Abstract
EcoP15I is the prototype of the Type III restriction enzyme family, comprised of two modification (Mod) subunits to which two (or one) restriction (Res) subunits are then added. The Mod subunits are responsible for DNA recognition and methylation, while the Res subunits are responsible for ATP hydrolysis and cleavage. Despite extensive biochemical and genetic studies, there is still no structural information on Type III restriction enzymes. We present here small angle X-ray scattering (SAXS) and analytical ultracentrifugation (AU) analysis of the EcoP15I holoenzyme and the Mod2 subcomplex. We show that the Mod2 subcomplex has a relatively compact shape with a radius of gyration (RG) of ~37.4 Å and a maximal dimension of ~ 110Å. The holoenzyme adopts an elongated crescent shape with an RG of ~65.3Å and a maximal dimension of ~ 218Å. From reconstructed SAXS envelopes, we postulate that Mod2 is likely docked in the middle of the holoenzyme with a Res subunit at each end. We discuss the implications of our model for EcoP15I action, whereby the Res subunits may come together and form a “sliding clamp” around the DNA.
Restriction-Modification (R-M) systems play crucial role in protecting bacterial and archaeal cells against invasion by foreign DNA. Based on their architecture, cofactors, and DNA recognition sequences, R-M systems can be broadly classified in three categories (I-III).1; 2 Type III restriction enzymes are large multimeric complexes that differ from the more common Type II enzymes (BamHI and EcoRI, for example) in requiring ATP for DNA hydrolysis. In addition, the DNA recognition, cleavage, methylation, and ATP dependent functions of Type III enzymes are all encoded within the same complex.3; 4 Despite decades of biochemical and genetic studies, there is still no structural information on Type III restriction enzymes.
EcoP15I is the prototype of the Type III restriction enzyme family, considered to contain two modification (Mod) and two restriction (Res) subunits in a heterotetrameric Mod2Res2 complex with a molecular mass of ~370 kDa.5 Interestingly, EcoP15I has also recently been suggested to exist as a hetrotrimeric Mod2Res1 complex.6 The Mod subunits are responsible for DNA recognition and methylation, while the Res subunits are responsible for ATP hydrolysis and cleavage. Also, whereas the Res subunit is unstable on its own, the Mod subunit is stable and can be purified as a Mod2 dimer.5 Cleavage of the duplex DNA occurs at 25 or 26 (on one strand) and 27 or 28 bases (on second strand) downstream to 3’end of an asymmetric recognition sequence, 5’ – CAGCAG – 3’.3; 5 Because of its unusual recognition sequence (two “glutamine codons”), EcoP15I has been used to count the CAG repeats in the Huntington’s disease gene.7 And, since the enzyme cleaves DNA far away from its recognition sequence, the enzyme is commonly used in serial analysis of gene expression (SAGE) to quantitate gene expression in human tumors and eukaryotic pathogens.8
For efficient DNA cleavage, EcoP15I requires two inversely oriented recognition sequences that can be either head-to-head or tail-to-tail.9 The sites can be separated by thousands of base pairs but ATP hydrolysis is absolutely necessary for cleavage.3 Several models have been proposed for this long-range communication between the sites and the requirement for ATP, including models based DNA translocation, DNA looping, and DNA sliding.10; 11; 12 To begin to understand the action mechanism by Type III restriction enzymes at a more structural level, we undertook small angle X-ray scattering (SAXS) and analytical ultracentrifugation (AU) analysis of the EcoP15I holoenzyme and the Mod2 subcomplex. We show here that the EcoP15I holoenzyme has an extended crescent shape, with the Mod subunits likely occupying the center and the Res subunits on the periphery.
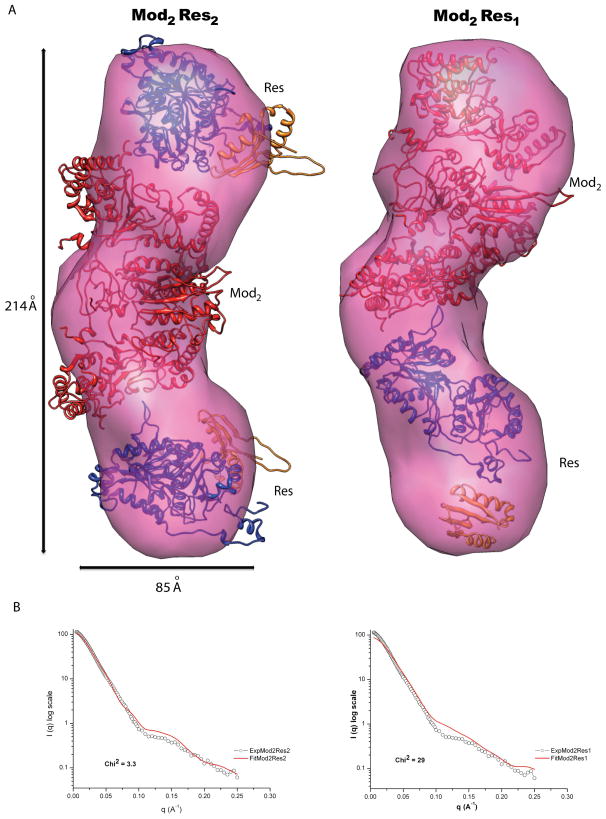
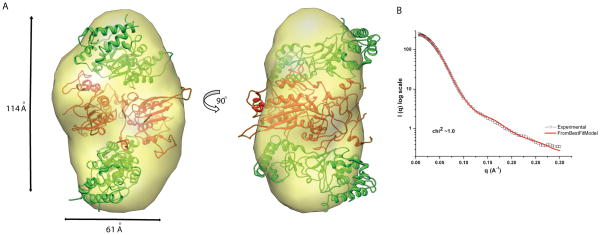
SAXS is an efficient method for investigating the overall size and shape of biological macromolecules in solution. Since the technique is highly sensitive to sample aggregation, we extensively purified both EcoP15I and the Mod subunit to achieve >95% purity and homogeneity (see legend of Figure 1 for details). Final samples were passed through a size exclusion column before SAXS data measurements. We observed single symmetrical peaks in the size exclusion profiles of EcoP15I and Mod2 in our experiments (Figure S1). We first measured SAXS data on the EcoP15I holoenzyme, wherein several dilutions in the range of 0.2 – 19.0 mg/ml were subjected to SAXS measurements. The buffer subtracted SAXS data from these samples were used for Guinier approximation to extrapolate radius of gyration (RG). An RG of ~ 65.3 Å and Dmax (maximum diameter of macromolecule) of ~ 218 Å were calculated for EcoP15I, which were consistent between independent samples of EcoP15I (Figure 1). Ab initio shape modeling was performed using DAMMIN, which employs simulated annealing based approach to restore solution structures in forms of dummy atoms defining overall shape.13 Models from 10 independent DAMMIN runs were averaged and filtered to yield the final shape. As shown in Figure 3, the averaged envelope of EcoP15I showed an elongated shaped molecule that is ~ 214 Å in length and ~ 85 Å in width. Significantly, although no symmetry was imposed during DAMMIN modeling, the final SAXS envelope of EcoP15I possessed an approximate two-fold symmetry. SAXS data were also collected and analyzed for the Mod2 subcomplex. Serial dilutions (0.14 to 11 mg/ml) of three independent samples of the Mod subunit yielded an RG of ~37.4 Å and a Dmax of ~110 Å. The averaged SAXS envelope, with imposed P2 symmetry, suggested a relatively compact shape for the Mod2 dimer that is ~ 114 Å in length and ~61 Å in width (Figure 2).
Figure 1. Small angle x-ray scattering (SAXS) data.
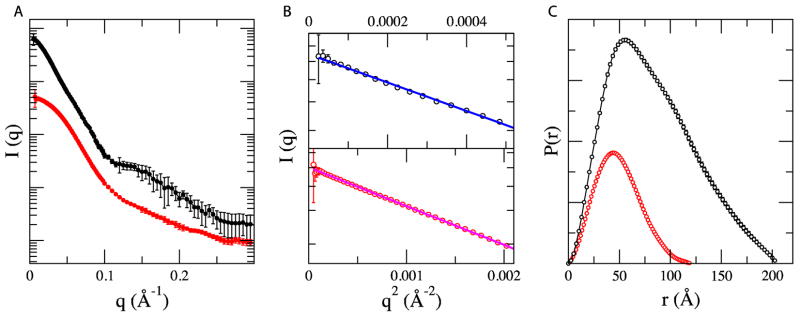
SAXS data were measured at beamline X9 of National Synchrotron Light Source at Brookhaven National Laboratory (BNL). Data processing, including azimuthal averaging and background subtraction, was performed using the pyXS software developed at beamline X9 (http://x9.nsls.bnl.gov/software/pyXS.htm). The wavelength of the x-ray beam and sample-to-detector distance was set to 0.918 Å and 2 meters respectively. A) Experimental SAXS profiles of holo enzyme (black line) and Mod2 subcomplex (red line). The two curves are offset for clarity. B) Corresponding Guinear fits for the experimental data with linear fit for EcoP15I (blue line) and Mod2 subcomplex (red line) in the limit qRg < 1.5. The radius of gyration (Rg) was derived by the Guinear approximation I(q) = I(0)exp(− q2Rg2/3). C) The intraparticle distance distribution (p (r)) functions of EcoP15I (black line) and Mod2 subcomplex (red line) computed for experimental data. The program GNOM28 was used to calculate the p (r) functions and corresponding maximum dimension of protein complexes, Dmax. The EcoP15I modification and restriction genes were PCR-amplified as a single 5.0 kb fragment from plasmid pSHI180 kindly provided by D.N. Rao (Indian Institute of Science, Bangalore). The PCR amplified mod-res DNA fragment were cloned in to an expression vector pRRS.29 The expression construct of EcoP15I (pRRS-ecoP15IRM) was transformed in to E. coli expression host NEB Express.Cells were harvested, lysed and the cleared soluble fraction was loaded onto a Heparin column and protein was eluted with a linear gradient of NaCl. Peak fractions containing EcoP15I activity were pooled and loaded onto a ceramic hydroxylapatite column (Bio-Rad) and eluted with a linear gradient of potassium phosphate. Peak fractions containing EcoP15I activity was pooled and was loaded onto a Source 15Q column (GE LifeSciences). Peak fractions from a linear gradient were collected, pooled and loaded on to a Superdex 200 column (GE lifesciences) in a buffer containing 25mM Tris-Cl, pH 7.5, 0.15M NaCl, 1mM DTT, 5% Glycerol. Mod2 subcomplex was purified similarly to EcoP15I holoenzyme.
Figure 3. Experimental SAXS envelops of EcoP15I and model building.
A) The upper panel shows overall shape of EcoP15I SAXS envelope in a surface representation with best-fit models of Mod2Res2 and Mod2Res1 complexes. Mod2 is shown in red ribbons whereas the helicase and endonuclease domains of Res subunit are shown in blue and orange ribbons respectively. B) Lower panel shows fitting of the experimental scattering of EcoP15I against scattering intensities computed from respective best-fit models in their SAXS envelopes.
Figure 2. SAXS envelope of Mod2 subcomplex.
A) The overall shape of Mod2 is shown. Two perpendicular views of envelopes, rotated by 90° around the y-axis are shown. The final experimental bead model was converted in to an EM style surface using the Situs package.30 A correlation based rigid body docking in program SASREF17 was performed to better-fit partial models of Mod (Mod-N in red; Mod-TRD in green) in the SAXS envelope. B) A comparison of experimental and computed scattering (from the best-fit model shown in A) resulted from program CRYSOL31 is shown.
There are no atomic structures for the Res or Mod subunits of Type III restriction enzymes. However, both Res and Mod subunits contain sequence motifs that can be used to derive partial homology models. The Mod subunit contains motifs characteristic of an adenine methyltransferase, while the Res subunit contains a helicase motif found commonly in members of superfamily 2 (SF2) helicases.14 Based on these motifs, we built partial models for the Mod and Res subunits using the protein modeling servers Phyre2 (http://www.sbg.bio.ic.ac.uk/~phyre2/), I-TASSER (http://zhanglab.ccmb.med.umich.edu/I-TASSER/), and the program Modeller.15 We built a model of the Mod subunit in two steps: Mod-N (aa34-285) was based on known N-6 methyltranseferase fold, whereas the C-terminal target recognition domain (TRD) Mod-TRD (aa 332-645) was based on the type IIG enzyme BpuSI (pdb id: 3S1S).16 The Res subunit model was built with two RecA-like folds (600 residues) as in SF2 DNA helicases and translocases. The endonuclease portion of Res subunit could only be built partially because of its low sequence homology to other proteins.
Next, we assembled these models in the SAXS envelopes by using a global rigid body modeling based protocol implemented in program SASREF.17 As shown in Figure 2B, two Mod subunit models, related by P2 symmetry, can be docked in the compact Mod2 subcomplex SAXS envelope with a low chi2 value of ~1.0. As docked, the Mod2 subcomplex is dimerized through the N6-methyltransferase fold (and not the TRD), analogous to that observed in other DNA methyltransferases homodimers, such as MboIIA (pdb id; 1G60)18 and TTHA0409 (pdb id; 2ZIG).19 Interestingly, this Mod2 orientation is maintained when, using SASREF, two substructures of Mod1, and Res subunits were treated as rigid bodies and allowed to fit in the EcoP15I SAXS envelope with P2 symmetry. The lowest chi2 value (~ 3.3) is obtained with Mod2 docked in the middle of the envelope and a Res subunit on each side. This is consistent with biochemical studies that show that a Res subunit is unstable on its own and requires interactions with the Mod subunit, whereas the Mod subunit is stable and purifies as a Mod2 dimer. A Ramachandran plot of this best fit Mod2Res2 model shows 87% of the residues in most favored and 8% in the allowed regions. We also attempted to fit Mod2Res1 in the SAXS envelope (Figure 3), but it gave a significantly poorer fit against the experimental scattering data with a chi2 > 20.
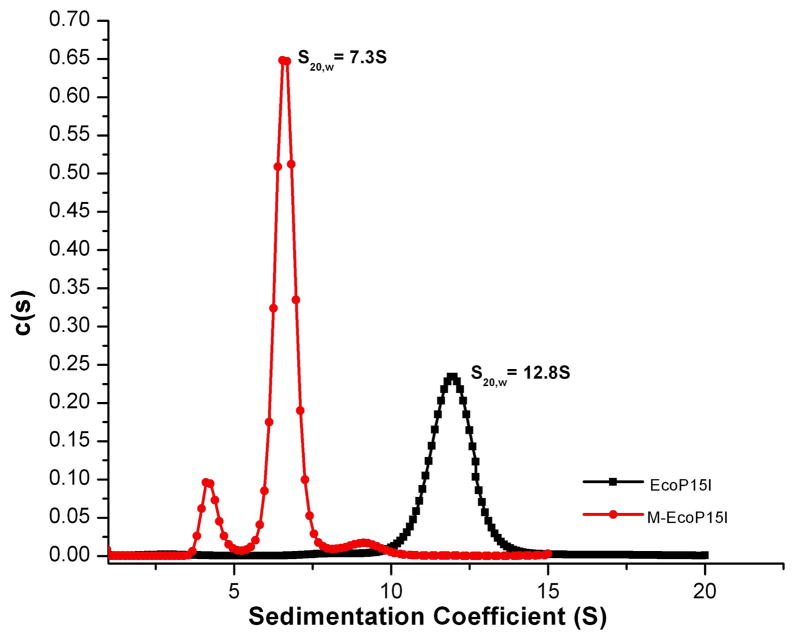
To further validate our SAXS models, we subjected the Mod2 subcomplex and the EcoP15I holoenzyme to analytical ultracentrifugation. The sedimentation velocity (SV) experiments were performed with centrifugation speeds in the range of 40000–48000 rpm. The data were analyzed by SEDFIT with a continuous sedimentation coefficients distribution (c (S)) Lamm equation model to retrieve sedimentation coefficients (S) values.20 The sedimentation velocity (SV) profile of Mod2 showed that the majority of protein (>85%) sediments as a single species (Figure 4) with an experimental sedimentation coefficient S20, w of ~7.3 S. A c(M) analysis performed in SEDFIT gives a molecular mass of ~140kDa for this species, which is in good agreement with the calculated molecular mass of a Mod2 dimer (~148kDa). Similarly, the EcoP15I holoenzyme sediments essentially as a single species with an experimental sedimentation coefficient S20,w of ~12.8.S (Figure 4). In a recent study6, EcoP15I was reported with S values ranging between 10.8S and 12.8S, where the 10.8S peak was characterized as Mod2Res1 and the 12.8S species was interpreted as an oligomeric form of this heterotrimeric complex (Mod2Res1)2. To more accurately estimate the molecular mass for the 12.8S species in our SV profile of the holoenzyme, we subjected EcoP15I to dynamic light scattering (DLS) and obtained experimental values of the translational diffusion coefficient (D) and the hydrodynamic radius (RH). The RH (~ 68 Å) of EcoP15I calculated from DLS measurements is consistent with Rg measured from SAXS data. A molecular mass of ~340kDa was calculated from the experimental values of the diffusion coefficient (3.7×10−7 cm2sec−1) and the sedimentation coefficient (12.8S). This suggests that the complex sedimenting at 12.8S in our experiments most likely contains two Mod and two Res subunits (calculated mass for Mod2Res2 is ~376kDa). To extend our analysis, we calculated theoretical sedimentation coefficients for both the SAXS Mod2 subcomplex and the EcoP15I holoenzyme using the SOMO bead modeling program in UltrascanII software package.21 The theoretical S values of ~7.7 S for the SAXS Mod2 subcomplex and ~13.3 S for the EcoP15I holoenzyme are in close agreement with the determined values ~7.3 S and ~12.8 S, respectively, and further corroborate the overall shapes and stoichiometry of the Mod2 subcomplex and the EcoP15I holoenzyme derived from our SAXS analysis. The sedimentation coefficient value we obtain for EcoP15I is in reasonable agreement with the values reported previously for a related Type III enzyme, EcoPI,5 which suggest that the overall shape is similar for members of the Type III R-M family. However, a recent finding that EcoP15I can assume the Mod2Res1 form at low protein concentration suggests a more dynamic assembly of Mod and Res subunits than previously considered.6 Indeed, the Type III enzyme PstII exists in Mod2Res1 and Mod2Res2 forms with differing enzymatic activities,22 and this may extend to other Type III enzymes.
Figure 4. Analytical Ultracentrifugation.
Sedimentation profiles of both EcoP15I and Mod2 from sedimentation velocity (SV) experiments are shown as a plot of c(s) (continuous sedimentation coefficient) distribution versus sedimentation coefficient (S). The sedimentation coefficients have been corrected to standard conditions and are reported in units of Svedbergs, where 1 S = 1 × 10−13 s. The SV experiments were carried out in an Optima XL-I analytical ultracentrifuge (Beckman Instruments Inc) using an An60Ti rotor. 0.5mg/ml Mod2 sample (~400μl) in 20mM Tris-Cl, pH 8.0, 0.2M NaCl was centrifuged at 40,000 rpm at 20°C. A total of 600 scans were collected at two wavelengths (270nm and 280nm) with 1 min intervals. For EcoP15I, we performed SV experiments at 48,000rpm at 25°C in 10mM Tris-Cl, pH 7.5, 0.15M NaCl, 1mM DTT. All samples were spun for ~10min at 13200 rpm prior to loading on to aluminum double sector cell. Sedimentation data were analyzed in SEDFIT20 program according to the c(s) distribution function of Lamm’s equation, which also yields an apparent weight-average frictional ratio (f/f0). The sedimentation coefficients from c(s) models and (f/f0) were then used to calculate the molecular weights for both proteins. In both cases, data correspond to monodisperse samples and were well fitted by single species model. SEDNTERP software (http://www.rasmb.bbri.org) was used to calculate the partial specific volume (v), the density (ρ) and the viscosity (η) of the solution.
Taken together, we present here the first structural analysis of a Type III restriction enzyme by SAXS. The analysis suggests that the EcoP15I enzyme has an elongated, slightly curved shape, wherein the Mod subunits are located towards the center and the Res subunits are at the edge. This arrangement of subunits favors the positioning of DNA towards the central region of the enzyme (Figure S2). Does the model provide any insights into how EcoP15I communicates between distant DNA sites? Type III restriction enzymes differ from most DNA helicases and translocases in their extremely low consumption of ATP.23; 24; 25 One suggestion, based on AFM studies,26 is that 3D DNA looping shortens the distance between the recognition sites, and thereby lowers the requirement for ATP for 1D translocation. Alternatively, based on single molecule and bulk solution cleavage assays, it is has been suggested that ATP is used primarily to catalyze a conformational switch in EcoP15I from a DNA recognition mode to a diffusive mode that is thermally driven and ATP independent.10; 11; 25 Based on the SAXS model, this would likely require the two Res subunits to come close together and form a “sliding clamp” around the DNA. Interestingly, an analogous open to closed transition has been proposed for the Mre11 nuclease in complex with Rad50 ATPase (MR complex) during double strand break repair.27 Significantly, the shape and dimensions of the apo MR complex (210Å × 70Å × 60Å) resemble those of EcoP15I, including the location of ATPase (Rad50) subunits at the tips of an extended crescent shape. The Rad50 ATPase subunits in MR are proposed to clamp onto the bound DNA upon ATP hydrolysis. Whether EcoP15I undergoes a similar clamping motion on ATP hydrolysis is unknown at present; however, the extended shape of the apo holoenzyme does raise this intriguing possibility and provides a basis for further experiments.
Supplementary Material
Highlights.
First small angle X-ray scattering (SAXS) analysis of a Type III restriction enzyme.
First low-resolution model for a Type III restriction enzyme.
-
Type III restriction enzymes adopt an elongated crescent shape.
Type III enzymes may form a “sliding clamp” around the DNA.
Acknowledgments
We thank the staff at BNL beamline X9 for the provision of X-ray facilities. This work was supported in part by grant GM044006 from the US National Institutes of Health. Use of the National Synchrotron Light Source, Brookhaven National Laboratory, was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-98CH10886.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev S, Dryden DT, Dybvig K, Firman K, Gromova ES, Gumport RI, Halford SE, Hattman S, Heitman J, Hornby DP, Janulaitis A, Jeltsch A, Josephsen J, Kiss A, Klaenhammer TR, Kobayashi I, Kong H, Kruger DH, Lacks S, Marinus MG, Miyahara M, Morgan RD, Murray NE, Nagaraja V, Piekarowicz A, Pingoud A, Raleigh E, Rao DN, Reich N, Repin VE, Selker EU, Shaw PC, Stein DC, Stoddard BL, Szybalski W, Trautner TA, Van Etten JL, Vitor JM, Wilson GG, Xu SY. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–12. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson GG, Murray NE. Restriction and modification systems. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- 3.Hadi SM, Bachi B, Shepherd JC, Yuan R, Ineichen K, Bickle TA. DNA recognition and cleavage by the EcoP15 restriction endonuclease. J Mol Biol. 1979;134:655–66. doi: 10.1016/0022-2836(79)90372-3. [DOI] [PubMed] [Google Scholar]
- 4.Reiser J, Yuan R. Purification and properties of the P15 specific restriction endonuclease from Escherichia coli. J Biol Chem. 1977;252:451–6. [PubMed] [Google Scholar]
- 5.Janscak P, Sandmeier U, Szczelkun MD, Bickle TA. Subunit assembly and mode of DNA cleavage of the type III restriction endonucleases EcoP1I and EcoP15I. J Mol Biol. 2001;306:417–31. doi: 10.1006/jmbi.2000.4411. [DOI] [PubMed] [Google Scholar]
- 6.Wyszomirski KH, Curth U, Alves J, Mackeldanz P, Moncke-Buchner E, Schutkowski M, Kruger DH, Reuter M. Type III restriction endonuclease EcoP15I is a heterotrimeric complex containing one Res subunit with several DNA-binding regions and ATPase activity. Nucleic Acids Res. 2011:1–13. doi: 10.1093/nar/gkr1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moncke-Buchner E, Reich S, Mucke M, Reuter M, Messer W, Wanker EE, Kruger DH. Counting CAG repeats in the Huntington's disease gene by restriction endonuclease EcoP15I cleavage. Nucleic Acids Res. 2002;30:e83. doi: 10.1093/nar/gnf082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsumura H, Kruger DH, Kahl G, Terauchi R. SuperSAGE: a modern platform for genome-wide quantitative transcript profiling. Curr Pharm Biotechnol. 2008;9:368–74. doi: 10.2174/138920108785915157. [DOI] [PubMed] [Google Scholar]
- 9.van Aelst K, Toth J, Ramanathan SP, Schwarz FW, Seidel R, Szczelkun MD. Type III restriction enzymes cleave DNA by long-range interaction between sites in both head-to-head and tail-to-tail inverted repeat. Proc Natl Acad Sci U S A. 2010;107:9123–8. doi: 10.1073/pnas.1001637107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szczelkun MD. Translocation, switching and gating: potential roles for ATP in long-range communication on DNA by Type III restriction endonucleases. Biochem Soc Trans. 2011;39:589–94. doi: 10.1042/BST0390589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szczelkun MD, Friedhoff P, Seidel R. Maintaining a sense of direction during long-range communication on DNA. Biochem Soc Trans. 2010;38:404–9. doi: 10.1042/BST0380404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raghavendra NK, Rao DN. Unidirectional translocation from recognition site and a necessary interaction with DNA end for cleavage by Type III restriction enzyme. Nucleic Acids Res. 2004;32:5703–11. doi: 10.1093/nar/gkh899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999;76:2879–86. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–24. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics. 2006;Chapter 5(Unit 5):6. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen BW, Xu D, Chan SH, Zheng Y, Zhu Z, Xu SY, Stoddard BL. Characterization and crystal structure of the type IIG restriction endonuclease RM.BpuSI. Nucleic Acids Res. 2011;39:8223–36. doi: 10.1093/nar/gkr543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J. 2005;89:1237–50. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osipiuk J, Walsh MA, Joachimiak A. Crystal structure of MboIIA methyltransferase. Nucleic Acids Res. 2003;31:5440–8. doi: 10.1093/nar/gkg713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morita R, Ishikawa H, Nakagawa N, Kuramitsu S, Masui R. Crystal structure of a putative DNA methylase TTHA0409 from Thermus thermophilus HB8. Proteins. 2008;73:259–64. doi: 10.1002/prot.22158. [DOI] [PubMed] [Google Scholar]
- 20.Schuck P, Perugini MA, Gonzales NR, Howlett GJ, Schubert D. Size-distribution analysis of proteins by analytical ultracentrifugation: strategies and application to model systems. Biophys J. 2002;82:1096–111. doi: 10.1016/S0006-3495(02)75469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brookes E, Demeler B, Rosano C, Rocco M. The implementation of SOMO (SOlution MOdeller) in the UltraScan analytical ultracentrifugation data analysis suite: enhanced capabilities allow the reliable hydrodynamic modeling of virtually any kind of biomacromolecule. Eur Biophys J. 2010;39:423–35. doi: 10.1007/s00249-009-0418-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sears A, Szczelkun MD. Subunit assembly modulates the activities of the Type III restriction-modification enzyme PstII in vitro. Nucleic Acids Res. 2005;33:4788–96. doi: 10.1093/nar/gki788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meisel A, Mackeldanz P, Bickle TA, Kruger DH, Schroeder C. Type III restriction endonucleases translocate DNA in a reaction driven by recognition site-specific ATP hydrolysis. EMBO J. 1995;14:2958–66. doi: 10.1002/j.1460-2075.1995.tb07296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saha S, Rao DN. ATP hydrolysis is required for DNA cleavage by EcoPI restriction enzyme. J Mol Biol. 1995;247:559–67. doi: 10.1016/s0022-2836(05)80137-8. [DOI] [PubMed] [Google Scholar]
- 25.Ramanathan SP, van Aelst K, Sears A, Peakman LJ, Diffin FM, Szczelkun MD, Seidel R. Type III restriction enzymes communicate in 1D without looping between their target sites. Proc Natl Acad Sci U S A. 2009;106:1748–53. doi: 10.1073/pnas.0807193106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crampton N, Roes S, Dryden DT, Rao DN, Edwardson JM, Henderson RM. DNA looping and translocation provide an optimal cleavage mechanism for the type III restriction enzymes. EMBO J. 2007;26:3815–25. doi: 10.1038/sj.emboj.7601807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lammens K, Bemeleit DJ, Mockel C, Clausing E, Schele A, Hartung S, Schiller CB, Lucas M, Angermuller C, Soding J, Strasser K, Hopfner KP. The Mre11:Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell. 2011;145:54–66. doi: 10.1016/j.cell.2011.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Cryst. 1992;25:495–503. [Google Scholar]
- 29.Skoglund CM, Smith HO, Chandrasegaran S. Construction of an efficient overproducer clone of HinfI restriction endonuclease using the polymerase chain reaction. Gene. 1990;88:1–5. doi: 10.1016/0378-1119(90)90052-s. [DOI] [PubMed] [Google Scholar]
- 30.Wriggers W. Using Situs for the integration of multi-resolution structures. Biophys Rev. 2010;2:21–27. doi: 10.1007/s12551-009-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Svergun DI, Barberato C, Koch MHJ. CRYSOL - a Program to Evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. J Appl Cryst. 1995;28:768–773. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.