Abstract
Superfamily I is a large and diverse group of monomeric and dimeric helicases defined by a set of conserved sequence motifs. Members of this class are involved in essential processes in both DNA and RNA metabolism in all organisms. In addition to conserved amino acid sequences, they also share a common structure containing two RecA-like motifs involved in ATP binding and hydrolysis and nucleic acid binding and unwinding. Unwinding is facilitated by a “pin” structure which serves to split the incoming duplex. This activity has been measured using both ensemble and single-molecule conditions. SF1 helicase activity is modulated through interactions with other proteins.
Introduction
Helicases are molecular motor proteins present in viruses, bacteria, and eukaryotes [1, 2]. They harness the chemical energy of ATP hydrolysis to break the energetically stable hydrogen bonding between the duplex DNA. By doing so, helicases allow access to the genetic information locked in the duplex DNA. Helicases participate in various aspects of nucleic acid metabolism such as DNA replication, recombination, repair, transcription, translation, and splicing of RNA transcripts [3–13].
Mutations in helicase genes involved in DNA repair processes have been linked to numerous human diseases [14–17] in which genomic instability, immunodeficiency, mental retardation, premature aging, and predisposition to cancer are common features [14, 15, 18–21]. Some of the diseases caused by defective helicases are xeroderma pigmentosum, Cockayne Syndrome, trichothiodystrophy, Werner’s syndrome, Bloom’s syndrome, and alpha-thalassemia mental retardation on the X chromosome [22–30]. A mutation in superfamily 1 (SF1) helicase Pif1 results in breast cancer predisposition [31]. Mutations in SetX helicase, involved in RNA splicing and termination, cause juvenile amyotrophic lateral sclerosis [32] and ataxia-ocular apraxia 2 [33], while mutations in IGHMBP2 (Smubp2), involved in translation [34], result in distal spinal muscular atrophy [35]. The diverse disease abnormalities caused by defective helicases suggest that multiple aspects of DNA and RNA metabolism are affected [18].
In some aggressive cancers, the activity of helicases in DNA repair reduces the efficacy of anticancer agents, because many of these agents are targeted to DNA. Studies have shown that the efficacy of chemotherapeutic agents could be increased by administering drugs that target helicases along with the anticancer drugs [36]. Helicases encoded by herpes simplex virus [37–39], West Nile virus, dengue virus, and hepatitis C virus [40] are targets for antiviral drug development [38, 41]. Some bacterial helicases such as Rep from Legionella pneumophilia are required for infection of mammalian cells [42]. The importance of helicases in the fundamental aspects of nucleic acid metabolism and the association of human, bacterial, and viral helicases in human diseases makes the study of helicases critical. This also makes it essential to understand the mechanisms by which helicases perform different biochemical functions so that the relationship between mutations and specific disease states can be understood at the molecular level.
Functions of Helicases
Unwinding of duplex or structured nucleic acids by helicases provides the ssNA intermediates required for metabolism of DNA and RNA [43–45]. In vivo, DNA unwinding is coupled to the action of many proteins such as primases, ssDNA-binding proteins, polymerases, and other factors depending on the functions of a particular helicase. Many of the biological functions of various helicases are listed in Table 2.1. Helicases are implicated in processes ranging from replication to translation [8, 10–12, 46–49] and also in ATP-dependent chromatin remodeling [50, 51]. Replicative helicases deal with the process of nucleic acid replication (initiation, elongation, and termination). Helicases play an important role in DNA repair, as these are frequently the first proteins that encounter DNA damage [13, 19, 52–54]. During DNA repair, the damaged area on the DNA has to be unwound before repair can proceed as most DNA repair processes require ssDNA. Helicases play roles in both initiation and branch migration during recombination [21, 25, 28, 55–60].
Table 2.1.
Biological functions carried out by various helicases
| Biological function | Helicase |
|---|---|
| Replicative helicases | PcrA1, RepA, UvrD, Dda, HSV UL5, HSV UL9, DnaB, PriA, T7gp4A and 4B, T4gp41, SV40, TAG, Polyoma TAG, BPV E1, MCM 4/6/7, Dna2, FFA-1, RecD, TraI, NS3, RecQL4 |
| Repair helicases | UvrD, UvrAB, PcrA, Rad3, helicase E, XPD, XPB, Dna2, RecD2, BACH1, HDH II, RecQ, WRN, Rtel1, BLM, RuvB, Mph 1, CHD4 |
| Recombination helicases | RecBCD, RecG, RecQ, RuvAB, PriA, UvrD, T4 UvsW, HDH II, HDH IV, WRN, Tra I, Rho, PDH65, BLM, Srs2, Sgs1, Rtel1 |
| Other functions of helicases | |
| Transcription | SWI2, SNF2, TFIIH, Rho, Factor 2, TRCF, RecQL5, ERCC6/RAD26 |
| Translation | HSV UL5, eIF4A, RHA, Ded1p, vasa |
| Chromatin remodeling | Rad54, ATRX, BLM, CHD4 |
| Maintenance of telomeres | Pif1, Dna2, Rtel 1, WRN, BLM, FANC |
| Okazaki fragment maturation | Dna2, Pif1, WRN |
Helicases alter DNA and RNA structures, remodel chromatin [24, 51, 61–65], and modulate access to the DNA template by transcriptional machinery. RNA polymerases that are involved in the elongation of RNA transcripts have been considered as helicases that unwind the dsDNA to expose the ssDNA strand that serves as the template for RNA synthesis [2]. Helicases thus play a role in most transcriptional processes including activation (TFIIH), initiation (TFIIH, SNF2), maintenance (SW1), DNA repair (TFIIH, ERCC6/RAD26), and termination (Factor 2, Rho) [49, 66–72].
Properties of Helicases
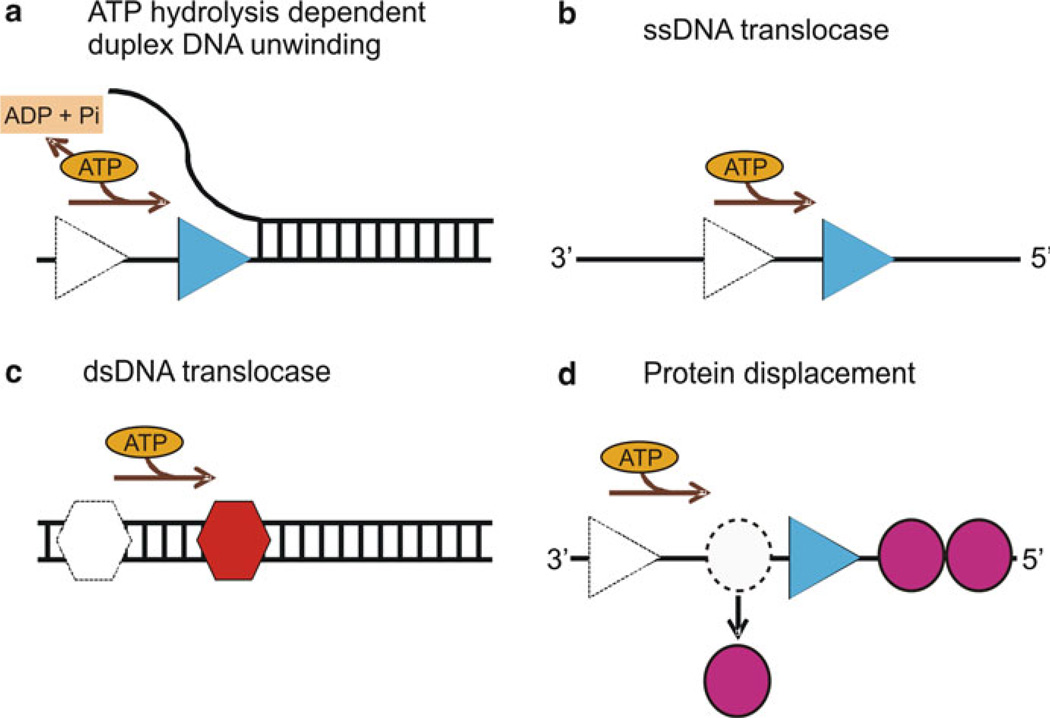
Some of the fundamental properties exhibited by helicases are nucleic acid binding, ATP binding and hydrolysis, translocation, unwinding of duplex nucleic acids, and displacement of proteins bound to the nucleic acid substrate (Fig. 2.1), although not all helicases are able to perform all of these activities. Helicases unwind DNA with a unique directionality (either from 5′ to 3′ or from 3′ to 5′) relative to the strand of DNA that is bound by the enzyme. RecBCD exhibits bipolar enzyme activity, where RecB and RecD components of the complex unwind DNA in 3′–5′ and 5′–3′ directions, respectively [73]. Other bipolar helicases are Bacillus anthracis PcrA [74] and Sulfolobus acidocaldarius HerA helicase [75].
Fig. 2.1.
Biochemical properties of helicases: (a) ATP hydrolysis-dependent unwinding of duplex DNA by helicase (blue triangle). The movement of helicase along the DNA utilizing the energy from ATP hydrolysis separates the duplex nucleic acid into single strands. (b) An ssDNA translocase (blue triangle) moves with biased directionality along the DNA powered by ATP binding and hydrolysis. The directionality of translocation can be 3′–5′ or 5′–3′. (c) A dsDNA translocase (red hexagon) moves along dsDNA. (d) Helicases can displace proteins (magenta circles) bound to the DNA strand as a result of their biased directional movement (adapted from ref. [45])
Most helicases require a short stretch of ssDNA as a loading strand in vitro, and they show a preference for binding to ssDNA over dsDNA. Many helicases (Rep, UvrD, PcrA, Dda, and HSV-UL5) are generally considered to be ssDNA translocases, while other helicases (eIF4A, RecG, and PriA) are dsDNA translocases [76, 77]. Some helicases need a replication fork-like structure on the substrate for optimum unwinding, whereas other DNA helicases can initiate unwinding from blunt-ended duplex DNA, such as RecBCD, UvrD, Rep, and RecQ [77–79]. While the ATPase activity of helicases is low in the absence of DNA, presence of ssDNA stimulates this activity [80].
Superfamily 1 Helicases
Helicases are divided into six superfamilies (SF1–6) based on the sequence identity among the conserved helicase motifs [1, 43, 47, 81]. Superfamily 1 is one of the largest classes of helicases with members that participate in virtually all steps in DNA or RNA metabolism [82–85]. SF1 includes three families (Rep/UrvD, Pif1/RecD, and Upf1 like) [86] and can be divided into groups based on the direction of translocation on ssDNA: 3′–5′ for SF1A helicases and 5′–3′ for SF1B helicases [43]. Some of the well-characterized SF1A helicases are PcrA, Rep, and UvrD T[87–92]. Well-characterized members of SF1B include RecD and Dda [93, 94] (He et al., in press). The biological functions, polarity, and active forms of some of the SF1 helicases are listed (Table 2.2).
Table 2.2.
Superfamily 1A (3′–5′) and SF1B (5′–3′) helicases
| Helicase | Source | Biological function | Active form | Polarity |
|---|---|---|---|---|
| PcrA | Bacillus stearothermophilus | DNA repair and rolling replication of plasmids | Monomer | 3′–5′ |
| Rep | Escherichia coli | DNA replication | Dimer | 3′–5′ |
| RecB | Escherichia coli | DNA recombination | Monomer | 3′–5′ |
| UvrD | Escherichia coli | DNA replication, recombination, and repair of UV damage and mismatched base pairs | Dimer | 3′–5′ |
| Pif1 | Saccharomyces cerevisiae | Maintenance of telomeres, ribosomal DNA replication, processing of Okazaki fragments | Monomer | 5′–3′ |
| Dna2 | Saccharomyces cerevisiae | Maintenance of telomeres, Okazaki fragment maturation, double strand break repair, aging | Monomer | 5′–3′ |
| Dda | Bacteriophage T4 | DNA replication initiation, and recombination | Monomer | 5′–3′ |
| UL5 | Herpes simplex virus | Viral DNA replication | Dimer | 5′–3′ |
| RecD2 | Deinococcus radiodurans | DNA repair | Monomer | 5′–3′ |
| TraI | Escherichia coli | DNA transfer during conjugation, DNA nicking | Monomer | 5′–3′ |
Helicase Motifs
A characteristic feature of helicases is the presence of highly conserved amino acid sequences termed the “helicase motifs” [46, 47, 95–98]. Based on their sequence similarity and organization, these motifs are useful in the grouping of helicases into different families. SF1 and SF2 contain at least seven conserved amino acid motifs whose sequences, organization, and secondary structures are, in general, very similar [43, 46]. SF1 (Rep and PcrA) and SF2 (NS3) helicases differ primarily in motifs III and IV. In NS3, motif IV makes contacts with the DNA backbone and is not in the same relative position as motif IV of Rep and PcrA. While motif III of Rep contacts the bound ssDNA molecule, this motif in NS3 does not [99, 100]. The SF1A and SF1B helicases also show differences with motifs Ia and III being particularly characteristic for each class. These motifs are usually clustered in a core region of 200–700 amino acids, separated by stretches of low sequence but high length conservation [79]. In contrast, the N-terminal and C-terminal regions of helicases are characterized by a high degree of sequence and length variability. The divergent regions are responsible for individual protein functions, whereas the highly conserved motifs are involved in ATP binding and hydrolysis or binding and unwinding of nucleic acids.
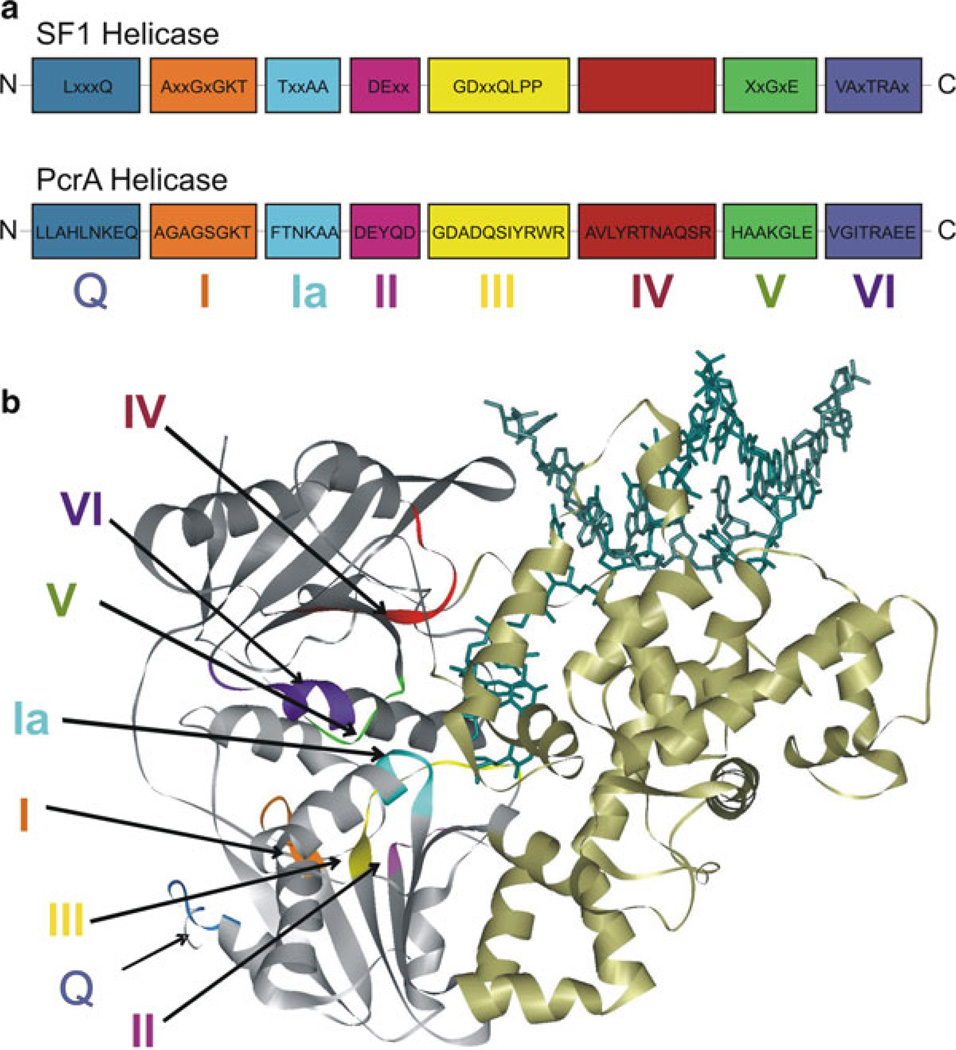
Helicase motifs involved in ATP binding are located at the interface between two RecA-like domains [43, 46, 47, 95, 96, 101] in the structures of SF1 and SF2 helicases [48, 102–104]. Figure 2.2 shows the conserved sequence of the motifs (Q, I, Ia, II, III, IV, V, and VI) of the SF1 helicase family and their location in PcrA. The biochemical functions of these motifs are described below.
Fig. 2.2.
Schematic representation of the motifs of SF1 helicases: (a) The consensus sequences for the conserved helicase motifs of SF1 helicases and PcrA are shown. The N-terminus of the protein is on the left and C-terminus is on the right side. Labels below the boxes are the names assigned to the motifs (motifs Q, I, Ia, II–VI). The relative positions of motifs and spacing between motifs are arbitrary. The consensus amino acid sequences of PcrA are taken from refs. [46, 89]. (b) The crystal structure of PcrA helicase (Protein Data Bank code 3PJR) [87, 89] bound to DNA (dark green) illustrating the different conserved motifs. The helicase motifs are in the cleft formed between the two RecA-like domains (grey). The colors of different motifs in the structure are as follows: motif Q, blue; motif I, orange; motif Ia, cyan; motif II, magenta; motif III, yellow; motif IV, red; motif V, green; motif VI, purple
Q Motif
The Q of the Q motif [101] is conserved among all SF1 helicases [86]. This motif coordinates the adenine base and is less conserved among those helicase families which do not show specificity for ATP [86]. Mutagenesis suggests that the Q motif is required for viability and plays a role in orienting ATP for hydrolysis [101, 105].
Motif I (Walker A)
The consensus sequence of this motif is AxxGxGKT [46]. It is present in many nucleotide-binding proteins and forms a phosphate-binding loop [106]. The residues “GKT” are required for the interaction of the protein with Mg2+ and ATP [107]. The conserved G in GKT helps to maintain the flexible loop conformation [46].
Motif Ia
The consensus sequence for motif Ia is TxxAA. It has been suggested that this motif is involved in ssDNA binding [89] and energy transduction from the ATP-binding site to the DNA-binding site [86]. For the SF1 helicases (UvrD, Rep, Pif1), motifs Ia and III have been proposed to play important roles in defining translocation polarity [94].
Motif II (Walker B) [108]
This motif, DExx, is involved in NTP hydrolysis [46, 76, 86]. The D and E residues coordinate the ATP-associated Mg2+ and activate the attacking water molecule, respectively [109, 110]. Mutation of these residues reduces ATPase and helicase reactivity [111–113].
Motif III
The consensus sequence for motif III is GDxxQLPP [86]. It functions in DNA binding through base stacking and hydrogen bonding with the bases [76]. The close proximity of some residues in motifs III to that in motif II suggests that motif III transduces the energy of ATP hydrolysis to the DNA [88]. Motif III mutants of UL5, UvrD, and eIF-4A exhibited uncoupling of ATPase and helicase activities [79, 110, 114]. The highly conserved Q in motif III contacts the γ-phosphate of the bound nucleotide in PcrA [89], UvrD, and UL5 [113, 115]. These results imply a role for motif III in coupling ATP hydrolysis with the unwinding of duplex DNA.
Motif IV
The consensus sequence for this motif varies among the three families within SF1. Motif IV supplies a stacking platform (conserved Y) for the adenine base [88] as well as direct contact with the γ-phosphate, suggesting that it may be involved directly in hydrolysis of the NTP [89].
Motif V
This motif interacts with the sugar-phosphate backbone of the DNA. Motif V mutants of UL5 exhibited reduced affinity for ssDNA and reduced rates of ATP hydrolysis [116].
Motif VI
The consensus sequence for this motif is VA(L/Y)TRA(K/R) [86]. It is proposed to be a part of the ATP-binding cleft and is involved in coupling the helicase and ATPase activities of the protein [88]. Several helicases exhibited nucleic acid binding defects when motif VI residues were altered [110]. Motif VI mutants of UvrD exhibited reduced ssDNA binding, ATP hydrolysis rate, and ligand-induced conformational changes [117]. In motif VI mutants of UL5, an uncoupling of ATPase and helicase activity was observed [113]. Studies on PcrA [89] and Upf1p [112, 113, 117] suggest that motif VI, by virtue of its close proximity to the NTP-and DNA-binding sites, mediates ligand-induced conformational changes, which are essential for the helicase to move along the nucleic acid substrate [88].
The conserved motifs bind and hydrolyze ATP and transduce the resulting energy to cause conformational changes in the helicase. These motifs function together to drive directional movement along ssDNA or dsDNA. They participate in the communication between nucleic acid and ATP-binding sites [118]. The ability to unwind dsDNA appears to be provided by additional protein domains which do not contain the helicase motifs [48].
Helicase Structure
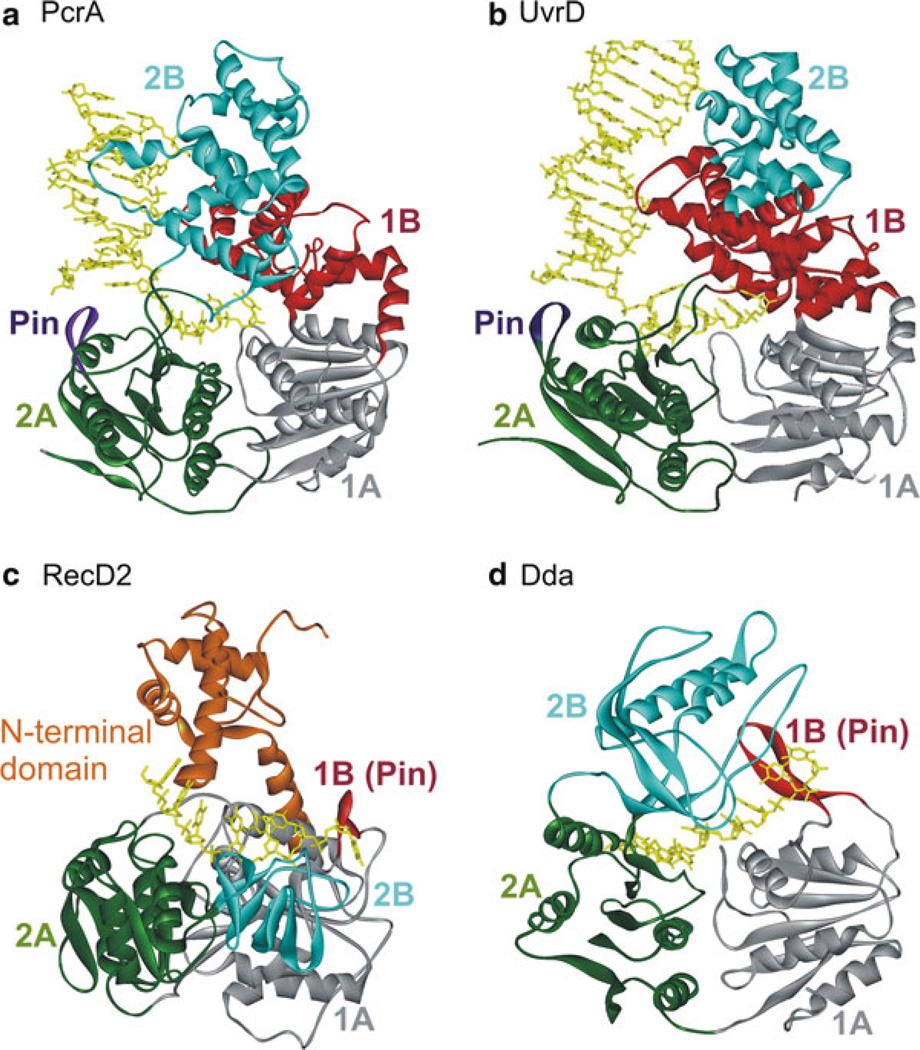
SF1 helicases constitute one of the best structurally characterized helicase families. The crystal structures have revealed that the helicase motifs are clustered together in two RecA-like domains, forming an ATP-binding pocket between them and a part of the nucleic acid-binding site. The nonconserved regions may contain specific domains such as protein–protein interaction domains, cellular localization domains, and DNA-recognition domains specific to individual helicases. Several helicase structures have been solved in the last decade contributing significantly to the overall understanding of the mechanism of SF1 helicases. Figure 2.3 shows the structures of two SF1A and two SF1B helicases [87–89, 93, 94, 103] (He et al., in press).
Fig. 2.3.
Crystal structures of SF1A (PcrA, UvrD) and SF1B (RecD2, Dda) helicases: (a) Ribbon diagram of PcrA helicase from Bacillus stearothermophilus (Protein Data Bank code 3PJR) [87, 89]. The RecA-like domains 1A and 2A are shown in grey and green colors, respectively. The structure shows the red 1B domain and the pin (purple) separating the strands of duplex DNA. The 2B domain is shown in cyan. (b) Structure of Escherichia coli UvrD helicase (PDB code 2IS1 [122]). Domains are colored as in (a). (c) Structure of RecD2 helicase from Deinococcus radiodurans (PDB code 3GPL [93]). Domains 1A, 2A, and 2B are colored as in (a). The beta-hairpin (1B) is red. The N-terminal domain is colored orange. (d) Structure of bacteriophage T4 Dda helicase bound to ssDNA (PDB id: 3UPU) (He et al., in press). The domains are colored as in (c). Nucleic acid is colored yellow in all structures
Structure of SF1A Helicases (PcrA and UvrD)
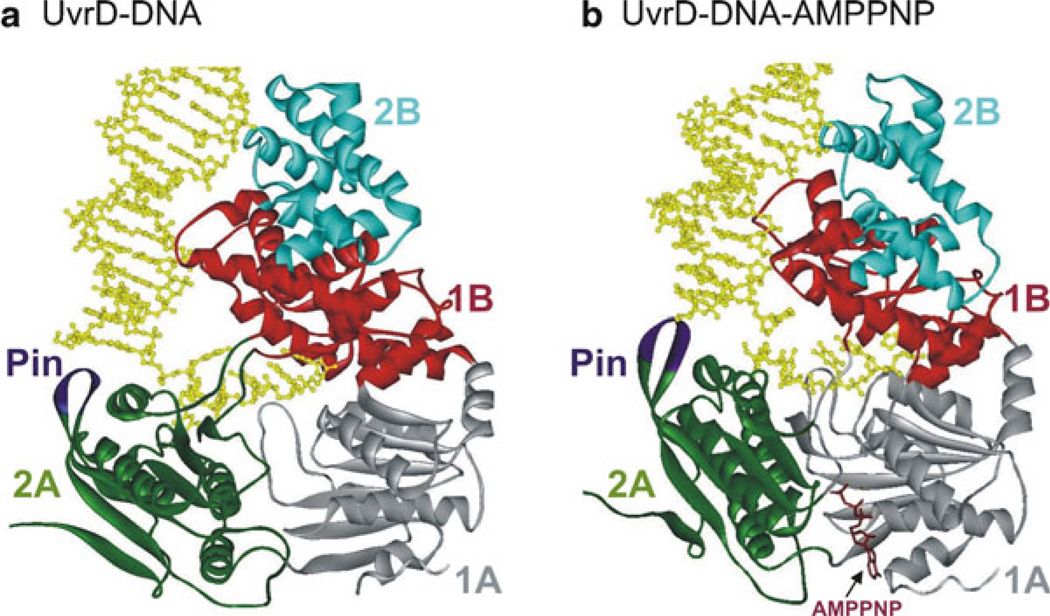
The first helicase structure to be solved was that of PcrA from B. stearothermophilus [87, 89]. PcrA is composed of four domains (1A, 2A, 1B, and 2B) (Fig. 2.3a), resembling other SF1 helicases [119–121]. The ATP-binding site is situated in a cleft between the RecA-like domains (1A and 2A). This cleft opens and closes in response to nucleotide binding and hydrolysis suggesting how translocation could occur [88, 89]. In UvrD (SF1A helicase), binding of an ATP analog, AMPPNP, in the cleft between domains 1A and 2A (Fig. 2.4) induces a 20° rotation between domain 2A and the remaining three domains (1A, 1B, and 2B). Upon AMPPNP binding, the duplex moves domains 1A/1B/2B towards 2A leading to untwisting of the duplex DNA as single-stranded DNA is pulled through the active site [122]. The structural data for UvrD, as for PcrA, predict one nt translocated and one bp unwound per ATP hydrolyzed.
Fig. 2.4.
Crystal structures of (a) UvrD–DNA and (b) UvrD–DNA–AMPPNP complexes [122]. The four domains (1A, 1B, 2A, and 2B) are colored grey, red, green, and cyan, respectively. The Pin region is shown in purple. The 3′-ssDNA tail is bound across domains 1A and 2A. Domains 1B and 2B interact with the DNA duplex. Binding of AMPPNP (brown) induces domains 2A and 1A to rotate towards each other by 20°
Structure of SF1B Helicases (RecD2 and Dda)
A crystal structure of Deinococcus radiodurans RecD2 helicase with ssDNA [93] is shown in Fig. 2.3c. RecD2 comprises five domains: the N-terminal, 1A, 1B, 2A, and 2B domains. Domains 1A and 2A have the RecA-like fold seen in all SF1 and SF2 helicases [48]. Domain 1B forms a rigid β-hairpin that protrudes from the surface of domain 1A, and the 2B domain has an SH3 fold [93]. The ssDNA-binding site runs in a 5′–3′ direction along a channel across the top of domains 2A and 1A. The DNA is also contacted by the 1B and 2B domains that form the sides of the channel.
Although the location of the ssDNA-binding site is similar in SF1A and SF1B helicases [88, 89], the contacts between the protein and the DNA and conformation of the DNA are different. PcrA interacts with the DNA through stacking of aromatic side chains with the DNA bases and there are relatively few contacts with the DNA backbone [89]. In contrast, the majority of protein–DNA contacts in RecD2 are via the phosphodiester backbone [93]. The bound ssDNA is in a configuration more similar to that found in a DNA duplex, with the bases stacked against one another, unlike the extended conformation observed in PcrA. Interestingly, the RecD2 mode of binding is more similar to that seen in SF2 enzymes such as NS3, Rad54, Vasa, and Hel308 [104, 123–125] rather than SF1A helicases [126].
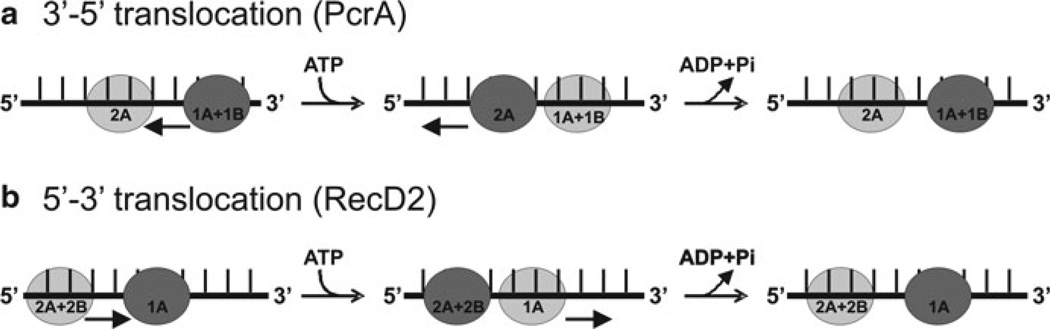
Even though there is significant structural similarity between SF1A and SF1B enzymes, they translocate in opposite directions. Comparison of the structures of PcrA and RecD2 shows that both SF1A and SF1B helicases bind the ssDNA in the same orientation (2A domain on the 5′ side of the DNA, 1A domain on the 3′ side of the DNA) [89, 94], and reveals how directionality is determined (Fig. 2.5a, b) [89, 94, 127]. Opening and closing of the cleft between the 1A and 1B domains in the presence and absence of ATP appears to provide the means of translocation [89]. The RecA-like domains bind to DNA and upon binding to ATP, the more weakly bound domain shifts towards the more tightly bound domain. For PcrA (SF1A), domain 1A moves towards domain 2A upon ATP binding, resulting in translocation in the 3′–5′ direction [94, 127]. For RecD2 (SF1B), ATP binding causes movement of domain 2A towards domain 1A, resulting in 5′–3′ translocation [94]. The net forward movement occurs in one nt physical steps with each ATP hydrolyzed. The efficiency of helicases may vary, with some ATP hydrolysis events being uncoupled from movement. Despite being in the same superfamily, SF1A and SF1B helicases exhibit significantly different mechanisms.
Fig. 2.5.
Comparison of the translocation mechanism of SF1A (PcrA) and SF1B (RecD2) helicases: In both enzyme classes, a cycle of ATP binding and hydrolysis induces conformational changes that result in translocation of the protein along DNA, but in opposite directions. Dark grey circles represent domains that have a tight grip on the ssDNA, and light grey circles represent domains that have a weaker grip and can slide along the DNA. Conversion between tight and weak grip (dark and light grey circles) is indicated by arrows. (a) The translocation mechanism of PcrA is shown in cartoon form demonstrating the change in affinity for ssDNA of domains 1A and 2A during translocation. Prior to ATP binding, ssDNA is bound to the enzyme spanning the 1A and 2A domains. Binding of ATP induces closure of the cleft between 1A and 2A domains. At this point, the grip is tightest on the 2A domain, causing the DNA to slide across the 1A/1B domains. Upon ATP hydrolysis, bind to ssDNA in 1A becomes tighter, whereas binding of ssDNA in 2A becomes weaker, releasing ssDNA. The domains also move apart, due to domain 2A sliding forward, causing the ssDNA to be pulled along the DNA-binding channel relative to the domain 2A. The result is translocation along the DNA in a 3′–5′ direction (indicated by black arrows). (b) Translocation mechanism of RecD2 helicase. When ATP binds to the enzyme, the cleft closes between 1A and 2A motor domains, causing domains 2A and 2B to slide along the DNA backbone (black arrows). The contacts between domain 1A and the DNA remain tight to anchor the DNA as domains 2A and 2B slide along it. When the conformational change is complete, the grip of domain 1A on the DNA is loosened. Then, ATP hydrolysis takes place, allowing the cleft to relax to the open conformation. The DNA is pulled back by domains 2A and 2B, which now have a tighter grip on bound DNA than domain 1A. This causes the DNA to slide across the surface of domain 1A as it moves away from domains 2A and 2B. The result is translocation by one base in a 5′–3′ direction (black arrows) during a single round of ATP binding and hydrolysis (adapted from ref. [94])
Duplex DNA Is Split by the Pin Region
In addition to the RecA-like 1A and 2A domains containing the conserved helicase motifs, all SF1 helicases contain accessory domains which can vary in structure. One common feature of these domains is the presence of a pin or wedge which functions to split the incoming DNA. The pin was first discovered in the SF2 helicase Hel308 from Archaeoglobus fulgidus [125]. A pin splitting the DNA was observed previously in RecC [103], the Chi recognition protein of the RecBCD helicase complex and in RuvA [128, 129], which along with RuvB and RuvC catalyzes branch migration resulting in Holliday junction resolution.
The location of the pin varies to correspond with the direction of translocation. For SF1A helicases, the 2A domain leads during unwinding in the 3′–5′ direction. In the structures of PcrA with DNA bound (Fig. 2.3a) [89] a pin is visible, positioned at the junction of the duplex and ssDNA. For SF1B helicases, 1A is the leading RecA-like domain, and an insertion in this domain, domain 1B, serves as the pin (Fig. 2.3c, d) [93]. In each case, the pin is positioned appropriately to split the incoming duplex during translocation along the ssDNA. Based on the structure of RecD2 bound to DNA, a mutant lacking the pin was designed which was completely devoid of unwinding activity although it hydrolyzed ATP at the same rate as wt RecD2 [93].
A recent report demonstrates that the mere presence of the pin is not sufficient for helicase activity (He et al., in press). Specific residues in the pin may be necessary for helicase activity. For the SF1B helicase Dda, mutation of a single F residue in the pin which stacks with the ssDNA completely eliminates strand separation although translocation on ssDNA is unaffected. In the case of RecD2 [93], the pin is short and it appears to function only in splitting the incoming duplex. Dda contains a long β-hairpin which is anchored at its tip by electrostatic interactions to domain 2B, allowing it to function not only directly in splitting the dsDNA but also in coupling ATP hydrolysis to unwinding (He et al., in press). Like Dda, the hepatitis C virus NS3 helicase (SF2) contains an extended pin [100], and mutation of conserved residues with this pin also uncouples ATP hydrolysis from unwinding [130].
Ensemble Kinetics for Helicase-Catalyzed DNA Unwinding of dsDNA and Translocation on ssDNA
SF1 helicases unwind the DNA in a stepwise manner so that more steps are required to unwind longer duplexes. Unwinding of duplexes of varying length has led to several descriptors of the kinetic and physical constants associated with helicases. One of the most discussed values relates to the “step size.” The kinetic step size refers to the number of base pairs unwound prior to a rate-limiting kinetic step. This value can be determined by measuring unwinding of increasing length duplexes [131]. The kinetic step sizes for some SF1 helicases are shown in Table 2.3. The physical step size refers to the number of base pairs that are unwound simultaneously. Single-molecule approaches have provided direct measures of the physical step size for a number of helicases (see below). The relationship between the kinetic step and the physical step size may be complex. A helicase might unwind one base pair at a time (physical step of one), but then proceed through a slow conformational change that occurs every three base pairs, resulting in a kinetic step size of three bps. The chemical step size refers to the number of base pairs unwound per ATP hydrolyzed. In the simplest case, all of these values are equal to one. However, there may be differences and care must be taken to distinguish between these values when comparing the activity of one helicase to another.
Table 2.3.
Kinetic constants for select SF1 helicases
| Helicase | Kinetic step size for unwind (bp) |
Kinetic step size for trans (nt) |
Unwind rate (bp/s) | Trans. rate (nt/s) | Unwinding processivity (bp) |
Trans. processivity (nt) |
|---|---|---|---|---|---|---|
| wtRep—monomer | N.D.a | N.D. | N.D. | 298 [155] | 0 [119] | 700 [155] |
| wtRep—EEb | 4c [119] | N.D. | 57 [119] | N.D. | N.D. | N.D. |
| ReΔ2B—monomer | 4c [155] | N.D. | 226 [155] | 530 [155] | <21 [155] | 800 [155] |
| UvrD—monomer | N.D. | 4–5 [92, 156, 234] | N.D. | 200 [92, 156, 234] | <18 bp [156] | 240 [92, 234] |
| UvrD—EE | 4.4 [131] | N.D. | 73.4 [131] | N.D. | 10 [131] | N.D. |
| RecBCD EE | 3–4 [235–238] | N.D.d | 700–1,350 [235–239] | N.D.d | 30,000 [183] | N.D. |
| PcrA—monomer | N.D. | 4.0 [145] | N.D. | 234 [145] | <18 [145] | 248 [145] |
| PcrA—EE | 4 [240] | N.D. | 31 [145, 240] | N.D. | N.D. | N.D. |
| Dda—monomer | 2.8 [241, 242] | N.D. | 262 [136, 241, 242] | 267 [136] | 19 [242] | N.D. |
| Dda—EE | 3.4 [242] | N.D. | 257 [242] | N.D. | 64 [242] | N.D. |
| TraI—monomer | 6.2 [159] | N.D. | 1,085 [159] | N.D. | N.D. | N.D. |
| TraI EE | 8.2 [159] | N.D. | 1,120 [159] | N.D. | >850 [243] | N.D. |
Not determined
Excess enzyme—enzyme concentration is in excess of substrate
Step size for ReΔ2B is assumed based on the duplex length and the number of steps in the kinetic mechanism
Kinetic step size and rates of translocation have been measured for RecB and RecBC, but not for RecBCD [244]
Similar to the unwinding studies, translocation can be examined by measuring the time needed to reach the end of varying lengths of ssDNA. The lag phase for these measurements provides kinetic information that can give rise to kinetic step sizes, rates of translocation, and coupling efficiencies when combined with ATP hydrolysis measurements. One assay that provided a breakthrough in helicase ATPase studies was the development of the phosphate-binding protein for measuring phosphate release kinetics [91]. This assay has been instrumental in relating the rates and efficiencies of ATPase activity to movement on ssDNA. However, it has not been generally applied, as yet, to understanding unwinding of dsDNA.
Helicases can be described as acting by an active or passive mechanism in reference to whether they actively separate the duplex or simply trap single-stranded intermediates that form as a result of thermal fraying [132, 133]. One suggestion for classifying active vs. passive helicases relies on comparing the ratio of the velocity for translocation on ssDNA to the velocity for unwinding of dsDNA [133]. If this ratio falls between 0.25 and 1, then a helicase can be considered as active. Most helicases likely fall between these extremes, and for some SF1 helicases, comparison of unwinding and translocation rates is complicated by oligomerization that occurs during unwinding [119, 134, 135]. However, bacteriophage T4 Dda has been shown to unwind duplex DNA and translocate on ssDNA at the same rate suggesting that it functions by a completely active mechanism [136].
Single-Molecule Methods Provide New Insights into SF1 Helicases
Breakthroughs in technology have resulted in corresponding breakthroughs in biology, and this theme has held true in understanding helicase mechanisms. Single-molecule Förster resonance energy transfer (smFRET) as well as laser tweezers or magnetic tweezers have been extensively applied to the study of helicases during the past decade [137, 138]. These techniques are particularly useful for visualizing kinetic events that are “hidden” within ensemble experiments. Recognition of the Chi sequence in DNA causes RecBCD to pause and reduce its translocation rate to approximately one-half the initial rate [139–141] resulting in a switch in motor usage with RecD being the lead motor prior to Chi and RecB after the Chi sites [142]. Magnetic tweezer analysis reported the average unwinding rate by RecBCD to be 900 bp/s [143] and the processivity of the complex to be ~1.
The physical step size can be directly observed in some cases using smFRET or laser tweezers. Single-molecule studies of PcrA reported the unwinding step size to be one nt [144], whereas a kinetic step size of four nts for translocation was estimated from ensemble experiments [145]. The larger kinetic step size determined from ensemble analysis could be an overestimation due to the presence of static disorder. In the case of Dda, single-molecule and ensemble experiments reported the unwinding rate to be ~250 bp/s and the rate varied little for forces ranging from 5 to 13 pN [136].
The mechanisms by which helicases catalyze protein displacement are beginning to be explored [146]. Single-molecule studies revealed the repetitive movements of Rep, PcrA, and UvrD helicases on the same stretch of DNA. The in vitro smFRET studies showed that the shuttling of the Rep monomer on ssDNA can prevent RecA filament formation [147], and that PcrA reeling in ssDNA can remove a preformed RecA filament [144]. Two other SF1 helicases, yeast Srs2 [148, 149] and UvrD [150], can displace Rad51 and RecA presynaptic filaments from ssDNA, respectively. Single-molecule studies offer insight into why many helicases display only limited unwinding processivity in vitro. For Rep, the reduced processivity in vitro is due to the relative instability of the functional complex [120]. The open and closed conformational states of Rep helicase undergoing ATP hydrolysis while bound to DNA were studied using smFRET [151]. The biological significance of having multiple conformations might be to regulate the helicase activity. Recent developments in three (or more)-color FRET [152, 153] should enable one to obtain simultaneous information on more than one activity, for example, ATP cycling and movement on DNA.
Protein–Protein Interactions That Regulate Helicase Activity
Helicases translocate along nucleic acids while separating dsDNA into single strands. Translocase activity alone is, in some cases, insufficient for helicase activity. In these cases, oligomerization and/or interactions with other proteins can regulate their translocase and helicase activities. Oligomerization can affect their NTPase, DNA-binding and -unwinding activities [45]. The monomeric forms of some SF1 enzymes, Rep [119, 120, 147, 154, 155], UvrD [121, 134, 156], and PcrA [89, 91, 127, 145], are processive translocases [91, 92, 156, 157], but do not display DNA unwinding activity in vitro [147]. On the other hand, the SF1 helicases Dda [158] and TraI [159] are able to function as monomeric helicases in vitro, b. But with the exception of the TraI, the SF1 helicases, when examined by themselves, generally unwind DNA with low processivity in vitro.
The nucleic acid unwinding processivity of some SF1 helicases can be increased significantly either through self-assembly or interactions with accessory proteins [119, 121, 134, 135, 160]. Rep helicase (SF1A) exists as a monomer in the absence of DNA [161]. However, Rep undergoes a DNA-induced dimerization upon binding either ss or dsDNA [132, 162], and the dimer appears to be the active form of the Rep helicase [119, 132, 161, 163–165]. Single turnover kinetic studies of UvrD-catalyzed DNA unwinding suggested that dimers are the minimal oligomeric form needed for optimal helicase activity [121, 134, 135]. In the case of Pif1, binding of ssDNA induces protein dimerization [166]. Oligomerization provides the active helicase with multiple DNA- and NTP-binding sites that are necessary for optimal DNA unwinding activity.
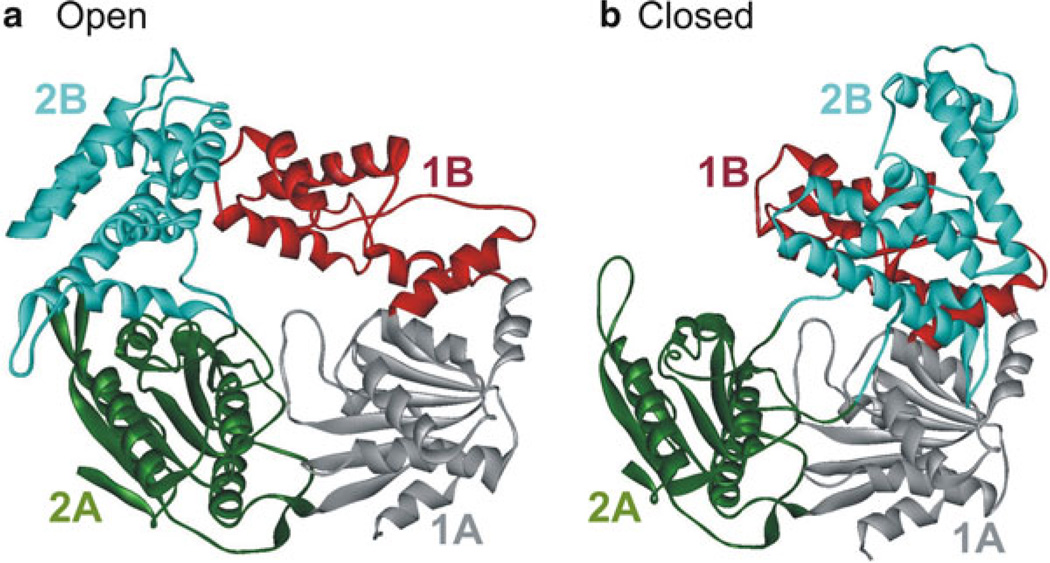
Regulation of helicase activity through protein–protein interactions may occur by altering conformations of the helicase. Crystal structures of Rep bound to ssDNA showed open and closed forms that differ in the 2B domain orientation [88] (Fig. 2.6). Deletion of the 2B domain in Rep was found to activate the helicase activity [154, 155]. Although, earlier smFRET studies have suggested the closed form of Rep to be an inhibited form [167], later detailed studies revealed two distinct Rep-partial-duplex DNA conformations in the ATPγS and ADP states. Here the primary conformation is found to be similar to the closed form, and in the secondary conformation the duplex DNA and 2B domain are rotated relative to the rest of the protein [151]. The multiple conformations may provide a mechanism of regulation of helicase activity whereby interactions between Rep and other proteins may determine the relative conformational states of domain 2B.
Fig. 2.6.
Crystal structure of E. coli Rep helicase (PDB code 1UAA [88]) in the open and closed conformations. Rep consists of four domains 1A, 1B, 2A, and 2B which are colored grey, red, green, and cyan, respectively. The open (a) and closed (b) conformations differ by rotations of around 130° of the 2B domain about a hinge region connecting it to the 2A domain [167]. The other domains are unchanged in both forms
In addition to helicases interacting among themselves, they can interact with other helicases to modulate their activity. It has been reported that Pif1 helicase activity is essential in top3 mutants in an Sgs1-dependent manner [168]. Pif1 has been suggested to strip Sgs1 from DNA, thereby downregulating the activity of Sgs1. Srs2 helicase has been reported to have a similar role in the disassembly of Rad51 filaments [148, 149]. Interaction between Rep and DnaB has been suggested to promote fork progression along protein-bound DNA [169].
Many SF1 enzymes are poor helicases in vitro; therefore, it is not surprising that their activities are enhanced through interactions with accessory proteins. Several helicases from prokaryotes and eukaryotes interact with other proteins to stimulate helicase activity [170–174]. The phage Φx174 gene A protein increases the helicase processivity of Rep [175–178]. PcrA is a nonprocessive helicase with difficulty unwinding even short lengths of duplex DNA [145], but the presence of plasmid replication initiator protein, RepD, enables PcrA to separate duplexes with high processivity [160, 179, 180].
The RecBCD holoenzyme contains two motors RecB (SF1A) and RecD (SF1B) in addition to the Chi recognition protein RecC [142]. RecBCD is compared to the world’s fastest supercar [181], since with the RecB and RecD helicase motors [73, 182], RecBCD is capable of moving along DNA at over 1,000 base pairs per second [183]. RecBCD is able to switch which of the two motors takes the lead, thereby regulating the translocation velocity of the complex [142] following the recognition of recombination hotspots called Chi sites [184, 185]. RecB helicase is activated through an interaction with RecC. RecB is a poor helicase by itself [186], but in complex with RecC is highly processive [139, 182, 183, 187]. Interaction with the accessory protein RecC is suggested to relieve an inhibitory function of the 2B subdomain of RecB [188]. Similarly, the inability of Rep monomers to function as helicases in vitro seems to be the result of an autoinhibitory effect of its subdomain 2B, and the deletion of this domain stimulates helicase activity of the Rep monomer [155].
Increased helicase processivity has been linked to helicase–SSB interactions [173, 189]. Rep and UvrD advance movement of replisomes blocked by nucleoprotein complexes in vitro [190]. Here the binding of successive monomers of Rep or UvrD at a blocked fork could facilitate protein displacement. Okazaki fragment processing in eukaryotes can occur either by the FEN1-only pathway or the two-nuclease pathway [191]. It has been reported that Dna2, Pif1, and RPA, the proteins of the two-nuclease pathway, stimulate FEN1 acting in the one-nuclease pathway [192]. Interactions with these proteins may change the conformation of FEN1 to optimally function on the substrate [192].
Saccharomyces Rrm3 helicase (SF1B) promotes replication fork progression through telomeric and subtelomeric DNA. Rrm3 is telomere-associated in vivo, suggesting a direct role in telomere replication [193]. Rrm3 is also needed for the timely replication of the entire genome, possibly through its role in promoting fork progression through difficult-to-replicate sites [194]. Rrm3 interacts with the catalytic subunit of DNA polymerase epsilon as it moves through both Rrm3p-dependent and -independent sites [194].
In addition to the stimulatory effects, some helicase-accessory protein interactions reduce the helicase activity. Through structural, biochemical, and functional studies, it was shown that the Srs2 helicase interacts with SUMO-PCNA thereby suppressing the Rad52-dependent recombinational repair pathway [195]. Also, Pif1 helicase negatively regulates telomere lengths by catalytically inhibiting telomerase activity [196–198].
Conclusion
The overall view from these studies is that SF1 helicases have multiple DNA-binding sites and that the entire enzyme does not move as a single unit, but instead different domains of the enzyme move at different times during the translocation–unwinding cycle as a function of ATP binding and hydrolysis. The description of this movement as an “inchworm” appears to hold true. Although the structures and helicase motifs are quite similar, the in vivo activities vary, which are likely achieved through structural differences outside of the 1A and 2A (RecA-like) domains. These variations can affect how the helicase interacts with the DNA substrate and how it interacts with other proteins which regulate its activity.
Despite the impressive progress made in understanding the kinetic and chemical mechanisms of helicases, there is much to be learned. The specific mechanism(s) by which most helicases actively pry apart the duplex remain to be determined. The specific step in the overall mechanism that limits the rate of the reaction is generally not known. Some data suggest that steps in the ATP hydrolysis cycle such as release of phosphate may limit the overall rate of DNA unwinding. The interactions between protein and DNA with the loading strand (or translocase strand) have been defined but interactions with the displaced strand largely remain a mystery. The role of protein–protein interactions in helicase mechanisms and other regulatory mechanisms remains to be uncovered. Finally, the specific mechanisms whereby helicases displace other proteins from DNA remain to be determined. There are certainly many unanswered questions that will require continued growth in the field.
Acknowledgements
Funding for this work was provided by NIH R01 GM098922 (K.D.R.).
Contributor Information
Kevin D. Raney, Email: raneykevind@uams.edu.
Alicia K. Byrd, Email: akbyrd@uams.edu.
Suja Aarattuthodiyil, Email: baarattuthodiyil@uams.edu.
References
- 1.Caruthers JM, McKay DB. Helicase structure and mechanism. Curr Opin Struct Biol. 2002;12:123–133. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 2.Delagoutte E, von Hippel PH. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part II: integration of helicases into cellular processes. Q Rev Biophys. 2003;36:1–69. doi: 10.1017/s0033583502003864. [DOI] [PubMed] [Google Scholar]
- 3.Matson SW, Kaiser-Rogers KA. DNA helicases. Annu Rev Biochem. 1990;59:289–329. doi: 10.1146/annurev.bi.59.070190.001445. [DOI] [PubMed] [Google Scholar]
- 4.Schmid SR, Linder P. D-E-A-D protein family of putative RNA helicases. Mol Microbiol. 1992;6:283–291. doi: 10.1111/j.1365-2958.1992.tb01470.x. [DOI] [PubMed] [Google Scholar]
- 5.Matson SW, Bean DW, George JW. DNA helicases: enzymes with essential roles in all aspects of DNA metabolism. Bioessays. 1994;16:13–22. doi: 10.1002/bies.950160103. [DOI] [PubMed] [Google Scholar]
- 6.Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- 7.Li J, Tang H, Mullen TM, Westberg C, Reddy TR, Rose DW, et al. A role for RNA helicase A in post-transcriptional regulation of HIV type 1. Proc Natl Acad Sci USA. 1999;96:709–714. doi: 10.1073/pnas.96.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linder P. Yeast RNA. helicases of the DEAD-box family involved in translation initiation. Biol Cell. 2003;95:157–167. doi: 10.1016/s0248-4900(03)00032-7. [DOI] [PubMed] [Google Scholar]
- 9.Bennett RJ, Keck JL. Structure and function of RecQ DNA helicases. Crit Rev Biochem Mol Biol. 2004;39:79–97. doi: 10.1080/10409230490460756. [DOI] [PubMed] [Google Scholar]
- 10.Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, Dunphy WG, et al. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell. 2005;121:887–898. doi: 10.1016/j.cell.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 11.Hartman TR, Qian S, Bolinger C, Fernandez S, Schoenberg DR, Boris-Lawrie K. RNA helicase A is necessary for translation of selected messenger RNAs. Nat Struct Mol Biol. 2006;13:509–516. doi: 10.1038/nsmb1092. [DOI] [PubMed] [Google Scholar]
- 12.Matsuno K, Kumano M, Kubota Y, Hashimoto Y, Takisawa H. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase alpha in the initiation of DNA replication. Mol Cell Biol. 2006;26:4843–4852. doi: 10.1128/MCB.02267-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Modrich P. Mismatch repair, genetic stability, and cancer. Science. 1994;266:1959–1960. doi: 10.1126/science.7801122. [DOI] [PubMed] [Google Scholar]
- 15.Sancar A. Mechanisms of DNA excision repair. Science. 1994;266:1954–1956. doi: 10.1126/science.7801120. [DOI] [PubMed] [Google Scholar]
- 16.Andressoo JO, Hoeijmakers JH, de Waard H. Nucleotide excision repair and its connection with cancer and ageing. Adv Exp Med Biol. 2005;570:45–83. doi: 10.1007/1-4020-3764-3_3. [DOI] [PubMed] [Google Scholar]
- 17.Stevnsner T, Muftuoglu M, Aamann MD, Bohr VA. The role of Cockayne syndrome group B (CSB) protein in base excision repair and aging. Mech Ageing Dev. 2008;129:441–448. doi: 10.1016/j.mad.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellis NA. DNA helicases in inherited human disorders. Curr Opin Genet Dev. 1997;7:354–363. doi: 10.1016/s0959-437x(97)80149-9. [DOI] [PubMed] [Google Scholar]
- 19.Andressoo JO, Hoeijmakers JH. Transcription-coupled repair and premature ageing. Mutat Res. 1997;577:179–194. doi: 10.1016/j.mrfmmm.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Brosh RM, Jr, Bohr VA. Human premature aging, DNA repair and RecQ helicases. Nucleic Acids Res. 2007;35:7527–7544. doi: 10.1093/nar/gkm1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ouyang KJ, Woo LL, Ellis NA. Homologous recombination and maintenance of genome integrity: cancer and aging through the prism of human RecQ helicases. Mech Ageing Dev. 2008;129:425–440. doi: 10.1016/j.mad.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 22.German J. Bloom’s syndrome. Dermatol Clin. 1995;13:7–18. [PubMed] [Google Scholar]
- 23.Shen JC, Gray MD, Oshima J, Kamath-Loeb AS, Fry M, Loeb LA. Werner syndrome protein. I. DNA helicase and dna exonuclease reside on the same polypeptide. J Biol Chem. 1998;273:34139–34144. doi: 10.1074/jbc.273.51.34139. [DOI] [PubMed] [Google Scholar]
- 24.Gibbons RJ, Bachoo S, Picketts DJ, Aftimos S, Asenbauer B, Bergoffen J, et al. Mutations in transcriptional regulator ATRX establish the functional significance of a PHD-like domain. Nat Genet. 1997;17:146–148. doi: 10.1038/ng1097-146. [DOI] [PubMed] [Google Scholar]
- 25.Prince PR, Emond MJ, Monnat RJ., Jr Loss of Werner syndrome protein function promotes aberrant mitotic recombination. Genes Dev. 2001;15:933–938. doi: 10.1101/gad.877001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saintigny Y, Makienko K, Swanson C, Emond MJ, Monnat RJ., Jr Homologous recombination resolution defect in werner syndrome. Mol Cell Biol. 2002;22:6971–6978. doi: 10.1128/MCB.22.20.6971-6978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hickson ID. RecQ helicases: caretakers of the genome. Nat Rev Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 28.Sharma S, Doherty KM, Brosh RM., Jr Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem J. 2006;398:319–337. doi: 10.1042/BJ20060450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ozgenc A, Loeb LA. Werner syndrome, aging and cancer. Genome Dyn. 2006;1:206–217. doi: 10.1159/000092509. [DOI] [PubMed] [Google Scholar]
- 30.Muftuoglu M, Oshima J, von Kobbe C, Cheng WH, Leistritz DF, Bohr VA. The clinical characteristics of Werner syndrome: molecular and biochemical diagnosis. Hum Genet. 2008;124:369–377. doi: 10.1007/s00439-008-0562-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chisholm KM, Aubert SD, Freese KP, Zakian VA, King MC, Welcsh PL. A genomewide screen for suppressors of Alu-mediated rearrangements reveals a role for PIF1. PLoS One. 2012;7:e30748. doi: 10.1371/journal.pone.0030748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen YZ, Bennett CL, Huynh HM, Blair IP, Puls I, Irobi J, et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4) Am J Hum Genet. 2004;74:1128–1135. doi: 10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreira MC, Klur S, Watanabe M, Nemeth AH, Le Ber I, Moniz JC, et al. Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat Genet. 2004;36:225–227. doi: 10.1038/ng1303. [DOI] [PubMed] [Google Scholar]
- 34.Guenther UP, Handoko L, Laggerbauer B, Jablonka S, Chari A, Alzheimer M, et al. IGHMBP2 is a ribosome-associated helicase inactive in the neuromuscular disorder distal SMA type 1 (DSMA1) Hum Mol Genet. 2009;18:1288–1300. doi: 10.1093/hmg/ddp028. [DOI] [PubMed] [Google Scholar]
- 35.Grohmann K, Schuelke M, Diers A, Hoffmann K, Lucke B, Adams C, et al. Mutations in the gene encoding immunoglobulin mu-binding protein 2 cause spinal muscular atrophy with respiratory distress type 1. Nat Genet. 2001;29:75–77. doi: 10.1038/ng703. [DOI] [PubMed] [Google Scholar]
- 36.Aggarwal M, Brosh RM., Jr Hitting the bull’s eye: novel directed cancer therapy through helicase-targeted synthetic lethality. J Cell Biochem. 2009;106:758–763. doi: 10.1002/jcb.22048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lehman IR, Boehmer PE. Replication of herpes simplex virus DNA. J Biol Chem. 1999;274:28059–28062. doi: 10.1074/jbc.274.40.28059. [DOI] [PubMed] [Google Scholar]
- 38.Kleymann G. Novel agents and strategies to treat herpes simplex virus infections. Expert Opin Investig Drugs. 2003;12:165–183. doi: 10.1517/13543784.12.2.165. [DOI] [PubMed] [Google Scholar]
- 39.Kleymann G. New antiviral drugs that target herpesvirus helicase primase enzymes. Herpes. 2003;10:46–52. [PubMed] [Google Scholar]
- 40.Kim DW, Gwack Y, Han JH, Choe J. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem Biophys Res Commun. 1995;215:160–166. doi: 10.1006/bbrc.1995.2447. [DOI] [PubMed] [Google Scholar]
- 41.De FR, Rice CM. New therapies on the horizon for hepatitis C: are we close? Clin Liver Dis. 2003;7:211–242. xi. doi: 10.1016/s1089-3261(02)00069-7. [DOI] [PubMed] [Google Scholar]
- 42.Harb OS, Abu KY. Essential role for the Legionella pneumophila rep helicase homologue in intracellular infection of mammalian cells. Infect Immun. 2000;68:6970–6978. doi: 10.1128/iai.68.12.6970-6978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 44.Enemark EJ, Joshua-Tor L. On helicases and other motor proteins. Curr Opin Struct Biol. 2008;18:243–257. doi: 10.1016/j.sbi.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Mol Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 46.Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989;17:4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure-function relationships. Curr Opin Struct Biol. 1993;3:419–429. [Google Scholar]
- 48.Singleton MR, Wigley DB. Modularity and specialization in superfamily 1 and 2 helicases. J Bacteriol. 2002;184:1819–1826. doi: 10.1128/JB.184.7.1819-1826.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von Hippel PH, Delagoutte E. Macromolecular complexes that unwind nucleic acids. Bioessays. 2003;25:1168–1177. doi: 10.1002/bies.10369. [DOI] [PubMed] [Google Scholar]
- 50.Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 51.Lusser A, Kadonaga JT. Chromatin remodeling by ATP-dependent molecular machines. Bioessays. 2003;25:1192–1200. doi: 10.1002/bies.10359. [DOI] [PubMed] [Google Scholar]
- 52.Petkovic M, Dietschy T, Freire R, Jiao R, Stagljar I. The human Rothmund-Thomson syndrome gene product, RECQL4, localizes to distinct nuclear foci that coincide with proteins involved in the maintenance of genome stability. J Cell Sci. 2005;118:4261–4269. doi: 10.1242/jcs.02556. [DOI] [PubMed] [Google Scholar]
- 53.Sharma S, Brosh RM., Jr Human RECQ1 is a DNA damage responsive protein required for genotoxic stress resistance and suppression of sister chromatid exchanges. PLoS One. 2007;2:e1297. doi: 10.1371/journal.pone.0001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Larsen DH, Poinsignon C, Gudjonsson T, Dinant C, Payne MR, Hari FJ, et al. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J Cell Biol. 2010;190:731–740. doi: 10.1083/jcb.200912135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raynard S, Bussen W, Sung P. A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIalpha, and BLAP75. J Biol Chem. 2006;281:13861–13864. doi: 10.1074/jbc.C600051200. [DOI] [PubMed] [Google Scholar]
- 56.Wu L, Bachrati CZ, Ou J, Xu C, Yin J, Chang M, et al. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci U S A. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barber LJ, Youds JL, Ward JD, McIlwraith MJ, O’Neil NJ, Petalcorin MI, et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh TR, Ali AM, Busygina V, Raynard S, Fan Q, Du CH, et al. BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes Dev. 2008;22:2856–2868. doi: 10.1101/gad.1725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bugreev DV, Brosh RM, Jr, Mazin AV. RECQ1 possesses DNA branch migration activity. J Biol Chem. 2008;283:20231–20242. doi: 10.1074/jbc.M801582200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uringa EJ, Youds JL, Lisaingo K, Lansdorp PM, Boulton SJ. RTEL1: an essential helicase for telomere maintenance and the regulation of homologous recombination. Nucleic Acids Res. 2011;39:1647–1655. doi: 10.1093/nar/gkq1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Picketts DJ, Tastan AO, Higgs DR, Gibbons RJ. Comparison of the human and murine ATRX gene identifies highly conserved, functionally important domains. Mamm Genome. 1998;9:400–403. doi: 10.1007/s003359900781. [DOI] [PubMed] [Google Scholar]
- 62.Travers A. Chromatin modification by DNA tracking. Proc Natl Acad Sci USA. 1999;96:13634–13637. doi: 10.1073/pnas.96.24.13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alexeev A, Mazin A, Kowalczykowski SC. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat Struct Biol. 2003;10:182–186. doi: 10.1038/nsb901. [DOI] [PubMed] [Google Scholar]
- 64.Zhang Z, Fan HY, Goldman JA, Kingston RE. Homology-driven chromatin remodeling by human RAD54. Nat Struct Mol Biol. 2007;14:397–405. doi: 10.1038/nsmb1223. [DOI] [PubMed] [Google Scholar]
- 65.Srivastava V, Modi P, Tripathi V, Mudgal R, De S, Sengupta S. BLM helicase stimulates the ATPase and chromatin-remodeling activities of RAD54. J Cell Sci. 2009;122:3093–3103. doi: 10.1242/jcs.051813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Troelstra C, van Gool A, de Wit J, Vermeulen W, Bootsma D, Hoeijmakers JH. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne’s syndrome and preferential repair of active genes. Cell. 1992;71:939–953. doi: 10.1016/0092-8674(92)90390-x. [DOI] [PubMed] [Google Scholar]
- 67.Evans E, Fellows J, Coffer A, Wood RD. Open complex formation around a lesion during nucleotide excision repair provides a structure for cleavage by human XPG protein. EMBO J. 1997;16:625–638. doi: 10.1093/emboj/16.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eisen A, Lucchesi JC. Unraveling the role of helicases in transcription. Bioessays. 1998;20:634–641. doi: 10.1002/(SICI)1521-1878(199808)20:8<634::AID-BIES6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 69.Moreland RJ, Tirode F, Yan Q, Conaway JW, Egly JM, Conaway RC. A role for the TFIIH XPB DNA helicase in promoter escape by RNA polymerase II. J Biol Chem. 1999;274:22127–22130. doi: 10.1074/jbc.274.32.22127. [DOI] [PubMed] [Google Scholar]
- 70.Fuller-Pace FV. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2008;34:4206–4215. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aygun O, Svejstrup J, Liu Y. A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc Natl Acad Sci U S A. 2008;105:8580–8584. doi: 10.1073/pnas.0804424105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Izumikawa K, Yanagida M, Hayano T, Tachikawa H, Komatsu W, Shimamoto A, et al. Association of human DNA helicase RecQ5beta with RNA polymerase II and its possible role in transcription. Biochem J. 2008;413:505–516. doi: 10.1042/BJ20071392. [DOI] [PubMed] [Google Scholar]
- 73.Dillingham MS, Spies M, Kowalczykowski SC. RecBCD enzyme is a bipolar DNA helicase. Nature. 2003;423:893–897. doi: 10.1038/nature01673. [DOI] [PubMed] [Google Scholar]
- 74.Naqvi A, Tinsley E, Khan SA. Purification and characterization of the PcrA helicase of Bacillus anthracis. J Bacteriol. 2003;185:6633–6639. doi: 10.1128/JB.185.22.6633-6639.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Constantinesco F, Forterre P, Koonin EV, Aravind L, Elie C. A bipolar DNA helicase gene, herA, clusters with rad50, mre11 and nurA genes in thermophilic archaea. Nucleic Acids Res. 2004;32:1439–1447. doi: 10.1093/nar/gkh283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hall MC, Matson SW. Helicase motifs: the engine that powers DNA unwinding. Mol Microbiol. 1999;34:867–877. doi: 10.1046/j.1365-2958.1999.01659.x. [DOI] [PubMed] [Google Scholar]
- 77.Tuteja N, Tuteja R. Prokaryotic and eukaryotic DNA helicases. Essential molecular motor proteins for cellular machinery. Eur J Biochem. 2004;271:1835–1848. doi: 10.1111/j.1432-1033.2004.04093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lohman TM. Escherichia coli DNA helicases: mechanisms of DNA unwinding. Mol Microbiol. 1992;6:5–14. doi: 10.1111/j.1365-2958.1992.tb00831.x. [DOI] [PubMed] [Google Scholar]
- 79.Tuteja N, Tuteja R. Unraveling DNA helicases. Motif, structure, mechanism and function. Eur J Biochem. 2004;271:1849–1863. doi: 10.1111/j.1432-1033.2004.04094.x. [DOI] [PubMed] [Google Scholar]
- 80.Bird LE, Subramanya HS, Wigley DB. Helicases: a unifying structural theme? Curr Opin Struct Biol. 1998;8:14–18. doi: 10.1016/s0959-440x(98)80004-3. [DOI] [PubMed] [Google Scholar]
- 81.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 82.Kadare G, Haenni AL. Virus-encoded RNA helicases. J Virol. 1997;71:2583–2590. doi: 10.1128/jvi.71.4.2583-2590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 84.Jankowsky E, Fairman ME. RNA helicases—one fold for many functions. Curr Opin Struct Biol. 2007;17:316–324. doi: 10.1016/j.sbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 85.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 86.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Subramanya HS, Bird LE, Brannigan JA, Wigley DB. Crystal structure of a DExx box DNA helicase. Nature. 1996;384:379–383. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- 88.Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 89.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 90.Soultanas P, Dillingham MS, Wiley P, Webb MR, Wigley DB. Uncoupling DNA translocation and helicase activity in PcrA: direct evidence for an active mechanism. EMBO J. 2000;19:3799–3810. doi: 10.1093/emboj/19.14.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dillingham MS, Wigley DB, Webb MR. Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry. 2000;39:205–212. doi: 10.1021/bi992105o. [DOI] [PubMed] [Google Scholar]
- 92.Tomko EJ, Fischer CJ, Niedziela-Majka A, Lohman TM. A nonuniform stepping mechanism for E. coli UvrD monomer translocation along single-stranded DNA. Mol Cell. 2007;26:335–347. doi: 10.1016/j.molcel.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Saikrishnan K, Griffiths SP, Cook N, Court R, Wigley DB. DNA binding to RecD: role of the 1B domain in SF1B helicase activity. EMBO J. 2008;27:2222–2229. doi: 10.1038/emboj.2008.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Saikrishnan K, Powell B, Cook NJ, Webb MR, Wigley DB. Mechanistic basis of 5′–3′ translocation in SF1B helicases. Cell. 2009;137:849–859. doi: 10.1016/j.cell.2009.03.036. [DOI] [PubMed] [Google Scholar]
- 95.Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. A novel superfamily of nucleoside triphosphate-binding motif containing proteins which are probably involved in duplex unwinding in DNA and RNA replication and recombination. FEBS Lett. 1988;235:16–24. doi: 10.1016/0014-5793(88)81226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. A conserved NTP-motif in putative helicases. Nature. 1988;333:22. doi: 10.1038/333022a0. [DOI] [PubMed] [Google Scholar]
- 97.Hodgman TC. A new superfamily of replicative proteins. Nature. 1988;333:22–23. doi: 10.1038/333022b0. [DOI] [PubMed] [Google Scholar]
- 98.Ilyina TV, Koonin EV. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Korolev S, Yao N, Lohman TM, Weber PC, Waksman G. Comparisons between the structures of HCV and Rep helicases reveal structural similarities between SF1 and SF2 super-families of helicases. Protein Sci. 1998;7:605–610. doi: 10.1002/pro.5560070309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kim JL, Morgenstern KA, Griffith JP, Dwyer MD, Thomson JA, Murcko MA, et al. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 101.Tanner NK. The newly identified Q motif of DEAD box helicases is involved in adenine recognition. Cell Cycle. 2003;2:18–19. doi: 10.4161/cc.2.1.296. [DOI] [PubMed] [Google Scholar]
- 102.Bernstein DA, Zittel MC, Keck JL. High-resolution structure of the E. coli RecQ helicase catalytic core. EMBO J. 2003;22:4910–4921. doi: 10.1093/emboj/cdg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- 104.Durr H, Korner C, Muller M, Hickmann V, Hopfner KP. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 2005;121:363–373. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 105.Rozen F, Pelletier J, Trachsel H, Sonenberg N. A lysine substitution in the ATP-binding site of eucaryotic initiation factor 4A abrogates nucleotide-binding activity. Mol Cell Biol. 1989;9:4061–4063. doi: 10.1128/mcb.9.9.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tanner NK, Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 107.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Venkatesan M, Silver LL, Nossal NG. Bacteriophage T4 gene 41 protein, required for the synthesis of RNA primers, is also a DNA helicase. J Biol Chem. 1982;257:12426–12434. [PubMed] [Google Scholar]
- 109.Story RM, Steitz TA. Structure of the recA protein-ADP complex. Nature. 1992;355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- 110.Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Brosh RM, Jr, Matson SW. Mutations in motif II of Escherichia coli DNA helicase II render the enzyme nonfunctional in both mismatch repair and excision repair with differential effects on the unwinding reaction. J Bacteriol. 1995;177:5612–5621. doi: 10.1128/jb.177.19.5612-5621.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Weng Y, Czaplinski K, Peltz SW. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol. 1996;16:5477–5490. doi: 10.1128/mcb.16.10.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Graves-Woodward KL, Gottlieb J, Challberg MD, Weller SK. Biochemical analyses of mutations in the HSV-1 helicase-primase that alter ATP hydrolysis, DNA unwinding, and coupling between hydrolysis and unwinding. J Biol Chem. 1997;272:4623–4630. doi: 10.1074/jbc.272.7.4623. [DOI] [PubMed] [Google Scholar]
- 114.Brosh RM, Jr, Matson SW. A point mutation in Escherichia coli DNA helicase II renders the enzyme nonfunctional in two DNA repair pathways. Evidence for initiation of unwinding from a nick in vivo. J Biol Chem. 1997;272:572–579. doi: 10.1074/jbc.272.1.572. [DOI] [PubMed] [Google Scholar]
- 115.Brosh RM, Jr, Matson SW. A partially functional DNA helicase II mutant defective in forming stable binary complexes with ATP and DNA. A role for helicase motif III. J Biol Chem. 1996;271:25360–25368. doi: 10.1074/jbc.271.41.25360. [DOI] [PubMed] [Google Scholar]
- 116.Graves-Woodward KL, Weller SK. Replacement of gly815 in helicase motif V alters the single-stranded DNA-dependent ATPase activity of the herpes simplex virus type 1 helicase-primase. J Biol Chem. 1996;271:13629–13635. doi: 10.1074/jbc.271.23.13629. [DOI] [PubMed] [Google Scholar]
- 117.Hall MC, Ozsoy AZ, Matson SW. Site-directed mutations in motif VI of Escherichia coli DNA helicase II result in multiple biochemical defects: evidence for the involvement of motif VI in the coupling of ATPase and DNA binding activities via conformational changes. J Mol Biol. 1998;277:257–271. doi: 10.1006/jmbi.1997.1614. [DOI] [PubMed] [Google Scholar]
- 118.Banroques J, Cordin O, Doere M, Linder P, Tanner NK. A conserved phenylalanine of motif IV in superfamily 2 helicases is required for cooperative, ATP-dependent binding of RNA substrates in DEAD-box proteins. Mol Cell Biol. 2008;28:3359–3371. doi: 10.1128/MCB.01555-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cheng W, Hsieh J, Brendza KM, Lohman TM. E. coli Rep oligomers are required to initiate DNA unwinding in vitro. J Mol Biol. 2001;310:327–350. doi: 10.1006/jmbi.2001.4758. [DOI] [PubMed] [Google Scholar]
- 120.Ha T, Rasnik I, Cheng W, Babcock HP, Gauss GH, Lohman TM, et al. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419:638–641. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 121.Maluf NK, Ali JA, Lohman TM. Kinetic mechanism for formation of the active, dimeric UvrD helicase-DNA complex. J Biol Chem. 2003;278:31930–31940. doi: 10.1074/jbc.M304223200. [DOI] [PubMed] [Google Scholar]
- 122.Lee JY, Yang W. UvrD helicase unwinds DNA bone base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yao N, Hesson T, Cable M, Hong Z, Kwong AD, Le HV, et al. Structure of the hepatitis C virus RNA helicase domain. Nat Struct Biol. 1997;4:463–467. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]
- 124.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 125.Buttner K, Nehring S, Hopfner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol. 2007;14:647–652. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- 126.Singleton MR, Scaife S, Wigley DB. Structural analysis of DNA replication fork reversal by RecG. Cell. 2001;107:79–89. doi: 10.1016/s0092-8674(01)00501-3. [DOI] [PubMed] [Google Scholar]
- 127.Soultanas P, Wigley DB. DNA helicases: ‘inching forward’. Curr Opin Struct Biol. 2000;10:124–128. doi: 10.1016/s0959-440x(99)00059-7. [DOI] [PubMed] [Google Scholar]
- 128.Hargreaves D, Rice DW, Sedelnikova SE, Artymiuk PJ, Lloyd RG, Rafferty JB. Crystal structure of E. coli RuvA with bound DNA Holliday junction at 6 A resolution. Nat Struct Biol. 1998;5:441–446. doi: 10.1038/nsb0698-441. [DOI] [PubMed] [Google Scholar]
- 129.Ariyoshi M, Nishino T, Iwasaki H, Shinagawa H, Morikawa K. Crystal structure of the Holliday junction DNA in complex with a single RuvA tetramer. Proc Natl Acad Sci USA. 2000;97:8257–8262. doi: 10.1073/pnas.140212997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lam AM, Keeney D, Frick DN. Two novel conserved motifs in the hepatitis C virus NS3 protein critical for helicase action. J Biol Chem. 2003;278:44514–44524. doi: 10.1074/jbc.M306444200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ali JA, Lohman TM. Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science. 1997;275:377–380. doi: 10.1126/science.275.5298.377. [DOI] [PubMed] [Google Scholar]
- 132.Wong I, Lohman TM. Allosteric effects of nucleotide cofactors on Escherichia coli Rep helicase-DNA binding. Science. 1992;256:350–355. doi: 10.1126/science.256.5055.350. [DOI] [PubMed] [Google Scholar]
- 133.Manosas M, Xi XG, Bensimon D, Croquette V. Active and passive mechanisms of helicases. Nucleic Acids Res. 2010;38:5518–5526. doi: 10.1093/nar/gkq273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ali JA, Maluf NK, Lohman TM. An oligomeric form of E. coli UvrD is required for optimal helicase activity. J Mol Biol. 1999;293:815–834. doi: 10.1006/jmbi.1999.3185. [DOI] [PubMed] [Google Scholar]
- 135.Maluf NK, Fischer CJ, Lohman TM. A dimer of Escherichia coli UvrD is the active form of the helicase in vitro. J Mol Biol. 2003;325:913–935. doi: 10.1016/s0022-2836(02)01277-9. [DOI] [PubMed] [Google Scholar]
- 136.Byrd AK, Matlock DL, Bagchi D, Aarattuthodiyil S, Harrison D, Croquette V, et al. Dda helicase tightly couples translocation on single-stranded dna to unwinding of duplex DNA: Dda is an optimally active helicase. J Mol Biol. 2012;420(3):141–154. doi: 10.1016/j.jmb.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Myong S, Ha T. Stepwise translocation of nucleic acid motors. Curr Opin Struct Biol. 2010;20:121–127. doi: 10.1016/j.sbi.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ha T, Kozlov AG, Lohman TM. Single-molecule views of protein movement on single-stranded DNA. Annu Rev Biophys. 2012;41:295–319. doi: 10.1146/annurev-biophys-042910-155351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bianco PR, Brewer LR, Corzett M, Balhorn R, Yeh Y, Kowalczykowski SC, et al. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature. 2001;409:374–378. doi: 10.1038/35053131. [DOI] [PubMed] [Google Scholar]
- 140.Dohoney KM, Gelles J. Chi-sequence recognition and DNA translocation by single RecBCD helicase/nuclease molecules. Nature. 2001;409:370–374. doi: 10.1038/35053124. [DOI] [PubMed] [Google Scholar]
- 141.Spies M, Bianco PR, Dillingham MS, Handa N, Baskin RJ, Kowalczykowski SC. A molecular throttle: the recombination hotspot chi controls DNA translocation by the RecBCD helicase. Cell. 2003;114:647–654. doi: 10.1016/s0092-8674(03)00681-0. [DOI] [PubMed] [Google Scholar]
- 142.Spies M, Amitani I, Baskin RJ, Kowalczykowski SC. RecBCD enzyme switches lead motor subunits in response to [chi] recognition. Cell. 2007;131:694–705. doi: 10.1016/j.cell.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rasnik I, Myong S, Ha T. Unraveling helicase mechanisms one molecule at a time. Nucleic Acids Res. 2006;34:4225–4231. doi: 10.1093/nar/gkl452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Park J, Myong S, Niedziela-Majka A, Lee KS, Yu J, Lohman TM, et al. PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell. 2010;142:544–555. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Niedziela-Majka A, Chesnik MA, Tomko EJ, Lohman TM. Bacillus stearothermophilus PcrA monomer is a single-stranded DNA translocase but not a processive helicase in vitro. J Biol Chem. 2007;282:27076–27085. doi: 10.1074/jbc.M704399200. [DOI] [PubMed] [Google Scholar]
- 146.Antony E, Tomko Q, Xiao L, Krejci L, Lohman TM, Ellengerger T. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol Cell. 2009;35:105–115. doi: 10.1016/j.molcel.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437:1321–1325. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- 148.Krejci L, Van KS, Li Y, Villemain J, Reddy MS, Klein H, et al. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- 149.Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le CE, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- 150.Veaute X, Delmas S, Selva M, Jeusset J, Le CE, Matic I, et al. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J. 2005;24:180–189. doi: 10.1038/sj.emboj.7600485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Balci H, Arslan S, Myong S, Lohman TM, Ha T. Single-molecule nanopositioning: structural transitions of a helicase-DNA complex during ATP hydrolysis. Biophys J. 2011;101:976–984. doi: 10.1016/j.bpj.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hohng S, Joo C, Ha T. Single-molecule three-color FRET. Biophys J. 2004;87:1328–1337. doi: 10.1529/biophysj.104.043935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Clamme JP, Deniz AA. Three-color single-molecule fluorescence resonance energy transfer. Chemphyschem. 2005;6:74–77. doi: 10.1002/cphc.200400261. [DOI] [PubMed] [Google Scholar]
- 154.Cheng W, Brendza KM, Gauss GH, Korolev S, Waksman G, Lohman TM. The 2B domain of the Escherichia coli Rep protein is not required for DNA helicase activity. Proc Natl Acad Sci U S A. 2002;99:16006–16011. doi: 10.1073/pnas.242479399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Brendza KM, Cheng W, Fischer CJ, Chesnik MA, Niedziela-Majka A, Lohman TM. Autoinhibition of Escherichia coli Rep monomer helicase activity by its 2B subdomain. Proc Natl Acad Sci U S A. 2005;102:10076–10081. doi: 10.1073/pnas.0502886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Fischer CJ, Maluf NK, Lohman TM. Mechanism of ATP-dependent translocation of E. coli UvrD monomers along single-stranded DNA. J Mol Biol. 2004;344:1287–1309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 157.Dillingham MS, Wigley DB, Webb MR. Direct measurement of single-stranded DNA translocation by PcrA helicase using the fluorescent base analogue 2-aminopurine. Biochemistry. 2002;41:643–651. doi: 10.1021/bi011137k. [DOI] [PubMed] [Google Scholar]
- 158.Nanduri B, Byrd AK, Eoff RL, Tackett AJ, Raney KD. Pre-steady-state DNA unwinding by bacteriophage T4 Dda helicase reveals a monomeric molecular motor. Proc Natl Acad Sci U S A. 2002;99:14722–14727. doi: 10.1073/pnas.232401899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sikora B, Eoff RL, Matson SW, Raney KD. DNA unwinding by Escherichia coli DNA helicase I (TraI) provides evidence for a processive monomeric molecular motor. J Biol Chem. 2006;281:36110–36116. doi: 10.1074/jbc.M604412200. [DOI] [PubMed] [Google Scholar]
- 160.Zhang W, Dillingham MS, Thomas CD, Allen S, Roberts CJ, Soultanas P. Directional loading and stimulation of PcrA helicase by the replication initiator protein RepD. J Mol Biol. 2007;371:336–348. doi: 10.1016/j.jmb.2007.05.050. [DOI] [PubMed] [Google Scholar]
- 161.Chao KL, Lohman TM. DNA-induced dimerization of the Escherichia coli Rep helicase. J Mol Biol. 1991;221:1165–1181. doi: 10.1016/0022-2836(91)90926-w. [DOI] [PubMed] [Google Scholar]
- 162.Wong I, Chao KL, Bujalowski W, Lohman TM. DNA-induced dimerization of the Escherichia coli rep helicase. Allosteric effects of single-stranded and duplex DNA. J Biol Chem. 1992;267:7596–7610. [PubMed] [Google Scholar]
- 163.Amaratunga M, Lohman TM. Escherichia coli rep helicase unwinds DNA by an active mechanism. Biochemistry. 1993;32:6815–6820. doi: 10.1021/bi00078a003. [DOI] [PubMed] [Google Scholar]
- 164.Bjornson KP, Amaratunga M, Moore KJ, Lohman TM. Single-turnover kinetics of helicase-catalyzed DNA unwinding monitored continuously by fluorescence energy transfer. Biochemistry. 1994;33:14306–14316. doi: 10.1021/bi00251a044. [DOI] [PubMed] [Google Scholar]
- 165.Bjornson KP, Moore KJ, Lohman TM. Kinetic mechanism of DNA binding and DNA-induced dimerization of the Escherichia coli Rep helicase. Biochemistry. 1996;35:2268–2282. doi: 10.1021/bi9522763. [DOI] [PubMed] [Google Scholar]
- 166.Barranco-Medina S, Galletto R. DNA binding induces dimerization of Saccharomyces cerevisiae Pif1. Biochemistry. 2010;49(39):8445–8454. doi: 10.1021/bi100984j. [DOI] [PubMed] [Google Scholar]
- 167.Rasnik I, Myong S, Cheng W, Lohman TM, Ha T. DNA-binding orientation and domain conformation of the E. coli rep helicase monomer bound to a partial duplex junction: single-molecule studies of fluorescently labeled enzymes. J Mol Biol. 2004;336:395–408. doi: 10.1016/j.jmb.2003.12.031. [DOI] [PubMed] [Google Scholar]
- 168.Wagner M, Price G, Rothstein R. The absence of Top3 reveals an interaction between the Sgs1 and Pif1 DNA helicases in Saccharomyces cerevisiae. Genetics. 2006;174:555–573. doi: 10.1534/genetics.104.036905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Atkinson J, Gupta MK, Rudolph CJ, Bell H, Lloyd RG, McGlynn P. Localization of an accessory helicase at the replisome is critical in sustaining efficient genome duplication. Nucleic Acids Res. 2011;39(3):949–957. doi: 10.1093/nar/gkq889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Korhonen JA, Gaspari M, Falkenberg M. TWINKLE Has 5′ -> 3′ DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J Biol Chem. 2003;278:48627–48632. doi: 10.1074/jbc.M306981200. [DOI] [PubMed] [Google Scholar]
- 171.Cadman CJ, McGlynn P. PriA helicase and SSB interact physically and functionally. Nucleic Acids Res. 2004;32:6378–6387. doi: 10.1093/nar/gkh980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Shereda RD, Bernstein DA, Keck JL. A central role for SSB in Escherichia coli RecQ DNA helicase function. J Biol Chem. 2007;282:19247–19258. doi: 10.1074/jbc.M608011200. [DOI] [PubMed] [Google Scholar]
- 173.Rajagopal V, Patel SS. Single strand binding proteins increase the processivity of DNA unwinding by the hepatitis C virus helicase. J Mol Biol. 2008;376:69–79. doi: 10.1016/j.jmb.2007.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sowd G, Wang H, Pretto D, Chazin WJ, Opresko PL. Replication protein A stimulates the Werner syndrome protein branch migration activity. J Biol Chem. 2009;284:34682–34691. doi: 10.1074/jbc.M109.049031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Scott JF, Eisenberg S, Bertsch LL, Kornberg A. A mechanism of duplex DNA replication revealed by enzymatic studies of phage phi X174: catalytic strand separation in advance of replication. Proc Natl Acad Sci U S A. 1977;74:193–197. doi: 10.1073/pnas.74.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Eisenberg S, Griffith J, Kornberg A. phiX174 cistron A protein is a multifunctional enzyme in DNA replication. Proc Natl Acad Sci U S A. 1977;74:3198–3202. doi: 10.1073/pnas.74.8.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yarranton GT, Gefter ML. Enzyme-catalyzed DNA unwinding: studies on Escherichia coli rep protein. Proc Natl Acad Sci U S A. 1979;76:1658–1662. doi: 10.1073/pnas.76.4.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Reinberg D, Zipursky SL, Weisbeek P, Brown D, Hurwitz J. Studies on the phi X174 gene A protein-mediated termination of leading strand DNA synthesis. J Biol Chem. 1983;258:529–537. [PubMed] [Google Scholar]
- 179.Soultanas P, Dillingham MS, Papadopoulos F, Phillips SE, Thomas CD, Wigley DB. Plasmid replication initiator protein RepD increases the processivity of PcrA DNA helicase. Nucleic Acids Res. 1999;27:1421–1428. doi: 10.1093/nar/27.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Slatter AF, Thomas CD, Webb MR. PcrA helicase tightly couples ATP hydrolysis to unwinding double-stranded DNA, modulated by the initiator protein for plasmid replication, RepD. Biochemistry. 2009;48(27):6326–6334. doi: 10.1021/bi900101h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wigley DB. RecBCD: the supercar of DNA repair. Cell. 2007;131:651–653. doi: 10.1016/j.cell.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 182.Taylor AF, Smith GR. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature. 2003;423:889–893. doi: 10.1038/nature01674. [DOI] [PubMed] [Google Scholar]
- 183.Roman LJ, Eggleston AK, Kowalczykowski SC. Processivity of the DNA helicase activity of Escherichia coli recBCD enzyme. J Biol Chem. 1992;267:4207–4214. [PubMed] [Google Scholar]
- 184.Lam ST, Stahl MM, McMilin KD, Stahl FW. Rec-mediated recombinational hot spot activity in bacteriophage lambda. II. A mutation which causes hot spot activity. Genetics. 1974;77:425–433. doi: 10.1093/genetics/77.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Anderson DG, Kowalczykowski SC. The recombination hot spot chi is a regulatory element that switches the polarity of DNA degradation by the RecBCD enzyme. Genes Dev. 1997;11:571–581. doi: 10.1101/gad.11.5.571. [DOI] [PubMed] [Google Scholar]
- 186.Boehmer PE, Emmerson PT. The RecB subunit of the Escherichia coli RecBCD enzyme couples ATP hydrolysis to DNA unwinding. J Biol Chem. 1992;267:4981–4987. [PubMed] [Google Scholar]
- 187.Bianco PR, Kowalczykowski SC. Translocation step size and mechanism of the RecBC DNA helicase. Nature. 2000;405:368–372. doi: 10.1038/35012652. [DOI] [PubMed] [Google Scholar]
- 188.Rigden DJ. An inactivated nuclease-like domain in RecC with novel function: implications for evolution. BMC Struct Biol. 2005;5:9. doi: 10.1186/1472-6807-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Marsh VL, McGeoch AT, Bell SD. Influence of chromatin and single strand binding proteins on the activity of an archaeal MCM. J Mol Biol. 2006;357:1345–1350. doi: 10.1016/j.jmb.2006.01.074. [DOI] [PubMed] [Google Scholar]
- 190.Guy CP, Atkinson J, Gupta MK, Mahdi A, Gwynn EJ, Rudolph CJ, et al. Rep provides a second motor at the replisome to promote duplication of protein-bound DNA. Mol Cell. 2009;36:654–666. doi: 10.1016/j.molcel.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bae SH, Seo YS. Characterization of the enzymatic properties of the yeast dna2 helicase/endonuclease suggests a new model for Okazaki fragment processing. J Biol Chem. 2000;275:38022–38031. doi: 10.1074/jbc.M006513200. [DOI] [PubMed] [Google Scholar]
- 192.Henry RA, Balakrishnan L, Ying-Lin ST, Campbell JL, Bambara RA. Components of the secondary pathway stimulate the primary pathway of eukaryotic Okazaki fragment processing. J Biol Chem. 2010;285:28496–28505. doi: 10.1074/jbc.M110.131870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ivessa AS, Zhou JQ, Schulz VP, Monson EK, Zakian VA. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 2002;16:1383–1396. doi: 10.1101/gad.982902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Azvolinsky AS, Dunaway S, Torres JZ, Bessler JB, Zakian VA. The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev. 2006;20:3104–3116. doi: 10.1101/gad.1478906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Armstrong AA, Mohideen F, Lima CD. Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2. Nature. 2012;483:59–63. doi: 10.1038/nature10883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Schulz VP, Zakian VA. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994;76:145–155. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- 197.Zhou J, Monson EK, Teng SC, Schulz VP, Zakian VA. Pif1p helicase, a catalytic inhibitor of telomerase in yeast. Science. 2000;289:771–774. doi: 10.1126/science.289.5480.771. [DOI] [PubMed] [Google Scholar]
- 198.Boule JB, Vega LR, Zakian VA. The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature. 2005;438:57–61. doi: 10.1038/nature04091. [DOI] [PubMed] [Google Scholar]
- 199.Jongeneel CV, Bedinger P, Alberts BM. Effects of the bacteriophage T4 dda protein on DNA synthesis catalyzed by purified T4 replication proteins. J Biol Chem. 1984;259:12933–12938. [PubMed] [Google Scholar]
- 200.Amundsen SK, Taylor AF, Chaudhury AM, Smith GR. recD: the gene for an essential third subunit of exonuclease V. Proc Natl Acad Sci U S A. 1986;83:5558–5562. doi: 10.1073/pnas.83.15.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lahue EE, Matson SW. Purified Escherichia coli F-factor TraY protein binds oriT. J Bacteriol. 1990;172:1385–1391. doi: 10.1128/jb.172.3.1385-1391.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Iordanescu S, Bargonetti J. Staphylococcus aureus chromosomal mutations that decrease efficiency of Rep utilization in replication of pT181 and related plasmids. J Bacteriol. 1989;171:4501–4503. doi: 10.1128/jb.171.8.4501-4503.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Dodson MS, Lehman IR. Association of DNA helicase and primase activities with a subassembly of the herpes simplex virus 1 helicase-primase composed of the UL5 and UL52 gene products. Proc Natl Acad Sci U S A. 1991;88:1105–1109. doi: 10.1073/pnas.88.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Rong L, Palladino F, Aguilera A, Klein HL. The hyper-gene conversion hpr5-1 mutation of Saccharomyces cerevisiae is an allele of the SRS2/RADH gene. Genetics. 1991;127:75–85. doi: 10.1093/genetics/127.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Gauss P, Park K, Spencer TE, Hacker KJ. DNA helicase requirements for DNA replication during bacteriophage T4 infection. J Bacteriol. 1994;176:1667–1672. doi: 10.1128/jb.176.6.1667-1672.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Bjornson KP, Wong I, Lohman TM. ATP hydrolysis stimulates binding and release of single stranded DNA from alternating subunits of the dimeric E. coli Rep helicase: implications for ATP-driven helicase translocation. J Mol Biol. 1996;263:411–422. doi: 10.1006/jmbi.1996.0585. [DOI] [PubMed] [Google Scholar]
- 207.Budd ME, Campbell JL. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol Cell Biol. 1997;17:2136–2142. doi: 10.1128/mcb.17.4.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Bird LE, Brannigan JA, Subramanya HS, Wigley DB. Characterisation of Bacillus stearothermophilus PcrA helicase: evidence against an active rolling mechanism. Nucleic Acids Res. 1998;26:2686–2693. doi: 10.1093/nar/26.11.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Dao V, Modrich P. Mismatch-, MutS-, MutL-, and helicase II-dependent unwinding from the single-strand break of an incised heteroduplex. J Biol Chem. 1998;273:9202–9207. doi: 10.1074/jbc.273.15.9202. [DOI] [PubMed] [Google Scholar]
- 210.Petit MA, Dervyn E, Rose M, Entian KD, McGovern S, Ehrlich SD, et al. PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication. Mol Microbiol. 1998;29:261–273. doi: 10.1046/j.1365-2958.1998.00927.x. [DOI] [PubMed] [Google Scholar]
- 211.Bruand C, Ehrlich SD. UvrD-dependent replication of rolling-circle plasmids in Escherichia coli. Mol Microbiol. 2000;35:204–210. doi: 10.1046/j.1365-2958.2000.01700.x. [DOI] [PubMed] [Google Scholar]
- 212.Ivessa AS, Zhou JQ, Zakian VA. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell. 2000;100:479–489. doi: 10.1016/s0092-8674(00)80683-2. [DOI] [PubMed] [Google Scholar]
- 213.Wang SW, Goodwin A, Hickson ID, Norbury CJ. Involvement of Schizosaccharomyces pombe Srs2 in cellular responses to DNA damage. Nucleic Acids Res. 2001;29:2963–2972. doi: 10.1093/nar/29.14.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Choe W, Budd M, Imamura O, Hoopes L, Campbell JL. Dynamic localization of an Okazaki fragment processing protein suggests a novel role in telomere replication. Mol Cell Biol. 2002;22:4202–4217. doi: 10.1128/MCB.22.12.4202-4217.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Lesur I, Campbell JL. The transcriptome of prematurely aging yeast cells is similar to that of telomerase-deficient cells. Mol Biol Cell. 2004;15:1297–1312. doi: 10.1091/mbc.E03-10-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ding H, Schertzer M, Wu X, Gertsenstein M, Selig S, Kammori M, et al. Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell. 2004;117:873–886. doi: 10.1016/j.cell.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 217.Opresko PL, Otterlei M, Graakjaer J, Bruheim P, Dawut L, Kolvraa S, et al. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol Cell. 2004;14:763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 218.Budd ME, Reis CC, Smith S, Myung K, Campbell JL. Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase delta. Mol Cell Biol. 2006;26:2490–2500. doi: 10.1128/MCB.26.7.2490-2500.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Liao S, Toczylowski T, Yan H. Identification of the Xenopus DNA2 protein as a major nuclease for the 5′ ->3′ strand-specific processing of DNA ends. Nucleic Acids Res. 2008;36:6091–6100. doi: 10.1093/nar/gkn616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Rossi ML, Pike JE, Wang W, Burgers PM, Campbell JL, Bambara RA. Pif1 helicase directs eukaryotic Okazaki fragments toward the two-nuclease cleavage pathway for primer removal. J Biol Chem. 2008;283:27483–27493. doi: 10.1074/jbc.M804550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.London TB, Barber LJ, Mosedale G, Kelly GP, Balasubramanian S, Hickson ID, et al. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J Biol Chem. 2008;283:36132–36139. doi: 10.1074/jbc.M808152200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wu Y, Shin-ya K, Brosh RM., Jr FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Pike JE, Burgers PM, Campbell JL, Bambara RA. Pif1 helicase lengthens some Okazaki fragment flaps necessitating Dna2 nuclease/helicase action in the two-nuclease processing pathway. J Biol Chem. 2009;284:25170–25180. doi: 10.1074/jbc.M109.023325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Ghosh A, Rossi ML, Aulds J, Croteau D, Bohr VA. Telomeric D-loops containing 8-oxo-2′-deoxyguanosine are preferred substrates for Werner and Bloom syndrome helicases and are bound by POT1. J Biol Chem. 2009;284:31074–31084. doi: 10.1074/jbc.M109.027532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem. 2009;284:4041–4045. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Chakraborty P, Grosse F. WRN helicase unwinds Okazaki fragment-like hybrids in a reaction stimulated by the human DHX9 helicase. Nucleic Acids Res. 2010;38:4722–4730. doi: 10.1093/nar/gkq240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.van de Putte P, van Sluis CA, van Dillewijn J, Rorsch A. The location of genes controlling radiation sensitivity in Escherichia coli. Mutat Res. 1965;2:97–110. doi: 10.1016/0027-5107(65)90041-2. [DOI] [PubMed] [Google Scholar]
- 228.Ogawa H, Shimada K, Tomizawa J. Studies on radiation-sensitive mutants of E. coli. I. Mutants defective in the repair synthesis. Mol Gen Genet. 1968;101:227–244. doi: 10.1007/BF00271625. [DOI] [PubMed] [Google Scholar]
- 229.Arthur HM, Lloyd RG. Hyper-recombination in uvrD mutants of Escherichia coli K-12. Mol Gen Genet. 1980;180:185–191. doi: 10.1007/BF00267368. [DOI] [PubMed] [Google Scholar]
- 230.Jongeneel CV, Formosa T, Alberts BM. Purification and characterization of the bacteriophage T4 dda protein. A DNA helicase that associates with the viral helix-destabilizing protein. J Biol Chem. 1984;259:12925–12932. [PubMed] [Google Scholar]
- 231.Jongeneel CV, Formosa T, Munn M, Alberts BM. Enzymological studies of the T4 replication proteins. Adv Exp Med Biol. 1984;179:17–33. doi: 10.1007/978-1-4684-8730-5_2. [DOI] [PubMed] [Google Scholar]
- 232.Bjornson KP, Hsieh J, Amaratunga M, Lohman TM. Kinetic mechanism for the sequential binding of two single-stranded oligodeoxynucleotides to the Escherichia coli Rep helicase dimer. Biochemistry. 1998;37:891–899. doi: 10.1021/bi9719307. [DOI] [PubMed] [Google Scholar]
- 233.Sikora B, Chen Y, Lichti CF, Harrison MK, Jennings TA, Tang Y, et al. Hepatitis C virus NS3 helicase forms oligomeric structures that exhibit optimal DNA unwinding activity in vitro. J Biol Chem. 2008;283:11516–11525. doi: 10.1074/jbc.M708125200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Tomko EJ, Fischer CJ, Lohman TM. Single-stranded DNA translocation of E. coli UvrD monomer is tightly coupled to ATP hydrolysis. J Mol Biol. 2012;418:32–46. doi: 10.1016/j.jmb.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Lucius AL, Wong CJ, Lohman TM. Fluorescence stopped-flow studies of single turnover kinetics of E. coli RecBCD helicase-catalyzed DNA unwinding. J Mol Biol. 2004;339:731–750. doi: 10.1016/j.jmb.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 236.Lucius AL, Lohman TM. Effects of temperature and ATP on the kinetic mechanism and kinetic step-size for E. coli RecBCD helicase-catalyzed DNA unwinding. J Mol Biol. 2004;339:751–771. doi: 10.1016/j.jmb.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 237.Lucius AL, Vindigni A, Gregorian R, Ali JA, Taylor AF, Smith GR, et al. DNA unwinding step-size of E. coli RecBCD helicase determined from single turnover chemical quenched-flow kinetic studies. J Mol Biol. 2002;324:409–428. doi: 10.1016/s0022-2836(02)01067-7. [DOI] [PubMed] [Google Scholar]
- 238.Wu CG, Lohman TM. Influence of DNA end structure on the mechanism of initiation of DNA unwinding by the Escherichia coli RecBCD and RecBC helicases. J Mol Biol. 2008;382:312–326. doi: 10.1016/j.jmb.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Dillingham MS, Webb MR, Kowalczykowski SC. Bipolar DNA translocation contributes to highly processive DNA unwinding by RecBCD enzyme. J Biol Chem. 2005;280:37069–37077. doi: 10.1074/jbc.M505520200. [DOI] [PubMed] [Google Scholar]
- 240.Yang Y, Dou SX, Ren H, Wang PY, Zhang XD, Qian M, et al. Evidence for a functional dimeric form of the PcrA helicase in DNA unwinding. Nucleic Acids Res. 2008;36:1976–1989. doi: 10.1093/nar/gkm1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Eoff RL, Raney KD. Intermediates revealed in the kinetic mechanism for DNA unwinding by a monomeric helicase. Nat Struct Mol Biol. 2006;13:242–249. doi: 10.1038/nsmb1055. [DOI] [PubMed] [Google Scholar]
- 242.Eoff RL, Raney KD. Kinetic mechanism for DNA unwinding by multiple molecules of Dda helicase aligned on DNA. Biochemistry. 2010;49:4543–4553. doi: 10.1021/bi100061v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Lahue EE, Matson SW. Escherichia coli DNA helicase I catalyzes a unidirectional and highly processive unwinding reaction. J Biol Chem. 1988;263:3208–3215. [PubMed] [Google Scholar]
- 244.Wu CG, Bradford C, Lohman TM. Escherichia coli RecBC helicase has two translocase activities controlled by a single ATPase motor. Nat Struct Mol Biol. 2010;17(10):1210–1217. doi: 10.1038/nsmb.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]