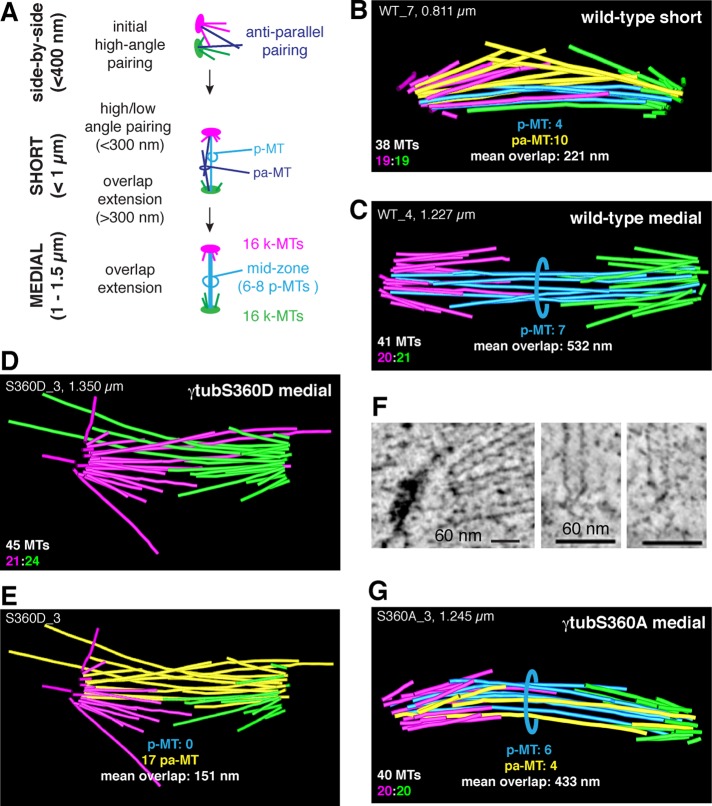
Correlation of spindle architecture with dynamic behavior shows that pairs of antiparallel microtubules are sufficient to form a bipolar spindle, whereas interpolar microtubules maintain the speed of pole displacement during spindle assembly. The number of interpolar microtubules formed is controlled in part through γ-tubulin phosphorylation.
Abstract
During spindle assembly, microtubules may attach to kinetochores or pair to form antiparallel pairs or interpolar microtubules, which span the two spindle poles and contribute to mitotic pole separation and chromosome segregation. Events in the specification of the interpolar microtubules are poorly understood. Using three-dimensional electron tomography and analysis of spindle dynamical behavior in living cells, we investigated the process of spindle assembly. Unexpectedly, we found that the phosphorylation state of an evolutionarily conserved Cdk1 site (S360) in γ-tubulin is correlated with the number and organization of interpolar microtubules. Mimicking S360 phosphorylation (S360D) results in bipolar spindles with a normal number of microtubules but lacking interpolar microtubules. Inhibiting S360 phosphorylation (S360A) results in spindles with interpolar microtubules and high-angle, antiparallel microtubule pairs. The latter are also detected in wild-type spindles <1 μm in length, suggesting that high-angle microtubule pairing represents an intermediate step in interpolar microtubule formation. Correlation of spindle architecture with dynamical behavior suggests that microtubule pairing is sufficient to separate the spindle poles, whereas interpolar microtubules maintain the velocity of pole displacement during early spindle assembly. Our findings suggest that the number of interpolar microtubules formed during spindle assembly is controlled in part through activities at the spindle poles.
INTRODUCTION
Mitotic spindles in animal and fungal cells share common design features and have a stereotyped architecture that is consistent with the requirement to ensure accurate chromosome segregation (Bouck et al., 2008). All spindles consist of kinetochore microtubules (k-MTs) and polar microtubules (p-MTs). During spindle assembly, the k-MTs attach to the centromeres of paired sister chromatids, whereas p-MTs contribute to separation of the poles and formation of a bipolar spindle. In anaphase, p-MTs both promote spindle elongation and stabilize the anaphase spindle. Proteins recruited to the p-MTs of the spindle midzone orchestrate spindle breakdown at the end of mitosis.
Microtubules have intrinsic structural and functional asymmetry, with a relatively stable (−) end and highly dynamic (+) end. The cap of the (+) end of microtubules undergoes cycles of polymerization, depolymerization (catastrophe), and repolymerization (rescue), allowing the microtubule to change its length (Mitchison and Kirschner, 1984). In cells, the dynamical properties of k-MTs and p-MTs are modulated by a plethora of proteins that bind along the contour length or at the ends of microtubules. These proteins make major contributions to the structure of the spindle, as they can stabilize or destabilize microtubule ends, couple spindle microtubules to centromeres, and cross-link microtubules into bundles.
The formation of p-MTs is promoted by the kinesin-5 orthologue Cin8 and to a lesser extent Kip1 (Hoyt et al., 1992). The homotetrameric structure of kinesin-5 motors is believed to enable multiple cross-links of antiparallel microtubules during spindle assembly followed by the sliding apart of p-MTs from opposite poles during anaphase (Straight et al., 1998). Kar3 (kinesin-14), Bim1 (EB1), and Ase1 (PRC-1) provide structural support to the anaphase spindle by cross-linking antiparallel microtubules. In the absence of Cin8, these proteins contribute to p-MT formation and pole separation during spindle assembly (Saunders and Hoyt, 1992; Kotwaliwale et al., 2007; Gardner et al., 2008b; Zimniak et al., 2009).
The budding yeast spindle has a relatively simple structure compared with spindles in animal cells (Bouck et al., 2008). At the completion of spindle assembly, the number of microtubules in the metaphase spindle of budding yeast is remarkably uniform, with one k-MT for each of the 32 sister chromatids and a core bundle composed of six to eight p-MTs (Winey et al., 1995). The number of centromeres defines the number of k-MTs. Intuitively, the number of p-MTs should be constrained by the abundance of proteins that promote and/or stabilize pairing, as well as by the number of single microtubules available for antiparallel pairing.
Spindle microtubules in budding yeast are nucleated by and project out of γ-tubulin ring complexes (γ-TURCs) composed of γ-tubulin and associated γ-tubulin–interacting proteins (GRIPs) located at spindle poles—centrosomes in animal cells and spindle pole bodies in fungal cells (Moritz et al., 1995; Zheng et al., 1995; Knop et al., 1997; Knop and Schiebel, 1997). Components of the spindle poles, specifically the γ-TURC nucleating complex, assemble microtubules and in this way can control the number of spindle microtubules. Whether γ-TURCs do enforce the number of spindle microtubules, thereby producing a spindle with a stereotyped core bundle, is not known.
We previously demonstrated that budding yeast γ-tubulin bound to purified spindle poles is phosphorylated at S360 and that S360 can be phosphorylated by Cdk1 in vitro (Keck et al., 2011). In this study, we perform a comprehensive characterization of the role of γ-tubulin S360 phosphorylation. Using electron tomography, we show that mimicking constitutive phosphorylation at S360 blocks the formation of p-MTs, whereas inhibiting S360 phosphorylation results in spindles containing two distinct types of paired antiparallel microtubules and 50% increase in the number of antiparallel microtubule pairs. Analysis of the dynamic behavior of spindles in living cells shows that inhibition of S360 phosphorylation increases the velocity of pole separation and dampens length fluctuations during assembly. Our findings suggest a model in which transient phosphorylation of γ-tubulin contributes to the formation of spindles with stereotyped structure and behavior.
RESULTS
Microtubule pairing and overlap are features of early steps in spindle assembly
The number and organization of microtubules in the yeast mitotic spindle have been studied using three-dimensional (3D) electron tomography, by which individual microtubules and their 3D organization can be determined (O'Toole et al., 1999, 2002). These studies provided insight into early events in the formation of a bipolar spindle in budding yeast (Figure 1A). Spindle assembly begins with events that lead to the separation of duplicated spindle poles. Subsequent to the nucleation of spindle microtubules from the new spindle pole, a subset of antiparallel microtubules pairs through the activity of cross-linking proteins. Because the duplicated poles are side by side, initial pairing occurs at a high angle. Short spindles, 0.8–1 μm, contain both long and short microtubules, some of which are paired and may contain p-MTs, which are defined by their continuous antiparallel overlap of ≥300 nm with a gap of ≤45 nm. Medial spindles 1–1.5 μm in length have a relatively stereotyped structure, with 32 k-MTs (16 per spindle pole) and a bundle of 6–8 p-MTs (Winey et al., 1995).
FIGURE 1:
Analysis of the number and organization of spindle microtubules in wild-type and S360 nonphosphorylatable (γ-tubS360A) and phosphor-mimetic (γ-tubS360D) spindles. (A) Summary of steps in the formation of the interpolar microtubules: antiparallel pairing is observed for spindles <400 nm in length (side by side); both pa-MTs and p-MTs are observed in wild-type spindles <1 μm in length. Wild-type spindles >1 μm have three or four pairs of p-MTs, each pair with a minimal overlap of 300 nm. The 3D structure and organization of k-MTs (pink, green), p-MTs (blue), and pa-MTs (yellow) in representative wild-type spindles during early (B) and late (C) phases of spindle assembly. The 3D structure and organization of k-MTs (pink, green), p-MTs (blue), and pa-MTs (yellow) in representative medial γ-tubS360A/D spindles. (D) A medial γ-tubS360D spindle (1.350 μm) contains pa-MTs (E) but lacks p-MTs. (F) Microtubule (−) ends in γ-tubS360D spindles are capped. (G) Both p-MTs and pa-MTs are observed in medial (>1 μm) γ-tubS360A spindles, and the MTs appear to be under compression.
We constructed seven models of wild-type spindles 0.8–1.35 μm in length and extracted the following information for each spindle: microtubule number, lengths, type, and pairing and contour overlap (Figure 2 and Supplemental Table S1). Representative models are shown in Figure 1. We observed that k-MTs and p-MTs could not be readily distinguished in short wild-type spindles <1 μm in length (Figure 1B and Supplemental Movie S1) in comparison with medial spindles >1 μm in length (Figure 1C). Short spindles contain few or no p-MTs but many paired antiparallel microtubules, which we called polar-approach MTs (pa-MTs). Similar to p-MTs, pa-MT pairs consist of two overlapping antiparallel microtubules with a gap of ≤45 nm. The length of contour overlap, however, is reduced (105 ± 77.9 nm) relative to p-MTs (585 ± 184.7; Figure 2). Because pa-MTs are abundant in short wild-type spindles but are rare (0.5 ± 1.2, n = 6) in medial wild-type spindles (Figure 2), pa-MTs may represent an intermediate step in the formation of p-MTs.
FIGURE 2:
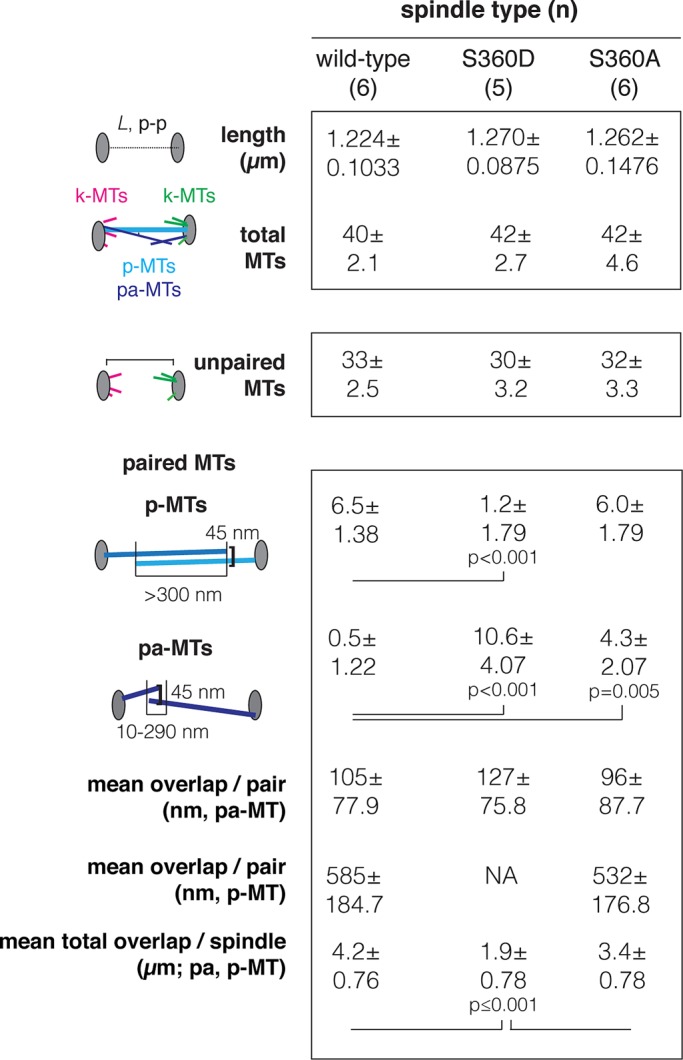
Summary of spindle length, microtubule number, type (k-MTs, p-MTs, and pa-MTs), and contour overlap in medial spindles. Data were extracted from wild-type (six), γ-tubS360D (five), and γ-tubS360A (six) tomographic models. The complete data set is provided in Supplemental Table S1. Spindle length is defined as pole–pole distance (in micrometers).
γ-Tubulin S360 phosphorylation state alters pairing and overlap but not microtubule number
We previously found that budding yeast γ-tubulin S360D/E mutations (γ-tubS360D/E), which mimic constitutive phosphorylation of S360, result in long anaphase spindles with an apparent defect in spindle breakdown (Keck et al., 2011). Overproduction of γ-tubS360D was reported to result in preanaphase spindles with a reduced number of spindle microtubules, presumably as a consequence of instability of the mutant protein (Lin et al., 2011). Given that S360 phosphorylation can be detected in γ-tubulin associated with purified spindle poles (Keck et al., 2011), we speculated that the physiological role of S360 phosphorylation is to limit the number of microtubules formed during spindle assembly. If this were the case, it would be expected that short and medial spindles in γ-tubS360D cells would have fewer microtubules per pole and an overall reduction in the number of both k-MTs and p-MTs relative to wild type. This would be particularly evident for microtubules projecting from the new spindle pole. Conversely, a mutation that inhibits phosphorylation of S360 (γ-tubS360A) might represent a basal state or increase in the number of microtubules per pole.
We sought to understand how the phosphorylation state of S360 relates to microtubule number and organization during spindle assembly. We used 3D electron tomography (O'Toole et al., 2002) to construct five γ-tubS360D models and six γ-tubS360A models, allowing us to determine microtubule number and 3D organization in medial (1–1.5 μm) spindles in the context of S360 phosphorylation state (Figure 2 and Supplemental Table S1). Figure 1, D and G, shows representative models. Our analysis revealed that the overall number of spindle microtubules is unaffected by S360 phosphorylation state, as there was no difference in microtubule number in γ-tubS360D spindles (n = 5, 42 ± 2.7) or γ-tubS360A spindles (n = 6, 42 ± 4.6) relative to wild-type spindles (n = 6, 40 ± 2.1; Figure 2). The number of unpaired microtubules (neither pa-MTs nor p-MTs) was also not significantly different among medial wild-type (33 ± 2.5), γ-tubS360D (30 ± 3.2), and γ-tubS360A (32 ± 3.3) spindles. Although the mean number of unpaired microtubules in γ-tubS360D spindles is statistically equivalent to that in wild type, based on the SE of the mean, γ-tubS360D is more likely to have <32 unpaired microtubules and as a result experience chromosome loss. The increased potential for chromosome loss in γ-tubS360D cells is consistent with a requirement for the spindle assembly checkpoint for viability of this mutant (Keck et al. 2011).
Mimicking constitutive phosphorylation of S360 inhibits the formation of p-MTs
p-MTs are clearly distinguishable from unpaired microtubules (k-MTs) in medial wild-type spindles (Winey et al., 2005), and all of our wild-type medial spindles (n = 6) had a well-organized bundle of ∼6 p-MTs (6.5 ± 1.38; Figure 2). p-MTs were conspicuously absent or reduced in number, however, in all medial γ-tubS360D spindles (n = 5, 1.2 ± 1.79; p = 0.001) relative to wild-type spindles (Figure 1D and Supplemental Movies S2 and S3). Numerous pa-MTs (10.6 ± 4.07, p < 0.001) were observed in γ-tubS360D spindles relative to wild-type spindles (Figure 1E and Supplemental Movie S3). The mean total contour overlap for all paired microtubules was decreased in γ-tubS360D spindles (1.89 ± 0.783 μm; p = 0.001) relative to wild type (4.15 ± 0.761 μm). These results suggest that S360 phosphorylation does not block antiparallel pairing that leads to the formation of pa-MTs but instead is defective in p-MT formation. Microtubule attachment to the spindle poles, however, did not appear to be perturbed in γ-tubS360D spindles. The (−) ends of microtubules in γ-tubS360D spindles were capped and closed (Figure 1F), which is inconsistent with the depletion or instability of γ-tubulin or the γ-TURC (O'Toole et al., 2012).
Inhibiting phosphorylation of S360 results in spindles with both pa-MTs and p-MTs
We next asked whether inhibiting S360 phosphorylation might increase the number of p-MTs and/or pa-MTs or increase total overlap without increasing the number of p-MTs. Like wild-type spindles, medial γ-tubS360A spindles (n = 6) have a bundle of p-MTs (6.0 ± 1.79) that are clearly distinguishable from short, unpaired microtubules (Figure 1G and Supplemental Movie S4). The number of unpaired microtubules in medial γ-tubS360A spindles (32 ± 3.3) is statistically equivalent to that in wild type, with a mean value equal to the number of centromeres after completion of DNA replication (Figure 2). All medial γ-tubS360A spindles contain pa-MTs (4.3 ± 2.07) that are uncharacteristic of medial wild-type spindles (Figures 1G and 2). The presence of p-MTs and pa-MTs results in 50% increase in paired microtubules relative to medial wild-type spindles. Surprisingly, the mean total pairing lengths contributed by the p-MTs in wild type (n = 6, 4.15 ± 0.761 μm) and the p-MTs + pa-MTs in γ-tubS360A spindles were not significantly different (n = 6, 3.43 ± 0.778 μm, p = 0.139). In the absence of S360 phosphorylation, the same amount of at least one of the proteins (Cin8, Kar3, Bim1) contributing to antiparallel cross-linking is likely distributed over a larger number of paired microtubules with the same total contour overlap as in wild type.
The mixture of p-MTs and pa-MTs in γ-tubS360A medial spindles offered an opportunity for a quantitative comparative analysis of their structure. We extracted several parameters from tomographic models of wild type and γ-tubS360A medial spindles: 1) the angle at which each microtubule projects from the spindle pole body, 2) the length of each microtubule, and 3) the contour overlap between the paired microtubules, and correlated pitch-length-pairing information for all antiparallel paired microtubules (Figure 3A). All medial wild-type and γ-tubS360A spindles contained p-MTs (blue dots, Figure 3, B and C). In wild-type spindles p-MTs were generally composed of two long microtubules (>500 nm) that project from the poles with a pitch of 2.7 ± 1.68° (n = 39). p-MTs in γ-tubS360A spindles project from the poles at an increased pitch (4.1 ± 2.58°, n = 35; p = 0.008) relative to wild-type p-MTs. In general, pa-MTs (yellow dots, Figure 3, B and C) were composed of at least one short microtubule projecting with a pitch of >5° (maximum 23°) paired with a longer, opposing microtubule (Figure 3C). The same organization was observed for the pa-MT pairing observed in one wild-type spindle (Figure 3B, inset). In most pairs, the increased pitch (>5°) of the short microtubule in the pa-MT pair was correlated with bending in the overlapping contour of one of the microtubules (Figure 3C, inset). In general, inhibiting S360 phosphorylation is correlated with an increase in the number of pole-biased antiparallel microtubule overlaps in the form of pa-MTs.
FIGURE 3:
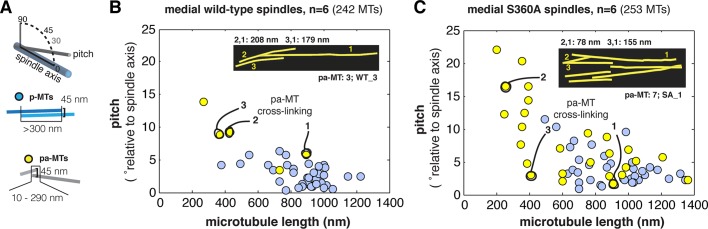
Topology of p-MTs and pa-MTs in wild-type and γ-tubS360A spindles. (A) Schematic representation of microtubule pitch–length pairing analysis. Pitch is defined by the angle of projection of the microtubule from its origin relative to the imaginary plane defined by the two spindle poles (spindle axis). The contour length (nanometers) of individual spindle microtubules and their intersection and spacing (gap) are extracted from the tomographic models using IMOD. Pairing events are represented by two types of antiparallel interactions: p-MTs (blue dots) and pa-MTs (yellow dots). p-MTs are defined as two microtubules (usually both >500 nm) projecting from opposite poles that interact over a contour length of >300 nm with gap of 45 nm. pa-MT pairs consist of two microtubules projecting from opposite poles that intersect over 10–290 nm of contour length with gap of <45 nm. (B) Distribution of p-MT pairing angles in wild-type spindles as a function of microtubule length (nanometers). The pitch of wild-type p-MTs is 2.7 ± 1.68° (n = 39). Three pa-MTs were identified in one wild-type spindle and are indicated with lines and shown in the inset. (C) Distribution of pa-MT and p-MT pairing angles in γ-tubS360A spindles as a function of microtubule length (nanometers). The pitch of γ-tubS360A p-MTs is 4.1 ± 2.58° (n = 35). pa-MT pairs are frequently composed of one long (>600 nm) microtubule projecting at <10° paired to at least one short (<500 nm) microtubule projecting at a pitch of >10°. High angle (>5°) antiparallel pairing is correlated with pa-MT bending. Bending is frequently observed in γ-tubS360A spindles (inset, C) and in one medial wild-type spindle that contained three pa-MTs (inset, B). p-MTs in γ-tubS360A spindles project from the poles at an increased pitch.
Analysis of the dynamic behavior of wild-type, γ-tubS360A, and γ-tubS360D spindles
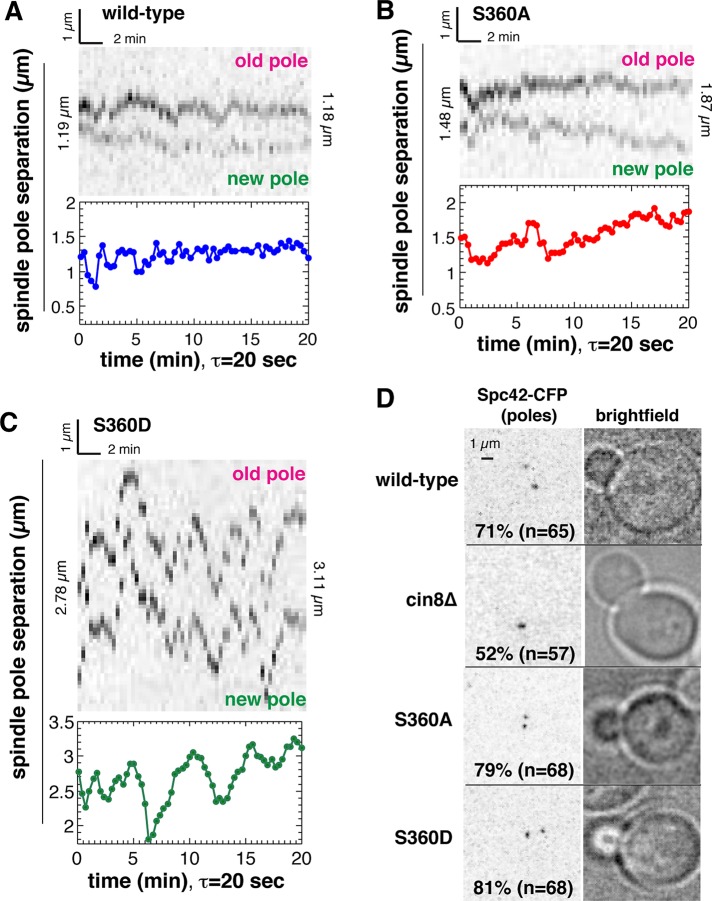
To investigate how the observed changes in spindle microtubule architecture in the presence of γ-tubS360A and γ-tubS360D mutations might affect the process of spindle assembly, we sought to accurately measure the dynamic behavior of spindles in living cells. The formation of a bipolar spindle and the transition to anaphase can be monitored in living yeast cells through the analysis of the displacements of fluorescently labeled spindle poles with respect to each other over time. Starting from initial pole separation, spindles slowly increase in length to ∼2.4 μm, followed by fast anaphase elongation to ∼6 μm (Figure 4A). The fluorescence intensity of a point-like diffraction-limited object can be fit to a Gaussian distribution in the x,y and z planes of an image stack (Crocker and Hoffman, 2007; Crocker and Grier, 1996), providing its position in space and total intensity (Figure 4B and Materials and Methods). This approach allows the accurate measurement of 3D displacements of individual spindle poles labeled with Spc42-CFP Cerulean over time at ∼20-nm precision (Figure 4C), with a resolution limit of ∼500 nm (Figure 4D). The total intensity of each pole of a bipolar spindle can be correlated with the pole's position relative to the bud neck plane (mean position over 20 min, τ = 20 s). We find that for the majority of spindles (∼80%) the brighter pole is correlated with proximity to the neck. Thus each pole can be assigned as either the old pole inherited from the previous cell cycle (high intensity) or as the new pole assembled in G1 (low intensity). Our method of pole assignment (Supplemental Figure S1) simplifies spindle pole tracking, as it does not require the use of a slow-maturing red fluorescent protein, which results in little or no signal from the new pole (Pereira et al., 2001) or color switching (Hotz et al., 2012).
FIGURE 4:
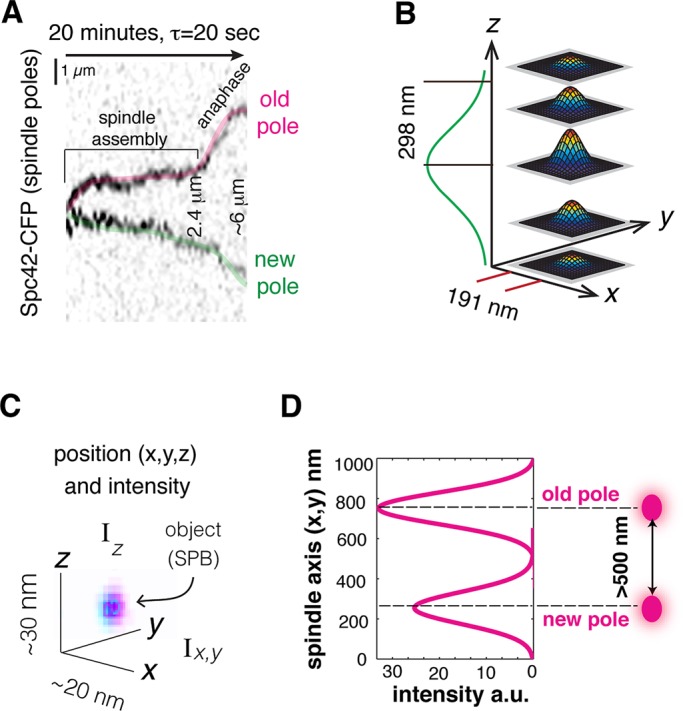
Spindle-pole tracking and methods of analysis of spindle dynamical behavior. (A) Spindle assembly and anaphase can be followed by tracking the 3D displacements of the spindle poles as a function of time. (B) Spindle poles, reported by Spc42-CFP Cerulean, are diffraction-limited, point-like objects whose fluorescence intensities in x, y, and z are fit to Gaussian distributions. (C) Subpixel measurements of pole position in coordinate space and fluorescence intensities are used for pole tracking. (D) Using 3D Gaussian fits to x, y, and z intensity distributions of each spindle pole provides a resolution limit of 500 nm.
The relative displacement of structures that remain within a single focal plane can be visualized in time and space using kymographs, which have been used for the analysis of the displacement of yeast centromeres (Pearson et al., 2001, 2003). We identified medial spindles (initial length 1–1.5 μm) for which both poles remained within a single 300-nm focal plane for the majority of the 20-min time lapse. These examples were used to create kymographs composed of extended projections of a time series of image stacks. The resolution limit and identity of the old and new poles can be easily distinguished in the kymograph. The kymograph of each spindle was then compared with the corresponding 3D tracking data.
We observed that all spindles undergo length fluctuations during their assembly. Length fluctuations indirectly report the stability of the spindle and, by extension, its structure. An example of a wild-type medial spindle, with an initial length of 1.19 μm, is shown in Figure 5A. In the first 8 min, the spindle undergoes large fluctuations in length, some of which are visible in the corresponding kymograph. The tracking data show that the amplitude of length fluctuations decreases over time; however, tilting of the spindle through more than one focal plane during the time lapse results in a lack of correspondence with the kymograph. Relative to the initial length of 1.19 μm, the spindle has no net increase in length at the end of the time lapse (1.18 μm; Figure 5A). An example of a γ-tubS360A medial spindle, with an initial length of 1.48 μm, is shown in Figure 5B. The spindle undergoes fluctuations in length within the first minute and at 7 min; however, spindle length increases steadily to a final length of 1.87 μm. In this example, the correspondence between the kymograph and tracking data reveals a phase of rapid growth (t = 8–18 min) corresponding to outward displacement of both old and new spindle poles relative to the spindle center.
FIGURE 5:
Analysis of spindle pole displacements in living cells. Spindle dynamical behavior was measured using 3D spindle-pole tracking and extended projection kymographs. (A) Both the kymograph and tracking data show that a wild-type spindle does not increase in net length, whereas pole tracking demonstrates that the amplitude of length fluctuations decreases over time. (B) Both the kymograph and tracking data show that a γ-tubS360A undergoes steady increase in length. The kymograph reveals that net increase in length correlates with outward displacement of both old and new spindle poles. (C) γ-tubS360D spindles are unstable, exhibiting very large fluctuations in length. Frequent tilting of the γ-tubS360D spindle through more than a single focal plane results in lack of correspondence between kymograph and pole tracking data and illustrates the need for pole tracking for accurate measurement of spindle dynamics. (D) Initial pole separation is delayed as a consequence of a cin8∆ loss-of-function mutation but is unaffected by γ-tubS360A and γ-tubS360D mutations.
Both the kymograph and pole-tracking data show that γ-tubS360D spindles are extremely unstable relative to wild-type and γ-tubS360A spindles. An example of a γ-tubS360D spindle, with an initial length of 2.78 μm, is shown in Figure 5C. The spindle undergoes large fluctuations in length throughout the time lapse. Of the 75 cells imaged, this is the only example of a spindle for which the kymograph and tracking data corresponded over portions of the time lapse. This example demonstrates that kymographs are of limited use for quantitative analysis of spindle behavior.
The instability of γ-tubS360D spindles led us to investigate whether this mutation blocks initial pole separation, as shown for cells lacking the kinesin-5 orthologue Cin8 (Crasta et al., 2006). Spindle poles first separate in S phase in budding yeast, soon after bud emergence (Winey and O'Toole, 2001). The majority (71%, n = 65) of wild-type cells with a small bud of ≤0.3 of the volume of the mother cell contain a bipolar spindle (Figure 5D). Initial pole separation is defective in cin8 mutants, and consistent with this, 52% of cells with a bud ∼0.5 of the volume of the mother cell (n = 57) contained unseparated spindle poles. Neither the γ-tubS360A nor the γ-tubS360D mutation perturbed initial pole separation, with, respectively, 79% (n = 68) and 81% (n = 68) of cells with a small bud ≤0.3 of the volume of the mother cell containing a bipolar spindle (Figure 5D).
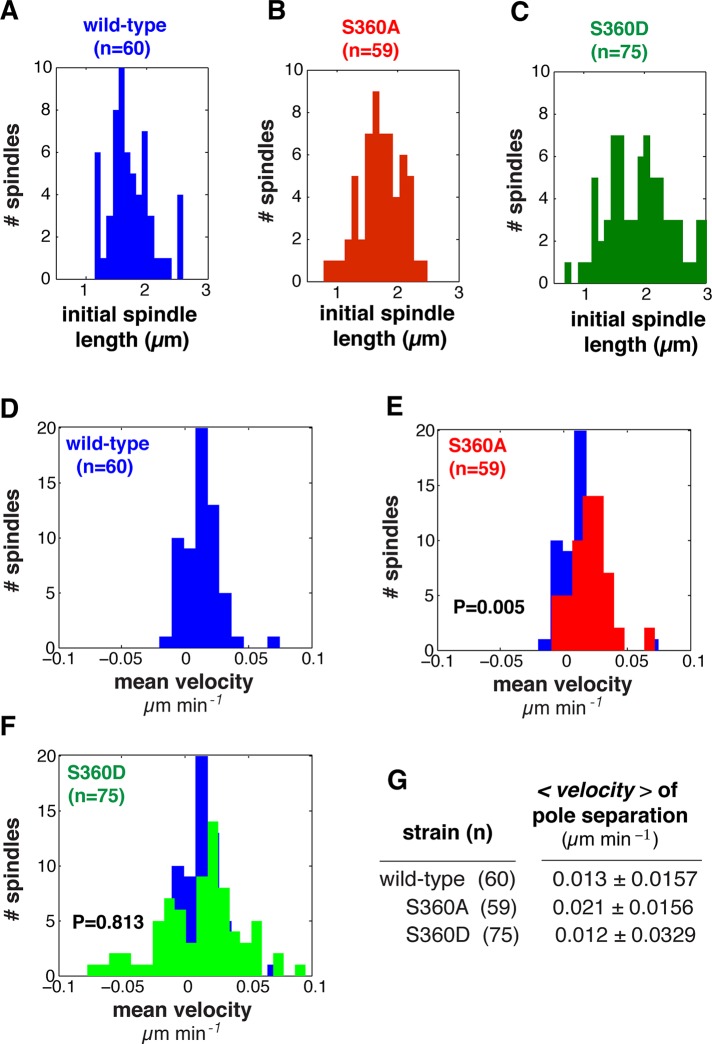
Inhibiting S360 phosphorylation increases pole separation velocity during spindle assembly
Little is known about how spindle organization might influence the separation of the poles before anaphase. The tracking and corresponding kymographs of spindle pole separation suggested that the velocity of pole separation is increased in the γ-tubS360A mutant (Figure 5B), which was unexpected. To determine whether this was the case, we measured the dynamic behavior of wild-type, γ-tubS360A, and γ-tubS360D spindles with initial lengths ranging from ∼1 to 3 μm (Figure 6, A–C). We found that the velocity of pole separation of γ-tubS360A spindles (Figure 6E; n = 59, 0.021 ± 0.0156 μm/min) increased 75% relative to that of wild-type spindles (Figure 6D; n = 60, 0.013 ± 0.0157 μm/min, p = 0.005). Whereas the mean velocity of pole separation in γ-tubS360D spindles (Figure 6F; n = 75, 0.012 μm/min) was similar to that of wild type, the variance (σ = 0.0329) of γ-tubS360D was much larger, consistent with large fluctuations in length characterized by rapid shrinking and elongation events.
FIGURE 6:
Inhibiting S360 phosphorylation increases the velocity of pole separation during spindle assembly. (A–C) Distribution of initial spindle lengths in wild-type, γ-tubS360A, and γ-tubS360D cells used for analysis of spindle assembly shown in Figures 5–7. (D–F) Distribution of mean pole separation velocities of individual wild-type (n = 60, blue), γ-tubS360A (n = 59, red), and γ-tubS360D (n = 75, green) spindles. (G) Mean velocity of the wild-type, γ-tubS360A, and γ-tubS360D spindle populations. The mean velocity of pole separation is significantly increased in γ-tubS360A spindles (0.021 ± 0.0156 μm/min) relative to wild type (0.013 ± 0.0145 μm/min; p = 0.005). Mean velocity of pole separation for γ-tubS360D (0.012 ± 0.0329 μm/min) is decreased relative to wild type but was not significantly different (p = 0.813).
The amplitude of length fluctuations decreases as spindle length increases
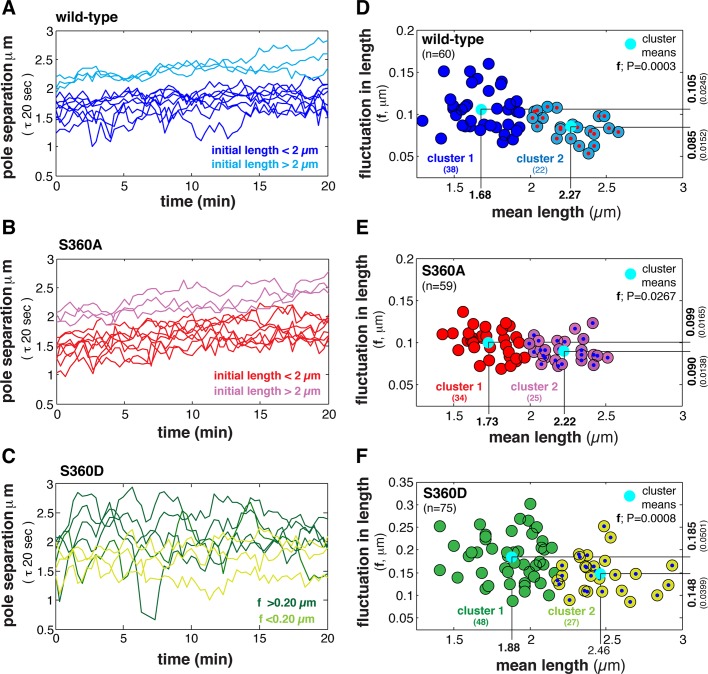
Spindle stability is expected to increase as the process of spindle assembly is completed, resulting in a compact bundle of p-MTs and the completion of biorientation. The amplitude of length fluctuations is therefore expected to decrease over time. Our analysis of individual spindles using kymographs suggested that length fluctuations are not isotropic throughout spindle assembly but instead tend to decrease in amplitude over time. In general, wild-type spindles with an initial length of >2 μm exhibited small/uniform length fluctuations and a net increase in length, whereas spindles with an initial length of <2 μm exhibited larger length fluctuations with little increase in length (Figure 7A). In contrast, γ-tubS360A spindles with initial lengths between 1 and 2.3 μm exhibited moderate length fluctuations, and most spindles with an initial length of <2 μm steadily increased in length (Figure 7B). This suggested that inhibiting S360 phosphorylation results in spindles <2 μm in length exhibiting dynamic behavior that is observed for wild-type spindles >2 μm in length. Conversely, γ-tubS360D spindles exhibited large length fluctuations regardless of initial spindle length (Figure 7C).
FIGURE 7:
Inhibiting S360 phosphorylation dampens fluctuations during early steps in spindle assembly. (A–C) Length dynamics of individual representative wild-type, γ-tubS360A, and γ-tubS360D spindles: blue and light blue (A), red and purple (B), and green and light green (C), respectively. Tracks represent spindles with initial length <2 or ≥2 μm, respectively. (D–F) Analysis of length fluctuations and their relationship to spindle length in wild-type (n = 60, blue), γ-tubS360A (n = 59, red), and γ-tubS360D (n = 75, green) cells. The mean fluctuation in length (<f>, in micrometers) is plotted as a function of the mean spindle length (<L>), using data collected over 20 min at a time step of 20 s. Spindles could be clustered using K-means into two populations based on the amplitude of length fluctuations (cluster 1, large; cluster 2, small), with a transition length of ∼1.9 μm for the wild-type (D) and γ-tubS360A (E) populations. Length fluctuations of wild-type cluster 1 spindles (<2 μm in length) are larger than fluctuations in γ-tubS360D cluster 1 spindles (F-test, p < 0.05). (F) Length fluctuations in the γ-tubS360D spindle population were larger relative to all preanaphase wild-type spindles (F-test, p < 0.01) but decrease in amplitude as spindle length increases.
We next analyzed the amplitude of length fluctuations as a function of mean length of populations of wild-type, γ-tubS360A, and γ-tubS360D preanaphase spindles (Figure 7, D–F). In general, the amplitude of fluctuations decreased as spindle length increased. Two distinct subpopulations of wild-type, γ-tubS360A, and γ-tubS360D spindles were identified by k-means clustering. Spindles <1.9 μm in length (cluster 1) underwent larger fluctuations. Cluster 2 (1.9–2.6 μm) contained spindles that were relatively stable.
S360 phosphorylation state can dampen (S360A) or increase (S360D) length fluctuations
Our analysis of length fluctuations during spindle assembly (Figure 6, A–C) yielded three outcomes. First, wild-type spindles <2 μm in length are less stable (cluster 1) than spindles ≥2 μm in length (p = 0.0003; Figure 7, A and D). Our analysis of individual wild-type spindles (Figures 5A and 7A) suggests that the dynamics of ∼2-μm spindles is characterized by steady growth. Second, fluctuations of all γ-tubS360A spindles are damped relative to wild-type and γ-tubS360D spindles, and although cluster 1 and cluster 2 populations exist (p = 0.0267), all γ-tubS360A spindles are surprisingly similar in behavior (Figure 6, B and E). A two-tailed F-test was used to compare the fluctuations of cluster 1 wild-type and cluster 1 γ-tubS360A spindles. It revealed that γ-tubS360A spindles were characterized by more uniform (damped) fluctuations relative to wild type (p = 0.001; Figure 7, D and E). Third, whereas the amplitude of length fluctuations decreased as γ-tubS360D spindle length increased, these spindles are unstable relative to wild-type spindles.
Damped fluctuations and steady growth are correlated with the completion of spindle assembly
Wild-type spindles exhibit stereotyped behavior during anaphase (Straight et al., 1997; Yang et al., 1997), characterized by three modes of pole separation; initial rapid pole separation of ∼0.7 μm/min, a mid-anaphase slowing of pole separation at ∼4 μm, followed by a slow-growth plateau phase (Supplemental Figure S2). Using this information, we determined the minimum and maximum values for spindle length at the transition to anaphase for wild-type (n = 60; 2.17, 2.79), γ-tubS360A (n = 73; 2.26, 2.93), and γ-tubS360D (n = 18; 2.08, 3.28) spindles (Supplemental Figure S2). These minimum and maximum lengths overlaps considerably with the distribution of mean lengths of cluster 2 spindles (Figure 7, D–F), which suggests that the majority of these spindles are completing the process of spindle assembly.
Inhibiting S360 phosphorylation increases outward displacement of the new spindle pole
Pole separation during spindle assembly is the outcome of both internal and external forces applied through the spindle microtubules and the cytoplasmic microtubules (c-MTs). This process has been extensively characterized in budding yeast. Three classes of motor proteins contribute to the forces applied through the c-MTs: cytoplasmic dynein, kinesins, and type V myosin (Carminati and Stearns, 1997; Lee et al., 2000; Yeh et al., 2000; Liakopoulos et al., 2003). Before anaphase, these forces are applied asymmetrically such that the old pole is pulled toward the plane of cleavage at the bud neck while the new pole is pushed. As a consequence, net increase in spindle length is expected to be the result of net outward displacement of the old pole, with little or no contribution from the displacements of the new pole.
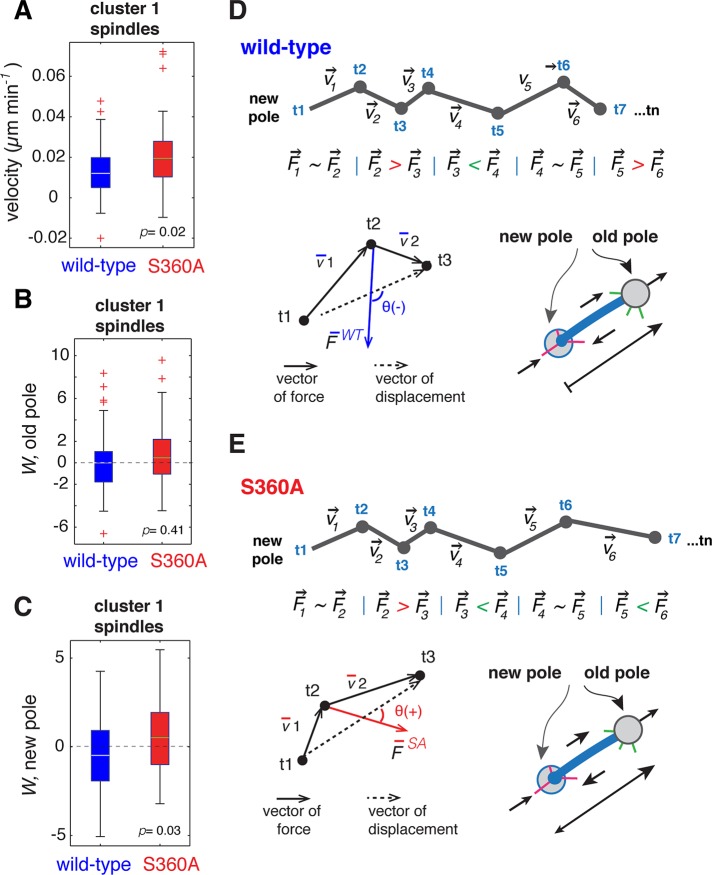
We found that inhibiting S360 phosphorylation increases the velocity of pole separation during the early phase of spindle assembly (Figure 6G). In an effort to understand the basis of the increased velocity of pole separation observed for γ-tubS360A spindles, we measured the individual displacements of the old and new spindle poles of wild-type and γ-tubS360A cluster 1 (<1.9 μm) spindles. We compared the displacements of oldWT versus oldS360A and newWT versus newS360A poles. Although it is not possible to directly measure forces applied to the spindle poles in living yeast cells, the displacement of each spindle pole (old, new) relative to the vector of total force indirectly reports the ensemble forces (including opposing force from c-MTs) in play at each spindle pole. The velocity of pole separation was increased in cluster 1 γ-tubS360A spindles relative to cluster 1 wild-type spindles (p = 0.02; Figure 8A). The displacement of the old pole (Wold) was characterized by overall acceleration in wild-type (0.065 ± 3.282) and γ-tubS360A spindles (0.10 ± 2.900; p = 0.41; Figure 8B). The displacement of the new pole in wild-type spindles (−0.31 ± 2.143) was characterized by overall deceleration, whereas outward displacement dominated at the new pole (Wnew) in γ-tubS360A spindles (0.70 ± 2.139; p = 0.03; Figure 8C). Thus, in wild-type spindles, there was an overall deceleration of the new pole (Figure 8D), whereas in γ-tubS360A spindles the displacement of both old and new poles was characterized by overall acceleration (Figure 8E). This is consistent with the additional acceleration of the new spindle pole contributing to the overall increase in the velocity of pole separation of γ-tubS360A spindles.
FIGURE 8:
Measurement of individual pole displacements during spindle assembly. (A) Mean velocity of pole separation for wild-type and γ-tubS360A cluster 1 spindles (Figure 7). Measurement of the vector of total force relative to the vector of displacement (W) for the old or new spindle poles. Each spindle pole moves from t1 to t2 to t3 over time with corresponding velocities v1 and v2. Movement (t1…t2…t3) results from the total force (F) applied to the pole relative to the vector of displacement. (B) W of the old pole in wild-type and γ-tubS360A spindles is positive (force is applied toward the vector of displacement; >90°) and not significantly different (p = 0.411). (C) W of new pole in the γ-tubS360A mutant (0.70 ± 2.139 μm2/min2) was significantly larger than in wild type (−0.31 ± 2.143 μm2/min2) and demonstrates that overall acceleration of the new pole occurs in γ-tubS360A spindles. The wild type had negative W and defines overall deceleration of the new spindle pole. For plots of velocity (A) and W (B, C), the edges of the boxes represent the 25th and 75th percentiles, whiskers represent extreme data points covering 99.3% of the data distribution, and + represent outliers. Green/yellow lines (boxes) are mean values. (D, E) Cartoon representation of relative displacements of the new pole in wild-type (D) and γ-tubS360A (E) spindles.
DISCUSSION
S360 phosphorylation state contributes to the formation of interpolar spindle microtubules
In this study, we demonstrate that the S360 phosphorylation state influences antiparallel pairing, with increased pairing when S360 phosphorylation is inhibited as a result of the γ-tubS360A mutation and a severe defect in the formation of p-MTs in the γ-tubS360D phosphomimetic mutant. The γ-tubS360A and γ-tubS360D mutations offered an opportunity to investigate how p-MTs form. In the case of the γ-tubS360A mutation, we detected antiparallel microtubule pairs, which we call pa-MTs, in medial spindles that contain a normal number (approximately six) of p-MTs. Because pa-MTs are infrequently observed in wild-type medial spindles (only one of six models) but are observed in short (<1 μm) wild-type spindles, they likely form before p-MTs and may represent a transient intermediate in p-MT formation.
The presence of both p-MTs and pa-MTs in γ-tubS360A medial spindles suggests that blocking phosphorylation of S360 does not inhibit either antiparallel pairing (e.g., formation of pa-MTs) or the extension of pairing overlap required to form p-MTs. If inhibiting S360 promotes pairing and p-MT formation, it follows that constitutive phosphorylation of S360 would be expected to inhibit the formation of pa-MTs and p-MTs. We found that, indeed, p-MT formation is defective in the γ-tubS360D mutant. γ-tubS360D spindles contain pa-MTs, however, suggesting that phosphorylation of S360 causes a specific defect in the extension of antiparallel microtubule overlap that is required to form a p-MT.
If a pairing event (pa-MT) generally leads to the formation of a p-MT in wild-type spindles, the combination of p-MTs and pa-MTs in γ-tubS360A medial spindles suggests that availability of cross-linking factors also contributes to limiting the number p-MTs formed during spindle assembly. Our observation that the total contour overlap in wild-type and γ-tubS360A medial spindles is equally distributed over a variable number of pairs (∼6 in wild type vs. ∼10 in γ-tubS360A) is consistent with a limiting amount of at least one cross-linker protein that is required for pairing. We propose that S360 phosphorylation, in combination with the availability and/or activity of cross-linking proteins, enforces the formation of pa-MTs that will efficiently convert to p-MTs during early steps in spindle assembly.
pa-MTs form early and are sufficient for initial pole separation
The formation of p-MTs is promoted by the kinesin-5 orthologue Cin8 and to a lesser extent Kip1 (Hoyt et al., 1992). The homotetrameric structure of kinesin-5 motors is believed to enable cross-linking of antiparallel microtubules during spindle assembly, followed by the antiparallel sliding of p-MTs during anaphase (Straight et al., 1998). Kar3 (kinesin-14), Bim1 (EB1), and Ase1 (PRC-1) also provide structural support to the anaphase spindle by cross-linking p-MTs. Deletion of Cin8 results in a significant defect in initial pole separation, indicating that its function in cross-linking antiparallel microtubules occurs early and is crucial for initial pole separation (Saunders and Hoyt, 1992; Kotwaliwale et al., 2007; Gardner et al., 2008b; Zimniak et al., 2009). It was reported that initial pole separation does not require the motor activity of Cin8 (Crasta et al., 2006). Side-by-side spindle poles each have a small number of long (400–600 nm) microtubules, which are paired at a very high angle (O'Toole et al., 1999). We propose that pa-MTs form at this time and are sufficient for initial pole separation. This model for pa-MT formation and function is consistent with our finding that pole separation is not defective in γ-tubS360D spindles, which contain numerous pa-MTs but rarely p-MTs.
p-MTs influence the velocity of pole separation before anaphase
Measurements of the length fluctuations of spindles provide an indirect yet quantitative means of measuring their relative stability, which then can be correlated with spindle structure (tomographic models). We found that γ-tubS360D spindles lacking p-MTs are unstable throughout spindle assembly, suggesting that p-MTs have an important role in maintaining spindle stability before anaphase. We find that fluctuations in spindle length, which indirectly report spindle stability, decrease as cells approach the metaphase–anaphase transition. Cross-linking proteins such as kinesin-5 are regulated by mitotic forms of Cdk1, which can increase or decrease the affinity of motor binding to microtubules (Cahu et al., 2008; Avunie-Masala et al., 2011). We observed that spindle dynamical behavior changes significantly at ∼2 μm (Figure 7), and this may reflect a change in the activity of proteins, such as kinesin-5, that dictate the behavior of the spindle through several activity modes: 1) microtubule depolymerization, 2) microtubule cross-linking, and finally 3) antiparallel sliding (Gardner et al., 2008a, b). We speculate that the activity of one or more proteins that perform antiparallel sliding required to increase antiparallel contour overlap may be sensitive to S360 phosphorylation—for example, may be inhibited by S360 phosphorylation. Correspondingly, antiparallel sliding may occur earlier if S360 cannot be phosphorylated. This model is consistent with our measurement of pole separation velocity, which is significantly increased in γ-tubS360A medial spindles regardless of their length.
The role of γ-tubulin in p-MT formation suggests that overlap extension requires (−)-end activity
Since its discovery in 1989 (Oakley and Oakley, 1989), γ-tubulin has been found to be a ubiquitous component of microtubule-organizing centers in animal, plant, and fungal cells. The nucleation function of γ-tubulin and the γ-TURC is essential for life, as null mutations or depletion of γ-tubulin or GRIPs are generally lethal. Although it is not surprising that γ-tubulin is required to assemble a functional spindle, S360 phosphorylation does not appear to contribute to the formation of microtubules. Our findings are surprising, given that formation of p-MTs during spindle assembly is believed to be governed through the activities of microtubule-associated proteins that interact with microtubules at their (+) ends and/or along their contour length. We propose that activities at microtubule (−) ends and the spindle pole also contribute to the specification of p-MTs. Numerous in vitro single-molecule measurements of kinesins, including Cin8/kinesin-5, have been performed with in vitro–assembled microtubules that have varying numbers of protofilaments (Cahu et al., 2008; Avunie-Masala et al., 2011; Roostalu et al., 2011), and there is no evidence that such changes in the microtubule lattice have a significant effect on Cin8 cross-linking or sliding activity. Thus, although formally possible, it is unlikely that the phosphorylation state of S360 promotes or inhibits pairing by changing the structure of the microtubule. The contribution of γ-tubulin is likely to be indirect, with S360 phosphorylation state perhaps influencing regulators of proteins that function in the formation of p-MTs and specifically in the extension of antiparallel overlap. Our findings suggest further investigation of regulatory kinases and phosphatases that associate with γ-tubulin and whether these interactions contribute to the proper function of microtubule-associated proteins.
Spindle assembly from the perspective of the (−) ends of microtubules
Components of the γ-TURC are evolutionarily conserved, with γ-tubulin, Spc97, and Spc98, the three principal proteins of the budding yeast γ-TURC, represented in unicellular eukaryotes and metazoans (Kollman et al., 2011). Although budding yeast γ-tubulin is divergent from the γ-tubulins of animals and plants (Marschall et al., 1996; Spang et al., 1996), the sequence containing S360 lies in one of several evolutionarily conserved regions (Keck et al., 2011). The sequence conservation of S360 and flanking residues suggests that S360 phosphorylation is coupled to a conserved function of γ-tubulin that contributes to early events in spindle assembly that are common to fungal and animal cells.
Much of our understanding of the process of spindle assembly and microtubule organization and dynamics during mitosis is based on analysis of the properties and behaviors of the (+) ends of spindle microtubules and activities of the proteins that bind to (+) ends and/or the contours of microtubules. Our findings suggest that the process of spindle assembly can be considered a series of events occurring at both ends of the spindle microtubules. From this perspective, γ-tubulin, and the γ-TURC, can be viewed as both a nucleation template and a launch pad that can receive regulatory inputs from the cell cycle machinery and use this information to appropriately deploy proteins that dictate microtubule dynamics and interactions that specify their fate as either k-MTs or p-MTs.
MATERIALS AND METHODS
Strain construction, genetic tests, and culture conditions
All strains used in this study are based in the haploid BY4741 wild-type strain (Brachmann et al., 1998) and are listed in Table 1. Budding yeast γ-tubulin mutations were introduced by PCR-directed homologous recombination into the genomic locus of the YBR4743 diploid strain expressing a Spc42-CFP fusion protein (pole marker; Cerulean variant) from the chromosomal locus. Isogenic haploid MATa strains expressing Spc42-CFP Cerulean that were wild type for γ-tubulin or bearing γ-tubS360A or γ-tubS360D mutations were isolated by tetrad dissection. For spindle analysis, data were collected from three (wild type, γ-tubS360D) or four independent haploid isolates (γ-tubS360A). Strain manipulation on solid media and growth of strains in liquid medium for microscopy and tomography were performed at 25°C.
TABLE 1:
Strains used in this study.
| Strain | Genotype | Notes | Source |
|---|---|---|---|
| YV856 | MATa SPC42-CFPCerulean-HygB; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0 | Microscopy; wild-type isolate 1 | This study |
| YV859 | MATa SPC42-CFPCerulean-HygB; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0 | Microscopy; wild-type isolate 2 | This study |
| YV860 | MATa SPC42-CFPCerulean-HygB; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0 | Microscopy; wild-type isolate 3 | This study |
| YV845 | MATa; tub4-S360A-NAT; SCP42-CFPCerulean-HygB; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0 | Microscopy; γ-TubS360A isolate 1 | This study |
| YV846 | MATa; tub4-S360A-NAT; SCP42-CFPCerulean-HygB; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0 | Microscopy; γ-TubS360A isolate 2 | This study |
| YV848 | MATa; tub4-S360A-NAT; SCP42-CFPCerulean-HygB; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0 | Microscopy; γ-TubS360A isolate 3 | This study |
| YV849 | MATa; tub4-S360A-NAT; SCP42-CFPCerulean-HygB; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0 | Microscopy; γ-TubS360A isolate 4 | This study |
| YV1607 | MATa; tub4-S360D-NAT; SCP42-CFPCerulean-HygB; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0 | Microscopy; γ-TubS360D isolate 1 | This study |
| YV1651 | MATa; tub4-S360D-NAT; SCP42-CFPCerulean-HygB; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0 | Microscopy; γ-TubS360D isolate 2 | This study |
| YV1652 | MATa; tub4-S360D-NAT; SCP42-CFPCerulean-HygB; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0 | Microscopy; γ-TubS360D isolate 3 | This study |
| YV447 | MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0 | Tomography; wild type | This study |
| YV1184 | MATa; tub4-S360A-NAT; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0 | Tomography; γ-TubS360A | This study |
| YV1209 | MATa; tub4-S360D-NAT; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0 | Tomography; γ-TubS360D | This study |
Tomography
Log-phase cells were prepared for electron microscopy as previously described (Winey et al., 1995). Electron tomography was carried out as previously described (O'Toole et al., 2002). Tomograms were reconstructed using the IMOD software package (Kremer et al., 1996; Mastronarde, 1997). Seven wild-type, five S360D, and six S360A complete spindles were reconstructed and spindle microtubules modeled. Microtubule lengths were measured from model contour data using the program IMODINFO, and the core bundle analysis was performed using the program MTPAIRING as previously described (Winey et al., 1995). The program FIBERPITCH was used to measure the angle between each microtubule and the spindle axis as described in Winey et al. (2005). Secondary pitch-pairing analysis was performed using MatLab (MathWorks, Natick, MA).
Live-cell imaging
Cells expressing Spc42-CFP Cerulean were imaged at 25°C as previously described (Rauch et al., 2010) using a custom spinning-disk confocal microscope, a Leica (Wetzlar, Germany) DM6000 inverted body with a 100×/1.46 numerical aperture plan-Apochromat objective, piezo stage (Mad City Labs, Madison, WI/Applied Scientific Instrumentation, Eugene, OR) and DAQ control board, Borealis head (conversion of a QLC100 Visitek unit; Quorum Technologies, Guelph, Canada), laser merge module (Spectral Applied Research, Richmond Hill, Canada), 441- and 493-nm solid-state lasers (Coherent, Santa Clara, CA), and a water-cooled, back-thinned electron-multiplying charge-coupled device camera with photon counting enabled (Hamamatsu, Hamamatsu, Japan). MetaMorph, version 7.7.3.0 (Universal Imaging, West Chester, PA), was used for image acquisition. Images were collected in a streaming regime as a stack (300-nm z-steps in 27–31 focal planes) with an exposure time for 50 ms/focal plane. For most studies, capture time was 20 min with 20-s interval (τ) between acquisitions.
Spindle analysis
Pole-tracking algorithm.
All data analysis was implemented in MatLab. The fluorescence signal from Spc42-CFP Cerulean reports the position of the spindle poles, which are tracked in successive time steps as point-like, diffraction-limited objects using methods described in Crocker and Hoffman (2007) and Crocker and Grier (1996). Spc42-CFP Cerulean was used to provide the coordinate position of each pole, as it is one of the first proteins recruited to the spindle pole body in G1 phase (Jaspersen and Winey, 2004). Fluorescence images were filtered using a 3D band-pass Gaussian kernel. By use of a spatial cut-off for the filter kernel close to the characteristic size of a spindle pole body (diameter, ∼100 nm; Winey et al., 1995), high-frequency noise was attenuated to a level below that of true features. The central part of the diffraction pattern, or Airy disk, can be approximated to a Gaussian profile. Fitting the profile of fluorescent signal of Spc42-CFP Cerulean to a Gaussian distribution in three dimensions detects the x, y, z position of each spindle pole with subpixel resolution (∼20 nm in x and y and ∼30 nm in z dimensions; Figure 4). Images were acquired over 20 min with 20-s intervals and 27–31 z-stacks. Each spindle pole is tracked independently.
To obtain the trajectory of each spindle pole, coordinates of the feature extracted from raw images are tracked in time, using a cut-off distance relative to the previous time step to identify the same feature displaced in space between adjacent time steps. Preanaphase cells undergoing spindle assembly and cells that went through anaphase were identified in the total sample population using the spindle-tracking algorithm shown in Supplemental Figure S2. Preanaphase cells were used for the analysis of spindle assembly as shown in Figure 4. We use this method for the analysis of populations of wild-type, S360A, and S360D cells to measure relative protein abundance at poles over time, extract instantaneous spindle length, quantify velocities of pole displacement and length fluctuations, and position of the poles in coordinate space relative to the bud neck (Rauch et al., 2010). Fluctuation of the spindle length was calculated for each spindle as <ν2> − <ν>2, where ν is a matrix of instantaneous velocities and <ν> is mean velocity averaged over 20 min for the population of cells. f = square root of the fluctuation (Figure 7).
Cells that transitioned to anaphase were used to determine the minimum and maximum values for the length at which wild-type or mutant cells transition to anaphase as follows: first, the curve of spindle elongation for each cell was fit to an eighth-order polynomial function. Local maxima and minima of the acceleration curve for the fitted function allow one to find biological transition points during anaphase spindle elongation that split spindle elongation into four modes: spindle assembly (preanaphase) and the fast elongation, slow elongation, and plateau phases of anaphase. Length and velocity parameters describing the metaphase-to-anaphase transition and early anaphase are automatically extracted and are provided in Supplemental Figure S2D.
Assignment of old and new spindle poles.
Old and new poles were assigned based on relative intensity of the Spc42-CFP Cerulean pole reporter. The maturation time of the Cerulean fluorophore is 49 min (Gordon et al., 2007), and the “new” pole has a lower fluorescence intensity than the old pole for most of the cell cycle. The integrated intensity of the fluorescent Spc42-CFP Cerulean–labeled poles was acquired by fitting each pole's intensity distribution to a Gaussian function. The integrated intensities were averaged over 20 min for each pole and compared with the position of that pole inside the cell relative to the neck plane (<Ip1> vs. <Ip2>). In 82.6% of cases in which spindles transitioned to anaphase (n = 150 cells; Figure 4D) the bright “old” pole approached and transited the neck plane, as expected based on previous studies (Pereira et al., 2001).
Measurement of new and old pole displacements during spindle assembly.
In a low–Reynolds number regime, viscous drag dominates the movement of particles (Purcell, 1977). In a living yeast cell the force applied to a spindle pole cannot be measured directly without perturbations that confound any analysis of the relationship between force and spindle dynamic behaviors. We simplify the indirect quantification of applied force by assuming that in this system force is proportional to velocity. On the basis of tomography measurements, we assume the shape and size of all spindle poles is equivalent for all conditions. On the basis of the detection and positions of the old and new spindle poles over time, we measured the vector of displacement, and based on this, we computed approximate speed and acceleration vectors. The vector of total force affecting the old/new poles is proportional to the magnitude of acceleration (Figure 8). Total force applied to the poles relative to the displacement of each pole was calculated as W = |F|⋅|d| cos θ, where W (arbitrary units) is total force relative to the displacement of the pole, F is the vector of total force, d is the vector of displacement, and θ is the angle between F and d. The angle of F and d can be <90° or >90°, with +W and −W representing acceleration or deceleration, respectively, of the pole relative to the previous time step.
Statistical analysis
All statistical analysis was performed using Matlab. Distributions were compared with the null hypothesis using Welch's unpaired t test. Spindles were clustered by K-means using mean fluctuation and spindle length. Two populations of spindles were optimal for minimizing the sum of point-to-centroid distances and are considered significantly different populations (based on fluctuation amplitude) if p < 0.05.
Note added in proof. Prior to publication, Ear et al. (2013)published an article describing Clb3-Cdk1 phosphorylation of gamma-tubulin.
Supplementary Material
Acknowledgments
We thank members of the Vogel and Winey labs, Gary Brouhard, Justin Kollman, Paul Lasko, and Stephen Michnick for comments on the manuscript; Yves Barrel and Paul Maddox for discussions; and Vincent Pelletier for initial contributions to the development of tracking algorithms. We also thank Tom Giddings and Christina Clarissa for electron microscope specimen preparation. E.N. is supported by a Collaborative Research and Training Experience–Cellular Dynamics of Macromolecular Complexes (Natural Sciences and Engineering Research Council of Canada) graduate training fellowship. E.O.T. is supported by Grants RR-000592 from the National Center for Research Resources of the National Institutes of Health and 8P41GM103431 from the National Institute of General Medical Sciences to Andreas Hoenger. This study was supported by National Institutes of Health Operating Grant R01 GM51312 (M.W.) and Canadian Institutes of Health Research Operating Grants MOP-64404 and MOP-123335 (J.V.).
Abbreviations used:
- c-MT
cytoplasmic microtubule
- GRIP
γ-tubulin interacting protein
- γ-TURC
γ-tubulin ring complex
- k-MT
kinetochore microtubule
- pa-MT
polar-approach microtubule (midzone)
- p-MT
polar microtubule (midzone)
- W
vector of total force relative to the vector of displacement
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-05-0232) on August 21, 2013.
REFERENCES
- Avunie-Masala R, Movshovich N, Nissenkorn Y, Gerson-Gurwitz A, Fridman V, Koivomagi M, Loog M, Hoyt MA, Zaritsky A, Gheber L. Phospho-regulation of kinesin-5 during anaphase spindle elongation. J Cell Sci. 2011;124:873–878. doi: 10.1242/jcs.077396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck DC, Joglekar AP, Bloom KS. Design features of a mitotic spindle: balancing tension and compression at a single microtubule kinetochore interface in budding yeast. Annu Rev Genet. 2008;42:335–359. doi: 10.1146/annurev.genet.42.110807.091620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Cahu J, Olichon A, Hentrich C, Schek H, Drinjakovic J, Zhang C, Doherty-Kirby A, Lajoie G, Surrey T. Phosphorylation by Cdk1 increases the binding of Eg5 to microtubules in vitro and in Xenopus egg extract spindles. PLoS One. 2008;3:e3936. doi: 10.1371/journal.pone.0003936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati JL, Stearns T. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta K, Huang P, Morgan G, Winey, Surana U. Cdk1 regulates centrosome separation by restraining proteolysis of microtubule-associated proteins. EMBO J. 2006;25:2551–2563. doi: 10.1038/sj.emboj.7601136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker JC, Grier DG. Methods of digital video microscopy for colloidal studies. J Colloid Interface Sci. 1996;179:298–310. [Google Scholar]
- Crocker J, Hoffman B. Multiple particle tracking and two-point microrheology in cells. Methods Cell Biol. 2007;83:141–178. doi: 10.1016/S0091-679X(07)83007-X. [DOI] [PubMed] [Google Scholar]
- Ear PH, Booth MJ, Abd-Rabbo D, Moreno JK, Hall C, Chen D, Vogel J, Michnick SW. Dissection of Cdk1–cyclin complexes in vivo. Proc Natl Acad Sci USA. 2013;110:15716–15721. doi: 10.1073/pnas.1305420110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MK, et al. Chromosome congression by kinesin-5 motor-mediated disassembly of longer kinetochore microtubules. Cell. 2008a;135:894–906. doi: 10.1016/j.cell.2008.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MK, et al. The microtubule-based motor Kar3 and plus end-binding protein Bim1 provide structural support for the anaphase spindle. J Cell Biol. 2008b;180:91–100. doi: 10.1083/jcb.200710164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon KD, et al. Absolute calibration and characterization of the Multiband Imaging Photometer for Spitzer. II. 70 micron imaging. Publ Astron Soc Pac. 2007;119:1019–1037. [Google Scholar]
- Hotz M, Lengefeld J, Barral Y. The MEN mediates the effects of the spindle assembly checkpoint on Kar9-dependent spindle pole body inheritance in budding yeast. Cell Cycle. 2012;11:3109–3116. doi: 10.4161/cc.21504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, He L, Loo KK, Saunders WS. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J Cell Biol. 1992;118:109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Winey M. The budding yeast spindle pole body: structure, duplication, and function. Annu Rev Cell Dev Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- Keck JM, et al. A cell cycle phosphoproteome of the yeast centrosome. Science. 2011;332:1557–1561. doi: 10.1126/science.1205193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Pereira G, Geissler S, Grein K, Schiebel E. The spindle pole body component Spc97p interacts with the gamma-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. EMBO J. 1997;16:1550–1564. doi: 10.1093/emboj/16.7.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Schiebel E. Spc98p and Spc97p of the yeast gamma-tubulin complex mediate binding to the spindle pole body via their interaction with Spc110p. EMBO J. 1997;16:6985–6995. doi: 10.1093/emboj/16.23.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Merdes A, Mourey L, Agard DA. Microtubule nucleation by gamma-tubulin complexes. Nat Rev Mol Cell Biol. 2011;12:709–721. doi: 10.1038/nrm3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotwaliwale CV, Frei SB, Stern BM, Biggins S. A pathway containing the Ipl1/aurora protein kinase and the spindle midzone protein Ase1 regulates yeast spindle assembly. Dev Cell. 2007;13:433–445. doi: 10.1016/j.devcel.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Lee L, Tirnauer JS, Li J, Schuyler SC, Liu JY, Pellman D. Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science. 2000;287:2260–2262. doi: 10.1126/science.287.5461.2260. [DOI] [PubMed] [Google Scholar]
- Liakopoulos D, Kusch J, Grava S, Vogel J, Barral Y. Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell. 2003;112:561–574. doi: 10.1016/s0092-8674(03)00119-3. [DOI] [PubMed] [Google Scholar]
- Lin TC, Gombos L, Neuner A, Sebastian D, Olsen JV, Hrle A, Benda C, Schiebel E. Phosphorylation of the yeast gamma-tubulin Tub4 regulates microtubule function. PLoS One. 2011;6:e19700. doi: 10.1371/journal.pone.0019700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall LG, Jeng RL, Mulholland J, Stearns T. Analysis of Tub4p, a yeast gamma-tubulin-like protein: implications for microtubule-organizing center function. J Cell Biol. 1996;134:443–454. doi: 10.1083/jcb.134.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN. Dual-axis tomography: an approach with alignment methods that preserve resolution. J Struct Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Sedat JW, Alberts B, Agard DA. Microtubule nucleation by gamma-tubulin-containing rings in the centrosome. Nature. 1995;378:638–640. doi: 10.1038/378638a0. [DOI] [PubMed] [Google Scholar]
- Oakley CE, Oakley BR. Identification of gamma-tubulin, a new member of the tubulin superfamily encoded by mipA gene of Aspergillus nidulans. Nature. 1989;338:662–664. doi: 10.1038/338662a0. [DOI] [PubMed] [Google Scholar]
- O'Toole E, Greenan G, Lange KI, Srayko M, Muller-Reichert T. The role of gamma-tubulin in centrosomal microtubule organization. PLoS One. 2012;7:e29795. doi: 10.1371/journal.pone.0029795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole ET, Winey M, McIntosh JR. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:2017–2031. doi: 10.1091/mbc.10.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole ET, Winey M, McIntosh JR, Mastronarde DN. Electron tomography of yeast cells. Methods Enzymol. 2002;351:81–95. doi: 10.1016/s0076-6879(02)51842-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Maddox PS, Salmon ED, Bloom K. Budding yeast chromosome structure and dynamics during mitosis. J Cell Biol. 2001;152:1255–1266. doi: 10.1083/jcb.152.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Maddox PS, Zarzar TR, Salmon ED, Bloom K. Yeast kinetochores do not stabilize Stu2p-dependent spindle microtubule dynamics. Mol Biol Cell. 2003;14:4181–4195. doi: 10.1091/mbc.E03-03-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G, Tanaka TU, Nasmyth, Schiebel E. Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J. 2001;20:6359–6370. doi: 10.1093/emboj/20.22.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell EM. Life at low Reynolds number. Am J Phys. 1977;45:3–11. [Google Scholar]
- Rauch A, Nazarova E, Vogel J. Analysis of microtubules in budding yeast. Methods Cell Biol. 2010;97:277–306. doi: 10.1016/S0091-679X(10)97016-7. [DOI] [PubMed] [Google Scholar]
- Roostalu J, Hentrich C, Bieling P, Telley IA, Schiebel E, Surrey T. Directional switching of the kinesin Cin8 through motor coupling. Science. 2011;332:94–99. doi: 10.1126/science.1199945. [DOI] [PubMed] [Google Scholar]
- Saunders WS, Hoyt MA. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- Spang A, Geissler S, Grein K, Schiebel E. gamma-Tubulin-like Tub4p of Saccharomyces cerevisiae is associated with the spindle pole body substructures that organize microtubules and is required for mitotic spindle formation. J Cell Biol. 1996;134:429–441. doi: 10.1083/jcb.134.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Marshall WF, Sedat JW, Murray AW. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- Straight AF, Sedat JW, Murray AW. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J Cell Biol. 1998;143:687–694. doi: 10.1083/jcb.143.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O'Toole ET, Mastronarde DN, Giddings TH, Jr, McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Morgan GP, Straight PD, Giddings TH, Jr, Mastronarde DN. Three-dimensional ultrastructure of Saccharomyces cerevisiae meiotic spindles. Mol Biol Cell. 2005;16:1178–1188. doi: 10.1091/mbc.E04-09-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, O'Toole ET. The spindle cycle in budding yeast. Nat Cell Biol. 2001;3:E23–E27. doi: 10.1038/35050663. [DOI] [PubMed] [Google Scholar]
- Yang SS, Yeh E, Salmon ED, Bloom K. Identification of a mid-anaphase checkpoint in budding yeast. J Cell Biol. 1997;136:345–354. doi: 10.1083/jcb.136.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Yang C, Chin E, Maddox P, Salmon ED, Lew DJ, Bloom K. Dynamic positioning of mitotic spindles in yeast: role of microtubule motors and cortical determinants. Mol Biol Cell. 2000;11:3949–3961. doi: 10.1091/mbc.11.11.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Wong ML, Alberts B, Mitchison T. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]
- Zimniak T, Stengl K, Mechtler K, Westermann S. Phosphoregulation of the budding yeast EB1 homologue Bim1p by Aurora/Ipl1p. J Cell Biol. 2009;186:379–391. doi: 10.1083/jcb.200901036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.