Abstract
Background and Aims
The olive (Olea europaea subsp. europaea) was domesticated in the Mediterranean area but its wild relatives are distributed over three continents, from the Mediterranean basin to South Africa and south-western Asia. Recent studies suggested that this crop originated in the Levant while a secondary diversification occurred in most westward areas. A possible contribution of the Saharan subspecies (subsp. laperrinei) has been highlighted, but the data available were too limited to draw definite conclusions. Here, patterns of genetic differentiation in the Mediterranean and Saharan olives are analysed to test for recent admixture between these taxa.
Methods
Nuclear microsatellite and plastid DNA (ptDNA) data were compiled from previous studies and completed for a sample of 470 cultivars, 390 wild Mediterranean trees and 270 Saharan olives. A network was reconstructed for the ptDNA haplotypes, while a Bayesian clustering method was applied to identify the main gene pools in the data set and then simulate and test for early generations of admixture between Mediterranean and Saharan olives.
Key Results
Four lineages of ptDNA haplotypes are recognized: three from the Mediterranean basin and one from the Sahara. Only one haplotype, primarily distributed in the Sahara, is shared between laperrinei and europaea. This haplotype is detected once in ‘Dhokar’, a cultivar from the Maghreb. Nuclear microsatellites show geographic patterns of genetic differentiation in the Mediterranean olive that reflect the primary origins of cultivars in the Levant, and indicate a high genetic differentiation between europaea and laperrinei. No first-generation hybrid between europaea and laperrinei is detected, but recent, reciprocal admixture between Mediterranean and Saharan subspecies is found in a few accessions, including ‘Dhokar’.
Conclusions
This study reports for the first time admixture between Mediterranean and Saharan olives. Although its contribution remains limited, Laperrine's olive has been involved in the diversification of cultivated olives.
Keywords: Admixture, domestication, Laperrine's olive, Mediterranean basin, microsatellite, Olea europaea, population genetic simulations, Sahara, secondary diversification, wild genetic resources
INTRODUCTION
The origins of the Mediterranean cultivated olive (Olea europaea subsp. europaea var. europaea) are hotly debated, but it is usually accepted that its primary domestication started in the Levant as attested to by archaeological, historical and molecular evidence (Kaniewski et al., 2012; Zohary et al., 2012; Besnard et al., 2013a). Multiple local selections of cultivars have also been reported by genetic analyses (e.g. Besnard et al., 2001a; Belaj et al., 2002; Baldoni et al., 2006; Breton et al., 2008), and secondary diversification of the crop followed the oleiculture diffusion over the whole Mediterranean basin (Terral. 1997; Besnard et al., 2001a). The contribution of the western wild olives in this diversification process remains, however, poorly understood. Some authors claimed that recurrent, ongoing crop–wild gene flow allowed maintenence of a high level of gene diversity in the cultivated compartment of perennial crops (Miller and Gross, 2011).
Hybrids between the Mediterranean olive (subsp. europaea) and other diploid wild olive subspecies (i.e. from South Africa, Iran and the Saharan Mountains) have been obtained under controlled conditions (e.g. Besnard et al., 2001b, 2008; Hannachi et al., 2009). This observation indicates that non-Mediterranean wild taxa could have also been involved in olive domestication or secondary diversification as hypothesized by numerous botanists since the 19th century (Oliver, 1868; Newberry, 1937; Chevalier, 1948; Turrill, 1951; Green, 2002). This assumption is usually no longer considered due to a lack of molecular evidence (e.g. Angiolillo et al., 1999; Besnard et al., 2007c). Yet, in a recent study, one unnamed cultivar from Morocco was shown to harbour a particular plastid DNA (ptDNA) haplotype, namely L1·1 (Besnard et al., 2011). In a brief report, this haplotype was further shown to be frequent in the Laperrine's olive tree (O. europaea subsp. laperrinei), a relictual endangered taxon endemic to the Central Saharan mountains (Besnard et al., 2012). This result strongly supports the hypothesis that crosses between O. europaea subsp. laperrinei and the Mediterranean olive could have contributed to cultivar diversification, but this hypothesis still needs to be properly tested with a detailed study of geographic patterns of genetic diversity (from nuclear and plastid genomes) on both Mediterranean and Saharan subspecies. In addition, such knowledge may have implications for the management of olive genetic resources.
According to Rubio de Casas et al. (2006), the genetic differentiation between subspecies europaea and laperrinei is highly significant [FST = 17·2 %; assessed with amplified fragment length polymorphisms (AFLPs)] as expected for allopatric taxa. Early generations of hybrids between two genetically differentiated taxa can be detected theoretically with genetic population analyses (e.g. admixture detection; Beaumont et al., 2001). Such a method has already been successfully used in olive to detect hybrids between europaea and cuspidata in the invasive Australian range (Besnard et al., 2007b, 2013b), and for detecting admixture between Mediterranean gene pools (Baldoni et al., 2006; Breton et al., 2006; Belaj et al., 2007). This approach was recently applied to document the secondary domestication history of the apple (Cornille et al., 2012) and almond trees (Delplancke et al., 2013).
In the present study, we describe patterns of genetic differentiation in the Mediterranean and Saharan olives, and we tested for admixture between these taxa. Nuclear microsatellites and plastid haplotypes from different studies were compiled and the genetic characterization of wild populations was completed. The maternal haplotype of all individuals was first used to detect possible seed–propagule exchanges between taxa. A Bayesian clustering method was applied for the detection of main gene pools, and simulations were used for assessing the power of our approach to detect admixture. Based on our results, the human-meditated diffusion of the oleiculture over the Mediterranean basin and the contribution of O. europaea subsp. laperrinei to the cultivated olive diversification are discussed.
MATERIALS AND METHODS
Plant material
In the present study, we analysed plastid and nuclear genetic data sets generated on 1130 olive accessions (see below). This sample encompasses Mediterranean cultivars and wild olives from the Mediterranean basin and the Saharan mountains. Mediterranean accessions were a priori classified according to three pre-defined zones (Supplementary Data Tables S1 and S2): East (from Croatia to the Levant, including Egypt), Central (from Italy to France and Libya to Algeria) and West (Iberian Peninsula and Morocco).
Olive cultivars (Olea europaea subsp. europaea var. europaea)
All cultivated accessions considered in this study were first characterized with nuclear microsatellite loci [or simple sequence repeats (SSRs)] by Besnard et al. (2007b), Khadari et al. (2008) and Haouane et al. (2011). When accessions were distinguished based on only one or two SSR alleles, the distinction was doubtful and could be attributed to mutations (Lopes et al., 2004; Baali-Cherif and Besnard, 2005; Khadari et al., 2008). To avoid considering variants of the same accession in the data analyses but also to avoid including families of very closely related individuals that can bias Bayesian clustering analyses (Rodríguez-Ramillo and Wang, 2012), we decided to keep (at random) only one tree for such closely related accessions (i.e. distinguished by one or two alleles per ten loci). A total of 470 accessions were analysed. Four hundred and thirty-five cultivated accessions were from the Olive World Germplasm Bank of Marrakech (OWGB-M). Additionally, 15 cultivated accessions were from the World Olive Germplasm Bank of Cordoba (WOGB-C) and 20 were sampled in the field (Supplementary Data Table S1). All these accessions have also been characterized with a plastid genomic profiling method (Besnard et al., 2013a).
Oleasters populations
Wild populations from the Mediterranean Basin (O. europaea subsp. europaea var. sylvestris) have been recently sampled for the study of ptDNA variation toward the Mediterranean basin (Besnard et al., 2013a). In the present study, a sub-set of 390 trees from 45 locations was considered for nuclear microsatellite characterization (Supplementary Data Table S2).
Laperrine's olive populations
Populations of O. europaea subsp. laperrinei (Batt. & Trab.) Cif. have been previously sampled in the Ahaggar (Hoggar and Tassili n'Ajjer, South Algeria) and the Aïr (Tamgak, Bagezane and Egalah, North Niger). Most individuals were characterized with nuclear SSR loci (seven of them shared with the present study) in order to identify genotypes and analyse the population dynamics (Baali-Cherif and Besnard, 2005; Besnard et al., 2007a). Here, 270 distinct genotypes were considered: 48 from the Bagezane-Egalah, 44 from the Tamgak, 45 from the Tassili n'Ajjer and 133 from the Hoggar.
Genetic characterization
Ten nuclear SSR loci were used to characterize all individuals: PA(ATT)2 (Saumitou-Laprade et al., 2000), ssrOeUA-DCA01, ssrOeUA-DCA05, ssrOeUA-DCA08, ssrOeUA-DCA09, ssrOeUA-DCA14, ssrOeUA-DCA15, ssrOeUA-DCA18 (Sefc et al., 2000), GAPU71A (Carriero et al., 2002) and EMO03 (de la Rosa et al., 2002). We completed the molecular analyses for all accessions (in particular for wild accessions) using the methods previously described (Baali-Cherif and Besnard, 2005; Haouane et al., 2011). The congruence between initial data sets was checked by comparing 35 genotypes (both wild and cultivated accessions; data not shown) that were independently characterized with the same set of markers in two different laboratories. We also showed that the unnamed cultivar from Morocco that harbours the ptDNA haplotype L1·1 (reported by Besnard et al., 2011) displays exactly the same nuclear SSR profile as ‘Dhokar’ (Tunisia; OWGB Marrakech) and can thus be considered as the same accession.
Laperrine's olive samples were analysed with ptDNA markers as described by Besnard et al. (2011). These data and available plastid profiles for 1797 Mediterranean olive trees (Besnard et al., 2013a) – including all Mediterranean accessions here analysed with nuclear SSRs – were compiled for further analyses.
Data analyses
Genetic diversity parameters were first estimated for each group (i.e. cultivars, oleasters and Laperrine's olive). In order to account for different sample sizes (minimum 270 individuals), allelic richness (RS) was estimated for all nuclear SSR loci and ptDNA haplotypes using FSTAT v.2.9.4 (Goudet 1995). The total genetic diversity (HT; Nei, 1987) was also estimated for each locus. A Wilcoxon singed-ranks test (one sided) was used to evaluate the significance of the difference of allelic richness and gene diversity at nuclear loci between cultivars, oleasters and Laperrine's olives.
Haplotype networks on the whole plastid data set or at a clade level (lineages E1 and L1) were reconstructed with the reduced-median method implemented in NETWORK V.4.112 (Bandelt et al., 1999). For the allele coding, we followed exactly the same procedure as previously described by Besnard et al., (2013a). A ptDNA haplotype was defined by the combination of alleles from the 45 polymorphic loci.
The inter- and intrasubspecies structure of the genetic diversity was then investigated. Pairwise FST between the three taxa were computed based on nuclear SSRs using FSTAT. The significance of pairwise differentiation was assessed using standard Bonferroni corrections. The number of genetically homogeneous clusters (K) in the whole data set and for each pair of taxa (i.e. cultivars/oleasters, cultivars/Laperrine's olive, oleasters/Laperrine's olive) was then determined using a model-based clustering method implemented in STRUCTURE v.2.3.4 (Pritchard et al., 2000). Bayesian analysis was run under the admixture model for 1 000 000 generations after a burn-in period of 200 000, assuming a correlation among allele frequencies. Analyses were run for K between one and ten clusters with ten iterations for each K value. The most likely number of clusters was determined using the ad-hoc measure ΔK (Evanno et al., 2005) with the R program (R Development Core Team, 2010), whereas the similarity index between ten replicates for the same K clusters (H′) was calculated with CLUMPP v1·1.2 (Greedy algorithm; Jakobsson and Rosenberg, 2007). For each K value that was retained, each accession was assigned to their respective clusters with a posterior membership coefficient (p).
The power of model-based clustering analyses to detect admixed individuals issued from intersubspecies hybridization or successive backcrosses was assessed by simulations based on the ten nuclear microsatellite loci used in our study. Two objectives were tackled: first, we assessed the level of assignment that allows a confident detection of admixture between Mediterranean and Saharan gene pools. We tested on pairs of simulated panmictic populations (that thus can be considered as non-admixed) the level of assignment to two genetic clusters: subspecies europaea (E) and laperrinei (L). In these simulated data sets, we thus estimated the level of assignment to the wrong subspecies that occurs by chance. In practice, panmictic populations of 1000 genotypes were generated for the Laperrine's olives (L) and oleasters (E) with the software HYBRIDLAB (Nielsen et al., 2006). Alleles were randomly sampled in each taxon assuming neutrality, linkage equilibrium and random mating. Allele frequencies in our simulated matrices were constrained to be similar to our observations. The procedure was replicated ten times, and simulated genotypes were used to carry out admixture analyses with STRUCTURE as described above (considering K = 2 clusters). The level of partial admixture between Mediterranean and Saharan olive was then investigated for each data set. The distribution of the posterior membership coefficient (p) values was plotted. The maximum value for a wrong assignment that may happen by chance was determined. For the second objective, we estimated the number of generations of backcrosses that occurred for accessions that were confidently detected as advanced generation of hybrids (see below). We randomly selected genotypes of 50 individuals of Laperrine's olive (from Tassili n'Ajjer and Hoggar), 50 oleasters (from the west-central Mediterranean gene pool) and 50 olive varieties from the central Mediterranean gene pool. Aiming to exclude recent hybrids, these genotypes were selected among individuals assigned to the Laperrine's olive or oleaster populations with p > 0·99 and to olive varieties with p > 0·95 (see below). We then generated 100 genotypes of each F1 and successive backcrossed genotypes (from BC1 to BC5) with HYBRIDLAB as described above. The procedure was replicated ten times and the simulated genotypes were used to carry out admixture analyses with STRUCTURE as described above. The level of partial admixture between Mediterranean and Saharan olive was then investigated and directly compared with the observed value on admixed individuals from our data set.
RESULTS
Comparison of genetic diversity parameters between Mediterranean and Saharan olives
No significant difference is observed between cultivars and the Laperrine's olive for the allelic richness and genetic diversity (Wilcoxon signed-ranks tests, P = 0·441 and 0·759, respectively; Table 1). In contrast, we observe a marginally significantly higher allelic richness in oleaster samples than in the Laperrine's olive (Wilcoxon signed-ranks test, P = 0·059), and a significantly higher genetic diversity in oleasters than in the Laperrine's olive (Wilcoxon signed-ranks test, P = 0·046), but these results could just reflect samplings on different geographic scales for these two taxa. Oleasters also show significantly higher allelic richness and genetic diversity than Mediterranean cultivars (Wilcoxon signed-ranks test, P<0·01 for RS and HT; Table 1). A similar trend is observed on ptDNA haplotypes (but not statistically testable; Table 1).
Table 1.
Total number of alleles (Na) observed on the whole data, allelic richness (RS; computed for 270 individuals) and genetic diversity (HT) computed on ten nuclear SSR loci and ptDNA haplotypes for each taxon
| Cultivars |
Oleasters |
Laperrine′s olive |
|||||
|---|---|---|---|---|---|---|---|
| Locus | Na | RS | HT | RS | HT | RS | HT |
| Nuclear SSR | |||||||
| ssrOeUA-DCA1 | 39 | 17·6 | 0·62 | 28·9 | 0·77 | 32·0 | 0·94 |
| ssrOeUA-DCA5 | 27 | 13·1 | 0·52 | 17·8 | 0·88 | 24·0 | 0·89 |
| ssrOeUA-DCA8 | 29 | 19·2 | 0·83 | 24·7 | 0·93 | 14·0 | 0·85 |
| ssrOeUA-DCA9 | 28 | 22·7 | 0·88 | 25·4 | 0·92 | 14·0 | 0·71 |
| ssrOeUA-DCA14 | 47 | 14·3 | 0·70 | 34·6 | 0·91 | 18·0 | 0·80 |
| ssrOeUA-DCA15 | 10 | 5·6 | 0·64 | 7·7 | 0·75 | 3·0 | 0·37 |
| ssrOeUA-DCA18 | 31 | 17·8 | 0·84 | 23·9 | 0·90 | 19·0 | 0·82 |
| PA(ATT)2 | 11 | 5·0 | 0·74 | 8·6 | 0·77 | 5·0 | 0·16 |
| GAPU71A | 30 | 12·5 | 0·47 | 23·3 | 0·74 | 7·0 | 0·65 |
| EMO03 | 25 | 13·0 | 0·81 | 16·5 | 0·89 | 17·0 | 0·82 |
| Mean | 27·7 | 14·1 | 0·71 | 21·1 | 0·85 | 15·3 | 0·70 |
| ptDNA | 51 | 12·1 | 0·34 | 31·7 | 0·90 | 18·0 | 0·79 |
Distinction of maternal lineages in Saharan and Mediterranean olives
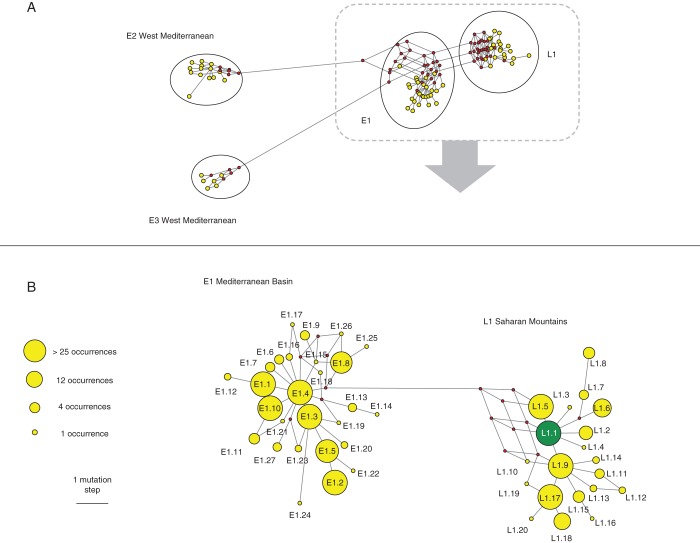
A total of 67 ptDNA haplotypes (Supplementary Data Table S3) are detected; 48 are found in the Mediterranean olive and 20 in the Laperrine's olive. The phylogenetic analysis reveals four clusters of closely related ptDNA haplotypes, or ptDNA lineages, on the whole data set (Fig. 1A). Lineages E1, E2 and E3 have been previously reported in the Mediterranean basin (Besnard et al., 2013a). Lineage L1 is sister to E1, from which it was previously not separated (Besnard et al., 2007c). A minimum of seven cpSSR mutation steps are detected between E1 and L1 haplotypes (Supplementary Data Table S3), attesting for a clear divergence of these two lineages. In the wild pool, three ptDNA lineages (namely E1, E2 and E3) are observed exclusively in oleasters, while the fourth lineage (L1) is only revealed in the Saharan populations. All cultivars, except one, show a Mediterranean haplotype. A Saharan haplotype (L1·1) is detected in ‘Dhokar’ (Fig. 1; Supplementary Data Table S2).
Fig. 1.
Plastid DNA haplotype networks reconstructed with the reduced-median method implemented in NETWORK (Bandelt et al., 1999). Haplotypes are represented by yellow or green circles, while the missing, intermediate nodes are indicated by small red dots. The length of branches is proportional to the number of mutation steps. (A) Network based on the whole data set. The Mediterranean lineages E1, E2 and E3 and the Saharan lineage L1 are indicated. (B) Haplotype network for the sister lineages E1 and L1. On this network, the name of each haplotype is indicated, and the circle size is relative to the number of observed occurrences for each haplotype. Haplotypes E1·1, E1·2 and E1·3 are the most frequent haplotypes in cultivars (Besnard et al., 2013a). Haplotype L1·1 (in green) is shared between Saharan and Mediterranean olives, but lineage L1 is primarily distributed in the Saharan Mountains. L1·1 was observed once in the Mediterranean basin (‘Dhokar’; Morocco–Tunisia).
Identification of genetic clusters in Saharan and Mediterranean olives
The genetic differentiation based on nuclear SSRs between cultivars, oleasters and the Laperrine's olives is highly significant (Table 2) and particularly high between the Mediterranean and Saharan olives (e.g. FST = 18·9 % between the Laperrine's olive and oleasters). The Bayesian clustering with STRUCTURE confirms these results, with a clear distinction of the Saharan and Mediterranean taxa (Figs 2 and 3).
Table 2.
Pairwise differentiation (FST) between the Mediterranean and Saharan olive taxa based on nuclear data
| Oleasters | Cultivars | |
|---|---|---|
| Cultivars | 7·36*** | |
| Laperrine's olive | 18·91*** | 26·61*** |
***P<0·001.
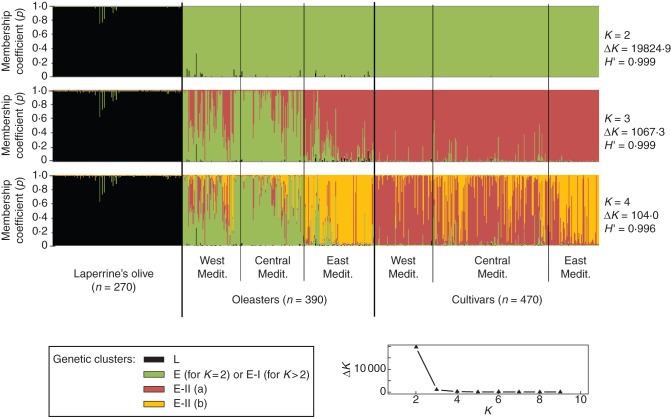
Fig. 2.
Inference of population structure based on ten nuclear SSRs using model-based Bayesian clustering implemented in STRUCTURE (Pritchard et al., 2000) on the whole data set (1130 individuals). Each vertical bar represents an individual. The membership coefficient of assignment (p) of each individual to different gene pools is shown for K = 2, K = 3 and K = 4 clusters. H′ represents the similarity coefficient between ten runs for each K, and ΔK is the ad-hoc measure of Evanno et al. (2005). The graph at the bottom right gives ΔK plotted against K. The three taxa (i.e. Laperrine's olive, oleasters and cultivars) are indicated, and three pre-defined regions are recognized for both oleasters and cultivars (Supplementary Data Tables S1 and S2). ‘West Mediterranean’ corresponds to accessions from Morocco and the Iberian Peninsula, ‘Central Mediterranean’ corresponds to accessions from Algeria to Libya and France to Continental Italy, and ‘East Mediterranean’ corresponds to accessions from Croatia to the Levant. The most likely genetic structure model is K = 2 clusters, according to ΔK and H' (ΔK = 19824·9 and H' = 0·999). At K = 2, each cluster corresponds to subspecies laperrinei and europaea (L and E, respectively), whereas at K= 3, numerous western and central oleasters (that mostly belong to cluster E-I) were shown to be distinguished from cultivars and eastern oleasters (that mostly belong to cluster E-II). At K = 4, most eastern oleasters and cultivars were distinguished from western/central cultivars (clusters E-IIa vs. E-IIb, respectively).
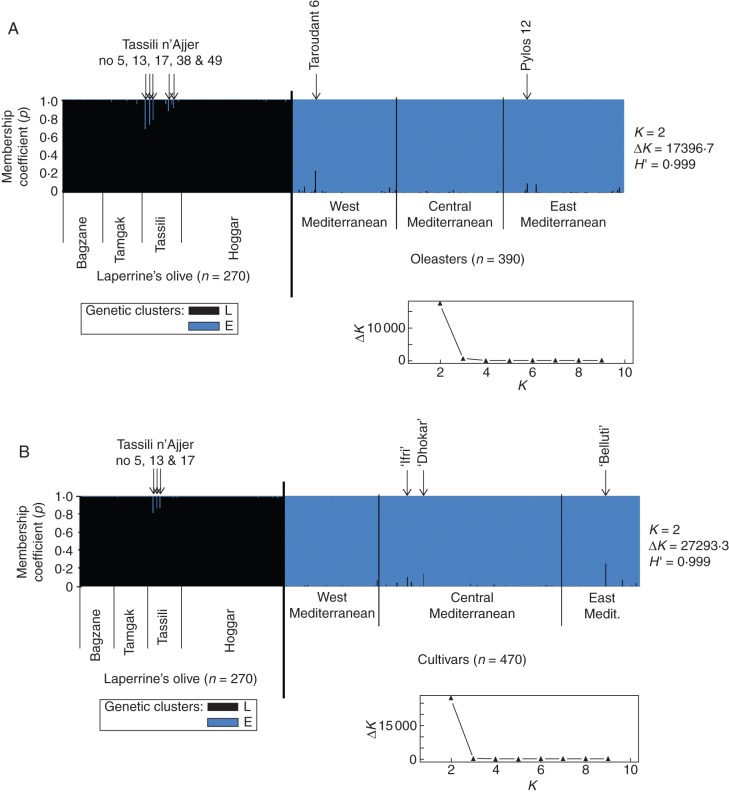
Fig. 3.
Inference of population structure based on ten nuclear SSRs using model-based Bayesian clustering implemented in STRUCTURE (Pritchard et al., 2000) for two pairwise comparisons: (A) Laperrine's olive–oleasters (660 individuals); and (B) Laperrine's olive–cultivars (740 individuals). Each vertical bar represents an individual. The membership coefficients of assignment (p) of each individual to the different clusters averaged over ten iterations is shown for K = 2 clusters (but see Supplementary Data Fig. S3 for K = 3 or 4). H′ represents the similarity coefficient between ten runs for each K, and ΔK is the ad-hoc measure of Evanno et al. (2005). The graph on the bottom right gives ΔK plotted against K. Most accessions of subspecies laperrinei and europaea are assigned to clusters L and E, respectively. The putative admixed individuals (p >0·1) are indicated by arrows.
The K value selected following the criterion defined by Evanno et al. (2005) is 2 (Fig. 2), but here we also show results for three and four clusters because they reflect sub-structures in the Mediterranean olive data set that are supported in separate analyses of Mediterranean accessions (see Supplementary Data Figs S1 and S2). At K = 2, the two clusters correspond to subspecies laperrinei and europaea (named L and E, respectively), and all individuals are assigned to their respective subspecies with a mean membership coefficient (p) >0·7. Assignments to clusters E and L is, however, not always categorical (p < 0·9 for ‘Taroudant no 6’ and a few accessions from the Tassili n'Ajjer). At K = 3, two clusters of oleasters (namely E-I and E-II) are distinguished (see also Supplementary Data Figs S1 and S2). Oleasters from the western and central Mediterranean are mainly assigned to cluster E-I, while cultivars and the remaining oleasters (from Croatia to the Levant) are mainly assigned to cluster E-II. Note that 463 (98·5 %) and 390 (83 %) cultivated olive accessions are assigned to cluster E-II with p>0·5 and 0·8, respectively. At K = 4, most of the western and central Mediterranean cultivars are distinguished from eastern Mediterranean accessions (both oleasters and cultivars; see also Supplementary Data Figs S1 and S2). A general trend for the assignment of genotypes from a given Mediterranean zone to a genetic cluster (e.g. EII-b for eastern accessions or E-I for western oleasters; see also Fig. 3) is thus observed. A separate analysis of all Mediterranean accessions also supports a soft distinction of cultivated and wild gene pools according to their geographical origin (Supplementary Data Fig. S1).
Nuclear evidence for a few admixture events between Mediterranean and Saharan olives
Based on the analysis of the whole data set, no obvious admixture between cultivars and the Laperrine's olive is revealed (see above), but the complex genetic structure in wild and cultivated Mediterranean olives could mask subtle relationships between these taxa and the Laperrine's olive (as recently shown in apple; Cornille et al., 2012). For this reason, we chose to analyse the genetic structure between oleasters and the Laperrine's olive (Fig. 3A) and between cultivars and the Laperrine's olive (Fig. 3B). In these two separate analyses, most accessions of subspecies laperrinei (263/270) and europaea (852/860) are assigned to clusters L and E, respectively, with a mean membership coefficient (p) >95 % (Fig. 3; see also Supplementary Data Fig. S3 for K = 3 or 4 clusters). This clear assignment was expected since the genetic differentiation between both subspecies is high (Table 2). Nevertheless, a few accessions were assigned to their respective subspecies with a mean membership coefficient (p) <0·90. Considering the high genetic differentiation between Mediterranean and Saharan olive populations, such patterns may reflect admixture between gene pools E and L, but this hypothesis needs to be statistically tested (see below).
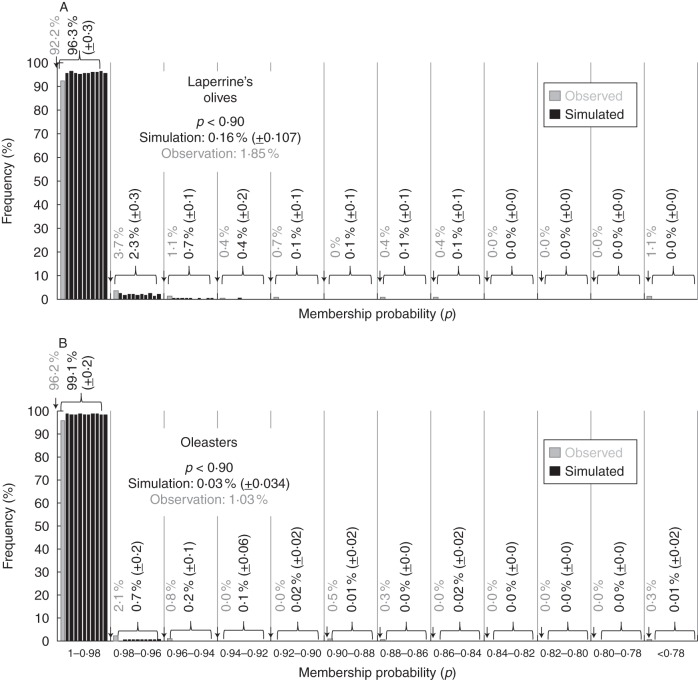
Because partial assignment to both subspecies could be not necessarily due to admixture and to some extent may arise by chance (Vähä and Primmer, 2006; Winkler et al., 2011), we analysed pairs of simulated panmictic populations of laperrinei and europaea based on observed allelic frequencies in each taxon. The assignment of genotypes to their respective cluster (i.e. L for the Laperrine's olive and E for oleasters) is correct using both observed and simulated data sets (Fig. 4). The mean membership probability (p) on the observed data is slightly lower than on simulated data. In particular, the assignment of simulated genotypes to their cluster is rarely below 0·9 (i.e. 0·16 and 0·03 % of simulated genotypes in the Laperrine's olives and oleasters, respectively) while this occurs in >1 % of observed genotypes (1·85 and 1·03 % of individuals in the Laperrine's olives and oleasters, respectively; Fig. 4). This means that when one individual is partially assigned to both subspecies with p values >0·1, this is probably not random and may reflects admixture between laperrinei and europaea.
Fig. 4.
Comparison of membership assignments (p) to the Laperrine's olive and oleaster genetic clusters on natural populations (observations) and simulated panmictic data sets (simulations; ten iterations of 1000 genotypes). A model-based Bayesian clustering implemented in STRUCTURE (Pritchard et al., 2000) was used considering K = 2 clusters. The frequency of genotypes assigned to their respective subspecies is reported for each class of values of p in the Laperrine's olives (A) and oleasters (B). Each simulated data set is individually treated. Frequencies of assignment of genotypes to their genetic cluster for p<0·9 are given on the right for simulated and observed data sets.
Based on an assignment threshold of 0·1, a few Laperrine's olive individuals from the Tassili n'Ajjer (South Algeria) show evidence of admixture with the Mediterranean cluster E (Fig. 3). Five individuals from this population are assigned to E with p >0·1, reaching a maximum of 0·317 in individual Tassili no 5 (Fig. 3A). We also find five Mediterranean accessions (three cultivars and two oleasters) that are similarly assigned to cluster L with p >0·1 (Fig. 3A, B): cultivars ‘Belluti’ (Turkey; assignment probability to cluster L, p = 0·255), ‘Ifri’ (Algeria; p = 0·105), ‘Dhokar’ (Tunisia–Morocco; p = 0·136), Oleasters ‘Pylos no 12’ (Greece; p = 0·101) and ‘Taroudant no 6’ (Morocco; p = 0·237). All putatively admixed individuals from the Sahara and Mediterranean basin are more assigned to subsp. laperrinei and subsp. europaea, respectively, suggesting they may result from early generations of backcrosses (e.g. BC1 or BC2) with the local taxon. Our simulations of backcrosses indicate that the observed level of admixture may indeed correspond to BC1 or BC2, but the confidence interval of assignment to each gene pool is large for BC generations (see details in Supplementary Data Figs S4 and S5). This is probably due to an insufficient number of loci to assess precisely the contribution of each gene pool in advanced generations of backcrosses (e.g. Vähä and Primmer, 2006). This also means that our approach may not allow detection of all BC2 generations and is inappropriate for detecting more advanced generations of hybrids.
DISCUSSION
In this study, we directly compared for the first time the genetic diversity in the cultivated olive, and wild populations of oleasters and Laperrine's olives. In the following section, patterns of genetic differentiation in the Mediterranean basin are first interpreted in the light of recent studies that confirmed a primary origin of cultivated olives in the Levant (Kaniewski et al., 2012; Zohary et al., 2012; Besnard et al., 2013a). The admixture between Mediterranean and Saharan gene pools is then discussed.
On patterns of genetic differentiation among Mediterranean olives
Both oleaster and cultivar samplings cover the whole Mediterranean area and are representative of the different gene pools present in this region (Haouane et al., 2011; Besnard et al., 2013a). We observe a reduction of 33 % of the allelic richness and 16 % of the total gene diversity for nuclear SSRs in cultivars compared with oleasters, while a reduction of 62 % of the allelic richness and total gene diversity is revealed for ptDNA haplotypes in the cultivated pool. Such a lower genetic diversity in cultivars compared with oleasters has also been revealed by Lumaret et al. (2004), Belaj et al. (2010) or Besnard et al. (2011) with different marker sets. This substantial reduction of diversity on both nuclear and plastid genomes may be interpreted as a signature of genetic erosion linked to domestication and diffusion, and, here, we can definitely conclude that all the oleaster diversity was not captured in the cultivated pool despite recurrent crop–wild gene flow (Miller and Gross, 2011).
A clear genetic differentiation between eastern and western Mediterranean oleasters is also observed in our study. This result has been reported by several authors (Angiolillo et al., 1999; Besnard et al., 2001a, 2013a; Breton et al., 2006; Rubio de Casas et al., 2006). The assignment of most cultivars (83 %) to gene pool E-II (Fig. 2; Supplementary Data Fig. S1) with p >0·8 confirms the strong eastern affiliation of the cultivated gene pool as indicated by plastid markers (Besnard et al., 2013a). Actually, no cultivar was strongly assigned to the western gene pool E-I (for p >0·8), emphasizing its secondary contribution to the olive domestication. It is usually accepted that olive domestication has primarily started in the eastern Mediterranean basin (Kaniewski et al., 2012; Zohary et al., 2012). The human-mediated dispersal of oleiculture from the East to the West with the main civilizations was then associated with secondary diversification of cultivars in the central and western Mediterranean, as reported by several authors (Besnard et al., 2001a; Belaj et al., 2002, 2012; Owen et al., 2005; Baldoni et al., 2006; Sarri et al., 2006; Breton et al., 2008; Haouane et al., 2011; Díez et al., 2012). This long story may explain the existence of a sub-structure among cultivated olives with three main gene pools (Supplementary Data Fig. S2), as recently shown by Haouane et al. (2011), Belaj et al. (2012) and Díez et al. (2012). Additional investigations are still necessary to depict processes of cultivated olive diversification, and particularly to infer the history of olive diffusion and to determine precisely the contribution of the western gene pool.
A high genetic differentiation between Mediterranean and Saharan olives
The genetic differentiation based on nuclear SSRs between the Mediterranean and Saharan olives is particularly high (Table 2), as observed by Rubio de Casas et al. (2006). In addition, no recent seed-mediated gene flow is detected between oleaster and Laperrine's olive populations that do not share any ptDNA haplotype. These patterns may result from a long isolation between the Mediterranean and Saharan regions due to a desert barrier that promoted vicariance since the Miocene–Pliocene boundary (Sepulchre et al., 2006; Besnard et al., 2007c; Migliore et al., 2012). Yet, a Saharan haplotype (L1·1) is detected in ‘Dhokar’, attesting to an indisputable contribution of the Laperrine's olive in the Maghreb cultivated gene pool.
Evidence for admixture between Mediterranean and Saharan olives
Based on ten nuclear SSR loci, we then used model-based clustering analyses to test for recent admixture between Saharan and Mediterranean olives (Figs 3 and 4). According to simulations on oleasters and the Laperrine's olives, the probability to obtain assignment of one genotype to its respective cluster inferior to 0·9 does not exceed 2 × 10−3. A partial assignment with p-values >0·1 to each cluster is thus very unlikely to be reached by chance but instead indicates admixture between these taxa (Fig. 4). Based on this very conservative threshold, five Laperrine's olive individuals and five Mediterranean accessions (two oleasters and three cultivars, namely ‘Dhokar’, ‘Ifri’ and ‘Belluti’) show assignment to both clusters L and E (Fig. 3A, B). This analysis thus confirms that the Maghreb cultivar ‘Dhokar’, that has a Saharan maternal origin (see above), results from admixture between Laperrine and Mediterranean olives, but other wild and cultivated admixed accessions are detected, and particularly in the Tassili n'Ajjer population. Such reciprocal gene flow was also strongly suspected on an AFLP profiling (Rubio de Casas et al., 2006). These ten admixed individuals may result from early generations of backcrosses (e.g. BC1 to BC2) within the local taxon as supported by additional simulations (Supplementary Data Figs S4 and S5).
The Laperrine's olive has thus already been involved in the cultivated olive diversification, confirming its potential use for the olive breeding (Lavee and Zohary, 2011; Besnard et al., 2012). Interestingly, due to their singular nuclear SSR profiles, cultivars ‘Dhokar’ and ‘Ifri’ were included in the core collections defined by Haouane et al. (2011), while ‘Belluti’ was not analysed in this study. Moreover, the level of admixture between subsp. europaea and subsp. laperrinei is relatively low, since no hybrid of the first generation is detected and only ten individuals out of 1130 (<1 %) are recognized as early generations of hybrids (for p >0·1). This suggests that hybrids of the first generation are relatively rare and/or that their fitness is poor. In addition, successive backcrosses also greatly reduce the contribution of a wild progenitor, hampering its identification based on a few molecular markers as in the present study. Our approach based on ten nuclear loci is inappropriate for detecting advanced generations of hybrids (Supplementary Data Figs S4 and S5) as previously demonstrated by Vähä and Primmer (2006). In order to assess precisely the importance of the laperrinei introgression in the cultivated olive gene pool, our results could be refined with a genome scan analysis (Scascitelli et al., 2010; Zhao et al., 2010).
We can also question the vectors of dispersal between the Mediterranean basin and the Saharan mountains. Both natural and human-mediated vectors could be involved. The aridity on the Sahara has greatly increased during the last five millennia (Maley, 2011), contributing to a high isolation of the Saharan olive populations during recent times. Ancient gene flow between the two investigated taxa could be still detectable due to the slow turnover of populations (Baali-Cherif and Besnard, 2005). Yet, the only evidence of maternal introgression was detected in a cultivar that has been collected from Tunisia to Morocco. This observation indicates that humans managed some hybrids and spread them over long distances.
Lastly, the Laperrine's olive is a highly restricted and endangered plant (Besnard et al., 2012). Evidence of gene flow between Saharan and Mediterranean olive taxa needs to be taken into account by genetic resource managers and breeders. The Laperrine's olive population is a putatively important wild genetic resource of the cultivated olive, and thus deserves to be protected in situ. This wild taxon should be also included in ex situ collections for further evaluation of agronomic traits.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
This work was funded by the fellowships PIEF-GA-2008-220813 and ANR-12-AGRI-0002 (ARIMNET 2011-PESTOLIVE). It has been conducted in Silwood Park, the UMR AGAP and the lab EDB, part of the LABEX entitled TULIP (ANR-10-LABX-41). We also thank two referees for their constructive comments.
LITERATURE CITED
- Angiolillo A, Mencuccini M, Baldoni L. Olive genetic diversity assessed using amplified fragment length polymorphisms. Theoretical and Applied Genetics. 1999;98:411–421. [Google Scholar]
- Baali-Cherif D, Besnard G. High genetic diversity and clonal growth in relict populations of Olea europaea subsp. laperrinei (Oleaceae) from Hoggar, Algeria. Annals of Botany. 2005;96:823–830. doi: 10.1093/aob/mci232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldoni L, Tosti N, Ricciolini C, et al. Genetic structure of wild and cultivated olives in the Central Mediterranean Basin. Annals of Botany. 2006;98:935–942. doi: 10.1093/aob/mcl178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Beaumont M, Barratt EM, Gottelli D, et al. Genetic diversity and introgression in the Scottish wildcat. Molecular Ecology. 2001;10:319–336. doi: 10.1046/j.1365-294x.2001.01196.x. [DOI] [PubMed] [Google Scholar]
- Belaj A, Satovic Z, Rallo L, Trujillo I. Genetic diversity and relationships in olive (Olea europaea L.) germplasm collections as determined by randomly amplified polymorphic DNA. Theoretical and Applied Genetics. 2002;105:638–644. doi: 10.1007/s00122-002-0981-6. [DOI] [PubMed] [Google Scholar]
- Belaj A, Muñoz-Diez C, Baldoni L, Porceddu A, Barranco D, Satovic Z. Genetic diversity and population structure of wild olives from the north-western Mediterranean assessed by SSR markers. Annals of Botany. 2007;100:449–458. doi: 10.1093/aob/mcm132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaj A, Muñoz-Diez C, Baldoni L, Satovic Z, Barranco D. Genetic diversity and relationships of wild and cultivated olives at regional level in Spain. Scientia Horticulturae. 2010;124:323–330. [Google Scholar]
- Belaj A, del Carmen Dominguez-García M, Atienza SG, et al. Developing a core collection of olive (Olea europaea L.) based on molecular markers (DArTs, SSRs, SNPs) and agronomic traits. Tree Genetics and Genomes. 2012;8:365–378. [Google Scholar]
- Besnard G, Baradat P, Breton C, Khadari B, Bervillé A. Olive domestication from structure of oleasters and cultivars using nuclear RAPDs and mitochondrial RFLPs. Genetics, Selection, Evolution. 2001a;33:S251–S268. [Google Scholar]
- Besnard G, Baradat P, Chevalier D, Tagmount A, Bervillé A. Genetic differentiation in the olive complex (Olea europaea) revealed by RAPDs and RFLPs in the rRNA genes. Genetic Resources and Crop Evolution. 2001b;48:165–182. [Google Scholar]
- Besnard G, Christin PA, Baali-Cherif D, Bouguedoura N, Anthelme F. Spatial genetic structure in the Laperrine's olive (Olea europaea subsp. laperrinei), a long-living tree from the central Saharan mountains. Heredity. 2007c;99:649–657. doi: 10.1038/sj.hdy.6801051. [DOI] [PubMed] [Google Scholar]
- Besnard G, Henry P, Wille L, Cooke D, Chapuis E. On the origin of the invasive olives (Olea europaea L. Oleaceae) Heredity. 2007d;99:608–619. doi: 10.1038/sj.hdy.6801037. [DOI] [PubMed] [Google Scholar]
- Besnard G, Rubio de Casas R, Vargas P. Plastid and nuclear DNA polymorphism reveals historical processes of isolation and reticulation in the olive tree complex (Olea europaea) Journal of Biogeography. 2007;34:736–752. [Google Scholar]
- Besnard G, García-Verdugo C, Rubio de Casas R, Treier U, Galland N, Vargas P. Polyploidy in the olive complex (Olea europaea): evidence from flow cytometry and nuclear microsatellite analyses. Annals of Botany. 2008;101:25–30. doi: 10.1093/aob/mcm275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard G, Hernández P, Khadari B, Dorado G, Savolainen V. Genomic profiling of plastid DNA variation in the Mediterranean olive tree. BMC Plant Biology. 2011;11:80. doi: 10.1186/1471-2229-11-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard G, Anthelme F, Baali-Cherif D. The Laperrine's olive tree (Oleaceae): a wild genetic resource of the cultivated olive and a model-species for studying the biogeography of the Saharan Mountains. Acta Botanica Gallica – Botany Letters. 2012;159:319–328. [Google Scholar]
- Besnard G, Khadari B, Navascués M, et al. The complex history of the olive tree: from Late Quaternary diversification of Mediterranean lineages to primary domestication in the northern Levant. Proceedings of the Royal Society B: Biological Sciences. 2013a;280:20122833. doi: 10.1098/rspb.2012.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard G, Dupuy J, Larter M, Cuneo P, Cooke D, Chikhi L. History of the invasive African olive tree in Australia and Hawaii: evidence for sequential bottlenecks and hybridizations with the Mediterranean olive. Evolutionary Applications. 2013b doi: 10.1111/eva.12110. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton C, Tersac M, Bervillé A. Genetic diversity and gene flow between the wild olive (oleaster, Olea europaea L.) and the olive: several Plio-Pleistocene refuge zones in the Mediterranean basin suggested by simple sequence repeats analysis. Journal of Biogeography. 2006;34:1916–1928. [Google Scholar]
- Breton C, Terral JF, Pinatel C, Médail F, Bonhomme F, Bervillé A. The origins of the domestication of the olive tree. Comptes Rendus Biologies. 2008;332:1059–1064. doi: 10.1016/j.crvi.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Carriero F, Fontanazza G, Cellini F, Giorio G. Identification of simple sequence repeats (SSRs) in olive (Olea europaea L.) Theoretical and Applied Genetics. 2002;104:301–307. doi: 10.1007/s001220100691. [DOI] [PubMed] [Google Scholar]
- Chevalier A. L'origine de l'Olivier cultivé et ses variations. Revue Internationale de Botanique Appliquée et d'Agriculture Tropicale. 1948;28:1–25. [Google Scholar]
- Cornille A, Gladieux P, Smulders MJM, et al. New insight into the history of domesticated apple: secondary contribution of the European wild apple to the genome of cultivated varieties. PLoS Genetics. 2012;8:e1002703. doi: 10.1371/journal.pgen.1002703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delplancke M, Alvarez N, Benoit L, et al. Evolutionary history of almond tree domestication in the Mediterranean basin. Molecular Ecology. 2013;22:1092–1104. doi: 10.1111/mec.12129. [DOI] [PubMed] [Google Scholar]
- Díez CM, Imperato A, Rallo L, Baranco D, Trujillo I. Worldwide core collection of olive cultivars based on simple sequence repeat and morphological markers. Crop Science. 2012;52:211–221. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Goudet J. FSTAT (Version 1.2): a computer program to calculate F-statistics. Journal of Heredity. 1995;86:485–486. [Google Scholar]
- Green PS. A revision of Olea L. Kew Bulletin. 2002;57:91–140. [Google Scholar]
- Hannachi H, Sommerlate H, Breton C, et al. Oleaster (var. sylvestris) and subsp. cuspidata are suitable genetic resources for improvement of the olive (Olea europaea subsp. europaea var. europaea) Genetic Resources and Crop Evolution. 2009;56:393–403. [Google Scholar]
- Haouane H, El Bakkali A, Moukhli A, et al. Genetic structure and core collection of the World Olive Germplasm Bank of Marrakech: towards the optimised management and use of Mediterranean olive genetic resources. Genetica. 2011;139:1083–1094. doi: 10.1007/s10709-011-9608-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Kaniewski D, Van Campo E, Boiy T, Terral JF, Khadari B, Besnard G. Primary domestication and early uses of the emblematic olive tree: palaeobotanical, historical and molecular evidences from the Middle East. Biological Reviews. 2012;87:885–899. doi: 10.1111/j.1469-185X.2012.00229.x. [DOI] [PubMed] [Google Scholar]
- Khadari B, Charafi J, Moukhli A, Ater M. Substantial genetic diversity in cultivated Moroccan olive despite a single major cultivar: a paradoxical situation evidenced by the use of SSR loci. Tree Genetics and Genomes. 2008;4:213–221. [Google Scholar]
- Lavee S, Zohary D. The potential of genetic diversity and the effect of geographically isolated resources in olive breeding. Israel Journal of Plant Sciences. 2011;59:3–13. [Google Scholar]
- Lopes MS, Mendonça D, Sefc KM, Gil FS, da Câmara Machado A. Genetic evidence of intra-cultivar variability within Iberian olive cultivars. HortScience. 2004;39:1562–1565. [Google Scholar]
- Lumaret R, Ouazzani N, Michaud H, Vivier G, Deguilloux MF, Di Giusto F. Allozyme variation of oleaster populations (wild olive tree) (Olea europaea L.) in the Mediterranean Basin. Heredity. 2004;92:343–351. doi: 10.1038/sj.hdy.6800430. [DOI] [PubMed] [Google Scholar]
- Maley J. Climate and palaeoenvironment evolution North Tropical Africa from the end of Tertiary to the upper Quaternary. Palaeoecology of Africa. 2011;30:227–278. [Google Scholar]
- Migliore J, Baumel A, Juin M, Médail F. From Mediterranean shores to central Saharan mountains: key phylogeographical insights from the genus. Myrtus. Journal of Biogeography. 2012;39:942–956. [Google Scholar]
- Miller AJ, Gross BL. From forest to field: perennial fruit crop domestication. American Journal of Botany. 2011;98:1389–1414. doi: 10.3732/ajb.1000522. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Newberry PE. On some African species of the genus Olea and the original home of the cultivated olive-tree. Proceedings of the Linnean Society of London. 1937;150:3–16. [Google Scholar]
- Nielsen EEG, Bach LA, Kotlicki P. HYBRIDLAB (version 1·0): a program for generating simulated hybrids from population samples. Molecular Ecology Notes. 2006;6:971–973. [Google Scholar]
- Oliver D. Flora of tropical Africa. Ashford, UK: Reeve L & Co; 1868. [Google Scholar]
- Owen CA, Bita EC, Banilas G, et al. AFLP reveals structural details of genetic diversity within cultivated olive germplasm from the Eastern Mediterranean. Theoretical and Applied Genetics. 2005;110:1169–1176. doi: 10.1007/s00122-004-1861-z. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure from multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2010. www.r-project.org . [Google Scholar]
- Rodríguez-Ramillo ST, Wang J. The effect of close relatives on unsupervised Bayesian clustering algorithms in population genetic structure analysis. Molecular Ecology Resources. 2012;12:873–884. doi: 10.1111/j.1755-0998.2012.03156.x. [DOI] [PubMed] [Google Scholar]
- de la Rosa R, James CM, Tobutt KR. Isolation and characterization of polymorphic microsatellites in olive (Olea europaea L.) and their transferability to other genera in the Oleaceae. Molecular Ecology Notes. 2002;2:265–267. [Google Scholar]
- Rubio de Casas R, Besnard G, Schönswetter P, Balaguer L, Vargas P. Extensive gene flow blurs phylogeographic but not phylogenetic signal in Olea europaea L. Theoretical and Applied Genetics. 2006;113:575–583. doi: 10.1007/s00122-006-0306-2. [DOI] [PubMed] [Google Scholar]
- Sarri V, Baldoni L, Porceddu A, et al. Microsatellite markers are powerful tools for discriminating among olive cultivars and assigning them to geographically defined populations. Genome. 2006;49:1606–1615. doi: 10.1139/g06-126. [DOI] [PubMed] [Google Scholar]
- Saumitou-Laprade P, Vassiliadis C, Epplen JT, Hardt C. Isolation of microsatellite loci for paternity in Phillyrea angustifolia L. (Oleaceae) Molecular Ecology. 2000;9:112–114. doi: 10.1046/j.1365-294x.2000.00764-4.x. [DOI] [PubMed] [Google Scholar]
- Scascitelli M, Whitney KD, Randell RA, King M, Buerkle CA, Rieseberg LH. Genome scan of hybridizing sunflowers from Texas (Helianthus annuus and H. debilis) reveals asymmetric patterns of introgression and small islands of genomic differentiation. Molecular Ecology. 2010;19:521–541. doi: 10.1111/j.1365-294x.2009.04504.x. [DOI] [PubMed] [Google Scholar]
- Sefc KM, Lopes MS, Mendonça D, et al. Identification of microsatellite loci in olive (Olea europaea) and their characterization in Italian and Iberian olive trees. Molecular Ecology. 2000;9:1171–1173. doi: 10.1046/j.1365-294x.2000.00954.x. [DOI] [PubMed] [Google Scholar]
- Sepulchre P, Ramstein G, Fluteau F, Schuster M, Tiercelin JJ, Brunet M. Tectonic uplift and Eastern Africa aridification. Science. 2006;313:1419–1423. doi: 10.1126/science.1129158. [DOI] [PubMed] [Google Scholar]
- Terral JF. France: Université Montpellier II; 1997. La domestication de l'olivier (Olea europaea L.) en Méditerranée nord-occidentale: approche morphométrique et implications paléoclimatiques. PhD. [Google Scholar]
- Turrill WB. Wild and cultivated olives. Kew Bulletin. 1951;3:437–442. [Google Scholar]
- Vähä JP, Primmer CR. Efficiency of model-based Bayesian methods for detecting hybrid individuals under different hybridization scenarios and with different numbers of loci. Molecular Ecology. 2006;15:63–72. doi: 10.1111/j.1365-294X.2005.02773.x. [DOI] [PubMed] [Google Scholar]
- Winkler KA, Pamminger-Lahnsteiner B, Wanzenböck J, Weiss S. Hybridization and restricted gene flow between native and introduced stocks of Alpine whitefish (Coregonus sp.) across multiple environments. Molecular Ecology. 2011;20:456–472. doi: 10.1111/j.1365-294X.2010.04961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Wright M, Kimball J, et al. Genomic diversity and introgression in O. sativa reveal the impact of domestication and breeding on the rice genome. PLoS One. 2010;5:e10780. doi: 10.1371/journal.pone.0010780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohary D, Hopf M, Weiss E. Domestication of plants in the Old World: the origin and spread of cultivated plants in Southwest Asia, Europe, and the Mediterranean basin. Oxford: Oxford University Press; 2012. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.