et al. Structure and specificity of lamprey monoclonal antibodies. Proc Natl Acad Sci USA. 2008;105:2040–2045. doi: 10.1073/pnas.0711619105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Han BW, Herrin BR, Cooper MD, Wilson IA. Antigen recognition by variable lymphocyte receptors. Science. 2008;321:1834–1837. doi: 10.1126/science.1162484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Velikovsky CA, et al. Structure of a lamprey variable lymphocyte receptor in complex with a protein antigen. Nature Struct Mol Biol. 2009;16:725–730. doi: 10.1038/nsmb.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Alder MN, et al. Antibody responses of variable lymphocyte receptors in the lamprey. Nature Immunol. 2008;9:319–327. doi: 10.1038/ni1562. [DOI] [PubMed] [Google Scholar]
- 107.Guo P, et al. Dual nature of the adaptive immune system in lampreys. Nature. 2009;459:796–801. doi: 10.1038/nature08068. This paper indicates that lampreys have two lineages of lymphoid cells that resemble T and B cells of jawed vertebrates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pancer Z, et al. Variable lymphocyte receptors in hagfish. Proc Natl Acad Sci USA. 2005;102:9224–9229. doi: 10.1073/pnas.0503792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kasamatsu J, Suzuki T, Ishijima J, Matsuda Y, Kasahara M. Two variable lymphocyte receptor genes of the inshore hagfish are located far apart on the same chromosome. Immunogenetics. 2007;59:329–331. doi: 10.1007/s00251-007-0200-3. [DOI] [PubMed] [Google Scholar]
- 110.Haruta C, Suzuki T, Kasahara M. Variable domains in hagfish: NICIR is a polymorphic multigene family expressed preferentially in leukocytes and is related to lamprey TCR-like. Immunogenetics. 2006;58:216–225. doi: 10.1007/s00251-006-0098-1. [DOI] [PubMed] [Google Scholar]
- 111.Yokoyama WM, Plougastel BF. Immune functions encoded by the natural killer gene complex. Nature Rev Immunol. 2003;3:304–316. doi: 10.1038/nri1055. [DOI] [PubMed] [Google Scholar]
- 112.Takahashi T, et al. Natural killer cell receptors in the horse: evidence for the existence of multiple transcribed LY49 genes. Eur J Immunol. 2004;34:773–784. doi: 10.1002/eji.200324695. [DOI] [PubMed] [Google Scholar]
- 113.Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–767. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- 114.Boehm T. One problem, two solutions. Nature Immunol. 2009;10:811–813. doi: 10.1038/ni0809-811. [DOI] [PubMed] [Google Scholar]
- 115.Wada H, et al. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–772. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- 116.Litman GW, Cannon JP, Dishaw LJ. Reconstructing immune phylogeny: new perspectives. Nature Rev Immunol. 2005;5:866–879. doi: 10.1038/nri1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Beutler B, et al. Genetic analysis of resistance to viral infection. Nature Rev Immunol. 2007;7:753–766. doi: 10.1038/nri2174. [DOI] [PubMed] [Google Scholar]
- 118.Hedrick SM. The acquired immune system: a vantage from beneath. Immunity. 2004;21:607–615. doi: 10.1016/j.immuni.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 119.Solem ST, Stenvik J. Antibody repertoire development in teleosts — a review with emphasis on salmonids and Gadus morhua L. Dev Comp Immunol. 2006;30:57–76. doi: 10.1016/j.dci.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 120.Matsunaga T, Rahman A. What brought the adaptive immune system to vertebrates? — The jaw hypothesis and the seahorse. Immunol Rev. 1998;166:177–186. doi: 10.1111/j.1600-065x.1998.tb01262.x. [DOI] [PubMed] [Google Scholar]
- 121.Kaufman J, Volk H, Wallny HJ. A ‘minimal essential Mhc’ and an ‘unrecognized Mhc’: two extremes in selection for polymorphism. Immunol Rev. 1995;143:63–88. doi: 10.1111/j.1600-065x.1995.tb00670.x. [DOI] [PubMed] [Google Scholar]
- 122.Flajnik MF. Churchill and the immune system of ectothermic vertebrates. Immunol Rev. 1998;166:5–14. doi: 10.1111/j.1600-065x.1998.tb01248.x. [DOI] [PubMed] [Google Scholar]
- 123.Tasumi S, et al. High-affinity lamprey VLRA and VLRB monoclonal antibodies. Proc Natl Acad Sci USA. 2009;106:12891–12896. doi: 10.1073/pnas.0904443106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Travis J. Origins. On the origin of the immune system. Science. 2009;324:580–582. doi: 10.1126/science.324_580. [DOI] [PubMed] [Google Scholar]
- 125.Fall-Ngai M. Adaptive immunity: care for the community. Nature. 2007;445:153. doi: 10.1038/445153a. [DOI] [PubMed] [Google Scholar]
- 126.Long JA, Trinajstic K, Johanson Z. Devonian arthrodire embryos and the origin of internal fertilization in vertebrates. Nature. 2009;457:1124–1127. doi: 10.1038/nature07732. [DOI] [PubMed] [Google Scholar]
- 127.Friedman R, Hughes AL. Pattern and timing of gene duplication in animal genomes. Genome Res. 2001;11:1842–1847. doi: 10.1101/gr.200601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hughes AL, Friedman R. 2R or not 2R: testing hypotheses of genome duplication in early vertebrates. J Struct Funct Genomics. 2003;3:85–93. [PubMed] [Google Scholar]
- 129.Gibson TJ, Spring J. Evidence in favour of ancient octaploidy in the vertebrate genome. Biochem Soc Trans. 2000;28:259–264. doi: 10.1042/bst0280259. [DOI] [PubMed] [Google Scholar]
- 130.Lundin LG, Larhammar D, Hallbook F. Numerous groups of chromosomal regional paralogies strongly indicate two genome doublings at the root of the vertebrates. J Struct Funct Genomics. 2003;3:53–63. [PubMed] [Google Scholar]
- 131.Putnam NH, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. This paper, written by core members of the amphioxus genome project, describes the salient features of the amphioxus genome, with emphasis on genome evolution. It provides strong evidence to support the 2R hypothesis. [DOI] [PubMed] [Google Scholar]
- 132.Kuraku S, Meyer A, Kuratani S. Timing of genome duplications relative to the origin of the vertebrates: did cyclostomes diverge before or after? Mol Biol Evol. 2009;26:47–59. doi: 10.1093/molbev/msn222. This paper proposes that, contrary to the commonly held view, both the first and second rounds of WGD occurred after the emergence of a common ancestor of vertebrates and before the divergence of jawed and jawless vertebrates. [DOI] [PubMed] [Google Scholar]
- 133.Karre K. NK cells, MHC class I molecules and the missing self. Scand J Immunol. 2002;55:221–228. doi: 10.1046/j.1365-3083.2002.01053.x. [DOI] [PubMed] [Google Scholar]
- 134.Jaillon O, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- 135.Vandepoele K, De Vos W, Taylor JS, Meyer A, Van de Peer Y. Major events in the genome evolution of vertebrates: paranome age and size differ considerably between ray-finned fishes and land vertebrates. Proc Natl Acad Sci USA. 2004;101:1638–1643. doi: 10.1073/pnas.0307968100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Blair JE, Hedges SB. Molecular phylogeny and divergence times of deuterostome animals. Mol Biol Evol. 2005;22:2275–2284. doi: 10.1093/molbev/msi225. [DOI] [PubMed] [Google Scholar]
- 137.Kasahara M. The chromosomal duplication model of the major histocompatibility complex. Immunol Rev. 1999;167:7–32. doi: 10.1111/j.1600-065x.1999.tb01379.x. [DOI] [PubMed] [Google Scholar]
- 138.Horton R, et al. Gene map of the extended human MHC. Nature Rev Genet. 2004;5:889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.