Abstract
Background and Aims
Non-S-ribonucleases (non-S-RNases) are class III T2 RNases constitutively expressed in styles of species with S-RNase-based self-incompatibility. So far, no function has been attributed to these RNases. The aim of this work is to examine if NnSR1, a non-S-RNase from Nicotiana alata, is induced under conditions of phosphate (Pi) deprivation. The hypothesis is that under Pi-limited conditions, non-S-RNase functions may resemble the role of S-like RNases. To date, the only RNases reported to be induced by Pi deficiency are class I and class II S-like RNases, which are phylogenetically different from the class III clade of RNases.
Methods
Gene and protein expression of NnSR1 were assayed in plants grown hydroponically with and without Pi, by combining RT-PCR, immunoblot and enzymatic activity approaches.
Key Results NnSR1
transcripts were detected in roots 7 d after Pi deprivation and remained stable for several days. Transcript expression was correlated based on Pi availability in the culture medium. Antiserum against a peptide based on a hypervariable domain of NnSR1 recognized NnSR1 in roots and stems but not leaves exposed to Pi shortage. NnSR1 was not detected in culture medium and was pelleted with the microsomal fraction, suggesting that it was membrane-associated or included in large compartments. The anti-NnSR1 inhibited selectively the enzymatic activity of a 31-kDa RNase indicating that NnSR1 was induced in an enzymatically active form.
Conclusions
The induction of NnSR1 indicates that there is a general recruitment of all classes of T2 RNases in response to Pi shortage. NnSR1 appears to have regained ancestral functions of class III RNases related to strategies to cope with Pi limitation and also possibly with other environmental challenges. This constitutes the first report for a specific function of class III RNases other than S-RNases.
Keywords: Class III RNases, gene induction, Nicotiana alata, phosphate deficiency, S-RNases, stress responses
INTRODUCTION
Ribonucleases (RNases) of the T2 family are secreted endoribonucleases highly conserved in all kingdoms of life which catalyse the cleavage of single-strand RNA. Despite this common biochemical function, T2 RNases have many different biological roles (Deshpande and Shankar, 2002; Luhtala and Parker, 2010). In plants, T2 RNases are classified into two subfamilies, S-RNases and S-like RNases, according to their function (Green, 1994). S-RNases are associated with the pollen rejection system in self-incompatible species of Solanaceae, Rosaceae and Plantaginaceae (McClure et al., 1989). They show high allelic polymorphism and pistil-specific expression, and are both the molecular determinant for pollen recognition (Lee et al., 1994; Murfett et al., 1994) and the cytotoxic factor for self-pollen rejection, mediated by growth inhibition and RNA degradation of incompatible pollen tubes (McClure et al., 1990). Supporting these findings, the catalytic active site of S-RNases is essential for self-incompatibility (SI) manifestation (Royo et al., 1994; Huang et al., 1994). In contrast, S-like RNases are widely distributed in self-compatible as well as in self-incompatible species of all plant families examined. They are expressed in many tissues and are not involved in the self-pollen recognition process. Plant S-like RNases were implicated in diverse biological responses, often related to stress induction, and to development and senescence (reviewed by MacIntosh, 2011). For instance, they are induced in several species in response to pathogenic fungi and bacteria (Galiana et al., 1997; Hugot et al., 2002; MacIntosh et al., 2010), virus infections (Kurata et al., 2002; Ohno and Ehara, 2005) and insect feeding (Bodenhausen and Reymond, 2007; MacIntosh et al., 2010). Many S-like RNases are also induced as a consequence of abiotic stress, such as phosphate (Pi) deficiency (see below), after mechanical wounding (LeBrasseur et al., 2002; Köck et al., 2004; Hillwig et al., 2008), during the development of tracheary elements (Ye and Droste, 1996; Lehmann et al., 2001) and in response to senescence stimuli (Lers et al., 2006). Conversely, other S-like RNases showed high constitutive expression; consequently, they were thought to perform a housekeeping role (MacIntosh et al., 2010). Arabidopsis RNS2 proved to be necessary for rRNA degradation to a level compatible with cellular homeostasis (Hillwig et al., 2011a). Constitutive expression also occurs in tomato RNase LER (Köthke and Köck, 2011) and the rice OsRNS2 gene (MacIntosh et al., 2010). In most of these cases, the expression of S-like RNases contributes to Pi mobilization from nucleic acids for nutritional, healing, recycling and defence functions (MacIntosh, 2011).
Further distinction between S-like RNases and S-RNases was established from evolutionary studies. Igic and Kohn (2001) classified plant T2 RNases into three classes, based on phylogenetic analysis and the number and position of gene introns: class I and class II include S-like RNases, while class III groups S-RNases and non-S-RNases. Non-S-RNases are structurally very similar to S-RNases and, like them, are specifically expressed in the pistil (Lee et al., 1992; Liang et al., 2003; Banović et al., 2009; Roldán et al., 2010). However, like S-like RNases, they are not polymorphic or functional in the SI recognition system (Roalson and McCubbin, 2003; Kao and Tsukamoto, 2004). Non-S-RNases probably originated by duplication from S-RNase genes and subsequent translocation away from the SI locus (Golz et al., 1998; Igic and Kohn, 2001). The physiological role of these groups of RNases, often called relic S-RNases, remains unknown. A putative role related to plant defence was suggested for some class III non-S-RNases from the genera Pisum, Luffa and Momordica (Igic and Kohn, 2001), as well as for non-S-RNases and S-RNases found in Petunia nectar (Hillwig et al., 2010, 2011b).
Five conserved regions have been established in the amino acids of plant T2 RNases, termed C1–C5 (Ioerger et al., 1991; Green, 1994). The C2 and C3 domains, also called CAS I and CAS II (conserved active site), are almost absolutely conserved in all T2 RNases and conform the active site (Irie, 1999). While C1, C4 and C5 domains are somewhat different between S-RNases and S-like RNases, Vieira et al. (2008) determined four amino acid patterns between CAS I and CAS II that allowed an accurate distinction between S-RNases and S-like RNases. Two of these patterns are exclusively found in S-RNases while the other two are included in the vast majority of S-like RNases but not in S-RNases.
Morphological and metabolic changes occur when plants grow in a Pi-deprived environment, a condition not unusual in natural ecosystems due to the low solubility of Pi in the soil (Yang and Finnegan, 2010; Péret et al., 2011). The induction of T2 RNase genes in several species of plants under Pi deprivation has been reported (reviewed by MacIntosh, 2011; Ivanov and Anderson, 2011). For instance, tomato LE and LX RNase and tobacco NE RNase were induced in cultivated cells or root seedlings grown in Pi-deficient media (Nürnberger et al., 1990; Köck et al., 1995, 2006; Dodds et al., 1996). Similarly, RNS1 and RNS2 were highly expressed in Arabidopsis seedlings in response to Pi limitation (Taylor et al., 1993; Bariola et al., 1994). Induced S-like RNases may contribute to recycling Pi both from extracellular and from intracellular RNA sources (Jost et al., 1991; Löffler et al., 1993; Bariola et al., 1999; Hillwig et al., 2011a). Thus far, most S-like RNases induced by low Pi belong to class I RNases, with a few exceptions belonging to class II RNases (MacIntosh et al., 2010). No class III RNase has been reported to be induced by Pi deficit.
Recently, a non-S-RNase gene sequence isolated from styles was identified in all the individuals examined of a natural population of Nicotiana alata (Roldán et al., 2010). This sequence, termed NnSR1 (Nicotiana non-S-RNase1) in this work, grouped into class III RNases with around 80 % identity to the functional S70-RNase sequence.
Here we examine whether NnSR1, a class III RNase, is induced under Pi deprivation, resembling the functionality of class I and II S-like RNases. We demonstrate that NnSR1 expression was mainly induced in roots in an enzymatically active form in N. alata plants subjected to Pi deprivation. To our knowledge, this is the first report of a specific function for a class III RNase, other than the role of S-RNases in the SI reaction. The induction of NnSR1 indicates that there is a general recruitment of RNases, including those of class III, to Pi mobilization from nucleic acids in plants exposed to Pi deficiency.
MATERIALS AND METHODS
Plant material and growth conditions
Nicotiana alata plants used in this study come from a natural population described by Roldán et al. (2010). Plants were grown in a chamber at 28 °C under white fluorescent and high-pressure sodium lights (150–200 µmol m−2 s−1) suspended 1 m above the plants with 16/8-h light/dark period. Seeds were germinated and grown for 20 d in Petri dishes on sterilized soil. Sets of nine selected plants were removed from soil and hydroponically cultivated in recipients containing 3 litres of complete Hoagland's solution containing 1·5 mm KNO3, 1·25 mm Ca(NO3)2, 0·75 mm MgSO4, 0·5 mm KH2PO4, 50 µm KCl, 10 µm MnSO4, 2 µm ZnSO4, 1·5 µm CuSO4, 72 µm Fe-EDTA, 75 nm (NH4)6Mo7O24, at pH 6. Nutrient solution was changed weekly and distilled water was added to replenish water loss every 2 d. After 15 d of acclimatization, healthy plants at the vegetative stage with 6–8 leaves were divided into two sets and cultured for different periods in complete Hoagland's solution or in Hoagland's solution containing no Pi. After culture plants were separated into different organs, frozen in liquid nitrogen and stored at –80 °C until use.
RNA extraction and RT-PCR conditions
For each culture period, roots from three representative plants cultivated with or without Pi were mixed and total RNA was extracted according to McClure et al. (1990) or using a SpectrumTM Plant RNA total Kit (Sigma, St Louis, MO, USA). RNA was treated with DNase (RQ1 RNase-free DNase; Promega, Madison, WI, USA) to remove contaminating genomic DNA. For RT-PCR amplification, 1 µg of total RNA and 5·5 µm oligo (dT)15 were incubated for 5 min at 70 °C, chilled in ice water for 5 min, and mixed with 6 mm MgCl2, 0·5 mm dNTPs, 1 µL of ImProm-II™ Reverse Transcriptase and ImProm-II™ buffer (Promega) in a final volume of 20 µL. Single-stranded cDNA synthesis was performed for 1 h at 42 °C, according to the manufacturer's instructions. A 2-μL aliquot of cDNA synthesis reaction was used as template for the amplification step, performed in a 20-μL reaction containing 0·4 units of GoTaq® Flexi DNA polymerase (Promega), GreenGoTaq Flexi buffer®, 1·5 mm MgCl2, 0·2 mm dNTPs and 12·5–33 pg DNA. All degenerate and specific primers used are detailed in Supplementary Data Table S1. Primers were used at 0·4 and 1·2 µm for specific and degenerate primers, respectively. The reaction was incubated at 95 °C for 5 min and then at 32–40 cycles of 1 min at 94 °C, 45 s at the annealing temperature indicated in Supplementary Data Table S1 and 1 min at 72 °C, followed by a final extension step of 5 min at 72 °C. Amplified fragments were analysed on agarose gels and assessed by densitometry with Gel-ProTM Analyzer 3·0 software. Transcript relative abundances were referred to actin transcript. For sequencing, amplified fragments were purified from agarose gels and cloned into pGEM-Teasy vector (Promega). Recombinant plasmids were analysed by restriction enzyme digestion and both DNA strands from two or more plasmids were sequenced with standard SP6 or T7 promoter primers (Macrogen Inc., MD, USA). Sequence alignment was performed using CLUSTAL W version 1·81 (Thompson et al., 1997).
Tissue extraction, subcellular fractionation and Pi content measurement
For each culture period, roots, stems, leaves and pistils from three representative plants cultivated with or without Pi were mixed. Samples were ground to a fine powder in liquid nitrogen using a pestle and mortar. The powder was extracted on ice by homogenizing 10 g with 1 mL of 10 mm Tris-HCl at pH 7·5 containing 1 mm MgCl, 2 mm KCl, 3 mm EDTA, 1 mm EGTA, 5 % (v/v) glycerol, 0·25 m sucrose, 0·1 mm PMSF, 2 µg mL−1 trypsin soybean inhibitor and 1 µg mL−1 aprotinin (buffer A). After 15 min centrifugation at 16 000g at 4 °C the pellet was discarded and the supernatant was stored at –80 °C until use. Protein content was estimated with the Bradford reagent (Bio-Rad, Richmond, CA, USA). Root extracts were usually 1 g L−1 in concentration.
Subcellular fractionation was performed on homogenates from roots exposed to Pi- deprivation for 14 d. All fractionation steps were carried out at 4 °C. The sample was centrifuged at 16 000g for 15 min and the pellet (P16), was washed, resuspended in buffer A and stored at –80 °C until use. The supernatant was subjected to additional centrifugation at 110 000g for 50 min. The supernatant (SN110) was saved and the pellet (P110) was washed and resuspended in buffer A. The three fractions, P16, P110 and SN110, were analysed by SDS-PAGE and Western blot, as detailed below.
The Pi content of roots, stems, leaves and pistils was analysed on tissue homogenates by the method of phosphomolybdenum blue reaction using a Shimadzu BioSpect-mini spectrophotometer (Shimadzu Corp., Kyoto, Japan). The amount of free available Pi in each tissue was calculated on a fresh weight basis.
In-gel ribonuclease activity
Electrophoresis for in-gel RNase activity staining was performed according to Yen and Green (1991), including torula yeast RNA (2·4 mg ml−1) in the separating gel and 0·1 and 2 % (w/w) SDS in the electrophoresis-running buffer and sample-loading buffer, respectively. After electrophoresis, gels were successively washed, incubated, stained and destained. SDS was removed from gels by two washes of 10 min each with 25 % (v/v) isopropanol in 10 mm Tris-HCl at pH 7·0. Isopropanol was then washed twice with 10 mm Tris-HCl at pH 7·0 and gels were incubated in 0·1 m Tris-HCl at pH 6·8 at 51 °C for 4 h. Following incubation, gels were rinsed twice with 10 mm Tris-HCl at pH 7·0 and stained for 10 min with 0·2 % (w/v) toluidine blue (Sigma) in 10 mm Tris-HCl. Finally, gels were destained with 10 mm Tris-HCl at pH 7·0 three times for 20 min each and kept in 10 mm Tris-HCl at pH 7·0 containing 10 % (v/v) glycerol. Band signals were visualized with a UVP EC3 bioimaging system (UVP Inc., Upland, CA, USA) and assessed by densitometry with Gel-ProTM Analyzer 3.0 software.
Antibody preparation, Western blot and analytical procedures
The peptide sequence GETFTKLREPREKKE corresponding to the hypervariable region A of NnSR1 (pNnSR1) was synthesized and coupled to KLH or BSA by Genbiotech (Buenos Aires, Argentina). Two rabbits were immunized with 0·6 mg of KLH- or BSA-pNnSR1 and boosted three times at intervals of 3 weeks. Both sera specifically recognized the pNnSR1 (Supplementary Data Fig. S1). Proteins from crude extracts were separated by 15 % SDS-PAGE of 1·5 mm thickness and stained with Coomassie blue R-250 or electrotransferred onto nitrocellulose membranes (AmershamTM HybondTM-ECL, GE Healthcare Bio-Sciences, Uppsala, Sweden) and blocked for 2 h at 4 °C with 4 % skimmed milk in PBS buffer. Membranes were then incubated overnight at 4 °C with anti-pNnSR1 serum (1 : 3000 unless otherwise indicated) in PBS containing 0·1 % Tween 20. After washing them five times with PBS–0·1 % Tween 20, membranes were reincubated with IRDye 800CW-conjugated anti-rabbit secondary antibody (LICOR Biosciences, Lincoln, NE, USA) in PBS containing 1 % skimmed milk. Following six washes with PBS, band signals were visualized with an Odyssey infrared imaging system (LICOR Biosciences) and then assessed by densitometry with Gel-ProTM Analyzer 3·0 software.
To confirm the identity of the immunodetected band in root extracts of Pi-deprived plants, the KLH-pNnSR1 antiserum (1 : 10 000) was preincubated overnight at 4 °C with 3 × 10−10 mol of BSA and 3 × 10−10 mol of BSA-conjugated pNnSR1 dotted onto nitrocellulose strips. Subsequently, the serum was incubated with root protein extracts electroblotted onto nitrocellulose as indicated above.
For in-gel inhibition of NnSR1 activity, root protein extracts were loaded onto 15 % SDS-PAGE of 1·0 mm thickness. After running, the gel was incubated overnight at 4 °C with (1 : 100) anti-pNnSR1, preimmune sera and PBS–0·5 % Tween 20. RNase activity staining was then assayed as indicated above.
To test root excretion of NnSR1, 350 mL of hydroponic culture medium was clarified by centrifugation for 10 min at 16 000g. The supernatant was passed through a 0·22-μm filter, concentrated in an Amicon Stirred Cell 8050 filter device (10k molecular weight cut-off, Millipore Corp., Bedford, MA, USA) and then dialysed overnight against PBS. Additional concentration was achieved using an Amicon Ultra-0·5 mL centrifugal filter device (10k molecular weight cut-off; Millipore). Aliquots representing 3 % of total culture medium (no protein detected by the Bradford assay) and root extract (20 µg protein) were assayed by Western blot as detailed above.
Statistical analysis
The data shown are the mean ± s.e.m. of at least three independent experiments. Statistical analysis were carried out through Student's t test for comparison between two groups of data and one-way ANOVA followed by Tukey's test to test significant differences in multiple comparisons. Statistical analyses were performed using the computer-based statistical GraphPad Prism V5·0.
RESULTS
Transcription of a class III non-S-RNase is induced in Nicotiana under Pi deprivation
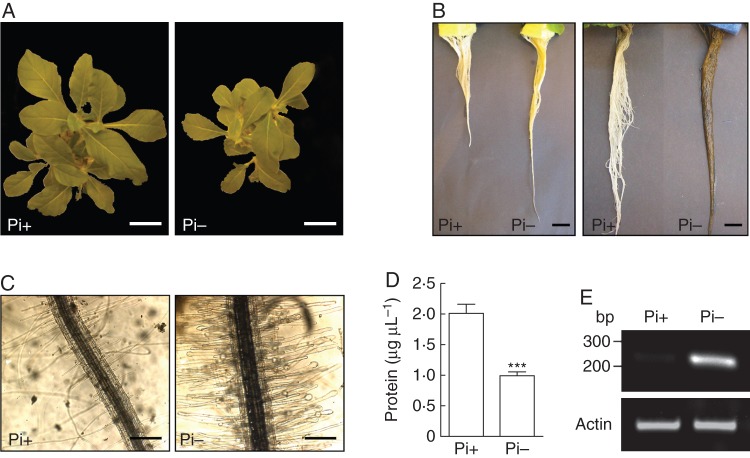
Nicotiana alata young plants were hydroponically cultivated in the absence of Pi for 14 d. Typical responses to Pi deprivation were observed in the stressed plants with respect to control plants supplied with Pi: (1) the aerial part growth was notably lower (Fig. 1A; Köck et al., 2006); (2) roots were longer and exhibited a dark red coloration, attributed to anthocyanin accumulation (Fig. 1B; Trull et al., 1997); (3) root hairs were much more abundant and showed frequent swollen tips (Fig. 1C; Ma et al., 2001); and (4) the protein content in root extracts decreased by almost 50 % (Fig. 1D). As expected, Pi deprivation strongly induced the transcript expression of class I S-like RNase NE (Fig. 1E), an RNase involved in Pi remobilization in N. alata previously reported (Dodds et al., 1996). Overall, these results confirmed that Nicotiana plants were clearly stressed by limited Pi supply.
Fig. 1.
Effects of Pi deprivation in Nicotiana alata. Young plants were hydroponically cultivated and subjected to Pi deficit for 14 d. Typical responses from Pi-starved plants (Pi-) compared with Pi-supplied plants (Pi+) confirmed the effectiveness of the treatment. (A) General growth of aerial part. Scale bar = 5 cm. (B) General root growth and dark red staining of roots, denoting anthocyanin accumulation. Scale bar = 2 cm. (C) Root hair density and morphology. Scale bar = 0·3 mm. (D) Protein content in root homogenates. Values are the mean ± s.e.m. of five independent experiments. Data were statistically analysed using the t-test. ***P < 0·001. (E) RT-PCR amplification from roots using specific primers for S-like RNase NE.
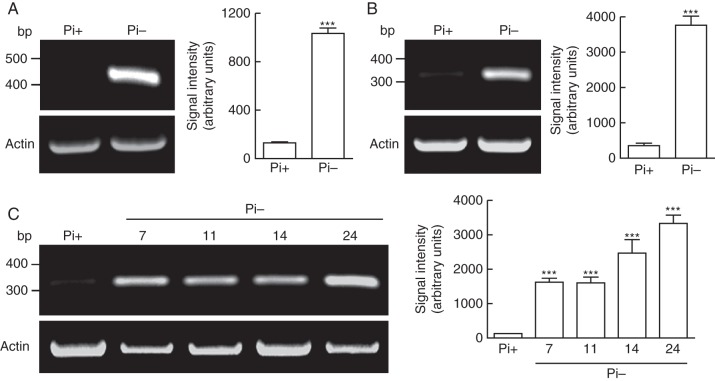
The only plant T2 RNases reported to be induced by Pi shortage were class I and class II S-like-RNases (MacIntosh, 2011). To test whether class III RNases are also induced by Pi deficit, we used RT-PCR with degenerate primers based on S-RNase conserved domains C2, C4 and C5 of N. alata (Supplementary Data Table S1; Ioerger et al., 1991; Roldán et al., 2010). Domains C4 and C5 of Nicotiana S-RNases are somewhat less conserved in S-like RNases (Green, 1994). Using two different combinations of primers (C2–C4 and C2–C5), a single band with the expected size was amplified in each case from roots grown without Pi, suggesting the involvement of class III RNases in the Pi response (Fig. 2A; Supplementary Data Fig. S2). The C2–C5 amplified band was cloned and analysed by sequencing. Only one species was identified, which was almost identical to a coding sequence for an RNase isolated from N. alata styles (GenBank accession no. D63887·1; Kuroda et al., 1994). This RNase was demonstrated not to be functional in the SI system (GenBank accession no. GQ850520·1; Roldán et al., 2010). Thus, we named this gene NnSR1 (Nicotiana non-S-RNase1). RT-PCR with NnSR1-specific primers and subsequent sequencing of the amplified fragment confirmed the selective induction of this gene in Pi-deprived roots, while the expression in roots grown in Pi-supplemented medium was hardly detectable (Fig. 2B). The induction of NnSR1 transcripts was detected after 7 d of Pi deprivation and was sustained for several days, with some increase on day 24 (Fig. 2C). Interestingly, another non-S-RNase cDNA sequence (GenBank accession no. GQ375151·1), termed NnSR2, was not induced by the absence of Pi (Supplementary Data Fig. S3), suggesting that these non-S-RNases had a different functional evolution. Deduced amino acid sequences of both NnSR1 and NnSR2 showed the typical amino acid patterns that distinguish class III S-RNases from class I and class II S-like-RNases (Supplementary Data Fig. S4; Vieira et al., 2008).
Fig. 2.
Expression of class III NnSR1 transcript induced by Pi deprivation in Nicotiana alata roots. (A) RT-PCR amplification from roots exposed to 14 d of Pi deprivation. Degenerate primers designed from C2 and C5 conserved domains of Nicotiana S-RNases were used. (B) Similar to (A) but using NnSR1-specific primers. (C) RT-PCR amplification using NnSR1-specific primers from roots. Numbers indicate the days of exposure to Pi starvation. Representative gels and the statistical analysis are shown. Signal intensity values represent the mean ± s.e.m. of three independent experiments. Data were analysed using the t-test (A, B) or one-way ANOVA followed by Tukey test (C). ***P < 0·001. Pi-, Pi-deficient medium; Pi+, Pi-sufficient medium.
The NnSR1 protein is induced in roots and stems in an enzymatically active form
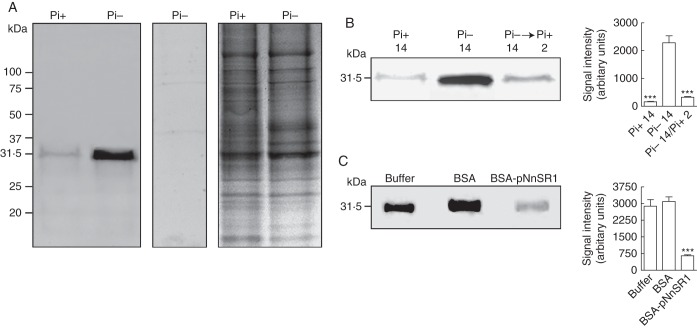
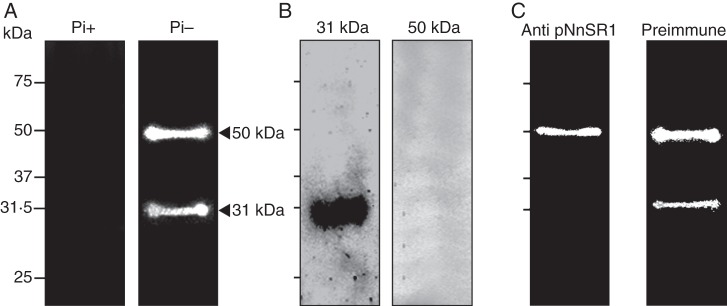
We developed a monospecific polyclonal antibody against a synthetic peptide based on the sequence of hypervariable region A of NnSR1 (heretofore abbreviated to pNnSR1). Hypervariable regions of S-RNases have been successfully used to generate highly specific antibodies recognizing only the corresponding allelic variant (Matton et al., 1999; Goldraij et al., 2006). The antisera generated against both BSA- or KLH-conjugated pNnSR1 contained antibodies that recognized the peptide sequence (Supplementary Data Fig. S1). Both antisera also recognized a single protein band of 31 kDa in root extracts of plants grown under Pi deprivation, while a very faint or no signal was detected in roots supplied with Pi-sufficient medium (Fig. 3A). The size of the protein band was in good agreement with the molecular weight of S-RNases, which ranges between 30 and 35 kDa. However, the predicted molecular weights of precursor and mature NnSR1 proteins are around 25 and 23 kDa, respectively (GenBank accession no. D63887·1). Thus, the apparent molecular weight estimated by Western blot may be caused by N-glycosylation at the Asn 49 site, close to the CAS I domain of the NnSR1 precursor. This site is commonly glycosylated in T2 class III RNases (MacIntosh, 2011).
Fig. 3.
Expression of class III NnSR1 protein induced in Nicotiana alata roots under Pi deprivation. NnSR1 induction was assayed by Western blot in roots subjected to Pi deprivation for 14 d. Anti KLH-pNnSR1 serum was used as probe. (A) Induction of a 31-kDa protein in root homogenates of Pi-starved plants. Controls using preimmune serum (central panel) and Coomassie blue staining (left panel) are shown. The result is representative of three independent experiments. Similar result was obtained using the anti-BSA-pNnSR1 serum. (B) Root homogenates assayed after Pi-starved plants were transferred to Pi-supplied medium for 2 d. (C) Root homogenates assayed after preincubation of anti KLH-pNnSR1 serum with buffer and with 3 × 10−10 mol of BSA and 3 × 10−10 mol of BSA-conjugated pNnSR1. Thirty micrograms of protein was loaded in each lane. Signal intensity values represent the mean ± s.e.m. of three independent experiments. The data in (B) and (C) were statistically analysed using one-way ANOVA followed by Tukey test: ***P < 0·001. Pi-, Pi-deficient medium; Pi+, Pi-sufficient medium.
Expression of the induced protein appeared to be dependent on the presence of Pi in the culture medium. The signal was decreased almost to basal values found in Pi-supplied plants by 2 d after the plants grown in Pi-deficient medium were moved to a Pi-sufficient medium (Fig. 3B). To confirm that the antibodies specifically generated against the pNnSR1 were involved in the recognition of the 31-kDa protein, the anti-KLH-pNnSR1 was separately incubated with BSA and BSA-pNnSR1 conjugate before Western blot analysis. The 31-kDa signal was reduced about 80 % when the serum was preincubated with the BSA-pNnSR1 conjugate before incubation with the Pi-deficient root extract (Fig. 3C). Taken together, these experiments provided compelling evidence that the 31-kDa protein induced in roots under Pi deficiency was NnSR1.
The spatial distribution of NnSR1 was analysed by comparing equal percentages of root tissue homogenate and plant growth medium in a protein gel blot. No signal was detected in the culture medium, indicating minor or no excretion of NnSR1 from the roots (Fig. 4A). Additional fractionation showed that NnSR1 was mostly excluded from the soluble cytosol/intercellular fluid fraction. NnSR1 was pelleted with the microsomal fraction, suggesting that it was associated with membranes or included in large compartments that were not broken during root homogenization (Fig. 4B).
Fig. 4.
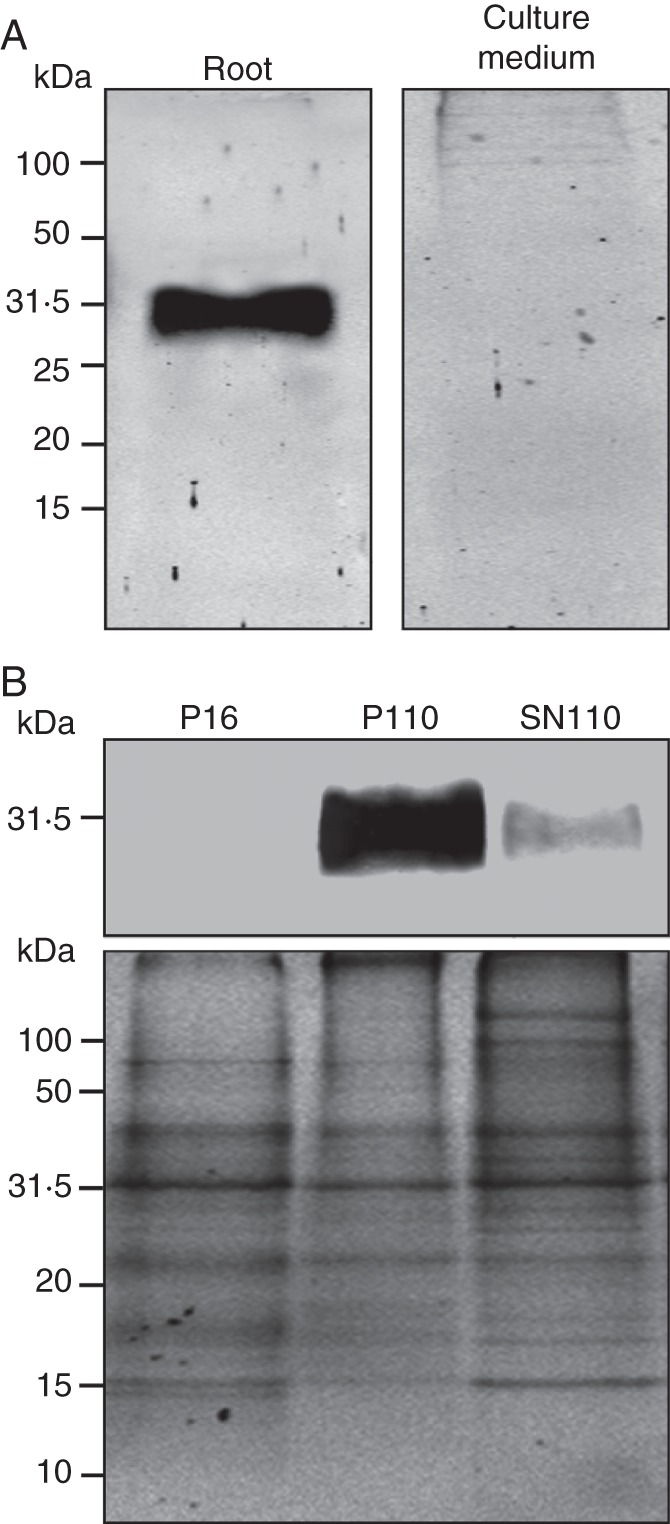
Fate of induced NnSR1. NnsR1 was assayed by protein gel blot in plants cultivated for 14 d in a Pi-deprived medium. Anti-KLH-pNnSR1 serum was used as probe. (A) Comparison of equal percentages of total root homogenate and culture medium. (B) Fractionation of root tissues by high-speed centrifugation. P16, 16 000g pellet; P110, 110 000g pellet; SN110, 110 000g supernatant. Coomassie blue staining is shown. Thirty micrograms of protein was loaded in each lane.
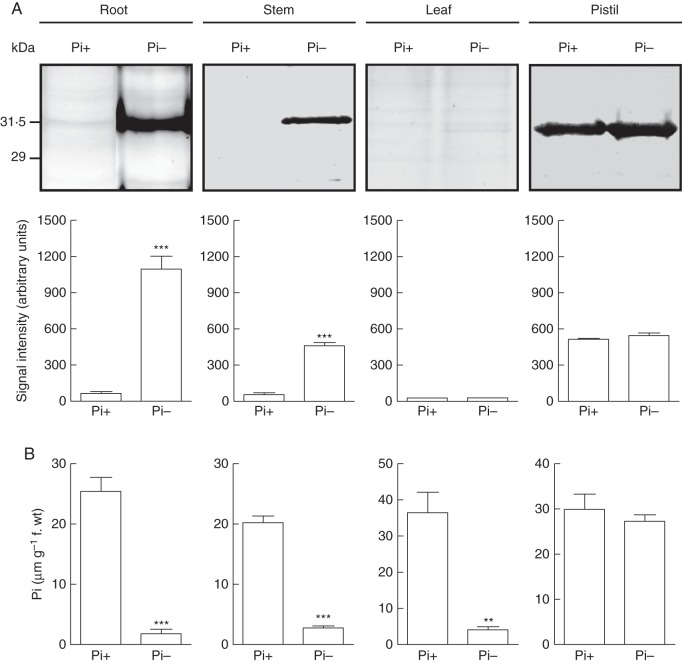
The induction of NnSR1 under Pi deprivation was evaluated in different tissues (Fig. 5). Besides roots, NnSR1 was induced in stems but not in leaves, where no signal was detected. The induction was not seen in the pistil either; however, consistent with the large abundance of NnSR1 expression in styles (Roldán et al., 2010), similar intensity signals of NnSR1 were detected in styles of both Pi-deprived and Pi-supplied plants. No signal of NnSR1 was detected in sepals or petals of plants cultivated with or without Pi (Supplementary Data Fig. S5). In parallel with NnSR1 induction, the tissue Pi content was determined. Except in pistils, where the Pi content was similar in plants cultured with and without Pi, all the tissues examined showed a pronounced decrease of the Pi content in plants exposed to Pi-deficient conditions.
Fig. 5.
Induction of NnSR1 and Pi content in different organs. Plants were grown for 14 d with or without Pi. (A) NnsR1 was assayed by protein gel blot in roots, stems, leaves and pistils. The anti-BSA-pNnSR1 serum was used as probe. Thirty micrograms of protein was loaded in each lane. Signal intensity values represent the mean ± s.e.m. of three independent experiments. (B) Pi content of roots, stems, leaves and pistils. The values represent the mean ± s.e.m. of three independent experiments. Data were analysed using the t-test. ***P < 0·001, **P < 0·05). Pi-, Pi-deficient medium; Pi+, Pi-sufficient medium.
It is thought that induction of T2 RNases under Pi limitation is related to Pi mobilization from RNA. Therefore, it was important to determine whether the induced NnSR1 was enzymatically active. To address this possibility, in-gel RNase activity was assayed in roots of N. alata. Two bands of 31 and 50 kDa were seen in Pi-deprived roots only, while no RNase activity was detected in roots grown in Pi-containing medium (Fig. 6A). To associate the RNase activity with NnSR1, a thin gel slice from the two bands was cut and loaded separately onto an SDS-PAGE. Electrophoresis followed by Western blot analysis revealed a single band in the lane containing the RNase activity of 31 kDa but not in the one loaded with the activity band of 50 kDa (Fig. 6B). This result strongly suggested that NnSR1 was induced in an enzymatically active form. Further confirmation was obtained from a complementary experiment in which a native gel loaded with Pi-deficient root extracts was incubated with the anti-pNnSR1 antibody. Subsequent in-gel RNase activity assay showed that the activity of the 31-kDa band was selectively inhibited by the antibody (Fig. 6C). This result confirmed that the activity band of the 31-kDa protein corresponded to the NnSR1.
Fig. 6.
RNase activity of NnSR1. (A) In-gel RNase activity assayed on roots cultivated for 14 d in a Pi-deprived medium. (B) Band slices of 31 and 50 kDa from A were separately loaded onto an SDS-PAGE, blotted onto nitrocellulose and probed with anti KLH-pNnSR1 serum. (C) In-gel RNase activity assayed as in A after preincubation of the gel with anti-KLH-pNnSR1 and preimmune sera. Thirty micrograms of protein was loaded in each lane. The result is representative of three independent experiments. Pi-, Pi-deficient medium; Pi+, Pi-sufficient medium.
DISCUSSION
NnSR1, a class III non-S-RNase, is induced under Pi limitation
We have demonstrated here the induction of T2 NnSR1, a class III non-S-RNase, under conditions of Pi deprivation in the self-incompatible species N. alata. It is well known that Pi deficiency promotes the induction of RNases in many plant species, presumably to recycle Pi from nucleic acid sources. However, all RNases described so far to be involved in this function have been class I and class II S-like RNases. Within T2 RNases, the class III S-RNase subfamily is phylogenetically divergent from the S-like RNase subfamily. The class III S-RNase subfamily includes the S-RNases and the non-S-RNases of self-incompatible species and the relic S-RNases of self-compatible species. Moreover, to the best of our knowledge, this is the first specific evidence of a physiological role for a class III non-S-RNase.
NnSR1 exhibited both structural and functional traits of S-RNases and S-like RNases. Like S-RNases, the deduced amino acid sequence of NnSR1 showed five conserved domains, C1–C5, and two hypervariable regions, Hva and Hvb (Kuroda et al., 1994; Roldán et al., 2010). Moreover, NnSR1 displayed the amino acid patterns attributed to S-RNases that distinguish them from S-like-RNases (Vieira et al., 2008; Supplementary Data Fig. S4). Despite these resemblances, however, NnSR1 was monomorphic and consequently not functional in the SI system (Roldán et al., 2010). Although the expression of this protein under normal growth conditions was similar to that of S-RNases, being exclusively restricted to the pistil (Fig. 5), it changed dramatically when plants were exposed to Pi deprivation. Under this condition, the expression of NnSR1 mimicked that of S-like RNases, especially class I RNases, which are typically induced in a variety of stress scenarios (Ivanov and Anderson, 2011; MacIntosh, 2011), mostly contributing to Pi mobilization from RNA. Exogenous RNA was effective in sustaining tomato cell growth (Abel et al., 2000) or growth of Arabidopsis seedlings (Chen et al., 2000), in parallel with a strong induction of class I S-like RNases (Nürnberger et al., 1990; Löffler et al., 1993). Consistently, the exogenous RNA (or DNA) was consumed from culture medium and ribonucleosides were generated, suggesting strongly that RNA degradation by RNase activity supported the growth of these cultured cells and seedlings. The NnSR1 also appeared to be induced to participate in Pi rescue, exhibiting a strong expression in roots and stems under Pi deprivation (Figs 2A, 3A and Fig. 5). This response was, in turn, almost fully reversed when Pi was resupplied to the culture medium (Fig. 3B). The fact that NnSR1 was expressed in an enzymatically active form (Fig. 6) reinforced the idea that it was induced specifically to recycle Pi from nucleic acids. Therefore, NnSR1 induction seems to be part of a general recruitment mechanism of RNases to cope with Pi limitation. Thus far, two RNases of different phylogenetic origins were reported to be induced in N. alata under Pi shortage: RNase NE, a class I S-like-RNase (Dodds et al., 1996); and NnSR1, the class III non-S-RNase reported in this work. Whether the induction of these two RNases is simultaneous or sequential will be the focus of future research, as well as the study of the Pi threshold triggering such induction.
As expected, the Pi content in roots, stems and leaves was significantly lower in plants grown for 14 d in a Pi-deficient medium. However, the NnSR1 induction was not uniform in these organs (Fig. 5). NnSR1 was detected mainly in roots and, to a lesser degree, in stems; however, it was not detectable in leaves. The latter is not surprising, given that an important fraction of genes modulated by Pi availability exhibited different or even contrasting expression in roots and leaves (Wu et al., 2003). Moreover, after 12–14 d of Pi deprivation, RNase NE and RNase LX were induced in roots but not in leaves of N. alata and Solanum lycopersicum plants, respectively (Dodds et al., 1996; Köck et al., 2006). Similarly, the expression of RNS1 was significantly up-regulated in Arabidopsis leaves but only marginally induced in shoots after 100 h of Pi deprivation (Hammond et al., 2003). Regardless, it is clear that distinct strategies may be used by different organs in response to a Pi shortage.
Like S-RNases, NnSR1 exhibited a constitutive expression in pistils that was not affected by Pi limitation (Fig. 5A). It has been proposed that expression of class III RNase genes in pistils may be related to defensive functions (Liang et al., 2003). Recently, two class III RNases genes from Petunia hybrida, Phy3 and Phy4, were also related to potential defence functions in pistils and nectar (Hillwig et al., 2010). In addition, given its resemblances to S-RNases and its pistil expression, it will be interesting to determine whether NnSR1 plays some role in pollen RNA degradation during the SI reaction.
In self-incompatible species, class III RNases with no functionality in the SI system probably originated from a duplication of S-RNase genes with a subsequent translocation from the S-locus (Golz et al., 1998; Kao and Tsukamoto, 2004). In particular, NnSR1 was derived from the S70-RNase, a functional S allele isolated from a natural population of N. alata. The two RNases shared around 81 % identity and phylogenetic analysis placed them together, supported by a maximal bootstrap value (Roldán et al., 2010). Importantly, the induction of NnSR1 as part of the response to Pi shortage implies that duplication from S-RNase was accompanied by a change of function. The acquired function resembles that of class I S-like RNases, which are typically induced by biotic and abiotic stress (MacIntosh et al., 2010; MacIntosh, 2011). This functional divergence could be reciprocal to the one that occurred ancestrally, as the S-RNase-based SI system probably originated from defence RNases of floral parts (Hiscock et al., 1996; Nasrallah, 2005; Hillwig et al., 2011b). If the SI system evolved by duplication and sub-functionalization from an ancestral RNase involved in biotic (and presumably abiotic) defence responses, then duplication of S-RNases to non-S-RNases may be conceived as the recovery of an ancestral function of class III RNases.
In conclusion, we report here that in self-incompatible Nicotiana a paralogous copy of S70-RNase termed NnSR1 is induced under Pi-deficient conditions, demonstrating a specific functional role for a class III RNase, other than the function of S-RNases in the SI reaction. NnSR1 was enzymatically active and presumably contributes to Pi mobilization from nucleic acids, as was described for S-like RNases. NnSR1 appears to have regained ancestral functions of class III RNases related to strategies to cope with environmental challenges. Further research will establish whether non-S-RNases are also induced in other developmental and stress scenarios in which S-like RNases are typically involved.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank Gabriela Diaz Cortez for editorial and language assistance. This work was supported by funds from Agencia Nacional de Promoción Científica y Tecnológica (PICT 32933), Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICET, PIP 11220090100265) and Secretaría de Ciencia y Técnica (SECyT) from Universidad Nacional de Córdoba (05/C466) to A.G. A.G. is a member of CONICET, Argentina. H.J.R. is a fellowship recipient of SECyT-Universidad Nacional de Córdoba, Argentina. J.A.R. is a fellowship recipient of CONICET, Argentina.
LITERATURE CITED
- Abel S, Nürnberger T, Ahnert V, Krauss G-J, Glund K. Induction of an extracellular cyclic nucleotide phosphodiesterase as an accessory ribonucleolytic activity during phosphate starvation of cultured tomato cells. Plant Physiology. 2000;122:543–552. doi: 10.1104/pp.122.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banović B, Šurbanovski N, Konstantinović M, Maksimović V. Basic RNases of wild almond (Prunus webbii): cloning and characterization of six new S-RNase and one “non-S RNase” genes. Journal of Plant Physiology. 2009;166:395–402. doi: 10.1016/j.jplph.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Bariola PA, Howard CJ, Taylor CB, Verburg MT, Jaglan VD, Green PJ. The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. Plant Journal. 1994;6:673–685. doi: 10.1046/j.1365-313x.1994.6050673.x. [DOI] [PubMed] [Google Scholar]
- Bariola PA, Macintosh GC, Green PJ. Regulation of S-like ribonuclease levels in Arabidopsis. Antisense inhibition of RNS1 or RNS2 elevates anthocyanin accumulation. Plant Physiology. 1999;119:331–342. doi: 10.1104/pp.119.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenhausen N, Reymond P. Signaling pathways controlling induced resistance to insect herbivores in Arabidopsis. Molecular Plant-Microbe Interactions. 2007;20:1406–1420. doi: 10.1094/MPMI-20-11-1406. [DOI] [PubMed] [Google Scholar]
- Chen DL, Delatorre CA, Bakker A, Abel S. Conditional identification of phosphate starvation-response mutants in Arabidopsis thaliana. Planta. 2000;211:13–22. doi: 10.1007/s004250000271. [DOI] [PubMed] [Google Scholar]
- Deshpande RA, Shankar V. Ribonucleases from T2 family. Critical Reviews in Microbiology. 2002;28:79–122. doi: 10.1080/1040-840291046704. [DOI] [PubMed] [Google Scholar]
- Dodds PN, Clarke AE, Newbigin E. Molecular characterisation of an S-like RNase of Nicotiana alata that is induced by phosphate starvation. Plant Molecular Biology. 1996;31:227–238. doi: 10.1007/BF00021786. [DOI] [PubMed] [Google Scholar]
- Galiana E, Bonnet P, Conrod S, et al. RNase activity prevents the growth of a fungal pathogen in tobacco leaves and increases upon induction of systemic acquired resistance with elicitin. Plant Physiology. 1997;115:1557–1567. doi: 10.1104/pp.115.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldraij A, Kondo K, Lee CB, et al. S-RNase compartmentalization and HT-B degradation in self-incompatible Nicotiana. Nature. 2006;439:805–810. doi: 10.1038/nature04491. [DOI] [PubMed] [Google Scholar]
- Golz JF, Clarke AE, Newbigin E, Anderson M. A relic S-RNase is expressed in the styles of self-compatible Nicotiana sylvestris. The Plant Journal. 1998;16:591–599. doi: 10.1046/j.1365-313x.1998.00331.x. [DOI] [PubMed] [Google Scholar]
- Green PJ. The ribonucleases of higher plants. Annual Reviews of Plant Physiology and Plant Molecular Biology. 1994;45:421–445. [Google Scholar]
- Hammond JP, Malcolm JB, Bowen HC, et al. Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiology. 2003;132:578–596. doi: 10.1104/pp.103.020941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillwig MS, LeBrasseur ND, Green PJ, MacIntosh GC. Impact of transcriptional, ABA-dependent, and ABA-independent pathways on wounding regulation of RNS1 expression. Molecular Genetics and Genomics. 2008;280:249–261. doi: 10.1007/s00438-008-0360-3. [DOI] [PubMed] [Google Scholar]
- Hillwig MS, Liu X, Liu G, Thornburg RW, MacIntosh GC. Petunia nectar proteins have ribonuclease activity. Journal of Experimental Botany. 2010;61:2951–2965. doi: 10.1093/jxb/erq119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillwig MS, Contento AL, Meyer A, Ebany D, Bassham DC, MacIntosh GC. RNS2, a conserved member of the RNase T2 family, is necessary for ribosomal RNA decay in plants. Proceedings of the National Academy of Sciences USA. 2011a;108:1093–1098. doi: 10.1073/pnas.1009809108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillwig MS, Kanobe C, Robert W., Thornburg RW, MacIntosh GC. Identification of S-RNase and peroxidase in Petunia nectar. Journal of Plant Physiology. 2011b;168:734–738. doi: 10.1016/j.jplph.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Hiscock SJ, Kües U, Dickinson HG. Molecular mechanisms of self-incompatibility in flowering plants and fungi: different means to the same end. Trends in Cell Biology. 1996;6:421–428. doi: 10.1016/s0962-8924(96)10037-4. [DOI] [PubMed] [Google Scholar]
- Huang S, Lee HS, Karunanandaa B, Kao T-h. Ribonuclease activity of Petunia inflata S proteins is essential for rejection of self-pollen. The Plant Cell. 1994;6:1021–1028. doi: 10.1105/tpc.6.7.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugot K, Ponchet M, Marais A, Ricci P, Galiana E. A tobacco S-like RNase inhibits hyphal elongation of plant pathogens. Molecular Plant-Microbe Interactions. 2002;15:243–250. doi: 10.1094/MPMI.2002.15.3.243. [DOI] [PubMed] [Google Scholar]
- Igic B, Kohn JR. Evolutionary relationships among self-incompatibility RNases. Proceedings of the National Academy of Sciences USA. 2001;98:13167–13171. doi: 10.1073/pnas.231386798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioerger TR, Gohlke JR, Xu B, Kao T-h. Primary structural features of the self-incompatbility protein in Solanaceae. Sexual Plant Reproduction. 1991;4:81–87. [Google Scholar]
- Irie M. Structure–function relationships of acid ribonucleases. Lysosomal, vacuolar, and periplasmic enzymes. Pharmacology and Therapeutics. 1999;81:77–89. doi: 10.1016/s0163-7258(98)00035-7. [DOI] [PubMed] [Google Scholar]
- Ivanov P, Anderson P. Stress-induced ribonucleases. In: Nicholson AW, editor. Ribonucleases, nucleic acids and molecular biology. Berlin: Springer-Verlag; 2011. pp. 115–119. [Google Scholar]
- Jost W, Bak H, Glund K, Terpstra P, Beintema JJ. Amino acid sequence of an extracellular, phosphate-starvation-induced ribonuclease from cultured tomato (Lycopersicon esculentum) cells. European Journal of Biochemistry. 1991;198:1–6. doi: 10.1111/j.1432-1033.1991.tb15978.x. [DOI] [PubMed] [Google Scholar]
- Kao T-h, Tsukamoto T. The molecular and genetic bases of S-RNase-based self-incompatibility. The Plant Cell. 2004;16:72–83. doi: 10.1105/tpc.016154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köck M, Löffler A, Abel S, Glund K. cDNA structure and regulatory properties of a family of starvation-induced ribonucleases from tomato. Plant Molecular Biology. 1995;27:477–485. doi: 10.1007/BF00019315. [DOI] [PubMed] [Google Scholar]
- Köck M, Groß N, Stenzel I, Hause G. Phloem-specific expression of the wound-inducible ribonuclease LE from tomato (Lycopersicon esculentum cv. Lukullus) Planta. 2004;219:233–242. doi: 10.1007/s00425-004-1227-4. [DOI] [PubMed] [Google Scholar]
- Köck M, Stenzel I, Zimmer A. Tissue-specific expression of tomato ribonuclease LX during phosphate starvation-induced root growth. Journal of Experimental Botany. 2006;57:3717–3726. doi: 10.1093/jxb/erl124. [DOI] [PubMed] [Google Scholar]
- Köthke S, Köck M. The Solanum lycopersicum RNaseLER is a class II enzyme of the RNase T2 family and shows preferential expression in guard cells. Journal of Plant Physiology. 2011;168:840–847. doi: 10.1016/j.jplph.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Kurata N, Kariu T, Kawano S, Kimura M. Molecular cloning of cDNAs encoding ribonuclease-related proteins in Nicotiana glutinosa leaves, as induced in response to wounding or to TMV-infection. Bioscience, Biotechnology, and Biochemistry. 2002;66:391–397. doi: 10.1271/bbb.66.391. [DOI] [PubMed] [Google Scholar]
- Kuroda S, Norioka S, Mitta M, Kato L, Sakiyama F. Primary structure of a novel stylar RNase unassociated with self-incompatibility in tobacco plant. Nicotiana alata. Journal of Protein Chemistry. 1994;13:438–439. [Google Scholar]
- LeBrasseur ND, MacIntosh GC, Pérez-Amador MA, Saitoh M, Green PJ. Local and systemic wound-induction of RNase and nuclease activities in Arabidopsis: RNS1 as a marker for a JA-independent systemic signaling pathway. The Plant Journal. 2002;29:393–403. doi: 10.1046/j.1365-313x.2002.01223.x. [DOI] [PubMed] [Google Scholar]
- Lee HS, Singh A, Kao T-h. RNase X2, a pistil-specific ribonuclease from Petunia inflata, shares sequence similarity with solanaceous S proteins. Plant Molecular Biology. 1992;20:1131–1141. doi: 10.1007/BF00028899. [DOI] [PubMed] [Google Scholar]
- Lee H-S, Huang S, Kao T-h. S proteins control rejection of incompatible pollen in Petunia inflata. Nature. 1994;367:560–563. doi: 10.1038/367560a0. [DOI] [PubMed] [Google Scholar]
- Lehmann K, Hause B, Altmann D, Köck M. Tomato ribonuclease LX with the functional endoplasmic reticulum retention motif HDEF is expressed during programmed cell death processes, including xylem differentiation, germination, and senescence. Plant Physiology. 2001;127:436–449. [PMC free article] [PubMed] [Google Scholar]
- Lers A, Sonego L, Green PJ, Burd S. Suppression of LX ribonuclease in tomato results in a delay of leaf senescence and abscission. Plant Physiology. 2006;142:710–721. doi: 10.1104/pp.106.080135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Huang J, Xue Y. Identification and evolutionary analysis of a relic S-RNase in Antirrhinum. Sexual Plant Reproduction. 2003;16:17–22. [Google Scholar]
- Löffler A, Glund K, Irie M. Amino acid sequence of an intracellular, phosphate-starvation-induced ribonuclease from cultured tomato (Lycopersicon esculentum) cells. European Journal of Biochemistry. 1993;214:627–633. doi: 10.1111/j.1432-1033.1993.tb17962.x. [DOI] [PubMed] [Google Scholar]
- Luhtala N, Parker R. T2 family ribonucleases: ancient enzymes with diverse roles. Trends in Biochemical Sciences. 2010;35:253–259. doi: 10.1016/j.tibs.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Bielenberg DG, Brown KM, Lynch JP. Regulation of root hair density by phosphorus availability in Arabidopsis thaliana. Plant, Cell and Environment. 2001;24:459–467. [Google Scholar]
- MacIntosh GC. RNase T2 family: enzymatic properties, functional diversity, and evolution of ancient ribonucleases. In: Nicholson AW, editor. Ribonucleases, nucleic acids and molecular biology. Berlin: Springer-Verlag; 2011. pp. 89–114. [Google Scholar]
- MacIntosh GC, Hillwig MS, Meyer A, Lex Flagel L. RNase T2 genes from rice and the evolution of secretory ribonucleases in plants. Molecular Genetics and Genomics. 2010;283:381–396. doi: 10.1007/s00438-010-0524-9. [DOI] [PubMed] [Google Scholar]
- Matton DP, Luu DT, Xike Q, et al. Production of an S RNase with dual specificity suggests a novel hypothesis for the generation of new S alleles. The Plant Cell. 1999;11:2087–2097. doi: 10.1105/tpc.11.11.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure BA, Haring V, et al. Style self-incompatibility gene products of Nicotlana alata are ribonucleases. Nature. 1989;342:955–957. doi: 10.1038/342955a0. [DOI] [PubMed] [Google Scholar]
- McClure BA, Gray JE, Anderson MA, Clarke AE. Self-incompatibility in Nicotiana alata involves degradation of pollen rRNA. Nature. 1990;347:757–760. [Google Scholar]
- Murfett J, Atherton TL, Mou B, Gasser CS, McClure BA. S-RNase expressed in transgenic Nicotiana causes S-allele-specific pollen rejection. Nature. 1994;367:563–566. doi: 10.1038/367563a0. [DOI] [PubMed] [Google Scholar]
- Nasrallah JB. Recognition and rejection of self in plant self-incompatibility: comparisons to animal histocompatibility. Trends in Immunology. 2005;26:412–418. doi: 10.1016/j.it.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Nürnberger T, Abel S, Jost W, Glund K. Induction of an extracellular ribonuclease in cultured tomato cells upon phosphate starvation. Plant Physiology. 1990;92:970–976. doi: 10.1104/pp.92.4.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno H, Ehara Y. Expression of ribonuclease gene in mechanically injured or virus-inoculated Nicotiana tabacum leaves. Tohoku Journal of Agricultural Research. 2005;55:99–109. [Google Scholar]
- Péret B, Clément M, Nussaume L, Desnos T. Root developmental adaptation to phosphate starvation: better safe than sorry. Trends in Plant Science. 2011;16:442–450. doi: 10.1016/j.tplants.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Roalson EH, McCubbin AG. S-RNases and sexual incompatibility: structure, functions, and evolutionary perspectives. Molecular Phylogenetics and Evolution. 2003;29:490–506. doi: 10.1016/s1055-7903(03)00195-7. [DOI] [PubMed] [Google Scholar]
- Roldán JA, Quiroga R, Goldraij A. Molecular and genetic characterization of novel S-RNases from a natural population of Nicotiana alata. Plant Cell Reports. 2010;29:735–746. doi: 10.1007/s00299-010-0860-6. [DOI] [PubMed] [Google Scholar]
- Royo J, Kunz C, Kowyama Y, Anderson MA, Clarke AE, Newbigin E. Loss of a histidine residue at the active site of S-locus ribonuclease is associated with self compatibility in Lycopersicon peruvianum. Proceedings of the National Academy of Sciences USA. 1994;91:6511–6514. doi: 10.1073/pnas.91.14.6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CB, Bariola PA, delCardayré SB, Raines RT, Green PJ. RNS2: a senescence-associated RNase of Arabidopsis that diverged from the S-RNases before speciation. Proceedings of the National Academy of Sciences USA. 1993;90:5118–5122. doi: 10.1073/pnas.90.11.5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmouguin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acid Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trull MC, Guiltinan MJ, Lynch JP, Deikman J. The responses of wild-type and ABA mutant Arabidopsis thaliana plants to phosphorus starvation. Plant, Cell and Environment. 1997;20:85–92. [Google Scholar]
- Vieira J, Fonseca NA, Vieira CP. An S-RNase-based gametophytic self-incompatibility system evolved only once in eudicots. Journal of Molecular Evolution. 2008;67:179–190. doi: 10.1007/s00239-008-9137-x. [DOI] [PubMed] [Google Scholar]
- Wu P, Ma L, Hou X, et al. Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiology. 2003;132:1260–1271. doi: 10.1104/pp.103.021022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Finnegan PM. Regulation of phosphate starvation responses in higher plants. Annals of Botany. 2010;105:513–526. doi: 10.1093/aob/mcq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye ZH, Droste DL. Isolation and characterization of cDNAs encoding xylogenesis-associated and wounding-induced ribonucleases in Zinnia elegans. Plant Molecular Biology. 1996;30:697–709. doi: 10.1007/BF00019005. [DOI] [PubMed] [Google Scholar]
- Yen Y, Green PJ. Identification and properties of the major ribonucleases of Arabidopsis thaliana. Plant Physiology. 1991;97:1487–1493. doi: 10.1104/pp.97.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.