Abstract
Chromatin insulators are DNA protein complexes situated throughout the genome capable of demarcating independent transcriptional domains. Previous studies point to an important role for RNA in gypsy chromatin insulator function in Drosophila; however, the identity of these putative insulator-associated RNAs is not currently known. Here we utilize RNA-immunoprecipitation and high throughput sequencing (RIP-seq) to isolate RNAs stably associated with gypsy insulator complexes. Strikingly, these RNAs correspond to specific sense-strand, spliced and polyadenylated mRNAs, including two insulator protein transcripts. In order to assess the functional significance of these associated mRNAs independent of their coding function, we expressed untranslatable versions of these transcripts in developing flies and observed both alteration of insulator complex nuclear localization as well as improvement of enhancer-blocking activity. Together, these data suggest a novel, noncoding mechanism by which certain mRNAs contribute to chromatin insulator function.
Keywords: RNA, chromatin, insulator, Drosophila , nuclear organization
Introduction
Chromatin insulator complexes define transcriptionally independent chromatin domains throughout the genome likely through alteration of higher order chromatin interactions. Consistent with the enhancer-blocking and barrier activities by which chromatin insulators are functionally defined, genome-wide chromatin conformation studies have demonstrated that chromatin domain boundaries preferentially correspond to certain insulator sites as reviewed in van Bortle and Corces [1]. The well-characterized Drosophila gypsy insulator complex comprises three core protein components, which interact directly. Its binding specificity is defined by the zinc-finger DNA binding protein Suppressor of Hairy-wing (Su(Hw)), whose partners include the 2.2 kb isoform of Modifier of mdg4 (Mod(mdg4)2.2) and Centrosomal protein 190 (CP190). Genome-wide mapping revealed thousands of Su(Hw)-binding sites, occurring most frequently in intergenic and intronic regions [2, 3]. Moreover, gypsy insulator complexes coalesce at a small number of large nuclear structures termed insulator bodies, which have been proposed to correspond to higher-order chromatin domains likely mediated by CP190 and Mod(mdg4)2.2 BTB-domain interactions [4, 5]. An alternative view is that insulator bodies are storage sites of reserve proteins not engaged in insulator activity [6]. Importantly, proper localization of insulator bodies are tightly correlated with gypsy insulator function and serves as a useful phenotypic readout [4, 7, 8].
A direct role for RNA in gypsy insulator function has been suggested by genetic and biochemical evidence implicating various RNA-binding proteins in the regulation of insulator activity. The putative RNA helicase Rm62 associates physically with CP190 complexes in an RNA-dependent manner [9]. In humans, the homologue of Rm62, p68, along with its associated noncoding RNA, SRA, interacts with the CTCF insulator protein and promotes its activity [10]. Recent work shows that the RNA-binding protein, Shep, interacts directly with gypsy insulator complexes and acts as a CNS-specific antagonist of insulator activity [11]. Therefore, utilization of RNA might be a generally conserved mechanism used to modulate the activity of chromatin insulators.
Here we report the unanticipated finding that certain messenger RNAs (mRNAs), and not other classes of RNA, specifically associate with gypsy insulator complexes. By native immunoaffinity purification and RNA sequencing (RIP-seq), we find that two of the most highly enriched transcripts are the su(Hw) and Cp190 mRNAs themselves. In order to study the functional significance of these transcripts outside their coding capacity, we ectopically expressed untranslatable versions in a subset of tissues within the developing fly. Remarkably, we found that expression of these untranslatable RNAs both alter insulator body localization and improve gypsy-dependent enhancer-blocking activity in certain tissues. These findings suggest a novel, noncoding capacity for some mRNA transcripts in chromatin regulation.
Results and discussion
Specific mRNAs copurify with gypsy insulator complexes
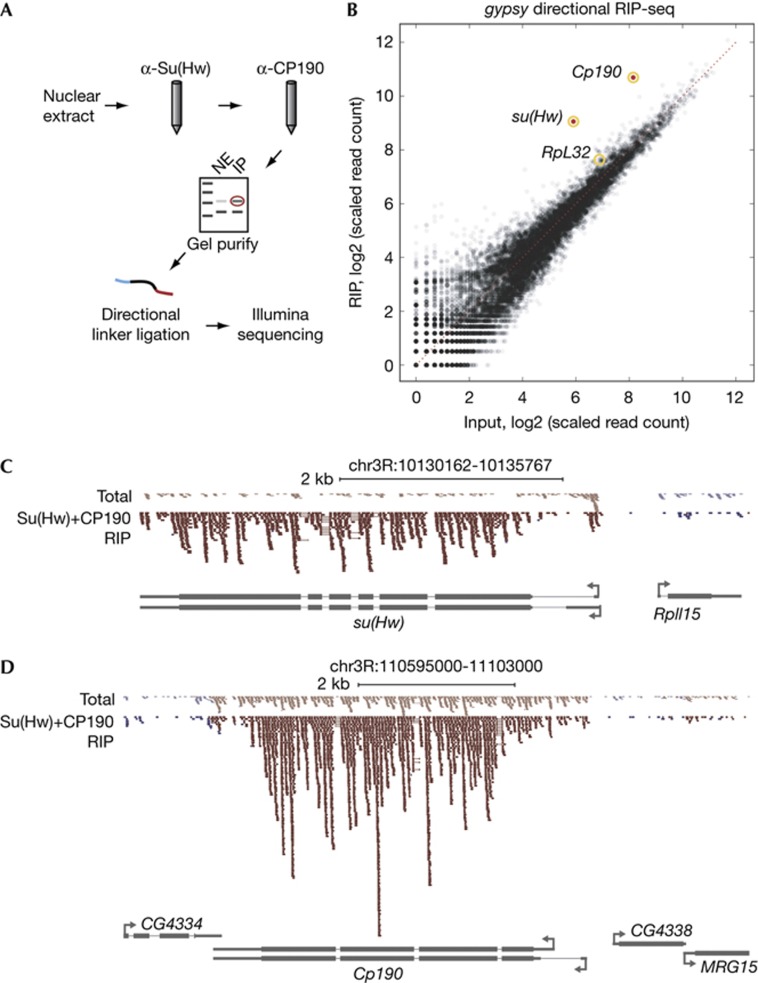
In order to identify RNAs stably associated with insulator complexes, we performed native sequential immunopurification of insulator complexes and high throughput sequencing. Nuclei from 40 g of mixed stage embryos were isolated by manual disruption and centrifugation through sucrose cushions. To achieve high specificity, nuclear extracts were first bound to an α-Su(Hw) column followed by mild salt elution, dilution and subsequent binding of the eluate to an α-CP190 column followed by high salt elution. Associated RNAs were 5′ end labelled and separated on a high percentage polyacrylamide gel. As certain RNA silencing mutants affect gypsy insulator activity [9], we were particularly interested to determine whether small RNAs associate with the gypsy insulator. However, we consistently observed 35–55 nt bands, considerably larger than known small RNA, enriched in IPs over unbound total nuclear extract (unpublished data). These products were gel extracted, directionally cloned and subjected to sequencing (Fig 1A). No products were obtained with α-Su(Hw) or α-CP190 preimmune sera used to preclear the nuclear extract, indicating specificity of the purification procedure.
Figure 1.
Messenger RNA associates with gypsy insulator complexes. (A) Purification scheme for gypsy insulator-associated RNA. Embryo nuclear extracts were passed sequentially over α-Su(Hw) and α-CP190 immunoaffinity columns. Purified RNA was 5′ end labelled and purified by electrophoresis. 35- and 55-nt bands were excised from NE and IP lanes, directionally cloned and sequenced. (B) Scatterplot of annotated genes corresponding to transcripts identified in total unbound and Su(Hw)/CP190 RIP. Mean scaled tag count across replicates of total unbound input (x-axis) and Su(Hw)/CP190 RIP (y-axis) for each gene is calculated as the mean number of tags across replicates mapping to the entire gene body, scaled by library size. Transcripts corresponding to su(Hw) and Cp190 (padj<0.02, red) are indicated. All other transcripts (black) correspond to padj=1. Note that genes displaying similar average fold-enrichment to su(Hw) and Cp190 show high variance across biological replicates, resulting in lack of statistical significance. The su(Hw), Cp190 and RpL32 transcripts are circled. (C) Reads mapping to the su(Hw) or neighbouring RpII15 locus from total unbound (2.4 M read alignments) or Su(Hw)/CP190 RIP (1.3 M read alignments) libraries. The height of each Watson (blue) and Crick (red) read is scaled to respective library size. Thin bars interrupting reads span splice junctions. (D) Reads mapping to the Cp190 locus as in (C). CP190, Centrosomal protein 190; IP, immunopurified; NE, nuclear extract; RIP, RNA immunoprecipitation; Su(Hw), Suppressor of Hairy wing.
A rigorous statistical approach was used to identify transcripts that specifically associate with gypsy insulator complexes. After clipping of adapter sequences, filtering of ribosomal RNA, mapping with TopHat [12], filtering of repetitive elements and removal of duplicate reads, we obtained ∼1.1 M unique reads in the IP and ∼2.3 M in the unbound total RNA samples. Together these libraries correspond to 11,523 annotated transcripts, including mRNA and other classes of noncoding RNA such as small nuclear RNA, small nucleolar RNA, transfer RNA and microRNA. We also considered reads that map to a set of ∼1,800 cis-regulatory modules such as enhancers and silencers annotated in the REDFly database [13], thereby also sampling putative unannotated noncoding RNA. To distinguish transcripts specifically associated with Su(Hw)/CP190 from nonspecific contaminants, we applied the DESeq algorithm [14] to results from two independent purifications. DESeq calculates the statistical probability of enrichment on the basis of fold-change as well as variance of read counts corresponding to transcripts across biological replicates, providing a stringent measure of reproducibility. Using this method, only two transcripts, su(Hw) and Cp190 mRNAs, emerge as significantly enriched (padj<2.0 × 10–6 and<1.9 × 10–2, respectively) compared with all other mRNA and annotated or putative noncoding RNA (padj=1) (Fig 1B; supplementary Table S1 online). Reads map across the entirety of su(Hw) and Cp190 transcripts, span known splice junctions and are primarily sense strand, suggesting that isolated RNAs are degradation products of full-length mRNA (Fig 1C,D, supplementary Fig S1 online), which were not directly sampled in this analysis. We examined the exon:intron ratio of all transcripts identified in these libraries and found that the Su(Hw)/CP190 RIP is enriched for exons compared with the unbound total RNA control (P=1.3 × 10–8, Fisher’s exact test). Due to the unavailability of more CP190 antibody, we validated the enrichment of spliced but not unspliced su(Hw) and Cp190 mRNAs by Su(Hw) RIP compared with preimmune serum followed by quantitative reverse transcription polymerase chain reaction (RT–PCR) (supplementary Fig S1 online). These results indicate that RNA present in the Su(Hw)/CP190 RIP is mainly spliced and likely polyadenylated, and enrichment of both transcripts is not an artifact caused by the use of two antibodies.
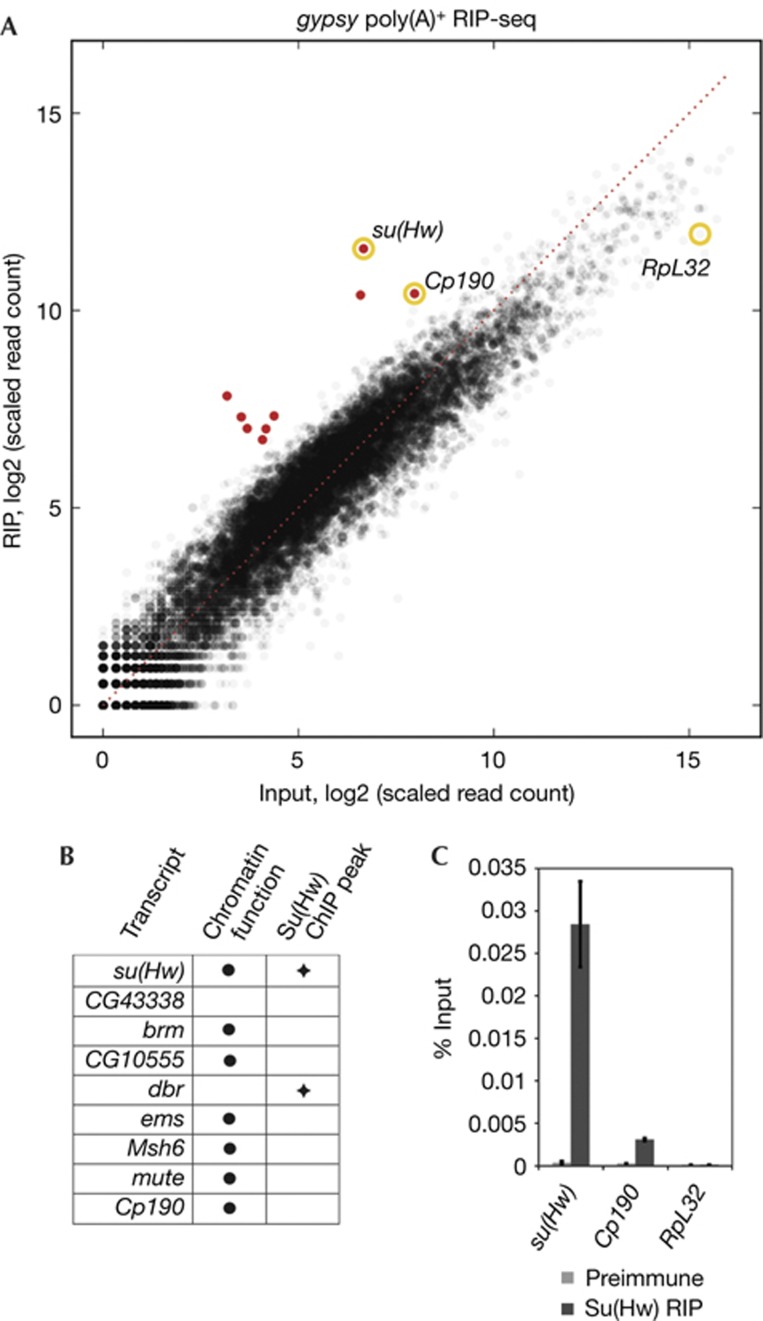
We confirmed that Su(Hw)/CP190 RIP transcripts are spliced and polyadenylated by performing sequential Su(Hw)/CP190 RIP followed by oligo-dT selection instead of gel purification, followed by non-directional cloning. We found that su(Hw), Cp190 as well as seven additional mRNA transcripts are highly enriched by this more efficient method (padj<0.02, Fig 2A, supplementary Table S2 and supplementary Fig S2 online). Only 2 of 9 transcripts are encoded by genes that coincide with a peak of Su(Hw) chromatin association defined by a genome-wide ChIP-chip study in embryos [3], similar to the frequency for all other transcripts (P=0.29, Fisher’s exact test). This result suggests that these mRNAs associate with insulator complexes in trans. We performed gene ontology analysis and found that 7 of 9 transcripts encode chromatin-associated proteins (Fig 2B). These transcripts could perhaps serve dual roles by also influencing insulator activity. In fact, mutation of brm, which encodes a Swi/Snf homologue involved in transcriptional activation, was shown to reduce gypsy insulator function [7], but given our results, this finding could reflect a requirement for either mRNA or protein. Enrichment of su(Hw) and Cp190 mRNAs in Su(Hw) RIP but not with preimmune serum was confirmed by quantitative RT–PCR using the abundant transcript RpL32 as a negative control, which is not enriched in the DESeq analysis (Fig 2A–C). These data demonstrate that gypsy insulator complexes associate with specific mRNAs mainly produced in trans.
Figure 2.
Oligo-dT selected RNA associated with gypsy insulator complexes. (A) Scatterplot of annotated genes corresponding to transcripts identified in total nuclear extracts and Su(Hw)/CP190 RIP. Transcripts corresponding to padj<0.02 are red. (B) Functional annotation of genes corresponding to significantly enriched transcripts (padj<0.02) using FlyBase annotations and correspondence of gene locus with Su(Hw) ChIP-chip peaks [3]. (C) Enrichment of transcripts in Su(Hw) RIP determined by quantitative RT–PCR. Amount of su(Hw), Cp190 and RpL32 in Su(Hw) IP or pre-immune control as a percentage of total. Parallel minus RT controls resulted in immeasurably low values. Error bars indicate standard deviation of quadruplicate measurements. ChIP, chromatin immunoprecipitation; IP, immunopurified; RIP, RNA immunoprecipitation; RT, reverse transcription; RT–PCR, reverse transcription polymerase chain reaction; Su(Hw), Suppressor of Hairy wing.
RIP-seq of Dorsal as a negative control
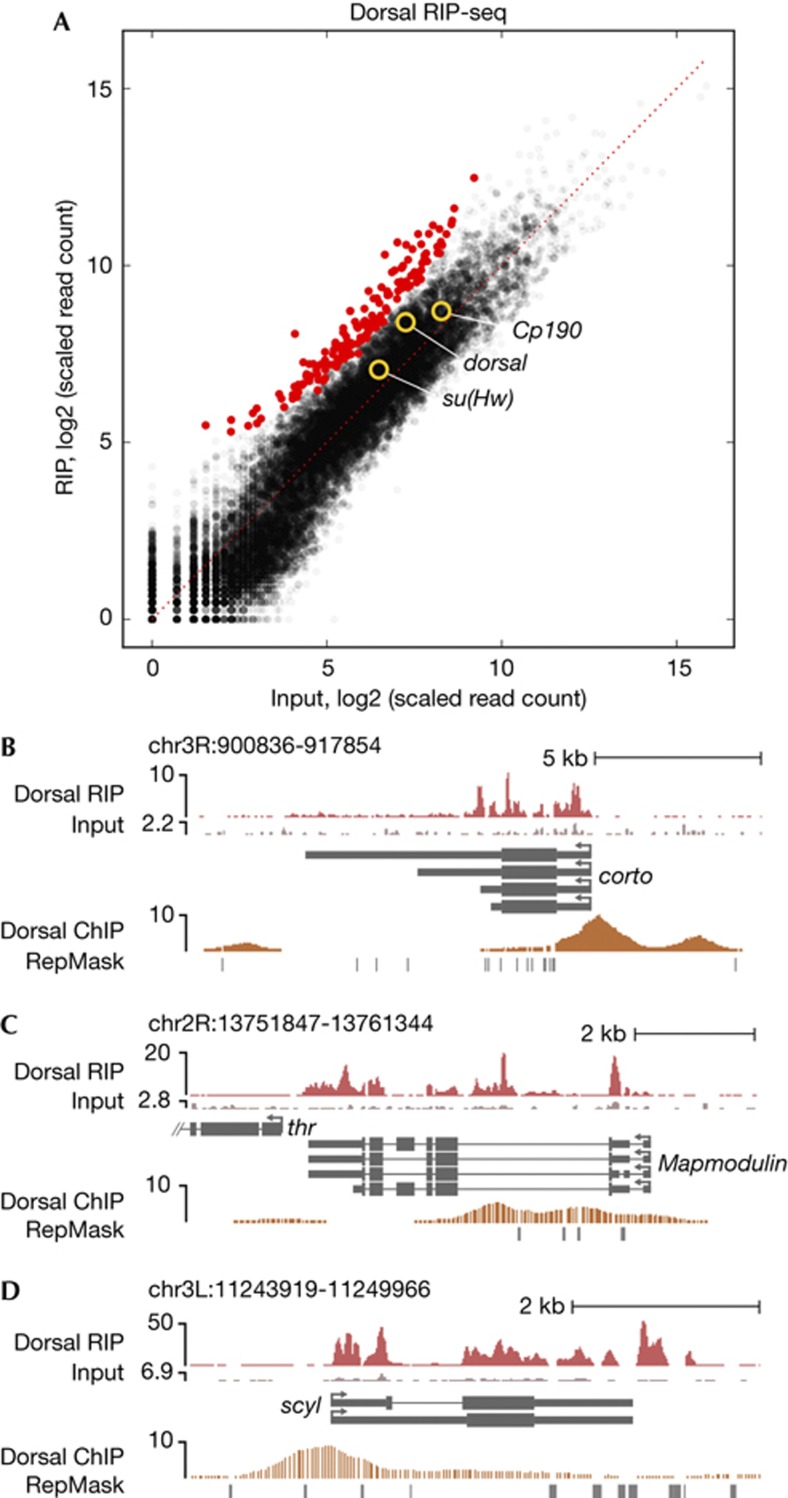
Given the unexpected result of su(Hw) and Cp190 transcript purification with insulator complexes, we considered the possibility that any chromatin-associated protein would copurify its own coding transcript using our protocol. We therefore performed RIP-seq analysis on the Dorsal (Dl) transcription factor, which is highly expressed in the embryo, is not known to be involved in insulator activity or to bind RNA directly, and has a well-defined embryonic chromatin association profile [15, 16]. We utilized antisera used previously for ChIP-chip analysis [16], ensuring access to a chromatin-associated pool. Although Dl was efficiently immunoprecipitated from nuclear extracts using α-Dl but not control serum, the resulting amounts of RNA isolated in the IP versus control fractions were almost equal, suggesting that RNA is not abundantly associated with Dl. Owing to the extremely low RNA yield, we were unable to perform oligo-dT selection and instead depleted ribosomal RNA, subjecting the remainder to sequencing. DESeq analysis of two biological replicates identified 161 transcripts as significantly enriched (padj<0.02) in Dl RIP compared with input (Fig 3A, supplementary Table S3 online). We suspect that the larger number of statistically significant enriched transcripts in the Dl RIP compared with Su(Hw)/CP190 RIP is mainly due to the higher consistency between biological replicates, and therefore higher sensitivity of detection, due to the simpler, single antibody purification. Importantly, the dl transcript is not significantly enriched in the Dl RIP (padj=0.37), arguing against our purifications being contaminated with translating ribosomes. Consistent with these results, mass spec of similarly purified insulator complexes did not detect ribosomal proteins [9].
Figure 3.
RIP-seq of Dorsal-associated RNA. (A) Scatterplot of annotated genes corresponding to transcripts identified in total nuclear extracts and Dl RIP. Transcripts corresponding to padj<0.02 are red. The su(Hw), Cp190 and dl transcripts are circled. (B–D) Reads mapping to the (B) corto, (C) Mapmodulin and (D) scylla loci from input (4.9 M read alignments) or Dl RIP (14.3 M read alignments) libraries. Input samples are shown on the same scale relative to respective IP, and the bottom of each scale bar corresponds to 0. Dl ChIP-chip signal at 1% FDR [15] is also shown. ChIP, chromatin immunoprecipitation; FDR, false discovery rate; IP, immunopurified; RIP, RNA immunoprecipitation; Su(Hw), Suppressor of Hairy wing.
Unlike Su(Hw)/CP190-associated transcripts, those copurified with Dl appear to be generated in cis from transcription factor binding sites. First, 142 or 88% of Dl-associated transcripts correspond to Dl ChIP peaks in the corresponding gene body, significantly more than expected compared with all other transcripts (P=1.2 × 10–10, Fisher’s exact test). Second, substantial numbers of Dl RIP reads map to introns or regions extending immediately 3′ beyond the annotated polyadenylation site (Fig 3B–D), consistent with some fraction of Dl-associated transcripts being nascent. The transcript association profile obtained for Dl contrasts starkly with that of the gypsy insulator, which mainly consists of processed mRNAs produced in trans to its binding sites. This important negative control also demonstrates that not all chromatin-associated proteins can associate with their own coding transcript.
In vivo expression of untranslatable transcripts using T7
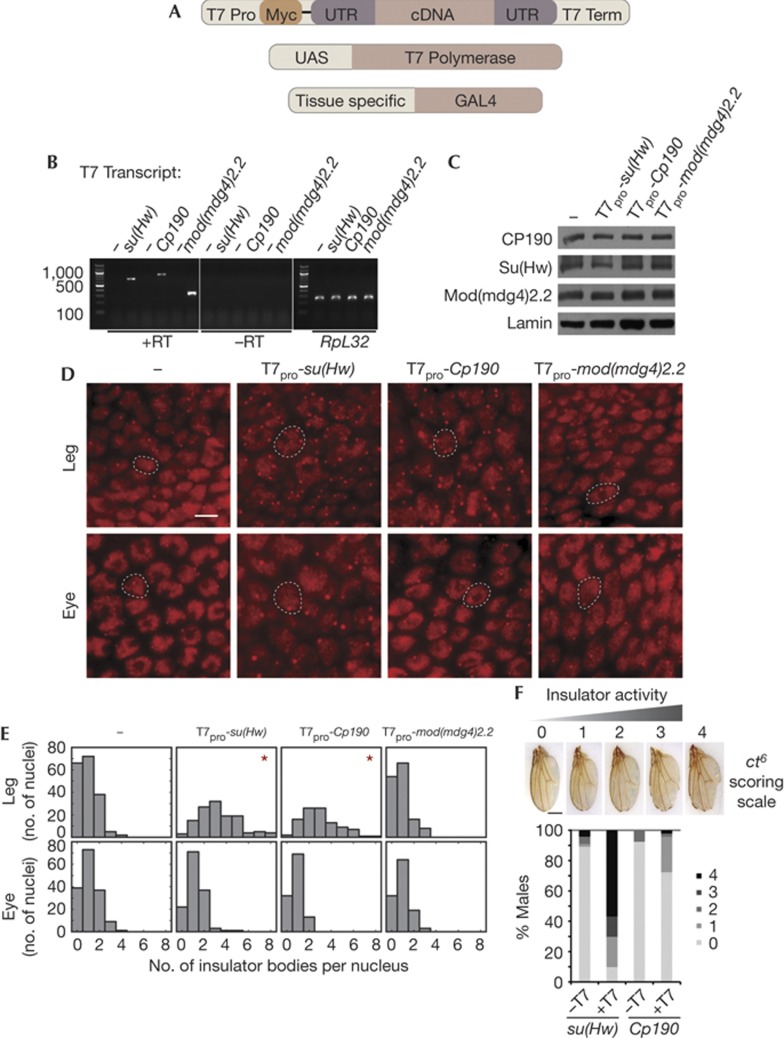
In order to address whether su(Hw) and Cp190 transcripts affect insulator activity, we expressed these transcripts uncoupled from RNA polymerase II transcription, processing, export and translation. This strategy avoids perturbing normal levels of Su(Hw) and CP190 proteins, which would confound subsequent analyses. The su(Hw) and Cp190 cDNAs along with a negative control were expressed using UAS-Gal4 inducible T7 polymerase [17, 18] (Fig 4A). The mod(mdg4)2.2 transcript serves as an ideal negative control as the endogenous transcript is expressed at a level similar to su(Hw) and Cp190 and shares sequence similarity to Cp190 but is not enriched in Su(Hw)/CP190 RIP purifications (supplementary Tables S1,S2 online). When T7-su(Hw), T7-Cp190 and T7-mod(mdg4)2.2 are expressed by inducing T7 polymerase with Ser::Gal4, the myc-tagged transcripts are readily detected by RT–PCR from total RNA at similar levels for each transgene (Fig 4B). We also verified that T7 transcripts are visible in the nucleus but not cytoplasm by in situ hybridization to myc in whole mount tissue in the expression patterns of Ser::Gal4, GMR::Gal4, or nrv2::Gal4, indicating transcripts are highly expressed in a Gal4 and T7 polymerase-dependent manner (unpublished data). No change in endogenous CP190, Su(Hw) or Mod(mdg4)2.2 protein levels is observed by western blotting on induction of T7 transcripts with Ser::Gal4 (Fig 4C; supplementary Fig S3 online). Furthermore, immunofluorescence of whole mount tissues and western blotting failed to detect Myc antigen from T7 transcript expressing larvae. These results indicate that T7 transcripts remain untranslated within the nucleus.
Figure 4.
Untranslatable versions of mRNA associated with the gypsy insulator affect insulator body localization and improve enhancer-blocking activity. (A) Schematic representation of three transgene system to produce untranslatable transcripts. Transcripts were cloned in their entirety including 5′ and 3′ UTRs; constructs contain a myc tag and are flanked by T7 promoter and T7 terminator hairpin sequence. Transcripts are induced by T7 polymerase under tissue-specific GAL4 control. (B) RT–PCR using myc and gene-specific primers to detect T7-su(Hw), T7-Cp190 and T7-mod(mdg4)2.2 transcripts from larvae expressing T7 polymerase with (+RT) or without (−RT) reverse transcription. Endogenous RpL32 transcript with RT serves as a positive control (right). (C) Western blotting of CP190, Su(Hw), Mod(mdg4)2.2 and Lamin in anterior larval extracts expressing T7 polymerase alone (−) or indicated T7 driven transcripts. (D) Indirect immunofluorescence of CP190 to detect insulator bodies in peripheral leg imaginal disc tissues expressing T7 polymerase alone or T7-su(Hw), T7-Cp190 or T7-mod(mdg4)2.2 using Ser::Gal4 (top row). Insulator bodies in eye imaginal discs, with no T7 expression, from corresponding samples (bottom row). Rabbit α-CP190 is detected by α-rabbit Alexa-594. White dotted lines outline an example nucleus in each field. Scale bar equals 5 μm. (E) Histograms of number of nuclei (y-axis, n>101) containing indicated number of insulator bodies per nucleus (x-axis) for samples shown in (D). Asterisks denote genotypes with distributions that show a statistically significant difference from the control (P<1.0 × 10–10, Kruskal–Wallace test). (F) Effects of T7-su(Hw) and T7-Cp190 on ct6 phenotype in males without T7 (−T7) or with (+T7) polymerase driven by Ser::Gal4. All flies are homozygous for mod(mdg4)u1. Percent of population scored on a 0–4 scale. 0, absence of notching; 1, light notch; 2, medium notch; 3, heavy notching; and 4, a single large notch in the distal wing margin, perhaps with notches in the wing tip. Scale bar is ∼0.5 mm. Example wings are of the following genotypes: y2wct6; Ser::Gal4/+; T7-Cp190, mod(mdg4)u1/+, mod(mdg4)u1 (0), y2wct6; pUAST7 25A/Ser::Gal4; T7-Cp190, mod(mdg4)u1/+, mod(mdg4)u1 (1 and 2), y2wct6; pUAST7 25A/Ser::Gal4; T7-su(Hw), mod(mdg4)u1/+, mod(mdg4)u1 (3 and 4). Note that all T7 transcript transgenes are paternally transmitted. See also supplementary Fig S5 online. mRNA, messenger RNA; RT, reverse transcription; RT-PCR, reverse transcription polymerase chain reaction; Su(Hw), Suppressor of Hairy wing.
Functional significance of gypsy-associated transcripts
We next tested whether T7-su(Hw) and T7-Cp190 transcripts affect insulator complex localization. We examined the distribution of insulator bodies by indirect immunofluorescence of endogenous CP190 protein in whole mount larval imaginal discs and brains. Compared with no T7 transcript or T7-mod(mdg4)2.2 expression using Ser::Gal4, T7-su(Hw) causes formation of ectopic and more pronounced insulator bodies relative to overall nucleoplasmic signal, particularly in peripheral cells of the leg discs (Fig 4D,E, top panel; observed in 10 of 12 experiments) as well as wing discs. Expression of T7-Cp190 causes a less penetrant and less pronounced but similar phenotype to T7-su(Hw) (observed in four of seven experiments). Importantly, no effect on overall development of disc or brain tissue is observed as a result of T7 polymerase expression with or without T7 transcript (supplementary Fig S4 online). No change in insulator bodies is observed in the eye disc, which does not express Ser::Gal4 (Fig 4D,E, bottom panel). Moreover, no change is detected in the brain, in which expression is much lower than the leg and wing discs (supplementary Fig S4 online). Similarly, no effects on insulator body localization in the eye or brain are observed when T7 transcripts are expressed by GMR::Gal4 or nrv2::Gal4, respectively (supplementary Fig S4 online). Resultant changes in insulator body morphology using Ser::Gal4 indicate that expression of untranslatable su(Hw) or Cp190 transcripts are sufficient to affect the overall nuclear distribution of insulator complexes in certain tissues. These RNAs might alter higher-order insulator interactions or those between insulators and factors that regulate their activities. An alternative explanation is that T7-su(Hw) or T7-Cp190 transcripts change the relative distribution of insulator proteins on chromatin versus storage sites either by competing with endogenous transcripts or nucleating ectopic insulator bodies.
Finally, we examined whether T7 transcripts also alter enhancer-blocking activity using gypsy-dependent phenotypic reporters. We scored enhancer-blocking activity of the well-characterized yellow2 (y2) and cut6 (ct6) alleles, which both result from a gypsy retrotransposon insertion between an enhancer and the promoter. Insertion results in loss of y or ct expression and corresponding loss of abdominal pigmentation or proper wing margin development, respectively [19]. In wild-type flies, ubiquitous expression of T7 polymerase using Act5C::Gal4 results in lethality, and expression in the wing using Ser::Gal4 causes blistering and subsequent necrosis of adult wings, preventing analysis of the y2 and ct6 phenotypes (supplementary Fig S4 online). However, in the sensitized mod(mdg4)u1 background, expression of T7-su(Hw) with Ser::Gal4 reduces wing blistering (supplementary Fig S4 online), allowing assessment of gypsy insulator activity at ct6. Compared with matched controls without T7 polymerase, expression of T7-su(Hw) results in dramatically increased wing notching whereas expression of T7-Cp190 produces a milder but similar phenotype, signifying improvement in insulator enhancer-blocking activity (Fig 4F). No change of y2 expression in the wing was observed using Ser::Gal4, and no effects on y2 expression in the wing or body were observed using a y::Gal4 driver that includes wing and body enhancers ([20]; supplementary Fig S5 online). Changes in both insulator activity and insulator body localization due to ectopic expression of these transcripts strongly suggest the functional importance of insulator-associated mRNA.
Core insulator proteins are not known to bind RNA directly, predicting the necessity for protein adapters to recruit RNAs to insulator complexes. On the basis of its antagonistic effect on insulator activity, it is unlikely that Shep mediates interaction between su(Hw) or Cp190 transcripts and insulator complexes. Future work to identify this putative adapter as well as the sequence specificity of its associated transcripts will provide important insights into this novel mechanism.
Insulator-associated mRNAs might be reminiscent of long noncoding RNA (lncRNA) that have been proposed to act as stabilizers of chromatin conformation or scaffolds for recruitment of chromatin-modifying factors to specific loci. Binding of a particular long noncoding RNA can provide a signal or induce a conformational change to alter the interaction potential of its associated protein [21]. Likewise, mRNA associated with gypsy insulator complexes could alter the likelihood of assembly at particular sites or affect insulator-dependent interactions that modulate their activities. Our results not only add to the growing list of dual coding/noncoding transcripts [22] but further suggest the potential for coevolution of bifunctionality.
Methods
Quantitative PCR. Transcripts from Su(Hw) RIP and preimmune RIP were reverse transcribed using gene-specific primers (supplementary Table S4 online) and quantified on an Applied Biosystems Real-Time PCR System using SYBR-green incorporation (Affymetrix/USB). PCR primers were tested for specificity and efficiency by electrophoresis and real-time PCR of serially diluted templates. Error is represented as the standard deviation of four technical replicates of each PCR reaction, removing statistical outliers. Transcript quantities were normalized to a 5-point genomic DNA serial dilution standard verified to be amplified in the linear range and expressed as percent of total nuclear RNA. Two independent biological replicates showed similar results.
Immunofluorescence. Brains and imaginal discs were dissected from at least five larvae of each genotype and stained as described previously [11] and were imaged using a Leica DM5000B epifluorescent microscope and captured using OpenLab software. See supplementary Methods online for antibody descriptions.
RIP-seq and computational analysis. See supplementary Methods online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank W. Bender and V. Corces for fly strains, M. Levine for α-Dl antisera and N. Perrimon for the pCa4B plasmid. We thank members of the Lei laboratory, J. Birchler, E. Clough, C. Kaplan, J. Kassis and V. Sartorelli for critical reading of the manuscript. This work was funded by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases.
Author contributions: L.H.M. and E.P.L. designed and performed the experiments, R.K.D. performed the computational analyses and all authors analysed the data and generated figures. L.H.M. and E.P.L. wrote the paper with input from R.K.D.
Footnotes
The authors declare that they have no conflict of interest.
References
- Van Bortle K & Corces VG (2012) Nuclear organization and genome function. Annu Rev Cell Dev Biol 28: 163–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey AM Ramos E & Corces VG (2009) Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev 23: 1338–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N et al. (2010) A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet 6: e1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova TI Byrd K & Corces VG (2000) A chromatin insulator determines the nuclear localization of DNA. Mol Cell 6: 1025–1035 [DOI] [PubMed] [Google Scholar]
- Byrd K & Corces VG (2003) Visualization of chromatin domains created by the gypsy insulator of Drosophila. J Cell Biol 162: 565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovnin A Volkov I & Georgiev P (2012) SUMO conjugation is required for the assembly of Drosophila Su(Hw) and Mod(mdg4) into insulator bodies that facilitate insulator complex formation. J Cell Sci 125: 2064–2074 [DOI] [PubMed] [Google Scholar]
- Dej KJ Gerasimova T Corces VG & Boeke JD (1998) A hotspot for the Drosophila gypsy retroelement in the ovo locus. Nucleic Acids Res 26: 4019–4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabler M et al. (2005) Trans-splicing of the mod(mdg4) complex locus is conserved between the distantly related species Drosophila melanogaster and D. virilis. Genetics 169: 723–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei EP & Corces VG (2006) RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat Genet 38: 936–941 [DOI] [PubMed] [Google Scholar]
- Yao H Brick K Evrard Y Xiao T Camerini-Otero RD & Felsenfeld G (2010) Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev 24: 2543–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzat LH Dale RK Moshkovich N & Lei EP (2012) Tissue-specific regulation of chromatin insulator function. PLoS Genet 8: e1003069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C Pachter L & Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo SM Gerrard DT Miner D Simich M Des Soye B Bergman CM & Halfon MS (2011) REDfly v3.0: toward a comprehensive database of transcriptional regulatory elements in Drosophila. Nucleic Acids Res 39: D118–D123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S & Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur S et al. (2009) Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol 10: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J Zinzen RP Stark A Kellis M Zhang H Young RA & Levine M (2007) Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev 21: 385–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall K & Bender W (1996) Probes of chromatin accessibility in the Drosophila bithorax complex respond differently to Polycomb-mediated repression. EMBO J 15: 569–580 [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald DP & Bender W (2001) Polycomb group repression reduces DNA accessibility. Mol Cell Biol 21: 6585–6597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gdula DA Gerasimova TI & Corces VG (1996) Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila. Proc Natl Acad Sci USA 93: 9378–9383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MK Tan YY & Hart CM (2006) The Drosophila boundary element-associated factors BEAF-32A and BEAF-32B affect chromatin structure. Genetics 173: 1365–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L Lin C Liu W Zhang J Ohgi KA Grinstein JD Dorrestein PC & Rosenfeld MG (2011) ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147: 773–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME Gascoigne DK & Mattick JS (2011) The evolution of RNAs with multiple functions. Biochimie 93: 2013–2018 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.