The Caulobacter polyphosphate synthesis enzyme, Ppk1, dynamically localizes to sites of polyphosphate granule production during the cell cycle by virtue of a C-terminal localization sequence. The processes of chromosome replication and segregation ensure that mother and daughter cells inherit a single granule upon cell division.
Abstract
Prokaryotes and eukaryotes synthesize long chains of orthophosphate, known as polyphosphate (polyP), which form dense granules within the cell. PolyP regulates myriad cellular functions and is often localized to specific subcellular addresses through mechanisms that remain undefined. In this study, we present a molecular-level analysis of polyP subcellular localization in the model bacterium Caulobacter crescentus. We demonstrate that biogenesis and localization of polyP is controlled as a function of the cell cycle, which ensures regular partitioning of granules between mother and daughter. The enzyme polyphosphate kinase 1 (Ppk1) is required for granule production, colocalizes with granules, and dynamically localizes to the sites of new granule synthesis in nascent daughter cells. Localization of Ppk1 within the cell requires an intact catalytic active site and a short, positively charged tail at the C-terminus of the protein. The processes of chromosome replication and segregation govern both the number and position of Ppk1/polyP complexes within the cell. We propose a multistep model in which the chromosome establishes sites of polyP coalescence, which recruit Ppk1 to promote the in situ synthesis of large granules. These findings underscore the importance of both chromosome dynamics and discrete protein localization as organizing factors in bacterial cell biology.
INTRODUCTION
Bacteria build a diverse array of specialized organelles and macromolecular structures that facilitate adaptation to and survival within complex environments. Exotic examples include gas vesicles, which function as cellular swim bladders (Klebahn, 1895; Pfeifer, 2012), and magnetosomes, which orient cells in magnetic fields (Blakemore, 1975). Some of these structures are inextricably linked to the basic metabolism of cells, including carboxysomes, whose polyhedral protein shells house carbon fixation machinery (Shively et al., 1973; Cannon et al., 2001), and polyhydroxybutyrate (PHB) granules, which are composed of carbon polymers analogous to fat stores of multicellular organisms (Stubbe et al., 2005). There is increasing evidence that bacteria employ dedicated mechanisms to organize these complex structures within the confines of the cytoplasmic compartment. Spatial organization is likely required for function in certain cases. For example, the membrane-bound magnetic crystals known as magnetosomes form a linear array, like a compass needle (Komeili et al., 2006). Regular cytosolic organization can also ensure each daughter inherits an equal number of structures upon cell division. Indeed, a functional role in inheritance has been proposed for carboxysomes in rod-shaped cells: when spacing is disrupted, daughter cells tend to inherit unequal numbers of organelles, hampering the fitness of cells receiving too few carboxysomes (Savage et al., 2010).
A ubiquitous subcellular structure whose organization and function in bacterial cells remains largely uncharacterized is the inorganic polyphosphate (polyP) granule. Unlike some of the more specialized structures just mentioned, polyP granules are found widely among Gram-positive and Gram-negative bacteria, suggesting that this structure is both evolutionarily ancient and functionally important. Known variously as metachromatic granules, Babes¸–Ernst bodies, or volutin granules, polyP inclusions were the first subcellular entities identified in bacteria (Babes¸, 1895). Granules are conserved in eukaryotes as well; in humans, polyP has been recognized as an important regulator of blood homeostasis (Morrissey et al., 2012), neuronal function (Holmstrom et al., 2013), and mitochondrial permeability (Abramov et al., 2007). Although the exact function of polyP in bacteria is still somewhat mysterious, it is known that loss of polyP production compromises cell survival under stress conditions in multiple species and attenuates virulence in pathogens such as Mycobacterium tuberculosis and Pseudomonas aeruginosa (Rashid et al., 2000; Sureka et al., 2007; Rao et al., 2009). It has been variously hypothesized that polyP serves as a cache of essential phosphorus, a store of energy, a buffer for cytosolic nucleoside triphosphates, and a signaling molecule promoting proteolysis and adaptation to nutrient stress (Harold, 1966; van Veen et al., 1994; Kuroda et al., 2001; Kulaev et al., 2004; Nocek et al., 2008; Boutte et al., 2012). Although some species coat polyP with a membrane to produce organelles known as acidocalcisomes (Docampo et al., 2005), most bacteria have no known proteins or accessory factors associated with polyP; granules are presumed to simply consist of long orthophosphate chains complexed with counterions. Indeed, granules can be easily recapitulated in vitro by mixing polyP and salts (Motomura et al., 2006). Despite our limited understanding of its physiological roles, its broad conservation across multiple kingdoms confirms that polyP is of fundamental importance to the biology of the cell and deserving of further study.
Like a number of other subcellular structures, polyP is nonrandomly distributed within the cytosol of many bacteria, forming discrete spherical granules, often with a definite and species-specific pattern of organization. Corynebacterium diphtheriae has historically been identified by its prominent polar granules (Breed et al., 1948). Helicobacter pylori localizes large polyP granules in its cytosol and small ones at the base of its flagellum (Bode et al., 1993), whereas Acetonema longum develops 8–12 dense storage granules, likely polyP, within developing spores (Tocheva et al., 2011). Transmission electron microscopy (TEM) studies of thin sections suggest that polyP may associate with the bacterial nucleoid (Voelz et al., 1966; Jensen, 1968; Takade et al., 1991). Despite these observations across diverse species, nothing is known regarding the mechanism by which these granules become localized within the cell. To develop our understanding of the cellular biology of polyP, we sought to dissect the chain of events that yields properly localized granules. To this end, we used Caulobacter crescentus, a robust model organism for the study of bacterial subcellular organization (Ausmees and Jacobs-Wagner, 2003). C. crescentus begins life as a motile swarmer cell, which differentiates into a replicative stalked cell and builds a new swarmer cell at the pole opposite the stalk. It is known to produce one or two polyP granules, found within both the stalked and nascent swarmer cell compartments (Poindexter, 1984; Comolli et al., 2006). Our studies provide a step-by-step model for the provisioning of each newborn cell with polyP. We define the positioning of granules throughout the cell cycle, elucidate molecular requirements for localization of the polyP biosynthetic enzyme, polyphosphate kinase 1 (Ppk1), and demonstrate the dependence of polyP localization on the replication state of the chromosome.
RESULTS
C. crescentus requires ppk1 to form polyP granules and survive in stationary phase
We previously showed by TEM that C. crescentus cells accumulate electron-dense granules in a polyphosphate kinase 1 (ppk1)-dependent manner (Boutte et al., 2012). Here we use 4′,6-diamidino-2-phenylindole (DAPI) staining to identify polyP granules by fluorescence microscopy (Aschar-Sobbi et al., 2008). Whereas DAPI-stained DNA fluoresces blue, DAPI-stained polyP granules fluoresce green. Bright green DAPI-fluorescent foci are evident in almost all C. crescentus cells growing in exponential phase (Figure 1A). Foci are entirely absent in a ∆ppk1-null strain, providing evidence that the fluorescently labeled structures we observe in the cell are bona fide polyP granules.
FIGURE 1:
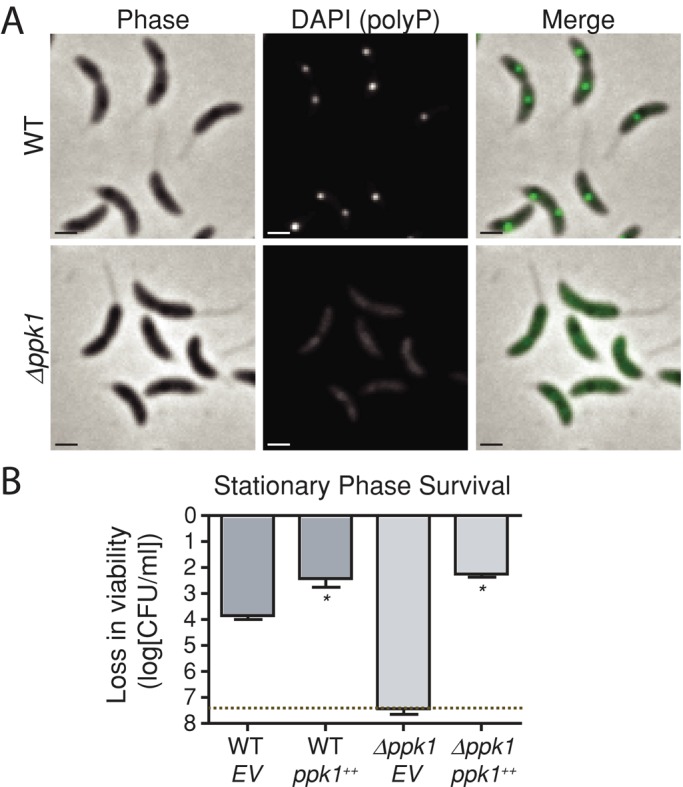
ppk1 is required for polyP granule production and survival in stationary phase. (A) Representative micrographs showing DAPI-stained log-phase WT C. crescentus and ∆ppk1 cells. Scale bars for this and subsequent micrographs, 1 μm. DAPI (polyP) images were taken with the same exposure and equally scaled for comparison. (B) Loss in viability of WT and ∆ppk1 complemented with xylose-inducible ppk1 (ppk1++) or an empty vector (EV) in M2X minimal medium after 24-h shaking in stationary phase. CFU/ml for three biological replicates of each strain plated upon entry to stationary phase and after 24-h culture in stationary phase; log change in CFU/ml from the initial plating to the 24-h plating is expressed on the y-axis. Dotted line, limit of detection (5 CFU/ml). Error bars, SE of the mean. *p < 0.001, Student's t test comparing the indicated strain with WT empty vector control (EV). Because zero CFU were isolated from ∆ppk1 EV cultures, error bars represent SE of the initial viability.
We next tested whether ppk1 has a functional role in cell survival under stress conditions, as in other species. After 24 h in stationary phase in minimal defined medium (M2-xylose, 108 colony-forming units [CFU]/ml starting culture), no viable CFU can be detected in ∆ppk1 cultures, whereas wild-type (WT) cells survive at a titer of 104 CFU/ml (Figure 1B). Expression of ppk1 from an inducible promoter complements this defect and even protects WT cells from death in stationary phase. Because ppk1 and polyP are important for C. crescentus stationary-phase physiology, we sought to leverage the strengths of C. crescentus as a developmental model organism to study the organization of polyP within bacterial cells.
PolyP granule production is cell cycle regulated, and granules are spatially organized within cells
The majority of newborn swarmer cells in a synchronized population contain a single DAPI-fluorescent granule (Figure 2A). As these cells progress through the cell cycle to the stalked and predivisional stages, a majority of cells develop a second granule. Indeed, when we quantify granules per cell over time, we observe a sharp increase in the proportion of cells with two granules, with a concomitant decrease in cells with a single granule (Figure 2B). Thus the number of polyP granules in a cell is a function of the cell cycle. Furthermore, these granules are nonrandomly distributed within cells. In swarmers, a single granule is positioned near mid-cell relative to the long cell axis. In predivisional cells, one granule resides in the stalked half of the cell and another in the nascent swarmer half (Figure 2A).
FIGURE 2:
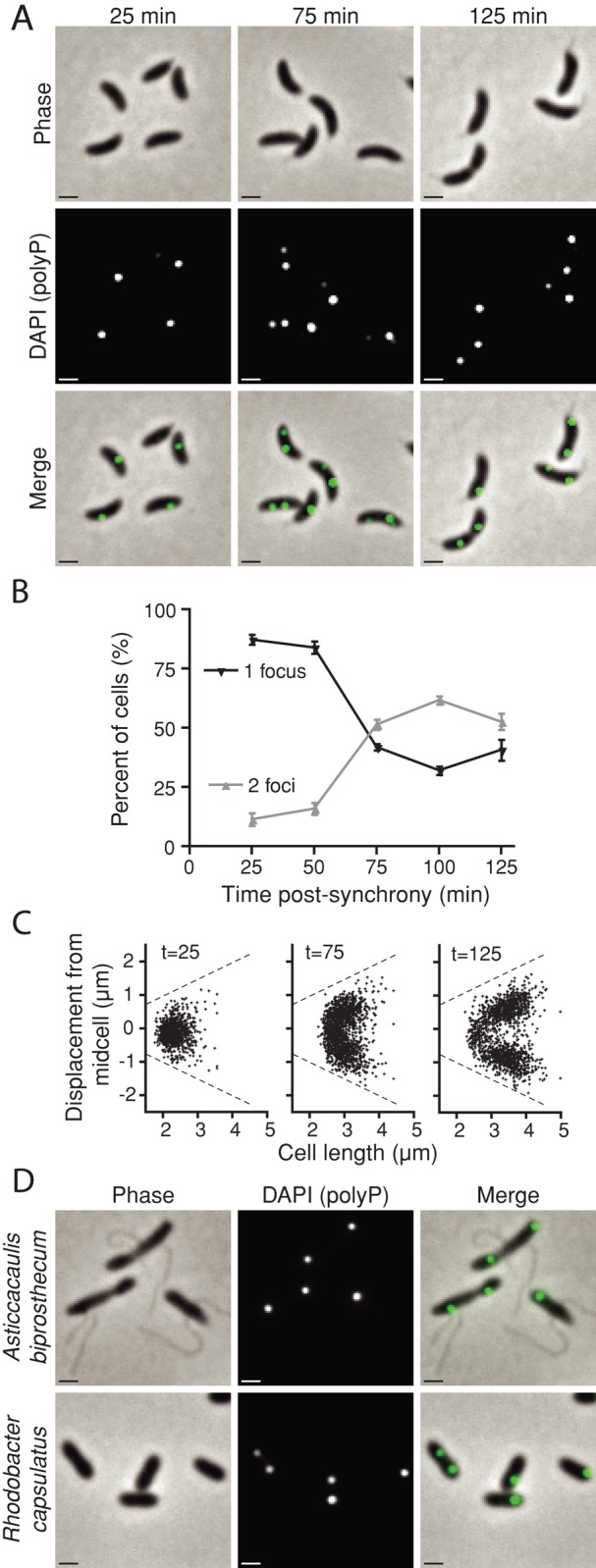
Granule production is cell cycle regulated and spatially organized. (A) Log-phase WT cells were synchronized, and samples were taken every 25 min and stained with DAPI for 25 min. Representative micrographs show cells at t = 25, 75, and 125 min postsynchrony, which includes staining time. (B) DAPI-staining foci per cell were manually enumerated from micrographs using ImageJ at each time point for two independent biological replicates. Error bars, SE of the mean. (C) MicrobeTracker was used to quantify the position of each granule within all cells in an image series. Each data point represents the position of a single DAPI-staining granule, plotted as a function of its displacement from the mid-cell along the cell's long axis vs. the total cell length. Dashed lines, the maximum position a granule can occupy, that is, the farthest possible distance from the mid-cell before leaving the detected cell boundary. Positive or negative displacements are arbitrary designations, as poles were not differentiated in this experiment. Times postsynchrony are given in minutes. For each frame, N = 1000 cells. (D) Representative micrographs of two additional DAPI-stained Alphaproteobacteria, A. biprosthecum and R. capsulatus.
To more fully define the spatial organization of polyP inside cells, we quantified the position of DAPI-fluorescent foci in 1000 synchronized cells at points across the cell cycle. Figure 2C depicts each polyP focus in a scatter plot by its position along the long axis of the cell versus the length of the cell. These data demonstrate that, on average, granules segregate away from the mid-cell at a fixed distance from each pole during cell growth (Figure 2C). In cells with two granules, this pattern can be described as one-fourth and three-fourths, or quarter-cell positioning. Thus both granule number and position are regulated cellular attributes. To assess conservation of polyP spatial organization in bacterial cells, we DAPI stained three related species in the class Alphaproteobacteria. We did not observe DAPI-staining granules in the plant symbiont Sinorhizobium meliloti (data not shown). In contrast, both the prosthecate bacterium Asticcacaulis biprosthecum and the purple photosynthetic bacterium Rhodobacter capsulatus display polyP organization similar to C. crescentus (Figure 2D). The related localization pattern observed across species suggests the existence of a conserved polyP-positioning mechanism.
Ppk1 is colocalized with polyP granules in cells
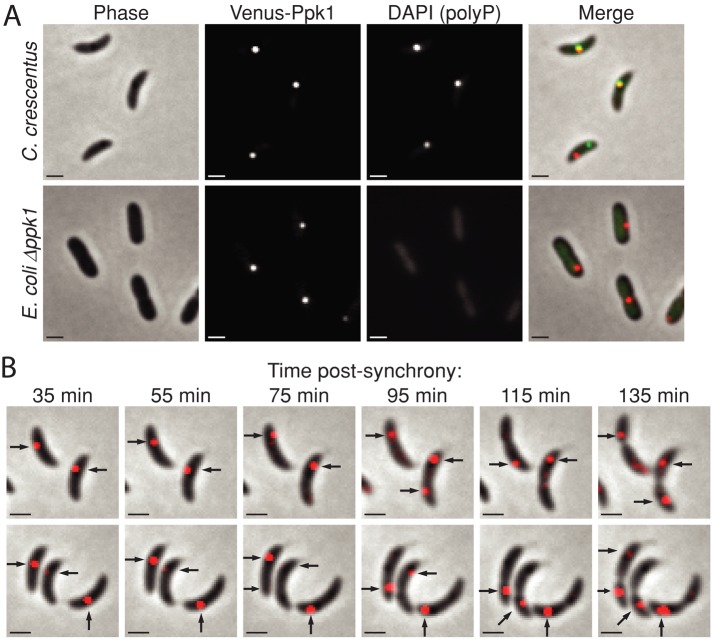
We sought to determine the site of granule biogenesis within cells and envisioned two possible patterns of Ppk1-dependent polyP production. Either 1) nascent granules are synthesized throughout the cytosol and localized postsynthesis or 2) polyP is produced in situ by a localized enzyme. To differentiate these models, we replaced the wild-type ppk1 with an allele encoding an N-terminally fluorescently tagged derivative, Venus-Ppk1. Venus-Ppk1 forms isolated fluorescent foci at the half and quarter positions within cells, a pattern easily distinguished from the diffuse fluorescence observed upon expression of Venus alone (Figure 3A and Supplemental Figure S1). To rule out Venus-dependent effects on Ppk1 localization, we also expressed mCherry-Ppk1 and confirmed that it too forms fluorescent foci at identical positions in the cell (Supplemental Figure S2). Of note, Venus-Ppk1 (and mCherry-Ppk1) foci colocalize with DAPI-stained polyP granules in most instances, consistent with a model in which granules are produced by local polyP biosynthesis. In cases in which a Venus-Ppk1 focus does not colocalize with a granule, it nevertheless occupies a quarter-cell position.
FIGURE 3:
Venus-Ppk1 forms clusters, colocalizes with polyP granules, and dynamically localizes over the cell cycle. (A) Representative micrographs of DAPI-stained C. crescentus expressing venus-ppk1 from the native chromosomal locus, as well as DAPI-stained E. coli ∆ppk1 expressing a plasmid-borne venus-ppk1(CC) fusion from an IPTG-inducible promoter (E. coli ∆ppk1 venus-ppk1++). In the merged image, Venus is depicted in the red channel, and DAPI (polyP) is depicted in green. (B) Time-lapse merged phase and fluorescence images of unstained venus-ppk1 growing on nutrient-agar pads. Black arrows, unambiguous Venus-Ppk1 foci, which are depicted in the red channel.
Ppk1 forms focal clusters in a heterologous host
To test the molecular requirements of Ppk1 localization, we assayed whether Ppk1 intrinsically forms focal clusters and localizes within cells, or whether clustering and localization require a C. crescentus–specific host factor. We expressed C. crescentus venus-ppk1 (venus-ppk1(CC)) in a heterologous host: an Escherichia coli K12 strain in which the native copy of ppk1 was deleted (E. coli Δppk1). Venus alone does not form fluorescent foci in E. coli Δppk1 (Supplemental Figure S2). We observe fluorescent foci, one per cell, in E. coli expressing C. crescentus venus-ppk1(CC) (Figure 3A). Ppk1(CC) does not, however, exhibit the predictable mid/quarter-position localization in E. coli that we observe in C. crescentus. We conclude that C. crescentus Ppk1 focal clustering is an inherent attribute of the enzyme but that the mechanism ensuring regular positioning of Ppk1 at the mid-cell early in the cell cycle and quarter positions late in the cell cycle is specific to C. crescentus. Wild-type E. coli does not typically produce discretely localized polyP granules, and, intriguingly, the E. coli fusion protein Venus-Ppk1(EC) is diffusely localized within cells (Supplemental Figure S2), indicating that Ppk1(CC) possesses specific attributes that promote the formation of discrete clusters.
Ppk1 dynamically relocalizes during the cell cycle
Having established that polyP granule synthesis and localization are cell cycle regulated, we sought to assess the dynamics of Ppk1/polyP localization in living cells across the cell cycle. Owing to DAPI toxicity, we were unable to monitor the development of stained granules in living cells, but we were able to visualize Venus-Ppk1 by time-lapse microscopy. Many Venus-Ppk1 foci are stable on a time scale of tens of minutes (Figure 3B). As the cell cycle progresses, we observe the development of new Venus-Ppk1 foci at previously unoccupied quarter-cell positions. This process varies from cell to cell. Appearance of a new Venus-Ppk1 focus can occur with the concomitant loss of the preexisting focus or can result in foci in both mother and nascent daughter compartments at the quarter positions. We increased the image acquisition rate to analyze movement of Venus-Ppk1 foci. As seen in Supplemental Movie S1, foci display three types of behavior on the time scale of minutes. Some cells show stable foci; others display rapid movement of foci from one quarter-position to the other; and some foci move erratically within cells and oscillate between mother and daughter compartments, homing intermittently to the quarter-positions. We conclude that Ppk1 dynamically relocalizes throughout the cell cycle to occupy the sites of granule production. These data might also explain why, at any given time, select Ppk1 foci are not colocalized with granules and vice versa.
Ppk1 forms foci independently of polyP catalysis, but localization requires the catalytic active-site residue, H434
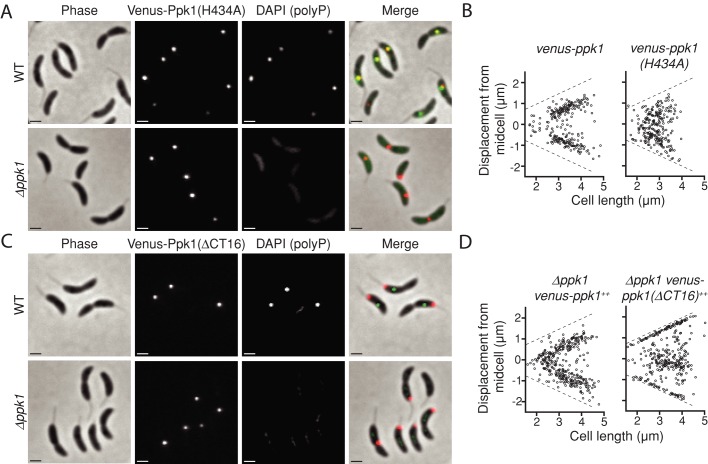
We next tested whether an intact Ppk1 catalytic active site is required for the proper localization of the enzyme within the cell. To this end, we expressed venus-ppk1(H434A), an allele for which the known catalytic histidine (Kumble et al., 1996) has been mutated to alanine. We have previously demonstrated that this variant is stably expressed but does not produce polyP granules in C. crescentus (Boutte et al., 2012). When expressed from an ectopic locus in a WT background, venus-ppk1(H434A) produces foci that colocalize with polyP granules and presumably wild-type Ppk1 (Figure 4A). Indeed, Ppk1-H434A colocalizes with wild-type Ppk1 in a heterologous host (Supplemental Figure S2), suggesting that these two proteins cooligomerize. We therefore expressed venus-ppk1(H434A) in the absence of wild-type ppk1 and confirmed that it cannot not complement granule production, although it still localizes to discrete foci (Figure 4A). This demonstrates that catalytic activity is not required for the formation of Ppk1 clusters within the cell. We quantified the position of Venus-Ppk1(H434A) fluorescent foci and plotted them as previously described for DAPI-fluorescent polyP granules. Whereas venus-ppk1 produces foci that consistently localize to the expected positions (i.e., mid- and quarter-cell positions), venus-ppk1(H434A) is randomly localized within cells (Figure 4B), indicating that catalysis is required for Ppk1 clusters to be properly addressed within the cell.
FIGURE 4:
Catalysis and a C-terminal tail are required for normal Venus-Ppk1 localization. (A) Representative micrographs of venus-ppk1(H434A)++ expressed in the WT or ∆ppk1 background. For merged images in A and C, Venus is depicted in the red channel and DAPI (polyP) in the green channel. (B) Positions of Venus-Ppk1 or Venus-Ppk1(H434A) expressed from the native locus in populations of cells 120 min postsynchrony. For A and B, N = 299 cells with detected foci. (C) Representative micrographs of venus-ppk1(∆CT16)++ expressed in the WT or ∆ppk1 background. (D) Positions of Venus-Ppk1(∆CT16) in populations of cells grown in M2G. Cells were imaged at 120 min postsynchrony. For C and D, N = 352 cells with detected foci.
These findings suggested that Ppk1 alone is not the target of the cell's localization mechanism and that polyP is required to localize the Ppk1/polyP complex. To separate Ppk1 localization from polyP localization, we attempted to complement granule production in C. crescentus Δppk1 by overexpressing venus-ppk1(EC). This protein was diffusely localized in C. crescentus and yet it was able to complement granule production and localization (Supplemental Figure S2). Thus it seems that polyP granules can be discretely localized in cells even when produced by a diffusely localized enzyme, supporting a model in which polyP is the target of the localization mechanism.
A C-terminal localization signal is required for proper Ppk1 positioning
Given that polyP localization likely precedes Ppk1 localization, we investigated the nature of Ppk1 colocalization with polyP granules. Discretely localized proteins commonly rely on specific localization sequences to find their intended subcellular addresses. To test whether segments of Ppk1 might serve a specific function in protein focal clustering or localization, we performed mutational analysis on the venus-ppk1 allele. The goal of these experiments was to discover variants that fail to form foci in the cell or that form foci but fail to properly localize. The crystal structure of the orthologous Ppk1 enzyme of E. coli (Zhu et al., 2005) aided this effort, facilitating construction of a C. crescentus Ppk1 homology model and design of structure-based mutations.
Ppk1 is composed of four conserved domains, labeled N, H, C1, and C2 (Supplemental Figure S1). Domain C1 contains the critical catalytic residue H434. We observed that C. crescentus Ppk1 contains a highly positively charged (pI = 11.8), 22-residue C-terminal tail not present in the E. coli enzyme. We systematically generated Venus fusions to Ppk1 variants lacking the N (∆N), N and H (∆NH), C2 (∆C2), or C1 and C2 (∆C1C2) domains, as well as a variant lacking 16 residues from the C-terminus (∆CT16).
Of the four whole-domain deletion constructs, ∆C1C2 and ∆C2 yield diffuse fluorescence signal throughout the cell similar to expression of venus alone (Supplemental Figure S1), indicating that at least domain C2 is required for focal clustering of Ppk1. ∆N and ∆NH yield very poor fluorescence signal and are likely unstable. No truncation construct complements granule production in a ∆ppk1 background, providing evidence that each domain is indispensable for in vivo granule production.
Unlike the whole-domain deletions, expression of venus-ppk1(∆CT16) produces discrete fluorescent foci (Figure 4C). Venus-Ppk1∆CT16 foci do not colocalize with polyP granules when expressed in a WT background, indicating that the C-terminal tail of Ppk1 is dispensable for focal cluster formation but required for proper subcellular enzyme localization. Moreover, when expressed in the ∆ppk1 background, this construct gives poor granule production, despite all known catalytic residues remaining intact.
At the population level, we quantified the position of fluorescent foci in cells expressing either venus-ppk1 or venus-ppk1(∆CT16). Whereas venus-ppk1 gives the expected pattern of localization, venus-ppk1(∆CT16) exhibits a dramatic mislocalization phenotype: rather than simply forming foci randomly throughout cells, this variant is excluded from the quarter-cell positions and redirected toward the poles and mid-cell (Figure 4D). Therefore the positively charged sequence at the C-terminus of C. crescentus Ppk1 is a key localization determinant that 1) is required for association with natively produced granules in a wild-type background and 2) is necessary for normal polyP granule production in the cell.
Inhibition of chromosome replication disrupts granule synthesis and localization
Typically, bacterial proteins are diffusely distributed throughout the cytosol or cell envelope. There are examples, however, of proteins that are addressed to one or both poles or to the mid-cell/division plane (Goley et al., 2007; Lindner et al., 2008). Localization to the quarter-position of the cell, as we observe for Ppk1 and polyP, is unusual. To our knowledge, no C. crescentus proteins with this localization pattern have been thoroughly characterized, despite genome-wide localization studies (Werner et al., 2009). Among the handful of known molecules displaying localization to the quarter-positions are certain low-copy plasmids (Gordon et al., 1997) and the cytoplasmic chemotaxis protein clusters of Rhodobacter sphaeroides (Wadhams et al., 2002), both of which associate with the chromosome to ensure an equal inheritance between mother and daughter cells (Roberts et al., 2012; Hwang et al., 2013). Note that carboxysomes and PHB granules also rely on the chromosome for proper localization (Savage et al., 2010; Galan et al., 2011; Pfeiffer et al., 2011). In considering the cellular basis of Ppk1/polyP localization, we hypothesized that that the C. crescentus chromosome may play an important role in this process. To test this hypothesis, we disrupted chromosome replication and chromosome segregation using both genetic and pharmacological approaches and measured the effects of these perturbations on synthesis and subcellular positioning of polyP granules.
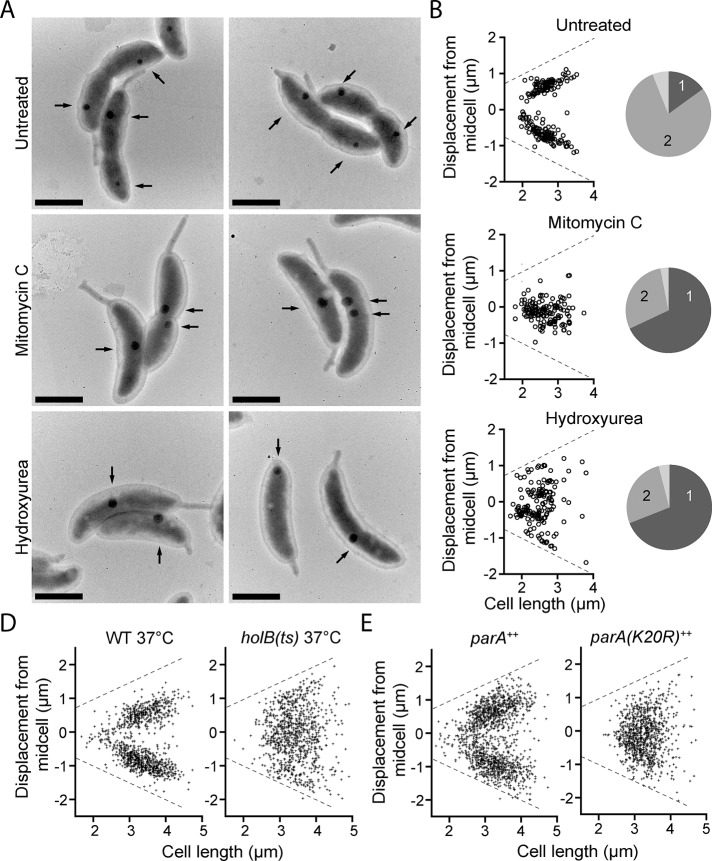
We first disrupted DNA synthesis in a synchronized population of wild-type swarmer cells with hydroxyurea (HU) or mitomycin C (MMC). Treatment with these drugs blocks chromosome replication, although swarmer cells continue to grow and differentiate into stalked cells (Degnen and Newton, 1972). Treated and untreated swarmer cells were permitted to develop into predivisional cells. We then quantified polyP granule position in the three groups by measuring the distance between the center of each granule to the stalked pole in TEM micrographs. The majority of untreated cells possess two polyP granules at quarter-cell positions, which is consistent with our DAPI fluorescence results (Figure 5, A and B). HU- and MMC-treated cells are generally defective in production of a second granule, and the positioning of the initial granule is disrupted. Despite the fact that treated and untreated cells grow to similar lengths, polyP granules did not regularly segregate into the mother and daughter cell compartments in treated cells. MMC, which cross-links DNA strands (Tomasz, 1995), caused granules to remain near mid-cell. HU treatment, which inhibits ribonucleotide reductase (Sinha and Snustad, 1972), left most cells with a single, randomly positioned granule that could occupy unusual positions such as extreme polar regions.
FIGURE 5:
Chromosome replication and segregation are required for normal granule biogenesis and positioning. (A) Representative TEM micrographs of synchronized WT cells treated with either MMC or HU or untreated. Black arrows, polyP granules. Scale bars, 1 μm. (B) Manually measured distances between the stalked pole and the center of each granule, following the center of the long axis of the cell. For the untreated group, N = 106 cells; MMC, N = 112 cells; HU, N = 118 cells. Pie graphs represent the percentage of cells in each treatment group containing one (dark gray), two (light gray), or three or more granules (off-white), as observed by TEM. (C) Position of DAPI-staining granules in synchronized populations of cells at t = 125 min postsynchrony, using a temperature-sensitive allele of holB to block DNA synthesis. Left, N = 764 cells; right, N = 742 cells. (D) Position of DAPI-staining granules in synchronized populations of cells at t = 125 min postsynchrony, using a dominant-negative allele of parA to block chromosome segregation. Left, N = 914 cells; right, N = 813 cells.
We repeated our polyP analysis using DAPI staining and fluorescence microscopy in a C. crescentus strain containing a temperature-sensitive allele of holB, the δ′ subunit of DNA polymerase (Quardokus and Brun, 2002). When shifted to the restrictive temperature of 37°C, DNA synthesis is blocked in this strain (Osley and Newton, 1977). Consistent with our previous findings, granule localization is disrupted in these strains at the restrictive temperature but not in WT cells grown at the same temperature (Figure 5C). Thus we conclude that chromosome replication governs the transition from the one- to two-granule state and that the mechanism by which DNA synthesis is perturbed has specific consequences for the localization of the initial granule.
Chromosome segregation governs polyP granule number and subcellular address
Blockade of DNA synthesis necessarily prevents chromosome segregation from mother to daughter. Thus experiments that inhibit new DNA synthesis cannot differentiate the effects of chromosome replication from chromosome segregation on polyP synthesis and localization. We hypothesized that segregation of the new chromosome to the nascent daughter cell determines localization of Ppk1/polyP to the quarter-cell positions. To test this hypothesis, we disrupted the parAB partitioning system, which is required for C. crescentus chromosome segregation (Ptacin et al., 2010). ParA forms filaments that guide the newly synthesized chromosome into the nascent daughter compartment. We expressed a dominant-negative parA(K20R) allele, which permits chromosome replication but blocks segregation (Toro et al., 2008; Supplemental Figure S3). As with blocked replication, we observe that blocked segregation disrupts polyP granule positioning. Rather than two granules per predivisional cell, each at quarter-position, we commonly observe cells with a single, randomly positioned granule in the strain expressing parA(K20R) (Figure 5D and Supplemental Figure S3). Control expression of the wild-type parA allele from an ectopic locus does not affect granule synthesis or positioning. Venus-Ppk1 continues to colocalize with polyP granules in both strains (Supplemental Figure S3).
These data provide evidence that quarter-position localization of polyP granules in each cell compartment requires that a second chromosome be not only synthesized, but also properly segregated. We conclude that polyP granules are capable of residing at any position within the cell, but that orderly progression of S-phase ensures that each mother and daughter cell inherit a polyP granule before cell division.
DISCUSSION
Despite the fact that polyP granules were first discovered in bacteria in the 19th century (Babes¸, 1895), the cellular factors that determine localization and inheritance of these important nutrient and energy stores have remained undefined. Our studies reveal many key elements of this process. PolyP itself, rather than Ppk1, is the target of the cell's partitioning mechanism (Supplemental Figure S2), and this mechanism requires proper chromosome replication and segregation (Figure 5). Although Ppk1 requires catalysis to localize to the quarter-positions (Figure 4A), it can be observed at these positions in the absence of detectible colocalizing DAPI-staining granules (Figure 3A). To accommodate these experimental observations, we propose a model in which chromosome segregation results in the partitioning of at least a small amount of polyP to the quarter-positions. Ppk1 is dynamically recruited to these positions by virtue of the C-terminal tail, produces additional polyP in situ, and generates detectable granules (Figure 6A). As cells grow, the polyP granules remain in place at fixed distances from the poles, consistent with tethering to the chromosome. When the cell divides, each daughter cell contains a polyP granule, and the process begins again (Figure 6B). Similar to carboxysomes, chemotaxis protein clusters, and PHB granules, we propose that the chromosome provides a necessary structure for proper partitioning of this nongenetic resource between cells.
FIGURE 6:
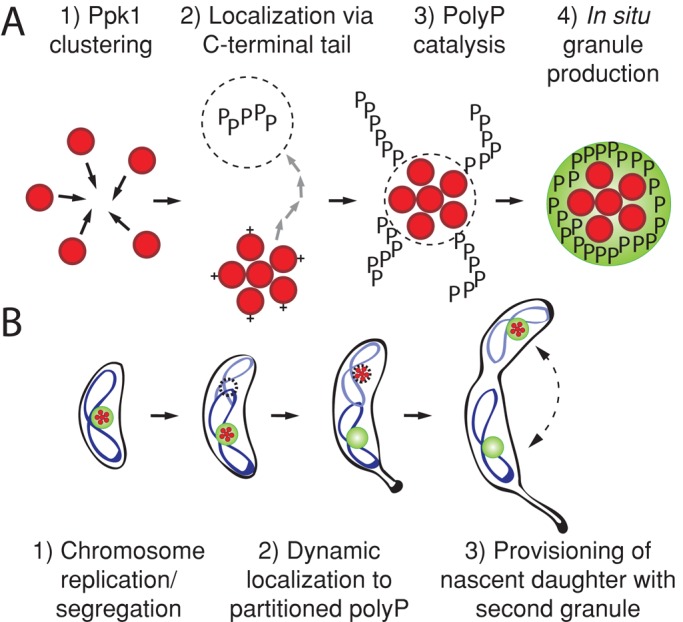
A model of polyP granule biogenesis in C. crescentus. (A) At the molecular level, Ppk1 enzyme (red circles) forms focal clusters. The positively charged Ppk1 C-terminal tail (+) and polyP catalytic activity are both required to properly localize these enzyme clusters, which in turn produce large, visible polyP granules (phosphate chains depicted as black Ps; PolyP granules depicted as green spheres). Groups of red circles represent Ppk1 clusters of unspecified stoichiometry. (B) At the cellular level, Ppk1 is localized to an existing granule in swarmer cells. As swarmers differentiate into stalked cells, the chromosome (blue ribbons) replicates, and one copy segregates to the nascent daughter cell. On segregation Ppk1 localizes to a new, chromosomally determined niche. A second granule is subsequently synthesized at this new subcellular address. Ppk1 can associate with either or both of these granules.
Although our top-down approach to the analysis of polyP provides a coarse-grained model of granule synthesis and partitioning in a bacterial cell, many molecular-level questions remain. For instance, if diffusely localized Ppk1(EC) produces discretely localized granules, is there a reason that Ppk1(CC) is discretely localized? It is possible that Ppk1, when not located at the quarter-positions, is inactive. This possibility is consistent with the finding that polar or mid-cell Ppk1(∆C16) cannot produce granules. Ppk1(EC) may complement granule localization by indiscriminately filling the cell with enzyme and thus occupying both the chromosomally determined polyP niche and all other subcellular addresses. This paradigm implies that Ppk1 forms discrete clusters to make the most use of the fewest enzymes per cell. Alternatively, it may be the case that Ppk1(CC) is more robustly attracted to cellular polyP than Ppk1(EC). It remains to be seen whether localized Ppk1 promotes efficient polyP accumulation, economizes enzyme production, or serves an as-yet-unidentified function.
Another related question centers on the biochemical/structural basis of Ppk1 clustering within cells, which occurs even in the absence of polyP. There is evidence from biochemical studies on orthologous versions of Ppk1 that formation of multimeric Ppk1 complexes is an inherent attribute of the enzyme. Specifically, Ppk1 from several species is known to form tetramers in vitro (Ahn and Kornberg, 1990; Ogawa et al., 2000; Jagannathan et al., 2010). Furthermore, upon addition of ATP, at least one Ppk1 orthologue forms large oligomers (Zhang et al., 2007); some Ppk1 orthologues may be capable of forming filaments (Gómez-García and Kornberg, 2004; Fraley et al., 2007). Indeed, the presence of high-order multimers has been put forward as an explanation for the lattice or string-like structures that are observed as part of polyP granules imaged by cryo-electron microscopy (Iancu et al., 2010). The role of Ppk1 quaternary structure in polyP synthesis, localization, and overall intracellular organization is an important future area of investigation.
The dynamics of individual Ppk1 proteins, as it relates to a localized model of polyP synthesis, is also of considerable interest. Our time-lapse microscopy data reveal both stable and highly dynamic Ppk1 fluorescent foci, the appearance of new foci, and the dissolution of others over the period of a cell cycle. We do not observe simple unidirectional movement of Ppk1 from mother to daughter, however, as is observed for tagged chromosomal loci (Viollier et al., 2004) and DNA-binding proteins (Thanbichler and Shapiro, 2006). It may be that Ppk1 clusters can diffuse throughout the cell but preferentially localize to polyP, as discussed later. Furthermore, we identified a previously uncharacterized localization signal at the Ppk1 C-terminus. An intriguing possibility is that this localization sequence functions to directly bind polyP. Alternatively, this localization sequence may simply enable Ppk1 to explore the nucleoid region, such that without it, Ppk1 is relegated to the poles and mid-cell.
The exact role of the chromosome in polyP granule localization and inheritance merits further discussion. Although we discovered that proper chromosome segregation is required for granule positioning, we do not yet understand the molecular details underlying this phenomenon. We envision three possible models that could explain our finding. The first and most parsimonious is that polyP directly associates with a specific chromosomal locus, perhaps by virtue of an as-yet-unidentified protein linker that binds both polyP and a specific DNA sequence or structure. A second model is that the cell uses a mechanism that promotes partitioning by binding polyP and nonspecific DNA, similar to ParA-family proteins (Roberts et al., 2012). Both models are supported by TEM studies that report polyP in proximity of the bacterial nucleoid. Finally, it is possible that some unknown factor determines granule localization and requires completion of S phase for its production or function. Because disruption of chromosome replication and segregation necessarily blocks downstream cell-cycle processes (Wortinger et al., 2000; Modell et al., 2011), we cannot formally rule out this possibility.
Although future study of this conserved partitioning mechanism will provide insight into these molecular details, our work provides the initial framework for understanding the mechanism of polyP localization and inheritance in bacterial cells. We have 1) demonstrated the highly regulated nature of polyP localization; 2) quantified the dynamics and subcellular localization of the polyP biosynthetic enzyme, Ppk1; 3) defined a novel, positively charged sequence at the Ppk1 C-terminus that is required for proper enzyme localization; and 4) demonstrated that chromosome replication and segregation govern the biogenesis and subcellular organization of these highly conserved nutrient and energy storage granules. We hope that our study is only the beginning of cellular analyses of this important and well-known but little-studied subcellular structure.
MATERIALS AND METHODS
Culture conditions and viability assessment
C. crescentus was grown in peptone–yeast extract (PYE) complex medium, M2G minimal medium with 0.2% glucose (Ely, 1991), or M2-xylose minimal medium with 0.2% xylose substituted for glucose. Unless otherwise specified, for xylose-inducible expression M2G was supplemented with 0.15% xylose for 4 h before imaging. Drugs were supplemented at the following concentrations in liquid/solid medium (μg/ml): kanamycin (5/25), nalidixic acid (–/20), hydroxyurea (5000/–; Alfa Aesar, Ward Hill, MA), and mitomycin C (3/–; A.G. Scientific, San Diego, CA). Cells were grown in a tube roller or rotary shaker at 30°C, except for temperature-shift experiments, for which cells were shaken at 37°C after synchrony. Cultures were maintained at an OD660 of <0.4 before each experiment, with the exception of stationary-phase survival experiments, for which cultures were grown in parallel until they reached an OD660 of >1.1, at which point serial dilutions were plated on PYE plus kanamycin to enumerate CFU/ml. Viability was assessed in the same way after 24 h of additional shaking at 30°C. E. coli grown for microscopy were cultured at 37°C in M9 minimal medium supplemented with 30 μg/ml kanamycin and induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and/or 0.2% arabinose for 2 h. Otherwise, E. coli Top10 and Mach1 strains (Invitrogen, Carlsbad, CA) used for cloning were cultured in Terrific Broth at 30°C. A. biprosthecum and R. capsulatus were grown in PYE at 30°C.
Strain construction
Supplemental Table S1 gives all strains used in this study. Truncation boundaries are denoted by protein residue numbers included in each construct name. ppk1 (CC_1710), truncations of this gene, or ppk1(H434A) were PCR amplified and ligated into pMT854 or pMT697. The H434A allele, based on current NA1000 annotation, corresponds to the H460A allele previously published (Boutte et al., 2012). venus-ppk1 and venus-ppk1(H434A) were amplified from pMT854-ppk1 and pMT854-ppk1(H434A), respectively and ligated into pNPTS138. Allelic replacements at the ppk1 locus were produced by a previously described double recombination method (Fiebig et al., 2010). venus and venus-ppk1 were also amplified from pMT854-ppk1 and ligated into pSRK-Kn. parA was amplified and ligated into pMT590; the parA(K20R) allele was generated by overlap-extension PCR.
Cell synchrony
Cells were synchronized as described previously. Synchronized cells are labeled by the total time elapsed between the isolation of swarmers and acquisition of images. DAPI-stained cells were incubated with DAPI for 25 min, and this time is included in the total time postsynchrony. For drug treatments, isolated swarmers were immediately incubated with the drug after synchrony; for temperature shifts, isolated swarmers were immediately shifted to 37°C. In the case of parA induction, cells were grown in M2G, and 0.03% xylose was added to the isolated swarmers.
Staining and fluorescence microscopy
To stain with DAPI (Sigma-Aldrich, St. Louis, MO), cells were treated with 12 μg/ml DAPI, added directly to the culture medium, and incubated at room temperature in the dark for 25 min. Cells were then spotted on 1% agarose pads for microscopy. For time-lapse microscopy, cells were spotted on 1% agarose M2G pads and grown at room temperature. Fluorescence microscopy was conducted with a Leica CTR5000 microscope (Leica, Wetzlar, Germany); images were acquired with a Hamamatsu ORCA-ER camera (Hamamatsu, Hamamatsu, Japan). A custom filter set was used to visualized DAPI-polyP, using a 390/70-nm excitation filter, a 488-nm dichroic, and a 515-nm long-pass emission filter (Chroma, Brattleboro, VT).
Electron microscopy
For transmission electron microscopy, log-phase cells were switched from M2G to M2 with no added carbon and collected after 4 h. Cells were fixed in a final concentration of 3.2% paraformaldehyde, 0.0001% glutaraldehyde, and 200 mM sodium phosphate buffer, pH 7.4, at room temperature for 8 min, then washed twice with distilled water. Unstained cells were spotted onto Formvar-carbon-coated 400 mesh copper grids, air dried, and visualized. Grids were imaged at ×16,800 using an FEI Tecnai F30 scanning transmission electron microscope equipped with an Ultrascan camera (Gatan, Pleasanton, CA).
Image analysis and bioinformatics
A model of C. crescentus Ppk1 was generated with SWISS-MODEL (Arnold et al., 2006) using Protein Data Bank entry 1XDO (Bernstein et al., 1977; Zhu et al., 2005). Protein isoelectric point was calculated using ExPASy ProtParam (Gasteiger et al., 2005). Measurements of TEM micrographs and image manipulation were performed using ImageJ (National Institutes of Health, Bethesda, MD; Collins, 2007). Automated cell outlining, spot detection, and coordinate output were performed by MicrobeTracker (Sliusarenko et al., 2011). Using Image-Pro Plus MDA (Media Cybernetics, Bethesda, MD), we applied a Gaussian filter to some images to sharpen diffraction-limited foci; when this operation was performed, it was applied uniformly to the entire image, and if that image was compared with other images in the text, we processed the others identically.
Supplementary Material
Acknowledgments
J.T.H. is supported by National Institutes of Health Medical Scientist National Research Service Award 5T32GM007281-38. We thank Justine Collier for key discussions and providing specifications for the DAPI (polyP) fluorescence filter set. We acknowledge Aretha Fiebig for thoughtful advice on experimental design and data presentation. We also thank Ariane Briegel and Grant Jensen for access to unpublished C. crescentus cryo-electron microscopy tomograms and K. C. Huang for helpful discussions at the outset of this project.
Abbreviations used:
- polyP
polyphosphate
- Ppk1
polyphosphate kinase 1
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-04-0182) on August 28, 2013.
REFERENCES
- Abramov AY, Fraley C, Diao CT, Winkfein R, Colicos MA, Duchen MR, French RJ, Pavlov E. Targeted polyphosphatase expression alters mitochondrial metabolism and inhibits calcium-dependent cell death. Proc Natl Acad Sci USA. 2007;104:18091–18096. doi: 10.1073/pnas.0708959104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn K, Kornberg A. Polyphosphate kinase from Escherichia coli. Purification and demonstration of a phosphoenzyme intermediate. J Biol Chem. 1990;265:11734–11739. [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp Jr, Schwede T. The SWISS-MODEL workspace: a Web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Aschar-Sobbi R, Abramov AY, Diao C, Kargacin ME, Kargacin GJ, French RJ, Pavlov E. High sensitivity, quantitative measurements of polyphosphate using a new DAPI-based approach. J Fluoresc. 2008;18:859–866. doi: 10.1007/s10895-008-0315-4. [DOI] [PubMed] [Google Scholar]
- Ausmees N, Jacobs-Wagner C. Spatial and temporal control of differentiation and cell cycle progression in Caulobacter crescentus. Annu Rev Microbiol. 2003;57:225–247. doi: 10.1146/annurev.micro.57.030502.091006. [DOI] [PubMed] [Google Scholar]
- Babes¸ V. Beobachtungen über die metachromatischen Körperchen, Sporenbildung, Verzwiegung, Kolben- und Kapsel-bildung pathogener Bakterien. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg. 1895;20:412–420. [Google Scholar]
- Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Blakemore R. Magnetotactic bacteria. Science. 1975;190:377–379. doi: 10.1126/science.170679. [DOI] [PubMed] [Google Scholar]
- Bode G, Mauch F, Ditschuneit H, Malfertheiner P. Identification of structures containing polyphosphate in Helicobacter pylori. J Gen Microbiol. 1993;139:3029–3033. doi: 10.1099/00221287-139-12-3029. [DOI] [PubMed] [Google Scholar]
- Boutte CC, Henry JT, Crosson S. ppGpp and polyphosphate modulate cell cycle progression in Caulobacter crescentus. J Bacteriol. 2012;194:28–35. doi: 10.1128/JB.05932-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breed RS, Bergey DH, Hitchens AP, Murray EGD. Bergey's Manual of Determinative Bacteriology. Baltimore: Williams & Wilkins; 1948. [Google Scholar]
- Cannon GC, Bradburne CE, Aldrich HC, Baker SH, Heinhorst S, Shively JM. Microcompartments in prokaryotes: carboxysomes and related polyhedra. Appl Environ Microbiol. 2001;67:5351–5361. doi: 10.1128/AEM.67.12.5351-5361.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins TJ. ImageJ for microscopy. Biotechniques. 2007;43:25–30. doi: 10.2144/000112517. [DOI] [PubMed] [Google Scholar]
- Comolli LR, Kundmann M, Downing KH. Characterization of intact subcellular bodies in whole bacteria by cryo-electron tomography and spectroscopic imaging. J Microsc. 2006;223:40–52. doi: 10.1111/j.1365-2818.2006.01597.x. [DOI] [PubMed] [Google Scholar]
- Degnen ST, Newton A. Dependence of cell division on the completion of chromosome replication in Caulobacter. J Bacteriol. 1972;110:852–856. doi: 10.1128/jb.110.3.852-856.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docampo R, de Souza W, Miranda K, Rohloff P, Moreno SNJ. Acidocalcisomes—conserved from bacteria to man. Nat Rev Microbiol. 2005;3:251–261. doi: 10.1038/nrmicro1097. [DOI] [PubMed] [Google Scholar]
- Ely B. Genetics of Caulobacter crescentus. Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- Fiebig A, Castro Rojas CM, Siegal-Gaskins D, Crosson S. Interaction specificity, toxicity and regulation of a paralogous set of ParE/RelE-family toxin-antitoxin systems. Mol Microbiol. 2010;77:236–251. doi: 10.1111/j.1365-2958.2010.07207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraley CD, Rashid MH, Lee SSK, Gottschalk R, Harrison J, Wood PJ, Brown MRW, Kornberg A. A polyphosphate kinase 1 (ppk1) mutant of Pseudomonas aeruginosa exhibits multiple ultrastructural and functional defects. Proc Natl Acad Sci USA. 2007;104:3526–3531. doi: 10.1073/pnas.0609733104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan B, Dinjaski N, Maestro B, de Eugenio LI, Escapa IF, Sanz JM, Garcia JL, Prieto MA. Nucleoid-associated PhaF phasin drives intracellular location and segregation of polyhydroxyalkanoate granules in Pseudomonas putida KT2442. Mol Microbiol. 2011;79:402–418. doi: 10.1111/j.1365-2958.2010.07450.x. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Walker JM. The Proteomics Protocols Handbook. Totowa, NJ: Humana Press; 2005. Protein identification and analysis tools on the ExPASy server; pp. 571–607. [Google Scholar]
- Goley ED, Iniesta AA, Shapiro L. Cell cycle regulation in Caulobacter: location, location, location. J Cell Sci. 2007;120:3501–3507. doi: 10.1242/jcs.005967. [DOI] [PubMed] [Google Scholar]
- Gómez-García MR, Kornberg A. Formation of an actin-like filament concurrent with the enzymatic synthesis of inorganic polyphosphate. Proc Natl Acad Sci USA. 2004;101:15876–15880. doi: 10.1073/pnas.0406923101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon GS, Sitnikov D, Webb CD, Teleman A, Straight A, Losick R, Murray AW, Wright A. Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell. 1997;90:1113–1121. doi: 10.1016/s0092-8674(00)80377-3. [DOI] [PubMed] [Google Scholar]
- Harold FM. Inorganic polyphosphates in biology: structure, metabolism, and function. Bacteriol Rev. 1966;30:772–794. doi: 10.1128/br.30.4.772-794.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmstrom KM, Marina N, Baev AY, Wood NW, Gourine AV, Abramov AY. Signalling properties of inorganic polyphosphate in the mammalian brain. Nat Commun. 2013;4:1362–1362. doi: 10.1038/ncomms2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang LC, Vecchiarelli AG, Han Y-W, Mizuuchi M, Harada Y, Funnell BE, Mizuuchi K. ParA-mediated plasmid partition driven by protein pattern self-organization. EMBO J. 2013;32:1238–1249. doi: 10.1038/emboj.2013.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iancu CV, Morris DM, Dou Z, Heinhorst S, Cannon GC, Jensen GJ. Organization, structure, and assembly of alpha-carboxysomes determined by electron cryotomography of intact cells. J Mol Biol. 2010;396:105–117. doi: 10.1016/j.jmb.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan V, Kaur P, Datta S. Polyphosphate kinase from M. tuberculosis: an interconnect between the genetic and biochemical role. PLoS One. 2010;5:e14336. doi: 10.1371/journal.pone.0014336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen TE. Electron microscopy of polyphosphate bodies in a blue-green alga, Nostoc pruniforme. Arch Mikrobiol. 1968;62:144–152. [Google Scholar]
- Klebahn H. Gasvakuolen, ein Bestandteil der Zellen der wasserblütenbildenden Phycochromaceen. Flora. 1895;80:241–282. [Google Scholar]
- Komeili A, Li Z, Newman DK, Jensen GJ. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science. 2006;311:242–245. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- Kulaev IS, Vagabov VM, Kulakovskaya TV. The Biochemistry of Inorganic Polyphosphates. Chichester, England: John Wiley & Sons; 2004. [Google Scholar]
- Kumble KD, Ahn K, Kornberg A. Phosphohistidyl active sites in polyphosphate kinase of Escherichia coli. Proc Natl Acad Sci USA. 1996;93:14391–14395. doi: 10.1073/pnas.93.25.14391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda A, Nomura K, Ohtomo R, Kato J, Ikeda T, Takiguchi N, Ohtake H, Kornberg A. Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science. 2001;293:705–708. doi: 10.1126/science.1061315. [DOI] [PubMed] [Google Scholar]
- Lindner AB, Madden R, Demarez A, Stewart EJ, Taddei F. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc Natl Acad Sci USA. 2008;105:3076–3081. doi: 10.1073/pnas.0708931105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modell JW, Hopkins AC, Laub MT. A DNA damage checkpoint in Caulobacter crescentus inhibits cell division through a direct interaction with FtsW. Genes Dev. 2011;25:1328–1343. doi: 10.1101/gad.2038911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey JH, Choi SH, Smith SA. Polyphosphate: an ancient molecule that links platelets, coagulation, and inflammation. Blood. 2012;119:5972–5979. doi: 10.1182/blood-2012-03-306605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motomura K, Takiguchi N, Ohtake H, Kuroda A. Polyamines affect polyphosphate accumulation in Escherichia coli. J Environ Biotechnol. 2006;6:41–46. [Google Scholar]
- Nocek B, Kochinyan S, Proudfoot M, Brown G, Evdokimova E, Osipiuk J, Edwards AM, Savchenko A, Joachimiak A, Yakunin AF. Polyphosphate-dependent synthesis of ATP and ADP by the family-2 polyphosphate kinases in bacteria. Proc Natl Acad Sci USA. 2008;105:17730–17735. doi: 10.1073/pnas.0807563105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa N, Tzeng CM, Fraley CD, Kornberg A. Inorganic polyphosphate in Vibrio cholerae: genetic, biochemical, and physiologic features. J Bacteriol. 2000;182:6687–6693. doi: 10.1128/jb.182.23.6687-6693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osley MA, Newton A. Mutational analysis of developmental control in Caulobacter crescentus. Proc Natl Acad Sci USA. 1977;74:124–128. doi: 10.1073/pnas.74.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer F. Distribution, formation and regulation of gas vesicles. Nat Rev Microbiol. 2012;10:705–715. doi: 10.1038/nrmicro2834. [DOI] [PubMed] [Google Scholar]
- Pfeiffer D, Wahl A, Jendrossek D. Identification of a multifunctional protein, PhaM, that determines number, surface to volume ratio, subcellular localization and distribution to daughter cells of poly(3-hydroxybutyrate), PHB, granules in Ralstonia eutropha H16. Mol Microbiol. 2011;82:936–951. doi: 10.1111/j.1365-2958.2011.07869.x. [DOI] [PubMed] [Google Scholar]
- Poindexter JS. The role of calcium in stalk development and in phosphate acquisition in Caulobacter crescentus. Arch Microbiol. 1984;138:140–152. doi: 10.1007/BF00413014. [DOI] [PubMed] [Google Scholar]
- Ptacin JL, Lee SF, Garner EC, Toro E, Eckart M, Comolli LR, Moerner WE, Shapiro L. A spindle-like apparatus guides bacterial chromosome segregation. Nat Cell Biol. 2010;12:791–798. doi: 10.1038/ncb2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quardokus EM, Brun YV. DNA replication initiation is required for mid-cell positioning of FtsZ rings in Caulobacter crescentus. Mol Microbiol. 2002;45:605–616. doi: 10.1046/j.1365-2958.2002.03040.x. [DOI] [PubMed] [Google Scholar]
- Rao NN, Gómez-García MR, Kornberg A. Inorganic polyphosphate: essential for growth and survival. Annu Rev Biochem. 2009;78:605–647. doi: 10.1146/annurev.biochem.77.083007.093039. [DOI] [PubMed] [Google Scholar]
- Rashid MH, Rumbaugh K, Passador L, Davies DG, Hamood AN, Iglewski BH, Kornberg A. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2000;97:9636–9641. doi: 10.1073/pnas.170283397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MAJ, Wadhams GH, Hadfield KA, Tickner S, Armitage JP. ParA-like protein uses nonspecific chromosomal DNA binding to partition protein complexes. Proc Natl Acad Sci USA. 2012;109:6698–6703. doi: 10.1073/pnas.1114000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage DF, Afonso B, Chen AH, Silver PA. Spatially ordered dynamics of the bacterial carbon fixation machinery. Science. 2010;327:1258–1261. doi: 10.1126/science.1186090. [DOI] [PubMed] [Google Scholar]
- Shively JM, Ball F, Brown DH, Saunders RE. Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science. 1973;182:584–586. doi: 10.1126/science.182.4112.584. [DOI] [PubMed] [Google Scholar]
- Sinha NK, Snustad DP. Mechanism of inhibition of deoxyribonucleic acid synthesis in Escherichia coli by hydroxyurea. J Bacteriol. 1972;112:1321–1324. doi: 10.1128/jb.112.3.1321-1334.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol Microbiol. 2011;80:612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbe J, Tian J, He A, Sinskey AJ, Lawrence AG, Liu P. Nontemplate-dependent polymerization processes: polyhydroxyalkanoate synthases as a paradigm. Annu Rev Biochem. 2005;74:433–480. doi: 10.1146/annurev.biochem.74.082803.133013. [DOI] [PubMed] [Google Scholar]
- Sureka K, Dey S, Datta P, Singh AK, Dasgupta A, Rodrigue S, Basu J, Kundu M. Polyphosphate kinase is involved in stress-induced mprAB-sigE-rel signalling in mycobacteria. Mol Microbiol. 2007;65:261–276. doi: 10.1111/j.1365-2958.2007.05814.x. [DOI] [PubMed] [Google Scholar]
- Takade A, Umeda A, Misumi T, Sawae Y, Amako K. Accumulation of phosphate-containing granules in the nucleoid area of Pseudomonas aeruginosa. Microbiol Immunol. 1991;35:367–374. doi: 10.1111/j.1348-0421.1991.tb01567.x. [DOI] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- Tocheva EI, Matson EG, Morris DM, Moussavi F, Leadbetter JR, Jensen GJ. Peptidoglycan remodeling and conversion of an inner membrane into an outer membrane during sporulation. Cell. 2011;146:799–812. doi: 10.1016/j.cell.2011.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasz M. Mitomycin C: small, fast and deadly (but very selective) Chem Biol. 1995;2:575–579. doi: 10.1016/1074-5521(95)90120-5. [DOI] [PubMed] [Google Scholar]
- Toro E, Hong S-H, McAdams HH, Shapiro L. Caulobacter requires a dedicated mechanism to initiate chromosome segregation. Proc Natl Acad Sci USA. 2008;105:15435–15440. doi: 10.1073/pnas.0807448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veen HW, Abee T, Kortstee GJ, Pereira H, Konings WN, Zehnder AJ. Generation of a proton motive force by the excretion of metal-phosphate in the polyphosphate-accumulating Acinetobacter johnsonii strain 210A. J Biol Chem. 1994;269:29509–29514. [PubMed] [Google Scholar]
- Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, McAdams HH, Shapiro L. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci USA. 2004;101:9257–9262. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelz H, Voelz U, Ortigoza RO. The “polyphosphate overplus” phenomenon in Myxococcus xanthus and its influence on the architecture of the cell. Arch Mikrobiol. 1966;53:371–388. doi: 10.1007/BF00409874. [DOI] [PubMed] [Google Scholar]
- Wadhams GH, Martin AC, Porter SL, Maddock JR, Mantotta JC, King HM, Armitage JP. TlpC, a novel chemotaxis protein in Rhodobacter sphaeroides, localizes to a discrete region in the cytoplasm. Mol Microbiol. 2002;46:1211–1221. doi: 10.1046/j.1365-2958.2002.03252.x. [DOI] [PubMed] [Google Scholar]
- Werner JN, Chen EY, Guberman JM, Zippilli AR, Irgon JJ, Gitai Z. Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc Natl Acad Sci USA. 2009;106:7858–7863. doi: 10.1073/pnas.0901781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortinger M, Sackett MJ, Brun YV. CtrA mediates a DNA replication checkpoint that prevents cell division in Caulobacter crescentus. EMBO J. 2000;19:4503–4512. doi: 10.1093/emboj/19.17.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gómez-García MR, Shi X, Rao NN, Kornberg A. Polyphosphate kinase 1, a conserved bacterial enzyme, in a eukaryote, Dictyostelium discoideum, with a role in cytokinesis. Proc Natl Acad Sci USA. 2007;104:16486–16491. doi: 10.1073/pnas.0706847104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Huang W, Lee SSK, Xu W. Crystal structure of a polyphosphate kinase and its implications for polyphosphate synthesis. EMBO Rep. 2005;6:681–687. doi: 10.1038/sj.embor.7400448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.