Abstract
Spinal muscular atrophy (SMA), a neurodegenerative disease with potentially devastating and even deadly effects on affected individuals, was first described in the late nineteenth century. Although the survival of motor neuron (SMN) gene was identified nearly 2 decades ago to be causative of the disease, neither an effective treatment nor a cure are currently available. Yet efforts are on-going to test a multitude of treatment strategies with the potential to alleviate disease symptoms in human and clinical trials. Among the most studied compounds for the treatment of SMA are histone deacetylase inhibitors. Several of these epigenetic modifiers have been shown to increase expression of the crucial SMN gene in vitro and in vivo, an effect linked to increased histone acetylation and remodeling of the chromatin landscape surrounding the SMN gene promoter. Here, we review the history and current state of use of histone deacetylase inhibitors in SMA, as well as the success of clinical trials investigating the clinical applicability of these epigenetic modifiers in SMA treatment.
Electronic supplementary material
The online version of this article (doi:10.1007/s13311-013-0209-2) contains supplementary material, which is available to authorized users.
Keywords: Epigenetic, Chromatin, Histones, Spinal muscular atrophy, Clinical trials
Introduction
Spinal muscular atrophy (SMA), a neurodegenerative autosomal recessive disease, is one of the worldwide leading genetic causes for death in early infancy and childhood. With an incidence of at least 1 in 10,000 live births and a carrier frequency of 1 in 35, its prevalence in Europeans is second only to cystic fibrosis [1–4]. The first description of the disease was recorded in two siblings more than a century ago (1891) by Guido Werdnig, and was quickly corroborated by Johan Hoffmann, who provided descriptions of several additional patients. Thus, one subtype of the disease is today still commonly known as Werdnig–Hoffmann disease. The clinical symptoms of SMA involve progressive weakness of voluntary muscles, eventually resulting in muscular atrophy. The proximal voluntary muscles are primarily affected, and patients lose or never acquire motor skills during disease progression. The observed muscle wasting is a result of deterioration of the alpha-motor neurons in the anterior horn of the spinal cord. Depending on the severity, SMA can lead to death within the first few years of life, primarily owing to respiratory insufficiency. A less severe, juvenile form of SMA, commonly referred to as Kugelberg–Welander disease, was described in 1956 [5]. Affected children were characterized with late age of onset, longer survival, and a higher level of acquired motor skills.
Age of onset of the first symptoms and the ability of patients to reach certain motor milestones, such as sitting, standing, and walking unaided, are today used as key clinical features to categorize SMA into a total of three major subgroups, as defined by the International Consortium on Spinal Muscular Atrophy 1991 [6–15]. Type I SMA (Werdnig–Hoffmann disease, Online Mendelian Inheritance in Man (OMIM) database access number #253300) is the most severe form of SMA, with onset of symptoms observed during the first 6 months of life. Patients are never able to sit or walk unaided, and usually die before the age of 2 years. Only 8 % of patients live more than 10 years. Type II SMA (OMIM #253550) patients have an age of onset between 6 and 18 months. They are able to sit unaided, but never achieve the ability to walk. Life expectancy is significantly higher than in patients with the severe type I form of SMA, with at least 70 % of patients exceeding a lifespan of 20 years. Type III (Kugelberg–Welander disease, OMIM # 253400) is a mild form of SMA that is subdivided into two classes, depending on the age of onset. In type IIIa, first symptoms occur before the age of 3 years, whereas the symptoms of type IIIb patient start between the age of 3 and 30 years. Patients are able to sit and walk unaided for at least part of their life, but regularly become wheelchair-bound with disease progression. Life expectancy is only slightly decreased compared with healthy individuals. More recently, two more forms of SMA were added (reviewed in [16, 17]). These are type IV (OMIM #271150), a very rare adult form of SMA that is characterized by the onset of symptoms after the 30th year of life and only slightly impaired motor skills, and type 0, which is used to characterize in utero onset of symptoms. Type 0 children require respiratory aid from birth.
SMA has long been thought to exclusively affect motor neurons, as patients generally retain normal facial and involuntary muscle function and full mental capabilities. Although motor neurons remain one of the foremost affected tissues, lately there has been on-going discussion about reclassifying SMA as a multisystem disorder, which, in severe cases, not only affects other neuronal tissues, such as sensory neurons and the brain, but also skeletal and heart muscle, vasculature, and other organs. For a detailed overview of the affected tissues please refer to a recent review by Hamilton and Gilligwater [18].
Genetic Basis of SMA
In 1990, the candidate region for SMA was mapped by linkage analysis to the long arm of chromosome 5 (5q11.2–5q13.2) [19, 20]. Through the use of positional cloning and newly discovered polymorphic microsatellite markers this region was narrowed down to an inverted and duplicated genomic section of 500 kilo base-pairs in length, which contained multiple copies of several genes [21–24]. Two genes, the neuronal inhibitory protein (NAIP) gene and the survival of motor neuron (SMN) gene, were subject to large deletions in the majority of SMA patients [24–26]. The NAIP gene has one functional copy (referred to as telomeric NAIP), which was found to be deleted in 50 % of SMA patients, but also in some asymptomatic carriers of the disease [25]. In contrast, SMN is present in 2, nearly identical, copies, referred to as SMN1 and SMN2. SMN1 was shown to map to the critical region on chromosome 5, and was deleted in 94 % of SMA patients [24, 27–32]. The remaining 6 % of patients were demonstrated to have different loss-of-function mutations in SMN1 [24, 32–34]. Consequently, SMA was classified as a monogenetic disease caused by the homozygous loss of function of the SMN1 gene. Presently, there are no reported cases where both SMN genes are deleted or mutated. It has been demonstrated in mice that the complete absence of murine SMN is embryonically lethal, which could explain the lack of human patients with complete functional loss of both SMN genes [35]. Interestingly, the SMA phenotype is specific to loss of SMN1, as deletion of all SMN2 copies, observed in approximately 5 % of the population, has no phenotypic effect [24, 36].
The SMN Gene
SMN1 spans a region of about 28 kilo base-pairs and contains an open reading frame of 885 base-pairs. It consists of 9 exons (also referred to as exons 1, 2a, 2b, and 3–8) that encode for a 294-amino acid protein [24, 37]. SMN2 is a near exact copy of SMN1 and differs only in 5 distinct base-pairs [24]. One of these nucleotide exchanges, a C to T transition at the 5’ end of exon 7 [c.840 C → T, nucleotide (nt.) position 27141], is located in the coding region. However, this nucleotide exchange is silent and has no functional effect on the amino acid sequence of the protein. Another exonic nucleotide exchange, a transition from G to A, is found in exon 8 (nt. position 27869). Because exon 8 belongs to the 3’ untranslated region, there is no effect on the amino acid sequence. The remaining three nucleotide exchanges are intronic, 1 G to A transition in intron 6 (nt. position 27092) and 2 A to G transitions in intron 7 (nt. positions 27289 and 27404), are intronic and do not directly affect the amino acid sequence of the encoded protein. The nucleotide exchange in exon 7, however, leads to a splice defect in SMN2 transcripts. The resulting exclusion of exon 7 during pre-messenger RNA (mRNA) processing is observed in 90 % of SMN2 transcripts [38].
Fully functional SMN1 pre-mRNA is spliced to an mRNA containing 9 exons, coding for the full-length, multidomain SMN protein with 294 amino acids (FL-SMN). In contrast, only 10 % of SMN2 mRNA transcripts are translated into FL-SMN. Owing to the above described splicing defect, the remaining 90 % of SMN2 mRNAs are translated into a truncated protein of 282 amino acids (SMNΔ7) [24, 39]. As the regular stop codon is located at the end of exon 7, transcripts lacking this exon force the utilization of an alternative stop codon in exon 8. Therefore, in addition to coding for a truncated protein, the exclusion of exon 7 results in a change of the last 4 amino acids of SMN [38]. SMNΔ7 is biochemically unstable and shows reduced self-oligomerization capability when compared with FL-SMN [40].
The SMN protein is highly abundant in the spinal cord, kidney, and liver, as well as the brain [41, 42]. Though expression levels of SMN genes vary in different tissues, there is no tissue specificity for the ratio of FL-SMN to SMNΔ7 proteins [38, 43]. Why the loss of SMN1 primarily affects the motor neurons of the spinal cord remains to be elucidated. Motor neurons appear to have a significantly higher demand for FL-SMN than other cells. Even though complete loss of both SMN genes is lethal, a single copy of SMN1 is sufficient to maintain a healthy individual; two copies of SMN2 alone produce enough FL-SMN protein to maintain the important nuclear functions of SMN in SMA patients, they are however insufficient to ensure survival of motor neurons. There is strong evidence that the role of SMN in axonal maintenance and RNA transport is the underlying cause for the development of SMA. It has been shown that the C-terminus of SMN, which is truncated in 90 % of SMN2 products, is important for axon outgrowth, which is mediated by actin metabolism in the growth cone [44].
SMN Copy Number
It long remained unclear how a genetic defect in a single gene could cause the broad variety in phenotypes observed in SMA. This variability today is attributed mainly to SMN2 copy number. The abundance of FL-SMN is decreased in SMA patients compared with healthy individuals, and levels of FL-SMN are inversely correlated with disease severity [24, 42, 43]. This is consistent with observations that SMN2 copy number is predictive of the SMA disease category [4, 34, 36, 45, 46]. The SMN2 gene encodes about 10 % of the full-length protein and is therefore unable to fully compensate for the loss of SMN1. However, an increased SMN2 copy number leads to more full-length transcript, lessening the severity of the disease. While 80 % of type I SMA patients possess only 1 or 2 copies of SMN2, 83 % of type II patients have 3 copies of SMN2. Type III patients have 3 or 4 copies of SMN2 in 96 % of cases [4]. In one case, a type IV patient with a very mild phenotype was reported to possess 6 copies of SMN2 [47]. Hence, the SMN2 copy number can be used to predict the severity of the SMA phenotype in children [4]. The observation that levels of FL-SMN protein are inversely correlated with the severity of SMA also provides much scope for the clinical treatment of the disease by aiming to increase FL-SMN levels.
Treatment of SMA
For a long time, treatment of SMA was restricted to supportive and palliative care. Although significant improvements have been made in the management of patients and disease-associated problems, culminating in a standard of care document in 2007 [48], to date there is still no effective treatment of the disease. However significant progress has been made in the field with the discovery of SMN as the disease-causing gene, yielding several promising options for treatment strategies in the last decade. As it is well established that the severity of SMA is inversely correlated with SMN protein abundance [42], many approaches to treat SMA are aimed at 1 of 2 targets: restoration of the SMN2 splice-pattern or an increase of the overall abundance of the FL-SMN protein.
SMN2 splice modulation has been achieved using several chemical compounds, including tetracyclines (Aclarubicin, hydroxyurea, PTK-SMA1) [49–51] and β2-adrenergic inhibitors (salbutamol/albuterol) [52, 53], some of which have been tested in human trials [50, 52, 53]. An alternative approach involves targeting of SMN2 pre-mRNA transcripts with RNA antisense oligonucleotides to facilitate exon 7 inclusion and/or exon 8 exclusion [54–59]. RNA-based strategies show encouraging results in pre-clinical experiments in mice [60, 61], and several clinical trials investigating the effects of antisense oligonucleotides in SMA are currently recruiting (clinical trial identifications: NCT01839656, NCT01780246, and NCT01703988).
Arguably the most desirable approach for treatment of SMA would be a permanent, long-term restoration of FL-SMN levels in neurons. Potentially, this could be achieved by the re-introduction of an intact copy of SMN1, or gene conversion from SMN2 to SMN1. Various approaches have been described using lentiviral and adeno-associated viral vectors to deliver an intact gene, and these approaches have shown success in mice [62–64]. Although no clinical trials appear to have started at this stage, major funding projects have recently been established to register clinical trials for systemic adeno-associated virus 9-SMN gene therapy in SMA [65, 66]. Similarly promising are very recent approaches using human-induced pluripotent stem cells in which the SMN2 gene has been altered ex vivo using vector-free oligonucleotide technology to change a single base and thus increase FL-SMN production [67]. Although this technology has not yet progressed to clinical trials, transplantation of these induced pluripotent stem cells into a mouse model of SMA significantly decreased SMA symptoms and increased survival of affected mice. For a detailed overview of the use of stem cells therapy in motor neuron disease see the review by Lunn et al. [68].
One of the most studied treatment options for SMA to date is oral drug delivery. Among the first chemical compounds identified in high-throughput assays for small molecules that are effective in increasing FL-SMN abundance were histone deacetylase (HDAC) inhibitors. Inhibition of HDAC enzymes is thought to increase SMN transcription levels by increasing histone acetylation levels in the respective gene promoters. They have been successfully applied in SMA mouse models, and several HDAC inhibitors have been or are currently being tested in human preclinical and clinical trials. In the next section, we will review in more detail the history and current state of application of HDAC inhibition in the treatment of SMA.
HDAC Inhibitors in SMA Treatment
The mechanism by which HDAC inhibitors increase the abundance of FL-SMN in SMA is most likely linked to a remodeling of the epigenetic landscape of the SMN2 promoter region, which results in increased gene expression. SMN protein levels fluctuate depending on the investigated tissue and developmental stage [41, 69–71], with a significant decline after birth [72, 73]. Interestingly, the highest levels of SMN are detected in the ventral horn of the spinal cord and motor neurons [41, 71], which are primarily affected in SMA. Accordingly, the strongest decline of SMN levels in SMA is also observed in these tissues [74]. Changes in gene expression patterns during development have previously been linked to epigenetic regulation [75]. The developmental changes in SMN protein levels, combined with a proposed differential regulation of SMN1 and SMN2 gene expression despite identical promoter sequences [76], and not least the described effects of various HDAC inhibitors on SMN gene expression, all indicate a strong epigenetic control of SMN promoter activity.
In 2001, sodium butyrate was the first HDAC inhibitor identified to elevate FL-SMN levels in vitro and in vivo [77]. However, sodium butyrate has a short half-life in human serum [78]. Alternative HDAC inhibitors increasing SMN gene expression were soon discovered (Table 1), led by the description of elevated SMN transcript and protein levels following valproic acid (VPA) treatment of SMA patient-derived fibroblast cells [79, 80]. VPA has a significantly longer half-life than sodium butyrate in human serum [1, 3, 7], and has been shown to increase SMN levels in the nervous system of rats [79].
Table 1.
List of histone deacetylase (HDAC) inhibitors used in spinal muscular atrophy models to date. Included are inhibitors that have been shown to increase survival of motor neuron (SMN) RNA and/or protein levels in vitro and/or in vivo, as well as their effective dosage range and HDAC targets [6, 8–15, 19, 20]
| Inhibitor name | Chemical class | Effective dose range | HDAC targets | Effect on SMN | Disadvantages |
|---|---|---|---|---|---|
| Phenylbutyrate | Short-chain fatty acid | mM | N/A | Yes | |
| Sodium butyrate | Short-chain fatty acid | mM | Class I, IIa | Yes | Short half-life |
| Valproic acid | Short-chain fatty acid | mM | Class I, IIa | Yes | Hepatotoxic, teratogen |
| m-Carboxycinnamic acid bishydroxamide | Hydroxamate | µM | N/A | Yes | High toxicity |
| Suberic bishydroxamic acid | Hydroxamate | µM | N/A | Yes | High toxicity |
| Suberoylanilide hydroxamic acid | Hydroxamate | µM | Class I, II | Yes | |
| Trichostatin A | Hydroxamate | nM | Class I, II | Yes | |
| LBH589 | Hydroxamate | nM | N/A | Yes | |
| M344 | Benzamide | µM | N/A | Yes | High toxicity |
| MS-275 | Benzamide | µM | HDAC 1, 2, 3, 9 | No |
NA not available
Also found to be effective in SMA patient-derived fibroblast cells was another short-chain fatty acid, phenylbutyrate (PB) [81]. To date, PB and VPA are the only 2 HDAC inhibitors that have been tested in randomized, double-blind clinical trials for treatment of SMA patients (see below). However, both PB and VPA require relatively high concentrations to effectively increase SMN levels in vitro and in vivo, concentrations that are difficult to achieve in the central nervous system. Hence, the search for more effective, second-generation HDAC inhibitors continues. Suberoylanilide hydroxamic acid (SAHA), which is approved by the US Food and Drug Administration for treatment of cutaneous T-cell lymphoma, as well as trichostatin A and M344 have been shown to be effective in elevating SMN levels at significantly lower levels than previously tested HDAC inhibitors [8, 9, 11], likely owing to the broader range of HDACs they inhibit [8]. More recently, LBH589 has been shown to strongly elevate SMN levels in human neuronal stem cells and SMA patient-derived fibroblast cells at nanomolar concentrations [15]. Intriguingly, LBH589 was able to elevate SMN levels in cells that were previously shown to be insensitive to VPA treatment [15].
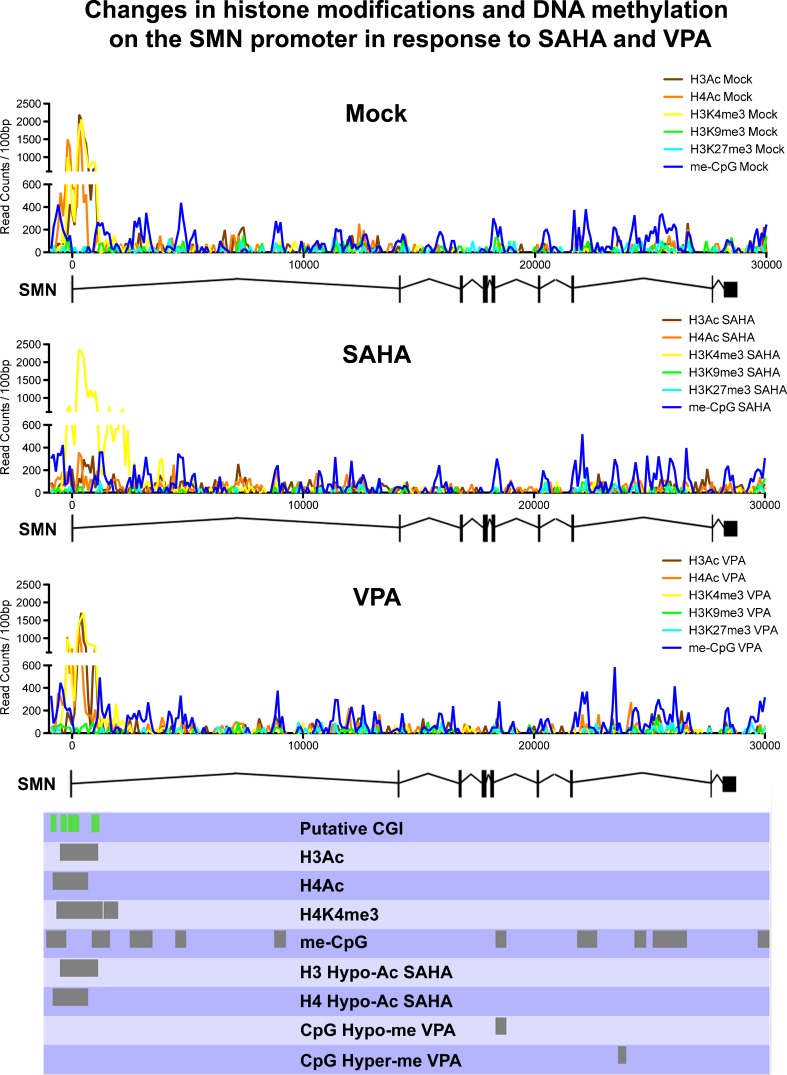
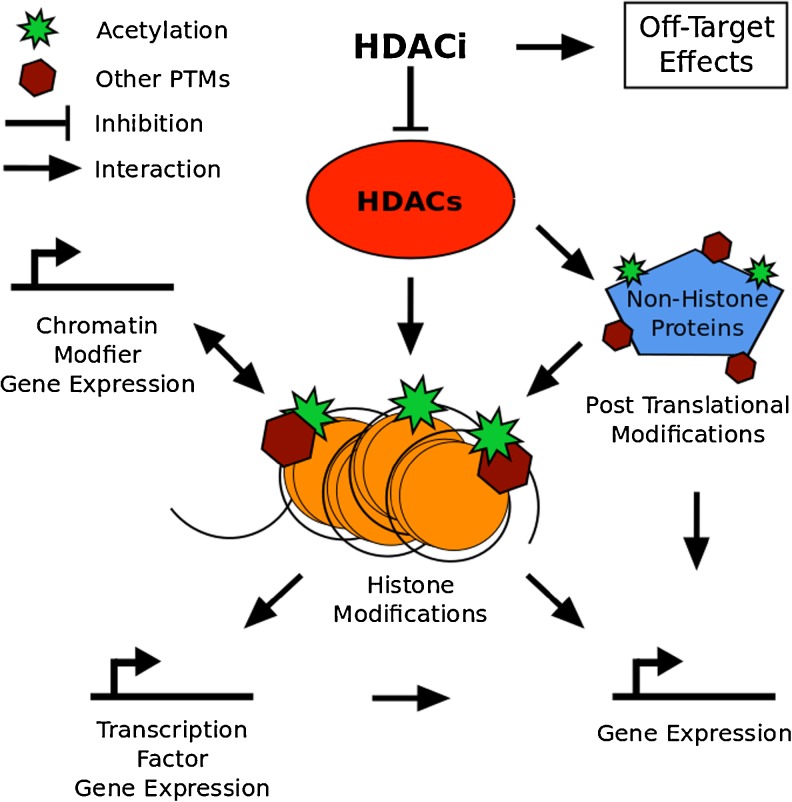
As mentioned above, a likely pathway by which HDAC inhibitors are increasing SMN gene expression and FL-SMN protein levels is through increased histone acetylation of the SMN promoter. It is well established that histone hyperacetylation can lead to a more permissive, transcriptionally-active chromatin structure [82, 83]. Accordingly, it has been shown using chromatin immunoprecipitation analyzed by polymerase chain reaction that the stimulation of human fibroblast cells with VPA increases SMN promoter acetylation [84, 85]. The underlying mechanism, however, might not be quite that simple. Supporting a more complex picture of regulatory changes on the SMN gene are experiments that we have recently performed in our laboratory using chromatin immunoprecipitation and massive parallel sequencing after exposure of human fibroblast cells to VPA or SAHA. Interestingly, in our hands, neither VPA nor SAHA significantly increased histone acetylation on the SMN2 transcription start site (TSS) (personal observations; Fig. 1), despite increasing SMN2 gene expression (data not shown). On the contrary, histone acetylation on the SMN2 TSS was significantly decreased after exposure to SAHA. Additionally, changes in DNA methylation were observed in the SMN gene body. The here-observed absence of increased histone acetylation on the SMN2 TSS following VPA exposure, as well as the decrease in histone acetylation on the SMN2 TSS in response to SAHA exposure, are in contrast to previous results linking increased SMN gene expression to increased histone acetylation on the SMN promoter [84, 85]. Albeit being preliminary, our observations were made using a higher resolution analysis than previous studies, and indicate a more complex mechanism behind the increased SMN2 gene expression in response to HDAC inhibitors than previously thought. This mechanism is likely to involve not only changes to other histone modifications, but also to DNA methylation. In addition to increasing histone acetylation, HDAC inhibitors have been proposed to facilitate increases in the histone 3 lysine 4 methylation mark [86–88], as well as to promote active DNA demethylation [41, 89–93]. It is unknown whether these changes are mediated by directly stimulating histone modifying enzymes or by other phenomena, such as chromatin cross-talk (reviewed in [94–96]). Another potential factor in SMN upregulation in response to HDAC inhibitors is increased access for other chromatin-modifying enzymes, and, indeed, transcription factors, through a more permissive chromatin structure. This has been proposed for DNA demethylases and their role in HDAC inhibitor-mediated DNA demethylation [41, 89–93], and might also be underlying the increased binding activity of AP1 and Sp1 transcription factors to the SMN promoter in response to VPA [79, 97, 98], as well as increased Serine/Arginine-rich (SR) and SR-like splicing factor activity on SMN pre-mRNA [79, 99]. Each of these events could elevate the abundance of FL-SMN by increasing gene-expression or restoring the SMN splice pattern [100]. Importantly, it has to be taken into consideration that HDAC inhibition affects the expression of a large number of genes, including transcription factors [101]. An altered transcription factor profile, and, indeed, altered transcription factor activity mediated by HDAC inhibitor induced differential modification of nonhistone proteins (reviewed in [102, 103]), may well be important in elevating FL-SMN protein levels independent of histone acetylation at the SMN promoter. Possible mechanisms of HDAC inhibition action are summarized in Fig. 2.
Fig. 1.
Chromatin landscape of the survival of motor neuron 2 (SMN2) gene in treated and untreated fibroblast cells. Read distribution for all analyzed datasets was calculated across the whole gene body plus 1 kilo base-pair (bp) up- and downstream using SeqMonk. Read counts were summed into 100 bp bins before plotting. Regions determined by Model-based Analysis for ChIP-Seq (MACS) to be significantly enriched in unexposed (Mock) cells or significantly differentially enriched after exposure were indicated in the bottom panel. Modifications not displaying significant enrichment were omitted to aid readability. Putative CpG islands (CGI) around the SMN2 transcription start site as determined by Hauke et al. [117] are indicated in green. VPA valproic acid, SAHA suberoylanilide hydroxamic acid
Fig. 2.
Possible mechanisms of histone deacetylase inhibitor (HDACi) action. Targets of inhibition are HDAC enzymes, which are associated with histone modifications that are implicated in altering chromatin structure and gene expression. Among the genes with altered expression are transcription factors, which also confer changes in expression of their respective target genes, as well as genes that code for proteins implicated in histone tail and chromatin modification. Lastly HDAC inhibition can alter activity and binding of important non-histone regulatory determinants, including transcription factors, thereby altering the expression of target genes. PTM: Post Translational Modification
Finally, a recent study using an in vitroSMN promoter activity assay and specific small interfering RNA knock-downs of single HDAC enzymes indicated that not all HDACs are equally involved in regulation of SMN gene expression [85]. More exhaustive experiments of this kind will establish in detail the merits of more targeted HDAC inhibition, as well as help unravel the mechanisms and response pathways behind the positive effects of HDAC inhibitors in SMA.
Use of HDAC Inhibitors in SMA Patients and Clinical Trials
A multitude of clinical trials studying the effectiveness of various HDAC inhibitors in the treatment of SMA have been conducted to date, although none have conclusively established the effectiveness of this class of compounds (Table 2). VPA treatment has been shown to increase SMN protein levels in blood of affected individuals [104]; however, a similar study shows increased blood SMN levels in only 7 out of 13 SMA patients [105]. Two further studies report increased muscle strength in a subset of SMA patients treated with VPA [106, 107]. A larger phase I clinical trial (NCT00374075) established the safe application of VPA in SMA patients; however, it also indicated carnitite depletion and weight gain as potential side effects. The at best moderate success of these initial studies was still seen as encouraging, and led to a larger phase II clinical trial employing co-treatment of SMA patients with VPA and L-carnitite (NCT00227266). This randomized, double-blind study was designed in 2 phases, investigating the results of treatment in SMA type I and SMA type II/III patients, respectively. Neither group, however, showed any significant improvements of muscle strength and other SMA-associated symptoms [108, 109]. This was, in part, attributed to the potential confounding factor of increased weight gain in the treatment group on the applied outcome measures [108, 109]. However, this trial did also fail to establish the previously reported significant increase of FL-SMN mRNA in the blood of treated patients [108, 109]. A second phase I/II trial employing VPA and L-carnitite in type I SMA patients (NCT00661453), as well as a phase II trial testing VPA in ambulant adults with SMA (NCT00481013), have been completed, yet the results have not yet been published. A new study assessing the efficacy of VPA and L-carnitite in type II and III SMA patients has been registered, but is not yet recruiting (NCT01671384).
Table 2.
List of patient/clinical trials employing histone deacetylase inhibition in the treatment of spinal muscular atrophy (SMA). ClinicalTrial.gov registration is provided where available
| Study/publication name | Trial ID | RCT | Status | Inhibitor | Clinically relevant conclusions |
|---|---|---|---|---|---|
| Randomized, double-blind, placebo-controlled trial of phenylbutyrate in spinal muscular atrophy | NA | Yes | Completed | PB | PB was not effective at the regimen, schedule, and duration used in this study [112] |
| Quantification of SMN protein in leucocytes from spinal muscular atrophy patients: Effects of treatment with Valproic acid | NA | No | Completed | VPA | VPA treatment resulted in significantly increased SMN protein levels in 5/6 SMA patients [104] |
| Valproic acid treatment in six patients with spinal muscular atrophy | NA | No | Completed | VPA | No effect in 1 type III adolescent and 2 type II/II adults, but muscle strength increase in 2 type II/III and 1 type III [106] |
| Pilot trial of phenylbutyrate in spinal muscular atrophy | NA | No | Completed | PB | PB might be beneficial to SMA patients without producing any major side effect [111] |
| Phenylbutyrate increases SMN gene expression in spinal muscular atrophy patients | NA | No | Completed | PB | PB significantly increases SMN expression in leukocytes of SMA patients [110] |
| In vivo activation of SMN in spinal muscular atrophy carriers and patients treated with valproate | NA | No | Completed | VPA | 7 of 10 carriers demonstrated increased SMN messenger RNA (mRNA) and protein levels. SMN2 mRNA levels were elevated in 7 patients and unchanged or decreased in 13 patients [105] |
| Valproate may improve strength and function in patients with type III/ IV spinal muscle atrophy | NA | No | Completed | VPA | VPA was followed by a sustained increase in function and strength in a group of patients with SMA III/IV [107] |
| Valproic Acid and Carnitine in Patients With Spinal Muscular Atrophy | NCT00227266 | Yes | Completed | VPA | VPA in combination with L-carnitine is not effective in improving strength or function in SMA children [108, 109] |
| Study of Safety and Dosing Effect on SMN Levels of Valproic Acid (VPA) in Patients With Spinal Muscular Atrophy | NCT00374075 | No | Completed | VPA | VPA appears safe and well-tolerated, but weight gain and carnitine depletion are likely to be significant confounding factors [114] |
| Clinical Trial of Sodium Phenylbutyrate in Children With Spinal Muscular Atrophy Type I (NPTUNE 02) | NCT00439218 | No | Terminated owing to slow recruitment | PB | NA |
| Clinical Trial of Sodium Phenylbutyrate in Children With Spinal Muscular Atrophy Types II or III (NPTUNE01) | NCT00439569 | No | Terminated owing to poor drug administration compliance | PB | NA |
| Valproic Acid in Ambulant Adults With Spinal Muscular Atrophy (VALIANTSMA) | NCT00481013 | No | Completed | VPA | NA |
| Study to Evaluate Sodium Phenylbutyrate in Pre-symptomatic Infants With Spinal Muscular Atrophy (STOPSMA) | NCT00528268 | No | Recruiting | PB | NA |
| CARNIVAL Type I: Valproic Acid and Carnitine in Infants With Spinal Muscular Atrophy (SMA) Type I | NCT00661453 | No | Completed | VPA | NA |
| Evaluation of the Muscle Strength and Motor Ability in Children With Spinal Muscle Atrophy(SMA) Treated With Valproic Acid | NCT01033331 | No | Completed | VPA | NA |
| Valproate and Levocarni ne in Children With Spinal Muscular Atrophy | NCT01671384 | No | Not yet recruiting | VPA | NA |
ID identification, RCT randomized controlled trial, SMN survival of motor neuron, VPA valproic acid, PB phenylbutyrate, NA not available
PB has also been used in several pilot studies establishing safe application in humans and indicating increased SMN levels in the blood of SMA patients [110, 111]. Two early clinical trials (NCT00439569 and NCT00439218), however, were terminated owing to poor compliance with the drug administration guidelines and slow recruitment. Similar to VPA, a larger, randomized, double-blind, placebo-controlled trial did not show any significant effects of PB in SMA patients using functional assessments of muscle strength [112]. Blood levels of FL-SMN protein were not assessed. This trial used an intermittent drug regimen, where patients were treated on a 7-day on, 7-day off schedule. The authors of the study argue that this arbitrarily chosen regimen may have weakened or negated any potential beneficial effect of the treatment [112]. A follow-up trial investigating the effects of PB in presymptomatic children that have been genetically confirmed as SMA patients using a continuous drug regimen (NCT00528268) is currently on-going.
For both HDAC inhibitors tested so far in clinical trials for treatment of SMA, pilot studies showed only moderate success and the larger randomized, double-blind, placebo-controlled trials did not show significant improvement of treatment over control groups at all. This has been partially attributed to the lack of adequate outcome measures [113], which might account for the observation that potential increases in SMN levels do not always seem to translate into tangible benefits for patients. Recent publications claim to have established more reliable measures for future trials [108, 109], which might increase their success. An additional complication arises, however, from the quick progression of the disease in its early stages, characterized by the loss of motor neurons. Once neurons are lost, most chemical-based treatments, like HDAC inhibitors, are highly unlikely to be able to restore their function. It therefore appears essential that treatment, and therefore clinical trials assessing the efficacy of HDAC inhibitors in SMA, are initiated before the onset of symptoms. In general, this applies to all forms of treatment aimed at restoring FL-SMN levels in SMA patients. At least 1 of the on-going clinical trials (NCT00528268) currently tries to address this problem by only recruiting presymptomatic SMA patients with confirmed molecular diagnosis. Still, the lack of consistent evidence for increases in FL-SMN in the blood of treated patients with both VPA and PB remains concerning, and it will have to be established whether second-generation HDAC inhibitors are more effective.
Conclusions
To date, VPA and PB are the only HDAC inhibitors investigated in clinical trials for SMA, with no conclusive evidence of success. The use of more potent, second-generation inhibitors in clinical trials may provide more promising results. Several second-generation HDAC inhibitors, including SAHA, trichostatin A, and LBH589, are either already US Food and Drug Administration-approved in other diseases or are frequently used in clinical trials in different disease settings. It is imperative, however, that those future clinical trials employ improved outcome measures to determine efficacy of the utilized treatment strategy, as well as aim to begin treatment before the critical motor neurons are irreversibly lost.
Overall, the application of HDAC inhibitors in the treatment of SMA is challenging. The mechanisms of action behind the increases in SMN protein levels remain poorly understood. It appears clear, however, that the mechanisms behind the increased FL-SMN levels in response to treatment with HDAC inhibitors go beyond the expected increases in histone acetylation at the SMN promoter, as they have been shown to also influence the binding of transcription factors to the SMN gene [15, 79, 115, 116] and alterations in splice patterns [15, 79, 99], as well as SMN post-translational ubiquitination and stability [15]. Understanding the molecular events that contribute to SMA will continue to provide new strategies and targets for the development of therapies to retard or reverse the symptoms of SMA in the future.
Electronic supplementary material
(PDF 123 kb)
Acknowledgments
The authors acknowledge grant and fellowship support from the National Health and Medical Research Council (NHMRC) and the National Heart Foundation of Australia (NHF). S.L. was awarded a Monash Graduate Scholarship and A.E-O. is NHMRC Senior Research Fellows. Supported in part by the Victorian Government’s Operational Infrastructure Support program.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
References
- 1.Loscher W. Basic pharmacology of valproate: a review after 35 years of clinical use for the treatment of epilepsy. CNS Drugs. 2002;16:669–694. doi: 10.2165/00023210-200216100-00003. [DOI] [PubMed] [Google Scholar]
- 2.Pearn J. Classification of spinal muscular atrophies. Lancet. 1980;1:919–922. doi: 10.1016/s0140-6736(80)90847-8. [DOI] [PubMed] [Google Scholar]
- 3.Perucca E. Pharmacological and therapeutic properties of valproate: a summary after 35 years of clinical experience. CNS Drugs. 2002;16:695–714. doi: 10.2165/00023210-200216100-00004. [DOI] [PubMed] [Google Scholar]
- 4.Feldkotter M, Schwarzer V, Wirth R, Wienker TF, Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet. 2002;70:358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kugelberg E, Welander L. Heredofamilial juvenile muscular atrophy simulating muscular dystrophy. AMA Arch Neurol Psychiatry. 1956;75:500–509. doi: 10.1001/archneurpsyc.1956.02330230050005. [DOI] [PubMed] [Google Scholar]
- 6.Andreassi C, Angelozzi C, Tiziano FD, et al. Phenylbutyrate increases SMN expression in vitro: relevance for treatment of spinal muscular atrophy. Eur J Hum Genet. 2004;12:59–65. doi: 10.1038/sj.ejhg.5201102. [DOI] [PubMed] [Google Scholar]
- 7.Munsat TL, Davies KE. International SMA consortium meeting (26–28 June 1992, Bonn, Germany) Neuromuscul Disord. 1992;2:423–428. doi: 10.1016/s0960-8966(06)80015-5. [DOI] [PubMed] [Google Scholar]
- 8.Hahnen E, Eyüpoglu IY, Brichta L, et al. In vitro and ex vivo evaluation of second-generation histone deacetylase inhibitors for the treatment of spinal muscular atrophy. J Neurochem. 2006;98:193–202. doi: 10.1111/j.1471-4159.2006.03868.x. [DOI] [PubMed] [Google Scholar]
- 9.Riessland M, Brichta L, Hahnen E, Wirth B. The benzamide M344, a novel histone deacetylase inhibitor, significantly increases SMN2 RNA/protein levels in spinal muscular atrophy cells. Hum Genet. 2006;120:101–110. doi: 10.1007/s00439-006-0186-1. [DOI] [PubMed] [Google Scholar]
- 10.Riessland M, Ackermann B, Förster A, et al. SAHA ameliorates the SMA phenotype in two mouse models for spinal muscular atrophy. Hum Mol Genet. 2010;19:1492–1506. doi: 10.1093/hmg/ddq023. [DOI] [PubMed] [Google Scholar]
- 11.Avila AM, Burnett BG, Taye AA, et al. Trichostatin A increases SMN expression and survival in a mouse model of spinal muscular atrophy. J Clin Invest. 2007;117:659–671. doi: 10.1172/JCI29562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darras BT, Kang PB. Clinical trials in spinal muscular atrophy. Curr Opin Pediatr. 2007;19:675–679. doi: 10.1097/MOP.0b013e3282f1884c. [DOI] [PubMed] [Google Scholar]
- 13.Swoboda KJ, Kissel JT, Crawford TO, et al. Perspectives on clinical trials in spinal muscular atrophy. J Child Neurol. 2007;22:957–966. doi: 10.1177/0883073807305665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khan N, Jeffers M, Kumar S, et al. Determination of the class and isoform selectivity of small-molecule histone deacetylase inhibitors. Biochem J. 2008;409:581–589. doi: 10.1042/BJ20070779. [DOI] [PubMed] [Google Scholar]
- 15.Garbes L, Riessland M, Hölker I, et al. LBH589 induces up to 10-fold SMN protein levels by several independent mechanisms and is effective even in cells from SMA patients non-responsive to valproate. Hum Mol Genet. 2009;18:3645–3658. doi: 10.1093/hmg/ddp313. [DOI] [PubMed] [Google Scholar]
- 16.Kolb SJ, Kissel JT. Spinal muscular atrophy: a timely review. Arch Neurol. 2011;68:979–984. doi: 10.1001/archneurol.2011.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewelt A, Newcomb TM, Swoboda KJ. New therapeutic approaches to spinal muscular atrophy. Curr Neurol Neurosci Rep. 2012;12:42–53. doi: 10.1007/s11910-011-0240-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamilton G, Gillingwater T. Spinal muscular atrophy: going beyond the motor neuron. Trends Mol Med. 2013;19:40–50. doi: 10.1016/j.molmed.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Melki J, Sheth P, Abdelhak S, et al. Mapping of acute (type I) spinal muscular atrophy to chromosome 5q12-q14. The French Spinal Muscular Atrophy Investigators. Lancet. 1990;336:271–273. doi: 10.1016/0140-6736(90)91803-i. [DOI] [PubMed] [Google Scholar]
- 20.Brzustowicz LM, Lehner T, Castilla LH, et al. Genetic mapping of chronic childhood-onset spinal muscular atrophy to chromosome 5q11.2-13.3. Nature. 1990;344:540–541. doi: 10.1038/344540a0. [DOI] [PubMed] [Google Scholar]
- 21.Melki J, Burlet P, Clermont O, et al. Refined linkage map of chromosome 5 in the region of the spinal muscular atrophy gene. Genomics. 1993;15:521–524. doi: 10.1006/geno.1993.1103. [DOI] [PubMed] [Google Scholar]
- 22.Wirth B. el-Agwany A, Baasner A, et al. Mapping of the spinal muscular atrophy (SMA) gene to a 750-kb interval flanked by two new microsatellites. Eur J Hum Genet. 1995;3:56–60. doi: 10.1159/000472274. [DOI] [PubMed] [Google Scholar]
- 23.Wirth B, Hahnen E, Morgan K, et al. Allelic association and deletions in autosomal recessive proximal spinal muscular atrophy: association of marker genotype with disease severity and candidate cDNAs. Hum Mol Genet. 1995;4:1273–1284. doi: 10.1093/hmg/4.8.1273. [DOI] [PubMed] [Google Scholar]
- 24.Lefebvre S, Burglen L, Reboullet S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 25.Roy N, Mahadevan MS, McLean M, et al. The gene for neuronal apoptosis inhibitory protein is partially deleted in individuals with spinal muscular atrophy. Cell. 1995;80:167–178. doi: 10.1016/0092-8674(95)90461-1. [DOI] [PubMed] [Google Scholar]
- 26.Thompson TG, DiDonato CJ, Simard LR, et al. A novel cDNA detects homozygous microdeletions in greater than 50% of type I spinal muscular atrophy patients. Nat Genet. 1995;9:56–62. doi: 10.1038/ng0195-56. [DOI] [PubMed] [Google Scholar]
- 27.Cobben JM, van der Steege G, Grootscholten P, de Visser M, Scheffer H, Buys CH. Deletions of the survival motor neuron gene in unaffected siblings of patients with spinal muscular atrophy. Am J Hum Genet. 1995;57:805–808. [PMC free article] [PubMed] [Google Scholar]
- 28.DiDonato CJ, Ingraham SE, Mendell JR, et al. Deletion and conversion in spinal muscular atrophy patients: is there a relationship to severity? Ann Neurol. 1997;41:230–237. doi: 10.1002/ana.410410214. [DOI] [PubMed] [Google Scholar]
- 29.Hahnen E, Forkert R, Marke C, et al. Molecular analysis of candidate genes on chromosome 5q13 in autosomal recessive spinal muscular atrophy: evidence of homozygous deletions of the SMN gene in unaffected individuals. Hum Mol Genet. 1995;4:1927–1933. doi: 10.1093/hmg/4.10.1927. [DOI] [PubMed] [Google Scholar]
- 30.Matthijs G, Schollen E, Legius E, et al. Unusual molecular findings in autosomal recessive spinal muscular atrophy. J Med Genet. 1996;33:469–474. doi: 10.1136/jmg.33.6.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodrigues NR, Owen N, Talbot K, Ignatius J, Dubowitz V, Davies KE. Deletions in the survival motor neuron gene on 5q13 in autosomal recessive spinal muscular atrophy. Hum Mol Genet. 1995;4:631–634. doi: 10.1093/hmg/4.4.631. [DOI] [PubMed] [Google Scholar]
- 32.Velasco E, Valero C, Valero A, Moreno F, Hernandez-Chico C. Molecular analysis of the SMN and NAIP genes in Spanish spinal muscular atrophy (SMA) families and correlation between number of copies of cBCD541 and SMA phenotype. Hum Mol Genet. 1996;5:257–263. doi: 10.1093/hmg/5.2.257. [DOI] [PubMed] [Google Scholar]
- 33.Parsons DW, McAndrew PE, Iannaccone ST, Mendell JR, Burghes AH, Prior TW. Intragenic telSMN mutations: frequency, distribution, evidence of a founder effect, and modification of the spinal muscular atrophy phenotype by cenSMN copy number. Am J Hum Genet. 1998;63:1712–1723. doi: 10.1086/302160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wirth B, Herz M, Wetter A, et al. Quantitative analysis of survival motor neuron copies: identification of subtle SMN1 mutations in patients with spinal muscular atrophy, genotype-phenotype correlation, and implications for genetic counseling. Am J Hum Genet. 1999;64:1340–1356. doi: 10.1086/302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schrank B, Götz R, Gunnersen JM, et al. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc Natl Acad Sci USA. 1997;94:9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McAndrew PE, Parsons DW, Simard LR, et al. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am J Hum Genet. 1997;60:1411–1422. doi: 10.1086/515465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Q, Baird SD, Mahadevan M, et al. Sequence of a 131-kb region of 5q13.1 containing the spinal muscular atrophy candidate genes SMN and NAIP. Genomics. 1998;48:121–127. doi: 10.1006/geno.1997.5141. [DOI] [PubMed] [Google Scholar]
- 38.Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gennarelli M, Lucarelli M, Capon F, et al. Survival motor neuron gene transcript analysis in muscles from spinal muscular atrophy patients. Biochem Biophys Res Commun. 1995;213:342–348. doi: 10.1006/bbrc.1995.2135. [DOI] [PubMed] [Google Scholar]
- 40.Lorson CL, Strasswimmer J, Yao JM, et al. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat Genet. 1998;19:63–66. doi: 10.1038/ng0598-63. [DOI] [PubMed] [Google Scholar]
- 41.Coovert DD, Le TT, McAndrew PE, et al. The survival motor neuron protein in spinal muscular atrophy. Hum Mol Genet. 1997;6:1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- 42.Lefebvre S, Burlet P, Liu Q, et al. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 43.Helmken C, Hofmann Y, Schoenen F, et al. Evidence for a modifying pathway in SMA discordant families: reduced SMN level decreases the amount of its interacting partners and Htra2-beta1. Hum Genet. 2003;114:11–21. doi: 10.1007/s00439-003-1025-2. [DOI] [PubMed] [Google Scholar]
- 44.van Bergeijk J, Rydel-Konecke K, Grothe C, Claus P. The spinal muscular atrophy gene product regulates neurite outgrowth: importance of the C terminus. FASEB J. 2007;21:1492–1502. doi: 10.1096/fj.06-7136com. [DOI] [PubMed] [Google Scholar]
- 45.Burghes AH. When is a deletion not a deletion? When it is converted. Am J Hum Genet. 1997;61:9–15. doi: 10.1086/513913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campbell L, Potter A, Ignatius J, Dubowitz V, Davies K. Genomic variation and gene conversion in spinal muscular atrophy: implications for disease process and clinical phenotype. Am J Hum Genet. 1997;61:40–50. doi: 10.1086/513886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wirth B, Brichta L, Schrank B, et al. Mildly affected patients with spinal muscular atrophy are partially protected by an increased SMN2 copy number. Hum Genet. 2006;119:422–428. doi: 10.1007/s00439-006-0156-7. [DOI] [PubMed] [Google Scholar]
- 48.Wang CH, Finkel RS, Bertini ES, et al. Consensus statement for standard of care in spinal muscular atrophy. J Child Neurol. 2007;22:1027–1049. doi: 10.1177/0883073807305788. [DOI] [PubMed] [Google Scholar]
- 49.Hastings ML, Berniac J, Liu YH, et al. Tetracyclines that promote SMN2 exon 7 splicing as therapeutics for spinal muscular atrophy. Sci Transl Med. 2009;1:5ra12. doi: 10.1126/scitranslmed.3000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen T-H, Chang JG, Yang Y-H, et al. Randomized, double-blind, placebo-controlled trial of hydroxyurea in spinal muscular atrophy. Neurology. 2010;75:2190–2197. doi: 10.1212/WNL.0b013e3182020332. [DOI] [PubMed] [Google Scholar]
- 51.Liang W-C, Yuo C-Y, Chang J-G, et al. The effect of hydroxyurea in spinal muscular atrophy cells and patients. J Neurol Sci. 2008;268:87–94. doi: 10.1016/j.jns.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 52.Kinali M, Mercuri E, Main M, et al. Pilot trial of albuterol in spinal muscular atrophy. Neurology. 2002;59:609–610. doi: 10.1212/wnl.59.4.609. [DOI] [PubMed] [Google Scholar]
- 53.Tiziano FD, Lomastro R, Pinto AM, et al. Salbutamol increases survival motor neuron (SMN) transcript levels in leucocytes of spinal muscular atrophy (SMA) patients: relevance for clinical trial design. J Med Genet. 2010;47:856–858. doi: 10.1136/jmg.2010.080366. [DOI] [PubMed] [Google Scholar]
- 54.Baughan TD, Dickson A, Osman EY, Lorson CL. Delivery of bifunctional RNAs that target an intronic repressor and increase SMN levels in an animal model of spinal muscular atrophy. Hum Mol Genet. 2009;18:1600–1611. doi: 10.1093/hmg/ddp076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coady TH, Shababi M, Tullis GE, Lorson CL. Restoration of SMN function: delivery of a trans-splicing RNA re-directs SMN2 pre-mRNA splicing. Mol Ther. 2007;15:1471–1478. doi: 10.1038/sj.mt.6300222. [DOI] [PubMed] [Google Scholar]
- 56.Dickson A, Osman E, Lorson C. A negatively-acting bifunctional RNA increases survival motor neuron in vitro and in vivo. Hum Gene Ther. 2008;19:1307–1315. doi: 10.1089/hum.2008.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hua Y, Vickers TA, Baker BF, Bennett CF, Krainer AR. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 2007;5:e73. doi: 10.1371/journal.pbio.0050073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Madocsai C, Lim SR, Geib T, Lam BJ, Hertel KJ. Correction of SMN2 Pre-mRNA splicing by antisense U7 small nuclear RNAs. Mol Ther. 2005;12:1013–1022. doi: 10.1016/j.ymthe.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 59.Marquis J, Meyer K, Angehrn L, Kampfer SS, Rothen-Rutishauser B, Schumperli D. Spinal muscular atrophy: SMN2 pre-mRNA splicing corrected by a U7 snRNA derivative carrying a splicing enhancer sequence. Mol Ther. 2007;15:1479–1486. doi: 10.1038/sj.mt.6300200. [DOI] [PubMed] [Google Scholar]
- 60.Passini MA, Bu J, Richards AM, et al. Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci Transl Med. 2011;3:72ra18. doi: 10.1126/scitranslmed.3001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hua Y, Sahashi K, Rigo F, et al. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011;478:123–126. doi: 10.1038/nature10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Foust KD, Wang X, McGovern VL, et al. Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat Biotechnol. 2010;28:271–274. doi: 10.1038/nbt.1610. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Valori CF, Ning K, Wyles M, et al. Systemic delivery of scAAV9 expressing SMN prolongs survival in a model of spinal muscular atrophy. Sci Transl Med. 2010;2:35ra42. doi: 10.1126/scitranslmed.3000830. [DOI] [PubMed] [Google Scholar]
- 64.Azzouz M, Le T, Ralph GS, et al. Lentivector-mediated SMN replacement in a mouse model of spinal muscular atrophy. J Clin Invest. 2004;114:1726–1731. doi: 10.1172/JCI22922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Families of SMA and nationwide children’s announce multi-million dollar award from NINDS to advance CNS gene therapy for spinal muscular atrophy. Available at: http://www.fsma.org/LatestNews/index.cfm?ID=7638&TYPE=1150. Accessed 7 Aug 2013.
- 66.FightSMA: history of gene therapy. Available at: http://www.fightsma.org/sma-research/gene-therapy/gene_therapy_history/. Accessed 7 Aug 2013.
- 67.Corti S, Nizzardo M, Simone C, et al. Genetic correction of human induced pluripotent stem cells from patients with spinal muscular atrophy. Sci Transl Med. 2012;4:165ra162. doi: 10.1126/scitranslmed.3004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lunn JS, Sakowski SA, Federici T, Glass JD, Boulis NM, Feldman EL. Stem cell technology for the study and treatment of motor neuron diseases. Regen Med. 2011;6:201–213. doi: 10.2217/rme.11.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.La Bella V, Cisterni C, Salaun D, Pettmann B. Survival motor neuron (SMN) protein in rat is expressed as different molecular forms and is developmentally regulated. Eur J Neurosci. 1998;10:2913–2923. doi: 10.1111/j.1460-9568.1998.00298.x. [DOI] [PubMed] [Google Scholar]
- 70.Kernochan LE, Russo ML, Woodling NS, et al. The role of histone acetylation in SMN gene expression. Hum Mol Genet. 2005;14:1171–1182. doi: 10.1093/hmg/ddi130. [DOI] [PubMed] [Google Scholar]
- 71.Battaglia G, Princivalle A, Forti F, Lizier C, Zeviani M. Expression of the SMN gene, the spinal muscular atrophy determining gene, in the mammalian central nervous system. Hum Mol Genet. 1997;6:1961–1971. doi: 10.1093/hmg/6.11.1961. [DOI] [PubMed] [Google Scholar]
- 72.Burlet P, Huber C, Bertrandy S, et al. The distribution of SMN protein complex in human fetal tissues and its alteration in spinal muscular atrophy. Hum Mol Genet. 1998;7:1927–1933. doi: 10.1093/hmg/7.12.1927. [DOI] [PubMed] [Google Scholar]
- 73.Jablonka S, Schrank B, Kralewski M, Rossoll W, Sendtner M. Reduced survival motor neuron (Smn) gene dose in mice leads to motor neuron degeneration: an animal model for spinal muscular atrophy type III. Hum Mol Genet. 2000;9:341–346. doi: 10.1093/hmg/9.3.341. [DOI] [PubMed] [Google Scholar]
- 74.Boda B, Mas C, Giudicelli C, et al. Survival motor neuron SMN1 and SMN2 gene promoters: identical sequences and differential expression in neurons and non-neuronal cells. Eur J Hum Genet. 2004;12:729–737. doi: 10.1038/sj.ejhg.5201217. [DOI] [PubMed] [Google Scholar]
- 75.Kiefer JC. Epigenetics in development. Dev Dyn. 2007;236:1144–1156. doi: 10.1002/dvdy.21094. [DOI] [PubMed] [Google Scholar]
- 76.Germain-Desprez D, Brun T, Rochette C, Semionov A, Rouget R, Simard LR. The SMN genes are subject to transcriptional regulation during cellular differentiation. Gene. 2001;279:109–117. doi: 10.1016/s0378-1119(01)00758-2. [DOI] [PubMed] [Google Scholar]
- 77.Chang JG, Hsieh-Li HM, Jong YJ, Wang NM, Tsai CH, Li H. Treatment of spinal muscular atrophy by sodium butyrate. Proc Natl Acad Sci USA. 2001;98:9808–9813. doi: 10.1073/pnas.171105098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pellizzaro C, Coradini D, Morel S, Ugazio E, Gasco MR, Daidone MG. Cholesteryl butyrate in solid lipid nanospheres as an alternative approach for butyric acid delivery. Anticancer Res. 1999;19:3921–3925. [PubMed] [Google Scholar]
- 79.Brichta L, Hofmann Y, Hahnen E, et al. Valproic acid increases the SMN2 protein level: a well-known drug as a potential therapy for spinal muscular atrophy. Hum Mol Genet. 2003;12:2481–2489. doi: 10.1093/hmg/ddg256. [DOI] [PubMed] [Google Scholar]
- 80.Sumner CJ, Huynh TN, Markowitz JA, et al. Valproic acid increases SMN levels in spinal muscular atrophy patient cells. Ann Neurol. 2003;54:647–654. doi: 10.1002/ana.10743. [DOI] [PubMed] [Google Scholar]
- 81.Andreassi C, Angelozzi C, Tiziano FD, et al. Phenylbutyrate increases SMN expression in vitro: relevance for treatment of spinal muscular atrophy. Eur J Hum Genet. 2003;12:59–65. doi: 10.1038/sj.ejhg.5201102. [DOI] [PubMed] [Google Scholar]
- 82.Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 83.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 84.Kernochan LE, Russo ML, Woodling NS, et al. The role of histone acetylation in SMN gene expression. Hum Mol Genet. 2005;14:1171–1182. doi: 10.1093/hmg/ddi130. [DOI] [PubMed] [Google Scholar]
- 85.Evans MC, Cherry JJ, Androphy EJ. Differential regulation of the SMN2 gene by individual HDAC proteins. Biochem Biophys Res Commun. 2011;414:25–30. doi: 10.1016/j.bbrc.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nightingale KP, Gendreizig S, White DA, Bradbury C, Hollfelder F, Turner BM. Cross-talk between histone modifications in response to histone deacetylase inhibitors: MLL4 links histone H3 acetylation and histone H3K4 methylation. J Biol Chem. 2007;282:4408–4416. doi: 10.1074/jbc.M606773200. [DOI] [PubMed] [Google Scholar]
- 87.Harikrishnan KN, Karagiannis TC, Chow MZ, El-Osta A. Effect of valproic acid on radiation-induced DNA damage in euchromatic and heterochromatic compartments. Cell Cycle. 2008;7:468–476. doi: 10.4161/cc.7.4.5405. [DOI] [PubMed] [Google Scholar]
- 88.Bradbury CA, Khanim FL, Hayden R, et al. Histone deacetylases in acute myeloid leukaemia show a distinctive pattern of expression that changes selectively in response to deacetylase inhibitors. Leukemia. 2005;19:1751–1759. doi: 10.1038/sj.leu.2403910. [DOI] [PubMed] [Google Scholar]
- 89.Milutinovic S, D'Alessio AC, Detich N, Szyf M. Valproate induces widespread epigenetic reprogramming which involves demethylation of specific genes. Carcinogenesis. 2007;28:560–571. doi: 10.1093/carcin/bgl167. [DOI] [PubMed] [Google Scholar]
- 90.Ou JN, Torrisani J, Unterberger A, et al. Histone deacetylase inhibitor Trichostatin A induces global and gene-specific DNA demethylation in human cancer cell lines. Biochem. Pharmacol. 2007;73:1297–1307. doi: 10.1016/j.bcp.2006.12.032. [DOI] [PubMed] [Google Scholar]
- 91.Detich N, Bovenzi V, Szyf M. Valproate induces replication-independent active DNA demethylation. J Biol Chem. 2003;278:27586–27592. doi: 10.1074/jbc.M303740200. [DOI] [PubMed] [Google Scholar]
- 92.Cervoni N, Szyf M. Demethylase activity is directed by histone acetylation. J Biol Chem. 2001;276:40778–40787. doi: 10.1074/jbc.M103921200. [DOI] [PubMed] [Google Scholar]
- 93.Cervoni N, Detich N, Seo SB, Chakravarti D, Szyf M. The oncoprotein Set/TAF-1beta, an inhibitor of histone acetyltransferase, inhibits active demethylation of DNA, integrating DNA methylation and transcriptional silencing. J Biol Chem. 2002;277:25026–25031. doi: 10.1074/jbc.M202256200. [DOI] [PubMed] [Google Scholar]
- 94.Lee J-S, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Verrier L, Vandromme M, Trouche D. Histone demethylases in chromatin cross-talks. Biol. Cell. 2011;103:381–401. doi: 10.1042/BC20110028. [DOI] [PubMed] [Google Scholar]
- 96.Suganuma T, Workman JL. Crosstalk among histone modifications. Cell. 2008;135:604–607. doi: 10.1016/j.cell.2008.10.036. [DOI] [PubMed] [Google Scholar]
- 97.Wlodarczyk BC, Craig JC, Bennett GD, Calvin JA, Finnell RH. Valproic acid-induced changes in gene expression during neurulation in a mouse model. Teratology. 1996;54:284–297. doi: 10.1002/(SICI)1096-9926(199612)54:6<284::AID-TERA3>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 98.Arinze IJ, Kawai Y. Sp family of transcription factors is involved in valproic acid-induced expression of Galphai2. J Biol Chem. 2003;278:17785–17791. doi: 10.1074/jbc.M209430200. [DOI] [PubMed] [Google Scholar]
- 99.Chang JG, Hsieh-Li HM, Jong YJ, Wang NM, Tsai CH, Li H. Treatment of spinal muscular atrophy by sodium butyrate. Proc Natl Acad Sci USA. 2001;98:9808–9813. doi: 10.1073/pnas.171105098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hofmann Y, Lorson CL, Stamm S, Androphy EJ, Wirth B. Htra2-beta 1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2) Proc Natl Acad Sci USA. 2000;97:9618–9623. doi: 10.1073/pnas.160181697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Peart MJ, Smyth GK, van Laar RK, et al. Identification and functional significance of genes regulated by structurally different histone deacetylase inhibitors. Proc Natl Acad Sci USA. 2005;102:3697–3702. doi: 10.1073/pnas.0500369102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 103.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 104.Piepers S, Cobben J-M, Sodaar P, et al. Quantification of SMN protein in leucocytes from spinal muscular atrophy patients: effects of treatment with valproic acid. J Neurol Neurosurg Psychiatr. 2011;82:850–852. doi: 10.1136/jnnp.2009.200253. [DOI] [PubMed] [Google Scholar]
- 105.Brichta L, Hölker I, Haug K, Klockgether T, Wirth B. In vivo activation of SMN in spinal muscular atrophy carriers and patients treated with valproate. Ann Neurol. 2006;59:970–975. doi: 10.1002/ana.20836. [DOI] [PubMed] [Google Scholar]
- 106.Tsai LK, Yang C-C, Hwu W-L, Li H. Valproic acid treatment in six patients with spinal muscular atrophy. Eur J Neurol. 2007;14:e8–9. doi: 10.1111/j.1468-1331.2007.01992.x. [DOI] [PubMed] [Google Scholar]
- 107.Weihl CC, Connolly AM, Pestronk A. Valproate may improve strength and function in patients with type III/IV spinal muscle atrophy. Neurology. 2006;67:500–501. doi: 10.1212/01.wnl.0000231139.26253.d0. [DOI] [PubMed] [Google Scholar]
- 108.Kissel JT, Scott CB, Reyna SP, et al. SMA CARNIVAL TRIAL PART II: a prospective, single-armed trial of L-carnitine and valproic acid in ambulatory children with spinal muscular atrophy. PLoS ONE. 2011;6:e21296. doi: 10.1371/journal.pone.0021296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Swoboda KJ, Scott CB, Crawford TO, et al. SMA CARNI-VAL trial part I: double-blind, randomized, placebo-controlled trial of L-carnitine and valproic acid in spinal muscular atrophy. PLoS ONE. 2010;5:e12140. doi: 10.1371/journal.pone.0012140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Brahe C, Vitali T, Tiziano FD, et al. Phenylbutyrate increases SMN gene expression in spinal muscular atrophy patients. Eur J Hum Genet. 2004;13:256–259. doi: 10.1038/sj.ejhg.5201320. [DOI] [PubMed] [Google Scholar]
- 111.Mercuri E, Bertini E, Messina S, et al. Pilot trial of phenylbutyrate in spinal muscular atrophy. Neuromuscul Disord. 2004;14:130–135. doi: 10.1016/j.nmd.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 112.Mercuri E, Bertini E, Messina S, et al. Randomized, double-blind, placebo-controlled trial of phenylbutyrate in spinal muscular atrophy. Neurology. 2007;68:51–55. doi: 10.1212/01.wnl.0000249142.82285.d6. [DOI] [PubMed] [Google Scholar]
- 113.Markowitz JA, Singh P, Darras BT. Spinal muscular atrophy: a clinical and research update. Pediatr Neurol. 2012;46:1–12. doi: 10.1016/j.pediatrneurol.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 114.Swoboda KJ, Scott CB, Reyna SP, et al. Phase II open label study of valproic acid in spinal muscular atrophy. PLoS ONE. 2009;4:e5268. doi: 10.1371/journal.pone.0005268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wlodarczyk BC, Craig JC, Bennett GD, Calvin JA, Finnell RH. Valproic acid-induced changes in gene expression during neurulation in a mouse model. Teratology. 1996;54:284–297. doi: 10.1002/(SICI)1096-9926(199612)54:6<284::AID-TERA3>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 116.Arinze IJ, Kawai Y. Sp family of transcription factors is involved in valproic acid-induced expression of Galphai2. J Biol Chem. 2003;278:17785–17791. doi: 10.1074/jbc.M209430200. [DOI] [PubMed] [Google Scholar]
- 117.Hauke J, Riessland M, Lunke S, et al. Survival motor neuron gene 2 silencing by DNA methylation correlates with spinal muscular atrophy disease severity and can be bypassed by histone deacetylase inhibition. Hum Mol Genet. 2009;18:304–317. doi: 10.1093/hmg/ddn357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 123 kb)