Abstract
Fluorine-19 is a spin-½ NMR isotope with high sensitivity and large chemical shift dispersion, which makes it attractive for high resolution NMR spectroscopy in solution. For studies of membrane proteins it is further of interest that 19F is rarely found in biological materials, which enables observation of extrinsic 19F labels with minimal interference from background signals. Today, after a period with rather limited use of 19F-NMR in structural biology, we witness renewed interest in this technology for studies of complex supramolecular systems. Here we report on recent 19F-NMR studies with the G-protein-coupled receptor family of membrane proteins.
Introduction
The favorable nuclear spin properties of fluorine-19 were exploited in 19F-NMR experiments with fluorinated organic compounds already early in NMR history. As a result, the principles of 19F-NMR have been extensively described in the literature, including serial coverage in the Annual Reports on NMR Spectroscopy (starting with [1]) and monographs (for example, [2]). It was also readily recognized that the absence of a natural 19F background and the high sensitivity of 19F-NMR observation favor the use of fluorine-19 labels for studies of proteins. 19F-NMR labels incorporated into a protein can provide information on the local environment of the labels, including solvent exposure, and on the conformational states and dynamics of the protein [3,4,5,6••]. 19F-NMR experiments with integral membrane proteins further enable characterization of the variations of motional and structural properties in different environments, such as lipid vesicles, organic solvents and detergents [7,8,9,10,11,12,13,14,15,16,17,18••,19•,20•,21•].
As can be seen from Table 1, references to 19F-NMR studies of membrane proteins in the 21st century, and specifically during the last two years are scarce, and therefore our reference list includes only a small number of featured articles. The renewed interest that we see today is closely linked with breakthroughs in membrane protein crystal structure determination, which enable formulation of new questions about structure/function correlations that might be investigated with the use of NMR in solution [22,23]. In this review we focus on recent crystal structure-guided applications of 19F-NMR for studies of G-protein-coupled receptors (GPCRs).
Table 1. Survey of 19F-NMR studies with membrane proteins.
| Membrane Protein | Expression System |
Milieua | Year b (Ref’s) |
|---|---|---|---|
| M13 phage coat protein | E. coli | Lipid vesicles | 1978 [7,8] |
| Gramicidin A | synthetic | Lipid vesicles | 1979 [9,10] |
| D-lactate dehydrogenase | E. coli | Triton X-100, Triton X- 100/SDS, C12E7, lipid vesicles |
1986 [11, 12] |
| Bacteriorhodopsin | E. coli | CH3OH/CHCl3, Triton X-100 |
1987 [13] |
| Rhodopsin | HEK293S (mammalian cell) |
DM, OG, OG/DMPC | 1999 [14, 15] |
| Diacylglycerol kinase | E. coli | DPC | 2002 [16] |
| CIC-ec1. Cl−/H+ antiporter | E. coli | DM | 2009 [17] |
| β2-adrenergic receptor | sf9 (insect cell) | DDM/CHS, MNG | 2012 [18••,21•] |
Abbreviations: SDS, sodium dodecyl sulfate; DM, n-decyl-β-D-maltopyranoside; OG, n-ocfyl-β-D-glucopyranoside; DMPC, 1,2-Dimyristoyl-sn-Glycero-3-Phosphocholine; DPC, n-dodecylphosphocholine; DDM, n-dodecyl-β-D-maltopyranoside; CHS,cholesteryl semisuccinate; MNG, maltose neopentyl glycol
The year of the first publication is shown. The references include only the first and the last publications.
Fluorine-19 NMR in a nutshell
Due to the previously mentioned high popularity during the early history of NMR, fundamental considerations on the use of fluorine-19 NMR in structural biology have been extensively covered in the literature [3,4,5,6••]. Here we summarize some features of special interest for studies of membrane proteins.
The spin-½ fluorine-19 has high sensitivity for NMR observation, corresponding to 83% of the 1H-NMR sensitivity. In spite of this high intrinsic sensitivity, recent 19F-NMR applications with membrane proteins were dependent on the availability of state-of-the-art cryoprobes (see figure 3 for details), due to the fact that membrane proteins must generally be studied in dilute solutions. For many applications it is also attractive that informative 19F-NMR data can be collected at field strengths corresponding to 1H resonance frequencies of 600 MHz or lower. Due to the large chemical shift anisotropy (CSA) of some 19F-labels used with proteins (see the next section), experiments at lower fields often yield superior results to those obtained with higher-field instruments [6••].
Figure 3.
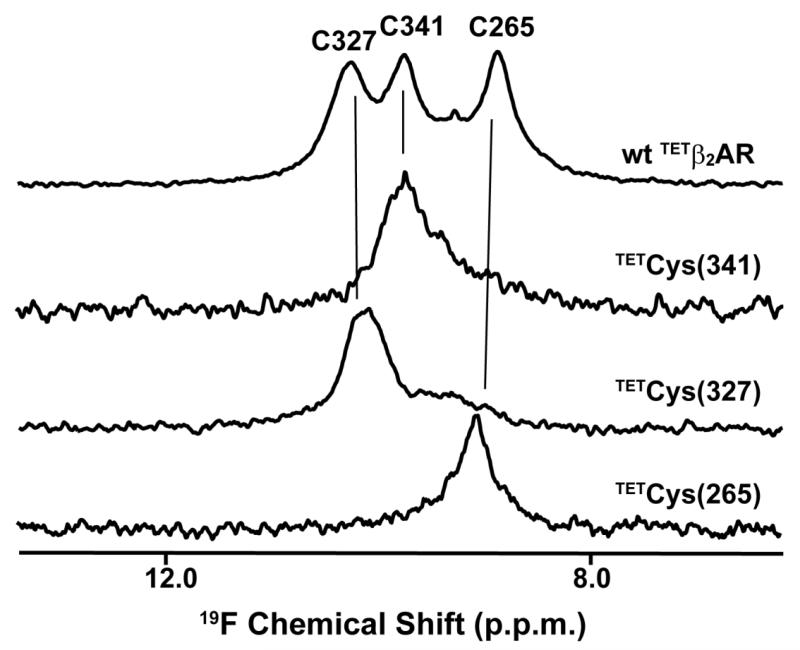
Mutagenesis-based sequence-specific 19F-NMR assignments in β2AR. 564 MHz 1D 19F-NMR spectra at 298K are shown of the carazolol complexes with the TET-labeled wild-type protein, β2AR (TETC265, TETC327, TETC341), and for three single-residue TET-labeled variants, β2AR (TETC265, C327S, C341A), β2AR (C265A, TETC327, C341A) and β2 AR (C265A, C327S,TET C341). The resonance assignments derived from comparison of the four spectra are indicated at the top. The spectra were recorded with a Bruker Avance 600 spectrometer (Bruker Biospin, Billerica, MA) equipped with a 5 mm 1H /19F, 13C/15N QCI cryoprobe (reproduced from [18••]).
As with other spin-½ NMR nuclei, multidimensional experiments have been developed, which are in common use in organic chemistry [24]. Heteronuclear and homonuclear two- and three-dimensional experiments with 19F have been applied also with soluble proteins, but few applications have been described with membrane proteins, probably mainly because of limited sensitivity [20•,25,26,27,28,29].
Fluorine-19 chemical shifts are outstandingly sensitive to the local environment characterized by van der Waals interactions, local electrostatic fields, and similar effects. 19F-labels are therefore highly sensitive reporters of changes in protein conformation and solvent environment. Direct applications are measurements of the immersion depth of membrane proteins into lipophilic environments by observation of isotope effects between H2O and D2O [30,31], or of effects from O2 as a paramagnetic relaxation agent [16,32]. In general, however, interpretation of 19F chemical shifts in terms of protein structure, or in terms of the nature of conformational changes is rather limited [33].
Introducing 19F-NMR labels into membrane proteins
Since 19F does not occur naturally in proteins, its use in structural biology typically relies on incorporation of extrinsic 19F-labels at structurally and/or functionally interesting locations of the target protein. Three methods of fluorine labeling are described here: chemical conjugation of fluorine-containing small molecules with reactive amino acids, biosynthetic introduction of fluorinated amino acid analogs which are compatible with a natural aminoacyl-tRNA synthetase, and site-specific incorporation of fluorinated amino acids which do not have a naturally compatible aminoacyl-tRNA synthetase.
Chemical modification of amino acid side chains
Fluorine can be postranslationally introduced by chemical conjugation of small 19F-containing molecules with reactive −SH or −NH moieties in proteins. It is an important advantage of the chemical modification approach that it can be applied with otherwise unlabeled proteins, which may either be obtained from natural sources or as recombinant proteins from cells grown in an optimized medium. This can be especially useful for membrane proteins, which are often poorly expressed even in eukaryotic cell lines. Two preferred reagents for membrane protein studies are 2,2,2-trifluoroethanethiol (TET) [14,15,18••,20•], and 3-bromo-1,1,1-trifluoroacetone (BTFA) [16,19•,21•], and 4-(perfluoro-tert-butyl)-phenyliodocetamide (PFP) [34] and S-ethyl-trifluorothioacetate (SETFA) [35] have also been used. TET and BTFA are commercially available, exhibit high labeling efficiency, and cause minimal structural perturbations. BTFA-protein conjugation is a one-step process that results in a stable thioester bond. Conjugation with TET starts with activation of cysteine −SH groups by 4,4-dithiodipyridine (4-DPS), and in a second step a disulfide linkage is formed. Therefore, TET-labeling may be reversible, especially at higher temperatures [18••,20•].
Trifluoromethyl probes have higher sensitivity for NMR detection and usually smaller chemical shift anisotropy (CSA) than monofluorinated probes, and are therefore the best option for studies of membrane proteins. Whereas, for example, NMR spectra of 3-fluorotyrosine-labelled alkaline phosphatase showed strong deteriorating effects due to CSA-induced line broadening at higher magnetic fields [36], trifluoromethyl groups may provide narrow signals even at high fields.
Biosynthetic incorporation of fluorinated amino acid analogs
Biosynthetic incorporation of fluorinated aliphatic or aromatic amino acid analogs has been popular in early 19F NMR studies of proteins [7,9,11]. For membrane proteins, fluorinated aromatic amino acids were primarily used, presumably because the relative scarcity of aromatic amino acids was expected to simplify the assignment process. Auxotrophic bacterial strains are available, and fluorinated aromatic amino acid analogs, such as m-monofluorotyrosine, 4-, 5- and 6- monofluorotryptophan, and o-, m- and p-monofluorophenylalanine are commercial reagents, which makes the labeling process quite efficient [37]. 13C- and 15N-enriched fluorinated amino acid analogs have been used to reduce spectral overlap in 19F-detected multidimensional heteronuclear correlation NMR experiments, and to obtain NMR-based assignments of the 19F signals [28,29,38]. Detailed biosynthetic labeling protocols can be found in earlier reviews [3,37].
Sequence-specific incorporation of fluorinated amino acid analogs
A potentially highly exciting approach is sequence-specific incorporation of 19F-labels into genetically encoded positions, which entails the use of an extrinsic orthogonal tRNA/aminoacyl-tRNA synthetase pair to incorporate 19F-labeled amino acids at positions defined by a TAG amber codon or a frameshift codon. Phenylalanine analogs such as p-OCF3-Phe and p-CF3-Phe have been incorporated into soluble proteins [39,40,41,42] and at least one membrane protein [43]. Genetically encoded labeling was also used for in-cell 19F-NMR measurements [39,44].
Three-dimensional protein structures guide 19F labeling
In principle, strategies for the introduction of 19F-labels into polypeptide chains can be developed based on the amino acid sequence, by taking the positions of reactive amino acids into account, such as cysteines and lysines, and by identifying interesting sequence locations for mutagenesis. However, reference to three-dimensional structures often enables more meaningful strategies. Examples may include the identification of sites near active centers or near binding sites for allosteric effectors. In principle, strategies based on three-dimensional structures can lead to triangulation of function-related interaction networks in larger systems, as will be illustrated below with studies of GPCRs.
19F-NMR applications with GPCRs
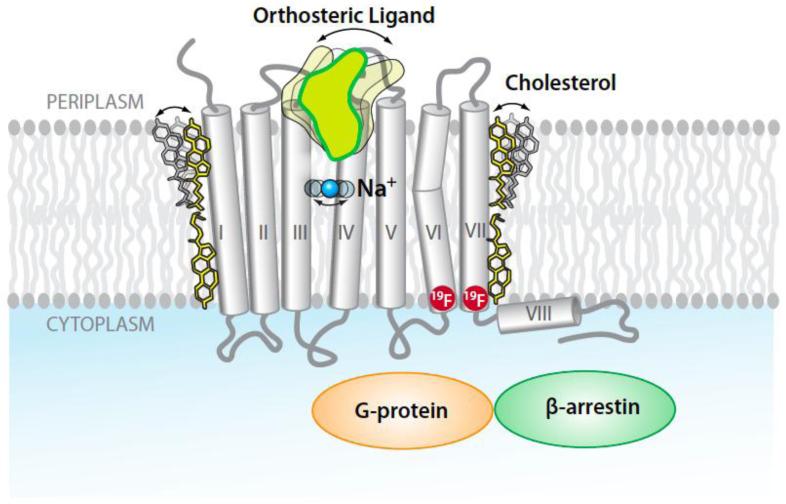
GPCRs constitute the largest membrane protein family in the human genome, with over 800 unique sequences [45,46]. They are also important drug targets, since over 30% of all prescription drugs on the market act via GPCRs [47,48,49]. GPCRs have a seven-transmembrane-helix topology and contain multiple binding sites for drugs and allosteric effectors (Figure 1). They recognize a diverse array of orthosteric ligands, including small organic compounds, peptides and small proteins. Upon ligand binding a signal is transmitted over a distance of about 30 Å across the cell membrane to intracellular partner proteins, such as a G-protein or β-arrestin (Figure 1). Other compounds, such as cholesterol or sodium, may act as allosteric effectors which modulate GPCR activity [50,51]. Depending on the chemical structure, different ligands have variable signaling efficacies [18••,52]. More than 20 GPCR crystal structures have been reported during the last seven years, which include structures of inactive and active states of some proteins, and a G-protein-bound state of the β2-adrenergic receptor (β2AR) [53].
Figure 1.
GPCR structure and function. GPCR structures are characterized by seven trans-membrane helices, which stretch across the lipid bilayer cell membrane. On the periplasmic surface there is an “orthosteric” ligand binding site (the arrow indicates mobility in the ligand binding site). Allosteric binding sites are located along the periphery of the seven-helix bundle (cholesterol) and in the center of the bundle (Na+). Binding of drug molecules to the orthosteric site elicits signaling to partner proteins in the cytoplasm, for example, G-protein and β-arrestin. The two red circles indicate locations for fluorine-19 labels that might be used for 19F-NMR observation of conformational changes associated with trans-membrane signaling between the orthosteric site and the cytoplasmic protein surface, which bridges a distance of about 30 Å.
GPCRs have so far almost exclusively been expressed in eukaryotic systems, such as yeast, insect or mammalian cells, which are not readily amenable to 15N and 13C labeling for NMR studies. Chemical conjugation of 19F-labels into GPCRs for 19F-NMR experiments is therefore an attractive approach for studies in non-crystalline milieus.
Questions for 19F-NMR with GPCRs
A key problem in GPCR research is to rationalize how the receptors transduce information encoded in the chemical structures of orthosteric ligands across the cell membrane to appropriate intracellular partners, such as the G-protein and β-arrestin (Figure 1). While crystal structures describe the molecular architecture in great detail and may identify structural differences between activated and inactive forms of a given GPCR, 19F-NMR can provide additional information on local conformational polymorphisms and the rate of associated conformational exchange processes.
19F labeling and sequence-specific NMR assignments of β2AR
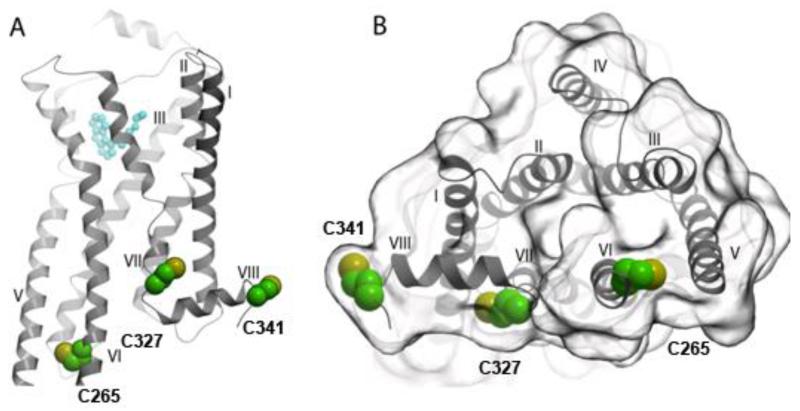
19F-NMR studies of the β2-adrenergic receptor (β2AR) have recently been reported by two different groups [18••,19•,20•,21•]. β2AR contains three native cysteines in positions 265, 327 and 341, which are located near the cytoplasmic protein surface (Figure 2). One project made use of the accessibility of these cysteines for covalent labeling with TET [18••,20•]. The resulting 1D 19F-NMR spectrum consists of three peaks (Figure 3, top trace). Site-specific mutagenesis was employed to obtain single-residue labeled β2AR variants. Comparison of the four spectra in Figure 3 then yielded sequence-specific assignments of the three 19F-NMR signals, and in functional studies the single-residue TET-labeled proteins enabled detailed line shape analyses of the individual signals [18••, 20•]. In the second project the residue Cys265 was selectively labeled with BTFA, and the observed 19F-NMR signal was assigned to this residue. [19•,21•].
Figure 2.
Locations of 19F-labels that were recently used for β2AR functional studies [18••,19•,20•,21•]. The green and yellow spheres indicate three TET-labeled cysteine residues in positions 265, 327 and 341. (A) Side view of β2AR. An agonist, Bl167107, in the orthosteric binding site is drawn in cyan. (B) View of the cytoplasmic surface of β2AR.
β2AR is a favorable molecule for the use of 19F-NMR, since reactive cysteine residues near the cytoplasmic surface are accessible for 19F-labeling. In the early work with rhodopsin [14,15] and in ongoing experiments with several human GPCRs [unpublished], cysteine residues had to be engineered into informative sequence positions, which then also requires special care to ascertain that such modifications preserve the biological activity.
19F-NMR studies of GPCR activation
19F-NMR studies of rhodopsin were started prior to the availability of high-resolution GPCR crystal structures [14,15]. In these seminal experiments, Khorana and co-workers introduced TET labels to endogenous as well as engineered cysteine residues. 19F-NMR spectra of various mutant rhodopsins containing only a single 19F-label were recorded in the dark and under light exposure. Chemical shift changes specific to receptor activation were thus observed, which elegantly demonstrated that 19F-NMR experiments can be used to monitor local conformational changes related to GPCR activation. In additional studies, close contacts in the three-dimensional structure were quantified by collecting 19F-19F nuclear Overhauser effects (NOEs) between pairs of TET labels, which had been incorporated by site-specific introduction of TET-accessible cysteines to obtain two-residue TET-labeled rhodopsin [15].
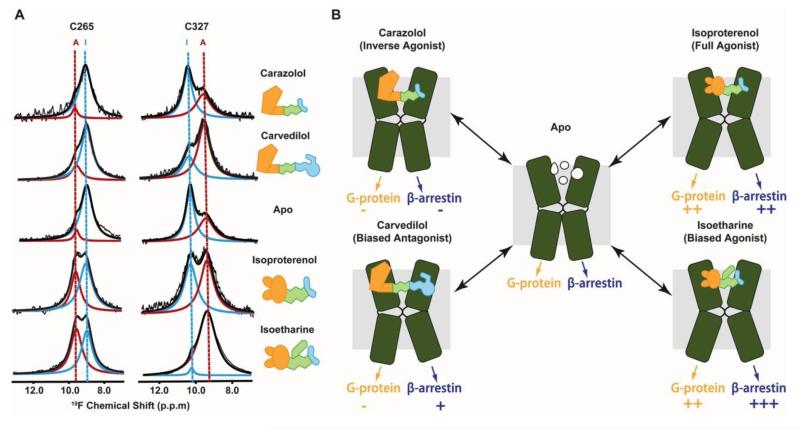
Unlike rhodopsin, β2AR is activated by binding to diffusible ligands, which exhibit a wide range of different efficacies in their receptor interactions, depending on their chemical structures. In the experiments of Figure 4A, 19F-NMR spectra of β2AR mutants containing single TET-labeled cysteine residues were recorded for complexes with different pharmacological ligands. Two largely independent equilibria were thus discovered by observation of Cys265 in helix VI, and Cys327 in helix VII, respectively. Exchange between the different conformations is slow on the chemical shift frequency scale, with k < 10 sec−1 [20•], so that these equilibria are manifested by two partially overlapped signals which have variable relative intensities in complexes with different ligands [18••]. Novel insights into correlations between the chemical structure of different orthosteric ligands and the resulting downstream signaling could thus be obtained. Specifically, ligands that shift the conformational equilibrium manifested in the 19F-NMR signal of TETCys265 to the activated state are likely to signal through the canonical G-protein pathway, whereas ligands that lead to activation observed on TETCys327 were found to signal primarily through the β-arrestin pathway (Figure 4).
Figure 4.
19F-NMR observation of conformational equilibria and signaling pathways in β2AR. (A) 19F-NMR signals of TETC265 and TETC327 in the apo-form and four drug complexes of single-residue TET-labeled β2AR recorded at 280K with the same equipment as the data in Figure 3. The experimental spectra (thin black line showing noise) have been deconvoluted into signals of an activated state (red) and an inactive state (blue) of β2AR. The thick black line represents the sum of the signals I and A. (B) Two β2AR signaling pathways suggested by the 19F-NMR experiments. The helices VI and VII of β2AR are shown as green kinked cylinders. The structures of the four bound ligands are schematically drawn and their functionalities are indicated. The arrows at the lower end of the cylinders indicate the signaling to the downstream effectors, with plus and minus signs indicating the signaling levels relative to the basal state (adapted from [18••]).
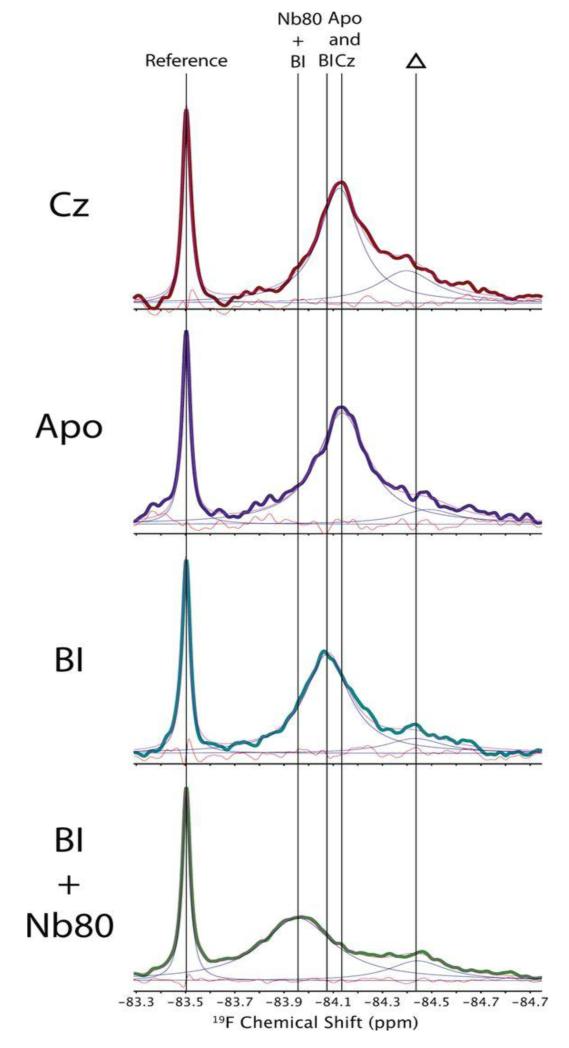
With the aforementioned single-residue BTFACys 265 variant of β2AR, a similar strategy to the experiments in Figure 4A was used. When the protein was solubilized in DDM micelles, evidence was obtained for the presence of two or multiple states of the protein in fast exchange on the chemical shift frequency scale [19•]. As a consequence, a single 19F-NMR signal was observed, which showed different chemical shifts when different ligands, and in one experiment a ligand and a nanobody were bound to β2AR (Figure 5). When the BTFA-labeled β2AR was solubilized with the detergent maltose-neopentyl-glycol (MNG-3), the authors concluded that the exchange between different states was slowed down, and that the 19F-NMR signals manifested at least three different states of β2AR in slow exchange on the chemical shift frequency scale.
Figure 5.
564 MHz 1D 19F NMR spectra of β2 AR (C77V, C275S, BTFAC265, C378A, C406A) at 298K. Cz, complex with the inverse agonist carazolol; Apo, apo-β2AR; BI, complex with the full agonist Bl-167107; BI + Nb80, complex with BI and a G-protein-mimicking nanobody. The sharp peak at −83.5 is from a small- molecule reference, and Δ indicates a background signal from partially 19F-labeled cysteines in other parts of the β2AR structure. The vertical lines indicate peak positions of BTFAC265 assigned to the complexes of β2AR with carazolol, Bl-167107, and Bl-167107/Nb80, as indicated at the top (reproduced from [19•]).
From a techniques viewpoint it is of interest that the aforementioned two studies with chemically different 19F-labels attached to the same residue, Cys265, provided complementary information. This comes rather as unexpected, since numerous earlier studies with a variety of different proteins indicated that replacement of natural amino acids with fluorinated analogs caused at most minor perturbations of the protein structures [54,55,56], and the introduction of TET- or BTFA-labels in GPCRs did not abolish their functions in the studies reported so far [14,15,18••,19•,20•,21•]. There is thus an indication that more comprehensive information may be obtained from repeating 19F-NMR experiments with chemically different 19F-containing groups even if they are attached to the same amino acids in membrane proteins.
Outlook
Due to recent advances in instrumentation, which ensured high sensitivity at moderate field strength, 19F-NMR is being recognized as an attractive technique for structural biology of complex systems, in particular membrane proteins. 19F-NMR can be used as a “probe method” to investigate conformational equilibria and associated rate processes, and can serve as a lead for the use of stable isotope labeling-based NMR studies of structural details associated with the plasticity of the system. With future improvements in membrane protein biochemistry, multidimensional NMR experiments with 19F-NMR may open additional new avenues for studies of membrane proteins.
Highlights.
Renewed interest in 19F-NMR for structural biology of complex molecular systems.
Advances in biochemistry and NMR techniques enable 19F-NMR studies of new classes of membrane proteins.
19F-NMR of GPCRs complements crystal structures with data on protein dynamics in long-range signal transfer.
Acknowledgments
This work was supported by NIH Common Fund grant P50 GM073197 and NIH PSI:Biology grant U54 GM094618. K.W. is the Cecil H. and Ida M. Green Professor of Structural Biology at The Scripps Research Institute. The authors thank K. Kadyshevskaya for the preparation of Figure 1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mooney E, Winson P. Fluorine-19 Nuclear Magnetic Resonance Spectroscopy. Annu Rep NMR Spectrosc. 1968;1:243–311. [Google Scholar]
- 2.Dolbier WR. Guide to fluorine NMR for organic chemists. Wiley; Hoboken, N.J.: 2009. [Google Scholar]
- 3.Gerig JT. Fluorine NMR of Proteins. Prog Nucl Mag Res Spectrosc. 1994;26:293–370. [Google Scholar]
- 4.Sykes BD, Hull WE. Fluorine nuclear magnetic resonance studies of proteins. Methods Enzymol. 1978;49:270–295. doi: 10.1016/s0076-6879(78)49015-9. [DOI] [PubMed] [Google Scholar]
- 5.Cobb SL, Murphy CD. F-19 NMR applications in chemical biology. J Fluorine Chem. 2009;130:132–143. [Google Scholar]
- 6••.Kitevski-LeBlanc JL, Prosser RS. Current applications of F-19 NMR to studies of protein structure and dynamics. Prog Nucl Mag Res Spectrosc. 2012;62:1–33. doi: 10.1016/j.pnmrs.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Hagen DS, Weiner JH, Sykes BD. Fluorotyrosine M13 coat protein: fluorine-19 nuclear magnetic resonance study of the motional properties of an integral membrane protein in phospholipid vesicles. Biochemistry. 1978;17:3860–3866. doi: 10.1021/bi00611a028. [DOI] [PubMed] [Google Scholar]
- 8.Wilson ML, Dahlquist FW. Membrane protein conformational change dependent on the hydrophobic environment. Biochemistry. 1985;24:1920–1928. doi: 10.1021/bi00329a018. [DOI] [PubMed] [Google Scholar]
- 9.Weinstein S, Wallace BA, Blout ER, Morrow JS, Veatch W. Conformation of gramicidin A channel in phospholipid vesicles: a 13C and 19F nuclear magnetic resonance study. Proc Natl Acad Sci U S A. 1979;76:4230–4234. doi: 10.1073/pnas.76.9.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weinstein S, Durkin JT, Veatch WR, Blout ER. Conformation of the gramicidin A channel in phospholipid vesicles: a fluorine-19 nuclear magnetic resonance study. Biochemistry. 1985;24:4374–4382. doi: 10.1021/bi00337a019. [DOI] [PubMed] [Google Scholar]
- 11.Ho C, Rule GS, Pratt EA. Nuclear magnetic resonance, biochemical, and molecular genetic studies of the membrane-bound d-lactate dehydrogenase of Escherichia coli. Biophys J. 1986;49:113–115. doi: 10.1016/S0006-3495(86)83615-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun ZY, Pratt EA, Simplaceanu V, Ho C. A 19F-NMR study of the equilibrium unfolding of membrane-associated D-lactate dehydrogenase of Escherichia coli. Biochemistry. 1996;35:16502–16509. doi: 10.1021/bi9620619. [DOI] [PubMed] [Google Scholar]
- 13.Arseniev AS, Kuryatov AB, Tsetlin VI, Bystrov VF, Ivanov VT, Ovchinnikov YA. F-19-NMR Study of 5-Fluorotryptophan-Labeled Bacteriorhodopsin. FEBS Lett. 1987;213:283–288. [Google Scholar]
- 14.Klein-Seetharaman J, Getmanova EV, Loewen MC, Reeves PJ, Khorana HG. NMR spectroscopy in studies of light-induced structural changes in mammalian rhodopsin: applicability of solution 19F NMR. Proc Natl Acad Sci U S A. 1999;96:13744–13749. doi: 10.1073/pnas.96.24.13744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loewen MC, Klein-Seetharaman J, Getmanova EV, Reeves PJ, Schwalbe H, Khorana HG. Solution 19F nuclear Overhauser effects in structural studies of the cytoplasmic domain of mammalian rhodopsin. Proc Natl Acad Sci U S A. 2001;98:4888–4892. doi: 10.1073/pnas.051633098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luchette PA, Prosser RS, Sanders CR. Oxygen as a paramagnetic probe of membrane protein structure by cysteine mutagenesis and F-19 NMR spectroscopy. J Am Chem Soc. 2002;124:1778–1781. doi: 10.1021/ja016748e. [DOI] [PubMed] [Google Scholar]
- 17.Elvington SM, Liu CW, Maduke MC. Substrate-driven conformational changes in ClC-ec1 observed by fluorine NMR. EMBO J. 2009;28:3090–3102. doi: 10.1038/emboj.2009.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18••.Liu JJ, Horst R, Katritch V, Stevens RC, Wüthrich K. Biased signaling pathways in β2-adrenergic receptor characterized by 19F-NMR. Science. 2012;335:1106–1110. doi: 10.1126/science.1215802. This paper demonstrates the use of 19F-NMR to probe structure-function relationships in GPCRs. The results revealed conformational equilibria associated with β2-adrenergic receptor signaling pathways, and provided novel insights into pathway-selective signal transduction.
- 19•.Chung KY, Kim TH, Manglik A, Alvares R, Kobilka BK, Prosser RS. Role of detergents in conformational exchange of a G protein-coupled receptor. J Biol Chem. 2012;287:36305–36311. doi: 10.1074/jbc.M112.406371. The authors investigated effects of various detergents on dynamic aspects of the β2-adrenergic receptor.
- 20•.Horst R, Liu JJ, Stevens RC, Wüthrich K. β2-adrenergic receptor activation by agonists studied with 19F-NMR. Angew Chemie. 2013 doi: 10.1002/anie.201305286. In Press. 19F NMR was used to characterize thermodynamic and kinetic parameters of conformational equilibria in β2-adrenergic receptor.
- 21•.Kim TH, Chung KY, Manglik A, Hansen AL, Dror RO, Mildorf TJ, Shaw DE, Kobilka BK, Prosser RS. The Role of Ligands on the Equilibria Between Functional States of a G Protein-Coupled Receptor. J Am Chem Soc. 2013 doi: 10.1021/ja404305k. 10.1021/ja404305k. Thermodynamic equilibria between functional states of the β2-adrenergic receptor were characterized using 1D 19F-NMR.
- 22.Stevens RC, Cherezov V, Katritch V, Abagyan R, Kuhn P, Rosen H, Wüthrich K. The GPCR Network: a large-scale collaboration to determine human GPCR structure and function. Nat Rev Drug Discov. 2013;12:25–34. doi: 10.1038/nrd3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White SH. Biophysical dissection of membrane proteins. Nature. 2009;459:344–346. doi: 10.1038/nature08142. [DOI] [PubMed] [Google Scholar]
- 24.Battiste J, Newmark RA. Applications of 19F multidimensional NMR. Prog Nucl Mag Res Spectrosc. 2006;48:1–23. [Google Scholar]
- 25.Li H, Frieden C. Fluorine-19 NMR studies on the acid state of the intestinal fatty acid binding protein. Biochemistry. 2006;45:6272–6278. doi: 10.1021/bi0602922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cistola DP, Hall KB. Probing internal water molecules in proteins using two-dimensional 19F-1H NMR. J Biomol NMR. 1995;5:415–419. doi: 10.1007/BF00182285. [DOI] [PubMed] [Google Scholar]
- 27.Feeney J, McCormick JE, Bauer CJ, Birdsall B, Moody CM, Starkmann BA, Young DW, Francis P, Havlin RH, Arnold WD. Oldfield E: F-19 nuclear magnetic resonance chemical shifts of fluorine containing aliphatic amino acids in proteins: Studies on Lactobacillus casei dihydrofolate reductase containing (2S,4S)-5-fluoroleucine. J Am Chem Soc. 1996;118:8700–8706. [Google Scholar]
- 28.Kitevski-LeBlanc JL, Al-Abdul-Wahid MS, Prosser RS. A Mutagenesis-Free Approach to Assignment of F-19 NMR Resonances in Biosynthetically Labeled Proteins. J Am Chem Soc. 2009;131:2054. doi: 10.1021/ja8085752. [DOI] [PubMed] [Google Scholar]
- 29.Kitevski-LeBlanc JL, Evanics F, Prosser RS. Approaches to the assignment of F-19 resonances from 3-fluorophenylalanine labeled calmodulin using solution state NMR. J Biomol NMR. 2010;47:113–123. doi: 10.1007/s10858-010-9415-y. [DOI] [PubMed] [Google Scholar]
- 30.Evanics F, Kitevski JL, Bezsonova I, Forman-Kay J, Prosser RS. 19F NMR studies of solvent exposure and peptide binding to an SH3 domain. Biochim Biophys Acta. 2007;1770:221–230. doi: 10.1016/j.bbagen.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 31.Hagen DS, Weiner JH, Sykes BD. Investigation of solvent accessibility of the fluorotyrosyl residues of M13 coat protein in deoxycholate micelles and phospholipid vesicles. Biochemistry. 1979;18:2007–2012. doi: 10.1021/bi00577a026. [DOI] [PubMed] [Google Scholar]
- 32.Prosser RS, Luchette PA, Westerman PW. Using O2 to probe membrane immersion depth by 19F NMR. Proc Natl Acad Sci U S A. 2000;97:9967–9971. doi: 10.1073/pnas.170295297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lau EY, Gerig JT. Origins of fluorine NMR chemical shifts in fluorine-containing proteins. J Am Chem Soc. 2000;122:4408–4417. [Google Scholar]
- 34.Kalbitzer HR, Rohr G, Nowak E, Goody RS, Kuhn W, Zimmermann H. A New High-Sensitivity F-19 Probe for Labeling Cysteine Groups of Proteins. Nmr Biomed. 1992;5:347–350. doi: 10.1002/nbm.1940050605. [DOI] [PubMed] [Google Scholar]
- 35.Mehta VD, Kulkarni PV, Mason RP, Constantinescu A, Antich PP. Fluorinated Proteins as Potential F-19 Magnetic-Resonance-Imaging and Spectroscopy Agents. Bioconjug Chem. 1994;5:257–261. doi: 10.1021/bc00027a011. [DOI] [PubMed] [Google Scholar]
- 36.Hull WE, Sykes BD. Fluorotyrosine alkaline phosphatase: internal mobility of individual tyrosines and the role of chemical shift anisotropy as a 19F nuclear spin relaxation mechanism in proteins. J Mol Biol. 1975;98:121–153. doi: 10.1016/s0022-2836(75)80105-7. [DOI] [PubMed] [Google Scholar]
- 37.Frieden C, Hoeltzli SD, Bann JG. The preparation of F-19-labeled proteins for NMR studies. Method Enzymol. 2004;380:400–415. doi: 10.1016/S0076-6879(04)80018-1. [DOI] [PubMed] [Google Scholar]
- 38.Alexeev D, Barlow PN, Bury SM, Charrier JD, Cooper A, Hadfield D, Jamieson C, Kelly SM, Layfield R, Mayer RJ, McSparron H, Price NC, Ramage R, Sawyer L, Starkmann BA, Uhrin D, Wilken J, Young DW. Synthesis, structural and biological studies of ubiquitin mutants containing (2S, 4S)-5-fluoroleucine residues strategically placed in the hydrophobic core. Chembiochem. 2003;4:894–896. doi: 10.1002/cbic.200300699. [DOI] [PubMed] [Google Scholar]
- 39.Jones DH, Cellitti SE, Hao X, Zhang Q, Jahnz M, Summerer D, Schultz PG, Uno T, Geierstanger BH. Site-specific labeling of proteins with NMR-active unnatural amino acids. J Biomol NMR. 2010;46:89–100. doi: 10.1007/s10858-009-9365-4. [DOI] [PubMed] [Google Scholar]
- 40.Cellitti SE, Jones DH, Lagpacan L, Hao X, Zhang Q, Hu H, Brittain SM, Brinker A, Caldwell J, Bursulaya B, Spraggon G, Brock A, Ryu Y, Uno T, Schultz PG, Geierstanger BH. In vivo incorporation of unnatural amino acids to probe structure, dynamics, and ligand binding in a large protein by nuclear magnetic resonance spectroscopy. J Am Chem Soc. 2008;130:9268–9281. doi: 10.1021/ja801602q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jackson JC, Hammill JT, Mehl RA. Site-specific incorporation of a (19)F-amino acid into proteins as an NMR probe for characterizing protein structure and reactivity. J Am Chem Soc. 2007;129:1160–1166. doi: 10.1021/ja064661t. [DOI] [PubMed] [Google Scholar]
- 42.Hammill JT, Miyake-Stoner S, Hazen JL, Jackson JC, Mehl RA. Preparation of site-specifically labeled fluorinated proteins for 19F-NMR structural characterization. Nat Protoc. 2007;2:2601–2607. doi: 10.1038/nprot.2007.379. [DOI] [PubMed] [Google Scholar]
- 43.Shi P, Wang H, Xi Z, Shi C, Xiong Y, Tian C. Site-specific (19)F NMR chemical shift and side chain relaxation analysis of a membrane protein labeled with an unnatural amino acid. Protein Sci. 2011;20:224–228. doi: 10.1002/pro.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li C, Wang GF, Wang Y, Creager-Allen R, Lutz EA, Scronce H, Slade KM, Ruf RA, Mehl RA, Pielak GJ. Protein (19)F NMR in Escherichia coli. J Am Chem Soc. 2010;132:321–327. doi: 10.1021/ja907966n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 46.Fagerberg L, Jonasson K, von Heijne G, Uhlen M, Berglund L. Prediction of the human membrane proteome. Proteomics. 2010;10:1141–1149. doi: 10.1002/pmic.200900258. [DOI] [PubMed] [Google Scholar]
- 47.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 48.Russ AP, Lampel S. The druggable genome: an update. Drug Discov Today. 2005;10:1607–1610. doi: 10.1016/S1359-6446(05)03666-4. [DOI] [PubMed] [Google Scholar]
- 49.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 50.Liu W, Chun E, Thompson AA, Chubukov P, Xu F, Katritch V, Han GW, Roth CB, Heitman LH, AP IJ, Cherezov V, Stevens RC. Structural basis for allosteric regulation of GPCRs by sodium ions. Science. 2012;337:232–236. doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien EY, Velasquez J, Kuhn P, Stevens RC. A specific cholesterol binding site is established by the 2.8 A structure of the human beta2-adrenergic receptor. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Urban JD, Clarke WP, von Zastrow M, Nichols DE, Kobilka B, Weinstein H, Javitch JA, Roth BL, Christopoulos A, Sexton PM, Miller KJ, Spedding M, Mailman RB. Functional selectivity and classical concepts of quantitative pharmacology. J Pharmacol Exp Ther. 2007;320:1–13. doi: 10.1124/jpet.106.104463. [DOI] [PubMed] [Google Scholar]
- 53.Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah ST, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunahara RK, Kobilka BK. Crystal structure of the β2-adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Campos-Olivas R, Aziz R, Helms GL, Evans JNS, Gronenborn AM. Placement of 19F into the center of GB1: effects on structure and stability. FEBS Letters. 2002;517:55–60. doi: 10.1016/s0014-5793(02)02577-2. [DOI] [PubMed] [Google Scholar]
- 55.Li HL, Frieden C. Observation of sequential steps in the folding of intestinal fatty acid binding protein using a slow folding mutant and F-19 NMR. Proc Natl Acad Sci U S A. 2007;104:11993–11998. doi: 10.1073/pnas.0705253104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiao GY, Parsons JF, Tesh K, Armstrong RN, Gilliland GL. Conformational changes in the crystal structure of rat glutathione transferase M1-1 with global substitution of 3-fluorotyrosine for tyrosine. J Mol Biol. 1998;281:323–339. doi: 10.1006/jmbi.1998.1935. [DOI] [PubMed] [Google Scholar]