Abstract
Precise and accurate quantification of protein expression levels in a complex biological setting is challenging. Here, we describe a method for absolute quantitation of endogenous proteins in cell lysates using an automated capillary immunoassay system (the size-based Simple Western system, ProteinSimple, CA). The method was able to accurately measure the absolute amounts of target proteins at picogram or sub-picogram levels per nanogram of cell lysates. The measurements were independent of the cell matrix or the cell lysis buffer and were not affected by different antibody affinities for their specific epitopes. We then applied this method to quantitate absolute levels of expression of protein kinase C (PKC) isoforms in LNCaP and U937 cells, two cell lines used extensively for probing the downstream biological responses to PKC targeted ligands. Our absolute quantitation confirmed the predominance of PKCδ in both cells, supporting the important functional role of this PKC isoform in these cell lines. The method described here provides an approach to accurately quantitate levels of protein expression and correlate protein level with function. In addition to enhanced accuracy relative to conventional western analysis, it circumvents the distortions inherent in comparison of signal intensities from different antibodies with different affinities.
Keywords: Protein kinase C (PKC), capillary immunoassay, Simple Western, quantative protein analysis
Introductory Statement
Information about protein expression is important for understanding the mechanism of protein function as well as for formulating strategies to develop biomedicines [1, 2]. While mRNA quantitation has been widely used as an indirect indication of protein expression, studies have found that mRNA and protein levels are not always correlated [3–5]. Protein quantitation methods have been established through both antibody-based and mass spectrometry based technologies. While those technologies provide good information regarding relative protein levels as well as protein modifications in response to stimuli, methods for precise and accurate quantitation of absolute protein levels in a complex biological matrix face many challenges [2, 6]. For example, quantification standards are difficult to establish in ELISA and antibody array assays due to endogenous signal interference, and poor assay reproducibility has been observed in conventional westerns. Though mass spectrometry provides a high precision analysis platform, it may have low accuracy due to the prevalence of interference from other peptides and small molecules in the sample matrix. Mass spectrometry methods also require complicated sample preparation procedures [2, 7].
In this manuscript we described a method to precisely and accurately quantitate absolute protein expression in cell lysates by using the size-based Simple Western system. Simple Western™ is a gel-free, blot-free, capillary-based, automated western blotting system recently developed by ProteinSimple (Santa Clara, CA). In Simple Western™ analysis, all steps following sample preparation are fully automated, including sample loading, size-based protein separation, immunoprobing, washing, detection and data analysis. The system greatly reduces the variability caused by manual processes in conventional western. Data generated by the Simple Western™ system are highly quantitative with good run to run reproducibility [8]. Using the Simple Western™ system, we developed a method for absolute quantitation of endogenous proteins in cell lysates by spiking the samples with GST-tagged recombinant proteins as a standard for comparison, and we demonstrated the reliability and accuracy of the method for measuring absolute levels of Erk1 and Erk2 in total cell lysates. To illustrate the power of this approach, we then applied this system to quantitate PKC isoforms in LNCaP and U937 cells. We were able to compare the relative abundance of PKC isoforms and determine the absolute protein levels of the different PKC isoforms in the two cell lines quantitatively. This latter information is helpful in understanding the relative contributions of these various PKC isoforms to the signaling network activated upon addition of PKC ligands such as the phorbol esters or bryostatin 1, a compound currently in cancer clinical trials (www.clinicaltrials.gov).
PKC is a validated therapeutic target for cancer [9, 10]. Intense efforts are underway to develop the next generation of bryostatin related drugs through structural simplification while maintaining its biological specificity for PKC proteins [11–13]. LNCaP and U937 are ideal cell lines for this investigation because they are well characterized systems that highlight the dramatic differences in biological response to the phorbol esters and bryostatin [11, 14]. A critical part of this effort is to understand which PKC isoforms in these cells are important for response and how these individual PKC isoforms are differentially modulated by phorbol ester, by bryostatin 1, and by various structural analogs of bryostatin 1 [14–16].
Materials and methods
Materials
Recombinant proteins GST-PKCα, GST-PKCβII, GST-PKCδ, GST-PKCε and GST-Erk1 were from SignalChem (Richmond, BC, Canada), and GST-Erk2 was from EMD Millipore (Billerica, MA). Primary antibodies anti-PKCβII (sc-13149), anti-PKCδ (sc-937), and anti-PKC (sc-214) were from Santa Cruz Biotechnology (Santa Cruz, CA), the anti-PKC (#1510-1) was from Epitomics (Burlingame, CA), and the anti-Erk1 (#05-957) and anti-Erk1/2 (#06-182) were from Millipore. The secondary antibodies, either goat-anti-rabbit or goat-anti-mouse horseradish peroxidase (HRP)-conjugated, were from Jackson ImmunoResearch (West Grove, PA) for Simple Western™ analysis and from Bio-Rad (Hercules, CA) for conventional western analysis. The human prostate cancer cell line LNCaP, the human monocytic leukemia cell line U937, the human breast cancer cell line MCF7, the human cervical cancer cell line HeLa, the fetal bovine serum (FBS), and RPM1-1640 medium were from ATCC (Manassas, VA).
Cell growth
The cells were grown at 37°C in a humidified atmosphere at 5% CO2 with growth media as recommended by the ATCC.
Cell Lysate Preparation for Simple Western Analysis
Cells were lysed with M-Per buffer (Thermo Scientific, Waltham, MA) or RIPA buffer (20 mM HEPES (pH 7.5), 150 mM NaCl, 1% NP40 alternative, 0.25% sodium deoxycholate (w/v) and 10 % glycerol) containing phosphatase and protease inhibitors (EMD Millipore, Billerica, MA). Protein concentration was measured on a SpectraMax Plus384 (Molecular Devices, Sunnyvale CA) microplate reader using the Pierce 660 protein assay and BSA standards (Thermo Scientific, Waltham, MA).
Real time RT-PCR analysis
RNA was isolated from cultured cells with TRIzol reagent following the manufacturer’s protocol (Invitrogen, Carlsbad, CA). For cDNA synthesis, 1.5 µg of total RNA was reverse transcribed using the iScript Advanced cDNA synthesis kit (Bio-Rad) as recommended by the manufacturer. Real time qPCR was performed on MyiQ or iQ5 instruments (Bio-Rad) in a volume of 20 µl using iQ SYBR Green Supermix (Bio-Rad) on 150 times diluted cDNA. The primers used were predesigned and validated primers from Qiagen (Valencia, CA) for PKC and GAPDH or from Origene (Rockville, MD) for the other genes. Relative gene expression levels were calculated using the 2−(Ct) formula, where Ct represents the cycle difference between the gene of interest and GAPDH, used as internal control. The efficiency of the qPCR reactions was between 99% and 109 % when tested on serially diluted (1:5) “universal” RNA samples prepared to contain all transcripts of interest.
Western blot analysis
Total cell lysates were prepared by vortexing and sonication in ice cold phosphate buffered saline solution (PBS) containing 1% Triton X-100 and supplemented with protease and phosphatase inhibitors (Roche, Branford, CT). Cell debris was removed by centrifugation at 2600 × g for 5 min in a cooled microcentrifuge (Eppendorf). Samples containing the same amounts of protein (protein concentration measured using the BIO-RAD DCTm protein assay) were added to SDS and beta-mercaptoethanol containing sample buffer (Quality Biological Inc), incubated at 100°C for 5 min, separated on 10% SDS-polyacrylamide gels (Invitrogen), and transferred to nitrocellulose membranes (GE Healthcare, Waukesha, WI) at 100 volts (constant) for 75 minutes using a Mini Trans-Blot® Electrophorectic Transfer Cell (Bio-Rad). The membranes were blocked with 5 % nonfat dry milk (Bio-Rad) diluted in PBS, incubated overnight at 4°C with primary antibodies (dilutions empirically determined to give optimal results), washed (3 times 10 min in PBS containing 0.5% Tween-20), and incubated for 1 hr at room temperature with secondary antibodies. After washing (3 times 10 min in PBS containing 0.5 % Tween 20), the signal was developed by ECL and detected on high performance chemiluminescence film (GE Healthcare). The scanned films were edited using Adobe Photoshop CS3 (Adobe Systems, McLean, VA).
Simple Western Analysis
Simple Western analyses were performed according to the ProteinSimple user manual. In brief, cell lysate samples were mixed with a master mix (ProteinSimple, Santa Clara, CA) to a final concentration of 1×sample buffer, 1×fluorescent molecular weight markers and 40mM DTT, then heated at 95 °C for 5 minutes. The samples, blocking reagent, primary antibodies, HRP conjugated second antibodies, chemiluminescent substrate, separation and stacking matrices were also dispensed to designated wells in a 384-well plate. After plate loading, the separation electrophoresis and immunodetection steps took place in the capillary system and were fully automated. Simon Simple Western analysis is carried out at room temperature, and instrument default settings were used except as specified below.. Capillaries were first filled with separation matrix followed by stacking matrix, and about 40nL sample loading. During electrophoresis, proteins were separated on the basis of molecular weight through the stacking and separation matrices at 250 volts for 40–50 minutes and then immobilized on the capillary wall using proprietary photo-activated capture chemistry. The matrices were then washed out. Capillaries were next incubated with a blocking reagent for 15 minutes, and target proteins were immunoprobed with primary antibodies followed by HRP-conjugated secondary antibodies. All antibodies were diluted in Immunobooster (Bioworld Consulting Laboratories, Mt. Airy, MD) or antibody diluent (ProteinSimple) with a 1:50 or 1:100 dilution. The antibody incubation time was 10–15 minutes with Immunnobooster or 60–120 minutes with antibody diluents. Luminol and peroxide (ProteinSimple) were then added to generate chemiluminescence which was captured by a CCD camera. The digital image was analyzed with Compass software (ProteinSimple), and the quantified data of the detected protein were reported as molecular weight, signal/peak intensity etc.
Results and discussion
Absolute Quantitation of Endogenous Protein in Cell Lysates
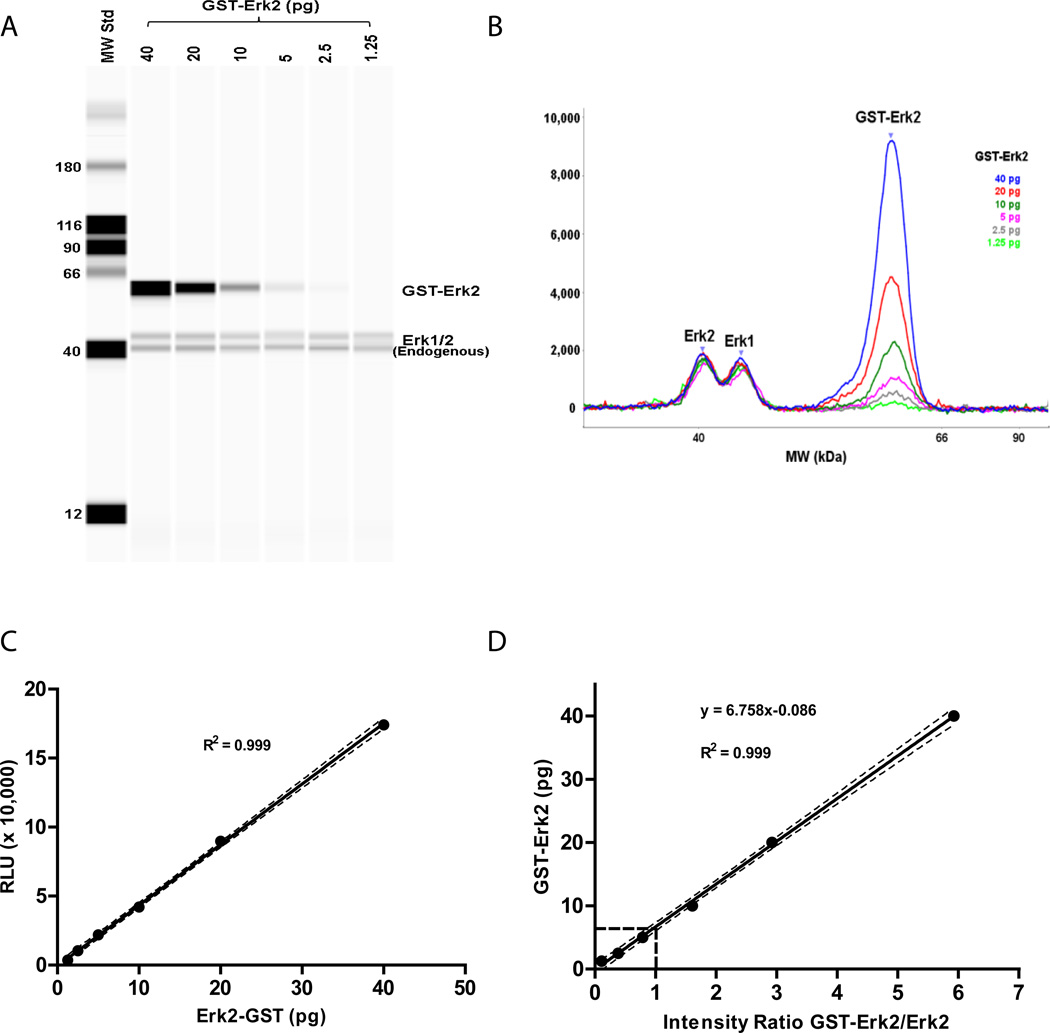
A method for absolute quantitation of endogenous proteins, using the Simple Western™ system, was developed using Erk2 as a prototype in LNCaP cell lysates. Serial dilutions of recombinant GST-Erk2, representing the standard, were spiked into LNCaP lysates. The recombinant protein / cell lysate mixes were separated on the Simple Western system and analyzed for Erk2. The GST-tag on the GST-Erk2 standard confers a change in size from the endogenous Erk2, permitting separation of the signals from the endogenous Erk2 and the exogenous Erk2 standard. Figures 1A/B illustrates a representative experiment, with data displayed in pseudo-gel and electropherogram formats, respectively, using an ERK1/2 antibody that recognizes both Erk1 and Erk2. A constant level of Erk1/2 can be seen with the serially decreasing GST-Erk2 standard. The peak areas in figure 1B represent target signal intensities and are precisely quantitated by the computer software. Linearity of the detected Erk2 signal was observed over the range of 1.25 to 40 pg GST-Erk2 protein (Figure 1C). Using the linear regression analysis approach described by Rudolph et al [17], the amount of endogenous Erk2 was calculated from the point at which the intensities of spiked-in GST-Erk2 standard and endogenous Erk2 were equivalent (intensity ratio = 1). In a single experiment as illustrated in Figure 1, we found that 6.67 pg of GST-Erk2 was predicted from the best-fit linear curve to yield a signal equivalent to that for the endogenous Erk2 present in 16.5 ng of LNCaP lysate (Figure 1D). Further conversion, accounting for the molecular weight difference between the two proteins (GST -Erk2 MW = 67.8 kDa, Erk2 MW = 42 kDa), reveals that 4.13 pg of endogenous Erk2 was present in 16.5 ng LNCaP lysate, indicating a concentration of 0.25 pg Erk2 / ng LNCaP lysate.
Figure 1. Absolute quantitation of endogenous Erk2 in LNCaP Lysate.
Serial dilutions of GST-Erk2 recombinant proteins were spiked into 16.5 ng LNCaP lysate, recombinant protein / cell lysate samples were mixed with sample loading master mix and analyzed with the Simple Western system as described under Methods. (A) Pseudo-gel and (B) electro-pherogram image of Simple Western results using anti-Erk1/2 antibody for blotting; (C) GST-Erk2 signal intensity is plotted versus spiked-in amount of GST-Erk2. Detection linearity is observed with 1.25 to 40 pg GST-Erk2; (D) Linear regression analysis for endogenous Erk2 determination. 6.67 pg of GST-Erk2 was predicted from the best-fit linearity curve when intensity ratio = 1. Data are of a representative experiment. Dash-lines indicate the 95% confidential intervals.
To test the robustness of the quantitation method, absolute amounts of endogenous Erk2 were measured as the total amount of LNCaP lysates was varied. As shown in Table 1, an average of 0.26 pg Erk2 per ng of total cell lysate protein was obtained for samples ranging from 7.6 to 60.8 ng. The CV% among the samples was 8.4%.
Table 1.
Absolute Quantitation of Endogenous Erk2 in LNCaP cells
| Lysate Amount (ng) |
Erk2 (pg in lysate) |
Erk2 (pg / ng of lysate) |
|---|---|---|
| 7.6 | 2.06 | 0.27 |
| 16.5 | 3.93 | 0.24 |
| 33 | 8.11 | 0.25 |
| 60.8 | 17.9 | 0.29 |
| mean | 0.26 | |
| %CV | 8.4 | |
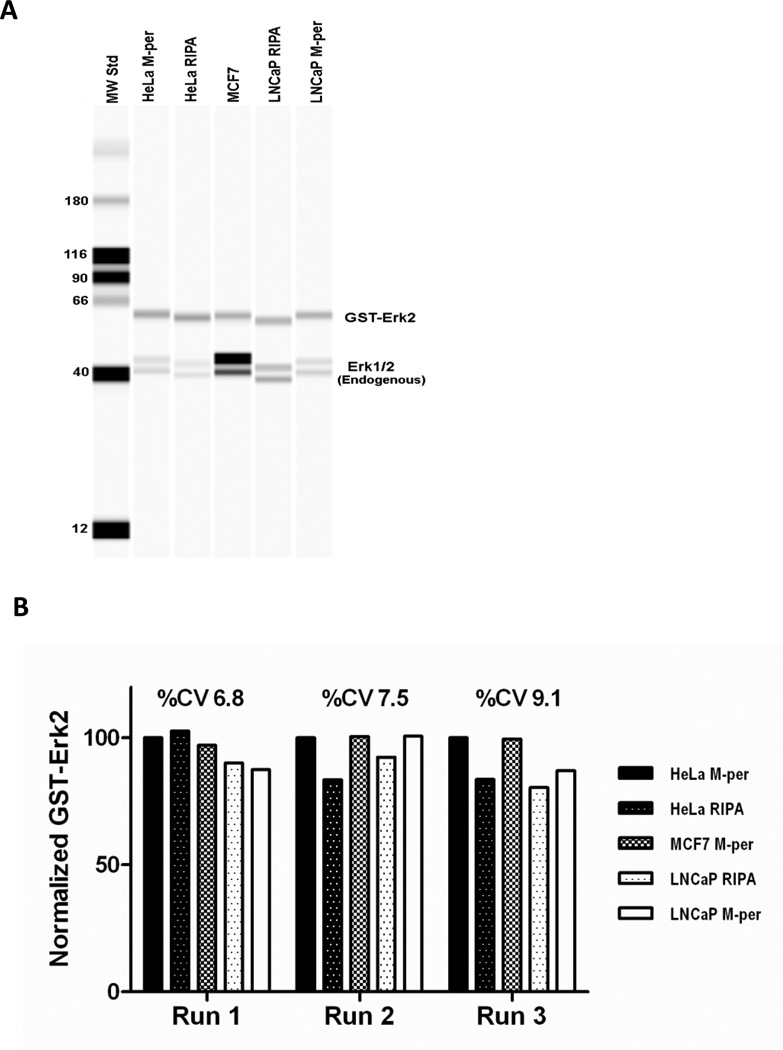
To examine if different lysates or different lysate preparation conditions may influence the quantitation of GST-Erk2, we compared the signals after adding equal amounts of GST-Erk2 protein to different lysates. The lysates were from three cell lines, either HeLa adenocarcinoma cells, MCF7 breast cancer cells, or LNCaP prostate cancer cells, and were prepared with either Bicine/CHAPS based M-per lysis buffer [18] or HEPES/NP-40 based RIPA lysis buffer (Figure 2A). Although the mobilities of GST-Erk2 and endogenous Erk1 and Erk2 were affected by the different buffer conditions (Figure 2A), the measured quantity of GST-Erk2 was not changed. A %CV of less than 10% was observed for its signal under the various conditions (Figure 2B). Although the purpose of this experiment was to examine the possible influence of different cell lysates or lysis conditions on the GST-Erk2 signal, it might be noted that higher Erk2 expression was observed in MCF7 cells compared to Hela and LNCaP cells both on the absolute expression level and the ratio to Erk1 signal. The method described in this manuscript can be used to further quantify and compare the expression levels of the Erk proteins to study the possible differential functions of the Erk proteins in different cell type or biological process. We further evaluated how the measured amounts of the endogenous protein compared if we use different primary antibodies that recognize different epitopes on the protein. Endogenous Erk1 was used as the target in this evaluation, and purified Erk1 was used as standard. One antibody was Erk1 specific (Millipore, #05-957) and the other antibody recognized both Erk1 and Erk2 proteins (Millipore, #06-182). Comparable results were obtained with the two antibodies (Table 2). Measured values of 0.27 ± 0.02 pg and 0. 31 ± 0.05 pg (mean ± SD, n = 3 experiments) Erk1 per ng of total cell lysate proteins from MCF7 cells were measured by the anti-Erk1 and anti-Erk1/2 antibodies, respectively. A somewhat higher standard deviation was observed with the anti-Erk1/2 antibody, perhaps due to the slight interference from the Erk2 signal.
Figure 2. Matrix effect on Erk2 signal.
10 pg (Run 1& 3) or 20 pg (Run 2) of GST-Erk2 was spiked into HeLa, MCF-7, and LNCaP cell lysates prepared with M-per or RIPA buffer and analyzed with the Simple Western system. (A) Pseudo-gel image of one representative run indicates almost identical GST-ERk2 signal strength in different cell lysates prepared with either M-per or RIPA buffer. (B) GST-Erk2 signals were normalized to the GST-Erk2 signal in HeLa M-per lysate and compared.
Table 2.
Endogenous Erk1 Quantitation with anti-Erk1 Specific and anti-Erk1/2 Antibody in MCF7 cells
| anti-Erk1 (pg Erk1 / ng lysate) |
anti-Erk1/2 (pg Erk1 / ng lysate) |
|
|---|---|---|
| Run1 | 0.24 | 0.25 |
| Run2 | 0.28 | 0.33 |
| Run3 | 0.28 | 0.35 |
| Mean | 0.27 | 0.31 |
| SD | 0.02 | 0.05 |
| %CV | 7.4 | 16.1 |
The above data demonstrate that, using the Simple Western™ system, we are able to precisely and accurately quantitate the absolute amounts of an endogenous protein in cell lysates. The measurement was not appreciably affected by cell matrix, sample lysis buffer or antibody affinity differences.
Absolute quantitation of PKC isoforms and a comparison in LNCaP and U937 cells
As part of our characterization of the differential mechanisms of action of phorbol esters, bryostatin 1, and synthetic bryostatin 1 analogs, we have found that the various ligands differentially interact with and down regulate the various endogenous PKC isoforms in the LNCaP and U937 cells [14–16]. They likewise have differential effects on cellular and molecular responses downstream of PKC. Given that PKC isoforms show only modest differences in substrate selectivity and absolute activity, it would be helpful to know the absolute levels of PKC isoform expression to better understand the contribution of the various isoforms to the downstream responses. For example, using siRNA to suppress the expression of individual PKC isoforms in the LNCaP cells, Caino et al concluded that PKCδ played the predominant role in the induction of gene response upon phorbol ester treatment [19]. Was this an effect of PKC isoform specificity or of expression level?
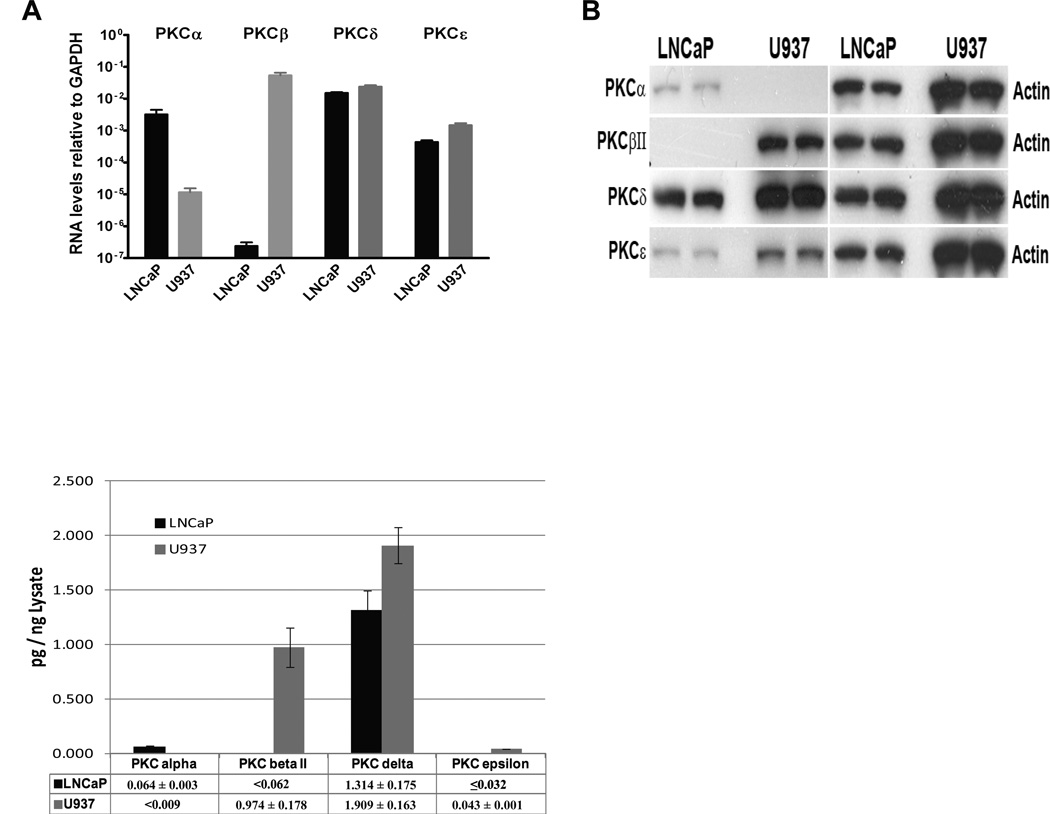
Using qPCR, we compared the levels of mRNA expression for the different PKC isoforms in the LNCaP and U937 cells. Differences in expression level of up to 6 orders of magnitude were found (Figure 3A). PKCδ and PKCα were the major isoforms in the LNCaP cells, with a lower level of PKCε. Relative levels of expression were 1, 0.25 and 0.03, respectively (note the Log10 scale in Figure 3A). The mRNA level of PKC II was orders of magnitude lower than that of PKCδ. In the U937 cells, PKCδ and PKCβII were the predominant isoforms, with a lower level of PKCε and PKCα. Relative levels of expression were 1, 2.2, 0.07 and 0.0005 for PKCδ, PKC II, PKCε and PKCα, respectively.
Figure 3. PKC isoform comparison in LNCaP and U937 cells.
(A) qPCR quantitation of PKC isoforms. Real time RT-PCR analysis was performed on RNA prepared from LNCaP and U937 cells using predesigned and validated primers for the indicated genes and GAPDH as control. Data represent average ± SEM values from three independent experiments. (B) Conventional western blot analysis. 15 g total cell lysates prepared from LNCaP and U937 cells were separated on SDS-PAGE, electro-transferred and immunoblotted with the indicated antibodies. Representative images using the same ECL exposure time for all PKC signals from three independent experiments are shown. The -actin signal was used as loading control. (C) Absolute quantitation of PKC isoform proteins using methods described in Figure 1. Data represent mean ± SD values from at least three independent experiments (see supplemental table for detail).
Conventional western blotting for protein expression of PKC isoforms gave a somewhat different image (Figure 3B). In the LNCaP cells, PKCδ was predominant and PKCα and PKCε appeared to be similar, contrasting with the appreciably lower level of PKCε mRNA expression, relative to PKCα. In the U937 cells, the band intensity for PKCδ was strongest, with decreasing intensities for PKC II and PKCε. Once again, the western blotting showed a particular difference with the qPCR for the relative expression of PKCε and PKCβII, which were 30 fold different by qPCR but much more similar by western blotting.
Of course, both measures of relative expression of PKC isoforms are only indirect indicators of the actual levels of PKC isoform protein expression. The levels of mRNA expression cannot incorporate efficiencies of translation or differential stability of the proteins. Comparison of band intensities by western blotting is very much subject to the differences in antibody affinities for the different proteins being compared and possible variations on image exposure time.
To determine the absolute levels of the PKC isoforms in the LNCaP and U937 cells, we used the approach described above with spiking of the cell lysates with individual purified GST-PKC isoforms as standards and quantitation with the Simple Western™ system (Figure 3C, also see supplemental material for representative linear regression analysis data). We could thus determine both the relative expression levels of the individual PKC isoforms in each cell line as well as their absolute levels relative to total protein. In the LNCaP cells relative levels of protein expression were 1, 0.05 and 0.02 for PKCδ, PKCα respectively, with the PKCε being barely detectable in the lowest PKCε antibody detection limit, which is about 0.032 pg per ng of total protein. No endogenous PKC II protein was detected at the lowest PKC II antibody detection limit, which was less than 0.062 pg per-ng of total protein. In the U937 cells relative levels of protein expression were 1, 0.51 and 0.02 for PKCδ, PKC II and PKCε, respectively. No endogenous PKCα protein was detected at the lowest PKCα antibody detection limit, which was less than 0.009 pg per ng of total protein. The data indicated that PKCδ is the dominant PKC isoform in LNCaP cells, while there is high PKCδ and PKCβII expression in U937 cells.
All three methods show similarities regarding relative PKC isoform expression levels between LNCaP and U937 cells. For example, expression levels of PKCβII, PKCδ and PKCε are higher in U937 cells, while LNCaP cells have higher PKCα expression than U937 cells. Comparing the absolute quantitation method with the indirect measures afforded by qPCR for mRNA expression or comparison on band intensities by western blotting, some discrepancies were found regarding the expression levels of different PKC isoforms in particular cells. In the U937 cells, qPCR had suggested a higher level of PKC II than of PKC, in contrast to the absolute protein quantitation or western blotting. In the LNCaP cells, western blotting would suggest that PKC was similar in level to PKC, in contrast to the absolute protein quantitation or qPCR which showed lower PKC expression than PKC (see Figure 3 for detail).
The predominance of the PKCδ isoform in the LNCaP cells is consistent with the contribution of this isoform to PMA induced gene expression [19]. The relatively low level of PKCε expression in these cells, as quantitated by Simple Western™, likewise helps to explain the limited contribution of this isoform to the effects of PMA on gene expression. In the U937 cells, the high level of PKCβII protein expression fits with the findings that bryostatin 1, which causes PKCβII to down regulate to a much greater extent than does PMA, fails to induce the same long term effects as does PMA for cell attachment and inhibition of proliferation [15].
Conclusions
The Simple Western™ system, with its high quality data quantitation and excellent assay reproducibility, allowed us to detect the relative abundance of the protein isoforms and to develop a method for precise quantitation of proteins at the picogram or sub-picogram level per nanogram cell lysate. The method of absolute quantitation of endogenous protein in cell lysates was demonstrated to be reliable and accurate as it was not affected by variations in sample matrix, sample concentration and detection antibody. Absolute quantitation of protein circumvents the problem caused by varying antibody affinities for different target proteins, which in many circumstance may yield a misleading impression of relative abundance. Although some approximation of levels of absolute expression could be obtained by traditional western blotting upon inclusion of purified protein standards as was done here, the Simple Western™ system combines ease of operation, linearity over a broad range of protein loads, tight quantitation, and good assay reproducibility. The method described here provides an approach to accurately correlate protein quantities with their function. This may be of particular importance in cases where relative levels of expression are central to their function, as would be case for postulated protein partners in a complex.
While the approach for standardization described here represents one approach, other alternatives could also be considered. Provided linearity of signal has been confirmed in a specific setting, then replicates of a single concentration of standard could be used for normalization. Likewise, standardization could be provided by purified, untagged standard protein loaded in parallel samples not including the experimental cell lysate. This latter approach would control for any distortions of antibody signal of the standard conferred by the GST or other protein tag, although it would reciprocally fail to control for any influence of the lysate or of loading. Given the robustness of the system as described here, all of these approaches would be expected to be powerful.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program, National Institutes of Health, Center for Cancer Research, National Cancer Institute (Project Z1A BC 005270, Z1C BC 011434).
ABBREVIATIONS
- PKC
Protein Kinase C
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brody EN, Gold L, Lawn RM, Walker JJ, Zichi D. High-content affinity-based proteomics: unlocking protein biomarker discovery. Expert Rev Mol Diagn. 2010;10(8):1013–1022. doi: 10.1586/erm.10.89. [DOI] [PubMed] [Google Scholar]
- 2.Gillette MA, Carr SA. Quantitative analysis of peptides and proteins in biomedicine by targeted mass spectrometry. Nat Methods. 2013;10(1):28–34. doi: 10.1038/nmeth.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst. 2009;5(12):1512–1526. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pascal LE, True LD, Campbell DS, Deutsch EW, Risk M, Coleman IM, Eichner LJ, Nelson PS, Liu AY. Correlation of mRNA and protein levels: cell type-specific gene expression of cluster designation antigens in the prostate. BMC Genomics. 2008;9:246. doi: 10.1186/1471-2164-9-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol. 2007;25(1):117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- 6.Ellington AA, Kullo IJ, Bailey KR, Klee GG. Antibody-based protein multiplex platforms: technical and operational challenges. Clin Chem. 2010;56(2):186–193. doi: 10.1373/clinchem.2009.127514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24(8):971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen USN, Boge A, Fung PA. The Simple Western [trade]: a gel-free, blot-free, hands-free Western blotting reinvention. Nat Methods. 2011;8(11):v–vi. [Google Scholar]
- 9.Griner EM, Kazanietz MG. Protein kinase C and other diacylglycerol effectors in cancer. Nat Rev Cancer. 2007;7(4):281–294. doi: 10.1038/nrc2110. [DOI] [PubMed] [Google Scholar]
- 10.Teicher BA. Protein kinase C as a therapeutic target. Clin Cancer Res. 2006;12(18):5336–5345. doi: 10.1158/1078-0432.CCR-06-0945. [DOI] [PubMed] [Google Scholar]
- 11.Keck GE, Kraft MB, Truong AP, Li W, Sanchez CC, Kedei N, Lewin NE, Blumberg PM. Convergent assembly of highly potent analogues of bryostatin 1 via pyran annulation: bryostatin look-alikes that mimic phorbol ester function. J Am Chem Soc. 2008;130(21):6660–6661. doi: 10.1021/ja8022169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hale KJ, Manaviazar S. New approaches to the total synthesis of the bryostatin antitumor macrolides. Chem Asian J. 2010;5(4):704–754. doi: 10.1002/asia.200900634. [DOI] [PubMed] [Google Scholar]
- 13.Wender PA, Miller BL. Synthesis at the molecular frontier. Nature. 2009;460(7252):197–201. doi: 10.1038/460197a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kedei N, Telek A, Czap A, Lubart ES, Czifra G, Yang D, Chen J, Morrison T, Goldsmith PK, Lim L, Mannan P, Garfield SH, Kraft MB, Li W, Keck GE, Blumberg PM. The synthetic bryostatin analog Merle 23 dissects distinct mechanisms of bryostatin activity in the LNCaP human prostate cancer cell line. Biochem Pharmacol. 2011;81(11):1296–1308. doi: 10.1016/j.bcp.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kedei N, Telek A, Michalowski AM, Kraft MB, Li W, Poudel YB, Rudra A, Petersen ME, Keck GE, Blumberg PM. Comparison of transcriptional response to phorbol ester, bryostatin 1, and bryostatin analogs in LNCaP and U937 cancer cell lines provides insight into their differential mechanism of action. Biochem Pharmacol. 2013;85(3):313–324. doi: 10.1016/j.bcp.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kedei N, Lewin NE, Geczy T, Selezneva J, Braun DC, Chen J, Herrmann MA, Heldman MR, Lim L, Mannan P, Garfield SH, Poudel YB, Cummins TJ, Rudra A, Blumberg PM, Keck GE. Biological Profile of the Less Lipophilic and Synthetically More Accessible Bryostatin 7 Closely Resembles That of Bryostatin 1. ACS Chem Biol. 2013 doi: 10.1021/cb300671s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rudolph C, Adam G, Simm A. Determination of copy number of c-Myc protein per cell by quantitative Western blotting. Anal Biochem. 1999;269(1):66–71. doi: 10.1006/abio.1999.3095. [DOI] [PubMed] [Google Scholar]
- 18.Yang-Boja E, DeFilippes F, Fales HM. Electrospray mass spectra of three proprietary detergents. Anal Biochem. 2000;285(2):205–210. doi: 10.1006/abio.2000.4734. [DOI] [PubMed] [Google Scholar]
- 19.Caino MC, von Burstin VA, Lopez-Haber C, Kazanietz MG. Differential regulation of gene expression by protein kinase C isozymes as determined by genome-wide expression analysis. J Biol Chem. 2011;286(13):11254–11264. doi: 10.1074/jbc.M110.194332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.