Abstract
The L-arabinose utilization pathway was established in Saccharomyces cerevisiae, by expressing the codon-optimized araA, araB, and araD genes of Lactobacillus plantarum. After overexpressing the TAL1, TKL1, RPE1, RKI1, and GAL2 genes and adaptive evolution, the L-arabinose utilization of the recombinant strain became efficient. The resulting strain displayed a maximum specific growth rate of 0.075 h−1, a maximum specific L-arabinose consumption rate of 0.61 g h−1 g−1 dry cell weight, and a promising ethanol yield of 0.43 g g−1 from L-arabinose fermentation.
1. Introduction
To reduce the dependence on fossil fuels, the worldwide production of bioethanol was increased from ~45 million liters in 2005 to ~113 billion liters in 2012 [1–3]. The future large-scale production of fuel ethanol will most likely be based on abundant lignocellulosic materials instead of sugar and grain, which are food for humans and animals [4]. Cost-effective fuel ethanol production from lignocellulosic materials requires the full use of the raw materials. One goal of bioethanol production is to endow the fermentation microorganism with the capacity to convert all of the sugars in lignocellulosic materials [5, 6]. Approximately 3–15% L-arabinose component can be recovered from lignocellulosic materials [7]. It is therefore necessary to construct an L-arabinose fermenting microorganism to increase the utilization of this sugar [8].
Two types of L-arabinose metabolic pathways exist in fungi and bacteria. The aldose reductase (AR), L-arabitol-4-dehydrogenase (LAD), L-xylulose reductase (LXR), and D-xylitol dehydrogenase (XDH) constitute the fungal L-arabinose metabolic pathway. The reaction catalyzed by AR and LXR is coupled with the oxidation of NADPH to NADP+, and the LAD and XDH use NAD+ as a cofactor [9]. The xylulose produced is phosphorylated and enters the pentose-phosphate pathway (PPP). The bacterial L-arabinose metabolic pathway is cofactor independent and consists of L-arabinose isomerase (AraA), L-ribulokinase (AraB), and L-ribulose-5-phosphate 4-epimerase (AraD). The D-xylulose-5-phosphate produced enters the PPP [9, 10]. Both L-arabinose metabolic pathways were established in Saccharomyces cerevisiae, which is the traditional ethanol-producing microorganism with excellent sugar fermenting capacity and tolerance to the harsh environment, but it cannot ferment L-arabinose [11]. Not surprisingly, a redox imbalance occurs in the recombinant S. cerevisiae strain containing the fungal L-arabinose metabolic pathway. The yield of the by-product L-arabitol was as high as 0.48 g g−1 of pentose sugar consumed in the D-xylose and L-arabinose cofermentation, although the strain expressing NADH preferred AR and LXR to decrease the redox imbalance [12].
Compared to the fungal L-arabinose metabolic pathway, the bacterial pathway is simpler and cofactor independent. However, because of the lack of effective activity assays for enzymes involved in the bacterial L-arabinose metabolic pathway, the optimization of this pathway in S. cerevisiae was not straightforward. The S. cerevisiae strain expressing the araA, araB, and araD genes of Escherichia coli could not utilize L-arabinose. However, after the E. coli L-arabinose isomerase gene was replaced with the araA cloned from Bacillus subtilis, the strain could grow and produce ethanol on L-arabinose after several circles of adaptive growth [13, 14]. Furthermore, the L-arabinose utilization was further improved by changing the codon usage of the bacterial araA, araB, and araD genes to the preferred yeast codons [15]. The L-arabinose metabolic genes of Lactobacillus plantarum matched the codon usage of S. cerevisiae more closely than the genes previously reported. Wisselink et al. [8] introduced multiple copies of araA and araD and a single copy of araB of L. plantarum into S. cerevisiae. After overexpressing the genes encoding the enzymes of nonoxidative PPP and extensive adaptive evolution, the resulting strain exhibited a high ethanol yield up to 0.43 g g−1 during anaerobic growth on L-arabinose, with a high arabinose consumption rate (0.70 g h−1 g−1 dry cell weight (DCW)) [8]. The metabolome, transcriptome, and metabolic flux analysis of a more evolved strain revealed that higher expression levels of the galactose transporter, transketolase, and transaldolase isoenzymes benefit the growth of S. cerevisiae on L-arabinose [16].
In the present work, the unique codon-optimized araA, araB, and araD genes of L. plantarum were expressed in the S. cerevisiae strain CEN.PK102-3A at different levels. Next, the genes TAL1, TKL1, RPE1, and RKI1 involved in PPP were overexpressed in this recombinant strain. The resulting strain was sequentially selected on L-arabinose under aerobic conditions and in oxygen-limited conditions. A strain with a significantly enhanced L-arabinose utilization capacity was obtained. The L-arabinose metabolic capacity of the evolved strains and the strain that also overexpressed the transporter gene GAL2 were investigated. The factors affecting L-arabinose metabolism efficiency are discussed.
2. Materials and Methods
2.1. Media and Culture Conditions
The yeast synthetic complete medium (SC) containing 1.7 g L−1 yeast nitrogen base (YNB, Sangon, China) and 5 g L−1 ammonium sulfate (Sangon, China), with additional carbon sources of glucose (Sangon, China) or L-arabinose (Sinopharm, China), was used for yeast cultivation. The complete supplement mixture, 0.77 g L−1 CSM-URA or 0.67 g L−1 CSM-LEU-URA (MP Biomedicals, Solon, OH), was added to maintain the required plasmids with auxotrophic selection when necessary. For strains with the KanMX4 marker, the medium was supplied with 200 μg mL−1 of the antibiotic G418 sulfate (Promega, Madison, WI, USA). All yeasts were cultivated at 30°C.
2.2. Codon Adaptation Index Analysis
The codon adaptation index (CAI) is used to illustrate the preference of codon usage in specific species [24]. For the CAI analysis, CODONW (http://mobyle.pasteur.fr/cgi-bin/MobylePortal/portal.py?form=codonw) [15] was used.
2.3. Plasmid and Strain Construction
E. coli DH5α [12] was used for subcloning. S. cerevisiae strains and plasmids used in this study are listed in Table 1. The primers used in this study are listed in Table 2.
Table 1.
S. cerevisiae strains and plasmids used in this study.
| Relevant genotype | Source/reference | |
|---|---|---|
| Strain | ||
| CEN.PK102-3A | MATα leu2-3, 112 ura3-52 | [17] |
| BSW1A1 | CEN.PK102-3A derivative; {YIp5-ara} | This work |
| BSW1AY | CEN.PK102-3A derivative; {YIp5-ara, pYX242} | This work |
| BSW1A7 | CEN.PK102-3A derivative; {YIp5-ara, pYX2422-HXT7araA} | This work |
| BSW1AT | CEN.PK102-3A derivative; {YIp5-ara, pYX2422-TEF1araA} | This work |
| BSW2AP | BSW1AT, gre3 (−241, +338):: TPI1p-RKI1-RKI1t-PGK1p-TAL1-TAL1t-FBA1p-TKL1-TKL1t-ADH1p-RPE1-RPE1t-loxP | This work |
| BSW3AP | BSW2AP, selected for oxygen-limited growth on L-arabinose | This work |
| BSW3AG | BSW3AP derivative; {pJFE318-GAL2} | This work |
| Plasmid | ||
| pUG6 | E. coli plasmid with segment LoxP-KanMX4-LoxP | [18] |
| pJPPP3 | pUC19-based yeast integration plasmid, containing GRE3-targeting recombinant arms, overexpression cassette of Sc-TAL1, Sc-TKL1, Sc-RPE1, Sc-RKI1, and selectable marker loxP-KanMX4-loxP | [19] |
| YEp24-PGKp | 2μURA3 | [20] |
| pHX | YEp24-PGKp PGK1p::HXT7p | This work |
| YIp5 | Integration plasmid, Ura3 | [21] |
| YIp5-ara | YIp5-HXT7p-araA-PGK1t-HXT7p-araB-PGK1t-HXT7p-araD-PGK1t, and selectable marker loxP-KanMX4-loxP | This work |
| pYX242 | 2μLEU2 | [22] |
| pYX242-WS | pYX242-PGK1t-TEF1p | This work |
| pYX2422-TEF1araA | pYX242-PGK1t-TEF1p-araA | This work |
| pYX2422-HXT7araA | pYX242-PGK1t-HXT7p-araA | This work |
| pJFE3 | 2μURA3 | [23] |
| pJFE3-GAL2 | pJFE3-TEF1p-GAL2-PGK1t | This work |
| pJFE318-GAL2 | pJFE3-GAL2 URA3::KanMX4 | This work |
Table 2.
Oligonucleotides used in this work.
| Primers | Sequence (5′-3′) | Purpose |
|---|---|---|
| Hxt7 upstream-HX | CATAGATCTCTCACAAATTAGAGCTTCAATTTAAT | Cloning the fragment of HXT7p-araA-PGKt1 |
| Pgk6 downstream-S | CATGTCGACAGCAATTTAACTGTGATAAACTACCG | Cloning the fragment of HXT7p-araA-PGKt1 |
| Hxt7 upstream-EEB | CATCGGCCGAGATCTCCTAGGCTCACAAATTAGAGCTTCAATTTAAT | Cloning the fragment of HXT7p-araB-PGKt1 |
| Pgk6 downstream-S | CATGTCGACAGCAATTTAACTGTGATAAACTACCG | Cloning the fragment of HXT7p-araB-PGKt1 |
| Hxt7 upstream | CATCCTAGGCTCACAAATTAGAGCTTCAATTTAAT | Cloning the fragment of HXT7p-araD-PGKt1 |
| Pgk6 downstream | CATCCTAGGAGCAATTTAACTGTGATAAACTACCG | Cloning the fragment of HXT7p-araD-PGKt1 |
| HXT7p-F | CCCAAGCTTCTCACAAATTAGAGCTTCAATT | Cloning HXT7p |
| HXT7p-R | ACGCGTCGACATTGGATCTAGATGCATTCGCG | Cloning HXT7p |
| TEF1 W up | CCCAAGCTTCACAATGCATACTTTGTACGTT | Cloning TEF1p |
| TEF1 W down | GCGCGTCGACTTGTAATTAAAACTTAGATTAG | Cloning TEF1p |
| AraA W up | ACGCGTCGACATGTTATCTGTTCCTGATTATG | Cloning araA |
| AraA W down-His | TACGAGTCTTTAGTGGTGGTGGTGGTGGTGTTTTAAAAATGCTTTTGTCA | Cloning araA |
| AraA-F | CAAGCAGGTGGTGGTCATCATAC | For quantitative real-time PCR of araA |
| AraA-R | TACCAACCATTGTAGCGTAATCTTCC | For quantitative real-time PCR of araA |
| AraB-1F | ATGCAGCATTCGCACCTTTG | For quantitative real-time PCR of araB |
| AraB-1R | CCTTCACCTGCTGTGGACAT | For quantitative real-time PCR of araB |
| AraD-1F | CCAGCTGCAGATGCATTAACT | For quantitative real-time PCR of araD |
| AraD-1R | ACAGCCTTAGCTGGTGTTGG | For quantitative real-time PCR of araD |
| Gal2 up | GCTCTAGAATGGCAGTTGAGGAGAACAATATGC | Cloning GAL2 |
| Gal2 down | ACGCGTCGACTTATTCTAGCATGGCCTTGTAC | Cloning GAL2 |
| pG418-Apa I up | AGTGGGCCCTAGGTCTAGAGATCTGTTTAGC | Cloning KanMX4 |
| pG418-Nde I down | GGAATTCCATATGATTAAGGGTTCTCGAGAGCTCG | Cloning KanMX4 |
The unique codon-optimized araA, araB, and araD genes encoding the L-arabinose isomerase (GenBank: CCC80517.1), L-ribulokinase (GenBank: CCC80519.1), and L-ribulose-5-phosphate 4-epimerase (GenBank: CCC80518.1) of L. Plantarum were artificially synthesized and ligated between the HXT7 promoter and PGK1 terminator sequences of plasmid pHX, which was constructed by substituting the PGK1p of plasmid YEp24-PGKp [20] with HXT7p, containing sites for the restriction enzymes Kpn I and Sma I. The HXT7p-araD-PGK1t fragment was amplified by PCR with terminal sites for the restriction enzymes Hind III and Bln I and then inserted into the Hind III and Nhe I sites of YIp5, resulting in plasmid YIp5-araD. The HXT7p-araA-PGK1t fragment containing terminal Bgl II and Sal I sites was inserted into the BamH I and Sal I sites of YIp5-araD, resulting in the plasmid YIp5-araAD. The HXT7p-araB-PGK1t fragment with Eag I and Stu I sites was inserted into the Eag I and Stu I sites of YIp5-araAD, resulting in the plasmid YIp5-ara (Figure 1(a)). The TEF1 promoter fragment (with terminal sites for Hind III and Sal I) and the PGK1 terminator fragment (with terminal sites for BamH I and Hind III) were cloned from the plasmids pJFE3 [23] and pYMIKP [25], respectively. These two fragments were ligated and inserted into the plasmid pYX242 to construct a vector, pYX242-WS, with two sites that can be used to express genes. Then, the gene araA was inserted between the Sal I and Sac I sites of this vector under control of the TEF1 promoter and the PloyA terminator for its expression, and the resulting plasmid was named pYX2422-TEF1araA (Figure 1(b)). The plasmid pYX2422-HXT7araA (Figure 1(b)) was constructed using the fragment of HXT7p to displace the TEF1p fragment of plasmid pYX2422-TEF1araA; the joints were BamH I and Sal I recognition sequences. The gene GAL2 was cloned from the chromosomal DNA of CEN.PK102-3A and then inserted into the Xba I and Sal I sites of plasmid pJFE3, resulting in plasmid pJFE3-GAL2. The URA3 fragment of plasmid pJFE3-GAL2 between the Nde I and Apa I sites was replaced by the KanMX4 gene cloned from pUG6 [18], resulting in plasmid pJFE318-GAL2 (Figure 1(c)).
Figure 1.
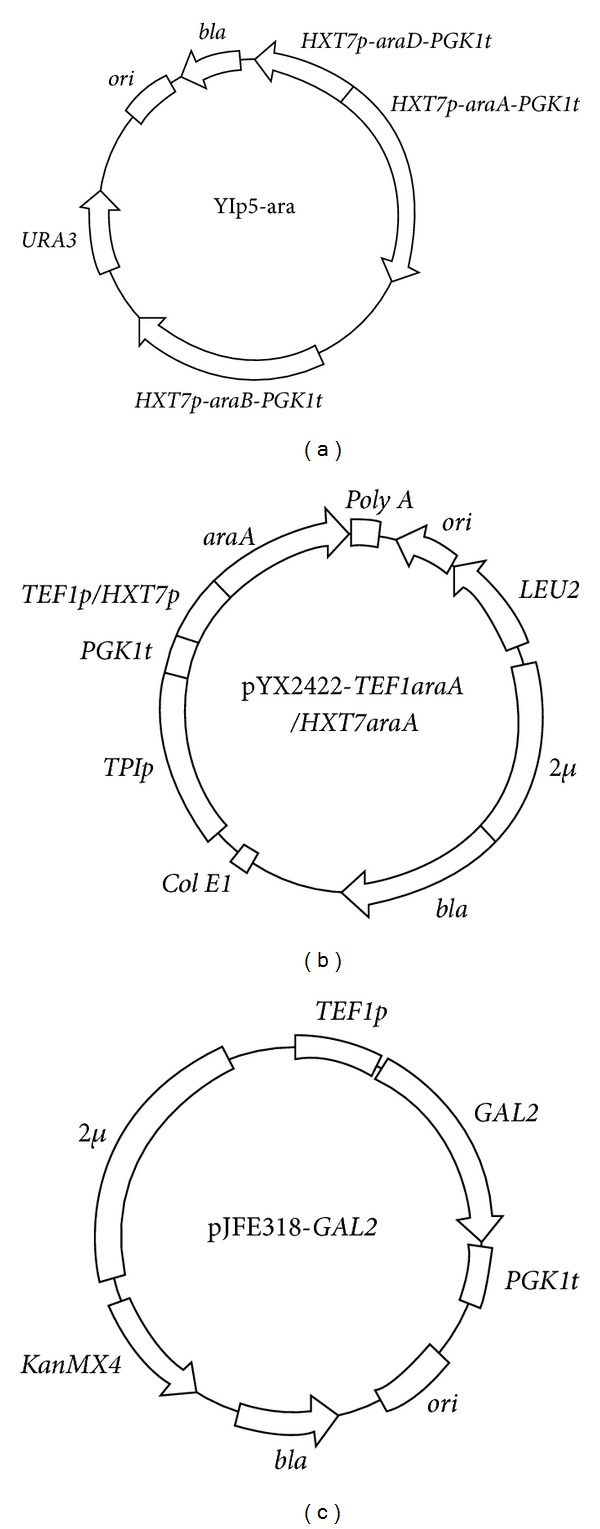
The physical maps of the plasmids (a) YIp5-ara, (b) pYX2422-TEF1araA/HXT7araA, and (c) pJFE318-GAL2.
The yeast transformation was performed using the lithium acetate transformation method [26]. The plasmid YIp5-ara was linearized at the Stu I site and then transformed into CEN.PK102-3A. The transformants with the araA, araB, and araD genes integrated into the chromosomal URA3 gene were selected in SC medium containing CSM-URA, and after being confirmed by sequencing, the desired transformant was named BSW1A1. Plasmids pYX242, pYX2422-HXT7araA, and pYX2422-TEF1araA were transformed into BSW1A1, resulting in BSW1AY, BSW1A7, and BSW1AT, respectively. The linearized pJPPP3, which contains the expression frames of genes TAL1, TKL1, RPE1, and RKI1 [19], was integrated into the chromosome of BSW1AT at the GRE3 gene locus, resulting in strain BSW2AP. The strain BSW2AP was adapted on 20 g L−1 L-arabinose under aerobic conditions and then under oxygen-limited conditions. Once the stationary phase was reached, a new batch was initiated by transferring the culture into fresh medium with an initial biomass of 0.15 g DCW L−1. When the doubling time of the strain stabilized, mutant BSW3AP was selected from the adapted mutants based on its excellent growth on L-arabinose. The plasmid pJFE318-GAL2 was then transformed into strain BSW3AP, resulting in strain BSW3AG.
2.4. Real-Time Quantitative PCR
The cells were cultured in SC medium containing 20 g L−1 glucose and collected when the OD600 of cultures reached 1. The total RNA was extracted using TRIzol reagent (Sangon, China). The first strand of cDNA was reverse transcribed from 1 μg of total RNA using PrimeScript RT reagent kits with gDNA Eraser (Takara, Japan). Diluted cDNA products were used for real-time quantitative PCR using the SYBR Green Real-time PCR Master Mix (TOYOBO, Japan) and the LightCycle PCR System (Roche Molecular Biochemicals, Germany). The actin-encoding gene ACT1 was used as the reference gene for normalization. The data of real-time PCR was calculated according to the 2−ΔΔCT method [19, 27]. The primers for these PCR were listed in Table 2.
2.5. Fermentation
A single colony was cultured overnight in SC medium containing 20 g L−1 glucose. A sample of the overnight culture was diluted to an initial OD600 of 0.5 in SC medium containing 10 g L−1 glucose and 10 g L−1 L-arabinose. After 10 h cultivation, the cells were collected and used for fermentation. All the shaker flask fermentations were performed at 30°C, 200 r min−1, in 200 mL shaker flasks containing 40 mL medium. The oxygen-limited condition was maintained by using a rubber stopper. The batch fermentations under anaerobic conditions were performed in 1.4 L fermentors (Infors AG, Switzerland) with a working volume of 900 mL. Anaerobic conditions were maintained by sparging with nitrogen (0.1 L min−1); the agitation rate was 500 r min−1. The pH was maintained at 5.0 by automatically pumping 1 mol L−1 NaOH and 1 mol L−1 H3PO4 [19]. The initial biomass was 0.2 g DCW L−1. The carbon source in the SC plus CSM-LEU-URA medium was 20 g L−1 L-arabinose; 200 μg mL−1 G418 was supplied in the fermentation of strain BSW3AG. The dry cell weight of evolved strains and the unevolved strains were calculated according to the formula of dry weight (mg mL−1) = 0.266 × OD600 − 0.0762 and dry weight (mg mL−1) = 0.2365 × OD600 + 0.1149, respectively.
2.6. Analysis of Fermentation Products
The high performance liquid chromatography (HPLC) Prominence LC-20A (Shimadzu, Japan) equipped with the refractive index detector RID-10A (Shimadzu, Japan) was used to determine the concentrations of sugars and metabolites. The Aminex HPX-87P ion exchange column (Bio-Rad, USA) was used to analyze L-arabinose, arabitol, and ethanol at 80°C with a mobile phase of water at a flow rate of 0.6 mL min−1. The Aminex HPX-87H ion exchange column (Bio-Rad, Hercules, USA) was used to analyze glycerol and acetate at 45°C using 5 mmol L−1 H2SO4 as the mobile phase [12].
3. Results
3.1. Expression of the Codon-Optimized Genes Involved in the L-Arabinose Pathway in S. cerevisiae
Based on the amino acid sequence of L-arabinose isomerase (GenBank accession no. CCC80517.1), L-ribulokinase (GenBank accession no. CCC80519.1), and L-ribulose-5-phosphate 4-epimerase (GenBank accession no. CCC80518.1) recorded in the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/), the araA, araB, and araD genes of L. plantarum were artificially synthesized using S. cerevisiae preferred codons. The CAIs of codon-optimized araA, araB, and araD were 0.599, 0.580, and 0.646, respectively, which were higher than those of the native sequences (0.324, 0.223, and 0.243, resp.).
The expression cassettes of codon-optimized araA, araB, and araD were integrated into the chromosome of strain CEN.PK102-3A, resulting in strain BSW1A1. However, BSW1A1 could not grow on L-arabinose, although the transcribed mRNAs of these genes were all detectable. Then, more copies of araA were introduced into BSW1A1, carried by the episomal plasmid pYX242 and expressed under control of the HXT7 and TEF1 promoters. The transcriptional levels of araA in the resulting strains, BSW1A7 and BSW1AT, were 12.9 ± 2.9-fold and 32.5 ± 0.7-fold higher than in the reference strain BSW1AY carrying only the integrated, expressed araA. The BSW1A7 and BSW1AT strains were aerobically incubated on L-arabinose, and the growth of strain BSW1AT was observed after ~150 h, whereas BSW1A7 could not grow even when cultured longer.
3.2. Improvement of the L-Arabinose Utilization in S. cerevisiae by Engineering and Evolution
The TAL1, TKL1, RPE1, and RKI1 genes involved in the nonoxidative pentose phosphate pathway were overexpressed in a single colony isolated from the BSW1AT 150 h culture by integrating the linearized plasmid pJPPP3 [19] into the chromosome. The resulting strain, BSW2AP, was evolved on L-arabinose. After 9 transfers in aerobic conditions and 12 transfers in oxygen-limited conditions, the doubling time of the culture decreased from 22 h to 4.5 h. The mutants were screened on L-arabinose plates, and a large colony was selected and named BSW3AP.
The transcriptional levels of genes in the recombinant strains BSW1AT, BSW2AP, and BSW3AP were determined by real-time quantitative PCR (Figure 2). The araA expression level in BSW2AP was 2-fold higher than in BSW1AT, whereas the expression levels of araB and araD in BSW2AP were lower. These changes might be due to mutations that occurred during the cultivation of BSW1AT on L-arabinose. In the evolved strain BSW3AP, all three genes were expressed at high levels. The araA, araB, and araD expression levels in BSW3AP were 4.1-fold, 1.6-fold, and 2.5-fold higher than those in strain BSW1AT, respectively.
Figure 2.
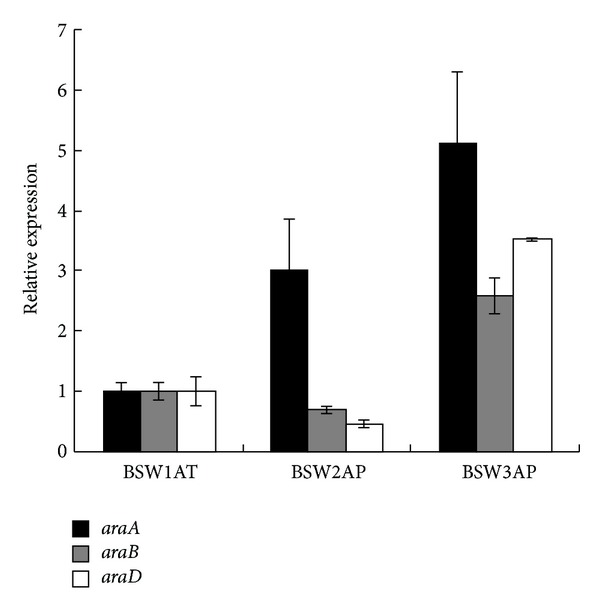
The expression of araA (black bars), araB (gray bars), and araD (blank bars) of strains BSW2AP and BSW3AP compared to strain BSW1AT. The fold-changes of mRNA levels of these genes are normalized to the expression of ACT1. The tested strains were cultivated on 20 g L−1 glucose. The values given are obtained from three independent measurements.
The L-arabinose utilization of strains BSW1AT, BSW2AP, and BSW3AP was compared in shaker-flasks under oxygen-limited conditions (Figure 3); the initial OD600 was 0.5. No growth of BSW1AT was observed within 120 h. The strain BSW2AP grew on L-arabinose with a maximum specific growth rate (μ max) of 0.011 h−1; 4.4 g L−1 L-arabinose was consumed, and 1.2 g L−1 ethanol was produced in 120 h of fermentation. In contrast, the μ max of the evolved strain BSW3AP increased to 0.23 h−1. After 120 h of fermentation, 18.6 g L−1 L-arabinose had been consumed with a maximum specific consumption rate of 0.7 g h−1 g−1 DCW; 6.9 g L−1 ethanol had been produced, and the ethanol yield was 0.43 g g−1; only 0.13 g L−1 L-arabitol had accumulated.
Figure 3.
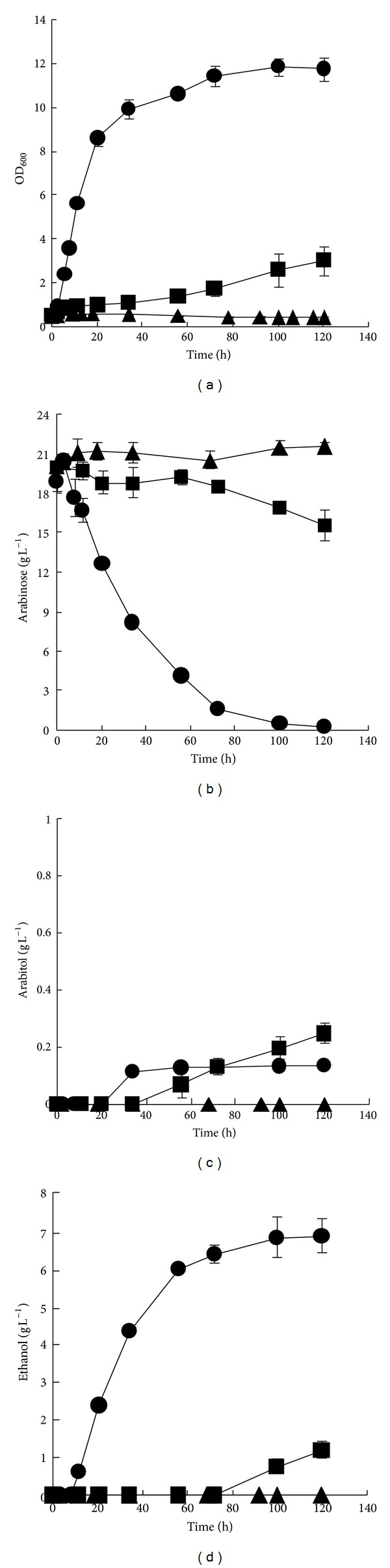
The L-arabinose fermentation of strains in shaker flasks. Growth capacity (a), L-arabinose consumption (b), arabitol formation (c), and ethanol formation (d) by BSW1AT (▲), BSW2AP (■), and BSW3AP (⚫). The strains were cultured in 40 mL SC medium with 20 g L−1 L-arabinose at 30°C, 200 r min−1 with an initial OD600 of 0.5. The data are the averages of three independent experiments.
3.3. Overexpression of GAL2 Improved the L-Arabinose Anaerobic Fermentation of the Evolved Strain
The galactose permease gene GAL2 was overexpressed in BSW3AP, resulting in strain BSW3AG. The anaerobic L-arabinose fermentation properties of strain BSW3AP and BSW3AG were studied (Figure 4 and Table 3) in bioreactors. Strain BSW3AP grew on L-arabinose with a maximum specific growth rate of 0.067 h−1. The maximum specific consumption rate of L-arabinose was 0.49 g h−1 g−1 DCW. Ethanol was produced at a maximum specific rate of 0.20 g h−1 g−1 DCW with a yield of 0.42 g g−1. The overexpression of GAL2 significantly improved the L-arabinose fermentation capacity. The maximum specific growth rate of BSW3AG was 0.075 h−1, which was 12% faster than that of BSW3AP. The L-arabinose specific consumption rate of BSW3AG was 0.61 g h−1 g−1 DCW, which was 24% faster than that of BSW3AP. The ethanol production rate was 0.27 g h−1 g−1 DCW, and the ethanol yield was 0.43 g g−1. Furthermore, both BSW3AP and BSW3AG produced small amounts of glycerol (1.4 g L−1 for both strains) and almost undetectable amounts of arabitol and acetate.
Figure 4.
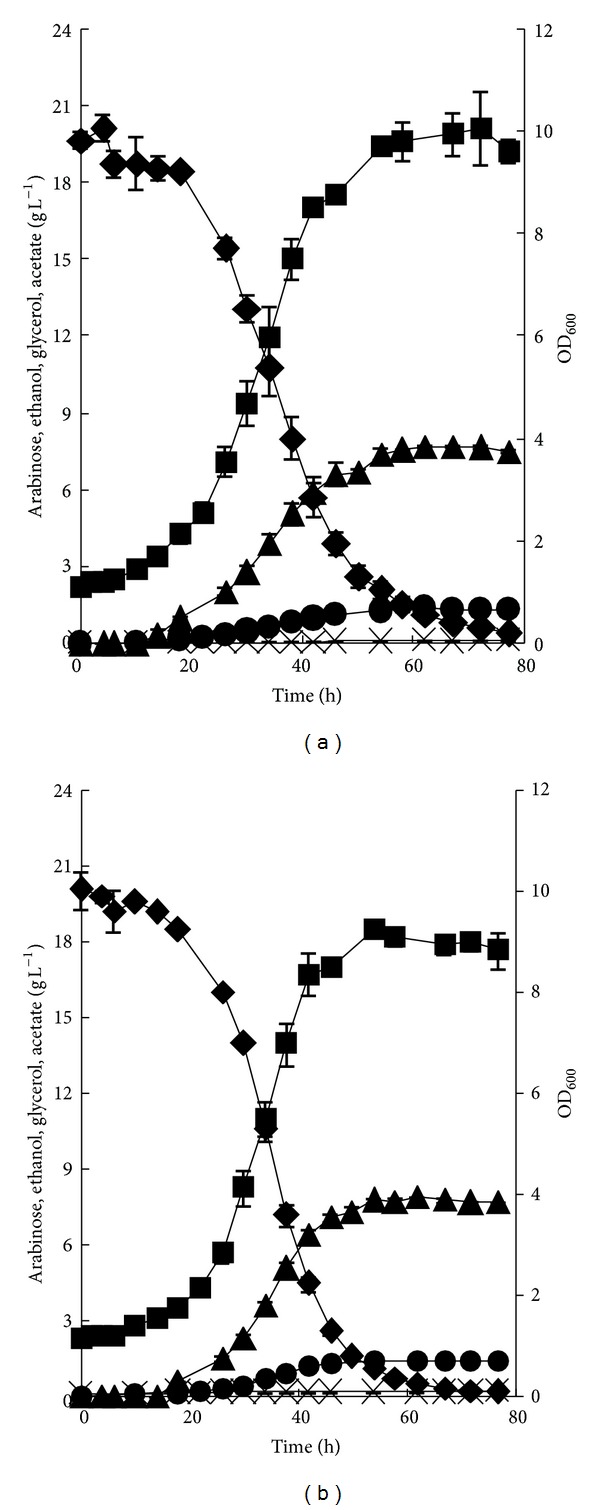
The anaerobic batch fermentation of BSW3AP (a) and BSW3AG (b) on 20 g L−1 arabinose. Levels of OD600 (■), arabinose (◆), ethanol (▲), Glycerol (⚫), and acetate (×). The fermentation was performed in 1.4 L fermentors with a working volume of 900 mL. Anaerobic conditions were maintained by sparging nitrogen (0.1 L min−1); the agitation rate was 500 r min−1. The pH was maintained at 5.0 by automatically pumping in 1 mol L−1 NaOH and 1 mol L−1 H3PO4. The initial biomass was 0. 2 g DCW L−1. The 20 g L−1 L-arabinose was used as the carbon source in SC plus CSM-LEU-URA medium, and 200 μg mL−1 G418 was supplied in the fermentation of strain BSW3AG. The data are the average of duplicate determinations.
Table 3.
The maximum specific growth rates (μ max), the maximum specific L-arabinose-consumption rate, the ethanol production rate, and the ethanol yield for BSW3AP and BSW3AG on 20 g L−1 L-arabinose.
| Strain | μ max (h−1) | The maximum specific L-arabinose consumption rate (g h−1 g−1 DCW) |
Ethanol production rate (g h−1 g−1 DCW) |
Ethanol yield (g g−1 L-arabinose consumed) |
|---|---|---|---|---|
| BSW3AP | 0.067 | 0.49 | 0.20 | 0.42 |
| BSW3AG | 0.075 | 0.61 | 0.27 | 0.43 |
4. Discussion
The complete conversion of sugars is important for efficient and cost-effective fuel ethanol production from lignocellulosic materials. Even small improvements in substrate utilization can significantly decrease the costs of the whole process [28]. L-arabinose is an important component of lignocellulosic materials. Expression of the L. plantarum L-arabinose pathway has proven to be effective in constructing L-arabinose utilizing S. cerevisiae [8]. Given that the codon-optimized genes might lead to increased expression of the proteins [15, 29], in the present work, the original araA, araB, and araD genes of L. plantarum were modified to match the codon usage of S. cerevisiae and then integrated into the chromosome of strain CEN.PK102-3A. However, this recombinant strain could not grow on L-arabinose. More copies of the araA gene were then introduced into the recombinant strain under the control of the HXT7 and TEF1 promoters. When the two resulting strains were cultured on L-arabinose, growth was only observed in cultures of the strain expressing araA under the control of the TEF1 promoter, in which the araA transcriptional level was 1.4-fold higher than in the strain expressing araA controlled by the HXT7 promoter. We suggest that only when the transcription level of araA is higher than a certain level can growth on L-arabinose occur. In contrast, only one copy of araB and araD was introduced into this recombinant strain, and the transcriptional levels of these genes were lower than in the parental strain. These phenomena indicated that araB and araD were less important for growth on L-arabinose because only one copy of these genes allowed the recombinant strain to grow on L-arabinose.
Adaptive evolution was proven to be a powerful method to enhance the strains' metabolic efficiency [8, 13]. In the present study, the evolved strain BSW3AP shows significantly improved L-arabinose metabolizing capacity. The increased transcription levels of all the three genes (araA, araB, and araD) might contribute to the enhancement. Compared to araA and araD, the expression level of araB was lower. Becker and Boles [13] reported that a mutant on L-arabinose decreased the L-ribulokinase activity expressed by araB. The relatively lower expression of araB avoids the overconsumption of ATP, which would benefit the growth of the strain on L-arabinose.
L-arabinose is a novel carbon source for S. cerevisiae. The uptake of L-arabinose in S. cerevisiae mainly depends on the nonspecific transport by the hexose transporter Gal2p. The Hxt9p and Hxt10p also can transport L-arabinose, but the efficiency is very low [30]. It was reported that overexpressing GAL2 improves the L-arabinose utilization [13, 16]. In this study, overexpressing GAL2 notably increased the growth rate and L-arabinose consumption rate of our evolved strain BSW3AP. This result suggested that the theoretical L-arabinose metabolic flux was higher than we detected in BSW3AP. The L-arabinose utilization of BSW3AP was limited by its absorption rate. When the GAL2 was overexpressed, more Gal2p in the plasma membrane lead to an increased L-arabinose uptake and then promote the L-arabinose utilization. Our result further confirmed the importance of transporters for L-arabinose utilization; however, the affinity of Gal2p for L-arabinose is low, and glucose competitively inhibited its binding to L-arabinose [30]. Improving the efficiency of the L-arabinose specific transporter remains to be conducted.
5. Conclusions
With multiple steps of genetic engineering and adaptive evolution, we obtained the strain BSW3AG, which grows on L-arabinose with a μ max of 0.075 h−1. The maximum specific L-arabinose consumption rate is 0.27 g h−1 g−1 DCW, and the maximum ethanol yield is 0.43 g g−1 L-arabinose consumed, which is 84.3% of the theoretical amount. A high level of araA expression is notably important in establishing an efficient L-arabinose pathway in S. cerevisiae, and more efficient transporters are necessary to improve the L-arabinose absorption capacity of the evolved strains.
Conflict of Interests
The authors declare that they have no conflict of interests.
Acknowledgments
This work was supported by the National Key Basic Research Program (2011CB707405), the National High-Technology Research and Development Program of China under Grant (2012AA022106), the National Natural Science Foundation of China (30970091, 31070096, and 31270151), and the International S&T Cooperation Program of China (2010DFA32560). The authors thank Dr. Peter Kötter from Johann Wolfgang Goethe-University Frankfurt for the strain CEN.PK102-3A.
References
- 1.den Haan R, Kroukamp H, Mert M, Bloom M, Görgens JF, van Zyl WH. Engineering Saccharomyces cerevisiae for next generation ethanol production. Journal of Chemical Technology & Biotechnology. 2013;88(6):983–991. [Google Scholar]
- 2.Mabee WE. Policy options to support biofuel production. Advances in Biochemical Engineering/Biotechnology. 2007;108:329–357. doi: 10.1007/10_2007_059. [DOI] [PubMed] [Google Scholar]
- 3.Farrell AE, Plevin RJ, Turner BT, Jones AD, O’Hare M, Kammen DM. Ethanol can contribute to energy and environmental goals. Science. 2006;311(5760):506–508. doi: 10.1126/science.1121416. [DOI] [PubMed] [Google Scholar]
- 4.Hahn-Hägerdal B, Galbe M, Gorwa-Grauslund MF, Lidén G, Zacchi G. Bio-ethanol—the fuel of tomorrow from the residues of today. Trends in Biotechnology. 2006;24(12):549–556. doi: 10.1016/j.tibtech.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Galbe M, Zacchi G. A review of the production of ethanol from softwood. Applied Microbiology and Biotechnology. 2002;59(6):618–628. doi: 10.1007/s00253-002-1058-9. [DOI] [PubMed] [Google Scholar]
- 6.Kim SR, Ha S, Wei N, Oh EJ, Jin Y. Simultaneous co-fermentation of mixed sugars: a promising strategy for producing cellulosic ethanol. Trends in Biotechnology. 2012;30(5):274–282. doi: 10.1016/j.tibtech.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Seiboth B, Metz B. Fungal arabinan and L-arabinose metabolism. Applied Microbiology and Biotechnology. 2011;89(6):1665–1673. doi: 10.1007/s00253-010-3071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wisselink HW, Toirkens MJ, Berriel MDRF, et al. Engineering of Saccharomyces cerevisiae for efficient anaerobic alcoholic fermentation of L-arabinose. Applied and Environmental Microbiology. 2007;73(15):4881–4891. doi: 10.1128/AEM.00177-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hahn-Hägerdal B, Karhumaa K, Jeppsson M, Gorwa-Grauslund MF. Metabolic engineering for pentose utilization in Saccharomyces cerevisiae . Advances in Biochemical Engineering/Biotechnology. 2007;108:147–177. doi: 10.1007/10_2007_062. [DOI] [PubMed] [Google Scholar]
- 10.Schleif R. Regulation of the L-arabinose operon of Escherichia coli . Trends in Genetics. 2000;16(12):559–565. doi: 10.1016/s0168-9525(00)02153-3. [DOI] [PubMed] [Google Scholar]
- 11.Fonseca C, Romão R, de Sousa HR, Hahn-Hägerdal B, Spencer-Martins I. L-arabinose transport and catabolism in yeast. FEBS Journal. 2007;274(14):3589–3600. doi: 10.1111/j.1742-4658.2007.05892.x. [DOI] [PubMed] [Google Scholar]
- 12.Bettiga M, Bengtsson O, Hahn-Hägerdal B, Gorwa-Grauslund MF. Arabinose and xylose fermentation by recombinant Saccharomyces cerevisiae expressing a fungal pentose utilization pathway. Microbial Cell Factories. 2009;8, article 40 doi: 10.1186/1475-2859-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Becker J, Boles E. A modified Saccharomyces cerevisiae strain that consumes L-arabinose and produces ethanol. Applied and Environmental Microbiology. 2003;69(7):4144–4150. doi: 10.1128/AEM.69.7.4144-4150.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sedlak M, Ho NWY. Expression of E. coliaraBAD operon encoding enzymes for metabolizing L-arabinose in Saccharomyces cerevisiae . Enzyme and Microbial Technology. 2001;28(1):16–24. doi: 10.1016/s0141-0229(00)00282-9. [DOI] [PubMed] [Google Scholar]
- 15.Wiedemann B, Boles E. Codon-optimized bacterial genes improve L-arabinose fermentation in recombinant Saccharomyces cerevisiae . Applied and Environmental Microbiology. 2008;74(7):2043–2050. doi: 10.1128/AEM.02395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wisselink HW, Cipollina C, Oud B, et al. Metabolome, transcriptome and metabolic flux analysis of arabinose fermentation by engineered Saccharomyces cerevisiae . Metabolic Engineering. 2010;12(6):537–551. doi: 10.1016/j.ymben.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Entian KD, Kötter P. 23 yeast mutant and plasmid collections. In: Alistair JPB, Mick T, editors. Methods in Microbiology. New York, NY, USA: Academic Press; 1998. pp. 431–449. [Google Scholar]
- 18.Güldener U, Heck S, Fiedler T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Research. 1996;24(13):2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng B, Shen Y, Li X, Chen X, Hou J, Bao X. Improvement of xylose fermentation in respiratory-deficient xylose-fermenting Saccharomyces cerevisiae . Metabolic Engineering. 2012;14(1):9–18. doi: 10.1016/j.ymben.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 20.Walfridsson M, Anderlund M, Bao X, Hahn-Hägerdal B. Expression of different levels of enzymes from the Pichia stipitis XYL1 and XYL2 genes in Saccharomyces cerevisiae and its effects on product formation during xylose utilisation. Applied Microbiology and Biotechnology. 1997;48(2):218–224. doi: 10.1007/s002530051041. [DOI] [PubMed] [Google Scholar]
- 21.Struhl K, Stinchcomb DT, Scherer S, Davis RW. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proceedings of the National Academy of Sciences of the United States of America. 1979;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, Zhang X, Wang C, Liu L, Lei M, Bao X. Genetic and comparative transcriptome analysis of bromodomain factor 1 in the salt stress response of Saccharomyces cerevisiae . Current Microbiology. 2007;54(4):325–330. doi: 10.1007/s00284-006-0525-4. [DOI] [PubMed] [Google Scholar]
- 23.Shen Y, Chen X, Peng B, Chen L, Hou J, Bao X. An efficient xylose-fermenting recombinant Saccharomyces cerevisiae strain obtained through adaptive evolution and its global transcription profile. Applied Microbiology and Biotechnology. 2012;96(4):1079–1091. doi: 10.1007/s00253-012-4418-0. [DOI] [PubMed] [Google Scholar]
- 24.Sharp PM, Li W. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Research. 1987;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji L, Shen Y, Xu L, Peng B, Xiao Y, Bao X. Enhanced resistance of Saccharomyces cerevisiae to vanillin by expression of lacA from Trametes sp. AH28-2. Bioresource Technology. 2011;102(17):8105–8109. doi: 10.1016/j.biortech.2011.06.057. [DOI] [PubMed] [Google Scholar]
- 26.Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11(4):355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.von Sivers M, Zacchi G. Ethanol from lignocellulosics: a review of the economy. Bioresource Technology. 1996;56(2-3):131–140. [Google Scholar]
- 29.Wu G, Zheng Y, Qureshi I, et al. SGDB: a database of synthetic genes re-designed for optimizing protein over-expression. Nucleic Acids Research. 2007;35(supplement 1):D76–D79. doi: 10.1093/nar/gkl648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subtil T, Boles E. Improving L-arabinose utilization of pentose fermenting Saccharomyces cerevisiae cells by heterologous expression of L-arabinose transporting sugar transporters. Biotechnology for Biofuels. 2011;4:p. 38. doi: 10.1186/1754-6834-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]


